Abstract
The understanding of the interaction between non-steroidal anti-inflammatory drugs and human serum albumin plays a fundamental role in the development of new drugs and new therapeutic strategies. Several studies have been performed, nevertheless, the interaction phenomena are still not fully understood. In this work, high-field solution Nuclear Magnetic Resonance (NMR) spectroscopy was applied to compare the strength of the interaction of diclofenac sodium salt, ketorolac tris salt and flurbiprofen sodium salt toward albumin. To this aim, mono- and bi-selective relaxation rate measurements were performed by applying selective π-pulses at the selected frequencies and by following magnetization recovery. On the basis of the dependence of relaxation parameters on albumin concentration, normalized affinity indexes were calculated for several protons of the drugs. Affinity indexes for diclofenac were about five-fold higher in comparison with ketorolac and flurbiprofen. Aromatic moieties of the three drugs and methine protons at the chiral centers of ketorolac and flurbiprofen were more involved in the interaction with albumin. In conclusion, NMR spectroscopy allows not only for the comparison of drug-to-protein affinities but also points out the nature of the drug sites that are more extensively involved in the interaction.
Keywords: protein/molecule interaction, NSAIDs, selective relaxation rates, affinity index, binding, NMR
1. Introduction
Nowadays non-steroidal anti-inflammatory drugs (NSAIDs) are among the most commonly used pharmaceuticals. These compounds are used for the treatment of several conditions in virtue of their anti-inflammatory, analgesic, antipyretic, and anti-aggregant properties [1]. Depending on the nature of the drug, oral, intraocular, intravenous, intramuscular, topical and rectal administration can be selected. In all cases, the drug reaches the site of action through the bloodstream, interacting with the proteins present in the blood plasma, especially human serum albumin (HSA), a globular protein that accounts for 60% of the total protein content. Efficient drug delivery systems, such as albumin-based nanoparticles [2], were proposed as well.
The remarkable flexibility together with the high concentration in the blood and the presence of multiple binding sites make HSA particularly inclined to interact with small molecules such as anticoagulants, anesthetics, steroids, amino acids and, indeed, NSAIDs [3]. The strength of this interaction has a direct effect on the bioavailability and on the metabolism: if the interaction with HSA is too strong, the drug cannot be released in the body, or a higher concentration is necessary to guarantee therapeutic efficacy. On the other hand, if the interaction is too weak the drug could be metabolized before reaching the site of action. In spite of the fact that the knowledge of the drug-to-HSA interaction mechanisms is fundamental for optimizing pharmacokinetic, pharmacodynamic, and toxicological profiles, a profound and complete comprehension of binding phenomena is still far from being achieved.
Several studies were performed, starting in the 1980s [4,5,6], to understand and rationalize the nature of the interactions between NSAIDs and HSA. Different analytical techniques were exploited to reach this goal: for example, isothermal titration calorimetry was combined with frontal analysis/capillary electrophoresis to shed light on the thermodynamic parameters of the interaction [7]. Fluorescence spectroscopy was employed for detecting the formation of the drug/albumin complex and obtaining the binding parameters [8]. Molecular modeling and spectroscopic techniques (fluorescence and UV) contributed to improving the comprehension of the interaction of HSA with diclofenac sodium salt, highlighting the pivotal role played by the hydrophobic interactions in the binding, with hydrogen bonds acting as support [9]. Such interactions were confirmed also via X-ray crystallography, which pointed out the protein residues involved in the interaction with the drug [10]. Competitive binding processes were investigated through circular dichroism and fluorescence spectroscopy [11] and better mastery of more complex binding processes, e.g., photo-induced ones, was obtained by UV spectroscopy [12].
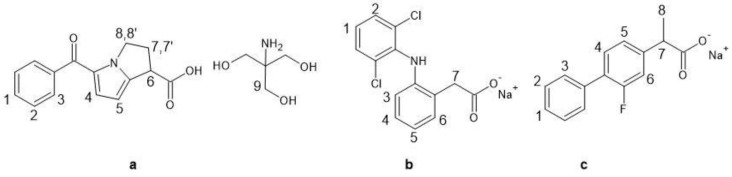
NMR constitutes an analytical technique largely employed in the investigation of drug/biomacromolecule interactions [13,14]. As a matter of fact, NMR spectroscopy allows determining not only the complexation stoichiometry and the association constants or, more generally, the affinity of the drug for the protein, but also the stereochemical parameters, thus providing information about the nature of the sites more extensively involved in the interaction. In this work, an NMR investigation was performed on the interaction between HSA and three drugs belonging to the group of arylalcanoic acid derivatives that are among the most used NSAIDs, known for their antipyretic and anti-inflammatory properties: ketorolac tris salt (KTR), diclofenac sodium salt (DCF) and flurbiprofen sodium salt (FBP), reported in Scheme 1. The interaction was investigated by measuring the mono-selective spin-lattice relaxation rates (R1ms) of KTR, DCF and FBP nuclei in the presence of HSA since this parameter is much more sensitive to the occurrence of interactions compared to the non-selective relaxation rate (R1ns) [15].
Scheme 1.
Chemical structures and numbering schemes for NMR analysis of (a) KTR, (b) DCF and (c) FBP.
The interaction between KTR and HSA has already been studied mainly via chromatographic methods [16] and, to the best of our knowledge, has not been investigated in detail via NMR, whereas there is only one NMR study focused on the low-affinity interaction between HSA and DCF, based on the analysis of chemical shift and linewidth of the 13C NMR signals of the drug in the presence of the protein [17]. Regarding FBP, its interaction with HSA was followed via NMR diffusion measurements, which were used for the calculation of the ligand–protein dissociation constant and the stoichiometry of binding [18]. The presence of a fluorine nucleus on the skeleton, moreover, made possible the use of 19F NMR spectroscopy as an investigation tool [19].
2. Results and Discussion
Proton mono-selective relaxation rates (R1ms) of KTR, DCF and FBP in their free state and in a mixture with HSA were calculated from the corresponding mono-selective relaxation times (T1ms) measured for each drug in D2O (pH 7.4, phosphate buffer, 0.1 M), according to Equation (1).
| R1ms = 1/T1ms | (1) |
This parameter was chosen as an interaction probe taking into consideration the experimental conditions in which the studies were carried out. The notable difference in the molecular weight of the species involved, in fact, does not allow working with solutions at the equimolar ratio, since the NMR signals of the small molecule (the drug) would be hardly superimposed on those belonging to the macromolecule. Therefore, it is necessary to exploit a parameter highly sensitive to the interaction even when the macromolecule is in strong defect with respect to the drug, such as the mono-selective relaxation rates.
It is important to underline that, rather than R1ms, the corresponding normalized relaxation rate (ΔR/Rf, where ΔR = Robs − Rf) constitutes a very efficient tool for comparing the entity of the interaction of different protons of the drug. The determination of R1ms of the drugs’ nuclei in mixtures containing different amounts of HSA allows extracting their normalized affinity indexes ([AN]), a measure of the drug–HSA global affinity and a useful parameter to compare the ability of different ligands to interact with the same macromolecule.
For protons at a fixed and known distance, the cross-relaxation terms (σ) can be obtained as the difference between bi- and mono-selective relaxation rates, allowing the bound molar fractions (xb) to be calculated, as discussed in Appendix A [15,20,21].
2.1. KTR/HSA Mixtures
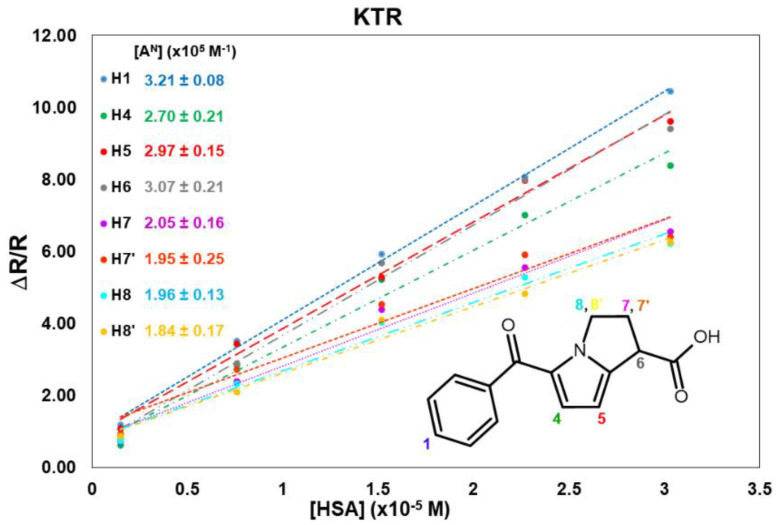
No superimposition was observed for KTR nuclei in the proton spectrum (Figure S1), hence it was possible to measure the relaxation parameters for almost all nuclei. All KTR protons increased their relaxation rate with an increasing HSA concentration, as reported in Table S1. The normalized values, reported in Table 1, indicate that the protons belonging to the two aromatic moieties (H1 and H4/H5) and the proton in α-position with respect to the carboxylate group (H6) are highly involved in the interaction. As an example, proton H1 undergoes more than a ten-fold increase in its mono-selective relaxation rate, which goes from 0.26 s−1 in the free state to 3.03 s−1 (Table S1) in the presence of the highest amount of protein (2 mg/mL).
Table 1.
1H mono-selective relaxation rates R1ms (s−1) (600 MHz, 25 °C, D2O, pH = 7.4) of some protons of KTR (2 mM) in its free state; normalized mono-selective relaxation rates (ΔR1ms/Rf, ΔR1ms = Robs − Rf) of KTR protons in mixtures with HSA at different molar ratios; cross-relaxation rate (σij, s−1) calculated for proton pair H4-H5 and corresponding bound molar fraction (xb).
| [HSA] mg/mL | KTR:HSA | H1 | H4 | H5 | H6 | H7 | H7′ | H8 | H8′ | σ4-5 | xb |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | / | 0.26 | 0.31 | 0.21 | 0.28 | 0.81 | 0.74 | 0.79 | 0.92 | 0.05 | |
| 0.1 | 1315:1 | 1.18 | 0.62 | 1.08 | 0.79 | 0.73 | 0.93 | 0.75 | 0.88 | 0.00 | 0.01 |
| 0.5 | 264:1 | 3.49 | 2.85 | 3.44 | 2.89 | 2.42 | 2.75 | 2.32 | 2.11 | −0.11 | 0.04 |
| 1.0 | 132:1 | 5.95 | 5.17 | 5.28 | 5.68 | 4.35 | 4.63 | 4.04 | 4.19 | −0.19 | 0.07 |
| 1.5 | 88:1 | 8.10 | 7.00 | 7.89 | 8.08 | 5.47 | 5.90 | 5.30 | 4.74 | −0.23 | 0.08 |
| 2.0 | 66:1 | 10.58 | 8.34 | 9.70 | 9.41 | 6.69 | 6.50 | 6.41 | 6.27 | −0.29 | 0.09 |
The linear fitting of normalized mono-selective relaxation rates vs. albumin concentration allowed calculating [AN] (see Appendix A) for KTR nuclei (Figure 1), and values of normalized affinity indexes ranging from 1.8 × 105 M−1 to 3.2 × 105 M−1 were obtained (see also Table S2 in Supplementary Material). Data confirm that the aromatic protons H1 and H4/H5 and proton H6, directly linked to the carboxyl group, are the nuclei more influenced by the protein.
Figure 1.
Normalized mono-selective relaxation rates plotted vs. HSA concentration and calculated affinity indexes for KTR protons.
By simultaneous inversion of aromatic protons H4 and H5, at a known and fixed distance, the bi-selective relaxation rate Rbs4-5 was measured to obtain the observed cross-relaxation rate σ4-5 (σ4-5 = R4-5bs − R4ms), listed in Table 1. The parameter assumes a positive value (0.05 s−1) for the free drug, as expected on the basis of its characteristics of motion, whereas the interaction with the protein determines a shift to negative values, typical of the slow motion region, where macromolecules and their complexes lie. The cross-relaxation term is highly sensitive to the variation of drug/protein molar ratio; in fact, it changes its values from 0 s−1 (KTR/HSA 1315:1) to −0.29 s−1(KTR/HSA 66:1).
On assuming that the reorientation time (τc) of the complex is controlled by albumin (τcHSA = 20 ns) [22], and by keeping in mind the distance r4-5 (2.61 Å) [23], a value of −3.69 s−1 is calculated for σb on the basis of Equation (A5) (Appendix A). Once σobs, σf and σb were obtained, the molar fraction in the bound state for each mixture KTR/HSA can be calculated from Equation (A6) (Appendix A). In the mixture with the higher KTR/HSA ratio (1315:1), about 1% of the drug is bound to the albumin and this amount slightly increases with decreasing of the molar ratio (9% for the mixture 66:1).
The plot of the normalized cross-relaxation terms in the function of the protein concentration gave a global normalized affinity index of 1.8 × 105 M−1 for KTR (the slope of the line in Figure S4 in Supplementary Material).
2.2. DCF/HSA Mixtures
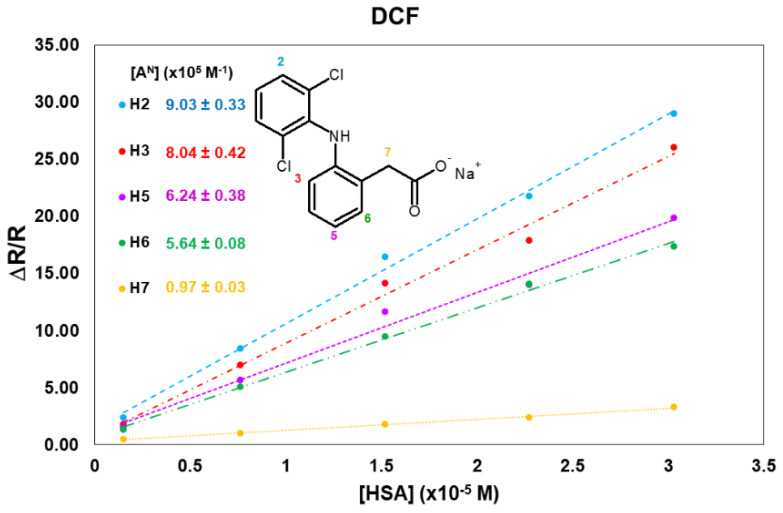
Greater variations in comparison to KTR were obtained for the aromatic nuclei of DCF in the presence of HSA (from a 17- to a 28-fold increase). The greatest variation was observed for H2, belonging to the chlorine-substituted aromatic ring: its mono-selective relaxation rate, equal to 0.17 s−1 in the free state, already increases up to 0.57 s−1 with a small amount of protein (0.1 mg/mL), and it remarkably undergoes almost a 29-fold increase in the mixture at the lowest molar ratio (66:1, Table 2 and Table S3 in Supplementary Material). A similar increment in the relaxation parameter is observed for the other aromatic proton H3.
Table 2.
1H mono-selective relaxation rates R1ms (s−1) (600 MHz, 25 °C, D2O, pH = 7.4) of some protons of DCF (2 mM) in its free state; normalized mono-selective relaxation rates (ΔR1ms/Rf, ΔR1ms = Robs − Rf) of DCF protons in mixtures with HSA at different molar ratios; cross-relaxation rate (σij, s−1) calculated for proton pair H6-H5 and corresponding bound molar fraction (xb).
| [HSA] mg/mL | DCF:HSA | H2 | H3 | H5 | H6 | H7 | σ6-5 | xb |
|---|---|---|---|---|---|---|---|---|
| 0 | / | 0.17 | 0.24 | 0.34 | 0.38 | 1.22 | 0.04 | |
| 0.1 | 1315:1 | 2.36 | 1.79 | 1.50 | 1.35 | 0.49 | −0.05 | 0.02 |
| 0.5 | 264:1 | 8.38 | 6.85 | 5.64 | 5.07 | 1.05 | −0.36 | 0.07 |
| 1.0 | 132:1 | 16.38 | 14.14 | 11.70 | 9.44 | 1.83 | −0.77 | 0.15 |
| 1.5 | 88:1 | 21.73 | 17.90 | 14.37 | 13.50 | 2.42 | −1.01 | 0.19 |
| 2.0 | 66:1 | 28.55 | 25.50 | 19.86 | 17.64 | 3.32 | −1.59 | 0.29 |
H5 and H6, belonging to the same aromatic ring of H3, are strongly involved in the interaction as well. Contrary to what was observed for KTR, methylene protons (H7) in the α-position to the carboxyl group showed the lowest effect (Table 2).
The normalized affinity indexes, obtained as the slope of the linear fittings shown in Figure 2, resulted very high and remarkably differentiated for DCF protons (Table S4), highlighting the differences in the binding of different groups of DCF with the HSA sites. This result is in agreement with the data obtained via fluorescence and crystallography measurements [9,10], where the important role of the hydrophobic interactions on the binding to the protein was pointed out.
Figure 2.
Normalized mono-selective relaxation rates plotted vs. HSA concentration, and calculated affinity indexes for DCF protons.
Similarly to KTR, aromatic protons H5 and H6, at a known and fixed distance (2.49 Å) [24], were simultaneously inverted to obtain R5-6bs and the corresponding cross-relaxation term (σ5-6). On the basis of Equation (A5) (Appendix A), a bound cross-relaxation term σb equal to −5.53 s−1 was obtained by assuming τc of the complex equal to τcHSA = 20 ns. As already discussed for KTR, the bound molar fractions at different molar ratios can be calculated from Equation (A6) (Appendix A) once σobs, σf and σb are given. The comparison of the calculated xb for DCF and KTR protons (Table 1 and Table 2, respectively) pointed out that the molar fraction of the two drugs is small and comparable (1% for KTR and 2% for DCF) in the solution with the lowest amount of protein (0.1 mg/mL), probably because the concentration of HSA is too low to observe significant differences in their activity. However, with the increase in the concentration of the protein, KTR and DCF show a very different affinity for the albumin, and this difference increases when increasing the protein concentration. At the lowest molar fraction, when the HSA concentration reaches 2 mg/mL, 9% of KTR is bound, in contrast to 29% of DCF.
The affinity index, calculated on the basis of the cross-relaxation term (Figure S5 in Supplementary Materials), is equal to 9.2 × 105 M−1, five times higher than that measured for KTR.
2.3. FBP/HSA Mixtures
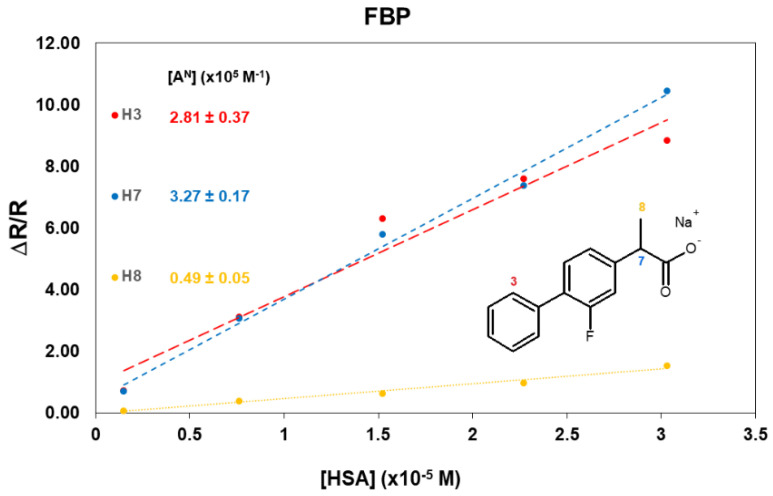
Unfortunately, the extensive superimposition between the aromatic resonances (Figure S3) did not allow for the implementation of cross-relaxation measurements for any proton pair of the drug. The mono-selective relaxation measurements performed pointed out remarkable differences in the involvement of different FBP nuclei in their interaction with HSA (Table 3 and Table S5), highlighting once again the primary role of the aromatic moiety of the drugs on the binding with HSA. While very small variations are observed for the methyl group of FBP, the relaxation rates of the aromatic proton H3 and methine proton (H7) in the α-position with respect to the carboxylate increase to a similar extent to what was observed for KTR.
Table 3.
1H mono-selective relaxation rates R1ms (s−1) (600 MHz, 25 °C, D2O, pH = 7.4) of some protons of FBP (2 mM) in its free state and normalized mono-selective relaxation rates (ΔR1ms/Rf, ΔR1ms = Robs − Rf) of FBP protons in mixtures with HSA at different molar ratios.
| [HSA] mg/mL | FBP:HSA | H3 | H7 | H8 |
|---|---|---|---|---|
| 0 | / | 0.34 | 0.34 | 1.33 |
| 0.1 | 1315:1 | 0.71 | 0.70 | 0.09 |
| 0.5 | 264:1 | 3.06 | 3.08 | 0.42 |
| 1.0 | 132:1 | 6.30 | 5.77 | 0.67 |
| 1.5 | 88:1 | 7.59 | 7.38 | 0.97 |
| 2.0 | 66:1 | 8.73 | 10.46 | 1.59 |
The different behavior of the methyl group is more evident in the mixture at the lowest molar ratio (66:1), where a value of 3.45 s−1 was measured for H8 (Table S3), corresponding to a moderate increase (normalized relaxation rate of 1.59 in Table 3), in contrast to an average ten-fold increase observed for protons H3 and H7 (Table 3). The affinity indexes calculated (Figure 3) confirm the involvement of the aromatic protons and the carboxyl group of FBP in the interaction with HSA.
Figure 3.
Normalized mono-selective relaxation rates plotted vs. HSA concentration, and calculated affinity indexes for FBP protons.
3. Materials and Methods
3.1. Materials
Ketorolac tris salt (≥99%), diclofenac sodium salt (≥98%), flurbiprofen sodium salt (pharmaceutical secondary standard), human serum albumin (66 kDa) and phosphate buffer powder, 0.1 M were purchased from Sigma Aldrich (St. Louis, MI, USA). Deuterated water (D2O, 99.9%) was purchased from Deutero GmbH (Kastellaun, Germany). All chemical compounds were used without any further purification.
3.2. NMR Measurements
The NMR experiments were carried out on a Varian INOVA600 spectrometer equipped with a 5 mm probe operating at 600 MHz for 1H nuclei; the temperature was controlled to ±0.1 °C.
Samples were degassed to remove paramagnetic dissolved oxygen. Degassing was carried out using five freeze–pump–thaw cycles.
The spin-lattice mono-selective relaxation times (T1ms) were measured by using the inversion recovery pulse sequence (180°-τ-90°-t)n and by applying a selective π-pulse at the selected frequency. Measurements were carried out for selected protons belonging to KTR, DCF and FBP, in the free state and in the presence of HSA; the drug concentration was kept equal to 2 mM for all three drugs, whereas the HSA concentration was increased from 0.1 mg/mL to 2 mg/mL.
The experimental error in the relaxation rate measurements was 5%. The errors relative to affinity indexes are reported in the Supplementary Material (Tables S2, S4 and S6).
In Appendix A, the theoretical discussion of the NMR approach with respect to mono-selective relaxation rates, cross-relaxation terms, and affinity indexes is reported.
3.3. NMR Characterization Data (Numbering Scheme Referred to Scheme 1)
KTR- 1H NMR (600 MHz, 2 mM, D2O pH 7.4 (0.1 mM), 298 K), δ (ppm) referred to the residual solvent (4.64 ppm): 7.64 (d, 3J3-2 = 7.6 Hz, 2H, H3), 7.52 (t, 3J1-2 = 7.6 Hz, 1H, H1), 7.42 (t, 3J2-1 = 3J2-3 = 7.6 Hz, 2H, H2), 6.85 (d, 3J4-5 = 4.2 Hz, 1H, H4), 6.03 (d, 3J5-4 = 4.2 Hz, 1H, H5), 4.32 (m, 1H, H8), 4.18 (m, 1H, H8′), 3.83 (dd, 3J6-7 = 8.9 Hz, 3J6-7′ = 6.0 Hz, 1H, H6), 3.57 (s, 6H, H9), 2.71 (m, 1H, H7), 2.57 (m, 1H, H7′).
DCF- 1H NMR (600 MHz, 2 mM, D2O pH 7.4 (0.1 mM), 298 K), δ (ppm) referred to the residual solvent (4.64 ppm): 7.35 (d, 3J2-1 = 8.2 Hz, 2H, H2), 7.14 (dd, 3J6-5 = 7.4 Hz, 4J6-4 = 1.3 Hz, 1H, H6), 7.03 (t, 3J1-2 = 8.2 Hz, 1H, H1), 6.99 (dt, 3J4-5 = 3J4-3 = 7.4 Hz, J44-6 = 1.4 Hz, 1H, H4), 6.84 (dt, 3J5-6 = 3J5-4 = 7.4 Hz, 4J5-3 = 1.3 Hz, 1H, H5), 6.35 (dd, 3J3-4 = 7.4 Hz, 4J3-5 = 1.3 Hz, 1H, H3), 3.53 (s, 2H, H7).
FBP- 1H NMR (600 MHz, 2 mM, D2O pH 7.4 (0.1 mM), 298 K), δ (ppm) referred to the residual solvent (4.64 ppm): 7.49 (d, 3J3-2 = 7.9 Hz, 2H, H3), 7.39 (t, 3J2-1 = 3J2-3 = 7.9 Hz, 2H, H2), 7.34 (t, 3J4-5 = 4J4-F = 8.0 Hz, 1H, H4), 7.32 (t, 3J1-2 = 7.9 Hz, 1H, H1), 7.09 (dd, 3J5-4 = 8.0 Hz, 4J5-6 = 1.4 Hz, 1H, H5), 7.05 (dd, 3J6-F = 12.2 Hz, 4J6-5 = 1.4 Hz, 1H, H6), 3.55 (q, 3J7-8 = 7.2 Hz, 1H, H7), 1.29 (d, 3J8-7 = 7.2 Hz, 3H, H8).
4. Conclusions
The affinity of ketorolac tris salt, diclofenac sodium salt and flurbiprofen sodium salt towards human serum albumin was evaluated via mono-selective relaxation rate measurements, thus upholding their reliability for a detailed investigation of drug/protein interactions. The exploitation of this NMR parameter, in fact, allows for identifying the moieties of the small molecule more extensively affected by the presence of the biomacromolecule, thus leading to reliable hypotheses about the interaction mechanisms that take place in solution and offering a tool for the design of tailored drugs.
A stronger affinity of DCF for HSA with respect to KTR and FBP, which behave similarly, was highlighted. The leading involvement of the aromatic moieties was clearly pointed out for all the analyzed compounds; in particular, in the case of DCF, the aromatic ring with the chlorine atoms is more affected by the interaction compared to the other one. No significant differences between the two aromatic groups of KTR were observed, whereas the strong superimposition observed for FBP resonances did not allow the analysis of the F-bearing ring, thus preventing the comparison between the aromatic moieties.
Bi-selective relaxation measurements, associated with the mono-selective ones, allowed for the determination of the cross-relaxation rates and of the bound molar fractions for KTR and DCF; the global affinity of the two drugs for HSA, calculated from the cross-relaxation terms in the bound state, confirmed that DCF interacts more strongly with the protein, with its affinity index being five times higher with respect to KTR.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/molecules27196647/s1, Figure S1: 1H NMR (600 MHz, D2O, pH 7.4, 298 K) spectrum of KTR (2 mM); Figure S2: 1H NMR (600 MHz, D2O, pH 7.4, 298 K) spectrum of DCF (2 mM); Figure S3: 1H NMR (600 MHz, D2O, pH 7.4, 298 K) spectrum of FBP (2 mM); Table S1: 1H mono-selective relaxation rates (ΔR1ms/Rf, ΔR1ms = Robs − Rf) (600 MHz, 25 °C, D2O, pH = 7.4) of some protons of KTR (2 mM) alone and in mixtures with HSA at different molar ratios; Table S2- Affinity indexes ([AN], M−1) and corresponding standard errors (ε, M-1) calculated for KTR protons from NMR data; Figure S4: Δσ/σf of KTR (2 mM) plotted in function of HSA concentration; Table S3: 1H mono-selective relaxation rates (ΔR1ms/Rf, ΔR1ms = Robs − Rf) (600 MHz, 25 °C, D2O, pH = 7.4) of some protons of DCF (2 mM) alone and in mixtures with HSA at different molar ratios; Table S4- Affinity indexes ([AN], M−1) and corresponding standard errors (ε, M-1) calculated for DCF protons from NMR data; Figure S5: Δσ/σf of DCF (2 mM) plotted in function of HSA concentration; Table S5: 1H mono-selective relaxation rates (ΔR1ms/Rf, ΔR1ms = Robs − Rf) (600 MHz, 25 °C, D2O, pH = 7.4) of some protons of FBP (2 mM) alone and in mixtures with HSA at different molar ratios; Table S6- Affinity indexes ([AN], M−1) and corresponding standard errors (ε, M−1) calculated for FBP protons from NMR data.
Appendix A. 1H Mono-Selective Relaxation Rates, Cross-Relaxation Terms, and Affinity Indexes
In the fast exchange regime, any NMR parameter (Pobs) is the weighted average of its value in the free (Pf) and bound state (Pb), according to Equation (A1):
| Pobs = Pfxf + Pbxb | (A1) |
where xf and xb are the molar fractions in the free and bound state, respectively.
When a drug interacts with a protein, the dynamics of the complex are driven by the latter, and it is possible to assume that the reorientation time of the complex (τc) corresponds to that of the macromolecule. As a consequence of the interaction, the drug goes from the fast motion region (ω2τc2 « 0.6; ω = Larmor frequency), characteristics of small molecules in their free state, to the slow-motion region (ω2τc2 » 0.6) where macromolecules and their complexes are found. The non-selective relaxation parameter R1ns increases with the increase in ω2τc2 up to 0.6, and then it reaches a maximum and starts decreasing (Equation (A2)); on the contrary, the function R1ms has a point of flex in correspondence of 0.6, and then keeps increasing with increasing of ω2τc2 (Equation (A3)) [25,26,27]. This high sensitivity to the change of motion regime explains why R1ms is usually preferred for investigations requiring a strong excess of drug with respect to the macromolecule [28].
| R1ns = 0.1 (γ4ħ2rij−6) [3τc/(1 + ω2τc2) + 12τc/(1 + 4 ω2τc2)] | (A2) |
| R1ms = 0.1 (γ4ħ2rij−6) [3τc/(1 + ω2τc2) + 6τc/(1 + 4 ω2τc2) + τc] | (A3) |
where γ is the gyromagnetic ratio and ħ is the reduced Plank’s constant. If two nuclei ij are simultaneously inverted, it is possible to measure the bi-selective relaxation rate R1bs; the difference between R1bs and R1ms gives the cross-relaxation term σij, a parameter that is dependent on the τc of the drug and from the proton distance rij. For small molecules in the fast-motion region, σij can be calculated according to Equation (A4), whereas for macromolecules and their complexes one must refer to Equation (A5).
| σij = 0.5γ4ħ2rij−6τc | (A4) |
| σij = −0.1γ4ħ2rij−6τc | (A5) |
In the case of a drug/protein complex, Equation (A5) can be used for calculating the bound cross-relaxation term (σb) for a proton pair at a known distance, when τc is known for the protein. Considering that a large excess of the ligand is used with respect to the macromolecule, the free molar fraction of the drug can be approximated to 1, and, from Equation (A1), the bound molar fraction of the drug can be obtained (Equation (A6)).
| xb = (σobs − σf)/σb | (A6) |
The extent to which different drug nuclei are involved in the interaction with the protein can be better evaluated by calculating the normalized mono-selective relaxation rate (Equation (A7)).
| ∆R/Rf = (Robs − Rf)/Rf | (A7) |
It is then possible to rewrite Equation (A1) in the form of Equation (A8), approximating the molar fraction of the free ligand to 1.
| Robs = Rf + Rbxb | (A8) |
In the occurrence of a 1:1 complexation equilibrium between the drug (L) and the macromolecule (M), the hetero association constant K can be expressed according to Equation (A9):
| K = [ML]/(([M0] − [ML])[L]) | (A9) |
where [ML] is the concentration of the drug/macromolecule complex, [M0] is the initial concentration of the macromolecule and [L] is the drug concentration. Therefore, xb (equal to [ML]/[L]) can be obtained from Equation (A9) and Robs can be expressed as follows (Equation (A10)):
| Robs = Rf + RbK[M0]/(1 + K[L]) | (A10) |
By plotting Robs in the function of [M0], a straight line should be obtained; the slope of the line is the affinity index [A] (Equation (A11)):
| [A] = KRb/(1 + K[L]) | (A11) |
which can be normalized to obtain [AN] (Equation (A12)).
| [AN] = [A]/Rf = KRb/(Rf(1 + K[L])) | (A12) |
For n binding sites of equal strength, the affinity index is (Equation (A13)):
| [A] = KRb[L]n−1/(1 + K[L]n) | (A13) |
For n binding sites, each with a different thermodynamic equilibrium constant, the affinity index takes the form of Equation (A14):
| (A14) |
Independently from the complexation stoichiometry, a linear relationship always correlates the relaxation rate and the macromolecule concentration (Equations (A15) and (A16)):
| Robs = Rf + [A][M0] | (A15) |
and
| ΔR/Rf = (Robs − Rf)/Rf = [AN][M0] | (A16) |
The linear fitting of the relaxation rates as a function of the macromolecule concentration [AN] can be obtained, providing a measure of ligand–macromolecule global affinity. The determination of this parameter represents a useful approach to compare the ability of different ligands to interact with the same macromolecule or to compare the ability of different macromolecules to bind the same drug.
Author Contributions
Conceptualization, F.A., G.U.-B. and F.B.; methodology, S.N.; software, C.P. and A.R.; validation, C.P., S.N. and A.R.; formal analysis, F.A. and C.P.; investigation, C.P. and S.N.; resources, G.U.-B. and F.B.; data curation, C.P. and A.R.; writing—original draft preparation, F.A. and A.R.; writing—review and editing, F.A., G.U.-B. and F.B.; visualization, F.A. and A.R.; supervision, F.A., G.U.-B. and F.B.; project administration, G.U.-B. and F.B.; funding acquisition, G.U.-B. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
Sample Availability
Samples of the compounds are commercially available.
Funding Statement
This research received no external funding.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.McPherson M.L., Cimino N.M. Topical NSAID Formulations. Pain Med. 2013;14:S35–S39. doi: 10.1111/pme.12288. [DOI] [PubMed] [Google Scholar]
- 2.Prasanthan P., Kishore N. HSA nanoparticles in drug recognition: Mechanistic insights with naproxen, diclofenac and methimazole. J. Biomol. Struct. Dynam. 2021:1–13. doi: 10.1080/07391102.2021.1953605. [DOI] [PubMed] [Google Scholar]
- 3.Czub M.P., Handing K.B., Venkataramany B.S., Cooper D.R., Shabalin I.G., Minor W. Albumin-Based Transport of Nonsteroidal Anti-Inflammatory Drugs in Mammalian Blood Plasma. J. Med. Chem. 2020;63:6847–6862. doi: 10.1021/acs.jmedchem.0c00225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chamouard J.-M., Barre J., Urien S., Houin G., Tillement J.-P. Diclofenac binding to albumin and lipoproteins in human serum. Biochem. Pharmacol. 1985;34:1695–1700. doi: 10.1016/0006-2952(85)90636-7. [DOI] [PubMed] [Google Scholar]
- 5.Takla P.G., Schulman S.G., Perrin J.H. Measurement of flurbiprofen—Human serum albumin interaction by fluorimetry. J. Pharm. Biomed. Anal. 1985;3:41–50. doi: 10.1016/0731-7085(85)80005-4. [DOI] [PubMed] [Google Scholar]
- 6.Honoré B., Brodersen R. Albumin binding of anti-inflammatory drugs. Utility of a site-oriented versus a stoichiometric analysis. Mol. Pharmacol. 1984;25:137–150. [PubMed] [Google Scholar]
- 7.Ràfols C., Zarza S., Bosch E. Molecular interactions between some non-steroidal anti-inflammatory drugs (NSAID׳s) and bovine (BSA) or human (HSA) serum albumin estimated by means of isothermal titration calorimetry (ITC) and frontal analysis capillary electrophoresis (FA/CE) Talanta. 2014;130:241–250. doi: 10.1016/j.talanta.2014.06.060. [DOI] [PubMed] [Google Scholar]
- 8.Amézqueta S., Beltrán J.L., Bolioli A.M., Campos-Vicens L., Luque F.J., Ràfols C. Evaluation of the Interactions between Human Serum Albumin (HSA) and Non-Steroidal Anti-Inflammatory (NSAIDs) Drugs by Multiwavelength Molecular Fluorescence, Structural and Computational Analysis. Pharmaceuticals. 2021;14:214. doi: 10.3390/ph14030214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cui Y., Hao E., Hui G., Guo W., Cui F. Investigations on the interactions of diclofenac sodium with HSA and ctDNA using molecular modeling and multispectroscopic methods. Spectrochim. Acta A. 2013;110:92–99. doi: 10.1016/j.saa.2013.01.093. [DOI] [PubMed] [Google Scholar]
- 10.Zhang Y., Lee P., Liang S., Zhou Z., Wu X., Yang F., Liang H. Structural Basis of Non-Steroidal Anti-Inflammatory Drug Diclofenac Binding to Human Serum Albumin. Chem. Biol. Drug Des. 2015;86:1178–1184. doi: 10.1111/cbdd.12583. [DOI] [PubMed] [Google Scholar]
- 11.Trynda-Lemiesz L., Wiglusz K. Interactions of human serum albumin with meloxicam: Characterization of binding site. J. Pharm. Biomed. Anal. 2010;52:300–304. doi: 10.1016/j.jpba.2009.12.025. [DOI] [PubMed] [Google Scholar]
- 12.Moser J., Hye A., Lovell W.W., Earl L.K., Castell J.V., Miranda M.A. Mechanisms of drug photobinding to proteins: Photobinding of suprofen to human serum albumin. Toxicol. Vitr. 2001;15:333–337. doi: 10.1016/S0887-2333(01)00033-9. [DOI] [PubMed] [Google Scholar]
- 13.Maity S., Gundampati R.K., Suresh Kumar T.K. NMR Methods to Characterize Protein-Ligand Interactions. Nat. Prod. Commun. 2019;14:1934578X19849296. doi: 10.1177/1934578X19849296. [DOI] [Google Scholar]
- 14.Gossert A.D., Jahnke W. NMR in drug discovery: A practical guide to identification and validation of ligands interacting with biological macromolecules. Prog. Nucl. Magn. Reson. Spectrosc. 2016;97:82–125. doi: 10.1016/j.pnmrs.2016.09.001. [DOI] [PubMed] [Google Scholar]
- 15.Valensin G., Sabatini G., Tiezzi E. Determination of Zero- and Double-Quantum Relaxation Transition Probabilities by Multiple-Selective Irradiation Methods. In: Niccolai N., Valensin G., editors. Advanced Magnetic Resonance Techniques in Systems of High Molecular Complexity. Birkhäuser; Boston, MA, USA: 1986. pp. 69–76. [Google Scholar]
- 16.Diaz-Perez M.J., Chen J.C., Aubry A.-F., Wainer I.W. The direct determination of the enantiomers of ketorolac and parahydroxyketorolac in plasma and urine using enantioselective liquid chromatography on a human serum albumin-based chiral stationary phase. Chirality. 1994;6:283–289. doi: 10.1002/chir.530060411. [DOI] [Google Scholar]
- 17.Ji Z.-S., Li C.-G., Mao X.-A., Liu M.-L., Hu J.-M. NMR Study on the Low-Affinity Interaction of Human Serum Albumin with Diclofenac Sodium. Chem. Pharm. Bull. 2002;50:1017–1021. doi: 10.1248/cpb.50.1017. [DOI] [PubMed] [Google Scholar]
- 18.Liu M., Nicholson J.K., Lindon J.C. Analysis of Drug–Protein Binding Using Nuclear Magnetic Resonance Based Molecular Diffusion Measurements. Anal. Commun. 1997;34:225–228. doi: 10.1039/a704268j. [DOI] [Google Scholar]
- 19.Jenkins B.G. Detection of site-specific binding and co-binding of ligands to macromolecules using 19F NMR. Life Sci. 1991;48:1227–1240. doi: 10.1016/0024-3205(91)90517-F. [DOI] [PubMed] [Google Scholar]
- 20.Martini S., Bonechi C., Corbini G., Rossi C. Determination of the modified ‘affinity index’ of small ligands and macromolecular receptors from NMR spin-lattice relaxation data. Chem. Phys. Lett. 2007;447:147–153. doi: 10.1016/j.cplett.2007.09.011. [DOI] [Google Scholar]
- 21.Rossi C., Bonechi C., Martini S., Ricci M., Corbini G., Corti P., Donati A. Ligand–macromolecule complexes: Affinity index determination by selective nuclear relaxation analysis. Magn. Reson. Chem. 2001;39:457–462. doi: 10.1002/mrc.876. [DOI] [Google Scholar]
- 22.Kamal A.J., Behere D.V. Spectroscopic studies on human serum albumin and methemalbumin: Optical, steady-state, and picosecond time-resolved fluorescence studies, and kinetics of substrate oxidation by methemalbumin. J. Biol. Inorg. Chem. 2002;7:273–283. doi: 10.1007/s007750100294. [DOI] [PubMed] [Google Scholar]
- 23.Momot K.I., Walker F.A. Proton NMR relaxation in six-coordinate low-spin Iron(III) tetraphenylporphyrinates: Temperature dependence of proton relaxation rates and interpretation of NOESY experiments. J. Phys. Chem. A. 1997;101:9207–9216. doi: 10.1021/jp972194j. [DOI] [Google Scholar]
- 24.Luy B., Frank A., Kessler H. Molecular Drug Properties: Measurement and Prediction. Wiley-VCH Verlag; Weinheim, Germany: 2008. Conformational Analysis of Drugs by Nuclear Magnetic. [Google Scholar]
- 25.Testa B., Vistoli G., Pedretti A., van de Waterbeemd H., Avdeef A., Ivanciuc O., Ertl P., Raevsky O., Sadowski J., Bostrom J., et al. In: Molecular Drug Properties: Measurement and Prediction. Mannhold R., editor. Wiley-VCH Verlag; Weinheim, Germany: 2007. pp. 207–254. [Google Scholar]
- 26.Martini S., Bonechi C., Casolaro M., Corbini G., Rossi C. Drug–protein recognition processes investigated by NMR relaxation data: A study on corticosteroid–albumin interactions. Biochem. Pharmacol. 2006;71:858–864. doi: 10.1016/j.bcp.2005.12.010. [DOI] [PubMed] [Google Scholar]
- 27.Corbini G., Martini S., Bonechi C., Casolaro M., Corti P., Rossi C. Synthetic polymers as biomacromolecular models for studying ligand–protein interactions: A nuclear spin relaxation approach. J. Pharm. Biomed. Anal. 2006;40:113–121. doi: 10.1016/j.jpba.2005.06.019. [DOI] [PubMed] [Google Scholar]
- 28.Zhai Y., Deng P., Wang X., Zhang C., Gan R., Gan N., Sun Q., Li H. Interaction mechanism of olaparib binding to human serum albumin investigated with NMR relaxation data and computational methods. RSC Adv. 2018;8:31555–31563. doi: 10.1039/C8RA05330H. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Not applicable.