Abstract
Depressive disorders (DDs) are an increasingly common health problem that affects all age groups. DDs pathogenesis is multifactorial. However, it was proven that stress is one of the most important environmental factors contributing to the development of these conditions. In recent years, there has been growing interest in the role of the glutamatergic system in the context of pharmacotherapy of DDs. Thus, it has become increasingly important to explore the functioning of excitatory synapses in pathogenesis and pharmacological treatment of psychiatric disorders (including DDs). This knowledge may lead to the description of new mechanisms of depression and indicate new potential targets for the pharmacotherapy of illness. An excitatory synapse is a highly complex and very dynamic structure, containing a vast number of proteins. This review aimed to discuss in detail the role of the key postsynaptic proteins (e.g., NMDAR, AMPAR, mGluR5, PSD-95, Homer, NOS etc.) in the excitatory synapse and to systematize the knowledge about changes that occur in the clinical course of depression and after antidepressant treatment. In addition, a discussion on the potential use of ligands and/or modulators of postsynaptic proteins at the excitatory synapse has been presented.
Keywords: PSD proteins, AMPAR, NMDAR, mGluR5, NOS, Homer, depression, animal models of depression, human study, excitatory synapse
1. Introduction
Depressive disorders (DDs) are widespread mental illnesses worldwide and pose a significant economic and psychosocial problem. The World Health Organization (WHO) estimates that depression affects about 3.8% of the human population (with a prevalence of 5% among adults), and it increases with age (in people over 60, its frequency is 5.7%) [1,2]. It has also been observed that the lifetime risk of developing DDs is twice as high in women as in men [3]. Moreover, mental illnesses (including DDs) are important health problem among adolescents and represent one of the leading causes of disease and disability in this age group [4,5,6]. Over 50 years ago, monoamine theory, assuming that symptoms of depression were related with deficiency or imbalances of monoamines systems, i.e., serotonin, norepinephrine, dopamine, was described. To this day, drugs able to modify (increase) the brain concentration of these neurotransmitters represent the most used type of pharmacotherapy [7,8]. Unfortunately, this therapy has many disadvantages, e.g., it takes weeks or months to achieve a therapeutic effect, low remission rate, and a high risk of relapse after responding to treatment [3,9]. An additional problem is the prevalence of treatment-resistant depression (TRD), characterized as the failure to achieve an inadequate response to at least two standard antidepressants. This condition is common in clinical practice. Up to 40% of major depressive disorder (MDD) patients suffer from TRD [10,11,12,13]. These factors can discourage patients from taking antidepressants regularly or even lead to their discontinuation, significantly reducing the chance of remission and triggering several complications including suicide [14]. A better understanding of the molecular mechanisms underlying DDs, including MDD and bipolar disorder (BD), seems to be necessary to obtain new, more effective drugs (both antidepressant and anxiolytic profile) [3,15,16,17].
Many environmental factors contribute to the development of DDs, among which stress is particularly important [18,19,20,21]. Numerous studies on depressed patients and animal models based on stress-related procedures have shown that specific brain areas (i.e., prefrontal cortex, hippocampus, amygdala, insula) have an altered volume [22,23,24,25,26,27,28]. It’s well documented that stress factors impair the expression of neurotrophins and cause an increase in the level of the pro-inflammatory cytokines, which may result in atrophy, depleted neurogenesis, and consequently changes in neuroplasticity [21,29,30,31,32,33,34]. Moreover, it was shown that acute stress causes an increase in extracellular glutamate (Glu) levels in the hippocampus and medial prefrontal cortex (mPFC), which may lead to excitotoxicity [35,36,37]. These results also suggest that imbalance between excitatory and inhibitory neurotransmission can be a potential substrate of DDs. The truth of the above evidence is confirmed by the ever-growing interest in the role of the glutamatergic system over the decades [15,38,39,40]. A thorough understanding of the mechanisms responsible for the Glu metabolism and its influence on postsynaptic proteins at the excitatory synapse is a new research direction for achieving effective pharmacotherapy of DDs [13,15,41,42], as evidenced by the growing number of clinical studies documenting the rapid and robust antidepressant effect of ketamine (N-methyl-D-aspartate receptors (NMDAR) antagonist with additional effects on α-Amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid receptors (AMPAR), hyperpolarization-activated cyclic nucleotide-gated (HCN) channels, L-type voltage-dependent calcium channel (L-VDCC), opioid receptors, and monoaminergic receptors) [43,44,45].
Glu acts through ionotropic and metabotropic receptors. However, NMDARs and AMPARs seem to be the most important for synaptic plasticity [46,47,48,49]. Synaptic plasticity, that is, changes in the onset or magnitude of long-term potentiation (LTP) or long-term depression (LTD), can be regulated by changing the number, types, or properties of these receptors in the postsynaptic membrane. AMPARs and NMDARs trafficking underlie activity-induced changes in synaptic transmission, and therefore their abundance at synapses can significantly enhance or weaken it [50,51,52,53]. The excitatory synapse is a highly dynamic structure in which receptors constantly circulate between the synaptic membrane and the cytoplasm as well as between the extra- and synaptic matrix, while postsynaptic density (PSD) proteins as well as post-translational modifications of the polypeptide chains of the receptors’ subunits play an important role in locating them and transmitting signals inside the cell [49,53,54,55,56,57]. PSD proteins modulate the signaling cascade by linking synaptic transmission from presynaptic neurons and neurotransmitter systems, mainly through by NMDARs, AMPARs, and group I metabotropic Glu receptors (especially mGluR5) [58,59,60,61]. A thorough understanding of the mechanisms responsible for the Glu turnover and its influence on postsynaptic proteins at the excitatory synapse is a new target of direction effective pharmacotherapy of DDs [41]. There is much evidence of changes in postsynaptic proteins at the excitatory synapse in depressive disorders, both in human and animal tissues. On the other hand, there are many inconsistencies in these findings. Taking this into account, the main goal of this review was to synthesize knowledge about changes in both PSD protein levels and post-translational modifications in MDD and BD patients, as well as in animal models of depression, and to determine whether these changes are characteristic of selected areas of the brain. In addition, we also reviewed the changes in these proteins following the administration of antidepressants (or compounds with antidepressant-like activity), and the expression of the genes encoding them.
2. The Role of Postsynaptic Density Proteins in the Excitatory Synapse
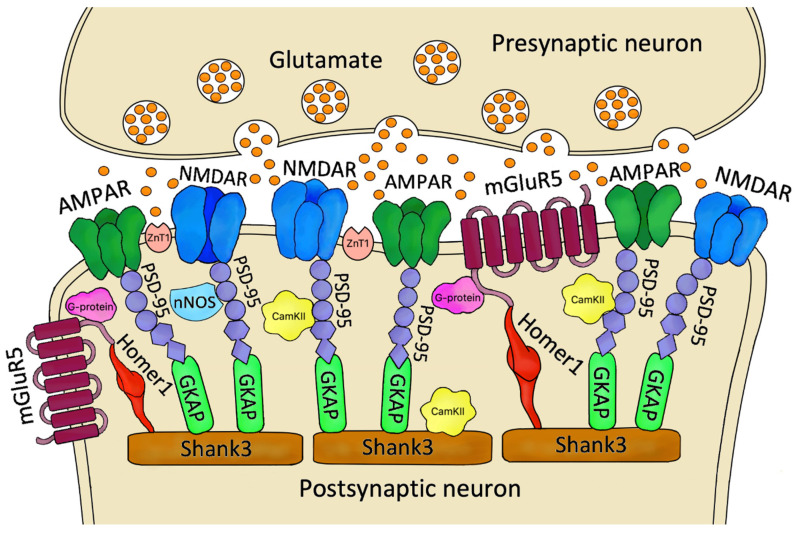
PSD is an electron-dense construction with a thickness of 20–50 nm (excitatory synapse in the hippocampus) located mainly on dendritic spines under the postsynaptic membrane, which contains many proteins that function as receptors, scaffolding and cytoskeletal elements, adhesion, signaling enzymes, and their regulators [62,63] (Figure 1). PSD is also a space whose components undergo dynamic changes, such as phosphorylation, diffusion, membrane insertion, as well as coupling and uncoupling in response to neuronal stimulation [62]. Numerous studies have shown that dysregulation within the PSD underlies many central nervous system diseases, e.g., DDs, schizophrenia, autism spectrum disorder, or neurodegenerative diseases [64,65,66].
Figure 1.
Schematic of the organization of selected PSD proteins. Released from synaptic vesicles Glu interacts with receptors on the postsynaptic neuron, specifically: AMPARs, NMDARs and mGluR5. The function of these receptors is modulated by various compounds, including nNOS, CamKII and transporters of many molecules, such as ZnT-1. An essential component of the PSD that maintains the proper proportions and alignment of the above elements are scaffold proteins, e.g., PSD-95, GKAP, Homer-1, Shank3, which bind to receptors and each other, thus performing a stabilizing function. Abbreviations: PSD—postsynaptic density; AMPAR—α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid receptor; NMDAR—N-methyl-D-aspartate receptor; mGluR5—metabotropic Glu receptor 5; nNOS—neuronal nitric oxide synthase; CamKII—calcium/calmodulin-dependent protein kinase II; ZnT1—Zn2+ transporter 1; PSD-95- postsynaptic density protein 95; GKAP- guanylate kinase-associated protein; Homer-1—Homer protein homolog 1; Shank3—SH3 and multiple ankyrin repeat domains 3.
NMDAR has a tetrameric structure and comprises two GluN1 subunits and two GluN2 subunits or, less frequently, GluN3 subunits [67] The GluN1 subunit is encoded by the GRIN1 gene. The GluN2 subunits occur in four forms: GluN2A-D are encoded sequentially by the GRIN2A-D genes, and the GluN3A and GluN3B subunits are encoded by the GRIN3A-B genes [67,68]. The occurrence of individual subunits is dependent on various factors, e.g., brain area and developmental stage. For example, GluN2B expression already occurs in fetal life in the brain. Still, with age in humans, there is a decline in GluN2B expression in favor of GluN2A in the CA1 and CA3 regions of the hippocampus, and finally, GluN2B in adults localizes in the forebrain. In contrast, GluN2A expression is widespread in the central nervous system (CNS). GluN2C expression is only postnatal and particularly abundant in the cerebellum, and GluN2D is predominantly embryonic in the midbrain [67,69]. Each NMDAR subunit comprises an extracellular N-terminal, three transmembrane domains (M1, M3, M4), a M2 re-entrant loop that forms part of the ion channel, and an intracellular C-terminal through which the subunit interacts with other PSD proteins [69,70]. Analysis of the GluN2 C-terminal revealed that it is five-fold longer in vertebrates than in invertebrates, which may significantly affect NMDAR-mediated signaling [70]. The action on NMDAR in the limbic system is modulated, among other things, by zinc ions (Zn2+), whose homeostasis is dependent on zinc transporters [71]. The ubiquitous Zn2+ transporter 1 (ZnT-1), encoded by the SLC30A1 gene, belongs to the solute carriers 30a (SLC30a) family and is the only transporter of this family located in the plasma membrane. ZnT-1 transports Zn2+ into extracellular space, thus exerting a protective effect and preventing toxicity by zinc accumulation in the cell [72]. Multiple zinc-deficiency animal models and postmortem studies have confirmed the significant role of zinc in the development of DDs [73,74].
Like NMDAR, AMPAR is also assembled as tetramer and consists of a combination of four subunits, GluA1–4 encoded by GRIA1–4 genes [75]. The expression of individual AMPAR subunits is function-dependent and region-specific. For example, GluA2 is the brain’s most widely distributed AMPAR subunit, which translates to the occurrence in mammals of mostly AMPAR complexes consisting of: GluA1/2, GluA2/3, and GluA1/2/3 [76]. GluA2 is essential for correct AMPAR function because its deletion results in increased calcium permeability and intracellular blockade by polyamines [75,77]. The presence of the GluA2 subunit in AMPAR prevents the influx of divalent cations, such as Ca2+ and Zn2+ [78]. Within the second transmembrane domain of the GluA2 subunit, there is positively charged arginine (R) at the Q/R site, which is not encoded at the genomic level, but is generated by RNA editing. Q/R editing is performed very efficiently in most of the GluA2 subunits of mammalian neurons while the equivalent position of the other AMPAR subunits is typically preserved as glutamine (Q) in its unmodified form [79]. The presence of a positively charged R residue in GluA-containing AMPARs makes the channel impermeable to Ca2+, slows down the kinetics of the channel and reduces its conductivity, increases the amplitude of synaptic events, enhances neuronal excitability, and regulates AMPAR trafficking and anchoring. Since the number of calcium-permeable AMPARs in the adult brain is very small, it is expected that even a slight change in their expression will have a significant impact on synaptic transmission and the functioning of neural circuits [80]. Functional proteomic analysis showed that GluA1 and GluA2 accounted for approximately 80% of all AMPAR subunits in the hippocampus, while in the cerebellum, the GluA4 subunit was the most significant (64%). In the cortex and striatum, GluA2 is the most abundant (45%), and the proportion of GluA1 and GluA3 is estimated to be about 21–27% [81]. Each AMPAR subunit consists of four domains: extracellular N-terminal domain (NTD), ligand-binding domain (LBD), transmembrane domain that forms the ion channel (TBD), and cytoplasmic C-terminal domain (CTD), through which other PSD proteins communicate [82,83]. Interestingly, the STAR*D study found a significant association between the occurrence of suicidal thoughts in MDD patients treated with citalopram and the presence of rs4825476 (single nucleotide polymorphism) in the GRIA3 gene [84].
AMPARs and NMDARs are the central receptors in the PSD that determine the proper functioning of the excitatory synapse. After an appropriately strong stimulus, Glu is released from the presynaptic neuron binds to binding sites on receptors and causes several conformational changes and ion flux, especially sodium Na+, potassium K+, and calcium Ca2+ [85]. NMDAR-mediated Ca2+ influx is responsible for a cascade of activation of many proteins, including calcium-dependent enzymes. One of these is calcium/calmodulin-dependent protein kinase II (CamKII), which acts as a signaling molecule and is crucial to modifying the actin cytoskeleton and synaptic plasticity. In mammals, CamKII is present in four isoforms: CamKIIα, CamKIIβ, CamKIIγ, CamKIIδ encoded by CAMK2A-D genes [86]. CamKIIα is the most common isoform in the forebrain, while CamKIIβ is particularly abundant in the cerebellum [67]. Activated CamKII contributes to the translocation of intracellular AMPARs to the PSD and phosphorylation of the GluA1 (S831) subunit, leading to enhanced synaptic transmission [75]. Additionally, CamKIIα, displaced from the cytoplasm to the PSD, also interacts with the GluN2B subunit, which is necessary for the induction of LTP [87].
In addition to rapid neurotransmission via ionotropic receptors, metabotropic receptors are also crucial at the excitatory synapse, among which metabotropic Glu receptor 5 (mGluR5) plays an important function in the development of DDs [88]. mGluR5 is encoded by GRM5 gene, belongs to the family of G-protein coupled receptors (GPCRs), and, together with mGluR1, is part of group 1 and mainly localized postsynaptically [89,90]. A characteristic feature of mGluRs is their occurrence in the form of homodimers and their structure because they contain a large extracellular domain at the N-terminus called a Venus flytrap (VFT), which possesses a ligand-binding site and, via a cystine-rich domain (CRD), binds to the 7-transmembrane domain (7TM). There is a C-terminal domain in the cytoplasm of mGluR, which allows the receptor to interact with other PSD proteins [91,92]. After Glu binds to mGluR5, a series of conformational changes occur, leading to activation of the phospholipase C (PLC) pathway and production of secondary messengers, e.g., inositol 1,4,5-triphosphate (IP3) and diacylglycerol (DAG) consequently responsible for slow neurotransmission [93]. The effect of IP3 is a Ca2+ influx, which affects calcium-dependent proteins such as CamKII [90,94]. Interestingly, mGluR5 also involves the NMDAR complex, as Jin et al. showed that the application of (RS)3,5-dihydroxyphenylglycine (3,5-DHPG, mGluR5 agonist) resulted in increased expression of membrane GluN1 and GluN2B subunits and reduced their levels intracellularly. This effect was abolished when 3-((2-methyl-1,3-thiazol-4-yl)ethynyl)pyridine hydrochloride (MTEP, mGluR5 selective antagonist) was used, indicating an essential role for mGluR5 as a molecule that determines the movement of NMDAR subunits to the surface [94].
To maintain the correct receptors composition, a balanced scaffold with many support proteins is necessary. One of them, especially widespread in forebrain, is postsynaptic density protein 95 (PSD-95), which is responsible for stabilizing and binding other PSD proteins [67]. PSD-95 is encoded by DLG-4 (discs large homolog 4) gene and belongs to the MAGUK (membrane-associated guanylate kinases) superfamily [65]. PSD-95 in its structure contains three PDZ (PSD-95/disc large/zonula occludens-1) domains followed sequentially by single SH3 (Src homology 3) and GK (guanylate kinase-like) domains [95]. PDZ domains allow PSD-95 to interact with many PSD proteins, including receptor (e.g., NMDAR, AMPAR, serotonin 5-HT2, dopamine D2) subunits, thus exerting a vast influence on glutamatergic, serotonergic, and dopaminergic transmission [96]. Chen et al. showed that RNA interference knockdown of PSD-95 caused PSD deficit and decreased AMPAR (but not NMDAR) levels in hippocampal neurons, thus confirming the crucial role of PSD-95 in maintaining correct protein architecture [97]. It has been observed that overexpression of PSD-95 can promote the formation of multi-innervated spines with up to seven presynaptic connections. The interaction of the PDZ2-domain of PSD-95 with nNOS (neuronal nitric oxide synthase) plays a vital role in this process [96,98]. nNOS is one of the three isoforms of the NOS enzyme and is widely distributed in the CNS and has also been demonstrated in human neutrophils [99]. Nikonenko et al. showed that both small interfering RNA (siRNA)-mediated knockdown of nNOS and pharmacological blockade by administration of L-NG-nitroarginine methyl ester (L-NAME, NOS inhibitor) result in the inhibition of multi-innervated spines formation, thus demonstrating the role of PSD-95 and nNOS as essential factors for synapse building [98].
The activity of individual elements included in the NMDAR-PSD-95-nNOS complex is modulated by various isoforms of the Homer 1 protein (Homer protein homolog 1) [100]. Homer 1 is widely distributed in the CNS and skeletal muscles and has nine isoforms, including Homer 1a-h and Ania-3 (Activity and neurotransmitter induced early gene 3) [101,102]. Homer-1 proteins consist of a Homer family specific EVH1 domain (enabled/vasodilator-stimulated phosphoprotein homology 1), a proline motif found in all Homer 1 proteins, and CC (coiled coil) domain, allowing the formation of homo- and heterooligomers. Short isoforms such as Homer 1a and Ania-3 lack a CC domain and function as negative modulators of Glu receptors [103]. Through the EVH1 domain, Homer 1 forms connections with other proteins, e.g., mGluRs (group 1), and can regulate their function. Wang et al. showed that Homer 1a is responsible for uncoupling between GluN2B, PSD-95, and nNOS, which was associated with the reduction of NMDAR-mediated transmission and thus had a neuroprotective effect. At the same time, Homer1 b/c facilitated mutual interactions among the NMDAR-PSD-95-nNOS complex [100]. Furthermore, an association between rs7713917 variant in the HOMER1 gene and an increased risk of suicide attempts and a worse response to antidepressant treatment with sleep deprivation and light therapy has been shown [104,105].
Homer 1 co-occurs with Shank (SH3 and multiple ankyrin repeat domain), forming a mesh-like structure that provides a scaffold for the other PSD proteins [106]. Shank3 (SH3 and multiple ankyrin repeat domains 3) is a member of the SHANK family, abundant at the excitatory synapse, and plays a vital role in the proper maturation and formation of dendritic spines [107]. Shank3 consists of a Shank/ProSAP N-terminal (SPN) domain followed by ankyrin repeats, src homology 3 (SH3) domain, PDZ domain, and sterile alpha motif (SAM) domain. PDZ domain binds to guanylate kinase-associated protein (GKAP). It thus indirectly allows Shank3 to interact with NMDARs and AMPARs [108], while the SAM domain is located at the C-terminal and binds zinc ions [109]. Duffney et al. showed that Shank-3 knockdown using si-RNA in cortical structures resulted in decreased NMDAR-mediated ionic current and decreased GluN1 subunit expression, significantly confirming the importance of Shank3 in correct excitatory synapse function [110]. Shank3 occurs in the Shank3a-f isoforms and is expressed in the nervous system, e.g., cortex, cerebellum, amygdala, hippocampus, spinal cord, striatum, dorsal root ganglia, but also in the heart, thymocytes, and spleen [107,111]. Interestingly, in a group of seven patients with Phelan-McDermid syndrome (22q13.3 deletion, which is associated with a deletion of the SHANK3 gene), four showed the presence of bipolar depression, which suggests a close relationship between Shank3 and this disorder [112].
3. Alterations in Postsynaptic Density Proteins in Depressive Disorders
Recent years, especially the last decade, have seen abundant research performed on postsynaptic proteins in the context of DDs. This review focuses only on the most important proteins, i.e., NMDAR, AMPAR, PSD-95, CamKII, Homer 1, Shank3, ZnT-1, and nNOS.
In the publications, we found clinical studies showing differences in PSD proteins expression in both depressed patients (Table 1) and preclinical studies, using animals (both mice and rats) showing depressive-like (mainly stress-induced) behaviors, as well as after treatment with drugs with antidepressant or antidepressant-like activity (Table 2, Table 3, Table 4 and Table 5). The observed alterations were sometimes varied and ambiguous, which may also be related to the diversity of the brain regions (hippocampus, prefrontal cortex, amygdala and locus coeruleus) in which the analyses were carried out. In addition, we found studies showing changes in peripheral blood cells (Table 1). Due to the large number of studies with the use of animal models, we divided our review according to the type of tested proteins (NMDAR and AMPAR, Table 2 and Table 3; other PSD proteins, Table 4 and Table 5) as well as by the species of animals used in studies (mice, Table 2 and Table 4; rats, Table 3 and Table 5). In this article, we included both research on protein and mRNA level changes.
Table 1.
Summary of clinical studies on the postsynaptic proteins in depressive disorders.
| Postsynaptic Proteins | Controls [N] | Patients [N] | Samples/Brain Region | Methods | Findings | Authors’ Names |
|---|---|---|---|---|---|---|
| NMDA receptor complex | ||||||
|
GluN1
GluN2A-D GluN3A |
N = 19 (male = 18, female = 1) | MDD = 18 male | Locus Coeruleus (LC) Prefrontal Cortex (PFC) |
qPCR | ↔GRIN1, GRIN2A mRNA in MDD-LC ↑GRIN2B mRNA in MDD-LC ↔GRIN1, GRIN2A, GRIN2B mRNA in MDD-PFC |
Chandley et al. [113] |
| GluN1 | N = 15 | MDD = 15 BD = 15 |
Prefrontal Cortex | Immunoautoradiography | ↔GluN1 protein in MDD and BD | Toro and Deakin [114] |
|
GluN1
GluN2A-D |
N = 15 | MDD = 15 BD = 15 |
Prefrontal Cortex | In situ hybridization | ↓GRIN1 mRNA in MDD and BD ↓GRIN2A mRNA in MDD ↔GRIN2B-D mRNA in MDD ↔GRIN2A-D mRNA in BD |
Beneyto and Meador-Woodruff [115] |
|
GluN1
GluN2A-D GluN3A |
N = 20 (male = 11, female = 9) N = 45 (male = 20, female = 25) |
MDD = 10 (male = 5, female = 4) BD = 10 (male = 5, female = 5) Suicide = 14 (male = 8, female = 6) |
Prefrontal cortex | In situ hybridization | ↑GRIN2D and ↔GRIN1, GRIN2A-C, and GRIN3A mRNA in MDD ↓GRIN2C and ↔GRIN1, GRIN2A, B, D, and GRIN3A mRNA in BD ↓GRIN2B and ↔GRIN1, GRIN2A, C, D and GRIN3A mRNA in Suicides |
Dean et al. [116] |
|
GluN1
GluN2A-B |
N = 32 (male = 19, female = 13) MDD-non-suicide (MDD-NS) = 19 (male, female) |
MDD = 53 (male = 26, female = 27) MDD-Suicide (MDD-S) = 34 (male, female) |
Dorsolateral Prefrontal Cortex |
qPCR | ↑GRIN2B, ↔GRIN1 and GRIN2A mRNA in MDD-S-both sexes ↑GRIN1, GRIN2A, GRIN2B mRNA in MDD-female ↑GRIN2B, ↔GRIN1 and GRIN2A mRNA in MDD-S-female ↔GRIN1, GRIN2A and GRIN2B mRNA in male MDD and MDD-S both sexes |
Gray et al. [68] |
| GluN2A | N = 10 (male) | MDD = 10 (male) | Prefrontal Cortex | Western Blot | ↓GluN2A protein in MDD | Rafalo-Ulinska et al. [74] |
|
GluN1
GluN2A-B |
N = 14 (male = 11, female = 3) | MDD = 14 (male =11, female = 3; 8 male and 2 female were suicides) | Prefrontal Cortex | Western Blot | ↔GluN1 protein in MDD ↓GluN2A, GluN2B protein in MDD |
Feyissa et al. [117] |
|
GluN1
GluN2A-B |
N = 14 (male = 13, female = 1) |
MDD = 14 (male = 13, female = 1; 12 subjects were suicides) |
Lateral amygdala | Western Blot | ↔GluN1, GluN2B protein in MDD ↑GluN2A protein in MDD |
Karolewicz et al. [118] |
| GluN2A-B | N = 6 | Suicide victims = 17 | Hippocampus | Western Blot | ↑GluN2A protein in suicides ↓GluN2B protein in suicides |
Sowa-Kućma et al. [119] |
|
GluN1
GluN2A-D |
N = 18 (female = 7, male = 11) |
MDD = 21 (female = 8, male = 13) | Hippocampus: CA1 region (CA1) and Dentate gyrus (DG) |
qPCR | ↔GRIN1, GRIN2A-D mRNA in CA1 and DG in MDD—both sexes | Duric et al. [120] |
| AMPA receptor complex | ||||||
| GluA1-2 | N = 19 (male = 18, female = 1) | MDD = 18 male | Locus Coeruleus (LC) Prefrontal cortex (PFC) |
qPCR | ↔GRIA1, GRIA2 mRNA in MDD—LC and PFC | Chandley et al. [113] |
| GluA1-2 | N = 32 (male = 19, female = 13) MDD-non-suicide (MDD-NS) = 19 (male, female) |
MDD = 53 (male = 26, female = 27) MDD-Suicide (MDD-S) = 34 (male, female) |
Dorsolateral Prefrontal Cortex |
qPCR | ↑GRIA2 and ↔GRIA1 mRNA in MDD—both sexes ↑GRIA2 and ↔GRIA1 mRNA in MDD-female ↔GRIA1 and GRIA2 mRNA in male MDD and MDD-S |
Gray et al. [68] |
| GluA1 | N = 10 (male) | MDD = 10 (male) | Prefrontal Cortex | Western Blot | ↓GluA1 protein in MDD | Rafalo-Ulinska et al. [74] |
| GluA1-4 | N = 18 (female = 7, male = 11) |
MDD = 21 (female = 8, male = 13) | Hippocampus: CA1 region (CA1) and Dentate gyrus (DG) | qPCR | ↓GRIA1 and GRIA3, ↔GRIA2 and GRIA4 mRNA in MDD-both sexes-CA1 and DG | Duric et al. [120] |
| Metabotropic glutamate receptor 5 | ||||||
| GluR5 | N = 32 (male = 19, female = 13) MDD-non-suicide (MDD-NS) = 19 (male, female) |
MDD = 53 (male = 26, female = 27) MDD-Suicide (MDD-S) = 34 (male, female) |
Dorsolateral Prefrontal Cortex |
qPCR | ↑GRM5 mRNA in MDD-female ↓GRM5 mRNA in MDD—male ↔GRM5 mRNA in MDD-both sexes and MDD-S-both sexes |
Gray et al. [68] |
| N = 19 (male = 18, female = 1) | MDD = 18 male | Locus Coeruleus (LC) Prefrontal Cortex (PFC) |
qPCR | ↑GRM5 mRNA in MDD—LC ↔GRM5 mRNA in MDD—PFC |
Chandley et al. [113] | |
| Postsynaptic density protein 95 | ||||||
| PSD-95 | N = 10 (male) | MDD = 10 (male) | Prefrontal Cortex | Western Blot | ↓PSD-95 protein in MDD | Rafalo-Ulinska et al. [74] |
| N = 14 (male = 11, female = 3) | MDD = 14 (male =11, female = 3; 8 male and 2 female were suicides) | Prefrontal Cortex | Western Blot | ↓PSD-95 protein level in MDD | Feyissa et al. [117] | |
| N = 14 (male = 13, female = 1) |
MDD = 14 (male = 13, female = 1; 12 subjects were suicides) |
Lateral amygdala | Western Blot | ↑PSD-95 protein in MDD | Karolewicz et al. [118] | |
| N = 6 | Suicide victims = 17 | Hippocampus | Western Blot | ↓PSD-95 protein in suicides | Sowa-Kućma et al. [119] | |
| N = 15 | MDD = 15 BD = 15 |
Prefrontal Cortex | In situ hybridization | ↔DLG4 mRNA in MDD ↓DLG4 mRNA in BD |
Kristiansen and Meador-Woodruff [121] | |
| N = 15 | MDD = 15 BD = 15 |
Prefrontal Cortex | Immunoautoradiography | ↓PSD-95 protein in BD vs. MDD | Toro and Deakin [114] | |
| N = 15 | MDD = 15 BD = 15 |
Prefrontal Cortex | In situ hybridization | ↔DLG4 mRNA in MDD and BD | Beneyto and Meador-Woodruff [115] | |
| N = 20 (male = 11, female = 9) N = 45 (male = 20, female = 25) |
MDD = 10 (male = 5, female = 4) BD = 10 (male = 5, female = 5) Suicide = 14 (male = 8, female = 6) |
Prefrontal cortex | Western Blot | ↔ PSD-95 protein in MDD and BD ↓PSD-95 protein in suicides |
Dean et al. [116] | |
| Zinc transporters | ||||||
| ZnT-1 | N = 10 (male) N = 8 (male, female) |
MDD = 10 (male) Suicide victims = 11 (male, female) |
Prefrontal Cortex | Western Blot | ↑ZnT-1 protein level in MDD and suicides | Rafalo-Ulinska et al. [74] |
| Nitric oxide synthases | ||||||
| nNOS | N = 27 (controls) N = 27 (first- degree relatives) |
MDD = 29 | Neutrophils (venous blood samples) | qPCR | ↑NOS1 mRNA in MDD ↔NOS1 mRNA in first-degree relatives |
Somani et al. [99] |
| N = 14 (male = 13, female = 1) |
MDD = 14 (male = 13, female = 1; 12 subjects were suicides) |
Lateral amygdala | Western Blot | ↔nNOS protein in MDD | Karolewicz et al. [118] | |
| N = 12 (male = 10, female = 2) | MDD = 12 (male = 9, female = 3; 12 subjects were suicides) | Locus coeruleus (LC) Cerebellum (CER) |
Western blot | ↓nNOS protein in MDD-LC ↔nNOS protein in MDD-CER |
Karolewicz et al. [122] | |
| NOS | N = 895 | MDD = 460 (drug-free = 104; antidepressant group = 356: SSRI = 138; SNRI = 137; another = 81) | Blood plasma | LC-MS | ↓NOS activity (L-Citrulline/L-Arginine ratio) in whole MDD group ↓NOS activity in drug-free MDD ↑NOS activity in whole MDD group at 3 months and 6 months ↑NOS activity in MDD responders group at 3 months and 6 months ↔NOS activity in MDD non-responders group at 3 months and 6 months |
Loeb et al. [123] |
| SH3 and multiple ankyrin repeat domains 3 | ||||||
| Shank3 | No control | MDD = 24 women BD = 32 women (All participants were treated with antidepressants and/or mood stabilizers of the 1st and 2nd generation) |
Peripheral blood mononuclear cells (PBMCs) | Microarray | ↑SHANK3 mRNA in MDD after treatment | Dmitrzak-Weglarz et al. [124] |
Abbreviations: qPCR —quantitative polymerase chain reaction; LC-MS—Liquid chromatography–mass spectrometry.
Table 2.
Summary of studies on the expression of postsynaptic Glu receptors (NMDAR, AMPAR and mGluR5) in mouse models of depression and/or after antidepressants treatment.
| Postsynaptic Proteins | Species/Strain | Model/Treatment/Groups | Samples/Brain Region | Methods | Findings | Authors’ Names |
|---|---|---|---|---|---|---|
| NMDA receptor complex | ||||||
|
NMDAR (no information on subunits) |
ICR male mice |
|
Hippocampus | qPCR | ↑NMDAR mRNA after CRS, CRS + Zn15, CRS + IMI5 and ↓NMDAR mRNA after CRS + Zn30, CRS + IMI20, and CRS + IMI5 + Zn15 |
Ding et al. [125] |
| GluN2A | C57BL/6J male mice |
|
Infralimbic Cortex (IL), Prelimbic cortex (PL), Basolateral Amygdala (BLA), Lateral Amygdala (LA), Central Amygdala (CEA) CA1 Strata Oriens (OR), CA1 Pyramidal (PYR), CA1 Radiatum (RAD) and CA1 Lacunosum-moleculare layers (LM) |
Fluorescence Immunohistochemistry | ↔ GluN2A protein in all brain region after FC + FLU ↑GluN2A protein in BLA, LA, CEA, CA1 PYR after FC+FLU+EXT |
Popova et al. [126] |
| GluN2A-B | C57Bl/6J male mice |
|
Hippocampus (HP) Prefrontal Cortex (PFC) |
Western Blot | ↓GluN2A, GluN2B protein after LUR10 and FLU20—HP and PFC | Stan et al. [127] |
| GluN2A-B | C57BL/6J male mice |
|
Hippocampus (HP) Prefrontal Cortex (PFC) |
Western Blot | ↓GluN2A, GluN2B and ↔GluN1 protein level after CUMS—HP ↔GluN1, GluN2A and GluN2B protein after CUMS—PFC ↔GluN2A protein and CUMS + 25 Hz HF-rTMS and ↔GluN2B protein after CUMS+15 Hz HF-rTMS and CUMS + 25 Hz HF-rTMS—HP ↔GluN1, GluN2A, GluN2B protein after CUMS+15 Hz HF-rTMS, CUMS + 25 Hz HF-rTMS |
Zuo et al. [128] |
| GluN2A-B | C57BL/6J male mice |
|
Basolateral Amygdala (BLA) Inferior-Limbic Prefrontal Cortex (IL-PFC) |
Western Blot | ↔GluN2A and ↓GluN2B protein in BLA and IL-PFC after KET | Asim et al. [129] |
|
pS1325-GluN2A
pS-1303-GluN2B GluN2A-B GluN1 |
C57BL/6J male mice |
|
Hippocampus | Western Blot qPCR |
↔GluN1, GluN2A, GluN2B, protein after KET1, 3, 5, 10 ↓GluN2A, GluN2B and ↔GluN1 protein after KET28 ↓GluN2A and GluN2B protein after KET_10 KET_20 and KET_30 pS1325-GluN2A, pS-1303-GluN2B, GluN2A, GluN2B (total, surface, intracellular) protein after KET_30 ↓Grin2A, Grin2B and ↔ Grin1 mRNA after KET_30 |
Luo et al. [130] |
| GluN2A-B | C57BL/6J female mice |
|
Hippocampus | qPCR | ↑Grin2A and Grin2B mRNA after CUS Grin2A and Grin2B mRNA after OVX + CUS ↔Grin2A and ↑Grin2B mRNA after OVX + CUS + P ↑Grin2A and Grin2B mRNA after OVX + CUS+ E |
Karisetty et al. [131] |
| GluN2B | Swiss male mice |
before each PTZ injection |
Prefrontal Cortex (PFC) Hippocampus (HP) | qPCR | ↔Grin2B mRNA level after PTE100, PTE200 + PTZ—PFC ↓Grin2B mRNA after VPA + PTZ—PFC ↔Grin2B mRNA after VPA, PTE100, 200 + PTZ in HP |
Nieoczym et al. [132] |
| AMPA receptor complex | ||||||
|
GluA1-pS845
GluA1-3 |
CD-1 male mice |
|
Prefrontal Cortex—whole tissue lysate Prefrontal Cortex—synaptic fraction |
Western Blot |
After 60 min of injection: ↔GluA1 and ↑GluA1-pS845 protein after PGM5, 8, 12.5 and IMI ↔GluA2 and GluA3 protein after PGM5, 8, 12.5 and IMI After 5 days of injection: ↑GluA1 protein after PGM 8 and 12.5 and ↑GluA1-pS845protein after PGM5, 8, 12.5 and IMI ↔GluA2 and GluA3 protein after PGM5, 8, 12.5 and IMI After 60 min of injection: ↔GluA1 and ↑GluA1-pS845 protein after PGM5, 8, 12.5 and IMI ↔GluA2 and GluA3 protein after PGM5, 8, 12.5 and IMI After 5 days of injection: ↑GluA1 and GluA1-pS845 protein after PGM5, 8, 12.5 and IMI ↑GluA2 protein after PGM8, 12.5 and IMI ↑GluA3 protein after PGM5, 8, 12.5 and IMI |
Bao et al. [133] |
| GluA1 | C57BL/6J male mice |
|
Hippocampus | Western Blot | ↑GluA1 protein after LPS+KET and LPS +YVAdD ↓GluA1 protein level after LPS ↔GluA1 protein level after LPS+YVAdD+KET |
Li et al. [134] |
|
GluA1
GluA1-pS818 GluA1-pS831 GluA1-pS845 GluA2 GluA2-pS880 |
C57BL/6J male mice |
|
Medial Prefrontal Cortex | Western Blot | ↔GluA1, ↓GluA1-pS831 and GluA2, and ↔GluA2-pS880 protein after CRS ↔ GluA1-pS818, ↑GluA1-pS831, ↔GluA2 and ↑ GluA2-pS880 after CRS+FLU |
Park et al. [135] |
| GluA1-2 | C57BL/6J male mice |
|
Infralimbic Cortex (IL), Prelimbic cortex (PL), Basolateral Amygdala (BLA), Lateral Amygdala (LA), Central Amygdala (CEA) CA1 Strata Oriens (OR), CA1 Pyramidal (PYR), CA1 Radiatum (RAD) and CA1 Lacunosum-moleculare layers (LM) |
Fluorescence Immunohistochemistry | ↓GluA1 protein in CA1 OR after FC+WAT+EXT ↓GluA1 protein in CA1 PYR after FC + FLU + EXT ↑GluA2 protein in all brain region after FC+WAT+EXT and FC+FLU+EXT |
Popova et al. [126] |
|
GluA1
GluA1-pS845 GluA2 |
C57BL/6J male mice |
|
Basolateral Amygdala |
Western Blot | ↑GluA1 and GluA1-pS845, and ↓GluA2 protein after CRS ↑GluA1, ↓GluA1-pS845 and ↑GluA2 protein after CRS + FLU |
Yi et al. [136] |
|
GluA1
GluA1-pS845 |
C57BL/6J male mice |
|
Western Blot | ↓GluA1 and GluA1-pS845 protein after CUS ↑GluA1 and GluA1-pS845 after CUS + SCO 25 and 50 |
Yu et al. [137] | |
| GluA1 | Swiss male mice |
|
Hippocampus (HP) Prefrontal Cortex (PFC) |
Western Blot | ↔ GluA1 protein in Hp after KET5, GUO5 ↓GluA1 protein in HP after CORT, CORT+GUO5 ↑GluA1 protein in HP after CORT+KET ↔GluA1 protein in PFC after KET5, GUO5, CORT, CORT +KET5, CORT + GUO5 ↔GluA1 protein in HP after FLU ↓GluA1 protein in HP after CORT ↓GluA1 protein in HP after CORT+FLU ↔GluA1 protein in PFC after FLU, CORT, CORT + FLU ↔GluA1 protein in HP after KET1+GUO5 ↓GluA1 protein in HP after CORT ↓GluA1 protein in HP after CORT+KET1+GUO5 ↔GluA1 protein in PFC after KET1 +GUO5, CORT, CORT+KET1+GUO5 |
Camargo et al. [138] |
| GluA1-2 | C57BL/6J male mice |
|
Hippocampus | Surface receptor cross-linking with BS3 and Western Blot |
↔GluA1, GluA2 protein in Susceptible and Resilient mice ↓GluA1, GluA2 protein after coss-linking in Susceptible mice |
Li et al. [139] |
| GluA1-pS845 | C57BL/6J male mice |
|
Prefrontal Cortex | Western Blot Fluorescence Immunohistochemistry |
↑GluA1-pS845 protein after NC shRNA+KET ↓GluA1-pS845 protein after VGF shRNA and VGF shRNA+KET ↑GluA1-pS845 protein after NC shRNA+KET ↓GluA1-pS845 protein after VGF shRNA and VGF shRNA+KET |
Shen et al. [140] |
| GluA1-2 | C57BL/6J male mice |
|
Hippocampus | Western Blot | ↓GluA1,GluA2 protein level after CRS, CRS+HNK, CRS+HNK+NBQX, CRS+HNK+ANA-12 | Ju et al. [141] |
|
pS831-GluA1
pS880-GluA2 GluA1-2 |
C57BL/6J male mice |
|
Hippocampus | Western BlotqPCR | ↔GluA1,GluA2 protein after KET1, 3, 5, 10 and KET_30 ↓GluA1, GluA2 protein after KET_10, 20 and 30 ↓GluA1,GluA2 after KET.10, KET.20, KET.30 ↓p-S831-GluA1 and p-S880-GluA2 (total, surface, intracellular) protein after KET_30 ↓Gria1, Gria2 mRNA after KET_30 |
Luo et al. [130] |
| Metabotropic glutamate receptor 5 | ||||||
| mGluR5 | C57BL/6J male mice |
|
Nucleus Accumbens (NAc) | Western Blot | ↓mGluR5 protein in Susceptible mice ↑mGluR5 protein in Resilient mice |
Xu et al. [142] |
| C57BL/6J male mice |
|
Hippocampus | qPCR | ↑Grm5 mRNA in Susceptible mice ↑Grm5 mRNA after CRS |
Li et al. [139] | |
Abbreviations: qPCR —quantitative polymerase chain reaction; Ac-YVAD-CMK—NLR family pyrin domain containing 3 (NLRP3) inflammasome inhibitor; BS3—bis(sulfosuccinimidyl)suberate.
Table 3.
Summary of studies on the expression of postsynaptic Glu receptors (NMDAR, AMPAR and GluR5) in rat models of depression and/or after antidepressants treatment.
| Postsynaptic Proteins | Species/Strain | Model/Treatment/Groups | Brain Region | Methods | Findings | Authors’ Names |
|---|---|---|---|---|---|---|
| NMDA receptor complex | ||||||
| GluN2A-B | Sprague Dawley male and female rats |
|
Hippocampus | Western blot Post-embedding Immunogold |
↓GluN2A/GluN2B protein in LH compared to NLH ↑GluN2B protein in LH compared to WT ↔GluN2B protein in NLH compared to WT |
Bieler et al. [143] |
|
NMDAR (no information on subunits) |
Sprague Dawley male rats |
|
Frontal cortex | qPCR | ↓NMDAR mRNA after CUMS ↑NMDAR mRNA n after CUMS+ PAR, CUMS+ Zn+ FA, CUMS+ Zn+ FA+ PAR |
Dou et al. [144] |
|
GluN1
GluN2A pT1325-GluN2A GluN2B pS1303-GluN2B |
Sprague Dawley male rats |
|
Medial prefrontal cortex (mPFC) | Western blot | ↑GluN1, GluN2B, p-GluN2B and ↔ GluN2A, p-GluN2A protein after CUS ↔GluN2A, p-GluN2A, GluN2B and ↓p-GluN2B protein after CUS+ IFE ↑GluN1 protein after CUS+ IFE ↔GluN1 protein level after CUS+TC-DAPK 6 ↔GluN2B and ↓p-GluN2B protein level after CUS+TC-DAPK 6 ↔GluN1 protein level after CUS+ AAV-shDAPK1 ↔GluN2B protein after CUS+ AAV-shDAPK1 ↓p-GluN2B protein after CUS+ AAV-shDAPK1 |
Li et al. [145] |
| GluN2A-B | Flinders Sensitive Line (FSL) male rats Flinders Resistant Line (FRL) male rats |
|
Hippocampus (HP) Prefrontal cortex (PFC) |
Western blot | ↓GluN2A and ↑GluN2B protein in FSL—HP ↔GluN2A and GluN2B protein in FSL—PFC |
Treccani et al. [146] |
| GluN2A-B | Sprague Dawley male rats |
|
Prefrontal cortex (PFC) Hippocampus (HP) Amygdala (AMY) |
Western blot qPCR |
↔GluN2A protein after OB in PFC vs. Sham ↔GluN2A protein after OB+ Mg15 in PFC vs. OB ↔GluN2B protein after OB in PFC vs. Sham ↔GluN2B protein after OB+ Mg15 in PFC vs. OB ↔GluN2A protein after OB in HP vs. Sham ↔GluN2A protein after OB+ Mg15 in HP vs. OB ↔GluN2B protein after OB in HP vs. Sham ↔GluN2B protein after OB+ Mg15 in HP vs. OB ↔GluN2A protein after OB in AMY vs. Sham ↔GluN2A protein after OB+ Mg15 in AMY vs. OB ↔GluN2B protein after OB in AMY vs. Sham ↑GluN2B protein after OB+ Mg15 in AMY vs. OB ↔Grin2A mRNA after OB in PFC vs. Sham ↔Grin2A mRNA after OB+ Mg15 in PFC vs. OB ↔Grin2B mRNA after OB in PFC vs. Sham ↑Grin2B mRNA after OB+ 15 Mg15 in PFC vs. OB ↓Grin2A mRNA after OB in HP vs. Sham ↔Grin2A mRNA after OB+ Mg15 in HP vs. OB ↓Grin2B mRNA after OB in HP vs. Sham ↑Grin2B mRNA after OB+ Mg15 in HP vs. OB |
Pochwat et al. [147] |
|
GluN1
GluN2A-B |
Sprague Dawley male rats |
|
Hippocampus | Western blot | ↑GluN1, GluN2A and GluN2B proteins after ZnD vs. ZnA ↓GluN1, GluN2A and GluN2B proteins after ZnD+ FLU vs. ZnD |
Doboszewska et al. [73] |
| GluN2A-B | Sprague Dawley male rats |
|
Hippocampus (whole tissue lysate) Hippocampus (synaptoneurosomes) |
Western blot | ↓GluN2A and ↔GluN2B protein after CRS ↔ GluN2A, ↔GluN2B protein after CRS+ FAS ↔GluN2A, GluN2B protein after CRS ↔GluN2A, GluN2B protein after CRS+ FAS |
Román-Albasini et al. [148] |
| GluN2B | Sprague Dawley male rats |
|
Hippocampus | Western blot | ↔ GluN2B protein after CUMS ↑GluN2B protein after CUMS+BDNF ↓GluN2B protein after proBDNF |
Qiao et al. [149] |
| GluN2B | Sprague Dawley male rats |
|
Medial prefrontal cortex | Western blot | ↓GluN2B protein after CUMS ↑GluN2B protein after CUMS+ YY-21 ↑GluN2B protein after CUMS+ FLU |
Guo et al. [150] |
| GluN2A-B | Sprague Dawley male rats |
|
Amygdala | Western blot | ↑GluN2A and GluN2B protein after CMS | Zhou et al. [151] |
| GluN2A-B | Dark Agouti male rats |
|
Frontal cortex | qPCR | ↑Grin2A and Grin2B mRNA after VLX | Tamási et al. [152] |
| GluN2B | Sprague Dawley male rats |
|
Hippocampus (HP) Nucleus accumbens (NAc) Amygdala (AMY) |
Western blot | ↓GluN2B protein after KET in HP ↑GluN2B protein after KET in NAc ↑GluN2B protein after KET in AMY |
Piva et al. [153] |
| AMPA receptor complex | ||||||
| GluA1 | Sprague Dawley male rats |
|
Hippocampus (HP) Medial prefrontal cortex (mPFC) |
Western blot | ↓GluA1 protein after CUMS and ↑GluA1 protein after CUMS+ IFE in HP ↓GluA1 protein after CUMS and ↑GluA1 protein after CUMS+ IFE in mPFC |
Yao et al. [154] |
| GluA1 | Sprague Dawley male rats |
|
Medial prefrontal cortex | Western blot | ↑GluA1 protein after NVP-AAM077 (only 30 min after administration) | Gordillo-Salas et al. [155] |
| GluA1 | Sprague Dawley male rats |
|
Medial prefrontal cortex | Western blot | ↓GluA1 protein after CUS ↑GluA1 protein after CUS+IFE |
Li et al. [145] |
|
GluA1
pS831-GluA1 pS845-GluA1 |
Sprague Dawley male rats |
|
Prefrontal cortex (PFC) Hippocampus (HP) Amygdala (AMY) |
Western blot | ↔GluA1, pS831-GluA1, pS845-GluA1 proteins after OB in PFC vs. Sham ↔GluA1,↑pS831-GluA1 and pS845-GluA1 protein after OB+ Mg15 in PFC vs. OB ↔GluA1, pS831-GluA1 and ↑pS845-GluA1 protein after OB in HP vs. Sham ↔GluA1, pS831-GluA1 and ↓pS845-GluA1 protein after OB+ Mg15 in HP vs. OB ↔GluA1, pS831-GluA1 and pS845-GluA1 protein after OB in AMY vs. Sham ↔GluA1, pS831-GluA1 and pS845-GluA1 protein after OB+ Mg15 in AMY vs. OB |
Pochwat et al. [147] |
|
GluA1
pS831-GluA1 pS845-GluA1 GluA2 |
Sprague Dawley male rats |
|
Hippocampus (whole tissue lysate) Hippocampus (synaptoneurosomes) |
Western blot | ↓GluA1 and ↔pS831-GluA1, pS845-GluA1, GluA2 protein level after CRS ↔GluA1, pS831-GluA1, pS845-GluA1, GluA2 protein after CRS+ FAS ↔GluA1, pS831-GluA1, pS845-GluA1, GluA2 protein after CRS ↔GluA1, pS845-GluA1, GluA2 and ↓pS831-GluA1 proteins after CRS+ FAS |
Román-Albasini et al. [148] |
| GluA3 | Dark Agouti male rats |
|
Frontal cortex | qPCR | ↑Gria3 mRNA after VLX | Tamási et al. [152] |
| GluA1 | Sprague Dawley male rats |
|
Hippocampus | Western blot | ↓GluA1 protein after CUMS ↑GluA1 protein after CUMS+S-KET ↔GluA1 protein after CUMS+ NSC23766 + S-KET ↔GluA1 protein after CUMS + NSC23766 |
Zhu et al. [156] |
| GluA1 | Sprague Dawley rats |
|
Amygdala (AMY) Prefrontal cortex (PFC) |
Western blot | ↑GluA1 protein after 3 CS-US in AMY ↔GluA1 protein after 6 CS-US in AMY ↑GluA1 protein after 10 CS-US in AMY ↓GluA1 protein after 10 CS-US+ KET in AMY ↔GluA1 protein after 10 CS-US + FLU in AMY ↓GluA1 protein after 3 CS-US in PFC ↓GluA1 protein after 6 CS-US in PFC ↓GluA1 protein after 10 CS-US in PFC ↑GluA1 protein after 10 CS-US+ KET in PFC ↔GluA1 protein after 10 CS-US + FLU in PFC |
Lee et al. [157] |
| GluA1 | Sprague Dawley male rats |
|
Hippocampus | Western blot | ↓GluA1 protein after CUMS ↑GluA1 protein after CUMS+ VLX ↑GluA1 protein after CUMS+ Dhmc |
Yang et al. [158] |
|
GluA1
pS845-GluA1 |
Sprague Dawley male rats |
|
Medial prefrontal cortex | Western blot | ↔GluA1 and ↓pS845-GluA1 proteins after CUMS ↔GluA1 and ↑pS845-GluA1 proteins after CUMS+ YY-21 ↔GluA1 and ↑pS845-GluA1 proteins after CUMS+ FLU |
Guo et al. [150] |
| GluA1 | Sprague Dawley male rats |
|
Prefrontal cortex | Western blot | ↔GluA1 protein 30 min after Zn5 treatment ↑GluA1 protein 3 h after Zn5 treatment ↑GluA1 protein 24h after Zn treatment |
Szewczyk et al. [159] |
|
GluA1
pS845-GluA1 |
Sprague Dawley male and female rats |
|
Hippocampus | Western blot | ↑GluA1 and pS845-GluA1 protein after KET ↔GluA1 and ↑pS845-GluA1 protein after MEM |
Zhang et al. [160] |
| GluA1 | Sprague Dawley male rats |
|
Hypothalamic paraventricular nucleus (hPVN) Hypothalamus (HY) |
Western blot qPCR |
↑ GluA1 protein after CUS in hPVN ↑GluA1 protein and ↑Gria1 mRNA after CUS in HY |
Li et al. [161] |
| GluA1-2 | Sprague Dawley male rats |
|
Hippocampus | Western blot | ↓GluA1 and GluA2 protein after CUMS ↑GluA1 protein after CUMS+ NaHS ↑GluA2 protein after CUMS+ NaHS |
Hou et al. [162] |
| GluA1 | Sprague Dawley male rats |
|
Hippocampus (HP) Nucleus accumbens (NAc) Amygdala (AMY) |
Western blot | ↓GluA1 protein after KET in HP ↓GluA1 protein after KET in NAc ↔GluA1 protein after KET in AMY |
Piva et al. [153] |
| Metabotropic glutamate receptor 5 | ||||||
| mGluR5 | Sprague Dawley male rats |
|
Hippocampus | Western blot Immunohistochemistry qPCR |
↓mGluR5 protein after CUMS ↑mGluR5 protein after CUMS+ FLU ↑mGluR5 protein after CUMS+ SSDH ↑mGluR5 protein after CUMS+ SSDL ↓mGluR5 protein after CUMS ↑mGluR5 protein after CUMS+ FLU ↑mGluR5 protein after CUMS+ SSDH ↑mGluR5 protein after CUMS+ SSDL ↓Grm5 mRNA after CUMS ↑Grm5mRNA after CUMS+ FLU ↑Grm5 mRNA after CUMS+ SSDH ↑Grm5 mRNA after CUMS+ SDDL |
Liu et al. [163] |
| mGluR5 | Sprague Dawley male rats |
|
Hippocampus (HP) Nucleus accumbens (NAc) Amygdala (AMY) |
Western blot | ↓mGluR5 protein after KET in HP ↓mGluR5 protein after KET in NAc ↑mGluR5 protein after KET in AMY |
Piva et al. [153] |
Abbreviations: YY-21—novel furostan skeleton secondary timosaponin, was obtained from timosaponin B-III via hydrolysis with hydrochloric acid; NVP-AAM07—[(S)-[[(1S)-1-(4-bromophenyl)ethyl]amino]-(2,3-dioxo-1,4-dihydroquinoxalin-5-yl)methyl]phosphonic acid; preferring GluN2A subunit antagonist; NSC23766—(N6-[2-(4-Diethylamino-1-methyl-butylamino)-6-methyl-pyrimidin-4-yl]-2-methyl-quinoline-4,6-diamine); Rac1 inhibitor; qPCR—quantitative polymerase chain reaction.
Table 4.
Summary of studies on the expression of postsynaptic density proteins in mouse models of depression and/or after antidepressants treatment.
| Postsynaptic Proteins | Species/Strain | Model/Treatment/Groups | Brain Region | Methods | Findings | Authors’ Names |
|---|---|---|---|---|---|---|
| Postsynaptic density protein 95 | ||||||
| PSD-95 | ICR male, female mice |
|
Hippocampus—CA1 | Immunofluorescence | ↓PSD-95 protein after KET ↑PSD-95 protein after BET100, KE+BET30 and KE+BET100 |
Chen et al. [164] |
| Swiss male mice |
|
Hippocampus (HP) Prefrontal Cortex (PFC) |
Western Blot | ↔PSD-95 protein in HP after KET5, GUO5↓PSD-95 protein in HP after CORT, CORT+GUO5 ↑PSD-95 protein in HP after CORT+KET5 ↔PSD-95 protein in PFC after KET5, GUO5, CORT, CORT+KET5, CORT+GUO5 ↔PSD-95 protein in HP after FLU ↓PSD-95 protein in HP after CORT ↓PSD-95 protein in HP after CORT +FLU ↔PSD-95 protein in PFC after FLU, CORT, CORT + FLU ↔PSD-95 protein in HP after KET1+GUO5 ↓PSD-95 protein in HP after CORT, CORT+KET1+GUO5 ↔PSD-95 protein in PFC after KET1 + GUO5, CORT, CORT + KET1 + GUO5 |
Camargo et al. [138] | |
| C57BL/6 male mice |
|
Basal nuclei (BLA) Lateral nuclei (LAT) Medial Prefrontal Cortex (mPFC) |
Immunohistochemistry | ↑PSD-95 protein in LAT and BLA after CRS ↓PSD-95 protein in LAT and BLA after CRS+IMI ↑PSD-95 expression in mPFC after CRS+IMI ↓PSD-95 expression in mPFC after CRS |
Leem et al. [165] | |
| C57BL/6 male mice |
|
Frontal Cortex Hippocampus |
Western Blot | ↔PSD-95 protein after CUMS, CUMS + FLU, CUMS+ASI10, 20, 40 ↓PSD-95 protein after CUMS ↑PSD-95 protein after CUMS+ASI10, CUMS + ASI20, CUMS +ASI40, CUMS+FLU |
Luo et al. [166] | |
| C57Bl/6J male mice |
|
Hippocampus (HP) Prefrontal Cortex (PFC) |
Western Blot | ↓PSD-95 protein after LUR10 and FLU20 ↓PSD-95 protein after LUR10 and FLU20 |
Stan et al. [127] | |
| C57Bl/6J male mice |
|
Basolateral Amygdala |
Western Blot | ↑PSD-95 protein after CRS ↓PSD-95 protein after CRS + FLU |
Yi et al. [136] | |
| C57BL/6J male mice |
|
Hippocampus | Western Blot | ↓PSD-95 protein after CRS, CRS+HNK, CRS+HNK+NBQX, CRS+HNK+ANA-12 | Ju et al. [141] | |
| KM (Kunming) male mice |
|
Prefrontal Cortex (PFC) Hippocampus (HP) Hypothalamus (HY) Olfactory Bulb (AL) |
qPCR Western Blot |
↓Dlg4 mRNA after UCMS, UCMS + Ala2+ Rapa in PFC, HP, Hy, AL. ↑Dlg4mRNA after UCMS+ Ala1 Ala2 in PFC, HP ↔Dlg4 mRNA after UCMS + Ala1 in HY, AL ↑Dlg4 mRNA after UCMS + Ala2 in HY, AL ↓PSD-95 protein after UCMS, UCMS+Ala2+Rapa in PFC ↑PSD-95 protein after UCMS+Ala2 in PFC ↔PSD-95 protein after UCMS+Ala1 in PFC ↓PSD-95 protein after UCMS, UCMS + Ala2 + Rapa in Hp ↑ PSD-95 protein after UCMS+Ala1, Ala2 in Hp ↓PSD-95 protein after UCMS in Hy ↔PSD-95 protein after UCMS + Ala1, Ala2, Ala2 + Rapa in HY ↓PSD-95 protein after UCMS in AL ↑PSD-95 protein after UCMS + Ala2 in AL ↔PSD-95 protein after UCMS + Ala1, Ala2+Rapa |
Zhuang et al. [167] | |
| Calcium/calmodulin-dependent protein kinase II | ||||||
| p-CaMKII | C57BL/6 male mice |
|
Basal nuclei (BA) Lateral nuclei (LAT) Medial Prefrontal Cortex (mPFC) |
Immunohistochemistry | ↑p-CamKII protein in LAT, BA after CRS and ↓after CRS+IMI ↓p-CamKII protein after CRS in mPFC ↑p-CamKII protein after CRS+IMI in mPFC |
Leem et al. [165] |
|
p-T286-CaMKII
t-CaMKII |
C57BL/6J male, female mice |
|
Prefrontal Cortex | Western Blot | ↓CaMKII-p-T286 and ↔ t-CaMKII protein after SD ↑CaMKII-p-T286 protein after CTM and CTM + SD |
Misrani et al. [168] |
| CaMKII | ICR male, female mice |
|
Hippocampus | Western Blot | ↔CaMKII protein after YG, ES, YG+LPS, ES+LPS (both sexes) ↓CaMKII (female) and ↑CaMKII (male) protein after CUS ↑CaMKII (female) and ↓CaMKII (male) protein after CUS+YG, CUS+ES |
Yin et al. [169] |
|
p-CaMKII (no information about the site of phosphorylation) |
ICR male, female mice |
|
Hippocampus | Western Blot | ↓p-CAMKII protein after cLH and cLH+YJ (at day 7) ↔ p-CAMKII protein after cLH+FLU (at day 7) ↓p-CAMKII protein after cLH and cLH+FLU (at day 26—after 5 days break in drug administration) |
Zou et al. [170] |
| p-T286- CaMKII | C57BL/6J male, female mice |
|
Hippocampus | Western Blot | ↓p-T286-CAMKII protein after SD ↑p-T286-CAMKII protein after CTM, CTM + SD |
Misrani et al. [171] |
| CaMKIIα | C57BL/6J male mice |
|
Basolateral Amygdala (BLA) Inferior-Limbic Prefrontal Cortex (IL-PFC) |
Western Blot | ↔CAMKIIα protein after KET in BLA and IL-PFC | Asim et al. [129] |
| Homer scaffold protein 1 | ||||||
| Homer 1a | C57BL/6J male mice |
|
Hippocampus (HP) Prefrontal Cortex (PFC) |
Western Blot | ↓Homer 1a protein in HP and PFC after CUMS ↔Homer 1a protein in HP and PFC after CUMS+15 Hz HF-rTMS, CUMS + 25 Hz HF-rTMS |
Zuo et al. [128] |
|
Homer 1a
Homer 1b/c |
C57BL/6 female mice |
|
Cortex | qPCR | ↓Homer1a and Homer1b/c mRNA expression after CDM ↑Homer1a and Homer1b/c mRNA expression after IMI, FLU, KET |
Sun et al. [172] |
|
Homer 1a
Homer 1b/c |
C57BL/6 J, CD1 male mice |
|
Medial Prefrontal Cortex (mPFC) Hippocampus (HP) Amygdala (AMY) |
qPCR Western Blot |
↔Homer1a mRNA in Susceptible, and Resilient mice after CSDS—all brain regions ↑Homer1b/c mRNA in Susceptible mice after CSDS—HP ↔Homer 1a protein in Susceptible and Resilient mice after CSDS—all brain regions ↑Homer 1b/c protein in Susceptible mice after CSDS—HP |
Li et al. [139] |
| Zinc transporters | ||||||
| ZnT-1 | Albino Swiss male mouse |
|
Prefrontal Cortex Hippocampus |
Western Blot | ↓ZnT-1 protein after Zn ↔ZnT-1 protein after IMI and Zn + IMI ↓ZnT-1 protein after Zn ↔ ZnT-1 protein level after IMI ↑ ZnT-1 protein level after Zn + IMI |
Rafało-Ulińska et al. [173] |
| Nitric oxide synthases | ||||||
| nNOS | ICR male, female mouse |
|
Hippocampus | Western Blot | ↔ nNOS protein after YG, ES, YG+LPS, ES+LPS (both sexes) ↓nNOS protein after CUS (female) ↑nNOS protein after CUS+YG, CUS+ES (female) ↑nNOS protein after CUS (male) ↓nNOS protein after CUS+YG, CUS+ES (male) |
Yin et al. [169] |
Abbreviations: Asiaticoside —product derived from the plant Centella asiatica. It has been shown to possess wound healing, anti-inflammatory and liver protective effects. In addition, studies demonstrated that asiaticoside could attenuate neurobehavioral andneurochemical; Alarin—neuropeptide with antidepressant activity; Yueju-Ganmaidazao (YG)–an herbal medicine prescribed for the treatment of mood disorders, consisting of two classic traditional Chinese herbal medicines Yueju and Ganmaidazao. Yueju and Ganmaidazao are used to treat depression.
Table 5.
Summary of studies on the expression of postsynaptic density proteins in rat models of depression and/or after antidepressants.
| Postsynaptic Proteins | Species/Strain | Model/Treatment | Brain Region | Methods | Findings | Authors’ Names |
|---|---|---|---|---|---|---|
| Postsynaptic density protein 95 | ||||||
| PSD-95 | Sprague Dawley male rats |
|
Medial prefrontal cortex | Western blot | ↓PSD-95 protein after CUS ↑PSD-95 protein after CUS+ IFE |
Li et al. [145] |
| Sprague Dawley male rats |
|
Hippocampus (CA1 and CA3 region) Dentate gyrus |
Western blot | ↓PSD-95 protein after CUMS in all regions ↑PSD-95 protein after CUMS+ NaHS in all regions |
Hou et al. [162] | |
| Sprague Dawley male rats |
|
Hypothalamus | Western blot qPCR |
↑PSD-95 protein after CUS ↑Dlg4 mRNA after CUS |
Li et al. [161] | |
| Sprague Dawley male rats |
|
Medial prefrontal cortex | Western blot | ↑PSD-95 protein after CX717 (only 2 h after administration) | Gordillo-Salas et al. [174] | |
| Sprague Dawley male rats |
|
Hippocampus | Western blot | ↓PSD-95 protein after CUMS ↑PSD-95 protein after CUMS+ S-KET ↑PSD-95 protein after CUMS+ NSC23766 + S-KET ↑PSD-95 protein after CUMS+ NSC23766 |
Zhu et al. [156] | |
| Sprague Dawley male rats |
|
Amygdala (AMY) Prefrontal cortex (PFC) |
Western blot |
↑PSD-95 protein after 3 CS-US in AMY ↔PSD-95 protein after 6 CS-US in AMY ↑PSD-95 protein after 10 CS-US in AMY ↔PSD-95 protein after 10 CS-US+ KET in AMY ↔PSD-95 protein after 10 CS-US+ FLU in AMY ↓PSD-95 protein after 3 CS-US in PFC ↓PSD-95 protein after 6 CS-US in PFC ↓PSD-95 protein after 10 CS-US in PFC ↑PSD-95 protein after 10 CS-US+ KET in PFC ↔PSD-95 protein after 10 CS-US + FLU in PFC |
Lee et al. [157] | |
| Sprague Dawley male rats |
|
Medial prefrontal cortex | Western blot | ↓PSD-95 protein after CUMS ↑PSD-95 protein after CUMS+ YY-21 ↔PSD-95 protein after CUMS+ FLU |
Guo et al. [150] | |
| Sprague Dawley male rats |
|
Prefrontal cortex | Western blot | ↔PSD95 protein 30min after Zn treatment ↑PSD95 protein level 3 h after Zn treatment ↔PSD95 protein level 24h after Zn treatment |
Szewczyk et al. [159] | |
| Wistar male rats |
|
Prefrontal cortex | qPCR | ↓Dlg4 mRNA after CMS (stress-reactive group) ↑Dlg4mRNA after CMS+ LUR |
Luoni et al. [175] | |
| PSD-95 | Sprague Dawley male rats |
|
Hippocampus | Western blot Immunohistochemistry qPCR |
↓PSD-95 protein after CUMS ↑PSD95 protein CUMS+ FLU ↑PSD95 protein CUMS+ SDDH ↔PSD95 protein CUMS+ SDDL ↓PSD-95 protein after CUMS ↑PSD95 protein CUMS+ FLU ↑PSD95 protein CUMS+ SDDH ↔PSD95 protein CUMS+ SDDL ↓Dlg4 mRNA after CUMS ↑Dlg4 mRNA after CUMS+ FLU ↑Dlg4 mRNA after CUMS+ SDDH ↑Dlg4 mRNA after CUMS+ SDDL |
Liu et al. [163] |
| Calcium/calmodulin-dependent protein kinase II | ||||||
|
CaMKII
p-T286-CaMKII p-T286-CaMKII α p-T286-CaMKII β |
Sprague Dawley male rats |
|
Hippocampus | Western blot | ↔CaMKII protein after CUMS ↔CaMKII protein after CUMS+ GXDSF in all groups ↑p-CaMKII protein after CUMS ↓p-CaMKII protein after CUMS+ GXDSF in all groups ↑p-CaMKIIα protein after CUMS ↓p-CaMKIIα protein after CUMS+ GXDSF in all groups ↑p-CaMKIIβ protein after CUMS ↓ p-CaMKIIβ protein after CUMS+ GXDSF in all groups |
Xie et al. [176] |
|
CaMKIIγ
CaMKII β |
Dark Agouti male rats |
|
Frontal cortex | qPCR | ↑Camk2g mRNA after VLX ↑Camk2b mRNA after VLX |
Tamási et al. [152] |
| Homer scaffold protein 1 | ||||||
| Homer 1 | Wistar male rats |
|
Prefrontal cortex | Western blot | ↑Homer1 protein in resilient group compared to anhedonic | Palmfeldt et al. [177] |
| Homer 1 | Sprague Dawley male rats |
|
Hippocampus (synaptoneurosomes) |
Western blot | ↔Homer1 protein after CRS ↔Homer1 protein after CRS+ FAS |
Román-Albasini et al. [148] |
| Homer 1 | Sprague Dawley male rats |
|
Hippocampus | Western blot Immunohistochemistry qPCR |
↑Homer1b/c protein after CUMS ↓Homer-1b/c protein after CUMS+ FLU ↓Homer1b/c protein after CUMS+ SSDH ↓Homer1b/c protein after CUMS+ SSDL ↑Homer1b/c protein after CUMS ↓Homer1b/c protein after CUMS+ FLU ↓Homer1b/c protein after CUMS+ SSDH ↓Homer1b/c protein after CUMS+ SSDL ↑Homer1 mRNA after CUMS ↓Homer1 mRNA after CUMS+ FLU ↓Homer1 mRNA after CUMS+ SDDH ↓Homer1 mRNA after CUMS+ SDDL |
Liu et al. [163] |
| Nitric oxide synthases | ||||||
|
nNOS
iNOS eNOS |
Wistar male rats |
|
Cerebellum (CER) Frontal cortex (FC) Midbrain (MID) Olfactory bulb (OB) Pons Striatum Temporal cortex (TC) Thalamus (TH) Hippocampus (HP) |
qPCR | ↔Nos1 mRNA after AR ↔Nos1 mRNA after AR+ FLU, AR+MIL, AR+MIR—all brain regions ↑Nos2 mRNA after AR—CER, FC, MID, OB, pons, striatum, TC, TH ↔Nos2 mRNA after AR—HP ↔Nos2 mRNA after AR+ FLU, AR+MIL, AR+MIR—all brain regions ↔Nos3 mRNA after AR ↑Nos3 mRNA after AR+ FLU—CER, Hp, midbrain, pons, striatum, thalamus, FC ↔Nos3 mRNA after AR+ FLU—OB, TC; after AR+ MIL and AR+ MIR—all brain regions |
Yoshino et al. [178] |
| nNOS | Sprague Dawley male rats |
|
Hippocampus | Western blot | ↑nNOS protein after CUS ↓nNOS protein after CUS + MEM |
Mishra et al. [179] |
Abbreviations: CX717—2,1,3-benzoxadiazol-5-yl(morpholin-4-yl)methanone; low-impact ampakine; NSC23766—(N6-[2-(4-Diethylamino-1-methyl-butylamino)-6-methyl-pyrimidin-4-yl]-2-methyl-quinoline-4,6-diamine), Rac1 inhibitor; YY-21—novel furostan skeleton secondary timosaponin obtained from timosaponin B-III via hydrolysis with hydrochloric acid; qPCR—quantitative polymerase chain reaction.
3.1. Human Studies
Based on the review, it can be concluded that among the PSD proteins, NMDAR analyses in post-mortem brains of patients with DDs constitute the vast majority. Most authors indicate no changes in both the protein and mRNA levels of the GluN1 subunit [113,114,116,117,118,120].
Interestingly, these observations apply to both MDD and BD patients [116]. Although slightly contrasting results (decrease in GluN1 mRNA level) were shown by Beneyto et al. [115], different gender ratios may be of importance. Similarly, according to Gray et al., GRIN1 gene expression was higher in women with MDD, while it did not change in men. Hence, we can conclude that an important factor influencing the NMDAR proteins level is, among other things, sex [68]. Analyses of different NMDAR subunits also confirm this. As shown by Gray et al., the expression of GRIN2A and GRIN2B was higher in the group of women with MDD, while it remained unchanged in men [68]. Other authors also indicate no changes in GRIN2A expression in the MDD group with predominantly males [113,115,116]. These observations seem to be consistent, despite the different areas of the brain studied [113] or the analytical methods used [68,113,116]. On the other hand, decreased expression of GRIN2B has been observed post-mortem in the locus coeruleus of the brain, which may indicate a multifactorial etiology of DDs and the participation of the noradrenergic-glutamatergic component [113]. GRIN2A and GRIN2B expression also appears to be relatively similar in MDD and BD patients. However, when we look at the levels of these proteins in different studies, the results are more varied and inconsistent with the expression of the genes that code for them. For example, two studies [74,117] showed a decrease in GluN2A level, while subsequent studies [118,119] showed its increase. The reason for these differences seems to stem from the dissimilarity of the studied structures (prefrontal cortex vs. lateral amygdala and hippocampus), which appeared to be of secondary importance in mRNA analysis. This hypothesis may not be entirely accurate when we analyze the changes in the GluN2B protein, which show a decrease in both the prefrontal cortex and the hippocampus [117,119] and no changes in the lateral amygdala [118]. The very cause of subjects’ death in the case of protein analysis may be irrelevant, which seemed to be essential for the study of gene expression [68]. Observed discrepancies may be explained by the different functional morphology and physiology of NMDARs in other parts of the CNS. As already mentioned above, NMDARs form heterotetramers, most often composed of two GluN1 subunits that bind to two additional subunits: GluN2 (A-D), which binds Glu, or less commonly GluN3 (A-B) with high affinity for glycine [180]. NMDARs most often consist of GluN1 and GluN2 subunits, particularly GluN2A and GluN2B [181,182]. Importantly, NMDARs containing GluN2A subunits show three times faster degradation times and reduced Glu affinity compared to GluN2B-containing receptors [183]. Mellone et al. showed that ZnT-1 in hippocampal neurons binds to GluN2A (1049-1464) C-terminal (but not GluN2B) and modulates PSD-95 (postsynaptic density protein 95) activity and dendritic spike morphology [184]. Importantly, studies on MDD subjects showed an increase in the level of ZnT-1 protein, which may suggest significant changes in NMDAR signaling caused by this transporter [74]. Recent studies have shown that activation of NMDA GluN2A receptors exerts a survival-promoting effect, while NMDA GluN2B receptors, which are mainly segregated into extrasynaptic sites, show deleterious effects [185]. Two further subunits, GluN2C and GluN2D, which are known to be positively involved in synaptic transmission and working memory, are also essential [186]. In addition, it is known that NMDARs in the CNS mainly comprise triheteromeric receptors consisting of the GluN1/GluN2A/GluN2B subunits. Other triheteromeric NMDARs, including GluN1/GluN2A/GluN2D and GluN1/GluN2B/GluN2D, have been observed in the human spinal cord as well as in the rat thalamus and midbrain. Much research to date has focused on diheteromeric NMDARs containing identical GluN2 or GluN3 subunits. However, the triheteromeric NMDAR containing a combination of GluN2 and/or GluN3 subunits has different channel gating kinetics and pharmacology from diheteromeric receptors [180]. With that in mind, extending protein research on NMDARs with additional subunits and studies showing their coexistence and variety in selected regions of the brain is essential.
Like NMDARs, AMPARs are abundantly located in the postsynaptic membrane and their effects are interdependent. During LTP, presynaptic Glu release activates AMPARs, and the triggered depolarization removes the Mg2+ blockade of the NMDA channel and allows Ca2+ influx. Strong activation of NMDARs triggers the Ca2+-calmodulin protein kinase II (CamKII) signaling cascade, which leads to LTP, brain-derived neurotrophic factor (BDNF) secretion and synaptic amplification [185]. Human post-mortem studies of AMPAR-building proteins show much greater variability compared to NMDAR analyses. The vast majority of these studies concern gene expression [68,113,120], while one study shows changes in protein levels [74]. Two papers show no change in the GRIA1 gene expression [68,113], and one of them shows its decrease [120]. In this situation, however, attention should be paid to the structural heterogeneity of the analyzed brain tissue. On the other hand, sexual differentiation is a likely cause of heterogeneous observations in the case of GRIA2 gene expression. Duric et al. [120] in the hippocampus and Chandley et al. [113] in the locus coeruleus showed no changes in the GRIA2 level, while Gray et al. [68] showed an increase in the expression of this gene in women and no changes in the group of men. Notably, most of the groups studied in the work of Chandley et al. and Duric et al. were men [113,120]. In addition, a decrease in GRIA3 expression and no change in GRIA4 level in MDD patients has been shown [120]. The observed AMPAR subunits changes have implications for the functionality of the entire receptor. It is indicated that AMPARs being a combination of GluA1 and GluA2 are essential for the plasticity of neurons and are rapidly recycled, therefore their number in the cell membrane reflects the balance between endo- and exocytosis processes. GluA1 subunits are delivered to the synapses in an activity-dependent manner, and GluA2/3 subunits are continuously provided to synapses independently of synaptic activity. Therefore, the trafficking process is an essential mechanism underlying synaptic plasticity since the recruitment of AMPAR to the postsynaptic membrane is positively correlated with LTP, and their endocytosis negatively correlated with LTP [185].
The most crucial PSD protein, necessary to maintain the molecular organization of postsynaptic density, anchors NMDARs and AMPARs, and mediates intracellular signaling is PSD-95 [187] (Figure 1). So, it is not surprising that it has been extensively studied in the context of DDs pathophysiology. Among the studies, almost all showed a reduced level of PSD-95 protein in both suicides and patients with MDD, including those who reported suicide [74,114,117,119]. Only one study indicates an increase in PSD-95 levels, but this may be due to a different brain structure (lateral amygdala) [118]. On the other hand, studies of DLG4 gene expression indicate its decrease, and this effect may vary depending on the diagnosis (MDD vs. BD) [121].
Peripheral blood is also a source of PSD protein mRNA. It has been observed that the level of SHANK3 in peripheral blood mononuclear cells (PBMCs) correlates with treatment response among women with MDD, indicating the potential employment of SHANK3 as a marker of antidepressant response [124]. Although some studies suggest a genetic predisposition to DDs, Somani et al. showed that neutrophils of drug-naïve MDD patients significantly have higher nNOS mRNA expression, but no such correlation was found among first-degree relatives of these patients [99]. In contrast, the locus coeruleus showed reduced nNOS protein immunoreactivity, which was absent in the cerebellum [122]. Different results were observed in the lateral amygdala as there were no significant differences in nNOS protein levels in patients with MDD and adjustment disorder with depressed mood [118]. In both nNOS analyses, post-mortem toxicology studies did not show antidepressant drugs use. This is a fundamental fact, because a case-control treatment study showed that the activity of NOS in the blood was initially lower in patients with depression than in healthy controls but increased in patients who responded to treatment. In this study, no changes in NOS activity were found in depressed patients who were not receiving drugs and were not responding to treatment. There is also no correlation between the class of antidepressants used and changes in NOS activity [123].
3.2. Animal Studies
In research on the pathophysiology of depression and amid the search for new, more effective antidepressants, animal models that initiate behavior similar to depression in humans are most often used. The models based on the use of various stressors are the best validated and most frequently applicated. Among them, we find: Chronic Unpredictable Stress (CUS), Chronic Unpredictable Mild Stress (CUMS), Chronic Restraint Stress (RST), Chronic Social Defeat Stress (CSDS), and Chronic Mild Stress (CMS) [188,189].
The stress response in different brain regions can vary dramatically. As is already known, one of the causes of depression is neurotransmission dysfunction in the brain. Available research suggests that the glutamatergic system is involved in the pathophysiology and treatment of depression [145]. NMDARs, especially GluN2A- and GluN2B-containing, are essential for regulating neuronal plasticity [128]. Molecular analysis in the hippocampus of female mice showed an increase in the expression of Grin2A and Grin2B genes after CUS and after administration of two hormonal compounds (estradiol, progesterone) [131]. Reports suggest estrogens’ role in structural and functional synaptic plasticity and long-term potentiation (LTP) in the hippocampus [131]. In the same brain region, the levels of GluN2A, GluN2B proteins were elevated after CUMS [128]. Interestingly, the increased levels of genes encoding NMDAR proteins were demonstrated by Tamasi et al. also after venlafaxine treatment in rats [152]. On the other hand, administration of lurasidone and fluoxetine to mice and infenprodil to rats also lowered the levels of these proteins in the hippocampus, prefrontal cortex, and medial prefrontal cortex (p-GluN2B), respectively [127,145]. Moreover, the analysis of NMDA genes and proteins after 28 days of ketamine administration showed a significant decrease in the hippocampus [130]. In contrast, in the basolateral and inferior limbic prefrontal cortex, ketamine decreased the expression of only GluN2B protein [129]. Other authors also indicate that GluN2A protein levels in the hippocampus were significantly lower than GluN2B in FSL rats [146]. In turn, in a rat model of zinc deficiency, GluN2A and GluN2B protein levels were reduced, while fluoxetine treatment had no significant effect [73]. Given the above, it can be concluded that changes in NMDAR expression levels may be largely dependent on factors inducing behavioral changes and appear more critical for the antidepressant response.
AMPARs for Glu, particularly those containing the GluA1 and GluA2 subunits, also contribute to normal plasticity of neurons, and various factors can modulate their in a multidirectional manner. For example, in rats, stress increased the GRIA1 expression in the paraventricular nucleus of the hypothalamus and the level of the GluA1 in the hypothalamus [161]. On the contrary, in stressed mice, the levels of AMPAR protein (and selected subunits) were significantly lower in the medial frontal cortex [135] and the prefrontal cortex [137]. The surface receptor crosslinking of BS3 showed a decrease in GluA1 and GluA2 levels associated with stress in the hippocampus [139]. Emerging data suggest that some fast-acting drugs in DDs, such as scopolamine, increase Glu release and induce neurotrophic factors through AMPAR activation [137]. It was observed that the administration of fluoxetine [135] and scopolamine [137] reversed stress-induced changes. In contrast, chronic stress increased in GluA1 and p-s845-GluA1 levels in the basolateral amygdala, while fluoxetine therapy decreased their levels [136]. Chronic ketamine administration caused a decrease in AMPARs, both gene and protein levels in the whole hippocampus and CA1 region [130]. Moreover, treatment with infenprodil, fluoxetine, and S-ketamine, venlafaxine, and NaHS normalized GluA1 protein stress-induced changes in the medial prefrontal cortex and hippocampus, respectively [145,150,154,156,162]. In its turn, the application of ketamine and memantine, induced a significant increase in the pS845-GluA1 protein subunit in the hippocampus [160]. Another interesting line of research turned out to be lipopolysaccharide-induced inflammation that caused a substantial decrease in GluA1 protein in the hippocampus while the administration of ketamine and Ac-YVAD-CMK (the selective NLRP3 inflammasome inhibitor) reversed its adverse effects [134].
PSD proteins, such as PSD-95, CamKII, Homer 1, and Shank3 may also be involved in the development of depressive disorders. Analysis of the Dlg4 gene of stressed animals revealed a decrease in this gene in the prefrontal cortex, hippocampus, and hypothalamus in mice [167] and gene and protein increases in the hypothalamus in rats [161]. On the other hand, in the hippocampus, it showed a decrease in protein in stressed animals [166]. Interestingly, PSD-95 protein levels were significantly higher in the basolateral amygdala after stress [136]. Application of fluoxetine, asioaticoside in mice, and ketamine or sodium hydrosulfide—NaHS (CA1, CA3 region) in rats increased PSD-95 protein levels in the hippocampus [156,162,166]. In contrast, in the basolateral amygdala, fluoxetine increased protein levels [136]. Chronic administration of ketamine decreased PSD-95 protein levels in the hippocampus, while in combination with betaine it increased them [164]. In stressed rats, administration of lurasidone increased Dlg4 gene levels in the prefrontal cortex [175]. In contrast, in male mice, lurasidone and fluoxetine induced a decrease in PSD-95 after both drugs in the hippocampus and prefrontal cortex [127]. It was also demonstrated that infenprodil and YY-21 decreased protein expression after CUMS in the medial prefrontal cortex in rats [145,150]. In the RST model, imipramine administration increased PSD-95 expression in lateral nuclei and basal nuclei and decreased in medial prefrontal cortex [165]. The application of CX717 therapy increased protein levels in mPFC (2h after administration) [174]. Leem et al. also showed a decrease in p-CAMKII expression in basolateral amygdala (BA) and an increase in medial prefrontal cortex (mPFC) after drug administration [165]. CAMKII dysfunction is involved in many neurological disorders, including depression. Short-term manipulation of CAMKII can result in long-term effects on disease-related behavior. On this basis, it can cause structural changes which in turn contribute to the progression of the disease and its duration [170]. Sleep deprivation caused a decrease in the CAMKII protein subunit in the prefrontal cortex and hippocampus and administration of citalopram reversed its effects in mice [168,171]. In addition, venlafaxine treatment increased the levels of Camk2g and Camk2b genes in the frontal cortex in rats [152].
Homer 1 also has its role in synaptic plasticity. Homer genes encode a family of proteins in the PSD, where they act as multimodal adaptors. Overexpression of Homer 1a protein may contribute to decreased density of postsynaptic proteins such as Shank and inhibit postsynaptic AMPAR and NMDAR currents. Moreover, it is also involved in Glu-induced changes in the distribution of pre- and postsynaptic proteins [96]. In stressed mice, a reduction in the Homer 1a gene expression in the prefrontal cortex and protein level in both the prefrontal cortex and hippocampus were noted [128,172]. Contrary observations were made in the prefrontal cortex of stressed rats in which an increase in the Homer 1 protein was shown [177]. Furthermore, the administration of imipramine, ketamine and fluoxetine increased Homer 1a, Homer 1b/c gene expression in the cortex of mice [172].
At the cellular level, zinc is one of the main enzymes involved in biochemical processes, and disturbance of homeostasis leads to physiological or pathological problems. Despite the high demand for zinc in cells, its levels must be kept low. Among others, the ZnT protein contributes to the maintenance of zinc balance. Rafalo-Ulinska et al. showed a decrease in this protein after zinc and imipramine supplementation in the prefrontal cortex, and an increase in the hippocampus. In contrast, supplementation with zinc alone contributed to a decreased ZnT-1 protein in the prefrontal cortex [173].
An important signaling molecule in CNS regulating anxiety behavior is nNOS. It has been shown that nNOS-derived free radical (NO) contributes to depression caused by chronic stress [190]. nNOS activity is also coupled to NMDAR activity via the membrane-bound postsynaptic density protein PSD-95. While PSD-95 expression is inhibited, calcium ion-activated NO production via NMDAR activation is blocked, and excitotoxicity is reduced [191]. In a rat model of CUS, memantine therapy reduced nNOS protein levels [179]. Interestingly, Yin et al. demonstrated the importance of gender on nNOS protein level. In a mouse stress model, female hippocampal nNOS protein levels decreased while male increased [169]. In addition, it seems of interest to use escitalopram and Yueju-Ganmaidazao (YG), which reversed the levels of this protein in stressed animals [169].
4. Postsynaptic Density Proteins as Therapeutic Targets for DDs
Among the discussed post-synaptic proteins of the excitatory synapse, the most promising seem to be research on the possibilities of modulating glutamatergic transmission by influencing Glu receptors. Because numerous preclinical studies have shown the function of various ligands or modulators of glutamate receptors, in this paper, the authors will focus mainly on clinical trials, which give more information about the safety and effectiveness of new antidepressant therapy. Targeting pharmacotherapy of DDs to disrupt glutamatergic signaling began with using low sub-anesthetic doses of ketamine, a drug commonly used in anesthesiology [192]. The first clinical trial in patients with MDD and BD using a 40-min infusion of racemic ketamine intravenously at a subanesthetic dose of 0.5 mg/kg was conducted by Berman et al. In the ketamine group, a sustained 72 h change in scores expressed by the 25-item Hamilton Depression Rating Scale (HDRS/HAM-D) have been noticed. A the same time, in the control group, there were no significant differences [193]. Zarate et al. used the same protocol of ketamine treatment (dose, route of administration) in TRD patients. Results indicate that ketamine induced a rapid antidepressant effect (measured by the 21-item HAM-D scale and Beck Depression Inventory (BDI) that remained for seven days [194]. This finding contributed to the subsequent studies on ketamine with multiple intravenous administration in a larger group of patients. Among 97 patients with TRD, 205 ketamine intravenous infusions (0.5 mg/kg/40 min; performed 2006 and 2012) showed antidepressant activity (improvement in Montgomery–Asberg Depression Rating Scale (MADRS) scores) in 67% of TRD individuals [195]. Ketamine was also administered by intramuscular (i.m.) injection with nearly 100% bioavailability. At 0.5 mg/kg dose, there is an improved antidepressant response in MDD subjects, similar to effects observed in the group receiving electroconvulsive therapy (ECT). Moreover, oral ketamine administration at a 1 mg/kg dose (despite its relatively low bioavailability on range 20–25%) for three weeks showed pharmacological efficacy (measured by the 17-item HAM-D and Beck Scale for Suicidal Ideation (BSSI) scales) compared to ECT [196]. On the other hand, sublingual ketamine shows a slightly higher bioavailability (30%), but even more importantly, indicates an improvement in mood, sleep, and cognitive functions in 26 patients with MDD and BD. Transient light-headedness was reported side-effects of this type of treatment [197]. Moreover, the Food and Drug Administration (FDA) approved esketamine nasal spray in 2019 and it is now a promising new therapeutic option for treating TRD [198].
In clinical trials, other NMDAR antagonists were also used, e.g., memantine (an uncompetitive, low-affinity, and selective open-channel blocker of NMDAR). A growing body of evidence shows that memantine significantly reduced depressive symptom scores in MDD patients [199]. Moreover, limited evidence shows its effectiveness in bipolar patients as an add-on treatment [200]. Recently, the increasing number of substances targeted to regulation-specific NMDAR subunits show potential antidepressant efficacy [201]. One of them is traxoprodil (CP-101,606), an antagonist of the GluN2B subunit, the use of which in monotherapy and in combination with escitalopram, imipramine, or fluoxetine in male Swiss mice resulted in a reduction of immobility time in a forced swim test (FST) [202]. Moreover, in a group of patients using paroxetine (but not responding to selective serotonin reuptake inhibitors; SSRIs), intravenous infusion of traxoprodil (0.75 mg/kg/h for 1.5 h and then 0.15 mg/kg/h for 6.5 h) resulted in a significant reduction in MADRS scores and a higher response rate in 17-item HAM-D compared to the placebo group [203]. Furthermore, a small clinical trial involving five patients with TRD showed that oral intake 4–8 mg/day of another GluN2B antagonist, rislenemdaz (MK-0657), resulted in significant changes in 17-item HDRS and BDI scale scores compared to placebo [204]. In addition, other GluN2B subunit antagonists such as EVT-101 (also known as ENS-101) or MIJ821 are the subject of clinical trials [201,205].
Another strategy of experimental pharmacology of DDs is based on the use of partial NMDAR agonists. [192]. The example is D-cycloserine (DCS), a partial agonist at NMDAR-associates glycine. Heresco-Levy et al. showed that oral administration of DCS at a gradually increasing dose (up to max 1000 mg/day) resulted in significant clinical improvement as measured by the 21-item HAM-D and BDI scales in TRD patients when receiving higher doses [206]. The following partial NMDAR agonist is rapastinel (GLYX-13), which affects the glycine site of this receptor. In male Sprague-Dawley rats subjected to 14 days of adrenocorticotropic hormone (ACTH) injection, it was observed that the administration of rapastinel (10 mg/kg, i.p.) significantly reduced immobilization time in the FST but did not affect the distance traveled in the open field test (OFT) [207]. A clinical trial with intravenous rapastinel in patients with MDD showed that 5 mg/kg and 10 mg/kg doses of GLYX-13 had a significant clinical effect as measured by the 17-item HAM-D scale, which lasted from 2 h to 7 days after administration [208].
Potential antidepressant effectiveness was found for “ampakines”, the compounds that potentiate AMPAR function. Intraperitoneal injection of 3 mg/kg/day of S47445 decreased olfactory bulbectomy-induced motor hyperactivity in male C57BL/6J mice. Still, oral intake of 400 mg of another AMPAR positive allosteric modulator (ORG 26576) by MDD patients did not significantly produce clinical antidepressant effects [209,210].
Due to the involvement of mGluRs in the pathogenesis of DDs, they are also targets for new pharmacological therapy. Many of them are focused on negative allosteric modulators of mGluR5. Among them, compounds, such as basimglurant (RG-7090, RO-4917523) and AZD2066, are in clinical trials, but their effects are inconclusive and further studies are required to determine their therapeutic benefits [192,201].
5. Conclusions
DDs represent a serious and growing health problem among both young and older adults, and if left untreated, they can lead to a suicide death. The appearance of a series of biochemical, electrophysiological, and behavioral studies in the 1990s indicated significant changes in the glutamatergic system in depressed people which, after antidepressant treatment or ECT, contributed to the formulation of the glutamatergic theory of depression and initiated intensive research in this field. It was essential to describe the changes taking place in the Glu synapse, both in depression and after effective antidepressant therapy. The first studies focused mainly on the NMDA and AMPA ionotropic receptors, while subsequent studies considered mGluRs. Despite the large variety of available studies, taking into account the research methodology, brain region, or heterogeneity of the studied groups, it can be concluded that changes in the GluN2A and GluN2B subunits in DDs were most often identified. In the case of the AMPARs, most studies show changes in the GluA1 subunit. Therefore, it seems that these should be the most important Glu therapeutic targets for potential new antidepressant therapies. This is confirmed by clinical studies of these receptors’ new ligands/modulators (e.g., traxoprodil). Over the years, the view of excitatory synapse function has changed dramatically, such that the static structure has become highly dynamic, in which postsynaptic density proteins play an important role in both the localization and function of Glu receptors and can modify intracellular signaling. The importance of these proteins, in the development of DDs is also evidenced by numerous studies that indicate a reduced level of PSD-95, CamKII, and Homer 1 proteins. Hence, apart from the receptor mentioned above, these proteins should also become an exciting direction in the searching for more effective drugs to combat DDs.
Author Contributions
Conceptualization: M.S.-K.; formal analysis: S.S., E.C. and M.S.-K.; funding acquisition: M.S.-K.; investigation (literature review and collection): S.S., E.C. and M.S.-K.; writing—original draft: S.S., E.C. and M.S.-K.; writing—review and editing: S.S., P.P.-T. and M.S.-K. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
Funding Statement
The study was supported by grants from the National Science Centre (contracts UMO-2016/21/B/NZ7/01623 to M. Sowa-Kućma). The funder had no role in the study design, data and literature collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.World Health Organization (WHO) Depression. [(accessed on 9 June 2022)]. Available online: https://www.who.int/news-room/fact-sheets/detail/depression.
- 2.Zenebe Y., Akele B., Woldeselassie Gebre M., Necho M. Prevalence and determinants of depression among old age: A systematic review and meta-analysis. Ann. Gen. Psychiatry. 2021;20:55. doi: 10.1186/s12991-021-00375-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Duman R.S., Sanacora G., Krystal J.H. Altered Connectivity in Depression: GABA and Glutamate Neurotransmitter Deficits and Reversal by Novel Treatments. Neuron. 2019;102:75–90. doi: 10.1016/j.neuron.2019.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.World Health Organization (WHO) Adolescent Mental Health. [(accessed on 9 June 2022)]. Available online: https://www.who.int/news-room/fact-sheets/detail/adolescent-mental-health.
- 5.Hossain M.M., Nesa F., Das J., Aggad R., Tasnim S., Bairwa M., Ma P., Ramirez G. Global burden of mental health problems among children and adolescents during COVID-19 pandemic: An umbrella review. Psychiatry Res. 2022;317:114814. doi: 10.1016/j.psychres.2022.114814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Thapar A., Eyre O., Patel V., Brent D. Depression in young people. Lancet. 2022;400:617–631. doi: 10.1016/S0140-6736(22)01012-1. [DOI] [PubMed] [Google Scholar]
- 7.Thompson S.M., Kallarackal A.J., Kvarta M.D., Van Dyke A.M., LeGates T.A., Cai X. An excitatory synapse hypothesis of depression. Trends Neurosci. 2015;38:279–294. doi: 10.1016/j.tins.2015.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tian H., Hu Z., Xu J., Wang C. The molecular pathophysiology of depression and the new therapeutics. MedComm. 2022;3:e156. doi: 10.1002/mco2.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Blackburn T.P. Depressive disorders: Treatment failures and poor prognosis over the last 50 years. Pharmacol. Res. Perspect. 2019;7:e00472. doi: 10.1002/prp2.472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Voineskos D., Daskalakis Z.J., Blumberger D.M. Management of Treatment-Resistant Depression: Challenges and Strategies. Neuropsychiatr. Dis. Treat. 2020;16:221–234. doi: 10.2147/NDT.S198774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Adu M.K., Shalaby R., Chue P., Agyapong V.I.O. Repetitive Transcranial Magnetic Stimulation for the Treatment of Resistant Depression: A Scoping Review. Behav. Sci. 2022;12:195. doi: 10.3390/bs12060195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Papalexi E., Galanopoulos A., Roukas D., Argyropoulos I., Michopoulos I., Douzenis A., Gkolia I., Fotiadis P., Kontis D., Zervas I.M. Residual cognitive and psychosocial functional impairment in outpatients in Greece who responded to conventional antidepressant monotherapy treatments for major depressive disorder (MDD) J. Affect. Disord. 2022;314:185–192. doi: 10.1016/j.jad.2022.07.009. [DOI] [PubMed] [Google Scholar]
- 13.Saez E., Erkoreka L., Moreno-Calle T., Berjano B., Gonzalez-Pinto A., Basterreche N., Arrue A. Genetic variables of the glutamatergic system associated with treatment-resistant depression: A review of the literature. World J. Psychiatry. 2022;12:884–896. doi: 10.5498/wjp.v12.i7.884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chevance A., Tomlinson A., Ravaud P., Touboul S., Henshall C., Tran V.T., Cipriani A. Important adverse events to be evaluated in antidepressant trials and meta-analyses in depression: A large international preference study including patients and healthcare professionals. Evid. Based Ment. Health. 2022 doi: 10.1136/ebmental-2021-300418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pilc A., Machaczka A., Kawalec P., Smith J.L., Witkin J.M. Where do we go next in antidepressant drug discovery? A new generation of antidepressants: A pivotal role of AMPA receptor potentiation and mGlu2/3 receptor antagonism. Expert Opin. Drug Discov. 2022;22:1–16. doi: 10.1080/17460441.2022.2111415. [DOI] [PubMed] [Google Scholar]
- 16.Hartig J., Nemeș B. BDNF-related mutations in major depressive disorder: A systematic review. Acta Neuropsychiatr. 2022;22:1–61. doi: 10.1017/neu.2022.22. [DOI] [PubMed] [Google Scholar]
- 17.Ramos-da-Silva L., Carlson P.T., Silva-Costa L.C., Martins-de-Souza D., de Almeida V. Molecular Mechanisms Associated with Antidepressant Treatment on Major Depression. Complex Psychiatry. 2021;7:49–59. doi: 10.1159/000518098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bijata M., Bączyńska E., Wlodarczyk J. A chronic unpredictable stress protocol to model anhedonic and resilient behaviors in C57BL/6J mice. STAR Protoc. 2022;3:101659. doi: 10.1016/j.xpro.2022.101659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Koert A., Ploeger A., Bockting C.L.H., Schmidt M.V., Lucassen P.J., Schrantee A., Mul J.D. The social instability stress paradigm in rat and mouse: A systematic review of protocols, limitations, and recommendations. Neurobiol. Stress. 2021;15:100410. doi: 10.1016/j.ynstr.2021.100410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim I.B., Lee J.H., Park S.C. The Relationship between Stress, Inflammation, and Depression. Biomedicines. 2022;10:1929. doi: 10.3390/biomedicines10081929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Willner P., Gruca P., Lason M., Tota-Glowczyk K., Litwa E., Niemczyk M., Papp M. Validation of chronic mild stress in the Wistar-Kyoto rat as an animal model of treatment-resistant depression. Behav. Pharmacol. 2019;30:239–250. doi: 10.1097/FBP.0000000000000431. [DOI] [PubMed] [Google Scholar]
- 22.Zhao Y., Zhang Q., Shao X., Ouyang L., Wang X., Zhu K., Chen L. Decreased Glycogen Content Might Contribute to Chronic Stress-Induced Atrophy of Hippocampal Astrocyte volume and Depression-like Behavior in Rats. Sci. Rep. 2017;7:43192. doi: 10.1038/srep43192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Evans J.W., Szczepanik J., Brutsché N., Park L.T., Nugent A.C., Zarate C.A., Jr. Default Mode Connectivity in Major Depressive Disorder Measured Up to 10 Days After Ketamine Administration. Biol. Psychiatry. 2018;84:582–590. doi: 10.1016/j.biopsych.2018.01.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim H., Han K.M., Choi K.W., Tae W.S., Kang W., Kang Y., Kim A., Ham B.J. Volumetric alterations in subregions of the amygdala in adults with major depressive disorder. J. Affect. Disord. 2021;295:108–115. doi: 10.1016/j.jad.2021.08.012. [DOI] [PubMed] [Google Scholar]
- 25.Belleau E.L., Treadway M.T., Pizzagalli D.A. The Impact of Stress and Major Depressive Disorder on Hippocampal and Medial Prefrontal Cortex Morphology. Biol. Psychiatry. 2019;85:443–453. doi: 10.1016/j.biopsych.2018.09.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chenani A., Weston G., Ulivi A.F., Castello-Waldow T.P., Huettl R.E., Chen A., Attardo A. Repeated stress exposure leads to structural synaptic instability prior to disorganization of hippocampal coding and impairments in learning. Transl. Psychiatry. 2022;12:381. doi: 10.1038/s41398-022-02107-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brosch K., Stein F., Schmitt S., Pfarr J.K., Ringwald K.G., Thomas-Odenthal F., Meller T., Steinsträter O., Waltemate L., Lemke H., et al. Reduced hippocampal gray matter volume is a common feature of patients with major depression, bipolar disorder, and schizophrenia spectrum disorders. Mol. Psychiatry. 2022 doi: 10.1038/s41380-022-01687-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yang Y., Li X., Cui Y., Liu K., Qu H., Lu Y., Li W., Zhang L., Zhang Y., Song J., et al. Reduced Gray Matter Volume in Orbitofrontal Cortex Across Schizophrenia, Major Depressive Disorder, and Bipolar Disorder: A Comparative Imaging Study. Front. Neurosci. 2022;16:919272. doi: 10.3389/fnins.2022.919272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Phillips C. Brain-Derived Neurotrophic Factor, Depression, and Physical Activity: Making the Neuroplastic Connection. Neural. Plast. 2017;2017:7260130. doi: 10.1155/2017/7260130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gałecki P., Talarowska M. Inflammatory theory of depression. Psychiatr. Pol. 2018;52:437–447. doi: 10.12740/PP/76863. [DOI] [PubMed] [Google Scholar]
- 31.Xu L., Sun H., Qu C., Shen J., Qu C., Song H., Li T., Zheng J., Zhang J. The environmental enrichment ameliorates chronic unpredictable mild stress-induced depressive-like behaviors and cognitive decline by inducing autophagy-mediated inflammation inhibition. Brain Res. Bull. 2022;187:98–110. doi: 10.1016/j.brainresbull.2022.07.001. [DOI] [PubMed] [Google Scholar]
- 32.Mograbi K.M., Suchecki D., da Silva S.G., Covolan L., Hamani C. Chronic unpredictable restraint stress increases hippocampal pro-inflammatory cytokines and decreases motivated behavior in rats. Stress. 2020;23:427–436. doi: 10.1080/10253890.2020.1712355. [DOI] [PubMed] [Google Scholar]
- 33.Koo J.W., Chaudhury D., Han M.H., Nestler E.J. Role of Mesolimbic Brain-Derived Neurotrophic Factor in Depression. Biol. Psychiatry. 2019;86:738–748. doi: 10.1016/j.biopsych.2019.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sowa-Kućma M., Styczeń K., Siwek M., Misztak P., Nowak R.J., Dudek D., Rybakowski J.K., Nowak G., Maes M. Lipid Peroxidation and Immune Biomarkers Are Associated with Major Depression and Its Phenotypes, Including Treatment-Resistant Depression and Melancholia. Neurotox. Res. 2018;33:448–460. doi: 10.1007/s12640-017-9835-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Popoli M., Yan Z., McEwen B.S., Sanacora G. The stressed synapse: The impact of stress and glucocorticoids on glutamate transmission. Nat. Rev. Neurosci. 2011;13:22–37. doi: 10.1038/nrn3138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Reagan L.P., Reznikov L.R., Evans A.N., Gabriel C., Mocaër E., Fadel J.R. The antidepressant agomelatine inhibits stress-mediated changes in amino acid efflux in the rat hippocampus and amygdala. Brain Res. 2012;1466:91–98. doi: 10.1016/j.brainres.2012.05.039. [DOI] [PubMed] [Google Scholar]
- 37.Reznikov L.R., Grillo C.A., Piroli G.G., Pasumarthi R.K., Reagan L.P., Fadel J. Acute stress-mediated increases in extracellular glutamate levels in the rat amygdala: Differential effects of antidepressant treatment. Eur. J. Neurosci. 2007;25:3109–3114. doi: 10.1111/j.1460-9568.2007.05560.x. [DOI] [PubMed] [Google Scholar]
- 38.Blacker C.J., Millischer V., Webb L.M., Ho A.M.C., Schalling M., Frye M.A., Veldic M. EAAT2 as a Research Target in Bipolar Disorder and Unipolar Depression: A Systematic Review. Mol. Neuropsychiatry. 2020;5((Suppl. S1)):44–59. doi: 10.1159/000501885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gruenbaum B.F., Zlotnik A., Frenkel A., Fleidervish I., Boyko M. Glutamate Efflux across the Blood-Brain Barrier: New Perspectives on the Relationship between Depression and the Glutamatergic System. Metabolites. 2022;12:459. doi: 10.3390/metabo12050459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sarawagi A., Soni N.D., Patel A.B. Glutamate and GABA Homeostasis and Neurometabolism in Major Depressive Disorder. Front. Psychiatry. 2021;27:637863. doi: 10.3389/fpsyt.2021.637863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Henter I.D., Park L.T., Zarate C.A., Jr. Novel Glutamatergic Modulators for the Treatment of Mood Disorders: Current Status. CNS Drugs. 2021;35:527–543. doi: 10.1007/s40263-021-00816-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jesus-Nunes A.P., Leal G.C., Correia-Melo F.S., Vieira F., Mello R.P., Caliman-Fontes A.T., Echegaray M.V.F., Marback R.F., Guerreiro-Costa L.N.F., Souza-Marques B., et al. Clinical predictors of depressive symptom remission and response after racemic ketamine and esketamine infusion in treatment-resistant depression. Hum. Psychopharmacol. Clin. Exp. 2022;37:e2836. doi: 10.1002/hup.2836. [DOI] [PubMed] [Google Scholar]
- 43.Abbar M., Demattei C., El-Hage W., Llorca P.M., Samalin L., Demaricourt P., Gaillard R., Courtet P., Vaiva G., Gorwood P., et al. Ketamine for the acute treatment of severe suicidal ideation: Double blind, randomised placebo controlled trial. BMJ. 2022;376:e067194. doi: 10.1136/bmj-2021-067194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hochschild A., Keilp J.G., Madden S.P., Burke A.K., Mann J.J., Grunebaum M.F. Ketamine vs midazolam: Mood improvement reduces suicidal ideation in depression. J. Affect. Disord. 2022;300:10–16. doi: 10.1016/j.jad.2021.12.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pochwat B., Nowak G., Szewczyk B. An update on NMDA antagonists in depression. Expert Rev. Neurother. 2019;19:1055–1067. doi: 10.1080/14737175.2019.1643237. [DOI] [PubMed] [Google Scholar]
- 46.Satarker S., Bojja S.L., Gurram P.C., Mudgal J., Arora D., Nampoothiri M. Astrocytic Glutamatergic Transmission and Its Implications in Neurodegenerative Disorders. Cells. 2022;11:1139. doi: 10.3390/cells11071139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chen Q.Y., Li X.H., Zhuo M. NMDA receptors and synaptic plasticity in the anterior cingulate cortex. Neuropharmacology. 2021;197:108749. doi: 10.1016/j.neuropharm.2021.108749. [DOI] [PubMed] [Google Scholar]
- 48.Micheli P., Ribeiro R., Giorgetti A. A Mechanistic Model of NMDA and AMPA Receptor-Mediated Synaptic Transmission in Individual Hippocampal CA3-CA1 Synapses: A Computational Multiscale Approach. Int. J. Mol. Sci. 2021;22:1536. doi: 10.3390/ijms22041536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Purkey A.M., Dell’Acqua M.L. Phosphorylation-Dependent Regulation of Ca2+-Permeable AMPA Receptors During Hippocampal Synaptic Plasticity. Front. Synaptic Neurosci. 2020;12:8. doi: 10.3389/fnsyn.2020.00008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lira M., Mira R.G., Carvajal F.J., Zamorano P., Inestrosa N.C., Cerpa W. Glutamatergic Receptor Trafficking and Delivery: Role of the Exocyst Complex. Cells. 2020;9:2402. doi: 10.3390/cells9112402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yang X., Gong R., Qin L., Bao Y., Fu Y., Gao S., Yang H., Ni J., Yuan T.F., Lu W. Trafficking of NMDA receptors is essential for hippocampal synaptic plasticity and memory consolidation. Cell Rep. 2022;40:111217. doi: 10.1016/j.celrep.2022.111217. [DOI] [PubMed] [Google Scholar]
- 52.Wu Q.L., Gao Y., Li J.T., Ma W.Y., Chen N.H. The Role of AMPARs Composition and Trafficking in Synaptic Plasticity and Diseases. Cell. Mol. Neurobiol. 2021 doi: 10.1007/s10571-021-01141-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sumi T., Harada K. Mechanism underlying hippocampal long-term potentiation and depression based on competition between endocytosis and exocytosis of AMPA receptors. Sci. Rep. 2020;10:14711. doi: 10.1038/s41598-020-71528-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Feng Z., Chen X., Zeng M., Zhang M. Phase separation as a mechanism for assembling dynamic postsynaptic density signalling complexes. Curr. Opin. Neurobiol. 2019;57:1–8. doi: 10.1016/j.conb.2018.12.001. [DOI] [PubMed] [Google Scholar]
- 55.Sathler M.F., Khatri L., Roberts J.P., Schmidt I.G., Zaytseva A., Kubrusly R.C.C., Ziff E.B., Kim S. Phosphorylation of the AMPA receptor subunit GluA1 regulates clathrin-mediated receptor internalization. J. Cell Sci. 2021;134:jcs257972. doi: 10.1242/jcs.257972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Giese K.P. The role of CaMKII autophosphorylation for NMDA receptor-dependent synaptic potentiation. Neuropharmacology. 2021;193:108616. doi: 10.1016/j.neuropharm.2021.108616. [DOI] [PubMed] [Google Scholar]
- 57.Pandey S., Ramsakha N., Sharma R., Gulia R., Ojha P., Lu W., Bhattacharyya S. The post-synaptic scaffolding protein tamalin regulates ligand-mediated trafficking of metabotropic glutamate receptors. J. Biol. Chem. 2020;295:8575–8588. doi: 10.1074/jbc.RA119.011979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhang Y., Matt L., Patriarchi T., Malik Z.A., Chowdhury D., Park D.K., Renieri A., Ames J.B., Hell J.W. Capping of the N-terminus of PSD-95 by calmodulin triggers its postsynaptic release. EMBO J. 2014;33:1341–1353. doi: 10.1002/embj.201488126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Thorsen. T.S., Madsen K.L., Rebola N., Rathje M., Anggono V., Bach A., Moreira I.S., Stuhr-Hansen N., Dyhring T., Peters D., et al. Identification of a small-molecule inhibitor of the PICK1 PD domain that inhibits hippocampal LTP and LTD. Proc. Natl. Acad. Sci. USA. 2010;107:413–418. doi: 10.1073/pnas.0902225107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bockaert J., Perroy J., Ango F. The Complex Formed by Group I Metabotropic Glutamate Receptor (mGluR) and Homer1a Plays a Central Role in Metaplasticity and Homeostatic Synaptic Scaling. J. Neurosci. 2021;41:5567–5578. doi: 10.1523/JNEUROSCI.0026-21.2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Fukaya M., Sugawara T., Hara Y., Itakura M., Watanabe M., Sakagami H. BRAG2a Mediates mGluR-Dependent AMPA Receptor Internalization at Excitatory Postsynapses through the Interaction with PSD-95 and Endophilin 3. J. Neurosci. 2020;40:4277–4296. doi: 10.1523/JNEUROSCI.1645-19.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kaizuka T., Takumi T. Postsynaptic density proteins and their involvement in neurodevelopmental disorders. J. Biochem. 2018;163:447–455. doi: 10.1093/jb/mvy022. [DOI] [PubMed] [Google Scholar]
- 63.Tao C.L., Liu Y.T., Sun R., Zhang B., Qi L., Shivakoti S., Tian C.L., Zhang P., Lau P.M., Zhou Z.H., et al. Differentiation and Characterization of Excitatory and Inhibitory Synapses by Cryo-electron Tomography and Correlative Microscopy. J. Neurosci. 2018;38:1493–1510. doi: 10.1523/JNEUROSCI.1548-17.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tomasetti C., Iasevoli F., Buonaguro E.F., De Berardis D., Fornaro M., Fiengo A.L., Martinotti G., Orsolini L., Valchera A., Di Giannantonio M., et al. Treating the Synapse in Major Psychiatric Disorders: The Role of Postsynaptic Density Network in Dopamine-Glutamate Interplay and Psychopharmacologic Drugs Molecular Actions. Int. J. Mol. Sci. 2017;18:135. doi: 10.3390/ijms18010135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wilkinson B., Coba M.P. Molecular architecture of postsynaptic Interactomes. Cell. Signal. 2020;76:109782. doi: 10.1016/j.cellsig.2020.109782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Moraes B.J., Coelho P., Fão L., Ferreira I.L., Rego A.C. Modified Glutamatergic Postsynapse in Neurodegenerative Disorders. Neuroscience. 2021;454:116–139. doi: 10.1016/j.neuroscience.2019.12.002. [DOI] [PubMed] [Google Scholar]
- 67.Baucum A.J., 2nd Proteomic Analysis of Postsynaptic Protein Complexes Underlying Neuronal Plasticity. ACS Chem. Neurosci. 2017;8:689–701. doi: 10.1021/acschemneuro.7b00008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Gray A.L., Hyde T.M., Deep-Soboslay A., Kleinman J.E., Sodhi M.S. Sex differences in glutamate receptor gene expression in major depression and suicide. Mol. Psychiatry. 2015;20:1057–1068. doi: 10.1038/mp.2015.91. [DOI] [PubMed] [Google Scholar]
- 69.Sanz-Clemente A., Nicoll R.A., Roche K.W. Diversity in NMDA receptor composition: Many regulators, many consequences. Neuroscientist. 2013;19:62–75. doi: 10.1177/1073858411435129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ryan T.J., Emes R.D., Grant S.G., Komiyama N.H. Evolution of NMDA receptor cytoplasmic interaction domains: Implications for organisation of synaptic signalling complexes. BMC Neurosci. 2008;9:6. doi: 10.1186/1471-2202-9-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Petrillil M.A., Kranz T.M., Kleinhaus K., Joe P., Getz M., Johnson P., Chao M.V., Malaspina D. The Emerging Role for Zinc in Depression and Psychosis. Front. Pharmacol. 2017;8:414. doi: 10.3389/fphar.2017.00414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sindreu C., Bayés Á., Altafaj X., Pérez-Clausell J. Zinc transporter-1 concentrates at the postsynaptic density of hippocampal synapses. Mol Brain. 2014;7:16. doi: 10.1186/1756-6606-7-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Doboszewska U., Szewczyk B., Sowa-Kućma M., Młyniec K., Rafało A., Ostachowicz B., Lankosz M., Nowak G. Antidepressant activity of fluoxetine in the zinc deficiency model in rats involves the NMDA receptor complex. Behav. Brain Res. 2015;287:323–330. doi: 10.1016/j.bbr.2015.03.064. [DOI] [PubMed] [Google Scholar]
- 74.Rafalo-Ulinska A., Piotrowska J., Kryczyk A., Opoka W., Sowa-Kucma M., Misztak P., Rajkowska G., Stockmeier C.A., Datka W., Nowak G., et al. Zinc transporters protein level in postmortem brain of depressed subjects and suicide victims. J. Psychiatr. Res. 2016;83:220–229. doi: 10.1016/j.jpsychires.2016.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Chater T.E., Goda Y. The role of AMPA receptors in postsynaptic mechanisms of synaptic plasticity. Front. Cell. Neurosci. 2014;8:401. doi: 10.3389/fncel.2014.00401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zhao Y., Chen S., Swensen A.C., Qian W.J., Gouaux E. Architecture and subunit arrangement of native AMPA receptors elucidated by cryo-EM. Science. 2019;364:355–362. doi: 10.1126/science.aaw8250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Swanson G.T., Kamboj S.K., Cull-Candy S.G. Single-channel properties of recombinant AMPA receptors depend on RNA editing, splice variation, and subunit composition. J. Neurosci. 1997;17:58–69. doi: 10.1523/JNEUROSCI.17-01-00058.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Man H.Y. GluA2-lacking, calcium-permeable AMPA receptors—Inducers of plasticity? Curr. Opin. Neurobiol. 2011;21:291–298. doi: 10.1016/j.conb.2011.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kawahara Y., Ito K., Sun H., Kanazawa I., Kwak S. w editing efficiency of GluR2 mRNA is associated with a low relative abundance of ADAR2 mRNA in white matter of normal human brain. Eur. J. Neurosci. 2003;18:23–33. doi: 10.1046/j.1460-9568.2003.02718.x. [DOI] [PubMed] [Google Scholar]
- 80.Guo C., Ma Y.Y. Calcium Permeable-AMPA Receptors and Excitotoxicity in Neurological Disorders. Front. Neural. Circuits. 2021;15:711564. doi: 10.3389/fncir.2021.711564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Schwenk J., Baehrens D., Haupt A., Bildl W., Boudkkazi S., Roeper J., Fakler B., Schulte U. Regional diversity and developmental dynamics of the AMPA-receptor proteome in the mammalian brain. Neuron. 2014;84:41–54. doi: 10.1016/j.neuron.2014.08.044. [DOI] [PubMed] [Google Scholar]
- 82.Kamalova A., Nakagawa T. AMPA receptor structure and auxiliary subunits. J. Physiol. 2021;599:453–469. doi: 10.1113/JP278701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Greger I.H., Watson J.F., Cull-Candy S.G. Structural and Functional Architecture of AMPA-Type Glutamate Receptors and Their Auxiliary Proteins. Neuron. 2017;94:713–730. doi: 10.1016/j.neuron.2017.04.009. [DOI] [PubMed] [Google Scholar]
- 84.Laje G., Paddock S., Manji H., Rush A.J., Wilson A.F., Charney D., McMahon F.J. Genetic markers of suicidal ideation emerging during citalopram treatment of major depression. Am. J. Psychiatry. 2007;164:1530–1538. doi: 10.1176/appi.ajp.2007.06122018. [DOI] [PubMed] [Google Scholar]
- 85.Smart T.G., Paoletti P. Synaptic neurotransmitter-gated receptors. Cold Spring Harb. Perspect. Biol. 2012;4:a009662. doi: 10.1101/cshperspect.a009662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Shor C., Zuo W., Eloy J.D., Ye J.H. The Emerging Role of LHb CaMKII in the Comorbidity of Depressive and Alcohol Use Disorders. Int. J. Mol. Sci. 2020;21:8123. doi: 10.3390/ijms21218123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Proietti Onori M., van Woerden G.M. Role of calcium/calmodulin-dependent kinase 2 in neurodevelopmental disorders. Brain Res. Bull. 2021;171:209–220. doi: 10.1016/j.brainresbull.2021.03.014. [DOI] [PubMed] [Google Scholar]
- 88.Terbeck S., Akkus F., Chesterman L.P., Hasler G. The role of metabotropic glutamate receptor 5 in the pathogenesis of mood disorders and addiction: Combining preclinical evidence with human Positron Emission Tomography (PET) studies. Front. Neurosci. 2015;9:86. doi: 10.3389/fnins.2015.00086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Nicoletti F., Bockaert J., Collingridge G.L., Conn P.J., Ferraguti F., Schoepp D.D., Wroblewski J.T., Pin J.P. Metabotropic glutamate receptors: From the workbench to the bedside. Neuropharmacology. 2011;60:1017–1041. doi: 10.1016/j.neuropharm.2010.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Bhattacharyya S. Inside story of Group I Metabotropic Glutamate Receptors (mGluRs) Int. J. Biochem. Cell Biol. 2016;77:205–212. doi: 10.1016/j.biocel.2016.03.003. [DOI] [PubMed] [Google Scholar]
- 91.Kim J.H., Marton J., Ametamey S.M., Cumming P.A. Review of Molecular Imaging of Glutamate Receptors. Molecules. 2020;25:4749. doi: 10.3390/molecules25204749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Koehl A., Hu H., Feng D., Sun B., Zhang Y., Robertson M.J., Chu M., Kobilka T.S., Laeremans T., Steyaert J., et al. Structural insights into the activation of metabotropic glutamate receptors. Nature. 2019;566:79–84. doi: 10.1038/s41586-019-0881-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Dickson E.J., Falkenburger B.H., Hille B. Quantitative properties and receptor reserve of the IP(3) and calcium branch of G(q)-coupled receptor signaling. J. Gen. Physiol. 2013;141:521–535. doi: 10.1085/jgp.201210886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Jin D.Z., Xue B., Mao L.M., Wang J.Q. Metabotropic glutamate receptor 5 upregulates surface NMDA receptor expression in striatal neurons via CaMKII. Brain Res. 2015;1624:414–423. doi: 10.1016/j.brainres.2015.07.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Laursen L., Karlsson E., Gianni S., Jemth P. Functional interplay between protein domains in a supramodular structure involving the postsynaptic density protein PSD-95. J. Biol. Chem. 2020;295:1992–2000. doi: 10.1074/jbc.RA119.011050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.de Bartolomeis A., Latte G., Tomasetti C., Iasevoli F. Glutamatergic postsynaptic density protein dysfunctions in synaptic plasticity and dendritic spines morphology: Relevance to schizophrenia and other behavioral disorders pathophysiology, and implications for novel therapeutic approaches. Mol. Neurobiol. 2014;49:484–511. doi: 10.1007/s12035-013-8534-3. [DOI] [PubMed] [Google Scholar]
- 97.Chen X., Nelson C.D., Li X., Winters C.A., Azzam R., Sousa A.A., Leapman R.D., Gainer H., Sheng M., Reese T.S. PSD-95 is required to sustain the molecular organization of the postsynaptic density. J. Neurosci. 2011;31:6329–6338. doi: 10.1523/JNEUROSCI.5968-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Nikonenko I., Boda B., Steen S., Knott G., Welker E., Muller D. PSD-95 promotes synaptogenesis and multiinnervated spine formation through nitric oxide signaling. J. Cell Biol. 2008;183:1115–1127. doi: 10.1083/jcb.200805132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Somani A., Singh A.K., Gupta B., Nagarkoti S., Dalal P.K., Dikshit M. Oxidative and Nitrosative Stress in Major Depressive Disorder: A Case Control Study. Brain Sci. 2022;12:144. doi: 10.3390/brainsci12020144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Wang Y., Rao W., Zhang C., Zhang C., Liu M.D., Han F., Yao L.B., Han H., Luo P., Su N., et al. Scaffolding protein Homer1a protects against NMDA-induced neuronal injury. Cell Death Dis. 2015;6:e1843. doi: 10.1038/cddis.2015.216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Shiraishi-Yamaguchi Y., Furuichi T. The Homer family proteins. Genome Biol. 2007;8:206. doi: 10.1186/gb-2007-8-2-206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Wang T., Zhang L., Shi C., Wei R., Yin C. Interaction of the Homer1 EVH1 domain and skeletal muscle ryanodine receptor. Biochem. Biophys. Res. Commun. 2019;514:720–725. doi: 10.1016/j.bbrc.2019.04.199. [DOI] [PubMed] [Google Scholar]
- 103.Clifton N.E., Trent S., Thomas K.L., Hall J. Regulation and Function of Activity-Dependent Homer in Synaptic Plasticity. Mol Neuropsychiatry. 2019;5:147–161. doi: 10.1159/000500267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Rao S., Siu C.O., Shi M., Zhang J., Lam M.H.B., Yu M., Wing Y.K., Waye M.M.Y. Associations of Homer Scaffolding Protein 1 gene and psychological correlates with suicide attempts in Chinese: A pilot study of multifactorial risk model. Gene. 2018;679:382–388. doi: 10.1016/j.gene.2018.09.025. [DOI] [PubMed] [Google Scholar]
- 105.Benedetti F., Poletti S., Locatelli C., Mazza E., Lorenzi C., Vitali A., Riberto M., Brioschi S., Vai B., Bollettini I., et al. A Homer 1 gene variant influences brain structure and function, lithium effects on white matter, and antidepressant response in bipolar disorder: A multimodal genetic imaging study. Prog. Neuropsychopharmacol. Biol. Psychiatry. 2018;81:88–95. doi: 10.1016/j.pnpbp.2017.10.011. [DOI] [PubMed] [Google Scholar]
- 106.Hayashi M.K., Tang C., Verpelli C., Narayanan R., Stearns M.H., Xu R.M., Li H., Sala C., Hayashi Y. The postsynaptic density proteins Homer and Shank form a polymeric network structure. Cell. 2009;137:159–171. doi: 10.1016/j.cell.2009.01.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Delling J.P., Boeckers T.M. Comparison of SHANK3 deficiency in animal models: Phenotypes, treatment strategies, and translational implications. J. Neurodev. Disord. 2021;13:55. doi: 10.1186/s11689-021-09397-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Roussignol G., Ango F., Romorini S., Tu J.C., Sala C., Worley P.F., Bockaert J., Fagni L. Shank expression is sufficient to induce functional dendritic spine synapses in aspiny neurons. J. Neurosci. 2005;25:3560–3570. doi: 10.1523/JNEUROSCI.4354-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Arons M.H., Lee K., Thynne C.J., Kim S.A., Schob C., Kindler S., Montgomery J.M., Garner C.C. Shank3 Is Part of a Zinc-Sensitive Signaling System That Regulates Excitatory Synaptic Strength. J. Neurosci. 2016;36:9124–9134. doi: 10.1523/JNEUROSCI.0116-16.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Duffney L.J., Wei J., Cheng J., Liu W., Smith K.R., Kittler J.T., Yan Z. Shank3 deficiency induces NMDA receptor hypofunction via an actin-dependent mechanism. J. Neurosci. 2013;33:15767–15778. doi: 10.1523/JNEUROSCI.1175-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Ortiz R., Niciu M.J., Lukkahati N., Saligan L.N., Nugent A.C., Luckenbaugh D.A., Machado-Vieira R., Zarate C.A., Jr. Shank3 as a potential biomarker of antidepressant response to ketamine and its neural correlates in bipolar depression. J. Affect. Disord. 2015;172:307–311. doi: 10.1016/j.jad.2014.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Denayer A., Van Esch H., de Ravel T., Frijns J.P., Van Buggenhout G., Vogels A., Devriendt K., Geutjens J., Thiry P., Swillen A. Neuropsychopathology in 7 Patients with the 22q13 Deletion Syndrome: Presence of Bipolar Disorder and Progressive Loss of Skills. Mol. Syndromol. 2012;3:14–20. doi: 10.1159/000339119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Chandley M.J., Szebeni A., Szebeni K., Crawford J.D., Stockmeier C.A., Turecki G., Kostrzewa R.M., Ordway G.A. Elevated gene expression of glutamate receptors in noradrenergic neurons from the locus coeruleus in major depression. Int. J. Neuropsychopharmacol. 2014;17:1569–1578. doi: 10.1017/S1461145714000662. [DOI] [PubMed] [Google Scholar]
- 114.Toro C., Deakin J.F. NMDA receptor subunit NRI and postsynaptic protein PSD-95 in hippocampus and orbitofrontal cortex in schizophrenia and mood disorder. Schizophr. Res. 2005;80:323–330. doi: 10.1016/j.schres.2005.07.003. [DOI] [PubMed] [Google Scholar]
- 115.Beneyto M., Meador-Woodruff J.H. Lamina-specific abnormalities of NMDA receptor-associated postsynaptic protein transcripts in the prefrontal cortex in schizophrenia and bipolar disorder. Neuropsychopharmacology. 2008;33:2175–2186. doi: 10.1038/sj.npp.1301604. [DOI] [PubMed] [Google Scholar]
- 116.Dean B., Gibbons A.S., Boer S., Uezato A., Meador-Woodruff J., Scarr E., McCullumsmith R.E. Changes in cortical N-methyl-D-aspartate receptors and post-synaptic density protein 95 in schizophrenia, mood disorders and suicide. Aust. N. Z. J. Psychiatry. 2016;50:275–283. doi: 10.1177/0004867415586601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Feyissa A.M., Chandran A., Stockmeier C.A., Karolewicz B. Reduced levels of NR2A and NR2B subunits of NMDA receptor and PSD-95 in the prefrontal cortex in major depression. Prog. Neuropsychopharmacol. Biol. Psychiatry. 2009;33:70–75. doi: 10.1016/j.pnpbp.2008.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Karolewicz B., Szebeni K., Gilmore T., Maciag D., Stockmeier C.A., Ordway G.A. Elevated levels of NR2A and PSD-95 in the lateral amygdala in depression. Int. J. Neuropsychopharmacol. 2009;12:143–153. doi: 10.1017/S1461145708008985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Sowa-Kućma M., Szewczyk B., Sadlik K., Piekoszewski W., Trela F., Opoka W., Poleszak E., Pilc A., Nowak G. Zinc, magnesium and NMDA receptor alterations in the hippocampus of suicide victims. J. Affect. Disord. 2013;151:924–931. doi: 10.1016/j.jad.2013.08.009. [DOI] [PubMed] [Google Scholar]
- 120.Duric V., Banasr M., Stockmeier C.A., Simen A.A., Newton S.S., Overholser J.C., Jurjus G.J., Dieter L., Duman R.S. Altered expression of synapse and glutamate related genes in post-mortem hippocampus of depressed subjects. Int. J. Neuropsychopharmacol. 2013;16:69–82. doi: 10.1017/S1461145712000016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Kristiansen L.V., Meador-Woodruff J.H. Abnormal striatal expression of transcripts encoding NMDA interacting PSD proteins in schizophrenia, bipolar disorder and major depression. Schizophr. Res. 2005;78:87–93. doi: 10.1016/j.schres.2005.06.012. [DOI] [PubMed] [Google Scholar]
- 122.Karolewicz B., Szebeni K., Stockmeier C.A., Konick L., Overholser J.C., Jurjus G., Roth B.L., Ordway G.A. Low nNOS protein in the locus coeruleus in major depression. J. Neurochem. 2004;91:1057–1066. doi: 10.1111/j.1471-4159.2004.02792.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Loeb E., El Asmar K., Trabado S., Gressier F., Colle R., Rigal A., Martin S., Verstuyft C., Fève B., Chanson P., et al. Nitric Oxide Synthase activity in major depressive episodes before and after antidepressant treatment: Results of a large case-control treatment study. Psychol. Med. 2020;52:80–89. doi: 10.1017/S0033291720001749. [DOI] [PubMed] [Google Scholar]
- 124.Dmitrzak-Weglarz M., Szczepankiewicz A., Rybakowski J., Kapelski P., Bilska K., Skibinska M., Reszka E., Lesicka M., Jablonska E., Wieczorek E., et al. Expression Biomarkers of Pharmacological Treatment Outcomes in Women with Unipolar and Bipolar Depression. Pharmacopsychiatry. 2021;54:261–268. doi: 10.1055/a-1546-9483. [DOI] [PubMed] [Google Scholar]
- 125.Ding Q., Li H., Tian X., Shen Z., Wang X., Mo F., Huang J., Shen H. Zinc and imipramine reverse the depression-like behavior in mice induced by chronic restraint stress. J. Affect. Disord. 2016;197:100–106. doi: 10.1016/j.jad.2016.03.017. [DOI] [PubMed] [Google Scholar]
- 126.Popova D., Ágústsdóttir A., Lindholm J., Mazulis U., Akamine Y., Castrén E., Karpova N.N. Combination of fluoxetine and extinction treatments forms a unique synaptic protein profile that correlates with long-term fear reduction in adult mice. Eur. Neuropsychopharmacol. 2014;24:1162–1174. doi: 10.1016/j.euroneuro.2014.04.002. [DOI] [PubMed] [Google Scholar]
- 127.Stan T.L., Sousa V.C., Zhang X., Ono M., Svenningsson P. Lurasidone and fluoxetine reduce novelty-induced hypophagia and NMDA receptor subunit and PSD-95 expression in mouse brain. Eur. Neuropsychopharmacol. 2015;25:1714–1722. doi: 10.1016/j.euroneuro.2015.07.007. [DOI] [PubMed] [Google Scholar]
- 128.Zuo C., Cao H., Ding F., Zhao J., Huang Y., Li G., Huang S., Jiang H., Jiang Y., Wang F. Neuroprotective efficacy of different levels of high-frequency repetitive transcranial magnetic stimulation in mice with CUMS-induced depression: Involvement of the p11/BDNF/Homer1a signaling pathway. J. Psychiatr. Res. 2020;125:152–163. doi: 10.1016/j.jpsychires.2020.03.018. [DOI] [PubMed] [Google Scholar]
- 129.Asim M., Hao B., Yang Y.H., Fan B.F., Xue L., Shi Y.W., Wang X.G., Zhao H. Ketamine Alleviates Fear Generalization Through GluN2B-BDNF Signaling in Mice. Neurosci. Bull. 2020;36:153–164. doi: 10.1007/s12264-019-00422-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Luo Y., Yu Y., Zhang M., He H., Fan N. Chronic administration of ketamine induces cognitive deterioration by restraining synaptic signaling. Mol. Psychiatry. 2021;26:4702–4718. doi: 10.1038/s41380-020-0793-6. [DOI] [PubMed] [Google Scholar]
- 131.Karisetty B.C., Maitra S., Wahul A.B., Musalamadugu A., Khandelwal N., Guntupalli S., Garikapati R., Jhansyrani T., Kumar A., Chakravarty S. Differential effect of chronic stress on mouse hippocampal memory and affective behavior: Role of major ovarian hormones. Behav. Brain Res. 2017;318:36–44. doi: 10.1016/j.bbr.2016.10.034. [DOI] [PubMed] [Google Scholar]
- 132.Nieoczym D., Socała K., Zelek-Molik A., Pieróg M., Przejczowska-Pomierny K., Szafarz M., Wyska E., Nalepa I., Wlaź P. Anticonvulsant effect of pterostilbene and its influence on the anxiety- and depression-like behavior in the pentetrazol-kindled mice: Behavioral, biochemical, and molecular studies. Psychopharmacology. 2021;238:3167–3181. doi: 10.1007/s00213-021-05933-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Bao H., Ran P., Zhu M., Sun L., Li B., Hou Y., Nie J., Shan L., Li H., Zheng S., et al. The Prefrontal Dectin-1/AMPA Receptor Signaling Pathway Mediates The Robust and Prolonged Antidepressant Effect of Proteo-β-Glucan from Maitake. Sci. Rep. 2016;6:28395. doi: 10.1038/srep28395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Li J.M., Liu L.L., Su W.J., Wang B., Zhang T., Zhang Y., Jiang C.L. Ketamine may exert antidepressant effects via suppressing NLRP3 inflammasome to upregulate AMPA receptors. Neuropharmacology. 2019;146:149–153. doi: 10.1016/j.neuropharm.2018.11.022. [DOI] [PubMed] [Google Scholar]
- 135.Park M.J., Seo B.A., Lee B., Shin H.S., Kang M.G. Stress-induced changes in social dominance are scaled by AMPA-type glutamate receptor phosphorylation in the medial prefrontal cortex. Sci. Rep. 2018;8:15008. doi: 10.1038/s41598-018-33410-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Yi E.S., Oh S., Lee J.K., Leem Y.H. Chronic stress-induced dendritic reorganization and abundance of synaptosomal PKA-dependent CP-AMPA receptor in the basolateral amygdala in a mouse model of depression. Biochem. Biophys. Res. Commun. 2017;486:671–678. doi: 10.1016/j.bbrc.2017.03.093. [DOI] [PubMed] [Google Scholar]
- 137.Yu H., Li M., Zhou D., Lv D., Liao Q., Lou Z., Shen M., Wang Z., Li M., Xiao X., et al. Vesicular glutamate transporter 1 (VGLUT1)-mediated glutamate release and membrane GluA1 activation is involved in the rapid antidepressant-like effects of scopolamine in mice. Neuropharmacology. 2018;131:209–222. doi: 10.1016/j.neuropharm.2017.12.028. [DOI] [PubMed] [Google Scholar]
- 138.Camargo A., Dalmagro A.P., de Souza M.M., Zeni A.L.B., Rodrigues A.L.S. Ketamine, but not guanosine, as a prophylactic agent against corticosterone-induced depressive-like behavior: Possible role of long-lasting pro-synaptogenic signaling pathway. Exp. Neurol. 2020;334:113459. doi: 10.1016/j.expneurol.2020.113459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Li M.X., Li Q., Sun X.J., Luo C., Li Y., Wang Y.N., Chen J., Gong C.Z., Li Y.J., Shi L.P., et al. Increased Homer1-mGluR5 mediates chronic stress-induced depressive-like behaviors and glutamatergic dysregulation via activation of PERK-eIF2α. Prog Neuropsychopharmacol. Biol. Psychiatry. 2019;95:109682. doi: 10.1016/j.pnpbp.2019.109682. [DOI] [PubMed] [Google Scholar]
- 140.Shen M., Lv D., Liu X., Li S., Chen Y., Zhang Y., Wang Z., Wang C. Essential roles of neuropeptide VGF regulated TrkB/mTOR/BICC1 signaling and phosphorylation of AMPA receptor subunit GluA1 in the rapid antidepressant-like actions of ketamine in mice. Brain Res. Bull. 2018;143:58–65. doi: 10.1016/j.brainresbull.2018.10.004. [DOI] [PubMed] [Google Scholar]
- 141.Ju L., Yang J., Zhu T., Liu P., Yang J. BDNF-TrkB signaling-mediated upregulation of Narp is involved in the antidepressant-like effects of (2R,6R)-hydroxynorketamine in a chronic restraint stress mouse model. BMC Psychiatry. 2022;22:182. doi: 10.1186/s12888-022-03838-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Xu X., Wu K., Ma X., Wang W., Wang H., Huang M., Luo L., Su C., Yuan T., Shi H., et al. mGluR5-Mediated eCB Signaling in the Nucleus Accumbens Controls Vulnerability to Depressive-Like Behaviors and Pain After Chronic Social Defeat Stress. Mol. Neurobiol. 2021;58:4944–4958. doi: 10.1007/s12035-021-02469-9. [DOI] [PubMed] [Google Scholar]
- 143.Bieler M., Hussain S., Daaland E.S.B., Mirrione M.M., Henn F.A., Davanger S. Changes in concentrations of NMDA receptor subunit GluN2B, Arc and syntaxin-1 in dorsal hippocampus Schaffer collateral synapses in a rat learned helplessness model of depression. J. Comp. Neurol. 2021;529:3194–3205. doi: 10.1002/cne.25155. [DOI] [PubMed] [Google Scholar]
- 144.Dou M., Gong A., Liang H., Wang Q., Wu Y., Ma A., Han L. Improvement of symptoms in a rat model of depression through combined zinc and folic acid administration via up-regulation of the Trk B and NMDA. Neurosci. Lett. 2018;683:196–201. doi: 10.1016/j.neulet.2018.07.036. [DOI] [PubMed] [Google Scholar]
- 145.Li S.X., Han Y., Xu L.Z., Yuan K., Zhang R.X., Sun C.Y., Xu D.F., Yuan M., Deng J.H., Meng S.Q., et al. Uncoupling DAPK1 from NMDA receptor GluN2B subunit exerts rapid antidepressant-like effects. Mol. Psychiatry. 2018;23:597–608. doi: 10.1038/mp.2017.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Treccani G., Gaarn du Jardin K., Wegener G., Müller H.K. Differential expression of postsynaptic NMDA and AMPA receptor subunits in the hippocampus and prefrontal cortex of the flinders sensitive line rat model of depression. Synapse. 2016;70:471–474. doi: 10.1002/syn.21918. [DOI] [PubMed] [Google Scholar]
- 147.Pochwat B., Sowa-Kucma M., Kotarska K., Misztak P., Nowak G., Szewczyk B. Antidepressant-like activity of magnesium in the olfactory bulbectomy model is associated with the AMPA/BDNF pathway. Psychopharmacology. 2015;232:355–367. doi: 10.1007/s00213-014-3671-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Román-Albasini L., Díaz-Véliz G., Olave F.A., Aguayo F.I., García-Rojo G., Corrales W.A., Silva J.P., Ávalos A.M., Rojas P.S., Aliaga E., et al. Antidepressant-relevant behavioral and synaptic molecular effects of long-term fasudil treatment in chronically stressed male rats. Neurobiol. Stress. 2020;13:100234. doi: 10.1016/j.ynstr.2020.100234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Qiao H., An S.C., Xu C., Ma X.M. Role of proBDNF and BDNF in dendritic spine plasticity and depressive-like behaviors induced by an animal model of depression. Brain Res. 2017;1663:29–37. doi: 10.1016/j.brainres.2017.02.020. [DOI] [PubMed] [Google Scholar]
- 150.Guo F., Zhang B., Fu Z., Ma Y., Gao Y., Shen F., Huang C., Li Y. The rapid antidepressant and anxiolytic-like effects of YY-21 involve enhancement of excitatory synaptic transmission via activation of mTOR signaling in the mPFC. Eur. Neuropsychopharmacol. 2016;26:1087–1098. doi: 10.1016/j.euroneuro.2016.05.006. [DOI] [PubMed] [Google Scholar]
- 151.Zhou M., Liu Z., Yu J., Li S., Tang M., Zeng L., Wang H., Xie H., Peng L., Huang H., et al. Quantitative Proteomic Analysis Reveals Synaptic Dysfunction in the Amygdala of Rats Susceptible to Chronic Mild Stress. Neuroscience. 2018;376:24–39. doi: 10.1016/j.neuroscience.2018.02.010. [DOI] [PubMed] [Google Scholar]
- 152.Tamási V., Petschner P., Adori C., Kirilly E., Ando R.D., Tothfalusi L., Juhasz G., Bagdy G. Transcriptional evidence for the role of chronic venlafaxine treatment in neurotrophic signaling and neuroplasticity including also Glutamatergic- and insulin-mediated neuronal processes. PLoS ONE. 2014;9:e113662. doi: 10.1371/journal.pone.0113662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Piva A., Caffino L., Padovani L., Pintori N., Mottarlini F., Sferrazza G., Paolone G., Fumagalli F., Chiamulera C. The metaplastic effects of ketamine on sucrose renewal and contextual memory reconsolidation in rats. Behav. Brain Res. 2020;379:112347. doi: 10.1016/j.bbr.2019.112347. [DOI] [PubMed] [Google Scholar]
- 154.Yao Y., Ju P., Liu H., Wu X., Niu Z., Zhu Y., Zhang C., Fang Y. Ifenprodil rapidly ameliorates depressive-like behaviors, activates mTOR signaling and modulates proinflammatory cytokines in the hippocampus of CUMS rats. Psychopharmacology. 2020;237:1421–1433. doi: 10.1007/s00213-020-05469-0. [DOI] [PubMed] [Google Scholar]
- 155.Gordillo-Salas M., Pilar-Cuéllar F., Auberson Y.P., Adell A. Signaling pathways responsible for the rapid antidepressant-like effects of a GluN2A-preferring NMDA receptor antagonist. Transl. Psychiatry. 2018;8:84. doi: 10.1038/s41398-018-0131-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Zhu X., Zhang F., You Y., Wang H., Yuan S., Wu B., Zhu R., Liu D., Yan F., Wang Z. S-Ketamine Exerts Antidepressant Effects by Regulating Rac1 GTPase Mediated Synaptic Plasticity in the Hippocampus of Stressed Rats. Cell. Mol. Neurobiol. 2022 doi: 10.1007/s10571-021-01180-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Lee C.W., Chen Y.J., Wu H.F., Chung Y.J., Lee Y.C., Li C.T., Lin H.C. Ketamine ameliorates severe traumatic event-induced antidepressant-resistant depression in a rat model through ERK activation. Prog. Neuropsychopharmacol. Biol. Psychiatry. 2019;93:102–113. doi: 10.1016/j.pnpbp.2019.03.015. [DOI] [PubMed] [Google Scholar]
- 158.Yang M., Luo C.H., Zhu Y.Q., Liu Y.C., An Y.J., Iqbal J., Wang Z.Z., Ma X.M. 7,8-Dihydroxy-4-methylcoumarin reverses depression model-induced depression-like behaviors and alteration of dendritic spines in the mood circuits. Psychoneuroendocrinology. 2020;119:104767. doi: 10.1016/j.psyneuen.2020.104767. [DOI] [PubMed] [Google Scholar]
- 159.Szewczyk B., Pochwat B., Rafało A., Palucha-Poniewiera A., Domin H., Nowak G. Activation of mTOR dependent signaling pathway is a necessary mechanism of antidepressant-like activity of zinc. Neuropharmacology. 2015;99:517–526. doi: 10.1016/j.neuropharm.2015.08.026. [DOI] [PubMed] [Google Scholar]
- 160.Zhang K., Yamaki V.N., Wei Z., Zheng Y., Cai X. Differential regulation of GluA1 expression by ketamine and memantine. Behav. Brain Res. 2017;316:152–159. doi: 10.1016/j.bbr.2016.09.002. [DOI] [PubMed] [Google Scholar]
- 161.Li R., Zhao X., Cai L., Gao W.W. Up-regulation of GluR1 in paraventricular nucleus and greater expressions of synapse related proteins in the hypothalamus of chronic unpredictable stress-induced depressive rats. Physiol. Behav. 2017;179:451–457. doi: 10.1016/j.physbeh.2017.07.024. [DOI] [PubMed] [Google Scholar]
- 162.Hou X.Y., Hu Z.L., Zhang D.Z., Lu W., Zhou J., Wu P.F., Guan X.L., Han Q.Q., Deng S.L., Zhang H., et al. Rapid Antidepressant Effect of Hydrogen Sulfide: Evidence for Activation of mTORC1-TrkB-AMPA Receptor Pathways. Antioxid. Redox Signal. 2017;27:472–488. doi: 10.1089/ars.2016.6737. [DOI] [PubMed] [Google Scholar]
- 163.Liu C.Y., Chen J.B., Liu Y.Y., Zhou X.M., Zhang M., Jiang Y.M., Ma Q.Y., Xue Z., Zhao Z.Y., Li X.J., et al. Saikosaponin D exerts antidepressant effect by regulating Homer1-mGluR5 and mTOR signaling in a rat model of chronic unpredictable mild stress. Chin Med. 2022;17:60. doi: 10.1186/s13020-022-00621-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Chen S.T., Hsieh C.P., Lee M.Y., Chen L.C., Huang C.M., Chen H.H., Chan M.H. Betaine prevents and reverses the behavioral deficits and synaptic dysfunction induced by repeated ketamine exposure in mice. Biomed. Pharmacother. 2021;144:112369. doi: 10.1016/j.biopha.2021.112369. [DOI] [PubMed] [Google Scholar]
- 165.Leem Y.H., Yoon S.S., Jo S.A. Imipramine Ameliorates Depressive Symptoms by Blocking Differential Alteration of Dendritic Spine Structure in Amygdala and Prefrontal Cortex of Chronic Stress-Induced Mice. Biomol. Ther. 2020;28:230–239. doi: 10.4062/biomolther.2019.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Luo L., Liu X.L., Mu R.H., Wu Y.J., Liu B.B., Geng D., Liu Q., Yi L.T. Hippocampal BDNF signaling restored with chronic asiaticoside treatment in depression-like mice. Brain Res. Bull. 2015;114:62–69. doi: 10.1016/j.brainresbull.2015.03.006. [DOI] [PubMed] [Google Scholar]
- 167.Zhuang F., Li M., Gao X., Wang Y., Wang D., Ma X., Ma T., Gu S. The antidepressant-like effect of alarin is related to TrkB-mTOR signaling and synaptic plasticity. Behav. Brain Res. 2016;313:158–171. doi: 10.1016/j.bbr.2016.06.057. [DOI] [PubMed] [Google Scholar]
- 168.Misrani A., Tabassum S., Wang M., Chen J., Yang L., Long C. Citalopram prevents sleep-deprivation-induced reduction in CaMKII-CREB-BDNF signaling in mouse prefrontal cortex. Brain Res. Bull. 2020;155:11–18. doi: 10.1016/j.brainresbull.2019.11.007. [DOI] [PubMed] [Google Scholar]
- 169.Yin Y., Qian S., Chen Y., Sun Y., Li Y., Yu Y., Li J., Wu Z., Yu X., Ge R., et al. Latent Sex Differences in CaMKII-nNOS Signaling That Underlie Antidepressant-Like Effects of Yueju-Ganmaidazao Decoction in the Hippocampus. Front. Behav. Neurosci. 2021;15:640258. doi: 10.3389/fnbeh.2021.640258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Zou Z., Huang J., Yang Q., Zhang Y., Xu B., Wang P., Chen G. Repeated Yueju, But Not Fluoxetine, Induced Sustained Antidepressant Activity in a Mouse Model of Chronic Learned Helplessness: Involvement of CaMKII Signaling in the Hippocampus. Evid.-Based Complement. Alternat. Med. 2022;2022:1442578. doi: 10.1155/2022/1442578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Misrani A., Tabassum S., Chen X., Tan S.Y., Wang J.C., Yang L., Long C. Differential effects of citalopram on sleep-deprivation-induced depressive-like behavior and memory impairments in mice. Prog. Neuropsychopharmacol. Biol. Psychiatry. 2019;88:102–111. doi: 10.1016/j.pnpbp.2018.07.013. [DOI] [PubMed] [Google Scholar]
- 172.Sun L., Verkaik-Schakel R.N., Biber K., Plösch T., Serchov T. Antidepressant treatment is associated with epigenetic alterations of Homer1 promoter in a mouse model of chronic depression. J. Affect. Disord. 2021;279:501–509. doi: 10.1016/j.jad.2020.10.040. [DOI] [PubMed] [Google Scholar]
- 173.Rafało-Ulińska A., Poleszak E., Szopa A., Serefko A., Rogowska M., Sowa I., Wójciak M., Muszyńska B., Krakowska A., Gdula-Argasińska J., et al. Imipramine Influences Body Distribution of Supplemental Zinc Which May Enhance Antidepressant Action. Nutrients. 2020;12:2529. doi: 10.3390/nu12092529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Gordillo-Salas M., Pascual-Antón R., Ren J., Greer J., Adell A. Antidepressant-Like Effects of CX717, a Positive Allosteric Modulator of AMPA Receptors. Mol. Neurobiol. 2020;57:3498–3507. doi: 10.1007/s12035-020-01954-x. [DOI] [PubMed] [Google Scholar]
- 175.Luoni A., Macchi F., Papp M., Molteni R., Riva M.A. Lurasidone exerts antidepressant properties in the chronic mild stress model through the regulation of synaptic and neuroplastic mechanisms in the rat prefrontal cortex. Int. J. Neuropsychopharmacol. 2014;18:pyu061. doi: 10.1093/ijnp/pyu061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Xie W., Meng X., Zhai Y., Ye T., Zhou P., Nan F., Sun G., Sun X. Antidepressant-like effects of the Guanxin Danshen formula via mediation of the CaMK II-CREB-BDNF signalling pathway in chronic unpredictable mild stress-induced depressive rats. Ann. Transl. Med. 2019;7:564. doi: 10.21037/atm.2019.09.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Palmfeldt J., Henningsen K., Eriksen S.A., Müller H.K., Wiborg O. Protein biomarkers of susceptibility and resilience to stress in a rat model of depression. Mol. Cell. Neurosci. 2016;74:87–95. doi: 10.1016/j.mcn.2016.04.001. [DOI] [PubMed] [Google Scholar]
- 178.Yoshino Y., Ochi S., Yamazaki K., Nakata S., Abe M., Mori Y., Ueno S. Antidepressant action via the nitric oxide system: A pilot study in an acute depressive model induced by arginin. Neurosci. Lett. 2015;599:69–74. doi: 10.1016/j.neulet.2015.05.043. [DOI] [PubMed] [Google Scholar]
- 179.Mishra S.K., Hidau M.K., Rai S. Memantine treatment exerts an antidepressant-like effect by preventing hippocampal mitochondrial dysfunction and memory impairment via upregulation of CREB/BDNF signaling in the rat model of chronic unpredictable stress-induced depression. Neurochem. Int. 2021;142:104932. doi: 10.1016/j.neuint.2020.104932. [DOI] [PubMed] [Google Scholar]
- 180.Benske T.M., Mu T.W., Wang Y.J. Protein quality control of N-methyl-D-aspartate receptors. Front. Cell. Neurosci. 2022;16:907560. doi: 10.3389/fncel.2022.907560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Paoletti P., Bellone C., Zhou Q. NMDA receptor subunit diversity: Impact on receptor properties, synaptic plasticity and disease. Nat. Rev. Neurosci. 2013;14:383–400. doi: 10.1038/nrn3504. [DOI] [PubMed] [Google Scholar]
- 182.Lee C.H., Lü W., Michel J.C., Goehring A., Du J., Song X., Gouaux E. NMDA receptor structures reveal subunit arrangement and pore architecture. Nature. 2014;511:191–197. doi: 10.1038/nature13548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Wyllie D.J., Livesey M.R., Hardingham G.E. Influence of GluN2 subunit identity on NMDA receptor function. Neuropharmacology. 2013;74:4–17. doi: 10.1016/j.neuropharm.2013.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Mellone M., Pelucchi S., Alberti L., Genazzani A.A., Di Luca M., Gardoni F. Zinc transporter-1: A novel NMDA receptor-binding protein at the postsynaptic density. J. Neurochem. 2015;132:159–168. doi: 10.1111/jnc.12968. [DOI] [PubMed] [Google Scholar]
- 185.Babaei P. NMDA and AMPA receptors dysregulation in Alzheimer’s disease. Eur. J. Pharmacol. 2021;908:174310. doi: 10.1016/j.ejphar.2021.174310. [DOI] [PubMed] [Google Scholar]
- 186.Suryavanshi P.S., Ugale R.R., Yilmazer-Hanke D., Stairs D.J., Dravid S.M. GluN2C/GluN2D subunit-selective NMDA receptor potentiator CIQ reverses MK-801-induced impairment in prepulse inhibition and working memory in Y-maze test in mice. Br. J. Pharmacol. 2014;171:799–809. doi: 10.1111/bph.12518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Stachowicz K. Is PSD-95 entangled in the side effects of antidepressants? Neurochem. Int. 2022;159:105391. doi: 10.1016/j.neuint.2022.105391. [DOI] [PubMed] [Google Scholar]
- 188.Song J., Kim Y.K. Animal models for the study of depressive disorder. CNS Neurosci Ther. 2021;27:633–642. doi: 10.1111/cns.13622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Planchez B., Surget A., Belzung C. Animal models of major depression: Drawbacks and challenges. J. Neural Transm. 2019;126:1383–1408. doi: 10.1007/s00702-019-02084-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Zhang J., Huang X.Y., Ye M.L., Luo C.X., Wu H.Y., Hu Y., Zhou Q.G., Wu D.L., Zhu L.J., Zhu D.Y. Neuronal nitric oxide synthase alteration accounts for the role of 5-HT1A receptor in modulating anxiety-related behaviors. J. Neurosci. 2010;30:2433–2441. doi: 10.1523/JNEUROSCI.5880-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Wiggins A.K., Shen P.J., Gundlach A.L. Neuronal-NOS adaptor protein expression after spreading depression: Implications for NO production and ischemic tolerance. J. Neurochem. 2003;87:1368–1380. doi: 10.1046/j.1471-4159.2003.02099.x. [DOI] [PubMed] [Google Scholar]
- 192.Jaso B.A., Niciu M.J., Iadarola N.D., Lally N., Richards E.M., Park M., Ballard E.D., Nugent A.C., Machado-Vieira R., Zarate C.A. Therapeutic Modulation of Glutamate Receptors in Major Depressive Disorder. Curr. Neuropharmacol. 2017;15:57–70. doi: 10.2174/1570159X14666160321123221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Berman R.M., Cappiello A., Anand A., Oren D.A., Heninger G.R., Charney D.S., Krystal J.H. Antidepressant effects of ketamine in depressed patients. Biol. Psychiatry. 2000;47:351–354. doi: 10.1016/S0006-3223(99)00230-9. [DOI] [PubMed] [Google Scholar]
- 194.Zarate C.A., Jr., Singh J.B., Carlson P.J., Brutsche N.E., Ameli R., Luckenbaugh D.A., Charney D.S., Manji H.K. A randomized trial of an N-methyl-D-aspartate antagonist in treatment-resistant major depression. Arch. Gen. Psychiatry. 2006;63:856–864. doi: 10.1001/archpsyc.63.8.856. [DOI] [PubMed] [Google Scholar]
- 195.Wan L.B., Levitch C.F., Perez A.M., Brallier J.W., Iosifescu D.V., Chang L.C., Foulkes A., Mathew S.J., Charney D.S., Murrough J.W. Ketamine safety and tolerability in clinical trials for treatment-resistant depression. J. Clin. Psychiatry. 2015;76:247–252. doi: 10.4088/JCP.13m08852. [DOI] [PubMed] [Google Scholar]
- 196.Kheirabadi D., Kheirabadi G.R., Mirlohi Z., Tarrahi M.J., Norbaksh A. Comparison of Rapid Antidepressant and Antisuicidal Effects of Intramuscular Ketamine, Oral Ketamine, and Electroconvulsive Therapy in Patients with Major Depressive Disorder: A Pilot Study. J. Clin. Psychopharmacol. 2020;40:588–593. doi: 10.1097/JCP.0000000000001289. [DOI] [PubMed] [Google Scholar]
- 197.Lara D.R., Bisol L.W., Munari L.R. Antidepressant, mood stabilizing and procognitive effects of very low dose sublingual ketamine in refractory unipolar and bipolar depression. Int. J. Neuropsychopharmacol. 2013;16:2111–2117. doi: 10.1017/S1461145713000485. [DOI] [PubMed] [Google Scholar]
- 198.Yavi M., Lee H., Henter I.D., Park L.T., Zarate C.A., Jr. Ketamine treatment for depression: A review. Discov. Ment. Health. 2022;2:9. doi: 10.1007/s44192-022-00012-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Hsu T.W., Chu C.S., Ching P.Y., Chen G.W., Pan C.C. The efficacy and tolerability of memantine for depressive symptoms in major mental diseases: A systematic review and updated meta-analysis of double-blind randomized controlled trials. J. Affect. Disord. 2022;306:182–189. doi: 10.1016/j.jad.2022.03.047. [DOI] [PubMed] [Google Scholar]
- 200.Krzystanek M., Surma S., Pałasz A., Romańczyk M., Krysta K. Possible Antidepressant Effects of Memantine-Systematic Review with a Case Study. Pharmaceuticals. 2021;14:481. doi: 10.3390/ph14050481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Vasiliu O. Investigational Drugs for the Treatment of Depression (Part 2): Glutamatergic, Cholinergic, Sestrin Modulators, and Other Agents. Front. Pharmacol. 2022;13:884155. doi: 10.3389/fphar.2022.884155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Poleszak E., Stasiuk W., Szopa A., Wyska E., Serefko A., Oniszczuk A., Wośko S., Świąder K., Wlaź P. Traxoprodil, a selective antagonist of the NR2B subunit of the NMDA receptor, potentiates the antidepressant-like effects of certain antidepressant drugs in the forced swim test in mice. Metab. Brain Dis. 2016;31:803–814. doi: 10.1007/s11011-016-9810-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Preskorn S.H., Baker B., Kolluri S., Menniti F.S., Krams M., Landen J.W. An innovative design to establish proof of concept of the antidepressant effects of the NR2B subunit selective N-methyl-D-aspartate antagonist, CP-101,606, in patients with treatment-refractory major depressive disorder. J. Clin. Psychopharmacol. 2008;28:631–637. doi: 10.1097/JCP.0b013e31818a6cea. [DOI] [PubMed] [Google Scholar]
- 204.Ibrahim L., Diaz Granados N., Jolkovsky L., Brutsche N., Luckenbaugh D.A., Herring W.J., Potter W.Z., Zarate C.A., Jr. A Randomized, placebo-controlled, crossover pilot trial of the oral selective NR2B antagonist MK-0657 in patients with treatment-resistant major depressive disorder. J. Clin. Psychopharmacol. 2012;32:551–557. doi: 10.1097/JCP.0b013e31825d70d6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Stroebel D., Buhl D.L., Knafels J.D., Chanda P.K., Green M., Sciabola S., Mony L., Paoletti P., Pandit J. A Novel Binding Mode Reveals Two Distinct Classes of NMDA Receptor GluN2B-selective Antagonists. Mol. Pharmacol. 2016;89:541–551. doi: 10.1124/mol.115.103036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Heresco-Levy U., Gelfin G., Bloch B., Levin R., Edelman S., Javitt D.C., Kremer I. A randomized add-on trial of high-dose D-cycloserine for treatment-resistant depression. Int. J. Neuropsychopharmacol. 2013;16:501–506. doi: 10.1017/S1461145712000910. [DOI] [PubMed] [Google Scholar]
- 207.Pereira V.S., Joca S.R.L., Harvey B.H., Elfving B., Wegener G. Esketamine and rapastinel, but not imipramine, have antidepressant-like effect in a treatment-resistant animal model of depression. Acta Neuropsychiatr. 2019;31:258–265. doi: 10.1017/neu.2019.25. [DOI] [PubMed] [Google Scholar]
- 208.Preskorn S., Macaluso M., Mehra D.O., Zammit G., Moskal J.R., Burch R.M. GLYX-13 Clinical Study Group. Randomized proof of concept trial of GLYX-13, an N-methyl-D-aspartate receptor glycine site partial agonist, in major depressive disorder nonresponsive to a previous antidepressant agent. J. Psychiatr. Pract. 2015;21:140–149. doi: 10.1097/01.pra.0000462606.17725.93. [DOI] [PubMed] [Google Scholar]
- 209.Pilar-Cuellar F., Castro E., Bretin S., Mocaer E., Pazos Á., Díaz Á. S 47445 counteracts the behavioral manifestations and hippocampal neuroplasticity changes in bulbectomized mice. Prog. Neuropsychopharmacol. Biol. Psychiatry. 2019;93:205–213. doi: 10.1016/j.pnpbp.2019.04.005. [DOI] [PubMed] [Google Scholar]
- 210.Nations K.R., Dogterom P., Bursi R., Schipper J., Greenwald S., Zraket D., Gertsik L., Johnstone J., Lee A., Pande Y., et al. Examination of Org 26576, an AMPA receptor positive allosteric modulator, in patients diagnosed with major depressive disorder: An exploratory, randomized, double-blind, placebo-controlled trial. J. Psychopharmacol. 2012;26:1525–1539. doi: 10.1177/0269881112458728. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.