Abstract
The histamine receptors belong to the G protein-coupled receptor (GPCR) superfamily, and play important roles in the regulation of histamine and other neurotransmitters in the central nervous system, as potential targets for the treatment of neurologic and psychiatric disorders. Here we report the crystal structure of human histamine receptor H3R bound to an antagonist PF-03654746 at 2.6 Å resolution. Combined with the computational and functional assays, our structure reveals binding modes of the antagonist and allosteric cholesterol. Molecular dynamic simulations and molecular docking of different antihistamines further elucidate the conserved ligand-binding modes. These findings are therefore expected to facilitate the structure-based design of novel antihistamines.
Subject terms: X-ray crystallography, Molecular modelling, G protein-coupled receptors
Crystal structure of human histamine receptor H3R bound to an antagonist PF-03654746 reveals the unexpected binding modes of the antagonist and allosteric cholesterol, which could facilitate the structure-based design of novel antihistamines.
Introduction
The biogenic amine histamine plays important pathophysiological roles in both the central nervous system (CNS) and periphery tissues, such as allergy, gastric acid secretion, neurotransmission, and immune response1. The action of histamine is mediated through four subtypes of G protein-coupled receptors (GPCRs), H1R, H2R, H3R, and H4R2. Antagonists of H1R and H2R have been clinically used for the treatment of allergies and gastric acid-related diseases, and the H3R inverse agonist Pitolisant (Wakix®) was approved for the treatment of narcolepsy3. While H4R antagonists are still in the clinical trials for their potential therapeutics in immune-related diseases4. Structures of H1R in complex with the agonist and antagonist have been determined5,6, providing the molecular mechanisms for ligand recognition and facilitating the structure-based design of novel drugs targeting H1R. However, the molecular mechanisms for ligand recognition with other histamine receptors were still elusive, due to the lacking of the H2R, H3R, and H4R structures.
H3R is expressed mainly in the brain and acts as an auto- or hetero-receptor in the histaminergic neurons7. As an auto-receptor, H3R modulates the histamine release by the negative feedback8. While, as a hetero-receptor, H3R regulates the release of various neurotransmitters such as dopamine, γ-aminobutyric acid (GABA), and acetylcholine9. It was suggested that H3R was associated with several physiological progresses such as sleeping and wakefulness, learning and memory, feeding, and cerebral ischemia10–12. Therefore, H3R is a potential target for the treatment of neurologic and psychiatric disorders, such as sleep disorders, Parkinson’s disease, schizophrenia, Alzheimer’s disease, and cerebral ischemia13,14. The imidazole antagonist of H3R showed poor penetration through the blood–brain barrier and unwanted interactions with hepatic cytochrome P45015. Thus, great efforts have been devoted to the development of non-imidazole H3R antagonists15. Here we determine the crystal structure of human H3R bound to a non-imidazole antagonist PF-03654746 at 2.6 Å resolution. The structure, together with the computational and functional assays, reveals the critical interactions for the ligand binding, as well as the unexpected cholesterol binding at the allosteric site, which could accelerate the structure-based design of novel antihistamines.
Results
Overall structure of H3R
To obtain the stable human H3R proteins for structure determination, the flexible regions of the N-terminal residues 1–26, intracellular loop 3 (ICL3) residues 242–346, and C-terminal residues 433–445 were truncated, and a thermostabilized apocytochrome b562RIL (BRIL) was inserted at the N-terminus. Additionally, a mutation of S1213.39K (superscript indicates residues numbers according to the Ballesteros–Weinstein scheme16) at the putative allosteric Na+ binding site was introduced to improve the homogeneity and thermostability of H3R as described in several GPCR structures determination17–22 (Supplementary Fig. 1b, c). In our calcium mobilization assays, the crystallized construct of H3R with S1213.39K mutation could be activated by histamine with ~3-fold lower efficacy but inhibited by PF-03654746 with ~18-fold higher efficacy (Supplementary Fig. 2, Supplementary Table 1), which was in consistent with our results that the crystallized H3R-PF-03654746 proteins showed significantly improved homogeneity and thermostability (Supplementary Fig. 1). The crystal structure of H3R in complex with the antagonist PF-03654746 was determined at 2.6 Å resolution (Fig. 1, Supplementary Fig. 1, Supplementary Table 3).
Fig. 1. Overall structure of H3R–PF-03654746 complex.
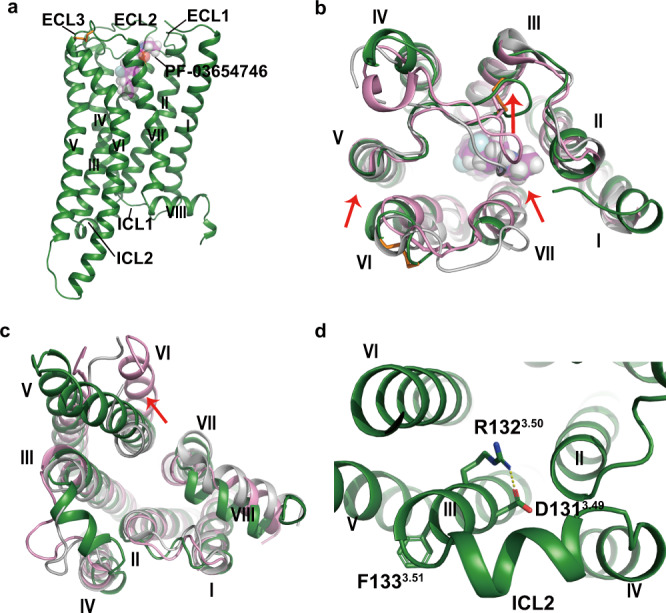
a Membrane view of H3R–PF-03654746 structure. H3R was shown in forest green ribbons. PF-03654746 was shown in a magenta sphere. The disulfide bond was shown as orange sticks. b, c Structural comparison of H3R (forest green) with inactive H1R (gray, PDB ID: 3RZE) and active H1R (pink, PDB ID: 7DFL) from extracellular view (b) and intracellular view (c). d Intracellular view showing a salt-bridge interaction (yellow dashed line) between D1313.49 and R1323.50. The red arrows indicated movements of TMs5/6 and ECL2 in the H3R structure compared to the H1R inactive structure.
The H3R structure consisted of the canonical seven transmembrane helical bundles (TMs1–7) connected by three extracellular loops (ECLs1–3) and three intracellular loops (ICLs1–3) with an amphipathic helix 8 (Fig. 1a). The ECL2 of H3R was stabilized by the conserved disulfide bridge between C1073.25 and C188ECL2, and the second disulfide bridge was found between C384ECL3 and C388ECL3 (Fig. 1a, b). Compared with the inactive H1R structure5, the extracellular tips of TM6 and TM7 in H3R moved inwards by 2.3 and 3.5 Å, respectively (Fig. 1b). Additionally, the first section of ECL2 shifted towards TM3 by 11 Å and extended from the receptor core, otherwise the antagonist PF-03654746 would clash with ECL2 if it adopted a similar conformation to that in H1R (Fig. 1b). At the intracellular side, the TM6 of H3R showed an outward movement of 2.8 Å compared to the inactive H1R, whereas the active H1R showed the TM6 outward movement of 12 Å (Fig. 1c). Moreover, the ICL2 of H3R was found to form an additional helix (Fig. 1c, d). Notably, the Y3.51 of D3.49–R3.50–Y3.51 motif in H3R was substituted by F1333.51, with the salt bridge formed between D1313.49 and R1323.50, which was a key feature of the inactive state of GPCRs23 (Fig. 1d).
PF-03654746 binding to H3R
In our H3R structure, PF-03654746 occupies a shallow pocket at the extracellular side, with clear densities for both the receptor and ligand (Fig. 2a). Although the orthosteric binding pocket of H3R is relatively shallow, an extended binding pocket (EBP) was found around TMs2/7 and ECL2 in H3R, compared to other aminergic receptors24,25 (Fig. 2a). The ligand-binding pocket of H3R is constituted by the residues mainly from TMs2/3/6/7 and ECL2 (Fig. 2b). At the extracellular side, the carbonyl and N-ethyl-carboxamide moieties of PF-03654746 extends into the EBP by forming hydrophobic and hydrogen interactions with E3957.36 and Y912.61, respectively (Fig. 2b). In our calcium mobilization assays, the E3957.36A mutant could fully abolish the PF-03654746 inhibition, while the Y912.61A mutant could significantly decrease the PF-03654746 inhibition by ~46-fold (Supplementary Fig. 3a, Supplementary Table 1). Both Y2.61 and E7.36 are located in the minor pocket of aminergic GPCRs, which were shown to determine the ligand affinity and selectivity26. Additionally, the 3-fluoro-phenyl moiety of PF-03654746 formed hydrophobic interaction with F193ECL2 (Fig. 2b). Mutating F193ECL2 to alanine could completely abolish the PF-03654746 inhibition (Supplementary Fig. 3a, Supplementary Table 1). This phenylalanine on ECL2 was suggested to determine the ligand specificity among the aminergic receptors27,28. Moreover, the hydrophobic interaction with PF-03654746 is seen with Y3746.51 (Fig. 2b). Mutagenesis of Y3746.51A could fully abolish the PF-03654746 inhibition (Supplementary Fig. 3a, Supplementary Table 1). Notably, the fluorine atom of 3-fluoro-cyclobutane of PF-03654746 engages a hydrogen bond with C18845.50, and the amine moiety of pyrrolidine of PF-03654746 forms a salt bridge with D1143.32 at the bottom of the pocket (Fig. 2b), which is highly conserved in the aminergic receptors28. Surprisingly, both D1143.32A and C18845.50A mutations displayed similar PF-03654746 inhibition on the histamine-induced calcium mobilization compared to the wild-type (Supplementary Fig. 3a, Supplementary Table 1). However, the D1143.32A and C18845.50A mutants showed ~6-fold and ~4-fold reduction of histamine activation, indicating these two residues might be involved in the binding of both histamine and PF-03654746 (Supplementary Fig. 3a, Supplementary Table 1). Indeed, D3.32 forms hydrogen bonds with histamine in H1R6.
Fig. 2. Binding modes of PF-03654746 and cholesterol to H3R.
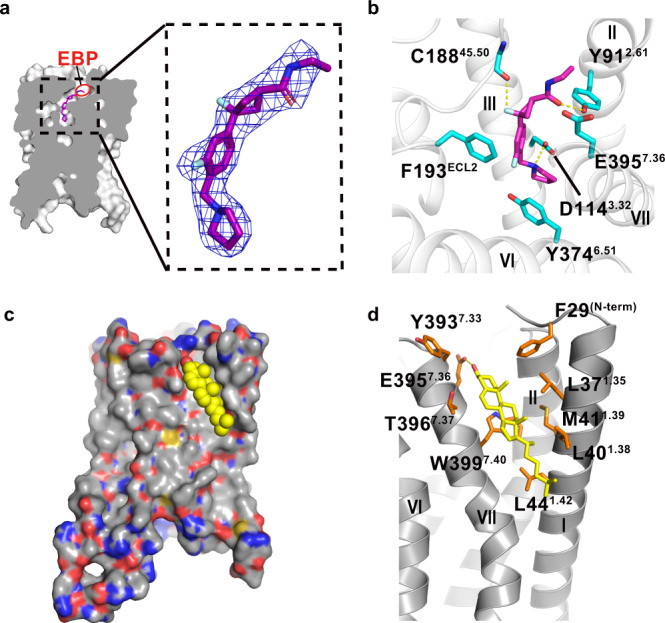
a Vertical cross section showing a shallow binding pocket in H3R. The extending binding pocket (EBP) of H3R-PF03654746 is shown in a red ellipse. |2Fo|−|Fc| electron density map for the PF-03654746 contoured at 1.0σ. b Detailed interactions of PF-03654746 in the H3R ligand-binding pocket. H3R was shown in gray ribbons, with critical residues for ligand-binding as cyan sticks and PF-03654746 as magenta sticks. Hydrogen bonds were shown as yellow dashed lines. c Surface representation of cholesterol-binding site with cholesterol shown in yellow spheres. d Detailed interactions of cholesterol with H3R. Residues critical for cholesterol binding were shown as orange sticks and cholesterol was shown as yellow sticks. The hydrogen bond was shown as yellow dashed lines.
Cholesterol binding to H3R
Cholesterol has been observed in many GPCR structures for its regulatory roles29–33, at the classical cholesterol consensus motif (CCM)34, as well as diverse binding sites35–40. In the adrenergic receptor β2AR, two cholesterols bound at the CCM stabilizing the receptor conformation34, while two other cholesterols were observed around helix 8 and TM1, modulating the β2AR dimerization39. In the histamine receptors, the cholesterol-binding site was not identified previously. In our structure, the electron density of a cholesterol molecule is observed around TM1 and TM7 of H3R (Fig. 2c). Cholesterol forms extensive hydrophobic interactions in the extrahelical pocket consisting of F29N-term, L371.35, M411.39, L401.38, L441.42, T3967.37, Y3937.33, and W3997.40. Especially, the β3-hydroxy head group of cholesterol interacts with E3957.36 through hydrogen bonding (Fig. 2d). Notably, E3957.36 also participates in the polar interactions with PF-03654736 (Fig. 2b). Our functional assays showed that mutating the negatively charged E3957.36 to uncharged alanine or positively charged arginine had little effects on the histamine activation, while completely abolishing the PF-03654746 inhibition, indicating that cholesterol binding to E3957.36 might not be critical for agonist binding and H3R activation, but might potentially to affect antagonist binding and H3R inhibition through an allosteric mode (Supplementary Fig. 3a, Supplementary Table 1).
To investigate the effects of cholesterol binding on H3R, molecular dynamics (MD) simulations were performed on H3R/PF-03654746 complex in the presence and absence of the crystal cholesterol molecule. Two systems, H3R/PF-03654746/cholesterol (hereafter referred to as CHL) and H3R/PF-03654746 (hereafter referred to as PF), were embedded in the palmitoyl oleoyl phosphatidylcholine (POPC) bilayer with a duration of 2000 ns, respectively, and each system was replicated to perform three independent simulations. A free-energy landscape was built to analyze the conformational changes in six 2-μs MD trajectories. RMSDresidues and RMSDPF, representing the root mean square deviations (RMSD) of orthosteric site residues and that of PF-03654746, respectively, were used as two collective variables of the landscape (Supplementary Fig. 4b). The small value of these parameters means the more approaching to the starting crystal conformation, while the larger value indicates obvious movements for both protein and PF-03654746.
The free-energy landscape showed three main minima corresponding to three states of the complexes: crystal-like state, state 2, and state 3 (Supplementary Fig. 4a). The crystal-like state contained snapshots from simulations CHL1, CHL2, and PF3 and displayed the smallest RMSDPF and RMSDresidues, representing the closest conformation to crystal structure. It is associated with the lowest free energy and is therefore the most stable. With larger RMSDPF and RMSDresidues, snapshots in simulation CHL3 formed state 2, and complexes from PF1 and PF2 fell into state 3. Both states were different from the crystal conformation and are characterized by higher free-energy values. In the crystal-like state, the PF-03654746-binding geometry was similar to that in the crystal structure, especially in the middle and bottom of the binding pocket (Supplementary Fig. 4a), where salt bridges with D1143.32 and hydrophobic interactions existed in every system. In the EBP, PF-03654746 was not that stable and adopted slightly different conformations, forming hydrogen bonds with Y912.61 in CHL1 and CHL2 systems or with Y942.64 in the PF3 system. Though PF-03654746 maintained the stable salt bridge with D1143.32 in state 2, its conformation changed in the middle and external parts of the pocket and only occasionally interacted with A190ECL2. For state 3, PF-03654746 totally lost its binding pose and rarely interacted with D1143.32, resulting in a random orientation in each MD trajectory. It’s noteworthy that the cholesterol molecule in CHL3 was not so stable as in CHL1 and CHL2 and eventually dissociated from its binding site at the TM1–TM7 interface (Supplementary Fig. 5a, c), so cholesterol-bound complexes only existed in simulations CHL1 and CHL2, and both of them were stabilized into the crystal-like conformations. Considering that one out of four cholesterol-unbound simulations also reproduced the crystal binding mode of PF-03654746, we came to the conclusion that cholesterol at the TM1–TM7 groove was not very stable and not the determining factor for complex stability, but bound cholesterol facilitated PF-03654746 present in the crystal pose at a higher frequency.
A significant phenomenon observed is that the conserved W3997.40 played an essential role in stabilizing the cholesterol binding and ligand–H3R interactions. W3997.40 predominantly maintained the original rotameric state (RI-I, χ1 ≈ −80° and χ2 ≈ 100°) in CHL1 and CHL2 (Supplementary Fig. 5b), and cholesterol resided stably in its site, forming a parallel π–π stacking with W3997.40 (Supplementary Fig. 5a, c). But in the CHL3 simulation, the side chain of W3997.40 flipped out of the TM1–TM7 cleft and pointed outward to the lipids at about 400 ns, resulting in a new rotamer conformation (RT-II, χ1 ≈ 175° and χ2 ≈ 100°) (Supplementary Fig. 5b, d). The side chain flipping reduced π–π stacking and caused a big steric hindrance for the bound cholesterol. As a result, cholesterol gradually dissociated from the cleft (Supplementary Fig. 5a, c). Lacking the stabilization of cholesterol, the side chain of W3997.40 turned to another conformation (RT-III) at about 1200 ns, and RMSDPF and RMSDresidues in CHL3 greatly increased at the same time (Supplementary Fig. 4b). The observations above predicted that cholesterol regulated the complex dynamics by stabilizing W3997.40 in RI-I state. To verify the role of W3997.40 in ligand binding, we further analyzed the rotameric states of W3997.40 in non-cholesterol system. As expected, W3997.40 in PF1 and PF2 underwent a certain conformational change, while W3997.40 of PF3 predominantly displayed RI-I state throughout the simulation, which should contribute to the stable conformation of H3R/PF-03654746 complex obtained in this trajectory (Supplementary Fig. 5f).
To explore how W3997.40 influenced the ligand binding, we examined its interactions with surrounding residues in the crystal structure. W3997.40 formed T-shape π–π stackings with Y912.61, which was important for the PF-03654746 binding (Supplementary Fig. 5e). Indeed, mutation of W3997.40A could completely abolish the PF-03654746 inhibition, while had little effects on the histamine activation (Supplementary Fig. 3a, Supplementary Table 1), indicating that cholesterol might affect the PF-03654746 binding mediated by the cholesterol–W3997.40–Y912.61–PF-03654746 interactions. W3997.40 and D1143.32 are completely conserved, and W4027.43 is highly conserved among monoamine receptors. Experiments have independently indicated the importance of W7.40 for the ligand binding in several GPCRs41,42. Therefore, our study provided additional support for this idea and suggested a relevance between cholesterol and the W3997.40–W4027.43–Y912.61 motif.
More importantly, cholesterol facilitated rearrangements of the TM1–TM7 interface and stabilized a polar network of cholesterol–E3957.36–R27N-term. By making extensive hydrophobic contacts with the extrahelical part of TM1 and TM7, cholesterol joined TM1 and TM7 tightly like a ‘glue’ and promoted the formation of E3957.36–R27N-term salt bridge (Supplementary Fig. 6a–c). Meanwhile, the hydroxy of cholesterol established a stable hydrogen bond with the carboxyl group of E3957.36 in our simulation, as indicated by the time dependences of their distance (Supplementary Fig. 5a). Hence, cholesterol–E3957.36–R27N-term polar network remained in CHL1 and CHL2, like in the crystal structure (Supplementary Fig. 6a). In the cholesterol-unbound simulations, only PF3 possessed the stable E3957.36–R27N-term salt bridge and similar compact conformation in TM1–TM7 interface. As for CHL3, PF1, and PF2, they showed declining stability of TM1 and TM7, as well as the E3957.36–R27N-term interaction (Supplementary Fig. 6b–e), consistent with their unstable complex states. Accordingly, the tight TM1–TM7–N-term contacts seemed to be favorable for ligand binding and cholesterol stabilized this receptor conformation through both hydrophobic and electrostatic interactions.
Conserved binding modes of H3R antagonists
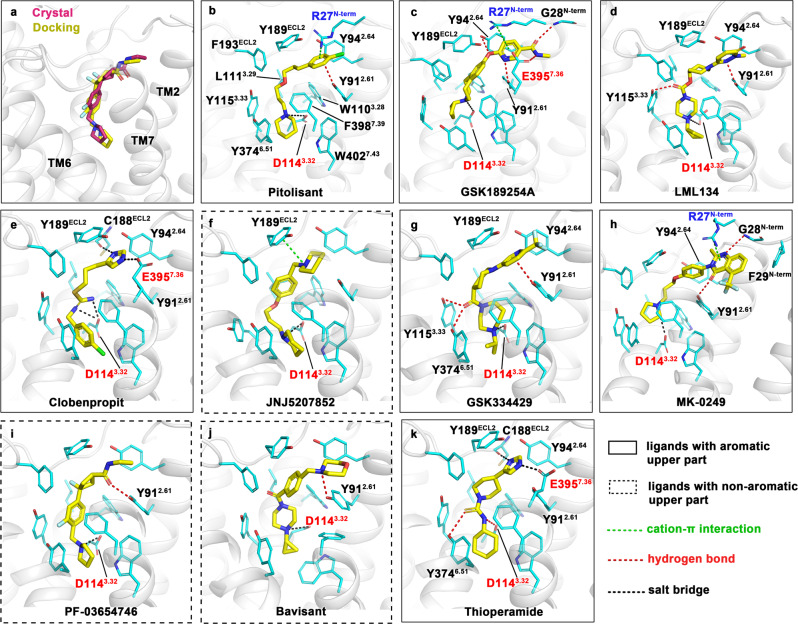
To explore the binding modes of different H3R antagonists, molecular docking studies were used to predict the binding conformations of other 9 H3R antagonists (Fig. 3). PF-03654746 was first re-docked into the protein to verify the reliability of the docking simulation, which showed the RMSD < 3.0 Å with the solved crystal structure (Fig. 3a). All ligands fit well in the binding pocket and all predicted docking scores were lower than −8.4 kcal/mol (Supplementary Table 4), which was inconsistent with the experimental Ki values of these ligands (Fig. 3b–k)1,13,43,44.
Fig. 3. Docking results of PF-03654746 and nine different H3R antagonists.
a Superimposition of PF-03654746 in the crystal structure (magenta) and the docked pose (yellow). b–k Binding modes of 10 H3R antagonists. The docked ligands were depicted as yellow sticks and arranged according to their Ki values from low to high as shown in Supplementary Table 2. Ligands with an aromatic moiety in the upper part of their binding poses were boxed with solid lines and ligands with non-aromatic upper parts were marked with dash lines. Interacting residues were presented as cyan sticks. In b, all residues involved in interactions were labeled. In c–k, only residues forming interactions with the external aromatic moiety or involved in polar contacts were labeled. Charged residues were marked in red (negative) or blue (positive). Polar interactions, including cation–π interactions, hydrogen bonds, and salt bridges, were represented by different colored dash lines, while π–π stackings were not shown.
The docking results showed a common binding pose for all ligands. Apart from the conserved salt bridges with D1143.32, docking studies revealed that favorable interactions between the aromatic upper part of ligands and residues in the EBP, as well as strong hydrophobic contacts at the bottom of the pocket, are of great importance for the ligand binding and efficacy. For all ligands, the downward heterocycle was in the hydrophobic pocket constituted by Y1153.33, Y3746.51, F3987.39, and W4027.43 (Fig. 3b). In the docked poses, conserved salt bridges were found between D1143.32 and protonated nitrogen atoms at the bottom of the binding pocket for each ligand except Thioperamide. As Thioperamide did not get protonated at the equivalent position, it only formed a hydrogen bond with D1143.32 (Fig. 3k), which might partly explain its worse inhibitive activity when compared with other antagonists (Supplementary Table 4). The middle part of the ligand was stabilized through hydrophobic interactions with L1113.29, W1103.28, F193ECL2, and Y189ECL2 (Fig. 3b), and the middle carbonyl group in GSK334429, LML134, and the carbothioamide group in Thioperamide formed additional hydrogen bonds with Y3746.51 and Y1153.33 (Fig. 3d, g, k).
In the EBP, there were two general patterns of receptor–ligand interactions. Except for PF-03654746, JNJ5207852, and Bavisant, the other seven ligands all possess an aromatic moiety in the upper part of their binding poses, which could establish favorable π–π stacking interactions and OH/π hydrogen bonds with a cluster of aromatic residues in the EBP that involve Y912.61 and Y189ECL2. This was further validated by our functional assays that the Y912.61A mutant significantly decreased the inhibition of GSK189254A and JNJ5207852 by ~25-fold and ~23-fold, respectively, and completely abolished the inhibition of Pitolisant (Supplementary Fig. 3b–d, Supplementary Table 2). While, the Y189ECL2A mutant decreased the inhibition of GSK189254A by ~88-fold and completely abolished the inhibition of Pitolisant (Supplementary Fig. 3b–d, Supplementary Table 2). On the other hand, ligands without aromatic moiety formed much fewer and weaker interactions in the EBP (Fig. 3f, i, j). Apparently, the extensive interactions benefit ligands binding and support the observation that most of the seven ligands with aromatic external moieties exert better inhibitive activity than ligands with non-aromatic groups (Fig. 3, Supplementary Table 4), highlighting the importance of aromatic rings in this part for H3R antagonists. Additionally, with larger aromatic groups, GSK189254A and MK-0249 extended to reach the TM1–TM7 interface and even interacted with G28N-term, E3957.36, and F29N-term (Fig. 3c, h). The imidazole moiety in Thioperamide and Clobenpropit also formed polar interactions with C188ECL2 and E3957.36 in H3R (Fig. 3e, k), in which the only non-conserved residue was R3417.36 in H4R (Supplementary Table 5), providing a structural basis for Thioperamide with similar affinity in H3R and H4R1. Indeed, the E3957.36A and E3957.36R mutants could fully abolish the inhibition of Thioperamide and Clobenpropit (Supplementary Fig 3e, f, Supplementary Table 2).
Further analysis suggested hydrophobic interactions at the bottom of the pocket play a role in ligand binding as well. Though JNJ5207852 and Bavisant showed similar contacts in the EBP through a protonated nitrogen atom (Fig. 3f, j), JNJ5207852 displays a much lower Ki value (Supplementary Table 4), which may be the result of stronger hydrophobic packings made by the piperidine of JNJ5207852 than the cyclopropane of Bavisant. This could also be the reason why JNJ5207852 has better activity than GSK334429 and MK-0249 in spite that they formed more contacts in the EBP (Fig. 3f, g, h). It is the same in the case of Clobenpropit and Thioperamide. With identical interactions in the EBP, Clobenpropit not only established more powerful salt bridges with D1143.32 as mentioned above but made more hydrophobic contacts through the fluorobenzene moiety at the bottom of the binding site (Fig. 3e, k).
Taken together, a combination of aromatic interactions in the EBP, salt bridges with D1143.32 and hydrophobic patterns at the bottom of the pocket stabilized H3R/antagonist complex. This exquisite binding feature rationalized the ability of Pitolisant, which possesses both a fluorobenzene group in the upper part and piperidine at the other end, to exhibit the best inhibitive activity among all ligands (Fig. 3b, Supplementary Table 4). In summary, the predicted poses of several H3R antagonists demonstrate a conserved binding feature targeting H3R, which could facilitate the future structure-based drug design.
Mechanism of H3R antagonism
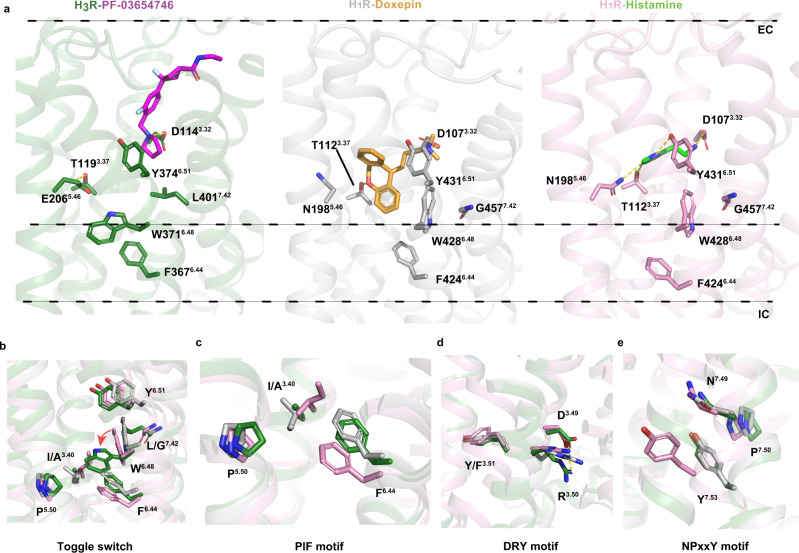
Structural comparison of our determined antagonist-bound H3R structure with the inactive doxepin-bound H1R5 and active histamine-bound H1R6 structures provides an opportunity to visualize how the antagonist inhibits H3R (Fig. 4a). A notable difference between H1R and H3R is the ligand-binding sites, where doxepin and histamine in H1R bound deeply in the ligand-binding pocket, without interactions with the extracellular part (Fig. 4a). While, in H3R, PF-03654746 occupies a shallow site near the extracellular part of the pocket, with only the pyrrolidine adopting a similar position to the primary amino group of doxepin and histamine in H1R (Fig. 4a). In the active structure of histamine-bound H1R6, three conserved residues D3.32, T3.37, and Y6.51 form extensive hydrogen bonds with histamine and pushes TM6 towards TM3 for H1R activation. In contrast, in the inactive structures of H1R5 and H3R, neither the inverse agonist doxepin in H1R nor the antagonist PF-03654746 in H3R form hydrogen bonds with Y6.51 (Fig. 4a). Y3746.51 of H3R forms hydrophobic interaction with PF-03654746 (Fig. 2b), and mutation of Y3746.51A could fully abolish the PF-03654746 inhibition, while showing little effects on histamine activation (Supplementary Fig. 3a, Supplementary Table 1), indicating Y3746.51 might be critical for PF-03654746 binding but not histamine binding to H3R. D1143.32 might be an overlapping binding site for both histamine and PF-03654746 since D1143.32A mutant showed similar PF-03654746 inhibition on the histamine-induced calcium mobilization compared to the wild-type, but a ~6-fold reduction of histamine activation (Supplementary Fig. 3a, Supplementary Table 1). T1193.37 in H3R forms two intramolecular hydrogen bonds with E2065.46, which is different from T1123.37 in H1R by forming hydrogen bonds with either doxepin or histamine (Fig. 4a). E2065.46 of H3R was suggested to form hydrogen bonds with the nitrogen atom in the imidazole ring of histamine and contribute to the binding of the selective H3R agonist with a similar imidazole ring13,45, indicating E2065.46 might be critical for the H3R activation.
Fig. 4. Mechanism of H3R antagonism.
a Superpositions of the ligand-binding pockets of H3R–PF-03654746 (H3R in forest green, PF-03654746 in violet), H1R–Doxepin (PDB ID: 3RZE, H1R in gray, Doxepin in yellow), and H1R–Histamine (PDB ID: 7DFL, H1R in pink, Histamine in green) from the membrane view. Structural comparisons of the toggle switch (b), PIF motif (c), DRY motif (d), and NPxxY motif (e) with the same colors as a.
Additionally, L4017.42 forming hydrophobic interaction with PF-03654746 in H3R corresponding to G4577.42 in H1R, which is likely to hinder the side chain of the toggle switch W3716.48 in H3R from forming a similar conformation in H1R (Fig. 4a, Supplementary Table 4). In H3R, the side chain of W3716.48 is rotated ~90° and exhibits a perpendicular conformation relative to that in the H1R structures (Fig. 4a, b). Consequently, the extracellular half of TM6 is pushed out by the outward displacement of W3716.48 and Y3746.51, thus expanding the ligand-binding pocket; contributing to the intracellular half of TM6 stabilizing an inactive state by forming the intramolecular hydrophobic interaction between W3716.48 and F3676.44 in the PIF motif. Indeed, the pocket volume of PF-03654746-bound H3R (calculated by the CASTp 3.0 server46) was similar to that of the doxepin-bound inactive H1R, but increased by ~3-fold in comparison with the histamine-bound active H1R, which is in agreement with the expansion of the extracellular binding pocket in the inactive state of H1R (Supplementary Fig. 7). Together with the intrahelical salt bridge observed between D3.49 and R3.50 in the DRY motif, and locked state of Y7.53 in the NP7.50xxY7.53 motif (Fig. 4d, e), these conformational changes resulted in an inactive state of H3R in complex with PF-03654746 (Fig. 4a, Supplementary Table 4).
Discussion
H3R plays a crucial role in controlling the release of histamine and other neurotransmitters, and many studies have shown the therapeutic potentials of H3R inverse agonists in CNS disorders10, despite its complex pharmacology13. Drug discovery targeting H3R was hampered by the lack of a three-dimensional structure to elucidate the molecular mechanisms for the ligand binding45. In this study, we reported a crystal structure of human H3R in complex with an antagonist PF-03654746, which was developed for the treatment of CNS diseases. Our structure revealed a ligand-binding mode distinct from that of the antagonist-bound H1R structure. Additionally, in combination with computational and functional assays, conserved binding modes of H3R antagonists were identified, highlighting the importance of the residues in the EBP and the hydrophobic contacts at the bottom of the pocket for the ligand binding and efficacy. Especially, a cholesterol-binding site was identified next to the ligand-binding pocket, which might be targeted by the allosteric modulators. Our results are therefore expected to facilitate the structure-based novel antihistamine drug discovery targeting H3R.
Methods
Protein engineering for structure determination
The codon-optimized human H3R gene was cloned into a modified pFastBac1 vector (Invitrogen) containing with N-terminal haemagglutinin (HA) signal sequence followed by a FLAG tag, a 10× His tag, and a tobacco etch virus (TEV) protease cleavage site. The H3R was modified by introducing S1213.39K mutation to improve the thermostability and expression. To facilitate crystallization, N terminal residues 1–26 were replaced by the thermostabilized apocytochrome b562RIL (BRIL) from Escherichia coli with mutations M7W, H102I, and R106L47. The ICL3 residues 242–346 and C terminal residues 433–445 were truncated.
Protein expression and purification
The engineered H3R protein was expressed in Spodoptera frugiperda (Sf9) insect cells (Invitrogen) using the Bac-to-Bac Baculovirus Expression System. Sf9 cells were infected at a density of 2–3 × 106 cells per ml with a multiplicity of infection 5. Cells were harvested 48 h post-infection and stored at −80 °C until use.
Frozen biomass was thawed and disrupted by extensive washing in hypotonic buffer (10 mM HEPES, pH 7.5, 10 mM MgCl2, 20 mM KCl) containing protease inhibitors (500 μM AEBSF, 1 μM E-64, 1 μM leupetain, 150 nM aprotinin) and high-osmotic buffer (10 mM HEPES, pH 7.5, 1.0 M NaCl, 10 mM MgCl2, 20 mM KCl). Purified membranes were resuspended in the hypotonic buffer with the presence of 2 mg/mL iodoacetamide at 4 °C for 30 min, and then solubilized in 50 mM HEPES, pH 7.5, 800 mM NaCl, 0.5% (w/v) n-dodecyl-β-d-maltopyranoside (DDM, Anatrace), 0.1% (w/v) cholesterol hemisuccinate (CHS, Sigma-Aldrich), and 10% (v/v) glycerol for 3 h at 4 °C. After high-speed centrifugation at 58,000×g for 1 h at 4 °C, the solubilized H3R proteins in the supernatants were incubated with TALON IMAC resin (TaKaRa) at 4 °C. After incubation overnight, the resin was then washed with 20 column volumes of washing buffer I (50 mM HEPES, pH 7.5, 800 mM NaCl, 10% (v/v) glycerol, 0.1% (w/v) lauryl maltose neopentyl glycol (LMNG, Anatrace), 0.01% (w/v) CHS, 20 mM imidazole), followed by 10 column volumes of wash buffer II (20 mM HEPES, pH 7.5, 500 mM NaCl, 5% (v/v) glycerol, 0.05% (w/v) LMNG, 0.005% (w/v) CHS, 40 mM imidazole). The protein was then eluted in 3 column volumes of elution buffer (10 mM HEPES, pH 7.5, 500 mM NaCl, 5% (v/v) glycerol, 0.01% (w/v) LMNG, 0.001% (w/v) CHS, 250 mM imidazole) and concentrated to 500 μL with a 100 kDa cutoff concentrator (Sartorius). Imidazole was removed by a PD MiniTrap G-25 column (GE Healthcare). Then, the sample was supplemented with 100 μM PF-03654746 and incubated with TEV protease overnight. The TEV protease, cleaved His-tag, and Flag-tag were removed by incubating with TALON IMAC resin (TaKaRa) at 4 °C for 2 h. The purified H3R–PF-03654746 complex protein was concentrated to ~40 mg/mL with a 100 kDa cutoff concentrator (Sartorius). The protein purity and monodispersity were tested by SDS–PAGE and analytical size-exclusion chromatography (aSEC).
Lipidic cubic phase crystallization
Purified protein was reconstituted in LCP by mixing 40% of protein with 60% of lipid (monoolein and cholesterol, 9:1, w/w) using a syringe lipid mixer. Crystallization trials were performed on a Gryphon LCP robot (ArtRobbins) by dispensing 40 nL of protein-loaded LCP on 96-well glass sandwich plates and overlaying with 800 nL precipitant solution per well. Crystals appeared after 1 day and grew to full size within 1 week in 0.1 M sodium cacodylate trihydrate, pH 6.4, 90 mM sodium citrate, 34% PEG400, and 0.005% dichloromethane. Crystals were collected directly from LCP using 50 μm micro-loops and flash-frozen in liquid nitrogen.
Data collection and structure determination
The X-ray diffraction data of crystals were collected at the BL18U1 beamline of Shanghai Synchrotron Radiation Facility, using 20 μm × 20 μm beams for 0.8 s and 1° oscillation per frame with a Pilatus3 6M detector at a wavelength of 1.0000 Å. Diffraction data were processed with HKL300048. Initial phase information was obtained by molecular replacement with CCP449 using M1R50 (PDB ID: 5CXV) and BRIL51 (PDB ID: 1M6T) as search models. Refinement was performed with COOT52 and Phenix53 using |2Fo|−|Fc| and |Fo|−|Fc| maps. Pymol (http://www.pymol.org) was used to generate all the structural images in this manuscript.
Molecular dynamics simulations
MD simulations on two systems (H3R/PF-03654746/cholesterol system, H3R/PF-03654746 system) were performed. Based on the crystal structure, we first built a complex model including H3R, PF-03654746, and cholesterol. BRIL in the crystal structure was removed and the S121K mutation was mutated back to serine. To investigate the influence of cholesterol, we removed the cholesterol molecule to build a complex model only including H3R and PF-03654746. These models were separately placed into a 110 Å × 110 Å palmitoyl oleoyl phosphatidylcholine (POPC) bilayer and the lipids located within 1 Å of the receptor were removed. Both systems were solvated in a box (110 Å × 110 Å × 110 Å) with TIP3P water molecules and 0.15 M NaCl. Each system was replicated to perform three independent simulations and each of the three simulations was run up to 2-μs.
MD simulations were carried out with GROMACS 202054 with an isothermal–isobaric (NPT) ensemble and periodic boundary conditions. The CHARMM36-CMAP force field55 was applied for protein, POPC phospholipids, cholesterol, ions, and water molecules. Ligand parameters were adapted from the CHARMM Generalized Force Field (CGenFF)56,57. For each system, stepwise energy minimizations were first performed to relieve unfavorable contacts with positional restraints imposed on i/protein, lipids, ligand, and cholesterol, ii/protein, ligand, and cholesterol, iii/mainchain atoms of protein, ligand, and cholesterol, iv/Cα atoms of protein, ligand, and cholesterol, v/no atoms. Subsequently, three parallel 50-ns equilibrations MD runs in the NPT ensemble were performed for each system with positional restraints applied in the same order as that in the energy minimization. During the equilibration, temperature and pressure were controlled using the v-rescale method58 and the Berendsen barostatv59, respectively. After equilibration, a 2-μs production run was carried out for each simulation. SETTLE constraints60 and LINCS constraints61 were applied to the hydrogen-involved covalent bonds in water molecules and in other molecules, respectively, and the time step was set to 2 fs. Electrostatic interactions were calculated with the particle-mesh Ewald (PME) algorithm62 with a real-space cutoff of 1.0 nm. The temperature was maintained at 310 K using the v-rescale method58 and the pressure was kept constant at 1 bar by semi-isotropic coupling to a Parrinello–Rahman barostat63 with τp = 2.5 ps and compressibility of 4.5 × 10−5 bar. Analysis of simulation data was conducted using PyMOL (http://www.pymol.org), tools implemented in GROMACS 2020, and in-house scripts.
Molecular docking
To investigate the interacting patterns between antagonists and H3R, we performed flexible molecular docking studies using AutoDock 464. The crystal structure of H3R reported here was used as the receptor and structures of 10 antagonists downloaded from the PubChem database were used as ligands. The receptor and ligands were respectively prepared by AutoDockTools to produce the corresponding low-energy three-dimensional conformation and the correct ionization state (pH 7.0). A 3D docking grid centered on PF-03654746 in the crystal structure was generated and residues around the pocket were treated as flexible. Then the processed antagonists were docked into the binding pocket of H3R, outputting the top 10 conformations for each ligand. The most reliable binding poses were selected according to the interaction energy and visual inspection. All results were analyzed and visualized using PyMOL (http://www.pymol.org).
Calcium mobilization assays
Calcium flux was performed as described in our previous studies. Briefly, CHO cells were co-transfected with wild-type or mutant H3R and Gqi5 using Lipofectamine 2000 according to the manufacturer’s manual. Transfected cells were seeded into a 96-well flat clear bottom black plate with a density of 25,000 cells per well and cultured overnight. Subsequently, cells were loaded with calcium dye solution from Calcium 5 assay kit (Molecular Devices) in Hanks’ balanced salt solution (20 mM HEPES, 2.5 mM probenecid in HBSS), and incubated at 37 °C for 45 min. Various concentrations of compounds were dispensed into the wells via a Flexstation III instrument (Molecular Devices). The intracellular calcium flux was detected immediately using the Flexstation III instrument (excitation at 485 nm, emission at 525 nm). Data were representative of three independent experiments and analyzed using GraphPad Prism 9.3.1.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Supplementary information
Acknowledgements
H.Z. is supported by the National Key R&D Program of China (2018YFA0508100), the National Natural Science Foundation of China (81722044, 91753115, 21778049, 81861148018), and the National Science and Technology Major Project of China (2018ZX09711002). We thank W.Q., Q.X., and other staff from the BL18U1 beamline of the National Facility for Protein Science in Shanghai (NFPS) at the Shanghai Synchrotron Radiation Facility, for assistance during data collection. We thank W.L. and M.L. from Shanghai Yuyao Biotech Ltd. for their assistance on the calcium mobilization assays.
Source data
Author contributions
X.P. designed, expressed, purified, and crystallized the protein, collected the X-ray diffraction data. L.Y. and M.L. performed the computational assays. Z.L., S.L., and S.M. assisted in protein expression, purification, and crystallization. Z.C. supervised the functional assays. H.Z. conceived and supervised the project, and determined the structures. X.P., L.Y., Z.C., and H.Z. wrote the manuscript with input from all other authors.
Peer review
Peer review information
Nature Communications thanks Giovanni Bottegoni, So Iwata and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Peer reviewer reports are available.
Data availability
The data that support this study are available from the corresponding author upon reasonable request. The structural data generated in this study have been deposited in the Protein Data Bank (http://www.pdb.org/) under accession code 7F61. The other data generated in this study are provided in the Supplementary Information and Source Data file. Source data are provided with this paper.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version contains supplementary material available at 10.1038/s41467-022-33880-y.
References
- 1.Panula P, et al. International union of basic and clinical pharmacology. XCVIII. Histamine receptors. Pharm. Rev. 2015;67:601–655. doi: 10.1124/pr.114.010249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Haas H, Panula P. The role of histamine and the tuberomamillary nucleus in the nervous system. Nat. Rev. Neurosci. 2003;4:121–130. doi: 10.1038/nrn1034. [DOI] [PubMed] [Google Scholar]
- 3.Panula P. Histamine receptors, agonists, and antagonists in health and disease. Handb. Clin. Neurol. 2021;180:377–387. doi: 10.1016/B978-0-12-820107-7.00023-9. [DOI] [PubMed] [Google Scholar]
- 4.Tiligada E, Ennis M. Histamine pharmacology: from Sir Henry Dale to the 21st century. Br. J. Pharm. 2020;177:469–489. doi: 10.1111/bph.14524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shimamura T, et al. Structure of the human histamine H1 receptor complex with doxepin. Nature. 2011;475:65–70. doi: 10.1038/nature10236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xia R, et al. Cryo-EM structure of the human histamine H(1) receptor/G(q) complex. Nat. Commun. 2021;12:2086. doi: 10.1038/s41467-021-22427-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nuutinen S, Panula P. Histamine in neurotransmission and brain diseases. Adv. Exp. Med. Biol. 2010;709:95–107. doi: 10.1007/978-1-4419-8056-4_10. [DOI] [PubMed] [Google Scholar]
- 8.Arrang JM, Garbarg M, Schwartz JC. Auto-inhibition of brain histamine release mediated by a novel class (H3) of histamine receptor. Nature. 1983;302:832–837. doi: 10.1038/302832a0. [DOI] [PubMed] [Google Scholar]
- 9.Haas HL, Sergeeva OA, Selbach O. Histamine in the nervous system. Physiol. Rev. 2008;88:1183–1241. doi: 10.1152/physrev.00043.2007. [DOI] [PubMed] [Google Scholar]
- 10.Schlicker E, Kathmann M. Role of the histamine H(3) receptor in the central nervous system. Handb. Exp. Pharm. 2017;241:277–299. doi: 10.1007/164_2016_12. [DOI] [PubMed] [Google Scholar]
- 11.Passani MB, Lin JS, Hancock A, Crochet S, Blandina P. The histamine H3 receptor as a novel therapeutic target for cognitive and sleep disorders. Trends Pharm. Sci. 2004;25:618–625. doi: 10.1016/j.tips.2004.10.003. [DOI] [PubMed] [Google Scholar]
- 12.Yan H, et al. Histamine H3 receptors aggravate cerebral ischaemic injury by histamine-independent mechanisms. Nat. Commun. 2014;5:3334. doi: 10.1038/ncomms4334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nieto-Alamilla G, Márquez-Gómez R, García-Gálvez AM, Morales-Figueroa GE, Arias-Montaño JA. The histamine H3 receptor: structure, pharmacology, and function. Mol. Pharm. 2016;90:649–673. doi: 10.1124/mol.116.104752. [DOI] [PubMed] [Google Scholar]
- 14.Hu W, Chen Z. The roles of histamine and its receptor ligands in central nervous system disorders: an update. Pharm. Ther. 2017;175:116–132. doi: 10.1016/j.pharmthera.2017.02.039. [DOI] [PubMed] [Google Scholar]
- 15.Berlin M, Boyce CW, Ruiz Mde L. Histamine H3 receptor as a drug discovery target. J. Med. Chem. 2011;54:26–53. doi: 10.1021/jm100064d. [DOI] [PubMed] [Google Scholar]
- 16.Ballesteros JA, Weinstein H. Integrated methods for the construction of three-dimensional models and computational probing of structure-function relations in G protein-coupled receptors. Methods Neurosci. 1995;25:366–428. doi: 10.1016/S1043-9471(05)80049-7. [DOI] [Google Scholar]
- 17.Toyoda Y, et al. Ligand binding to human prostaglandin E receptor EP(4) at the lipid-bilayer interface. Nat. Chem. Biol. 2019;15:18–26. doi: 10.1038/s41589-018-0131-3. [DOI] [PubMed] [Google Scholar]
- 18.Kimura KT, et al. Structures of the 5-HT(2A) receptor in complex with the antipsychotics risperidone and zotepine. Nat. Struct. Mol. Biol. 2019;26:121–128. doi: 10.1038/s41594-018-0180-z. [DOI] [PubMed] [Google Scholar]
- 19.Shao Z, et al. Structure of an allosteric modulator bound to the CB1 cannabinoid receptor. Nat. Chem. Biol. 2019;15:1199–1205. doi: 10.1038/s41589-019-0387-2. [DOI] [PubMed] [Google Scholar]
- 20.Yan W, et al. Structure of the human gonadotropin-releasing hormone receptor GnRH1R reveals an unusual ligand binding mode. Nat. Commun. 2020;11:5287. doi: 10.1038/s41467-020-19109-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Im D, et al. Structure of the dopamine D(2) receptor in complex with the antipsychotic drug spiperone. Nat. Commun. 2020;11:6442. doi: 10.1038/s41467-020-20221-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Qin J, et al. Molecular mechanism of agonism and inverse agonism in ghrelin receptor. Nat. Commun. 2022;13:300. doi: 10.1038/s41467-022-27975-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Palczewski K, et al. Crystal structure of rhodopsin: a G protein-coupled receptor. Science. 2000;289:739–745. doi: 10.1126/science.289.5480.739. [DOI] [PubMed] [Google Scholar]
- 24.Chen X, et al. Molecular mechanism for ligand recognition and subtype selectivity of α(2C) adrenergic receptor. Cell Rep. 2019;29:2936–2943.e2934. doi: 10.1016/j.celrep.2019.10.112. [DOI] [PubMed] [Google Scholar]
- 25.Qu L, et al. Structural basis of the diversity of adrenergic receptors. Cell Rep. 2019;29:2929–2935.e2924. doi: 10.1016/j.celrep.2019.10.088. [DOI] [PubMed] [Google Scholar]
- 26.Kooistra AJ, Kuhne S, de Esch IJ, Leurs R, de Graaf C. A structural chemogenomics analysis of aminergic GPCRs: lessons for histamine receptor ligand design. Br. J. Pharm. 2013;170:101–126. doi: 10.1111/bph.12248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Michino M, et al. What can crystal structures of aminergic receptors tell us about designing subtype-selective ligands? Pharm. Rev. 2015;67:198–213. doi: 10.1124/pr.114.009944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vass M, et al. Aminergic GPCR-ligand interactions: a chemical and structural map of receptor mutation data. J. Med. Chem. 2019;62:3784–3839. doi: 10.1021/acs.jmedchem.8b00836. [DOI] [PubMed] [Google Scholar]
- 29.Yao Z, Kobilka B. Using synthetic lipids to stabilize purified beta2 adrenoceptor in detergent micelles. Anal. Biochem. 2005;343:344–346. doi: 10.1016/j.ab.2005.05.002. [DOI] [PubMed] [Google Scholar]
- 30.Muth S, Fries A, Gimpl G. Cholesterol-induced conformational changes in the oxytocin receptor. Biochem. J. 2011;437:541–553. doi: 10.1042/BJ20101795. [DOI] [PubMed] [Google Scholar]
- 31.Qiu Y, Wang Y, Law PY, Chen HZ, Loh HH. Cholesterol regulates micro-opioid receptor-induced beta-arrestin 2 translocation to membrane lipid rafts. Mol. Pharm. 2011;80:210–218. doi: 10.1124/mol.110.070870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Taghon GJ, Rowe JB, Kapolka NJ, Isom DG. Predictable cholesterol binding sites in GPCRs lack consensus motifs. Structure. 2021;29:499–506.e493. doi: 10.1016/j.str.2021.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Duncan AL, Song W, Sansom MSP. Lipid-dependent regulation of ion channels and G protein-coupled receptors: insights from structures and simulations. Annu. Rev. Pharm. Toxicol. 2020;60:31–50. doi: 10.1146/annurev-pharmtox-010919-023411. [DOI] [PubMed] [Google Scholar]
- 34.Hanson MA, et al. A specific cholesterol binding site is established by the 2.8 A structure of the human beta2-adrenergic receptor. Structure. 2008;16:897–905. doi: 10.1016/j.str.2008.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jazayeri A, et al. Extra-helical binding site of a glucagon receptor antagonist. Nature. 2016;533:274–277. doi: 10.1038/nature17414. [DOI] [PubMed] [Google Scholar]
- 36.Song G, et al. Human GLP-1 receptor transmembrane domain structure in complex with allosteric modulators. Nature. 2017;546:312–315. doi: 10.1038/nature22378. [DOI] [PubMed] [Google Scholar]
- 37.Zhang Y, et al. Cryo-EM structure of the activated GLP-1 receptor in complex with a G protein. Nature. 2017;546:248–253. doi: 10.1038/nature22394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lu J, et al. Structural basis for the cooperative allosteric activation of the free fatty acid receptor GPR40. Nat. Struct. Mol. Biol. 2017;24:570–577. doi: 10.1038/nsmb.3417. [DOI] [PubMed] [Google Scholar]
- 39.Cherezov V, et al. High-resolution crystal structure of an engineered human beta2-adrenergic G protein-coupled receptor. Science. 2007;318:1258–1265. doi: 10.1126/science.1150577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang D, et al. Two disparate ligand-binding sites in the human P2Y1 receptor. Nature. 2015;520:317–321. doi: 10.1038/nature14287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Roth BL, Shoham M, Choudhary MS, Khan N. Identification of conserved aromatic residues essential for agonist binding and second messenger production at 5-hydroxytryptamine2A receptors. Mol. Pharmacol. 1997;52:259–266. doi: 10.1124/mol.52.2.259. [DOI] [PubMed] [Google Scholar]
- 42.Rivail L, et al. New insights into the human 5-HT4 receptor binding site: exploration of a hydrophobic pocket. Br. J. Pharm. 2004;143:361–370. doi: 10.1038/sj.bjp.0705950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Troxler T, et al. The discovery of LML134, a histamine H3 receptor inverse agonist for the clinical treatment of excessive sleep disorders. ChemMedChem. 2019;14:1238–1247. doi: 10.1002/cmdc.201900176. [DOI] [PubMed] [Google Scholar]
- 44.Leurs R, Vischer HF, Wijtmans M, de Esch IJ. En route to new blockbuster anti-histamines: surveying the offspring of the expanding histamine receptor family. Trends Pharm. Sci. 2011;32:250–257. doi: 10.1016/j.tips.2011.02.004. [DOI] [PubMed] [Google Scholar]
- 45.Kiss R, Keserű GM. Structure-based discovery and binding site analysis of histamine receptor ligands. Expert Opin. Drug Discov. 2016;11:1165–1185. doi: 10.1080/17460441.2016.1245288. [DOI] [PubMed] [Google Scholar]
- 46.Tian W, Chen C, Lei X, Zhao J, Liang J. CASTp 3.0: computed atlas of surface topography of proteins. Nucleic Acids Res. 2018;46:W363–w367. doi: 10.1093/nar/gky473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chun E, et al. Fusion partner toolchest for the stabilization and crystallization of G protein-coupled receptors. Structure. 2012;20:967–976. doi: 10.1016/j.str.2012.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Minor W, Cymborowski M, Otwinowski Z, Chruszcz M. HKL-3000: the integration of data reduction and structure solution—from diffraction images to an initial model in minutes. Acta Crystallogr. D Biol. Crystallogr. 2006;62:859–866. doi: 10.1107/S0907444906019949. [DOI] [PubMed] [Google Scholar]
- 49.Usón I, Ballard CC, Keegan RM, Read RJ. Integrated, rational molecular replacement. Acta Crystallogr. D Struct. Biol. 2021;77:129–130. doi: 10.1107/S2059798321001339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Thal DM, et al. Crystal structures of the M1 and M4 muscarinic acetylcholine receptors. Nature. 2016;531:335–340. doi: 10.1038/nature17188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chu R, et al. Redesign of a four-helix bundle protein by phage display coupled with proteolysis and structural characterization by NMR and X-ray crystallography. J. Mol. Biol. 2002;323:253–262. doi: 10.1016/S0022-2836(02)00884-7. [DOI] [PubMed] [Google Scholar]
- 52.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Adams PD, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Abraham MJ, et al. GROMACS: high performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX. 2015;1-2:19–25. doi: 10.1016/j.softx.2015.06.001. [DOI] [Google Scholar]
- 55.Huang J, MacKerell AD., Jr. CHARMM36 all-atom additive protein force field: validation based on comparison to NMR data. J. Comput. Chem. 2013;34:2135–2145. doi: 10.1002/jcc.23354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Vanommeslaeghe K, MacKerell AD. Automation of the CHARMM General Force Field (CGenFF) I: bond perception and atom typing. J. Chem. Inf. Model. 2012;52:3144–3154. doi: 10.1021/ci300363c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Vanommeslaeghe K, Raman EP, MacKerell AD. Automation of the CHARMM General Force Field (CGenFF) II: assignment of bonded parameters and partial atomic charges. J. Chem. Inf. Model. 2012;52:3155–3168. doi: 10.1021/ci3003649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bussi, G., Donadio. D. & Parrinello, M. Canonical sampling through velocity rescaling. J. Chem. Phys.126, 014101 (2007). 10.1063/1.2408420. [DOI] [PubMed]
- 59.Berendsen HJC, Postma JPM, Vangunsteren WF, Dinola A, Haak JR. Molecular-dynamics with coupling to an external bath. J. Chem. Phys. 1984;81:3684–3690. doi: 10.1063/1.448118. [DOI] [Google Scholar]
- 60.Miyamoto S, Kollman PA. Settle—an analytical version of the shake and rattle algorithm for rigid water models. J. Comput. Chem. 1992;13:952–962. doi: 10.1002/jcc.540130805. [DOI] [Google Scholar]
- 61.Hess B, Bekker H, Berendsen HJC, Fraaije JGEM. LINCS: a linear constraint solver for molecular simulations. J. Comput. Chem. 1997;18:1463–1472. doi: 10.1002/(SICI)1096-987X(199709)18:12<1463::AID-JCC4>3.0.CO;2-H. [DOI] [Google Scholar]
- 62.Essmann U, et al. A smooth particle mesh Ewald method. J. Chem. Phys. 1995;103:8577–8593. doi: 10.1063/1.470117. [DOI] [Google Scholar]
- 63.Parrinello M, Rahman A. Polymorphic transitions in single-crystals—a new molecular-dynamics method. J. Appl. Phys. 1981;52:7182–7190. doi: 10.1063/1.328693. [DOI] [Google Scholar]
- 64.Morris GM, et al. AutoDock4 and AutoDockTools4: automated docking with selective receptor flexibility. J. Comput. Chem. 2009;30:2785–2791. doi: 10.1002/jcc.21256. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support this study are available from the corresponding author upon reasonable request. The structural data generated in this study have been deposited in the Protein Data Bank (http://www.pdb.org/) under accession code 7F61. The other data generated in this study are provided in the Supplementary Information and Source Data file. Source data are provided with this paper.