Abstract
The hepatitis G virus (HGV) polyprotein was scanned by computer-aided prediction of antigenicity to search for B-cell epitopes. Four polypeptide sequences, V37D (amino acids [aa] 1685 to 1721), V36S (aa 2102 to 2137), P37R (aa 2156 to 2192), and C40P (aa 2280 to 2319), were identified and synthesized for use in immunoassays. Antibodies to these peptides were searched for in a panel of 239 serum samples, which were also tested for anti-E2 antibodies and HGV RNA. Furthermore, the course of HGV markers was studied prospectively in four patients who had been transfused with HGV RNA-positive blood. There was a negative association between immunoreactivity to V37D and P37R and presence of HGV RNA (2 of 53 and 1 of 53, respectively; P < 0.05); none of the subjects with dual antibody positivity was HGV RNA positive. Anti-V37D and anti-P37R antibodies compared favorably with anti-E2 antibodies as markers of recovery from HGV infection. These results might be useful for the development of new, more sensitive diagnostic assays.
A new hepatitis virus, named hepatitis G virus (HGV), has recently been isolated (7, 10). Studies based on serially obtained specimens from infected patients have shown that HGV can persist over the long term in the host; however, there is also evidence in the literature that many HGV infections are cleared, with development of protective immunity.
An assay that detects antibodies to the putative HGV envelope (E2) protein has been developed and is now commercially available. The appearance of anti-E2 antibodies is usually taken as an indication of recovery from HGV infection (2), although some patients are simultaneously positive for circulating HGV RNA and anti-E2 antibodies. Likely, other epitopes in the HGV polyprotein could be able to elicit a humoral immune response.
In the present study, new putative B-cell epitopes, located in two nonstructural proteins of HGV, were identified by computer-aided prediction of antigenicity. The corresponding synthetic peptides were used as antigens in enzyme-linked immunosorbent assays (ELISAs). To verify the performance of these new assays in comparison to those of existing tests, antibodies to these peptides were searched for in a large panel of serum samples, which were also tested for anti-E2 antibodies and HGV RNA. Furthermore, the course of HGV markers was studied prospectively in four patients who had been transfused with HGV RNA-positive blood.
MATERIALS AND METHODS
Subjects.
In a first, cross-sectional study, a panel of serum samples obtained from 239 subjects (169 males and 70 females; mean age, 57.5 ± 11.9 years; age range, 14 to 85 years) were studied. The population studied could be divided into the following five categories: patients with liver cirrhosis (n = 45), patients with hepatocellular carcinoma (n = 62), patients with extrahepatic malignancies (n = 45), patients with coagulopathy (n = 34), and asymptomatic control subjects (n = 53). Data regarding patients belonging to the first three groups have been reported previously (11); their characteristics are summarized in Table 1. None of them had autoimmune hepatitis. The group of patients with coagulopathy included 16 patients with hemophilia A, 7 patients with hemophilia B, 7 patients with Von Willebrand’s disease, 2 patients with factor XI deficiency, 1 patient with factor V deficiency, and 1 patient with factor X deficiency; these diagnoses were based on the identification of the specific coagulation defects.
TABLE 1.
Demographic and clinical characteristics of study groups
| Characteristic | Cirrhosis | Hepatocellular carcinoma | Extrahepatic malignancies | Coagulopathy | Controls |
|---|---|---|---|---|---|
| No. of subjects | 45 | 62 | 45 | 34 | 53 |
| No. of males:no. of females | 29:16 | 48:14 | 24:21 | 28:6 | 40:13 |
| Age (yr) | |||||
| Mean (SD) | 59.7 (6.6) | 63.3 (9.5) | 62.6 (11.1) | 39.2 (11.8) | 56.5 (4.7) |
| Range | 46–75 | 34–84 | 35–85 | 14–61 | 45–67 |
| No. of subjects: | |||||
| HBsAg positive | 5 | 5 | 2 | 3 | 0 |
| Anti-HCV positive | 22 | 28 | 3 | 30 | 0 |
| HCV RNA positive | 19 | 23 | 0 | 18 | 0 |
| % Subjects with abnormal ALTa levels | 70 | 70 | 38 | 47 | 4 |
ALT, alanine aminotransferase.
A second study was conducted prospectively with four patients who were identified during a survey of posttransfusion hepatitis conducted in our institution. For a period of 4 months, a sample from each blood unit transfused and a baseline blood sample from each recipient were collected for HGV RNA testing. Four patients (three males and one female; age range, 39 to 71 years) who were exposed to HGV RNA-positive units were monitored for a minimum of 6 months after the transfusion. Two other patients who were also exposed to HGV RNA-positive blood units could not be studied because they died shortly after the transfusion from causes related to their illnesses. Four control patients who had been transfused with HGV RNA-negative blood were also monitored. The rate of HGV RNA positivity in blood donors, as determined in the survey mentioned above, was 4%.
The study protocols were conducted with strict adherence to the Principles of the Declaration of Helsinki. Informed consent was obtained from the study subjects prior to their participation.
Antibodies to synthetic peptides derived from HGV genome.
The selection of putative B-cell epitopes of the HGV polyprotein was performed by alignment of four published sequences of HGV isolates (GenBank accession nos. HGU 36380, HGU 44402, HGU 45966, and HGU 75356). The consensus sequences were studied by computerized prediction of antigenicity by the method of Welling et al. (15), thus identifying one putative B-cell epitope in the nonstructural region 4 (NS4) and three putative B-cell epitopes in the nonstructural region 5 (NS5A) of the HGV polyprotein. The corresponding synthetic peptides V37D (VLS LAQ AKT AEA YTA TAK WLA GCY TGT RAV PTV SIVD; amino acids [aa] 1685 to 1721), V36S (VYG IGQ SVT IDG ERY TLP HQL RMR NVA PSE VSS EVS; aa 2102 to 2137), P37R (PAA AAL QAI ENA ARI LEP HID VXM EDC STP SLC GSSR; aa 2156 to 2192), and C40P (CVE KSV TRF FSL GLT VAD VAS LCE MEI QNH TAY CDK VRTP; aa 2280 to 2319) were synthesized with a peptide synthesizer (432A Peptide Synthesizer; Applied Biosystems, Perkin-Elmer) by using 9-fluorenylmethoxycarbonyl chemistry and were purified by reverse-phase high-performance liquid chromatography. These peptides were used as antigens in ELISAs by following a methodology previously described by our group (3). Briefly, the peptides were diluted to 5 μg/ml in sodium phosphate (10 mM; pH 7.4) coating buffer. One hundred microliters of this solution was added to each well of a Bio-Hit 96-well plate, and peptides were absorbed overnight at 37°C. After being washed five times with phosphate-buffered saline (PBS)–0.25% Tween 20, the wells were saturated with 2% bovine serum albumin in PBS–Tween 20. The plate was washed five times, dried, and stored at 4°C with silica gel as the drying agent. Serum samples were diluted 1:50 in PBS supplemented with 10% normal goat serum. One hundred microliters of this final solution was added to each well and the plate was incubated for 60 min at room temperature. After the plate was washed, 100 μl of horseradish peroxidase-conjugated anti-human immunoglobulin G (Bio-Rad, Richmond, Calif.) was added to the wells at room temperature. The plates were again washed five times and were incubated with a tetramethylbenzidine solution for 15 min in the dark at room temperature. The reaction was stopped with 100 μl of 2 N sulfuric acid; the optical density was read in a spectrophotometer equipped with a 450 to 620-nm filter. The background ELISA reactivity was estimated by examining sera from 200 healthy voluntary blood donors who were negative for HGV RNA. The analysis of optical density (OD) distribution was carried out with log-transformed values to approximate the normal distribution. To remove the outliers, the Dixon method was used (8), and cutoff values were calculated by adding 3 standard deviations to the mean OD values. All tests were performed twice. The results were expressed as sample/cutoff ratios. The sensitivity of the ELISA was 0.015 OD unit. The specificity of each positive ELISA was confirmed as follows: 100 μl of 1:100 serum dilution of the samples found to be reactive by the ELISA was mixed with 5 μl of 1 mg of a synthetic peptide solution per ml, and the mixture was kept for 1 h at 37°C. Both treated and untreated samples were then retested by the ELISA, and only those samples in which more than a 50% reduction between the two OD values was observed were considered truly positive.
Determination of other biohumoral responses.
Hepatitis B virus (HBV) surface antigen (HBsAg) was detected in sera by a commercially available ELISA (Ortho Diagnostics, Raritan, N.J.). Antibodies to hepatitis C virus (HCV) were detected by a third-generation ELISA (Ortho Diagnostics). Antibodies to the E2 protein of HGV were also detected by means of a commercially available ELISA (Boerhinger Mannheim, Mannheim, Germany). Circulating HGV RNA and HCV RNA sequences were detected in frozen serum samples by in-house reverse transcriptase PCR assays as described previously (13, 14).
Statistical analysis.
Statistical analysis of the data was performed with the BMDP statistical software package (release 7.0; Statistical Solution Ltd., Cork, Ireland). The associations between categorical variables were explored by chi-square tests (BMDP program 4F).
RESULTS
Cross-sectional study.
Circulating HGV RNA sequences were detected in serum samples from 53 patients. In detail, 8 of 45 (18%) patients with cirrhosis, 23 of 62 (37%) patients with hepatocellular carcinoma, 14 of 45 (31%) patients with extrahepatic cancers, 3 of 34 (9%) patients with coagulopathy, and 5 of 53 (9%) control subjects were HGV RNA positive.
The rates of reactivity to the synthetic peptides were as follows: C40P, 29 of 238 (12%); V36S, 64 of 238 (27%); V37D, 27 of 239 (11%); and P37R, 23 of 239 (10%). The rate of reactivity to anti-E2 antibodies was 62 of 239 (26%). Table 2 presents the rates of reactivity to synthetic peptides and to anti-E2 antibodies in relationship to the HGV RNA status of the patients. V36S represented the immunodominant epitope among the four peptides. Although antibodies to this peptide were found more commonly in HGV RNA-positive patients, they were also found in HGV RNA-negative patients, thus discriminating poorly between these two groups. In contrast, anti-V37D and anti-P37R antibodies were rarely detected in the presence of HGV RNA. In fact, sera from 196 of 239 subjects were found to be nonreactive to V37D and P37R, whereas sera from 36 of 239 subjects were found to be reactive to one of the two peptides and sera from 7 of 239 subjects had dual reactivities. HGV RNA was detected in 50 of 196 (26%) nonreactive subjects, 3 of 36 (8%) subjects with single reactivity, and none of 7 (0%) subjects with dual reactivities (P = 0.008 by the chi-square test for trend).
TABLE 2.
Reactivities to synthetic peptides C40P, V36S, V37D, and P37R and to recombinant E2 protein in relationship to serum HGV RNA status of patients
| HGV RNA status | No. of patients
reactive/total no. tested (%)
|
||||
|---|---|---|---|---|---|
| C40P | V36S | V37D | P37R | E2 | |
| Positive | 5/53 (9) | 20/53 (38) | 2/53 (4) | 1/53 (2) | 9/53 (17) |
| Negative | 24/185 (13) | 44/185 (24) | 25/186 (13) | 22/186 (12) | 53/186 (29) |
| P valuea | 0.487 | 0.043 | 0.049 | 0.030 | 0.091 |
Pearson chi-square tests.
The sensitivities of currently available tests for HGV would be increased by the inclusion of assays based on C40P, V36S, V37D, and P37R. In fact, among the population studied, 106 subjects were HGV RNA and/or anti-E2 positive, leading to an overall estimate of the rate of exposure to HGV of 44%. Fifteen further subjects, although HGV RNA and anti-E2 negative, tested positive by at least two of the four assays with synthetic peptides, leading to a new overall estimate of the rate of exposure to HGV of 51%.
Among the HBsAg-positive subjects, there was an increased frequency of positive results by the anti-C40P assay (5 of 15 versus 24 of 224; P = 0.01), the anti-V36D assay (8 of 15 versus 56 of 224; P = 0.017), and the anti-P37R assay (5 of 15 versus 18 of 224; P = 0.001). Among the anti-HCV antibody and/or HCV RNA-positive subjects, there was a significantly increased frequency of positive results by the anti-P37R assay (14 of 95 versus 9 of 144; P = 0.029) and the anti-E2 assay (36 of 95 versus 26 of 144; P < 0.001).
Longitudinal study.
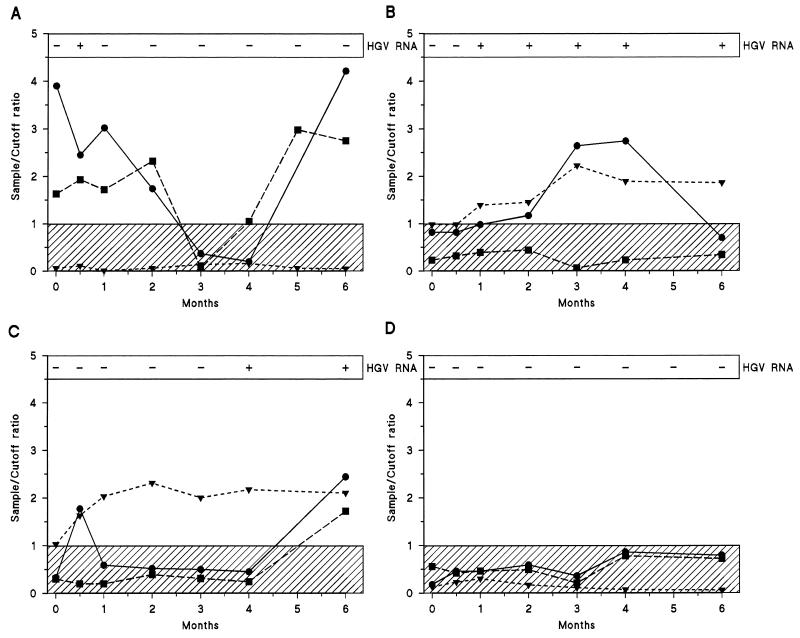
Figure 1 shows the course of the levels of HGV RNA, anti-E2 antibodies, anti-P37R antibodies, and anti-V37R antibodies in the four patients transfused with HGV RNA-positive blood units. The minimum follow-up period after blood transfusion was 6 months. One subject who was anti-P37R and anti-V37D positive at the baseline became transitorily HGV RNA positive early (2 weeks) after transfusion but rapidly cleared the virus. Anti-E2 antibodies remained consistently negative (Fig. 1A). The other three patients were anti-P37R and anti-V37D negative at the baseline. One patient became HGV RNA positive after 2 months; the detection of HGV viremia was followed by elevations in anti-V37D and anti-P37R antibody titers, but subsequently these antibodies became undetectable. The patient was still HGV RNA positive and anti-E2 positive at the end of the follow-up (Fig. 1B). Another patient became HGV RNA positive after 4 months; again, the appearance of viremia was followed by an elevation in anti-P37R and anti-V37D antibody titers. After 6 months, positivity for all serologic markers of HGV could be detected (Fig. 1C). Finally, the fourth patient remained negative for all serologic markers of HGV, including HGV RNA, throughout the entire follow-up period (Fig. 1D). Throughout the entire follow-up period the four control patients transfused with HGV RNA-negative blood remained negative for HGV RNA and anti-HGV antibodies (data not shown).
FIG. 1.
Serologic profiles of HGV infection in four persons (A to D) exposed to HGV by blood transfusion (day 0). At each time point, the results of HGV RNA and anti-HGV antibody testing (anti-E2 antibodies, solid triangles; anti-P37R antibodies, solid circles; anti-V37D antibodies, solid squares) are indicated. The shaded areas indicate the sample/cutoff ratio for negative test results.
DISCUSSION
Three of the synthetic peptides on which we based the immunoassays described here (C40P, P37R, and V36S) were derived from nucleotide sequences located in the NS5A region, whereas the fourth peptide (V37D) was derived from nucleotide sequences located in the NS4 region. The functional roles of the proteins encoded in these two regions of the viral genome are unknown. In any case, they appear to elicit a detectable antibody response. This is similar to what occurs in HCV infection, in which antibodies to nonstructural proteins are easily detected. However, the great sequence variability and the quasispecies nature of HCV lead to the escape of the virus from the immune system, but the virus remains detectable in blood, despite the presence of high titers of antibodies to many epitopes along the entire length of the HCV genome. In contrast, the appearance of antibodies to HGV seems to be most commonly (although not invariably) associated with recovery from infection, as demonstrated by the present study as well as by other studies (2, 5). Indeed, even in the setting of immunosuppression, as in liver transplant recipients, the presence of antibodies to the E2 glycoprotein seems to represent a marker of protective immunity to HGV (11).
Interestingly, as markers of recovery from HGV infection, antibodies to V37D and P37R compare favorably with anti-E2 antibodies, a fact which could be exploited to improve the sensitivities of currently available tests. An overall estimate of the rate of exposure to HGV made by combining together the results of the different assays indicates that traces of current or previous HGV infection may be found in up to 50% of subjects in selected populations. It should be noted, however, that assays based on combinations of antigens guarantee adequate sensitivity only for the detection of past exposure to HGV. Considerable need remains for new serologic markers of current HGV infection so that they can be used in alternatives to PCR-based assays for the detection of HGV RNA.
Even though HGV and HCV are two phylogenetically closely related viruses, we are confident that the reactivity of HGV to nonstructural proteins shown in the present study was not due to cross-reactivity with HCV epitopes. Overall, there is little primary sequence homology between the two viruses, which share approximately 25% identity at the amino acid level. As a consequence, there is little chance that antibodies directed against epitopes of HCV may cross-react with epitopes of HGV, or vice versa. The occasional patient who shows reactivity only to antigens derived from the HCV NS5B region but who has negative results for HCV infection by PCR may be the exception (5). However, we have tested sera from nine such patients and did not find any HGV RNA- or HGV antibody-positive patients (unpublished data). Moreover, scanning of the open reading frames of HBV and HCV demonstrates that these viruses do not have significant amino acid homology with any of the four synthetic peptides (maximum number of consecutive amino acids in common, 4).
A final comment regarding the association which we found between positive results of HGV assays and positive results of serologic assays for HBV and HCV is deserved. The pathogenic role and the hepatotropism of HGV are the subject of much debate (1, 9); nevertheless, the association of HGV with HBV and HCV confirms that transmission of HGV is likely to be by routes similar to those of these truly pathogenic viruses. Vertical transmission of HGV has already been demonstrated (4); because of the high prevalence of HGV in the general population, other main routes (including sexual intercourse) must also be assumed. Indeed, there is a high HGV RNA prevalence among non-drug-injecting homosexual and bisexual men (12) and prostitutes (6).
In summary, four new epitopes have been identified in the HGV polyprotein. These expanding data on the immune response to HGV might be helpful in the development of more sensitive assays that can be used for epidemiological surveys.
ACKNOWLEDGMENTS
This work was supported by grant 9502438.CT04 from the Consiglio Nazionale delle Ricerche and by a grant from the Associazione Italiana per la Ricerca sul Cancro, Rome, Italy.
REFERENCES
- 1.Cotler S J, Gretch D R, Bronner M P, Tateyama H, Emond M J, dela Rosa C, Perkins C D, Carithers R L., Jr Hepatitis G virus co-infection does not alter the course of recurrent hepatitis C virus infection in liver transplantation recipients. Hepatology. 1997;26:432–436. doi: 10.1002/hep.510260225. [DOI] [PubMed] [Google Scholar]
- 2.Dille B J, Surowy T K, Gutierrez R A, Coleman P F, Knigge M F, Carrick R J, Aach R D, Hollinger F B, Stevens C E, Barbosa L H, Nemo G J, Mosley J W, Dawson G J, Mushahwar I K. An ELISA for detection of antibodies to the E2 protein of GB virus C. J Infect Dis. 1997;175:458–461. doi: 10.1093/infdis/175.2.458. [DOI] [PubMed] [Google Scholar]
- 3.Ferroni P, Mascolo G, Zaninetti M, Colzani D, Pregliasco F, Pirisi M, Barbone F, Gasparini V. Identification of four epitopes in hepatitis C virus core protein. J Clin Microbiol. 1993;31:1586–1591. doi: 10.1128/jcm.31.6.1586-1591.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Feucht H H, Zöllner B, Polywka S, Laufs R. Vertical transmission of hepatitis G. Lancet. 1996;347:615–616. [PubMed] [Google Scholar]
- 5.Feucht H H, Zöllner B, Polywka S, Knödler B, Schröter M, Nolte H, Laufs R. Distribution of hepatitis G viremia and antibody response to recombinant proteins with special regard to risk factors in 709 patients. Hepatology. 1997;26:491–494. doi: 10.1002/hep.510260234. [DOI] [PubMed] [Google Scholar]
- 6.Kao J H, Chen W, Chen P J, Lai M Y, Lin R Y, Chen D S. GB virus-C/hepatitis G virus infection in prostitutes: possible role of sexual transmission. J Med Virol. 1997;52:381–384. [PubMed] [Google Scholar]
- 7.Linnen J, Wages J, Zhang-Keck Z Y, Fry K E, Krawczynski K Z, Alter H, Koonin E, Gallagher M, Alter M, Hadziyannis S, Karayiannis P, Fung K, Nakatsuji Y, Shih J W K, Young L, Piatak M, Hoover C, Fernandez J, Chen S, Zou J C, Morris T, Hyams K C, Ismay S, Lifsaon J D, Hess G, Foung S K H, Thomas H, Bradley D, Margolis H, Kim J P. Molecular cloning and disease association of hepatitis G virus: a transfusion-transmissible agent. Science. 1996;271:505–508. doi: 10.1126/science.271.5248.505. [DOI] [PubMed] [Google Scholar]
- 8.Linnet K. Two stage transformation systems for normalization of reference distribution evaluated. Clin Chem. 1987;33:381–386. [PubMed] [Google Scholar]
- 9.Petrik J, Guella L, Wight D G D, Pearson G M, Hinton J, Parker H, Allain J-P, Alexander G J M. Hepatic histology in hepatitis C virus carriers coinfected with hepatitis G virus. Gut. 1998;42:103–106. doi: 10.1136/gut.42.1.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Simons J N, Leary T P, Dawson G J, Pilot-Matias T J, Muerhoff A S, Schlauder G G, Desai S M, Mushawar I K. Isolation of novel virus-like sequences associated with human hepatitis. Nat Med. 1995;1:564–569. doi: 10.1038/nm0695-564. [DOI] [PubMed] [Google Scholar]
- 11.Silini E, Belli L, Alberti A B, Asti M, Cerino A, Bissolati M, Rondinara G, De Carlis L, Forti D, Mondelli M U, Ideo G. HGV/GBV-C infection in liver transplant recipients: antibodies to the viral E2 envelope glycoprotein protect from de novo infection. J Hepatol. 1998;29:533–540. doi: 10.1016/s0168-8278(98)80147-5. [DOI] [PubMed] [Google Scholar]
- 12.Stark K, Bienzle U, Hess G, Engel A M, Hegenscheid B, Schlüter V. Detection of the hepatitis G virus genome among injecting drug users, homosexual and bisexual men, and blood donors. J Infect Dis. 1996;174:1320–1323. doi: 10.1093/infdis/174.6.1320. [DOI] [PubMed] [Google Scholar]
- 13.Toniutto P, Pirisi M, Fabris C, Bardus P, Soardo G, Vitulli D, Tisminetzky S G, Pacco P, Gasparini V, Baralle F, Bartoli E. High prevalence of infection by hepatitis G virus in patients with hepatic and extrahepatic malignancies. J Hepatol. 1998;28:550–555. doi: 10.1016/s0168-8278(98)80277-8. [DOI] [PubMed] [Google Scholar]
- 14.Toniutto P, Pirisi M, Tisminetzsky S G, Fabris C, Chinellato E, Gerotto M, Falleti E, Ferroni P, Lombardelli T, Bartoli E, Baralle F. Discordant results from hepatitis C virus genotyping by procedures based on amplification of different genomic regions. J Clin Microbiol. 1996;34:2382–2385. doi: 10.1128/jcm.34.10.2382-2385.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Welling G W, Weijer W J, van der Zee R, Welling-Wester S. Prediction of sequential antigenic regions in proteins. FEBS Lett. 1985;188:215–218. doi: 10.1016/0014-5793(85)80374-4. [DOI] [PubMed] [Google Scholar]