Abstract
Variation in gene expression arises from cis- and trans-regulatory mutations, which contribute differentially to expression divergence. Here, we compare the impacts on gene expression and fitness for cis- and trans-regulatory mutations in Saccharomyces cerevisiae, with a focus on the TDH3 gene. We use the effects of cis-regulatory mutations to infer effects of trans-regulatory mutations attributable to impacts beyond the focal gene, revealing a distribution of pleiotropic effects. Cis- and trans-regulatory mutations had different effects on gene expression, with pleiotropic effects of trans-regulatory mutants impacting expression of genes both in parallel to and downstream of the focal gene. The more widespread and deleterious effects of trans-regulatory mutations we observed are consistent with their decreasing relative contribution to expression differences over evolutionary time.
One-Sentence Summary:
This study quantifies the pleiotropic impacts of trans-regulatory mutations on fitness and gene expression relative to a focal gene.
Heritable variation in gene expression is widespread within and between species and often contributes to phenotypic diversity (1). This variation arises from mutations that alter activity of the regulatory networks that control gene expression. Each regulatory mutation can act in cis or in trans with respect to a specific gene. cis-regulatory mutations tend to be located close to the focal gene and often impact non-coding sequences that regulate the focal gene’s expression, such as promoters or enhancers. trans-regulatory mutations can be located anywhere in the genome and can impact either coding or non-coding sequences of genes that influence the focal gene’s expression through activity of a diffusible molecule such as a protein or RNA. Prior work has shown that trans-regulatory variants appear to be the primary source of mRNA expression differences within a species but the relative contribution of cis-regulatory variants often increases with evolutionary time (2-6). Understanding how and why these classes of regulatory mutations contribute differently to variation in gene expression is important for understanding how gene expression evolves.
Differences in the way cis- and trans-regulatory mutations affect gene expression might contribute to a preferential fixation of cis-regulatory variants relative to trans (6-9). A cis-regulatory mutation alters expression of a focal gene, which can in turn have effects on expression of downstream genes (orange box in Figure 1). By contrast, a trans-regulatory mutation affecting expression of the same focal gene might have effects comparable to the cis-regulatory mutation plus independent effects on expression of other genes, each with its own downstream consequences (blue box in Figure 1). Trans-regulatory mutations are thus predicted to have more wide-spread effects on gene expression than mutations altering expression of the focal gene in cis. Consequently, trans-regulatory mutations might be fixed less often than cis-regulatory mutations because mutations that are more pleiotropic (i.e., affect more traits) are predicted to be more deleterious (10). This hypothesis has been difficult to test directly, however, because it is notoriously difficult to disentangle the effects of a pleiotropic mutation on one trait from its effects on others (11-13).
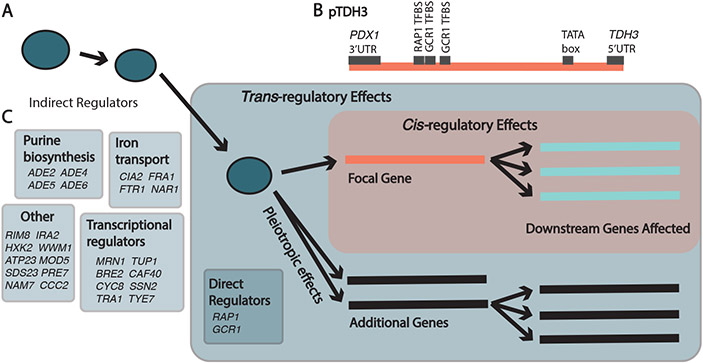
Fig. 1. cis- and trans-regulatory mutations have different effects on gene expression.
(A) Trans-regulatory mutations (blue), in either indirect or direct regulators, influence expression of a focal gene (orange), which in turn influences expression of downstream genes (light blue) either directly or indirectly, and can also impact additional genes (black). (B) Schematic shows the 678 bp cis-regulatory sequence (promoter) for the S. cerevisiae TDH3 gene (pTDH3) used as a focal gene for this work (see Fig S1, Data S1). (C) Previously identified (16) trans-regulators of TDH3 expression harboring mutations tested in this work (see Fig S1 for detail).
Here, we examine the pleiotropic effects of trans-regulatory mutations by using cis-regulatory mutations to separate the effects of a trans-regulatory mutation caused by its impact on a focal gene from its effects caused by impacts on other genes. We separate these mutational effects for fitness and gene expression by measuring relative growth rate and expression profiles (using RNA-seq) for strains of S. cerevisiae carrying either a cis- or trans-regulatory mutation affecting expression of the focal gene. We focus on mutations affecting expression of the TDH3 gene in the baker’s yeast Saccharomyces cerevisiae, which encodes a glyceraldehyde-3-phosphate dehydrogenase (GAPDH), because prior studies have systematically identified and isolated individual cis- and trans-regulatory mutations that affect its expression (14-16). To the best of our knowledge, comparable data are not available for other genes in S. cerevisiae or other eukaryotic species.
We analyzed the effects of 5 cis-regulatory mutants on fitness and gene expression that caused expression of TDH3 to vary from 0% to ~135% of wild-type levels (Data S1). These mutants were selected to cover the range of effects observed previously for 348 mutant alleles of the 678 bp TDH3 promoter (14, 17, 18). The five cis-regulatory mutants were genetically identical except that the 0% TDH3 expression strain carried a deletion of the TDH3 gene, the 20%, 50%, and 85% TDH3 expression strains each carried a single point mutation in the TDH3 promoter, and the 135% TDH3 expression strain carried a duplication of the TDH3 allele harboring a point mutation with a copy of URA3 separating the two copies of TDH3 (Fig. S1). The impact of each of these mutations on gene expression was determined by comparing expression in these strains to expression in the progenitor strain lacking any mutations in TDH3, either without (for the 0, 20, 50, and 85% strains) or with (for the 135% strain) the extra URA3 gene (Fig. S1, see Methods). In parallel, we analyzed the effects of 35 trans-regulatory mutant strains carrying mutations that caused TDH3 expression to vary from ~6% to ~130% of wild-type levels (Data S1). 26 of these 35 strain carried single mutations with a range of effects similar to those captured by the 5 cis-regulatory mutants as well as those observed for >1400 trans-regulatory mutations introduced randomly by ethyl methanesulfonate (EMS) (15). The remaining 9 strains each carried 1 to 6 mutations in either RAP1 and GCR1, which directly regulate TDH3 (16) (Fig. S1, Data S1).
To separate the fitness effects of trans-regulatory mutations attributable to changes in TDH3 expression from the fitness effects attributable to the pleiotropic impacts of these mutations on other genes, we first defined the relationship between TDH3 expression and fitness using only the cis-regulatory mutants. Relative fitness was estimated for each mutant based on measures of growth rate under the same conditions used to grow cells for expression profiling (see Methods). To predict the fitness effects of any change in TDH3 expression between 0 and 135% of wild-type expression, we fit a local polynomial regression (LOESS) curve to these data. The continuous relationship between fitness and TDH3 expression level observed (Fig. 2A) is consistent with prior work using competitive growth to estimate fitness of 43 cis-regulatory mutations in TDH3 (17, 18).
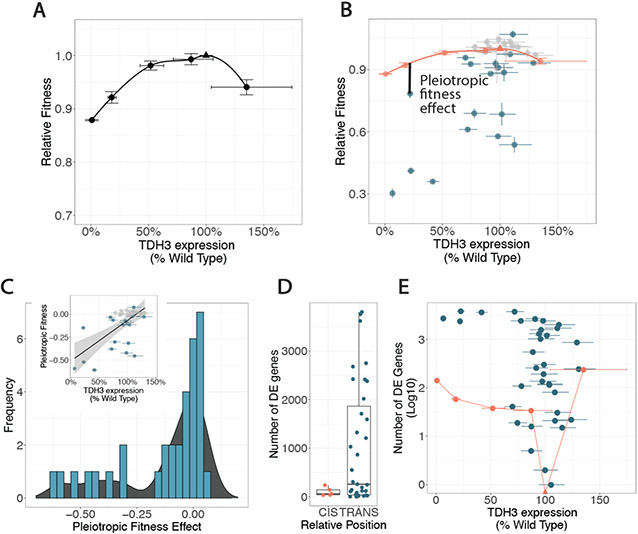
Fig. 2. Pleiotropic effects of trans-regulatory mutations on fitness and gene expression.
(A) Relative fitness is shown for cis-regulatory mutants and a wild-type strain (triangle) with different levels of TDH3 expression. Line fitted with a local polynomial regression (LOESS). (B) Relative fitness and TDH3 expression of trans-regulatory mutants is shown, with the cis-regulatory mutants and fitted LOESS curve from (A) in orange. Pleiotropic fitness effect of one trans-regulatory mutant indicated with solid black line. Mutants with significant pleiotropic fitness effects are blue, not significant are grey. (C) Histogram summarizes pleiotropic fitness effects of all trans-regulatory mutants. A smoothed density distribution derived from this histogram is underlaid in black. Inset shows the same pleiotropic fitness effects plotted vs the effect on TDH3 expression including a robust linear regression (black line). Grey shaded area is the 95% confidence interval. (D) Number of significantly differentially expressed (DE) genes at a 10% FDR is shown for cis-regulatory mutants (orange) and trans-regulatory mutants (blue). Boxplots show median and quartile values. (E) The log10 number of significantly differentially expressed genes at a 10% FDR is shown for each mutant, plotted according to the mutant’s impact on TDH3 expression. cis-regulatory mutants (circles) and wild type strain (triangle) are connected by straight line segments. In all panels, error bars represent one standard error of the mean.
Using these inferred effects of changes in TDH3 expression on fitness, we estimated the pleiotropic fitness effects of each trans-regulatory mutant by comparing its measured fitness to the fitness predicted for a cis-regulatory mutant with the same change in TDH3 expression (Fig. 2B). Excluding 2 flocculant trans-regulatory mutants for unreliable estimates of growth rate, 52% (17/33) of mutants had significant deleterious pleiotropic effects based on the LOESS regression curve falling above their 99% confidence intervals for fitness. Only 1 mutant had a significant beneficial pleiotropic effect; this mutant had a mutation in IRA2, which has also been found to harbor beneficial mutations in other studies (19-21). The remaining 15 trans-regulatory mutants (45%) showed fitness effects comparable to cis-regulatory mutants with similar impacts on TDH3. Overall, this empirical distribution of pleiotropic fitness effects was skewed toward deleterious effects (Fig. 2C). Mutations with the largest deleterious fitness effects tended to also cause the largest decreases in TDH3 expression, although some mutations with large deleterious effects had little impact on TDH3 expression (Fig. 2C, insert). An additional 1106 gene deletions that affected TDH3 expression in trans showed a similar distribution of pleiotropic effects (Fig. S2, (22, 23))
To test the idea that trans-regulatory mutations are more likely to be deleterious because they tend to be more pleiotropic, we compared the number of genes considered significantly differentially expressed in the cis- and trans-regulatory mutants at a false discovery rate (FDR) of 10%. Overall, we found no statistically significant difference in the median number of differentially expressed genes between the 5 cis- and 35 trans-regulatory mutants (Fig. 2D, permutation test p-value: 0.14, Fig. S3A) but observed significantly more variable effects for the trans-regulatory mutants (permutation test p-value = 0.01, Fig. S3B). Although the small number of cis-regulatory mutants included limited the power of this test, the absence of a larger median effect of trans-regulatory mutants might be due to differences in the severity of mutational effects between the sets of cis- and trans-regulatory mutants examined. To test this possibility, we examined the effects of cis- and trans-regulatory mutants on the number of differentially expressed genes while taking their impact on TDH3 expression into account. We found that most trans-regulatory mutants showed more widespread effects on gene expression than cis-regulatory mutants with the same effect on TDH3 expression (Fig. 2E).
As shown in Figure 1, trans-regulatory mutants are assumed to have effects on expression of genes downstream of the focal gene similar to cis-regulatory mutations but to also have additional pleiotropic effects on expression of other genes in parallel. To test this assumption, we sought to separately analyze the effects of trans-regulatory mutants on expression of genes downstream of and in parallel to TDH3. To identify the set of genes downstream of TDH3, we identified genes whose expression was significantly altered in the TDH3 null mutant. We found 140 such genes (10% FDR, p-value=0.002, Data S3), excluding TDH3 itself. 49 (35%) of these genes were under-expressed in the null mutant relative to the wild-type strain, and 91 (65%) were over-expressed (Data S3). This gene set was significantly enriched for genes encoding proteins involved in glycolytic processes (Fig. S4), suggesting that many expression changes observed in the TDH3 null mutant might be due to a homeostatic response of the cells to maintain metabolism in the absence of the TDH3p enzymatic activity involved in glycolysis and gluconeogenesis (24). These downstream genes were also enriched for genes associated with the gene ontology terms DNA biosynthesis, integration, and transposition (Fig. S4), which might result from non-metabolic functions of TDH3p in processes such as transcriptional silencing and rDNA recombination (25).
The median absolute log2 fold expression changes observed for this set of 140 genes downstream of TDH3 decreased monotonically as TDH3 expression approached wild type and was smallest in the cis-regulatory mutant overexpressing TDH3 (Fig. 3A). 122 (87%) of these 140 genes showed a significant linear relationship with TDH3 expression in the 5 cis-regulatory mutants (10% FDR, p-value = 0.09, Fig. S5A), with 44 positively correlated and 78 negatively correlated (Fig. 3B, Fig. S5B). For example, the GPD2 gene, which encodes an enzyme two steps away from TDH3 in the metabolic network, increased linearly when TDH3 expression was decreased by cis-regulatory mutations (Fig. 3C).
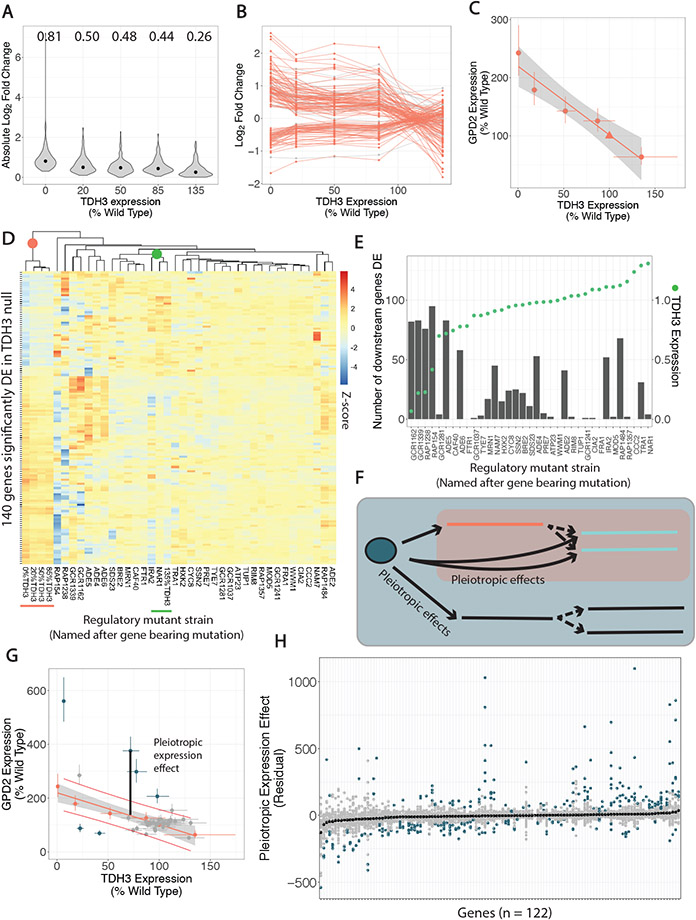
Fig. 3. cis- and trans-regulatory mutants have distinct effects on expression of genes downstream of TDH3.
(A) Violin plots show absolute log2 fold changes in the cis-regulatory mutants for the 140 genes identified as downstream of TDH3 because they were significantly differentially expressed (DE) in the TDH3 null mutant. Median absolute log2 fold changes are shown above each plot and indicated with black dots. (B) Log2 fold changes in cis-regulatory mutants are shown for the same 140 genes downstream of TDH3 connected by line segments. Genes whose expression was not significantly linearly correlated with TDH3 expression are shown in grey. (C) Expression of TDH3 and GPD2 is shown for the cis-regulatory mutants (circles) and the wild type strain (triangle). The best fit linear regression line and 95% confidence interval (grey shaded area) are shown. (D) A heatmap of the 140 genes downstream of TDH3 shows log2 fold changes in all cis- and trans-regulatory mutants in which genes are rows and mutants, named after the gene bearing the mutation in that mutant, are columns. Color intensity is scaled by row (by gene) and represents z-scores. Mutants are hierarchically clustered as shown by the dendrogram. (E) The number of downstream genes that are also significantly differentially expressed in each trans-regulatory mutant is shown as a column relating to the left y-axis. The expression level of TDH3 in that mutant is shown as a green point relating to the right y-axis. (F) Schematic shows that trans-regulatory mutants can have pleiotropic effects on genes downstream of TDH3 not mediated by their impact on TDH3 as well as pleiotropic effects on genes in parallel to TDH3. (G) Expression of GPD2 and TDH3 is shown for the trans-regulatory mutants, with the expression and linear regression from cis-regulatory mutants from (C) included in orange. Red lines delineate 95% prediction intervals for the cis-regulatory mutant relationship between TDH3 and GPD2. Effects of trans-regulatory mutants on GPD2 that are not explained by their impact on TDH3 (pleiotropic expression effects, example illustrated by black line) are colored blue. (H) For each of the 122 genes downstream of TDH3 with a significant linear relationship to TDH3 expression in the cis-regulatory mutants (x-axis), the pleiotropic expression effect (y-axis), as illustrated in (G), is shown. For each gene, each point represents a different trans-regulatory mutant. Genes are ordered on the x-axis by median residual (black points). Blue points indicate mutants with significant pleiotropic expression effects, while grey points indicate non-significant pleiotropic expression effects.
A heatmap showing expression with hierarchical clustering for these 140 genes downstream of TDH3 in the 40 cis- and trans-regulatory mutants also visually showed the correlation with TDH3 expression in the cis-regulatory mutants (Fig. 3D, orange cluster). The cis-regulatory mutant overexpressing TDH3 (135%TDH3) had opposing effects on expression of these genes that caused it to cluster separately, with expression most similar to two trans-regulatory mutants (bearing mutations in NAR1 and IRA2) that also caused overexpression of TDH3 (Fig. 3D, green cluster). The other 33 trans-regulatory mutants had more distinct patterns of expression for these genes (Fig. 3D), with some mutants (e.g., RAP1484 and IRA2) showing significant changes in expression for many genes despite minimal impacts on TDH3 expression (Fig. 3E). These observations suggest that the pleiotropic effects of trans-regulatory mutants include impacts on expression of genes downstream of TDH3 that are not explained by their impact on TDH3 (Fig. 3F).
To further explore this possibility, we used the linear models fitted to the cis-regulatory mutant data to predict the change in expression expected for each downstream gene due to the impact of the trans-regulatory mutant on TDH3 expression alone. Deviations from these expectations indicate pleiotropic effects of trans-regulatory mutants on the expression of genes downstream of TDH3. For example, 6 of the 35 trans-regulatory mutants showed evidence of a pleiotropic effect on expression of GPD2, as indicated by a change in GPD2 expression further than one standard error outside of the 95% prediction interval for the expression change expected based on cis-regulatory mutants (Fig. 3G). Such pleiotropic effects were observed for at least one trans-regulatory mutant for 111 of the 122 genes downstream of TDH3, with the magnitude of the pleiotropic effects (measured as residuals from the gene-specific regression models based on the cis-regulatory mutants) varying among trans-regulatory mutants and genes (Fig. 3H).
These pleiotropic effects on expression of genes downstream of TDH3 are in addition to the pleiotropic effects of trans-regulatory mutants in parallel to TDH3. A heatmap of expression with hierarchical clustering for the 5806 genes not classified as downstream of TDH3 shows these other effects (Fig. 4A). 14 of the trans-regulatory mutants showed minimal effects on expression of these genes (purple cluster in Fig. 4A). The four hypomorphic cis-regulatory mutants clustered together (orange cluster in Fig. 4A) and showed additional effects that appeared to scale with TDH3 expression level (consistent with Fig. S5), suggesting that there might be more genes downstream of TDH3 than were called differentially expressed in the TDH3 null mutant with the statistical thresholds used. The trans-regulatory mutants with larger effects tended to cluster according to related phenotype or function, such as the two flocculant strains (bearing mutations in CYC8 and SSN2), genes involved in adenine biosynthesis (ADE4, ADE5, and ADE6), and large-impact mutations in GCR1. Finally, the two RAP1 mutants with the largest impacts on TDH3 expression (RAP154 and RAP1238) showed the most different expression patterns from the other mutants and each other.
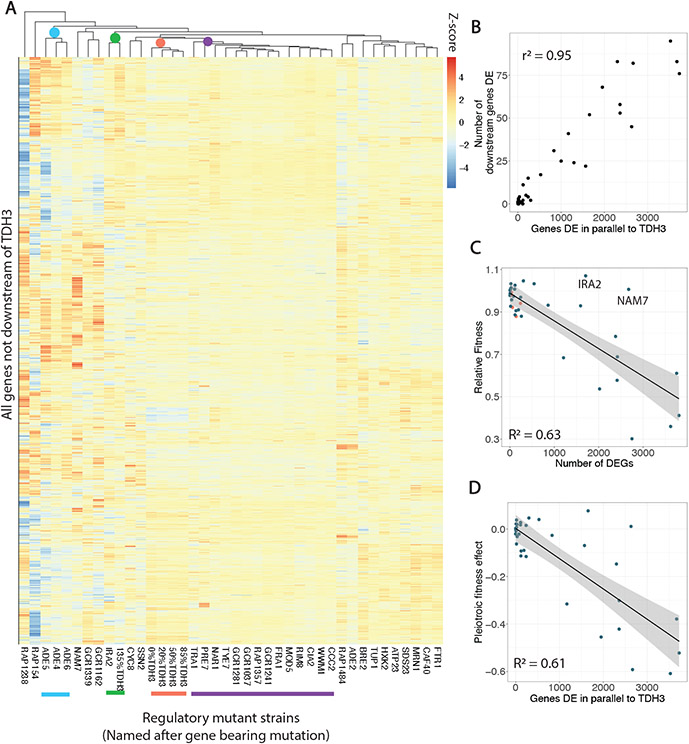
Fig. 4. Larger pleiotropic expression effects of trans-regulatory mutants correlate with negative fitness effects.
(A) A heatmap shows log2 fold changes for all genes not downstream of TDH3 as estimated by DESeq2, in which rows are genes and columns are mutants. Color intensity is scaled by row (by gene) and corresponds to z-scores. Hierarchical clustering of mutants is shown by the dendrogram above. Three of the four trans-regulatory mutants bearing mutations in the adenine biosynthesis pathway cluster together and are marked by a blue dot and line. The cis-regulatory mutant overexpressing TDH3 clusters with IRA2 (green), while the cis-regulatory mutants with reduced TDH3 expression cluster together (orange). A large cluster of trans-regulatory mutants with small effects across the genome are marked by a purple dot and line. (B) For all trans-regulatory mutants, the number of downstream genes differentially expressed is positively correlated with the number of genes differentially expressed in parallel to TDH3 in that mutant. (C) For both cis- (orange) and trans- (blue) regulatory mutants, the total number of differentially expressed genes is a strong predictor of relative fitness. A linear model is shown with grey shading representing 95% confidence intervals and two outliers, IRA2 and NAM7 are indicated. (D) For all trans-regulatory mutants with fitness data (i.e., excluding the two flocculant strains), the pleiotropic fitness effect, as defined in Fig. 2B, is plotted vs the number of genes differentially expressed in parallel to TDH3 in that trans-regulatory mutant. A linear regression is shown in black with 95% confidence intervals in grey.
Comparing the impacts of trans-regulatory mutations on gene expression downstream of and in parallel to TDH3 showed that mutants affecting expression of many genes in parallel to TDH3 tended to also affect expression of many genes downstream of TDH3 (Fig. 4B). This observation indicates that some trans-regulatory mutants had larger or smaller overall effects on the transcriptome. However, the impact of a mutation on TDH3 expression was not a strong predictor for this overall effect: some mutants with minimal effects on TDH3 expression altered expression of thousands of other genes, whereas other mutants changed TDH3 expression up to 30% but altered expression of less than 100 other genes (Fig. 2E). The overall number of genes whose expression was affected by a trans-regulatory mutation was a strong predictor of the relative fitness of the mutant (Fig. 4C, consistent with (26)), although two trans-regulatory mutants (IRA2 and NAM7) were notable exceptions (Fig. 4C). Directly comparing the pleiotropic effects of trans-regulatory mutations inferred for fitness and expression of genes in parallel to TDH3 showed a very similar relationship (Fig. 4D) because the cis-regulatory mutants had much smaller effects on both fitness and expression of other genes than trans-regulatory mutants with similar effects on TDH3 expression (Fig. 2B,E).
In summary, we developed a framework to decompose the effects of trans-regulatory mutations attributable to their impact on expression of a focal gene from their impact on other genes in the genome. These data support the widely held but rarely tested assumptions that trans-regulatory mutations tend to be more pleiotropic and more deleterious than cis-regulatory mutations, but also provide a more nuanced view of these differences at the level of specific mutations and show that the pleiotropic effects of trans-regulatory mutations might often affect expression of genes both in parallel to and downstream of the focal gene. Studies such as this one that empirically determine and compare the properties of different types of mutations affecting gene expression are critical for understanding how regulatory systems evolve.
Supplementary Material
Acknowledgments:
We thank Brian Metzger, Fabien Duveau, Mo Siddiq, Henry Ertl, Anna Redgrave, and other members of the Wittkopp lab for helpful discussions and feedback on drafts of this manuscript. We also thank Bin Z. He (University of Iowa) for sharing the protocol used for RNA-seq. The University of Michigan Advanced Genomics Core, Center for Statistical Consultation and Research, and High-Performance Computing Cluster provided services used to conduct this work. Current address for P.V.Z. is Department of Microbiology and Immunology, University of Minnesota, Minneapolis, Minnesota, USA. Current address for M.S.H. is Cancer Evolution and Genome Instability Laboratory, University College London Cancer Institute and The Francis Crick Institute, London, United Kingdom.
Funding:
National Institutes of Health grants R35GM118073 and R01GM108826 to P.J.W. and T32GM07544 to P.V.Z.
Footnotes
Publisher's Disclaimer: This manuscript has been accepted for publication in Science. This version has not undergone final editing. Please refer to the complete version of record at http://www.sciencemag.org/. The manuscript may not be reproduced or used in any manner that does not fall within the fair use provisions of the Copyright Act without the prior, written permission of AAAS.
Competing interests: Authors declare that they have no competing interests.
Data and materials availability: RNA-seq data is available at GEO accession GSE175398. Code used for data analysis, as well as supporting data files, are available from GitHub at https://github.com/pvz22/Trans-reg_pleiotropy with the version of record at time of publication available at Zenodo under DOI 10.5281/zenodo.6567260 (27). All other data is included with the manuscript or supplementary material. Mutant strains of S. cerevisiae available upon request.
References and Notes
- 1.Stern DL, Orgogozo V, The loci of evolution: how predictable is genetic evolution? Evolution. 62, 2155–2177 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Coolon JD, McManus CJ, Stevenson KR, Graveley BR, Wittkopp PJ, Tempo and mode of regulatory evolution in Drosophila. Genome Res. 24, 797–808 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Metzger BPH, Wittkopp PJ, Coolon JD, Evolutionary Dynamics of Regulatory Changes Underlying Gene Expression Divergence among Saccharomyces Species. Genome Biol. Evol 9, 843–854 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gokhman D, Agoglia RM, Kinnebrew M, Gordon W, Sun D, Bajpai VK, Naqvi S, Chen C, Chan A, Chen C, Petrov DA, Ahituv N, Zhang H, Mishina Y, Wysocka J, Rohatgi R, Fraser HB, Human-chimpanzee fused cells reveal cis-regulatory divergence underlying skeletal evolution. Nat. Genet (2021), doi: 10.1038/s41588-021-00804-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Signor SA, Nuzhdin SV, The Evolution of Gene Expression in cis and trans. Trends Genet. 34, 532–544 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hill MS, Vande Zande P, Wittkopp PJ, Molecular and evolutionary processes generating variation in gene expression. Nat. Rev. Genet (2020), doi: 10.1038/s41576-020-00304-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wray GA, The evolutionary significance of cis-regulatory mutations. Nat. Rev. Genet 8, 206–216 (2007). [DOI] [PubMed] [Google Scholar]
- 8.Wittkopp PJ, Haerum BK, Clark AG, Regulatory changes underlying expression differences within and between Drosophila species. Nat. Genet 40, 346–350 (2008). [DOI] [PubMed] [Google Scholar]
- 9.Schaefke B, Emerson JJ, Wang TY, Lu MYJ, Hsieh LC, Li WH, Inheritance of gene expression level and selective constraints on trans-and cis-regulatory changes in yeast. Mol. Biol. Evol 30, 2121–2133 (2013). [DOI] [PubMed] [Google Scholar]
- 10.Kimura M, Ohta T, On some principles governing molecular evolution. Proc. Natl. Acad. Sci. U. S. A 71, 2848–2852 (1974). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Paaby AB, Rockman MV, The many faces of pleiotropy. Trends Genet. 29, 66–73 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang J, Wagner GP, On the definition and measurement of pleiotropy. Trends Genet. 29 (2013), pp. 383–384. [DOI] [PubMed] [Google Scholar]
- 13.Paaby AB, Rockman MV, Pleiotropy: what do you mean? Reply to Zhang and Wagner. Trends Genet. 29 (2013), p. 384. [DOI] [PubMed] [Google Scholar]
- 14.Metzger BPH, Yuan DC, Gruber JD, Duveau F, Wittkopp PJ, Selection on noise constrains variation in a eukaryotic promoter. Nature. 521, 344–347 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Metzger BPH, Duveau F, Yuan DC, Tryban S, Yang B, Wittkopp PJ, Contrasting Frequencies and Effects of cis- and trans-Regulatory Mutations Affecting Gene Expression. Mol. Biol. Evol 33, 1131–1146 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Duveau F, Vande Zande P, Metzger BP, Diaz CJ, Walker EA, Tryban S, Siddiq MA, Yang B, Wittkopp PJ, Mutational sources of trans-regulatory variation affecting gene expression in Saccharomyces cerevisiae. Elife. 10 (2021), doi: 10.7554/eLife.67806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Duveau F, Toubiana W, Wittkopp PJ, Fitness Effects of Cis-Regulatory Variants in the Saccharomyces cerevisiae TDH3 Promoter. Mol. Biol. Evol 34, 2908–2912 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Duveau F, Hodgins-Davis A, Metzger BP, Yang B, Tryban S, Walker EA, Lybrook T, Wittkopp PJ, Fitness effects of altering gene expression noise in Saccharomyces cerevisiae. Elife. 7 (2018), doi: 10.7554/eLife.37272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sezmis AL, Malerba ME, Marshall DJ, McDonald MJ, Beneficial Mutations from Evolution Experiments Increase Rates of Growth and Fermentation. J. Mol. Evol 86, 111–117 (2018). [DOI] [PubMed] [Google Scholar]
- 20.Venkataram S, Dunn B, Li Y, Agarwala A, Chang J, Ebel ER, Geiler-Samerotte K, Hérissant L, Blundell JR, Levy SF, Fisher DS, Sherlock G, Petrov DA, Development of a Comprehensive Genotype-to-Fitness Map of Adaptation-Driving Mutations in Yeast. Cell. 166, 1585–1596.e22 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li Y, Venkataram S, Agarwala A, Dunn B, Petrov DA, Sherlock G, Fisher DS, Hidden Complexity of Yeast Adaptation under Simple Evolutionary Conditions. Curr. Biol 28, 515–525.e6 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kemmeren P, Sameith K, van de Pasch LAL, Benschop JJ, Lenstra TL, Margaritis T, O’Duibhir E, Apweiler E, van Wageningen S, Ko CW, van Heesch S, Kashani MM, Ampatziadis-Michailidis G, Brok MO, Brabers NACH, Miles AJ, Bouwmeester D, van Hooff SR, van Bakel H, Sluiters E, Bakker LV, Snel B, Lijnzaad P, van Leenen D, Groot Koerkamp MJA, Holstege FCP, Large-Scale Genetic Perturbations Reveal Regulatory Networks and an Abundance of Gene-Specific Repressors. Cell. 157, 740–752 (2014). [DOI] [PubMed] [Google Scholar]
- 23.Maclean CJ, Metzger BPH, Yang J-R, Ho W-C, Moyers B, Zhang J, Hernandez R, Deciphering the Genic Basis of Yeast Fitness Variation by Simultaneous Forward and Reverse Genetics. Molecular Biology and Evolution. 34, 2486–2502 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McAlister L, Holland MJ, Differential expression of the three yeast glyceraldehyde-3-phosphate dehydrogenase genes. J. Biol. Chem 260, 15019–15027 (1985). [PubMed] [Google Scholar]
- 25.Ringel AE, Ryznar R, Picariello H, Huang K-L, Lazarus AG, Holmes SG, Yeast Tdh3 (glyceraldehyde 3-phosphate dehydrogenase) is a Sir2-interacting factor that regulates transcriptional silencing and rDNA recombination. PLoS Genet. 9, e1003871 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Featherstone DE, Broadie K, Wrestling with pleiotropy: genomic and topological analysis of the yeast gene expression network. Bioessays. 24, 267–274 (2002). [DOI] [PubMed] [Google Scholar]
- 27.Code used for data analysis, as well as supporting datasets, are available from GitHub in the repository title “Trans-reg_pleiotropy” at https://github.com/pvz22/Trans-reg_pleiotropy, with the version of record at time of publication available at Zenodo; under DOI 10.5281/zenodo.6567260. [DOI] [Google Scholar]
- 28.Cherry JM, Hong EL, Amundsen C, Balakrishnan R, Binkley G, Chan ET, Christie KR, Costanzo MC, Dwight SS, Engel SR, Fisk DG, Hirschman JE, Hitz BC, Karra K, Krieger CJ, Miyasato SR, Nash RS, Park J, Skrzypek MS, Simison M, Weng S, Wong ED (2012) Saccharomyces Genome Database: the genomics resource of budding yeast. Nucleic Acids Res. Jan;40(Database issue):D700–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sprouffske K, Wagner A, Growthcurver: an R package for obtaining interpretable metrics from microbial growth curves. BMC Bioinformatics. 17, 172 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Martin M, Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.journal. 17, 10–12 (2011). [Google Scholar]
- 31.Patro R, Duggal G, Love MI, Irizarry RA, Kingsford C, Salmon provides fast and bias-aware quantification of transcript expression. Nat. Methods 14, 417–419 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Love MI, Huber W, Anders S, Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Smyth GK, Michaud J, Scott HS, Use of within-array replicate spots for assessing differential expression in microarray experiments. Bioinformatics. 21, 2067–2075 (2005). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.