Abstract
Pasture-based milk presents several advantages over milk from intensive industrial farming in terms of human health, the environment, animal welfare, and social aspects. This highlights the need for reliable methods to differentiate milk according to its origin on the market. Here, we explored whether miRNA profiles could serve as a marker of milk production systems. We compared levels of previously described miRNAs in milk from four production systems (altogether 112 milk samples): grazing, zero grazing, grass silage or corn silage. Total RNA was extracted from the fat phase, and miRNAs levels were quantified by real-time quantitative PCR. The levels of the miRNAs bta-miR-155 and bta-miR-103 were higher in the grazing system than in corn silage farms. The levels of bta-miR-532, bta-miR-103 and bta-miR-7863 showed differences between different farm managements. The miRNAs bta-miR-155 and bta-miR-103 were predicted to participate in common functions related to fat metabolism and fatty acid elongation. All four differentially expressed miRNAs were predicted to participate in transport, cell differentiation, and metabolism. These results suggest that the dairy production system influences the levels of some miRNAs in milk fat, and that bta-miR-155 and bta-miR-103 may be potential biomarkers to identify milk from pasture-managed systems.
Keywords: milk, microRNA, biomarker, dairy production systems
1. Introduction
Milk production systems vary in how extensive or intensive they are, ranging from one extreme of extensive pastoral livestock farming to the other extreme of intensive industrial farming [1]. In pasture-based production systems, animals with a genotype appropriate to the area feed on pastures or rangelands, external feed inputs are minimized, the use of pastoral resources is optimized, and the stocking rate is low [1,2,3,4]. At the other extreme, intensive farms rely on a high stocking rate and other measures to maximize milk production per cow. The animals are permanently housed and eat a diet based on silage and large amounts of concentrates [5]. Many farms combine certain characteristics between these two extremes, where animals may graze, receive supplements with conserved forages and concentrates and, depending on the climatic conditions, be stabled or on pastures [4,6].
Pasture-based milk production has environmental benefits over intensive production [2,7], as the milk contains higher levels of functional nutrients [8,9] and the marginal milk cost may be lower [10,11,12]. Pasture-based milk production is also more animal friendly [13] and it reduces workload, improving farmer lifestyle and creating a positive image of livestock farming [4].
Except for some regional or sectoral initiatives, no national regulations clearly define the differences among this continuum of milk production systems [1]. This makes it difficult for more or less extensive farms to certify the advantages of their milk to consumers who demand socially and environmentally responsible products [14,15]. A traceability system to identify the milk production system based on markers in the milk itself could support the certification and accurate marketing of milk from less intensive production systems.
The present study explored whether levels of microRNAs (miRNAs) in milk might serve as indicators of how extensively or intensively it was produced. As small, endogenous non-coding RNAs of 21–25 nucleotides, miRNAs bind to specific targets in mRNA to regulate their expression and thereby control various processes within cells [16]. Beyond their functions in the cells that produce them, microRNAs can also be transferred to other cells, or to other species, in protein complexes or through extracellular vesicles [17,18]. There is also compelling evidence that humans use microRNAs from cow’s milk in gene regulation [19,20], highlighting the bioactive characteristic of milk.
Various body fluids contain miRNAs, including tears, colostrum, plasma, and seminal fluids [21] as well as milk [22]. Milk miRNAs have already shown potential as biomarkers of mammary gland diseases [23,24], the cow’s physiological state [25] and stress [26], diet [27,28,29], and breed [30]. Such miRNAs can also serve as a quality assurance indicator to verify labeling on milk powder [31].
Given that miRNAs expression varies according to cow genotype [30,32] and environment [26,33], we wondered whether miRNAs profiles might differ reliably among milk samples from different production systems. In support of this idea, gene expression in cow mammary glands has been shown to depend on diet [34], exercise [35], and stress [36]. If so, milk miRNAs could be an ideal biomarker of the production system, given that miRNAs are stable to high temperature, freeze/thaw cycles, RNase digestion and low pH [37]. In addition, they can be sampled in a non-invasive or minimally invasive manner [38]. Hence, in the current study, the quantification of the abundance of 12 selected miRNAs was performed in tank milk from four dairy farming systems: grazing, zero grazing, grass silage or corn silage. The miRNAs were selected from a previous sequencing work [39], and others were selected from the literature for being associated with feeding and metabolism. By comparing the abundance levels of these miRNAs, we aim to highlight potential non-invasive biomarkers of the milk production systems, which may contribute to the authentication of socially and environmentally responsible dairy products.
2. Results
2.1. MiRNAs with Differential Levels in Cow Milk According to Production System
Total RNA concentrations in milk fat from the four types of milk production system varied between 84 and 144 ng/µL, and the RNA was of good quality: the absorbance ratio was 1.67–1.98 in all samples.
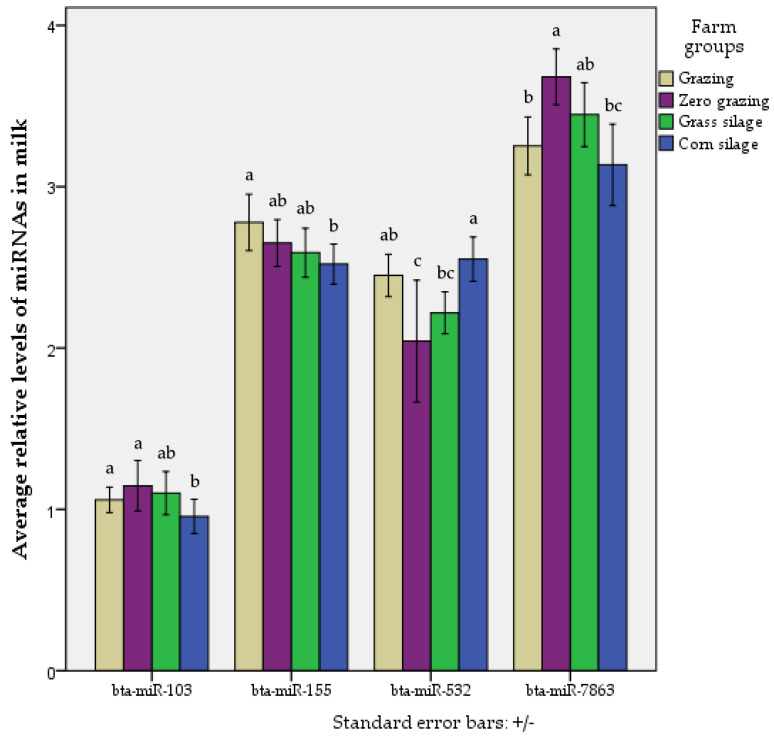
MiRNA levels in milk fat across the four dairy production systems for the 12 chosen miRNAs were estimated. We found that levels of the following four miRNAs differed significantly between at least two dairy production systems: bta-miR-155, bta-miR-103, bta-miR-532, and bta-miR-7863 (Figure 1). Post hoc analysis showed that miRNAs bta-miR-103 and bta-miR-155 showed significant differences between the grazing and corn silage groups, being more abundant in the grazing farms. The bta-miR-532 was significantly more abundant in grazing farms than in zero grazing, while on the contrary bta-miR-7863 was more abundant in zero-grazing farms than in grazing. Bta-miR-103, bta-miR-532 and bta-miR-7863 showed significant differences between zero grazing and corn silage groups, with bta-miR-103 and bta-miR-7863 being more abundant in the zero-grazing group, but bta-miR-532 was more abundant in the corn silage group. The miRNA bta-miR-532 was significantly more abundant in corn silage farms than in grass silage (Figure 1).
Figure 1.
Average relative levels of the miRNAs bta-miR-103, bta-miR-155, bta-miR-532, and bta-miR-7863 in raw milk from grazing (n = 44), zero-grazing (n = 13), grass silage, (n = 10), or corn silage (n = 45) milk production systems. The bar chart shows the average of miRNA levels in each farm group, and the standard error bars. Different letters show significant difference between groups.
2.2. miRNA Functionality and Pathway Analyses
To determine the possible implications of the studied miRNAs in the biological response to different production systems, we predicted the target genes of the four miRNAs as well as the functional pathways in which those target genes may participate. The 707 targets of bta-miR-103 were associated with 71 KEGG pathways (The Kyoto Encyclopedia of Genes and Genomes), 8 biological processes, and 15 molecular functions. The 460 targets of bta-miR-155 were associated with 106 KEGG pathways, 5 biological processes, and 9 molecular functions. The 2266 targets of bta-miR-7863 were associated with 76 KEGG pathways, 12 biological processes, and 7 molecular functions. The 208 targets of bta-miR-532 were associated with 29 KEGG pathways, 3 biological processes, and 22 molecular functions.
Among these target processes, we identified 15 KEGG pathways (Table 1), 8 biological processes (Table 2), and 12 molecular functions (Table 3) that were related to milk production and metabolism. In particular, the four miRNAs were all predicted to be involved in the MAPK signaling pathway (mitogen-activated protein kinases signaling pathway) and the molecular functions of transferases and serine/threonine-protein kinases. Two metabolic pathways stand out for their relationship with milk production and secretion, the oxytocin and prolactin signaling pathways.
Table 1.
KEGG pathways that are associated with milk production and metabolism and that are predicted to be regulated by milk miRNAs with differential levels across production systems.
| KEGG Signaling Pathway | Biomarker | No. of Target Genes | p-Value | |||
|---|---|---|---|---|---|---|
| bta-miR-103 | bta-miR-155 | bta-miR-532 | bta-miR-7863 | |||
| AMPK 1 | x | x | x | 38 | 4.6 × 10−7 | |
| MAPK | x | x | x | x | 67 | 3.2 × 10−6 |
| PI3K-Akt 2 | x | x | x | 27 | 5.6 × 10−5 | |
| Oxytocin | x | x | x | 38 | 1.1 × 10−4 | |
| Prolactin | x | x | 25 | 1.2 × 10−4 | ||
| Insulin | x | x | 35 | 1.4 × 10−4 | ||
| Ras 3 | x | x | x | 53 | 1.6 × 10−4 | |
| Growth hormone synthesis, secretion and action | x | x | 31 | 1.8 × 10−4 | ||
| TGF-beta 4 | x | x | 25 | 6.2 × 10−4 | ||
| Calcium | x | x | 52 | 7.3 × 10−4 | ||
| Glucagon | x | 24 | 6.0 × 10−3 | |||
| Lipid and atherosclerosis | x | x | 43 | 2.5 × 10−2 | ||
| cGMP-PKG 5 | x | 32 | 3.3 × 10−2 | |||
| Mineral absorption | x | 13 | 8.3 × 10−2 | |||
| Lysine degradation | x | 14 | 9.7 × 10−2 | |||
1 Adenosine monophosphate-activated protein kinase (AMPK), 2 phosphoinositide 3-kinases- protein kinase B (PI3Ks-Akt), 3 rat sarcoma virus (Ras), 4 transforming growth factor beta (TGF-beta), 5 cyclic guanosine monophosphate- protein kinase G (cGMP-PKG). x implies pathway associated.
Table 2.
Biological processes that are associated with milk production and metabolism and that are predicted to be regulated by milk miRNAs with differential levels across production systems.
| Biological Process | Biomarker | No. of Target Genes | p-Value | |||
|---|---|---|---|---|---|---|
| bta-miR-103 | bta-miR-155 | bta-miR-532 | bta-miR-7863 | |||
| Growth regulation | x | 11 | 2.5 × 10−3 | |||
| Transport | x | 249 | 1.4 × 10−2 | |||
| Ion transport | x | 79 | 1.6 × 10−2 | |||
| Calcium transport | x | 15 | 1.7 × 10−2 | |||
| Amino- acid transport | x | 7 | 2.4 × 10−2 | |||
| Protein transport | x | x | 67 | 2.7 × 10−2 | ||
| Differentiation | x | 45 | 6.0 × 10−2 | |||
| Sodium transport | x | 13 | 9.5 × 10−2 | |||
x implies biological processes associated.
Table 3.
Molecular functions that are associated with milk production and metabolism and that are predicted to be regulated by milk miRNAs with differential levels across production systems.
| Molecular Function | Biomarker | No. of Target Genes | p-Value | |||
|---|---|---|---|---|---|---|
| bta-miR-103 | bta-miR-155 | bta-miR-532 | bta-miR-7863 | |||
| Transferase | x | x | x | x | 279 | 1.4 × 10−20 |
| Activator | x | x | x | 64 | 1.3 × 10−8 | |
| Ion channel | x | x | 65 | 3.0 × 10−8 | ||
| Serine/threonine-protein kinase | x | x | x | x | 55 | 7.1 × 10−8 |
| Developmental protein | x | x | x | 64 | 3.0 × 10−6 | |
| Glycosyltransferase | x | 31 | 2.5 × 10−3 | |||
| Hydrolase | x | x | x | 182 | 2.8 × 10−3 | |
| Growth factor | x | 17 | 9.0 × 10−3 | |||
| Protein phosphatase | x | x | 19 | 1.3 × 10−2 | ||
| Guanine-nucleotide releasing factor | x | x | 16 | 1.6 × 10−2 | ||
| Calcium channel | x | 8 | 4.0 × 10−2 | |||
| Potassium channel | x | 12 | 4.8 × 10−2 | |||
x implies molecular functions associated.
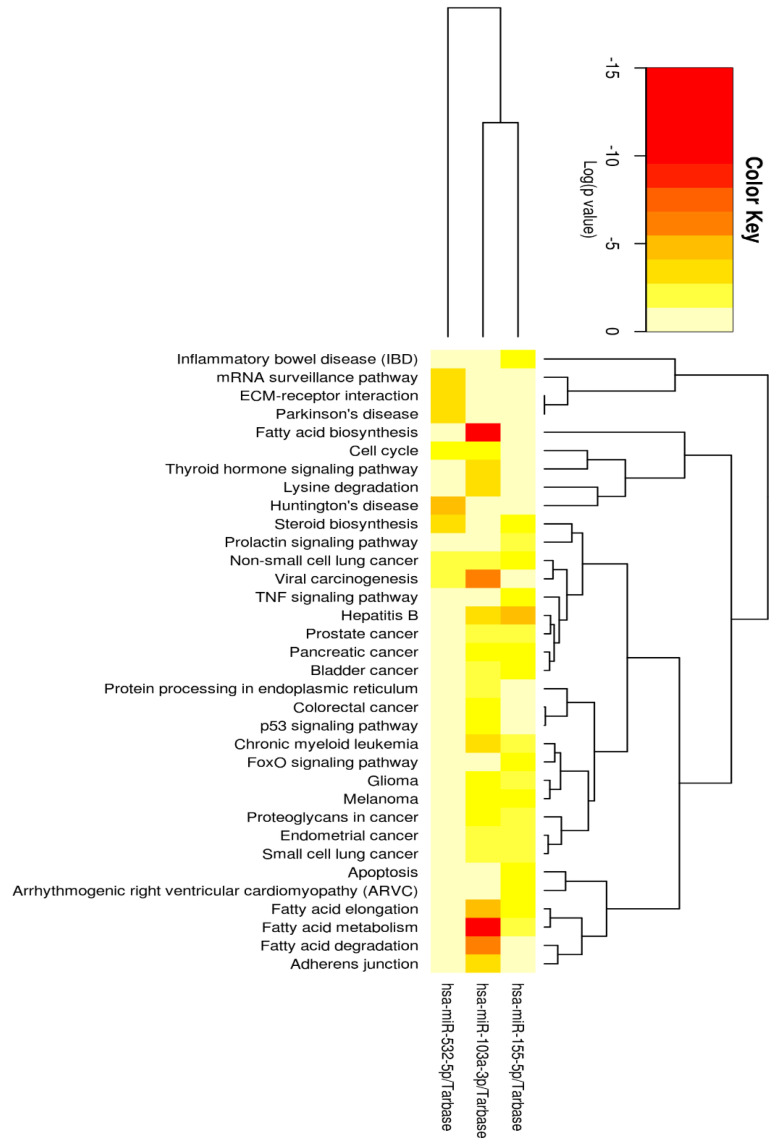
The miRNAs bta-miR-103, bta-miR-155 and bta-miR-532 showed high homology to human miRNAs, so we used DIANA miRPath for a second functionality analysis. Using Tarbase, we found that human target genes were experimentally validated for miR-103a-3p (2156 genes), miR-155 (1117 genes), and miR-532 (306 genes). These targets (using the option union for KEGG and intersection for GO (gene ontology) to merge results were associated with 34 KEGG pathways (Figure 2) and 62 GO categories (Figure S1). These analyses allowed us to predict pathways regulated by the three miRNAs (available in the Tarbase database) (Figure 2 and Figure S1). These results predicted miR-103a-3p and miR-155 to be involved in fatty acid elongation and metabolism.
Figure 2.
Heatmap of hierarchical clustering of miR-103, miR-155 and miR-532 based on mRNA target pathways, identified in DIANA using the Tarbase and KEGG pathway union representation. Darker colors represent lower p-values.
GO analysis revealed nine categories common to miR-103a-3p, miR-155, and miR-532: protein binding transcription factor activity, cellular protein modification processes, catabolic processes, biosynthetic processes, cellular nitrogen compound metabolic processes, cellular protein metabolic processes, small-molecule metabolic processes, mRNA metabolic processes, generation of precursor metabolites and energy, and regulation of glucose transport.
3. Discussion
This work aimed to identify miRNAs that vary according to milk production system in order to differentiate milk from farms managed extensively. In the present study, among the 12 miRNAs analyzed, the levels of 4 miRNAs differed significantly among the types of dairy production system. These findings suggest the potential of profiling miRNAs in milk in order to certify whether it came from an extensive grazing production system.
Previous work from our group showed that the levels of the miRNA bta-miR-215 in milk fat differed between the two extremes of extensiveness/intensiveness [40]. The present work is not limited to extreme production systems but compares grazing livestock farms with intensive and mixed managements.
The levels of bta-miR-103 were highest in farms including fresh grass in the diet, either grazed or harvested. Previous work showed that the expression of bta-miR-103 in blood [41] and subcutaneous fat [42] was similar between cows fed on pasture and those fed in a free-stall barn with fresh grass harvested every morning. The fresh grass delivery mode does not affect the expression of bta-miR-103.
In a study aiming to evaluate the effect of pasture during 3 months on stearoyl-CoA desaturase (SCD) and miR-103 expression in milk dairy goats, the SCD was significantly higher in grazing animals compared to housed animals consuming conserved forages, whereas miR-103 tended to be higher but not significantly [43]. Additionally, Lin et al., (2013) [44] showed that miR-103 and SCD gene expression had similar trends, and that the overexpression of miR-103 in mammary gland has been linked to the increased synthesis of milk fat, which is in line with the results obtained in our study linking grazing with higher milk fat content [45], and conversely, relating higher proportion of concentrate in ration with lower milk fat content (intensive farms) [46].
However, another study showed that when the grain-fed cattle were compared to the grazing cattle, the bta-miR-103 content in plasma tended to be higher in the first group (p = 0.057), although the difference was not significant [27]. Differences between the previous studies and the present work may reflect differences in the type of tissues and to the fact of studying the cows individually, which may introduce individual variability, such as lactation stage and number of lactations [43]. These and other factors can even mask differences in the expression of miRNAs according to production system [47].
Bta-miR-155 was abundant in milk from grazing farms compared with corn silage farms. This miRNA was implicated in aspects of energy balance regulation: feed restriction upregulates it in the mammary gland tissue of dairy cows [30], while oxidative stress upregulates it in a mouse model [48]. Negative energy balance in dairy cows activates the production of reactive oxygen metabolites, which in large quantities can create oxidative stress [49]. Grazing may improve immune function and oxidative status [50] due to the high amounts of antioxidants in fresh grass and the exercise involved [51]. On the contrary, other studies have reported the opposite results, with grazing favoring an increase in free radicals without a concomitant increase in the amounts of antioxidants [52]. In grazing cows, the energy input may be lower than in intensive farming [53], which is probably at the origin of the underlying bta-miR-155 levels in our study.
Studies linked miR-155 levels to a proinflammatory response in humans [54] and dairy cattle [24]. In dairy cattle, the upregulation of bta-miR-155 was related to significantly higher risk of mastitis [24]. On grazing farms, it is more likely to have higher levels of somatic cells in milk compared to housed animals [55]; an SSC level above a certain level indicates inflammatory risks, such as mastitis [56], but below these levels may indicate resilience capacity [57]. Therefore, SSC levels may be linked to the differences in bta-miR-155 levels between grazing or housed cattle.
Furthermore, bta-miR-7863 is more abundant in zero-grazing farms compared to grazing and corn silage animals. Bta-miR-7863 has been studied as a mammary biomarker of mastitis caused by Staphylococcus aureus and Escherichia coli [58].
To our knowledge, no studies have investigated bta-miR-532 expression in milk so far.
The four miRNAs identified in this study are related to the MAPK pathway, which regulates cell cycle entry and proliferation [59] and the molecular function of serine/threonine-protein kinases, which regulate cell proliferation, programmed cell death (apoptosis), cell differentiation, and embryonic development [60]. Thus, miRNAs bta-miR-155, bta-miR-103, bta-miR-532 and bta-miR-7863 may participate in cell differentiation in the mammary gland and thereby regulate milk production. Our GO-enrichment analysis identified 249 target genes of the 4 studied miRNAs that are related to transport activity, which may indicate the great involvement of the 4 studied miRNAs in the milk synthesis precursor transport process.
Using DIANA miRPath, experimentally validated human targets were identified, such as metabolic pathways related to fatty acid elongation and metabolism for miR-103 and miR-155. This suggests that the high levels of these two miRNAs in cow’s milk are related to the increased fat synthesis due to the consumption of fresh grass, especially during grazing [45]. In parallel this leads us to think about the importance of confirming these results and to study the expression of these miRNA according to the fatty acid profile of milk.
We observed variations in four miRNAs among the four types of farms grouped according to the presence or absence of grazing and different ration ingredients. We did not consider quantitative factors, such as daily grazing hours, pasture management, vegetation, animal density, and amounts of ration ingredients. This could the reason for the large variations in miRNA levels in the groups observed in this study. It would be interesting to investigate, in further studies, the variation of miRNA levels in milk by including these quantitative factors. Within the groups, the farms are not perfect replicates, as the samples belong to commercial farms, which increases the variability. In some groups, we were unable to obtain a large number of samples, as in the case of zero grazing and grass silage. This is due to the fact that few farms adopt this management system. In fact, this is the first investigation of miRNA variation in cow’s milk on commercial dairy farms representing a wide range of production systems.
4. Materials and Methods
4.1. Study Farms
The sampled farms are located in different parts of Asturias (Spain) and are representative of the characteristic production systems in the north of Spain. For each farm, the following data about feed for lactating cows were requested at three days before site visits: diet composition, whether fresh grass was consumed, and whether grass was consumed as grazed or cut. Data were also collected about the number of lactating cows, breed, and average milk production during the three days prior to the site visit.
4.2. Farm Classification
Given the continuum of farm extensiveness and lack of clear regulatory definitions [1], we defined four milk production systems for the present study based on what we considered the most relevant factors and based on the approach of Abou el qassim (2017) [39]. We considered only the management system and the presence or absence of certain ingredients in feed ration. We did not take into account quantitative variables, such as hours spent grazing, or the levels or proportions of certain ingredients in the food ration. In this way, we divided farms into the following four groups (Table 4): grazing (n = 44 farms), where animals had access to grazing and ate fresh grass and concentrated feed (grazing represents the extensive system); zero grazing (n = 13), where animals received a ration of fresh grass and concentrated feed in the stable, without grazing; grass silage (n = 10), where animals received ration based on grass silage and concentrated feed, without grazing or corn silage (zero grazing and grass silage represent intermediate systems); and corn silage (n = 45), where animals received ration of corn silage and concentrated feed, without grazing (corn silage represents intensive system).
Table 4.
Classification of the farms in the study based on milk production system.
| Ration Composition | |||||
|---|---|---|---|---|---|
| Production System | Grazing | Fresh Grass in the Stable | Grass Silage | Corn Silage | Concentrated Feed |
| Grazing (n = 44) | + | - | - | - | + |
| + | + | - | - | + | |
| + | - | + | - | + | |
| + | - | + | - | + | |
| + | - | - | - | + | |
| + | + | + | - | + | |
| + | - | + | + | + | |
| + | - | + | + | + | |
| Zero grazing (n = 13) | - | + | + | - | + |
| - | + | - | - | + | |
| - | + | + | - | + | |
| - | + | - | - | + | |
| Grass silage (n = 10) | - | - | + | - | + |
| Corn silage (n = 45) | - | - | + | + | + |
+ implies presence in the diet, - implies no presence in the diet.
4.3. Sample Collection and Processing
A total of 112 raw tank milk samples were collected, representing 112 farms, with 10 Holstein cows in the smaller farms and 250 Holstein cows in the bigger one, during fall 2016, spring 2017, and both fall and spring, 2019 and 2021. Samples included two milking sessions: afternoon and morning.
Tubes containing 50 mL of each milk sample were centrifuged at 1900× g for 20 min. The fat layer was transferred to fresh 50 mL RNase-free tubes, then Qiazol lysis reagent (Qiagen, Barcelona, Spain) was added (1 mL per milk fat gram). Tubes were vortexed until the fat was thoroughly dispersed, and samples were stored at −80 °C until RNA extraction.
4.4. Total RNA Extraction
Total RNA was extracted from 2 mL of milk fat with Qiazol using the mirVana isolation Kit (Ambion) following the manufacturer’s instructions. RNA was eluted in 100 µL of nuclease-free deionized water. The Nano-Drop spectrophotometer (ND-1000, Thermo Fisher Scientific, Madrid, Spain) was used to assess RNA concentration and purity (A260/280 ratio).
4.5. Quantitative Real-Time PCR
The isolated total RNA was reverse transcribed using the TaqMan Advanced miRNA cDNA Synthesis Kit (Thermo Fisher Scientific, Madrid, Spain). Levels of miRNAs were determined by quantitative real-time PCR (qRT-PCR) using the TaqMan Advanced miRNA Assay (ThermoFisher Scientific, Madrid, Spain) and a StepOne thermocycler (Applied Biosystems, Foster City, CA, USA) under the following conditions: 95 °C for 20 s, followed by 40 cycles of 95 °C for 1 s and 60 °C for 20 s. Template cDNA (5 μL of a 1:10 dilution) were added to 15 μL of a mix comprising 10 μL of 2× TaqMan Fast Advanced Master mix, 1 μL of 20× TaqMan Advanced miRNA Assay, and 4 μL of RNase-free water.
Specific miRNAs were quantified after selection based on previous sequencing studies [39]. Quantified miRNAs included bta-miR-215, bta-miR-369-5p, bta-miR-6520, bta-miR-7863, and bta-miR-532, all of which have been identified as the most highly expressed in milk [40]. We also quantified several miRNAs that have been associated with feeding and metabolism: bta-miR-148, bta-miR-155, bta-miR-451-5p, bta-miR-103, bta-miR-181, bta-miR-21-5p, and bta-miR-29 [27,28,41,61]. The relative abundance of miRNAs was quantified using the △△Ct method after normalization with bta-miR-30 and bta-miR-151 as described by Abou el qassim et al., (2022) [40]. All reactions were performed in duplicate. Negative controls lacking cDNA were included in all experiments.
4.6. Prediction of Potential Functions and Pathways of Genes Targeted by Milk miRNAs
Targets of miRNAs with different levels across farms were identified using TargetScan 7.2 [62] in the cow database. Target genes were selected based on cumulative weighted context++ score > 0. Functional enrichment analysis of signaling pathways involving the miRNAs target genes was performed using DAVID Bioinformatics tools (v.6.8). The Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways, biological process, and molecular function were analyzed [63].
The targets of the studied milk miRNAs and their associated pathways were also analyzed using DIANA miRPath 3.0 [64]. The validated miRNA targets were identified using DIANA TarBase 7.0 (http://diana.imis.athena-innovation.gr/DianaTools/ accessed on 1 May 2022). Gene Ontology (GO) categories and KEGG were assessed. As bovine genes are not included in DIANA miRPath, target prediction and pathways analysis were performed based on human miRNA annotations.
Statistical analyses were carried out using the integrated Fischer’s exact test followed by FDR (false discovery rate) adjustment.
4.7. Statistical Analysis
Data were analyzed using SPSS 22 software. One-way ANOVA was used to compare miRNA levels among the four study groups as independent biological types. When the analysis of variance gave a significant difference for the main effect, the Bonferroni post hoc test was applied (multiple comparison of means). When the assumption of equal variances was not met, Welch’s ANOVA was used as an alternative to one-way ANOVA. When the normality of the residues and homogeneity of variance was not verified, means were compared using the Kruskal–Wallis non-parametric test; when significant differences were attained, the Games–Howell post hoc test (multiple comparison of means) was performed. Significance was defined as p ≤ 0.05.
5. Conclusions
This study confirms that the dairy production system could influence miRNA levels in milk fat. In particular, the miRNAs bta-miR-103 and bta-miR-155 are significantly abundant in grazing farms and may be related to different factors intrinsic to grazing, such as diet and fresh grass consumption, as well as exercise and other aspects, such as immune response and oxidative status. These miRNAs emerge as potential biomarkers for the tracing of pasture-based milk.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/ijms231911681/s1.
Author Contributions
Conceptualization, L.J.R. and L.A; methodology, L.J.R., L.A.e.q. and S.L.G.; formal analysis, L.A.e.q., S.L.G. and L.J.R.; investigation, L.A.e.q., L.J.R.; resources, L.A.e.q.; data curation, L.J.R. and L.A.e.q.; writing—original draft preparation, L.A.e.q. and L.J.R.; visualization, L.A and L.J.R.; supervision, L.J.R..; project administration, L.J.R.; funding acquisition, L.J.R. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.
Conflicts of Interest
The authors declare no conflict of interest.
Funding Statement
This research was funded by Principado de Asturias Regional Government (project IDI/2021/000102) co-financed by the European Union through the ERDF (European Regional Development Fund). Loubna Abou el qassim was founded by FICYT Severo Ochoa Grant (BP17-49).
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Ruiz J., Herrera M.P., Barba R., Busqué J. Definición y Caracterización de La Extensividad En Las Explotaciones Ganaderas En España. Situación de La Ganadería Extensiva En España (I) Minist. Agric. Y Pesca Aliment. Y Medio Ambiente. 2017:1–98. [Google Scholar]
- 2.Vicente Mainar F., Martínez-Fernández A., Soldado A., De la Roza Delgado B., Argamentería Gutiérrez A. 3er Simposio Internacional del Producción Animal. Univ. Autónoma del Estado México; Ciudad de Mexico, Mexico: 2013. Producción Sostenible de Leche de Vaca Mediante El Aprovechamiento de Los Recursos Naturales y Su Impacto Sobre El Medio Ambiente; pp. 76–89. [DOI] [Google Scholar]
- 3.Rojas-Downing M.M., Nejadhashemi A.P., Elahi B., Cassida K.A., Daneshvar F., Hernandez-Suarez J.S., Abouali M., Herman M.R., Dawood Al Masraf S.A., Harrigan T. Food Footprint as a Measure of Sustainability for Grazing Dairy Farms. Environ. Manag. 2018;62:1073–1088. doi: 10.1007/s00267-018-1101-y. [DOI] [PubMed] [Google Scholar]
- 4.Villar Bonet A., Quintana Ruíz M. “Leche de Pastoreo” y “Leche de Pasto”. Es Urgente Una Figura de Calidad Reconocida Oficialmente. Vaca Pint. 2021;26:154–163. [Google Scholar]
- 5.Knaus W. Perspectives on Pasture versus Indoor Feeding of Dairy Cows. J. Sci. Food Agric. 2016;96:9–17. doi: 10.1002/jsfa.7273. [DOI] [PubMed] [Google Scholar]
- 6.Bienestar Animal y Sistemas de Producción de Vacas Lecheras. World Organisation for Animal Health; Paris, France: 2014. [(accessed on 1 May 2022)]. Comisión de Normas Sanitarias de la OIE para los Animales Terrestres. Proyecto de capítulo 7. X. Available online: https://www.woah.org/fileadmin/Home/esp/Internationa_Standard_Setting/docs/pdf/E_TAHSC_Feb_2014_Parte_B.pdf. [Google Scholar]
- 7.Marshall C.J., Beck M.R., Garrett K., Barrell G.K., Al-Marashdeh O., Gregorini P. Grazing Dairy Cows with Low Milk Urea Nitrogen Breeding Values Excrete Less Urinary Urea Nitrogen. Sci. Total Environ. 2020;739:139994. doi: 10.1016/j.scitotenv.2020.139994. [DOI] [PubMed] [Google Scholar]
- 8.Lopez C., Briard-Bion V., Ménard O. Polar Lipids, Sphingomyelin and Long-Chain Unsaturated Fatty Acids from the Milk Fat Globule Membrane Are Increased in Milks Produced by Cows Fed Fresh Pasture Based Diet during Spring. Food Res. Int. 2014;58:59–68. doi: 10.1016/j.foodres.2014.01.049. [DOI] [Google Scholar]
- 9.Veskoukis A.S., Kerasioti E., Sidiropoulos K., Maragou I., Skaperda Z.O.I., Kouretas D. Nutritional Habits and Free Grazing Regimen of Productive Animals along with Specific Ingredients Are Influential Factors for the Antioxidant Properties of Milk: From Farm to Market. Biomed. Rep. 2020;13:31–36. doi: 10.3892/br.2020.1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Heard J.W., Hannah M.C., Ho C.K.M., Wales W.J. Predicting Immediate Marginal Milk Responses and Evaluating the Economics of Two-Variable Input Tactical Feeding Decisions in Grazing Dairy Cows. Animals. 2021;11:1920. doi: 10.3390/ani11071920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Soder K.J., Rotz C.A. Economic and Environmental Impact of Four Levels of Concentrate Supplementation in Grazing Dairy Herds. J. Dairy Sci. 2001;84:2560–2572. doi: 10.3168/jds.S0022-0302(01)74709-1. [DOI] [PubMed] [Google Scholar]
- 12.Dartt B.A., Lloyd J.W., Radke B.R., Black J.R., Kaneene J.B. A Comparison of Profitability and Economic Efficiencies between Management-Intensive Grazing and Conventionally Managed Dairies in Michigan. J. Dairy Sci. 1999;82:2412–2420. doi: 10.3168/jds.S0022-0302(99)75492-5. [DOI] [PubMed] [Google Scholar]
- 13.Nakajima N., Yayota M. Grazing and Cattle Health: A Nutritional, Physiological, and Immunological Status Perspective. Anim. Behav. Manag. 2019;55:143–153. [Google Scholar]
- 14.Croissant A.E., Washburn S.P., Dean L.L., Drake M.A. Chemical Properties and Consumer Perception of Fluid Milk from Conventional and Pasture-Based Production Systems. J. Dairy Sci. 2007;90:4942–4953. doi: 10.3168/jds.2007-0456. [DOI] [PubMed] [Google Scholar]
- 15.Stampa E., Schipmann-Schwarze C., Hamm U. Consumer Perceptions, Preferences, and Behavior Regarding Pasture-Raised Livestock Products: A Review. Food Qual. Prefer. 2020;82:103872. doi: 10.1016/j.foodqual.2020.103872. [DOI] [Google Scholar]
- 16.He L., Hannon G.J. MicroRNAs: Small RNAs with a Big Role in Gene Regulation. Nat. Rev. Genet. 2004;5:522–531. doi: 10.1038/nrg1379. [DOI] [PubMed] [Google Scholar]
- 17.Zhou G., Zhou Y., Chen X. New Insight into Inter-Kingdom Communication: Horizontal Transfer of Mobile Small RNAs. Front. Microbiol. 2017;8:768. doi: 10.3389/fmicb.2017.00768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang J., Li S., Li L., Li M., Guo C., Yao J., Mi S. Exosome and Exosomal MicroRNA: Trafficking, Sorting, and Function. Genom. Proteom. Bioinforma. 2015;13:17–24. doi: 10.1016/j.gpb.2015.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.del Pozo-Acebo L., López de las Hazas M., Margollés A., Dávalos A., García-Ruiz A. Eating MicroRNAs: Pharmacological Opportunities for Cross-kingdom Regulation and Implications in Host Gene and Gut Microbiota Modulation. Br. J. Pharmacol. 2021;178:2218–2245. doi: 10.1111/bph.15421. [DOI] [PubMed] [Google Scholar]
- 20.Zempleni J., Baier S.R., Howard K.M., Cui J. Gene Regulation by Dietary MicroRNAs. Can. J. Physiol. Pharmacol. 2015;93:1097–1102. doi: 10.1139/cjpp-2014-0392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Weber J.A., Baxter D.H., Zhang S., Huang D.Y., Huang K.H., Lee M.J., Galas D.J., Wang K. The MicroRNA Spectrum in 12 Body Fluids. Clin. Chem. 2010;56:1733–1741. doi: 10.1373/clinchem.2010.147405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Carrillo-Lozano E., Sebastián-Valles F., Knott-Torcal C. Circulating Micrornas in Breast Milk and Their Potential Impact on the Infant. Nutrients. 2020;12:3066. doi: 10.3390/nu12103066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tzelos T., Ho W., Charmana V.I., Lee S., Donadeu F.X. MiRNAs in Milk Can Be Used towards Early Prediction of Mammary Gland Inflammation in Cattle. Sci. Rep. 2022;12:5131. doi: 10.1038/s41598-022-09214-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lai Y.C., Fujikawa T., Maemura T., Ando T., Kitahara G., Endo Y., Yamato O., Koiwa M., Kubota C., Miura N. Inflammation-Related MicroRNA Expression Level in the Bovine Milk Is Affected by Mastitis. PLoS ONE. 2017;12:e0177182. doi: 10.1371/journal.pone.0177182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li Z., Liu H., Jin X., Lo L., Liu J. Expression Profiles of MicroRNAs from Lactating and Non-Lactating Bovine Mammary Glands and Identification of MiRNA Related to Lactation. BMC Genom. 2012;13:731. doi: 10.1186/1471-2164-13-731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Colitti M., Sgorlon S., Licastro D., Stefanon B. Differential Expression of MiRNAs in Milk Exosomes of Cows Subjected to Group Relocation. Res. Vet. Sci. 2019;122:148–155. doi: 10.1016/j.rvsc.2018.11.024. [DOI] [PubMed] [Google Scholar]
- 27.Muroya S., Shibata M., Hayashi M., Oe M., Ojima K. Differences in Circulating MicroRNAs between Grazing and Grain-Fed Wagyu Cattle Are Associated with Altered Expression of Intramuscular MicroRNA, the Potential Target PTEN, and Lipogenic Genes. PLoS ONE. 2016;11:e0162496. doi: 10.1371/journal.pone.0162496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li R., Beaudoin F., Ammah A.A., Bissonnette N., Benchaar C., Zhao X., Lei C., Ibeagha-Awemu E.M. Deep Sequencing Shows MicroRNA Involvement in Bovine Mammary Gland Adaptation to Diets Supplemented with Linseed Oil or Safflower Oil. BMC Genom. 2015;16:884. doi: 10.1186/s12864-015-1965-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mobuchon L., Le Guillou S., Marthey S., Laubier J., Laloë D., Bes S., Le Provost F., Leroux C. Sunflower Oil Supplementation Affects the Expression of MiR-20a-5p and MiR-142-5p in the Lactating Bovine Mammary Gland. PLoS ONE. 2017;12:e0185511. doi: 10.1371/journal.pone.0185511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Billa P.A., Faulconnier Y., Ye T., Chervet M., Le Provost F., Pires J.A.A., Leroux C. Deep RNA-Seq Reveals MiRNome Differences in Mammary Tissue of Lactating Holstein and Montbéliarde Cows. BMC Genom. 2019;20:621. doi: 10.1186/s12864-019-5987-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen X., Gao C., Li H., Huang L., Sun Q., Dong Y., Tian C., Gao S., Dong H., Guan D., et al. Identification and Characterization of MicroRNAs in Raw Milk during Different Periods of Lactation, Commercial Fluid, and Powdered Milk Products. Cell Res. 2010;20:1128–1137. doi: 10.1038/cr.2010.80. [DOI] [PubMed] [Google Scholar]
- 32.Le Guillou S., Leduc A., Laubier J., Barbey S., Rossignol M.N., Lefebvre R., Marthey S., Laloë D., Le Provost F. Characterization of Holstein and Normande Whole Milk MiRNomes Highlights Breed Specificities. Sci. Rep. 2019;9:20345. doi: 10.1038/s41598-019-56690-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Guan J., Long K., Ma J., Zhang J., He D., Jin L., Tang Q., Jiang A., Wang X., Hu Y., et al. Comparative Analysis of the MicroRNA Transcriptome between Yak and Cattle Provides Insight into High-Altitude Adaptation. PeerJ. 2017;2017:e3959. doi: 10.7717/peerj.3959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bauman D.E., Harvatine K.J., Lock A.L. Nutrigenomics, Rumen-Derived Bioactive Fatty Acids, and the Regulation of Milk Fat Synthesis. Annu. Rev. Nutr. 2011;31:299–319. doi: 10.1146/annurev.nutr.012809.104648. [DOI] [PubMed] [Google Scholar]
- 35.Padovani M., Lavigne J.A., Chandramouli G.V.R., Perkins S.N., Barrett J.C., Hursting S.D., Bennett L.M., Berrigan D. Distinct Effects of Calorie Restriction and Exercise on Mammary Gland Gene Expression in C57BL/6 Mice. Cancer Prev. Res. 2010;12:1076–1087. doi: 10.1158/1940-6207.CAPR-09-0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tao S., Bubolz J.W., do Amaral B.C., Thompson I.M., Hayen M.J., Johnson S.E., Dahl G.E. Effect of Heat Stress during the Dry Period on Mammary Gland Development. J. Dairy Sci. 2011;94:5976–5986. doi: 10.3168/jds.2011-4329. [DOI] [PubMed] [Google Scholar]
- 37.Izumi H., Kosaka N., Shimizu T., Sekine K., Ochiya T., Takase M. Bovine Milk Contains MicroRNA and Messenger RNA That Are Stable under Degradative Conditions. J. Dairy Sci. 2012;95:4831–4841. doi: 10.3168/jds.2012-5489. [DOI] [PubMed] [Google Scholar]
- 38.Buschmann D., Haberberger A., Kirchner B., Spornraft M., Riedmaier I., Schelling G., Pfaffl M.W. Toward Reliable Biomarker Signatures in the Age of Liquid Biopsies—How to Standardize the Small RNA-Seq Workflow. Nucleic Acids Res. 2016;44:5995–6018. doi: 10.1093/nar/gkw545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Abou el qassim L. Master’s Thesis. Universidad de Zaragoza; Zaragoza, Spain: 2017. Variaciones de Los Perfiles de MicroARN En La Leche Cruda de Vaca Según El Sistema de Alimentación. [Google Scholar]
- 40.Abou el qassim L., Alonso J., Zhao K., Le Guillou S., Diez J., Vicente F., Fernández-Sanjurjo M., Iglesias-Gutiérrez E., Guan L.L., Royo L.J. Servicio Regional de Investigación y Desarrollo Agroalimentario (SERIDA); Villaviciosa, Spain: 2022. Differences in the MicroRNAs Levels of Raw Milk from Dairy Cattle Raised under Extensive or Intensive Production Systems. Manuscript under review. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Muroya S., Ogasawara H., Hojito M. Grazing Affects Exosomal Circulating MicroRNAs in Cattle. PLoS ONE. 2015;10:e0136475. doi: 10.1371/journal.pone.0136475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Muroya S., Ogasawara H., Nohara K., Oe M., Ojima K., Hojito M. Coordinated Alteration of MRNA-MicroRNA Transcriptomes Associated with Exosomes and Fatty Acid Metabolism in Adipose Tissue and Skeletal Muscle in Grazing Cattle. Asian-Australasian J. Anim. Sci. 2020;33:1824–1836. doi: 10.5713/ajas.19.0682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tudisco R., Morittu V.M., Addi L., Moniello G., Grossi M., Musco N., Grazioli R., Mastellone V., Pero M.E., Lombardi P., et al. Influence of Pasture on Stearoyl-CoA Desaturase and MiRNA 103 Expression in Goat Milk: Preliminary Results. Animals. 2019;9:606. doi: 10.3390/ani9090606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lin X., Luo J., Zhang L., Wang W., Gou D. MiR-103 Controls Milk Fat Accumulation in Goat (Capra Hircus) Mammary Gland during Lactation. PLoS ONE. 2013;8:e79258. doi: 10.1371/journal.pone.0079258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.De La Torre-Santos S., Royo L.J., Martínez-Fernández A., Menéndez-Miranda M., Rosa-García R., Vicente F. Influence of the Type of Silage in the Dairy Cow Ration, with or without Grazing, on the Fatty Acid and Antioxidant Profiles of Milk. Dairy. 2021;2:716–728. doi: 10.3390/dairy2040055. [DOI] [Google Scholar]
- 46.Sanh M.V., Wiktorsson H., Ly L.V. Effects of Natural Grass Forage to Concentrate Ratios and Feeding Principles on Milk Production and Performance of Crossbred Lactating Cows. Asian-Australasian J. Anim. Sci. 2002;15:650–657. doi: 10.5713/ajas.2002.650. [DOI] [Google Scholar]
- 47.Avril-Sassen S., Goldstein L.D., Stingl J., Blenkiron C., Le Quesne J., Spiteri I., Karagavriilidou K., Watson C.J., Tavaré S., Miska E.A., et al. Characterisation of MicroRNA Expression in Post-Natal Mouse Mammary Gland Development. BMC Genom. 2009;10:548. doi: 10.1186/1471-2164-10-548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vallanat B., Anderson S.P., Brown-Borg H.M., Ren H., Kersten S., Jonnalagadda S., Srinivasan R., Corton J.C. Analysis of the Heat Shock Response in Mouse Liver Reveals Transcriptional Dependence on the Nuclear Receptor Peroxisome Proliferator-Activated Receptor α (PPARα) BMC Genom. 2010;11:16. doi: 10.1186/1471-2164-11-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pedernera M., Celi P., García S.C., Salvin H.E., Barchia I., Fulkerson W.J. Effect of Diet, Energy Balance and Milk Production on Oxidative Stress in Early-Lactating Dairy Cows Grazing Pasture. Vet. J. 2010;186:352–357. doi: 10.1016/j.tvjl.2009.09.003. [DOI] [PubMed] [Google Scholar]
- 50.Braghieri A., Pacelli C., De Rosa G., Girolami A., De Palo P., Napolitano F. Podolian Beef Production on Pasture and in Confinement. Animal. 2011;5:927–937. doi: 10.1017/S1751731110002685. [DOI] [PubMed] [Google Scholar]
- 51.Celi P. Biomarkers of Oxidative Stress in Ruminant Medicine. Immunopharmacol. Immunotoxicol. 2011;33:233–240. doi: 10.3109/08923973.2010.514917. [DOI] [PubMed] [Google Scholar]
- 52.Di Grigoli A., Di Trana A., Alabiso M., Maniaci G., Giorgio D., Bonanno A. Effects of Grazing on the Behaviour, Oxidative and Immune Status, and Production of Organic Dairy Cows. Animals. 2019;9:371. doi: 10.3390/ani9060371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Vicente Mainar F., Morales-Almaraz E., Martínez-Fernández A. El Uso de Forrajes Para La Mejora de Producción y de Calidad de La Leche de Vacuno Lechero. Ganadería. 2012;82:30–33. [Google Scholar]
- 54.Tomé-Carneiro J., Larrosa M., Yáñez-Gascón M.J., Dávalos A., Gil-Zamorano J., Gonzálvez M., García-Almagro F., Ruiz Ros J.A., Tomás-Barberán F., Espín J.C., et al. One-Year Supplementation with a Grape Extract Containing Resveratrol Modulates Inflammatory-Related MicroRNAs and Cytokines Expression in Peripheral Blood Mononuclear Cells of Type 2 Diabetes and Hypertensive Patients with Coronary Artery Disease. Pharmacol. Res. 2013;72:69–82. doi: 10.1016/j.phrs.2013.03.011. [DOI] [PubMed] [Google Scholar]
- 55.De La Torre-Santos S. Ph.D. Thesis. Universidad de Zaragoza; Zaragoza, Spain: 2021. Identificación de Biomarcadores Específicos Para Autentificar El Origen y El Sistema de Alimentación Del Vacuno Lechero. [Google Scholar]
- 56.Lamarche A., Martin B., Hauwuy A., Coulon J.B., Poutrel B. Evolution of Milk Somatic Cell Count of Cows Grazing an Alpine Pasture According to the Infection of Udder by Pathogens. Anim. Res. 2000;49:45–54. doi: 10.1051/animres:2000107. [DOI] [Google Scholar]
- 57.Bishop S.C., Woolliams J.A. Genomics and Disease Resistance Studies in Livestock. Livest. Sci. 2014;166:190–198. doi: 10.1016/j.livsci.2014.04.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Luoreng Z.M., Wang X.P., Mei C.G., Zan L. Sen Comparison of MicroRNA Profiles between Bovine Mammary Glands Infected with Staphylococcus Aureus and Escherichia Coli. Int. J. Biol. Sci. 2018;14:87–99. doi: 10.7150/ijbs.22498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wei Z., Liu H.T. MAPK Signal Pathways in the Regulation of Cell Proliferation in Mammalian Cells. Cell Res. 2002;12:9–18. doi: 10.1038/sj.cr.7290105. [DOI] [PubMed] [Google Scholar]
- 60.Cross T.G., Scheel-Toellner D., Henriquez N.V., Deacon E., Salmon M., Lord J.M. Serine/Threonine Protein Kinases and Apoptosis. Exp. Cell Res. 2000;256:34–41. doi: 10.1006/excr.2000.4836. [DOI] [PubMed] [Google Scholar]
- 61.Wang D., Liang G., Wang B., Sun H., Liu J., Guan L.L. Systematic MicroRNAome Profiling Reveals the Roles of MicroRNAs in Milk Protein Metabolism and Quality: Insights on Low-Quality Forage Utilization. Sci. Rep. 2016;6:21194. doi: 10.1038/srep21194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Agarwal V., Bell G.W., Nam J.-W., Bartel D.P. Predicting Effective MicroRNA Target Sites in Mammalian MRNAs. Elife. 2015;4:e05005. doi: 10.7554/eLife.05005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Dennis G., Sherman B.T., Hosack D.A., Yang J., Gao W., Lane H.C., Lempicki R.A. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 2003;4:R60. doi: 10.1186/gb-2003-4-9-r60. [DOI] [PubMed] [Google Scholar]
- 64.Vlachos I.S., Zagganas K., Paraskevopoulou M.D., Georgakilas G., Karagkouni D., Vergoulis T., Dalamagas T., Hatzigeorgiou A.G. DIANA-MiRPath v3.0: Deciphering MicroRNA Function with Experimental Support. Nucleic Acids Res. 2015;43:W460–W466. doi: 10.1093/nar/gkv403. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.