Abstract
The role of the microbiome in hair follicle (HF) growth represents a growing field of research. Here, we studied the bacterial population in the scalp hair follicles of subjects with alopecia areata (AA). Two Healthy and two AA subjects, respectively (20–60 years old), were enrolled and studied regarding the microbial community in the subepidermal scalp compartments by means of a 4-mm biopsy punch. Samples were examined by 16S sequencing, histochemical staining (Gram’s method), and transmission electron microscopy (TEM). Bacterial foci were observed in the AA subjects’ follicles with both the two adopted complementary approaches (electron microscopy and Gram staining). Significant (p < 0.05) differences were also found in the three-layer biopsy samples (p < 0.05) regarding the bacterial population. In particular, in the deep epidermis and dermis levels, a significant (p < 0.05) lower abundance of Firmicutes and a higher abundance of Proteobacteria were found in AA samples compared to the healthy control. Firmicutes also showed a significant (p < 0.05) lower abundance in hypodermis in AA subjects. In addition, Enterobacteriaceae and the genera Streptococcus, Gemella, Porphyromonas, and Granulicatella were relatively more abundant in AA groups at the deep epidermis level. The Staphylococcus and Flavobacterium genera were significantly less abundant in AA samples than in controls in all three-layer biopsy samples (p < 0.05). In contrast, Veillonella and Neisseriaceae were relatively more abundant in the healthy control group compared to the AA sample. Therefore, higher alpha diversity was observed in all three-layer biopsy samples of AA patients compared to the control. In conclusion, our data suggest that tAA could be defined as a “hair disease associated with dysregulated microbiome-immunity axis of hair follicles”.
Keywords: alopecia areata, microbiota, immunity, bacteria, transmission electron microscopy
1. Introduction
The alopecia areata (AA), noncicatricial alopecia, is an autoimmune disease targeting hair follicles (HF) in the anagen phase without predilection for sex or race. The AA incidence ranges from 0.57% to 3.8% [1].
The exact pathogenesis of AA remains still unknown, but increasing evidence suggests the implication of the HF immune privilege (HFIP) collapse from the bulge downwards to the bulb [2,3,4] as a possible stepping stone. However, how perifollicular memory T cells and innate lymphoid cells are implicated in HFIP collapse remains an unexplored research frontier [5]. In addition, another suggestive and intricate aspect concerns the involvement of the HFs’ microbiome in the hair cycle in AA and especially in the HFIP process [6,7]. Other autoimmune diseases, such as vitiligo, Hashimoto’s thyroiditis, psoriasis, diabetes mellitus, lupus erythematosus, and celiac disease, have been associated with comorbidities in AA patients [8]. Among these, hypothyroidism has the strongest association [9,10,11].
Thousands of microorganisms, including bacteria, fungi, archaea, and viruses, inhabit the skin and scalp ecosystem; they represent the so-called microbiota [12]. The theatre of activities and interactions with the human host of the microbiota by molecules (metabolites, DNA, genes, etc.) is called a “microbiome” [13]. There is growing evidence of the presence of bacteria below the surface of the skin and alongside the HFs [14,15,16,17]. It is also becoming increasingly clear that the dialogue between bacteria and the underlying tissue is a dynamic interplay [18,19]. It has also been hypothesized that external microbial stimulation creates immune responses in the HF epithelium [20] and, contextually, immunological events around the HF could shape microbial composition [21].
HF microenvironment, including lipid-rich and hypoxic conditions, could contribute to the establishment of resident microbes. For example, the hair shaft environment may be unfavorable to the growth of genera such as Propionibacterium, which would prefer a low level of oxygen and a high level of sebum like that of HF [22]. Indeed, exactly like the skin, the resident microbiota varies among different hair environments [14].
Recently, we reported the scalp and gut microbial shift in AA patients compared to healthy controls [23,24]. In addition, the constant crosstalk between the microorganisms living on the scalp and the skin’s immune cells is well documented [13]. Notably, the failure of the fine regulation between host immunity and “tolerance” to the resident microbiota is critical and might contribute to inflammatory and autoimmune skin diseases, including AA [25,26,27,28].
Finally, it has been suggested that the keratinocytes, in response to alteration of the resident microflora near and around HF, could unbalance the chemokines’ secretion and IP guardian, playing a role in both AA development and response to therapy [5].
Recently, we highlighted the differences in microbial populations on the scalp of AA and healthy subjects [11] and, in detail, by analyzing the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways. We documented that the pathways related to bacterial chemotaxis and cellular antigens were predominant in AA samples. The bacterial chemotaxis allows different strains to access specific host niches, delivering proapoptotic host cell factors [29]. Interestingly, microbial-derived antigens have been reported to be responsible for the susceptibility of HFs to HFIP [30].
This provides initial evidence of how our resident skin flora could interact with the immunological part of the HF ecosystem.
So, to further explore the strict relationship between microbiota changes and pathogenesis and development of AA, we performed and compared observations by electron microscopy and gram staining for in situ analysis of the spatial arrangement of bacteria around the HF of AA patients and healthy control, respectively. Finally, the identified bacterial foci were then compared with taxonomic analysis obtained using 16S rRNA analysis.
2. Results
We used three different methodologic approaches to observe, define, and compare the bacterial foci in scalp biopsy patients affected by alopecia areata and healthy controls.
2.1. Microbiota Profiling of the Scalp in AA Patients
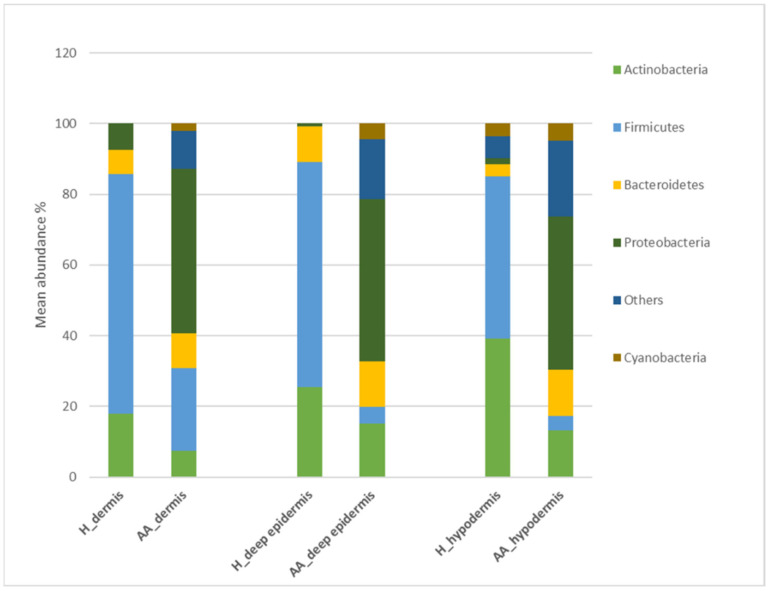
The composition of human scalp bacterial composition of controls (n = 2) and AA (N = 2) subjects have been analyzed by IlluminaSeq (Figure 1). A bacterial shift was found based on three-layer biopsy samples from AA subjects. Moreover, we found significant (p < 0.05) differences in the microbiota composition between healthy and AA subjects based on all three-layer biopsy samples (Table 1).
Figure 1.
Bacterial profiling in control and AA subjects. Percentage of bacteria at phylum level in the healthy controls (H) and patients with alopecia areata (AA) groups. Results are presented as the percentage (%) of total sequences.
Table 1.
Statistically different taxa emerged between microbial taxa healthy abundances versus alopecia areata (AA) at sub-epidermal layers (deep epidermis, dermis, hypodermis).
| Layer | Taxa Level | Feature | Healthy | AA | p-Value |
|---|---|---|---|---|---|
| deep epidermis | family | Enterobacteriaceae | 0.00 | 17.09 | 0.500 |
| deep epidermis | genus | Streptococcus | 2.45 | 16.30 | 0.551 |
| deep epidermis | genus | Gemella | 0.00 | 2.40 | 0.500 |
| deep epidermis | genus | Porphyromonas | 0.00 | 2.14 | 0.500 |
| deep epidermis | genus | Granulicatella | 0.00 | 2.50 | 0.500 |
| deep epidermis | genus | Staphylococcus | 49.60 | 0.00 | 0.227 |
| deep epidermis | genus | Flavobacterium | 8.00 | 0.00 | 0.156 |
| dermis | phylum | Firmicutes | 71.92 | 22.50 | 0.009 |
| dermis | phylum | Proteobacteria | 7.48 | 46.55 | 0.027 |
| dermis | family | Micrococcaceae | 0.00 | 6.41 | 0.137 |
| dermis | genus | Staphylococcus | 48.35 | 1.35 | 0.278 |
| dermis | genus | Flavobacterium | 5.00 | 0.00 | 0.275 |
| hypodermis | phylum | Firmicutes | 45.75 | 4.05 | 0.033 |
| hypodermis | phylum | Proteobacteria | 1.8 | 43.35 | 0.009 |
| hypodermis | family | Bacteroidetes | 2.96 | 12.74 | 0.015 |
| hypodermis | family | Micrococcaceae | 0.00 | 4.85 | 0.06 |
| hypodermis | family | Neisseriaceae | 0.00 | 1.00 | 0.500 |
| hypodermis | genus | Staphylococcus | 10.20 | 0.90 | 0.021 |
| hypodermis | genus | Flavobacterium | 7.10 | 0.00 | 0.269 |
| hypodermis | genus | Veillonella | 0.00 | 4.30 | 0.500 |
Regarding the deep epidermis level and the dermis level, Firmicutes showed significantly lower abundances in AA samples compared to those in healthy samples. In contrast, Proteobacteria was associated with significantly higher abundances (p < 0.05).
Notably, at the hypodermis level, Firmicutes showed significantly lower abundances in AA samples compared to those in healthy samples. Finally, the Proteobacteria and Bacteroidetes phyla were relatively more abundant in AA samples than in controls (p < 0.05).
In addition, the family Enterobacteriaceae and the genera Streptococcus, Gemella, Porphyromonas, and Granulicatella were relatively more abundant in AA groups at the deep epidermis level. Therefore, Neisseria subflava was absent in healthy controls at the species level.
The Staphylococcus and Flavobacterium genera were significantly less abundant in AA samples than in controls in all three-layer biopsy samples (p < 0.05).
The genus Pseudomonas and the family Micrococcaceae were more abundant in the dermis and hypodermis layers of the AA subjects. In contrast, Veillonella and Neisseriaceae were relatively more abundant in the healthy control group compared to AA samples.
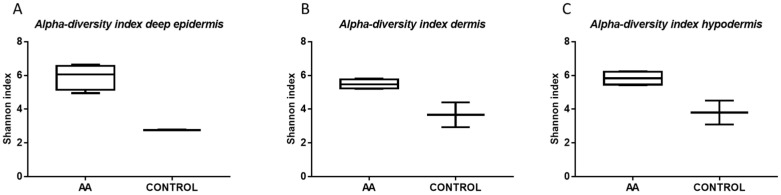
The Shannon index values were indicative of differences in diversity among the analyzed biopsy layers (Figure 2). In particular, in all three-layer biopsy samples of AA patients, we observed a higher alpha diversity.
Figure 2.
Alpha diversity index. All the diversity indices for (A) deep epidermis, (B) dermis, and (C) hypodermis in AA and healthy control group are expressed as Shannon index.
2.2. Bacterial Infection in AA Subjects
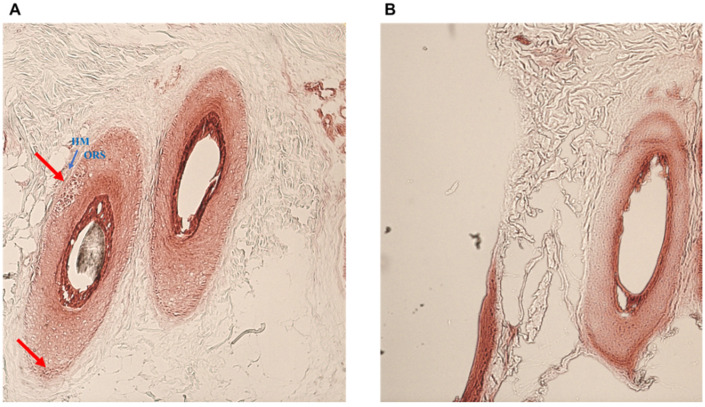
Both the two adopted complementary approaches (electron microscopy and Gram staining) have allowed the detection of bacterial foci in the follicle of AA subjects. By the histochemical analysis, we observed that Gram-positive and negative bacteria foci were around the hair follicle (Figure 3).
Figure 3.
Gram staining. Hair follicles of AA patients (A) and control subjects (B) at the level of the isthmus showing bacterial foci (red arrow). HM: hyaline membrane (blue arrow); ORS: outer root sheet.
2.3. Histology and Ultrastructure of Normal and Alopecia Aerate Scalp Hair Bulb
Before describing histologic and ultrastructural alterations in AA, the normal growth of hair follicles will be described briefly as a good starting point for explaining the morphological changes in several pathological conditions.
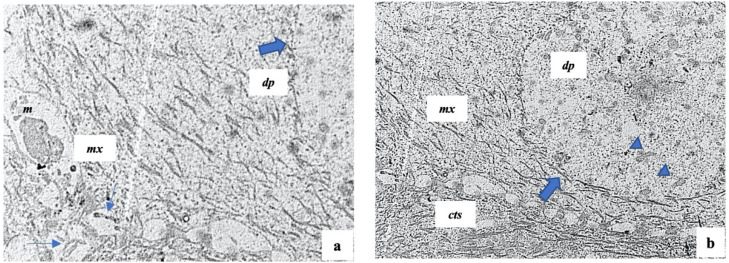
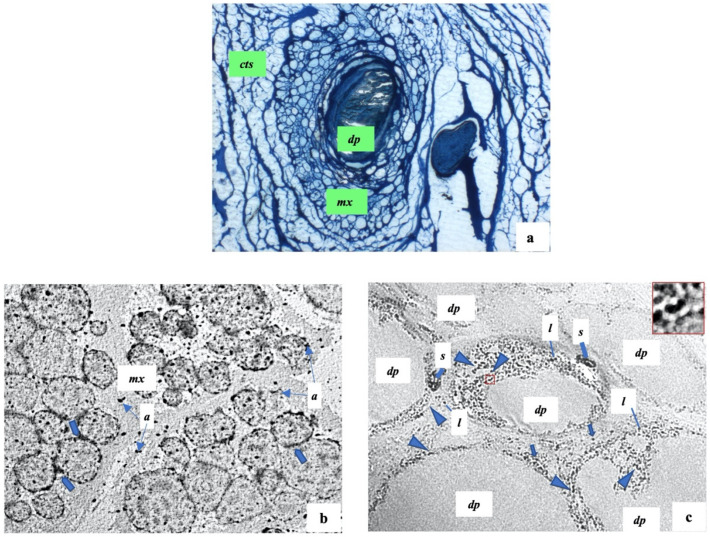
The hair bulb shows, from outside to inside, three major components: the epithelial hair matrix showing mainly the keratinocytes and melanocytes; the connective tissue sheath—a mesenchymal sheath wrapping the entire hair follicles; the follicular papilla—a vascularized mesenchymal structure which presents a fine basal lamina. We also recognized some keratinocytes in the epithelia hair matrix in the mitotic stage, indicating a proliferative moment linked to the hair phase (Figure 4a,b). The follicular papilla shows many fibroblasts and some fibrocytes embedded in ground substance; they are separated from the surrounding hair matrix (Figure 4b).
Figure 4.
Representative electron microscopical overview of normal scalp hair follicles. cts: connective tissue sheath, mx: hair matrix; dp: follicular papilla; m: melanosomes; thin arrows: keratinocytes in mitotic stage; thick arrows: fine basal lamina of follicular papilla; arrowheads: fibroblasts. (a) 6200×; (b) 6200×.
As previously reported, the hair follicles are altered in AA, and we confirmed, via semithin and electron microscopy sections, that these alterations involve both hair follicles and their matrix; we showed both these changes.
In this regard, by semithin sections, we showed many changes in the hair matrix, the basal lamina, and the area around the follicular papilla (Figure 5a). The basal lamina of the follicular papilla seems very thick, while the area around the papilla shows many not correctly organized cells.
Figure 5.
Scalp hair follicles in alopecia areata. (a) A representative semithin section shows: a follicular papilla (dp) with abundant flocculent substance; a hair matrix (mx) with many cells not well organized; cts: connective tissue sheath. (b,c) The representative ultrastructural sections of hair matrix and follicular papilla respectively show: mx: hair matrix; a: apoptotic debris; thick arrows: beaded membrane of keratinocytes; dp: follicular papilla; head arrows: bacteria infiltrations; s: melanosome structures; l: lymphocytes; red square: area of inset magnification. (a) 200×; (b) 1750×; (c) 12,500×.
Based on these results, we performed an electron microscopical analysis. We observed that the hair matrix showed many keratinocytes with cellular injuries, such as a marked thickening of their membrane that could be defined “beaded” membrane. Moreover, these cells and the other part of the hair matrix showed much apoptotic debris (Figure 5b,c).
Of note, as documented in detail in Figure 6, the follicular papilla and its surrounding area showed the presence of elongated-shaped electron-dense particles such as the bacteria infections (Figure 5c). In healthy controls, rare black particles were present, while in AA patients, the black particles were clearly present in a widespread manner. Moreover, it is important to highlight that these bacterial infections were surrounded by well-recognized lymphocytic infiltrates thanks to their rounded nucleus and their size. Further, the latter were also scattered and in the hair matrix (Figure 5c).
Figure 6.
Microbiota and immunity crosstalk in alopecia areata.
Finally, in addition to the bacterial infections and lymphocytic infiltrates, giant spherical melanin structures occurred in the altered follicular papilla (Figure 5c); these structures contained many melanosomes and were located between the follicular papilla at the hair matrix. Furthermore, the follicular papilla showed many fibrocytes and abundant flocculent substances concerning healthy control.
3. Discussion
The best and most crucial result of the study is that, for the first time, we documented the presence of bacterial foci around hair follicles in patients with alopecia areata.
We analyzed the microbiota composition considering three skin biopsy layers (deep epidermis, dermis, and hypodermis) that is in agreement with our previous works [10,11]. We confirmed the differences in microbial populations inhabiting the scalp of AA patients compared to those of healthy controls.
We observed significant differences (p < 0.05) in all three-layer biopsy samples, documenting an increase in some species, such as Neisseria subflava and Pseudomonas, and a decrease in Veillonella genus at the dermis level. We confirmed that specific microbiota characteristics are associated with damaged AA tissue. Interestingly, Neisseria sicca/subflava, a bacterium generally considered commensal inhabitants of the human oropharynx, was reported to cause occasional infections [31,32].
In addition, we found an increase in gram-positive cocci, such as Micrococcaceae. Usually, species from this family are common noninfectious bacteria of the human skin; however, recently, Boldock and colleagues [33] suggested that some pathogens might recruit them (e.g., S. aureus) to support and initiate infection.
In addition, we observed differences between different layers and confirmed that skin location is the main impacting factor on the composition of bacterial communities. Therefore, an unexpected increase in α-diversity was found in AA patients (compared to the healthy controls), suggesting a higher susceptibility of the HF to be colonized by microorganisms.
Finally, we used both optical and electron microscopy techniques to localize the microbes in the deep layers; in AA samples, the gram staining showed the presence of bacteria incorporated inside the outer root sheet at the isthmus level. Moreover, at a deep level, we identified in the follicular papilla and its surrounding area, the presence of widespread electron-dense particles such as bacterial infections (rarely observed in healthy controls). This relevant finding represents the most important result of our study. In contrast, many studies have confirmed the presence of numerous bacteria and fungi within the follicular opening and upper part of the HF (the infundibulum). Only some experimental studies have located microbes deeply in the follicle [21].In other skin pathology, such as acne vulgaris [33] and folliculitis decalvans [34], but anyone in AA. Furthermore, we found that the bacterial outbreak was surrounded by infiltrating lymphocytes suggesting an active and dysregulated role of the host immune response in this pathology.
In this scenario, it is irrelevant to remember that, like the gut microbiome, the skin microbiome has critical and crucial crosstalk with the host immune system modulating the immune responses (Figure 6) [21,35]. However, while the bacterial role in HF-associated inflammatory diseases has been well established [36], poor information is currently available on how alterations in the HF microbiota can affect the growth and immunological status of hair follicles.
Previously [11], we correlated the HF microbiota modifications, especially at the dermis level, with changes in pathways related to immune responses such as bacterial chemotaxis and glycosaminoglycan (GAG) degradation; GAGs can be used by bacteria to evade the host immunosurveillance. Therefore, we also found a positive correlation between microbial HF changes in AA patients and nucleotide-binding oligomerization domain containing 2 (NOD2), an innate immune receptor [37].
Regarding these last considerations, it is important to remember that AA is classified as an autoimmune disease. However, an increased prevalence in other forms of inflammatory skin diseases (e.g., psoriasis, vitiligo, and atopic dermatitis) were reported in AA patients, suggesting that they have an increased risk of developing T cell-driven inflammatory skin diseases [38]. Moreover, in physiological conditions, the hair follicle is defined as an “immune-privileged site” that prevents autoimmune responses [39], and the breakdown of this homeostasis is usually considered one of the major AA drivers. It has been proposed that follicular keratinocytes have the ability to release chemotactic for the T cells’ enrollment into the perifollicular area [40], thereby triggering a self-replicating process.
Additionally, the electron microscopy images showed the ultrastructural complex change in which the follicle undergoes AA pathology. In detail, we observed an altered hair follicle matrix with much apoptotic debris that affects early cortical differentiation epithelium and the presence of infiltrating lymphocytes. These data suggest that the matrix seems to be the primary target of an immune attack on the hair follicle [41,42]. In addition, these degenerative changes lead to a localized region of weakness in the hair shaft, which causes hair breaking when it emerges from the ostium at the skin surface.
Finally, we observed giant spherical melanin structures in the altered follicular papilla that is in line with previous data, showing [43,44] that the hair bulb melanocytes were damaged in acute AA, resulting in impaired melanogenesis and the loss of the ellipsoidal shape. The main limitation of the study was the small sample size. Further studies consisting of a larger number of patients are needed to support the provided evidence better.
4. Materials and Methods
4.1. Subjects’ Recruitment
The study was in the form of a monocentric study involving both healthy and AA subjects.
Healthy subjects have been screened in the absence of any history of dermatological or scalp disorders and enrolled if confirmed after clinical examination.
AA patients were diagnosed clinically and confirmed as having AA by biopsy, according to the WHO criteria. Essential clinical data were collected at baseline under dermatological control according to the guidelines of the National Alopecia Areata Foundation [45].
We adopted the following exclusion criteria: (i) the administration of antibiotics in the last 30 days before the sampling; (ii) the probiotics’ use in the last 15 days; (iii) women on pregnancy or lactation; (iv) the documented presence of other dermatological diseases; (vi) therapy with antitumor, immunosuppressant, or radiation therapy in the last 3 months; (vii) treatment with topical or hormonal therapy on the scalp in the last 3 months. The study started after the approval of the Ethical Independent Committee for Clinical, not a pharmacological investigation in Genoa (Italy). All patients were evaluated and enrolled after signing informed consent.
4.2. Sample Collection
Two healthy and two AA subjects, respectively (20–60 years old), were enrolled under dermatological control at the Italian private dermatological clinic, Studio Rinaldi (Milan, Italy).
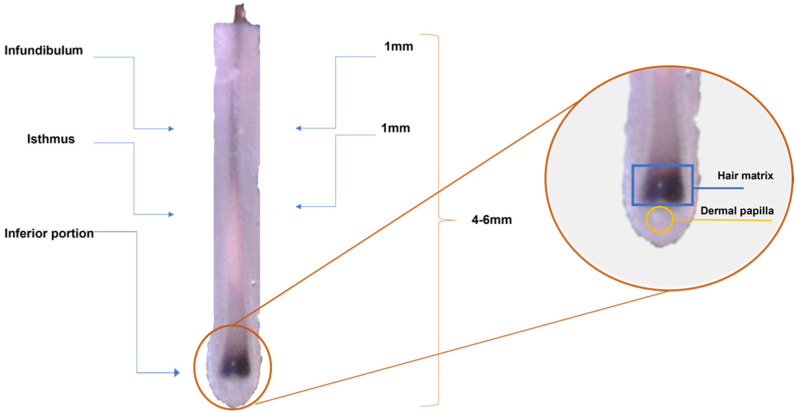
The subjects were sampled using a 4-mm biopsy punch to assess the microbial community in the subepidermal scalp compartments, as previously described [10] (Figure 7). For each subject, two biopsy samples were collected from the occipital area of the scalp and, for AA subjects, in correspondence with an affected area. After collection, samples were stored in a preservative medium (Allprotect® Tissue reagent, Qiagen, Milan-Italy) at +4 °C until further analysis. One biopsy sample was used for the metataxonomic profile. These samples were already analyzed in a previously published cohort [10], and sequences were deposited into the National Centre for Biotechnology Information (NCBI) BioProject database under the project number PRJNA510206. The other biopsy sample was divided into two halves, one half used for resident gram-positive and gram-negative bacteria by means of chemical staining (Gram’s method) and the other for electron microscopy.
Figure 7.
Stereomicroscopic view of the anatomy of an anagen hair follicle.
4.3. DNA Extraction and 16S Amplicon Generation, Sequencing, and Analysis-Illumina Sequencing
Bacterial DNA was extracted through the QIAamp Dneasy Tissue kit (Qiagen, Milan, Italy) according to manufacturer protocol, with minor modifications [46] followed by quantification with the QIAexpert system (Qiagen, Milan, Italy) before sequencing.
Following universal prokaryotic primers were used for the V3-V4 variable region: 341 F CTGNCAGCMGCCGCGGTAA [47,48] and 806bR GGACTACNVGGGTWTCTAAT [49,50,51] at Personal Genomics (Verona, Italy) following the method of Caporaso and colleague [52] and Kozich and colleagues [53], with minor modifications. The 300PE instrument (Illumina, San Diego, CA, USA) was used for library generation.
Following bacterial 16S rRNA gene sequences, FastQC v0.11.5. was used for quality control of fastq reads and the quality trimming made by Cutadapt, v. 1.14 [54] and Sickle v. 1.33 software toolkits. OTU tables were generated by QIIME v1.9. [55]. The Greengenes database v13_8 was used as a reference for the bacterial taxonomic assignment [56]. Alpha diversity was inspected using the Shannon index.
4.4. Transmission Electron Microscopy (TEM)
Small skin samples obtained from the enrolled subjects were fixed in 2.5% glutaraldehyde in 0.1 M sodium cacodylate buffer, pH 7.4 (Electron Microscopy Sciences, Hatfield, PA, USA) overnight at 4 °C. They were rinsed in buffer, postfixed in osmium tetroxide in cacodylate buffer for one four at room temperature, rinsed in buffer again, and dehydrated through a graded series of acetone to 100%. Then, they were infiltrated with Araldite-Epon resin (Electron Microscopy Sciences, Hatfield, PA, USA) in a 1:1 solution of Araldite-Epon: acetone for one hour at room temperature. After, they were placed in fresh Araldite-Epon resin for one hour at 37 °C and then embedded in Araldite-Epon overnight at 60 °C. Semithin sections (1 μm) were cut on a Reichert-Jung UltraCutE ultramicrotome, collected, and stained with methylene blue. After, thin sections (70–80 nm) were cut on the same ultramicrotome, collected onto formvar-coated grids, and stained with UranyLess and lead citrate (EMS, Hatfield, PA, USA). Finally, the thin sections were observed in a transmission electron microscope (Tecnai G2 Spirit; FEI Company, Eindhoven, the Netherlands) at 80 kV. Images were collected using a digital imaging system. Two blinded observers evaluated 10 images for each sample according to previous work for morphology and the presence of electron-dense particles ascribable to bacteria or lymphocytes [42,57].
4.5. Tissue Gram Staining
Cryosections (6 μm) were fixed in Acetone and stained for Gram stain (Sigma-Aldrich S.r.l., Milan, Italy) by procedures described by the manufacturer.
4.6. Statistical Analysis
Statistically significant differences in bacterial community determined by AA condition were determined by Student’s t with Welch’s correction. Analyses were performed with GraphPad Prism 7.0 (GraphPad Software, Inc., San Diego, CA, USA). p–values equal to or less than 0.05 were considered significant.
5. Conclusions
In conclusion, our data suggest that the alopecia areata is not a single clinic-pathological entity. However, it has to be managed in a holistic way considering its complexity and the alterations of microbiome homeostasis, so that it could be defined as a “hair disease associated with dysregulated microbiome-immunity axis of hair follicles”.
Acknowledgments
The authors acknowledge Maria De Angelis for her scientific support.
Author Contributions
Conceptualization, F.R., D.P. and R.R.; methodology, D.P., E.B. and S.C.; investigation, F.R., D.P., E.B. and S.C.; resources, F.R. and R.R.; data analysis and curation, F.R., D.P., E.B., S.C. and R.R.; writing—original draft preparation, F.R., D.P., E.B. and R.R.; writing—review and editing, F.R., A.A. and R.R.; supervision, F.R. and R.R.; project administration, F.R.; funding acquisition, F.R. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
The study was conducted in accordance with the Declaration of Helsinki and approved by the Ethical Independent Committee for Clinical, not a pharmacological, investigation in Genoa (Italy), protocol code n. 2018/3, date of approval 27 February 2018.
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study.
Data Availability Statement
The datasets used during and/or analyzed during the current study are available from the corresponding author upon reasonable request.
Conflicts of Interest
The authors have conflicts of interest to declare concerning this study. F.R. serves as a consultant for Giuliani S.P.A., and D.P. is employed by Giuliani S.P.A. All named authors meet the International Committee of Medical Journal Editors (ICMJE) criteria for authorship for this manuscript, take responsibility for the integrity of the work as a whole, and have given final approval for the version to be published.
Funding Statement
This research and the APC were funded by Giuliani SPA, Milan, Italy.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Villasante Fricke A.C., Miteva M. Epidemiology and burden of alopecia areata: A systematic review. Clin. Cosmet. Investig. Dermatol. 2015;8:397–403. doi: 10.2147/CCID.S53985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Harries M.J., Meyer K.C., Chaudhry I.H., Griffiths C.E., Paus R. Does collapse of immune privilege in the hair-follicle bulge play a role in the pathogenesis of primary cicatricial alopecia? Clin. Exp. Dermal. 2010;35:637–644. doi: 10.1111/j.1365-2230.2009.03692.x. [DOI] [PubMed] [Google Scholar]
- 3.Paus R., Bertolini M. The role of hair follicle immune privilege collapse in alopecia areata: Status and perspectives. J. Investig. Dermatol. Symp. Proc. 2013;16:S25–S27. doi: 10.1038/jidsymp.2013.7. [DOI] [PubMed] [Google Scholar]
- 4.Bertolini M., McElwee K., Gilhar A., Bulfone-Paus S., Paus R. Hair follicle immune privilege and its collapse in alopecia areata. Exp. Dermatol. 2020;29:703–725. doi: 10.1111/exd.14155. [DOI] [PubMed] [Google Scholar]
- 5.Paus R., Bulfone-Paus S., Bertolini M. Hair Follicle Immune Privilege Revisited: The Key to Alopecia Areata Management. J. Investig. Dermatol. Symp. Proc. 2018;19:S12–S17. doi: 10.1016/j.jisp.2017.10.014. [DOI] [PubMed] [Google Scholar]
- 6.Constantinou A., Kanti V., Polak-Witka K., Blume-Peytavi U., Spyrou G.M., Vogt A. The Potential Relevance of the Microbiome to Hair Physiology and Regeneration: The Emerging Role of Metagenomics. Biomedicines. 2021;9:236. doi: 10.3390/biomedicines9030236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rinaldi F., Pinto D., Marzani B., Rucco M., Giuliani G., Sorbellini E. Human microbiome: What’s new in scalp diseases. J. Transl. Sci. 2018;4:1–4. [Google Scholar]
- 8.Naik P.P., Farrukh S.N. Association between alopecia areata and thyroid dysfunction. Postgrad. Med. 2021;133:895–898. doi: 10.1080/00325481.2021.1974689. [DOI] [PubMed] [Google Scholar]
- 9.Puavilai S., Puavilai G., Charuwichitratana S., Sakuntabhai A., Sriprachya-Anunt S. Prevalence of thyroid diseases in patients with alopecia areata. Int. J. Dermatol. 1994;33:632–633. doi: 10.1111/j.1365-4362.1994.tb02921.x. [DOI] [PubMed] [Google Scholar]
- 10.Lewiński A., Broniarczyk-Dyła G., Sewerynek E., Zerek-Mełeń G., Szkudliński M. Abnormalities in structure and function of the thyroid gland in patients with alopecia areata. J. Am. Acad. Dermatol. 1990;23:768–769. doi: 10.1016/S0190-9622(08)81090-0. [DOI] [PubMed] [Google Scholar]
- 11.Thomas E.A., Kadyan R.S. Alopecia areata and autoimmunity: A clinical study. Indian J. Dermatol. 2008;53:70–74. doi: 10.4103/0019-5154.41650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cogen A.L., Nizet V., Gallo R.L. Skin microbiota: A source of disease or defence? Br. J. Dermatol. 2008;158:442–455. doi: 10.1111/j.1365-2133.2008.08437.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Byrd A., Belkaid Y., Segre J. The human skin microbiome. Nat. Rev. Microbiol. 2018;16:143–155. doi: 10.1038/nrmicro.2017.157. [DOI] [PubMed] [Google Scholar]
- 14.Lousada M.B., Lachnit T., Edelkamp J., Rouillé T., Ajdic D., Uchida Y., Di Nardo A., Bosch T.C.G., Paus R. Exploring the human hair follicle microbiome. Br. J. Dermatol. 2021;184:802–815. doi: 10.1111/bjd.19461. [DOI] [PubMed] [Google Scholar]
- 15.Constantinou A., Polak-Witka K., Tomazou M., Oulas A., Kanti V., Schwarzer R., Helmuth J., Edelmann A., Blume-Peytavi U., Spyrou G.M., et al. Dysbiosis and Enhanced Beta-Defensin Production in Hair Follicles of Patients with Lichen Planopilaris and Frontal Fibrosing Alopecia. Biomedicines. 2021;9:266. doi: 10.3390/biomedicines9030266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ho B.S., Ho E.X.P., Chu C.W., Ramasamy S., Bigliardi-Qi M., de Sessions P.F., Bigliardi P.L. Microbiome in the hair follicle of androgenetic alopecia patients. PLoS ONE. 2019;14:e0216330. doi: 10.1371/journal.pone.0216330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Carmona-Cruz S., Orozco-Covarrubias L., Sáez-de-Ocariz M. The Human Skin Microbiome in Selected Cutaneous Diseases. Front. Cell. Infect. Microbiol. 2022;12:834135. doi: 10.3389/fcimb.2022.834135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zheng D., Liwinski T., Elinav E. Interaction between microbiota and immunity in health and disease. Cell Res. 2020;30:492–506. doi: 10.1038/s41422-020-0332-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Racine P.J., Janvier X., Clabaut M., Catovic C., Souak D., Boukerb A.M., Groboillot A., Konto-Ghiorghi Y., Duclairoir-Poc C., Lesouhaitier O., et al. Dialog between skin and its microbiota: Emergence of “Cutaneous Bacterial Endocrinology”. Exp. Dermatol. 2020;29:790–800. doi: 10.1111/exd.14158. [DOI] [PubMed] [Google Scholar]
- 20.Kabashima K., Honda T., Ginhoux F., Egawa G. The immunological anatomy of the skin. Nat. Rev. Immunol. 2019;19:19–30. doi: 10.1038/s41577-018-0084-5. [DOI] [PubMed] [Google Scholar]
- 21.Belkaid Y., Segre J.A. Dialogue between skin microbiota and immunity. Science. 2014;346:954–959. doi: 10.1126/science.1260144. [DOI] [PubMed] [Google Scholar]
- 22.Sanford J.A., O’Neill A.M., Zouboulis C.C., Gallo R.L. Short-Chain Fatty Acids from Cutibacterium acnes Activate Both a Canonical and Epigenetic Inflammatory Response in Human Sebocytes. J. Immunol. 2019;202:1767–1776. doi: 10.4049/jimmunol.1800893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pinto D., Sorbellini E., Marzani B., Rucco M., Giuliani G., Rinaldi F. Scalp bacterial shift in Alopecia areata. PLoS ONE. 2019;14:e0215206. doi: 10.1371/journal.pone.0215206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pinto D., Calabrese F.M., De Angelis M., Celano G., Giuliani G., Gobbetti M., Rinaldi F. Predictive Metagenomic Profiling, Urine Metabolomics, and Human Marker Gene Expression as an Integrated Approach to Study Alopecia Areata. Front. Cell. Infect. Microbiol. 2020;10:146. doi: 10.3389/fcimb.2020.00146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Naik S., Bouladoux N., Wilhelm C., Molloy M.J., Salcedo R., Kastenmuller W., Deming C., Quinones M., Koo L., Conlan S., et al. Compartmentalized control of skin immunity by resident commensals. Science. 2012;337:1115–1119. doi: 10.1126/science.1225152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Campbell D.J., Koch M.A. Living in Peace: Host-Microbiota Mutualism in the Skin. Cell Host Microbe. 2017;4:419–420. doi: 10.1016/j.chom.2017.03.012. [DOI] [PubMed] [Google Scholar]
- 27.Scharschmidt T.C., Vasquez K.S., Pauli M.L., Leitner E.G., Chu K., Truong H.A., Lowe M.M., Sanchez Rodriguez R., Ali N., Laszik Z.G., et al. Commensal Microbes and Hair Follicle Morphogenesis Coordinately Drive Treg Migration into Neonatal Skin. Cell Host Microbe. 2017;21:467–477.e5. doi: 10.1016/j.chom.2017.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Polak-Witka K., Rudnicka L., Blume-Peytavi U., Vogt A. The role of the microbiome in scalp hair follicle biology and disease. Exp. Dermatol. 2020;29:286–294. doi: 10.1111/exd.13935. [DOI] [PubMed] [Google Scholar]
- 29.Rolig A.S., Carter J.E., Ottemann K.M. Bacterial chemotaxis modulates host cell apoptosis to establish a T-helper cell, type 17 (Th17)-dominant immune response in Helicobacter pylori infection. Proc. Natl. Acad. Sci. USA. 2011;108:19749–19754. doi: 10.1073/pnas.1104598108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Paus R., Slominski A., Czarnetzki B.M. Is alopecia areata an autoimmune-response against melanogenesis-related proteins, exposed by abnormal MHC class I expression in the anagen hair bulb? Yale J. Biol. Med. 1993;66:541–554. [PMC free article] [PubMed] [Google Scholar]
- 31.Crew P.E., McNamara L., Waldron P.E., McCulley L., Jones S.C., Bersoff-Matcha S.J. Unusual Neisseria species as a cause of infection in patients taking eculizumab. J. Infect. 2019;78:113–118. doi: 10.1016/j.jinf.2018.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Humbert M.V., Christodoulides M. Atypical, Yet Not Infrequent, Infections with Neisseria Species. Pathogens. 2019;9:10. doi: 10.3390/pathogens9010010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Boldock E., Surewaard B.G.J., Shamarina D., Na M., Fei Y., Ali A., Williams A., Pollitt E.J.G., Szkuta P., Morris P., et al. Human skin commensals augment Staphylococcus aureus pathogenesis. Nat. Microbiol. 2018;3:881–890. doi: 10.1038/s41564-018-0198-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jahns A.C., Lundskog B., Ganceviciene R., Palmer R.H., Golovleva I., Zouboulis C.C., McDowell A., Patrick S., Alexeyev O.A. An increased incidence of Propionibacterium acnes biofilms in acne vulgaris: A case-control study. Br. J. Dermatol. 2012;167:50–58. doi: 10.1111/j.1365-2133.2012.10897.x. [DOI] [PubMed] [Google Scholar]
- 35.Matard B., Meylheuc T., Briandet R., Casin I., Assouly P., Cavelier-balloy B., Reygagne P. First evidence of bacterial biofilms in the anaerobe part of scalp hair follicles: A pilot comparative study in folliculitis decalvans. J. Eur. Acad. Dermatol. Venereol. 2013;27:853–860. doi: 10.1111/j.1468-3083.2012.04591.x. [DOI] [PubMed] [Google Scholar]
- 36.Thaiss C.A., Zmora N., Levy M., Elinav E. The microbiome and innate immunity. Nature. 2016;535:65–74. doi: 10.1038/nature18847. [DOI] [PubMed] [Google Scholar]
- 37.Lavrinienko A., Tukalenko E., Mappes T. Skin and gut microbiomes of a wild mammal respond to different environmental cues. Microbiome. 2018;6:209. doi: 10.1186/s40168-018-0595-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Negroni A., Pierdomenico M., Cucchiara S., Stronati L. NOD2 and inflammation: Current insights. J. Inflamm. Res. 2018;11:49–60. doi: 10.2147/JIR.S137606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Garzorz N., Alsisi M., Todorova A., Atenhan A., Thomas J., Lauffer F., Ring J., Schmidt-Weber C., Biedermann T., Eyerich S., et al. Dissecting susceptibility from exogenous triggers: The model of alopecia areata and associated inflammatory skin diseases. J. Eur. Acad. Dermatol. Venereol. 2015;29:2429–2435. doi: 10.1111/jdv.13325. [DOI] [PubMed] [Google Scholar]
- 40.Tharumanathan S. Understanding the biological mechanism of alopecia areata. Am. J. Dermatol. Venereol. 2015;4:1–4. doi: 10.5923/j.ajdv.20150401.01. [DOI] [Google Scholar]
- 41.Hull C.M., Nickolay L.E., Estorninho M., Richardson M.W., Riley J.L., Peakman M., Maher J., Tree T.I. Generation of human islet-specific regulatory T cells by TCR gene transfer. J. Autoimmun. 2017;79:63–73. doi: 10.1016/j.jaut.2017.01.001. [DOI] [PubMed] [Google Scholar]
- 42.Messenger A.G., Bleehen S.S. Alopecia areata: Light and electron microscopic pathology of the regrowing white hair. Br. J. Dermatol. 1984;110:155–162. doi: 10.1111/j.1365-2133.1984.tb07461.x. [DOI] [PubMed] [Google Scholar]
- 43.Pratt C.H., King L.E., Jr., Messenger A.G., Christiano A.M., Sundberg J.P. Alopecia areata. Nat. Rev. Dis. Prim. 2017;3:17011. doi: 10.1038/nrdp.2017.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Coroaba A., Chiriac A.E., Sacarescu L., Pinteala T., Minea B., Ibanescu S.A., Pertea M., Moraru A., Esanu I., Maier S.S., et al. New insights into human hair: SAXS, SEM, TEM and EDX for Alopecia Areata investigations. PeerJ. 2020;8:e8376. doi: 10.7717/peerj.8376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Olsen E.A., Hordinsky M.K., Price V.H., Roberts J.L., Shapiro J., Canfield D., Duvic M., King L.E., Jr., McMichael A.J., Randall V.A., et al. National Alopecia Areata Foundation. Alopecia areata investigational assessment guidelines–Part II. national alopecia areata foundation. J. Am. Acad. Dermatol. 2004;51:440–447. doi: 10.1016/j.jaad.2003.09.032. [DOI] [PubMed] [Google Scholar]
- 46.Gao Z., Perez-Perez G.I., Chen Y., Blaser M.J. Quantitation of major human cutaneous bacterial and fungal populations. J. Clin. Microbiol. 2010;48:3575–3581. doi: 10.1128/JCM.00597-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Klindworth A., Pruesse E., Schweer T., Peplies J., Quast C., Horn M., Glöckner F.O. Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res. 2013;41:e1. doi: 10.1093/nar/gks808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Takahashi S., Tomita J., Nishioka K., Hisada T., Nishijima M. Development of a prokaryotic universal primer for simultaneous analysis of Bacteria and Archaea using next-generation sequencing. PLoS ONE. 2014;9:e105592. doi: 10.1371/journal.pone.0105592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Apprill A., McNally S., Parsons R., Weber L. Minor revision to V4 region SSU rRNA 806R gene primer greatly increases detection of SAR11 bacterioplankton. Aquat. Microb. Ecol. 2015;75:129–137. doi: 10.3354/ame01753. [DOI] [Google Scholar]
- 50.Parada A.E., Needham D.M., Fuhrman J.A. Every base matters: Assessing small subunit rRNA primers for marine microbiomes with mock communities, time series and global field samples. Environ. Microbiol. 2016;18:1403–1414. doi: 10.1111/1462-2920.13023. [DOI] [PubMed] [Google Scholar]
- 51.Walters W., Hyde E.R., Berg-Lyons D., Ackermann G., Humphrey G., Parada A. Improved Bacterial 16S rRNA Gene (V4 and V4-5) and Fungal Internal Transcribed Spacer Marker Gene Primers for Microbial Community Surveys. mSystems. 2016;1:e00009-15. doi: 10.1128/mSystems.00009-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Caporaso J.G., Lauber C.L., Walters W.A., Berg-Lyons D., Lozupone C.A., Turnbaugh P.J., Fierer N., Knight R. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc. Natl. Acad. Sci. USA. 2011;108:4516–4522. doi: 10.1073/pnas.1000080107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kozich J.J., Westcott S.L., Baxter N.T., Highlander S.K., Schloss P.D. Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq Illumina sequencing platform. Appl. Environ. Microbiol. 2013;79:5112–5120. doi: 10.1128/AEM.01043-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. J. 2011;17:10–12. doi: 10.14806/ej.17.1.200. [DOI] [Google Scholar]
- 55.Bolyen E., Rideout J.R., Dillon M.R., Bokulich N.A., Abnet C.C., Al-Ghalith G.A., Alexander H., Alm E.J., Arumugam M., Asnicar F., et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 2019;37:852–857. doi: 10.1038/s41587-019-0209-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Desantis T.Z., Hugenholtz P., Larsen N., Rojas M., Brodie E.L., Keller K., Huber T., Dalevi D., Hu P., Andersen G.L. Greengenes, a chimerachecked 16S rRNA gene database and workbench compatible with ARB. Appl. Environ. Microbiol. 2006;72:5069–5072. doi: 10.1128/AEM.03006-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tobin D.J. Morphological analysis of hair follicles in alopecia areata. Microsc. Res. Tech. 1997;38:443–451. doi: 10.1002/(SICI)1097-0029(19970815)38:4<443::AID-JEMT12>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used during and/or analyzed during the current study are available from the corresponding author upon reasonable request.