Abstract
The tripartite partition defect (PAR) polarity complex, which includes the proteins PAR3, atypical protein kinase C (aPKC), and PAR6, is a major regulator of cellular polarity. It is highly conserved and expressed in various tissues. Its largest component, PAR3, controls protein–protein interactions of the PAR complex with a variety of interaction partners, and PAR3 self-association is critical for the formation of filament-like structures. However, little is known about the structure of the PAR complex. Here, we purified non-filamentous PAR3 and the aPKC–PAR6 complex and characterized them by single-particle electron microscopy (EM). We expressed and purified an oligomerization-deficient form of PAR3, PAR3V13D,D70K, and the active aPKC–PAR6 dimer. For PAR3, engineering at two positions is sufficient to form stable single particles with a maximum dimension of 20 nm. aPKC–PAR6 forms a complex with a maximum dimension of 13.5 nm that contains single copies of aPKC. Thus, the data present a basis for further high-resolution studies of PAR proteins and PAR complex formation.
Supplementary Information
The online version contains supplementary material available at 10.1007/s12033-022-00504-1.
Keywords: PAR3, PAR6, aPKC, PAR complex, Polarity, Single-particle electron microscopy
Introduction
Polarity is a hallmark of cellular development such as anterior–posterior polarity in zygotes, apical-basal and planar cell polarity in epithelia, axon-dendrite differentiation in neurons, and transient polarity in migrating cells [1–7]. Polarity is established and regulated by a set of evolutionarily highly conserved proteins including a set of PAR (for partitioning defective) proteins. PAR proteins have been originally identified as factors required for polarization in C. elegans zygotes [8]. The PAR protein family includes a polarity complex (PAR complex) comprising PAR3, atypical protein kinase C (aPKC), and PAR6 [9, 10]. In vertebrates, the PAR complex has been studied extensively in epithelia, where the proteins localize in the apical compartment near tight junctions and have been demonstrated to be central for the establishment and maintenance of apical-basal polarity [11]. The PAR complex is also critical for neural development, where it has been suggested to be essential for the differentiation of neurites into dendrites and the axon [12–14]. Moreover, growing evidence suggests a link between deregulation of PAR3 and cancer development favoring cell proliferation, epithelial–mesenchymal transition (EMT) and metastatic spread in a number of tumor types [15].
In human, its major component, PAR3, is a large (> 1300 amino acids) protein rich in interaction domains. PAR3 binds PAR6 through PDZ (PSD-95, Discs-large, ZO-1) domains [9], and interacts with aPKC through an aPKC-binding domain [16], while PAR6 and aPKC interact with each other through their PB1 (Phox and Bem1) domains [17]. PAR3 has been suggested to bind to two PAR6 proteins via two of its three PDZ domains [18]. The first PDZ domain of PAR3 can also bind to membrane proteins including junctional adhesion molecules (JAM) and the p75 neurotrophin receptor [19–21].
Moreover, PAR3 exhibits a microtubule binding and bundling activity [22], and the PAR complex has been implicated in the regulation of the microtubule and actin cytoskeleton as a critical step in neuronal development [12, 13, 23]. The semi-CRIB (Cdc42- and Rac-interactive binding) domain of PAR6 can bind to the active Rho GTPase CDC42 [9], which activates aPKC to phosphorylate PAR3 and cause PAR3 dissociation from the PAR complex [24].
Major attention has been paid to the ability of the proteins to enrich in certain cellular compartments as a hallmark of polarization [13, 14, 25–27]. Particularly, PAR3 self-association may represent the molecular basis for the enrichment of the PAR complex at target sites. The first approximately 83 amino acids of PAR3 form an N-terminal domain (NTD) that has been demonstrated to exhibit a critical role in self-association of PAR3 [25, 28]. Crystallization and cryo-EM studies of the rat PAR3 NTD fragment have shown that the isolated NTD forms protein helices with a regular pitch through a number of residues including T4, V13, and D70 providing lateral interactions, as well as R9 providing longitudinal interactions [29]. Mutation studies of the isolated PAR3 NTD furthermore suggested that mutations of V13 and D70 prevent oligomerization of NTD fragments [29]. However, the structural organisation of PAR3 clusters in vivo remains unknown.
Although the ability of the PAR complex to function in diverse cellular contexts such as epithelia and neurons would clearly be explained by knowing its structure, still little is known about its architecture. Here, we aimed at characterizing non-polymerized PAR complexes, the heterodimeric aPKC–PAR6 complex, and PAR3 alone.
Results
Identification of PAR3, aPKC, and PAR6 Isoforms Expressed in Human Neural Cells
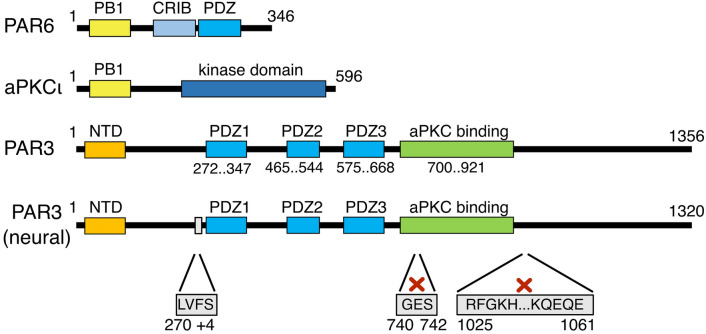
The genes of PAR6, PAR3, and aPKC subtype iota were amplified from human neural cells [30], and their identity was confirmed by sequencing (Fig. 1). The sequences of the encoded aPKC and PAR6 proteins are identical to the canonical human isoforms (PKCι, GenBank accession code: NP_002731.4; PAR6α; GenBank accession code: NP_001032358.1), respectively. PAR3 is a novel isoform as a result of minor changes in alternative splicing. In comparison to the human PAR3 isoform 1 (GenBank accession code: NP_062565.2), the following differences were observed: (i) the neural PAR3 isoform possessed four additional amino acids after residue 269 with D269 changed to E immediately N-terminal to the PDZ1 domain; (ii) three amino acids were omitted between positions 739–743 in the aPKC-binding domain; and (iii) 37 residues were omitted between positions 1024–1062 C-terminal to the aPKC-binding domain. The omission of these 37 residues is also seen in isoform 4 of PAR3 (NCBI accession no. NP_001171716). All these modifications in the PAR3 isoform can be explained by alternative splice sites of the transcript. Besides, we observed a K312R change in the PDZ1 domain.
Fig. 1.
Protein domains of human PAR6, aPKC, and PAR3. PAR6 and aPKC interact via their PB1 domains. aPKC harbors a kinase domain that can phosphorylate PAR3 followed by PAR3 release from the complex. PAR3 binds to PAR6–aPKC via a PDZ/aPKC-binding domain. The neural isoform analyzed herein showed alternative splicing leading to an extra mini-exon at amino acid (aa) 270, an omission of 3 amino acids at aa 739–743, and a skipped exon encoding aa 1024–1062 (positions compared to the canonical PAR3 sequence, accession no. NP_062565.2)
aPKC and PAR6 form a Defined Complex
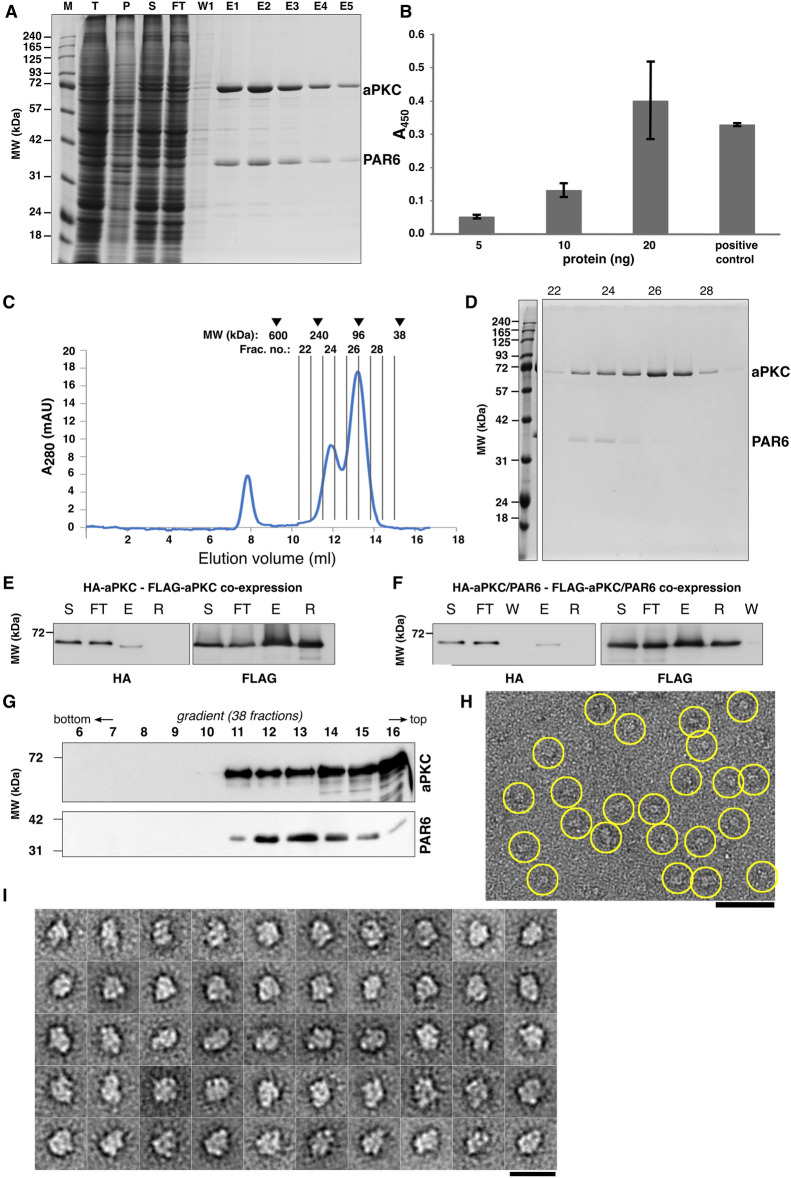
Wild-type tagged aPKC and untagged PAR6 were co-expressed in their full-length forms using a baculovirus/insect cell system. Upon immuno-purification of aPKC via its N-terminal 3 × FLAG tag (Fig. 2A), a dimeric complex was obtained. We confirmed by using a kinase assay that the purified aPKC–PAR6 complex was functionally active in phosphorylation (Fig. 2B). By size exclusion chromatography (SEC), the aPKC–PAR6 complex (theoretical molecular weight of 106 kDa assuming a 1:1 complex) and co-eluting unbound aPKC (theoretical molecular weight of monomer, 68 kDa) showed peaks in fraction 24 and 26, respectively, at elution volumes somewhat higher than expected for near spherical proteins (Fig. 2C, D).
Fig. 2.
aPKC and PAR6 associate as a heterodimeric complex functional in phosphorylation. A Anti-FLAG-affinity selected aPKC–PAR6 complex visualized by SDS-PAGE and Coomassie staining. Proteins are indicated to the right. M, T, P, S, FT, W1, and E1-E5 correspond to marker, total cell lysate, pellet, supernatant, flow through, first wash, and elution fractions 1–5, respectively. B Kinase activity of the purified aPKC–PAR6 complex measured as absorption at 450 nm for protein concentrations of 5 ng, 10 ng, and 20 ng, and a positive control (n = 3). As positive control, 36 ng of control protein was used. C Gel-filtration chromatography profile of the aPKC–PAR6 complex. Protein fractions (500 µl) were collected, and the absorption was monitored at 280 nm wavelength. Running behaviors of standard proteins (in kDa) are indicated at the top. D The peak fractions 22–29 (fraction numbers indicated in C) are visualized by a Coomassie-stained SDS-PAGE (theoretical MW: aPKC, 71 kDa; PAR6, 37 kDa). The aPKC–PAR6 complex peaks in fraction 24, and aPKC alone peaks in fractions 26. Proteins are indicated to the right. E, F Recovery of HA-aPKC in FLAG pulldown assays upon 3 × FLAG-aPKC,HA-aPKC co-expression (E) and 3 × FLAG-aPKC,HA-aPKC,PAR6 co-expression (F) (S, supernatant; FT, flow through; E, eluate; R, FLAG affinity resin; W, wash). G Western blot of the aPKC–PAR6 heterodimer after sedimentation in a 5–20% glycerol gradient. Protein samples were collected in a total of 38 fractions, of which fractions 6–16 are shown (fraction numbering from the bottom; molecular weight marker indicated on the left). H A representative negative stain EM image of the aPKC–PAR6 complex using peak fractions 12–14 shows a monodisperse particle population (scale bar: 50 nm). I 2D class averages of aPKC–PAR6 demonstrate particles with a compact, moderately elongated shape and maximum dimensions of 13.5 nm (scale bar: 20 nm)
aPKC Does Not Exhibit Self-Interaction
To assess whether or not the main component of the heterodimeric aPKC–PAR6 complex, aPKC occurs in monomeric form, we co-expressed aPKC with two different tags by replacing the 3 × FLAG sequence in the aPKC construct with an HA tag sequence, followed by co-expression of the two distinguishable tagged forms of aPKC and purification via the 3 × FLAG-tag. We expressed HA-aPKC together with 3 × FLAG-aPKC in the absence of PAR6, and we also expressed HA-aPKC and 3 × FLAG-aPKC in the presence of PAR6 to test whether the presence of PAR6 had an influence on the stoichiometry of the protein complex (Supplementary Fig. S1A, B). We measured the recovery of HA-aPKC in FLAG pulldown assays upon 3 × FLAG-aPKC,HA-aPKC co-expression (Fig. 2E) and 3 × FLAG-aPKC,HA-aPKC,PAR6 co-expression (Fig. 2F) using anti HA western blotting and anti FLAG western blotting as control. Only a minor fraction of HA-aPKC was recovered irrespective of the absence or presence of PAR6: when normalized to the input (supernatant), the elution yielded 0.5% recovery of HA-aPKC and 0.14% of HA-aPKC–PAR6 (Fig. 2E, F; for SDS-PAGE, see Fig. S1A, B) consistent with no noteworthy self-interaction and predominantly monomeric aPKC alone and in complex with PAR6. Thus, the predominant form of the aPKC–PAR6 complex is an assembly with a single copy of aPKC.
aPKC–PAR6 Forms Moderately Elongated Particles
For all particles, we employed gradient ultracentrifugation as final purification step, which provides an optimum sample quality for EM [31]. When aPKC–PAR6 was run on a 5–20% glycerol gradient, the peak of the protein complex (Fig. 2G and Supplementary Fig. S1C) occurred in fraction 12 – 14 (out of 38 fractions), which corresponds to an apparent Svedberg value of about 4.5S. By EM, raw images showed a monodisperse particle population (Fig. 2H). The maximum dimension of the particles is approximately 13.5 nm, and class averages with about 24 images per class show well-discernible fine-structural details indicating a well-defined structure (Fig. 2I). In particular, the particles reveal an asymmetrical, compact, moderately elongated structure.
We also combined the Stoke’s radii and sedimentation data to a molecular weight estimate (MW) using Erickson’s approximation [32] based on Siegel and Monty [33], where S is the sedimentation in Svedberg Units and Rs is the radius in nm. For the aPKC–PAR6 complex, this estimation yields a predicted MW of max. ~ 100 kDa (Table 1), which is consistent with a monomeric stoichiometry of the largest protein in the complex, aPKC, in addition to PAR6.
Table 1.
Characteristics of the protein complexes
| PAR3V13D,D70K | aPKC | aPKC–PAR6 | |
|---|---|---|---|
| Theoretical monomeric MW [kDa] | 141 | 71 | 108 |
| SEC Rs [nm] | 6.1 | 4.0 | 4.7 |
| Sedimentation coefficient [S] | 5.2–7.1 | 3.5–4.2 | 4.4–5.3 |
| EM max. diameter [nm] | 20 | N/A | 13.5 |
| MWSM [kDa] | 133–182 | 59–71 | 87–105 |
MW molecular weight, SEC size exclusion column, Rs Stoke’s radius measured by SEC, S Svedberg, EM electron microscopy, MWSM molecular weight estimate derived from Rs and S using the Siegel-Monty estimation
PAR3V13D,D70K Forms a Stable Elongated Particle
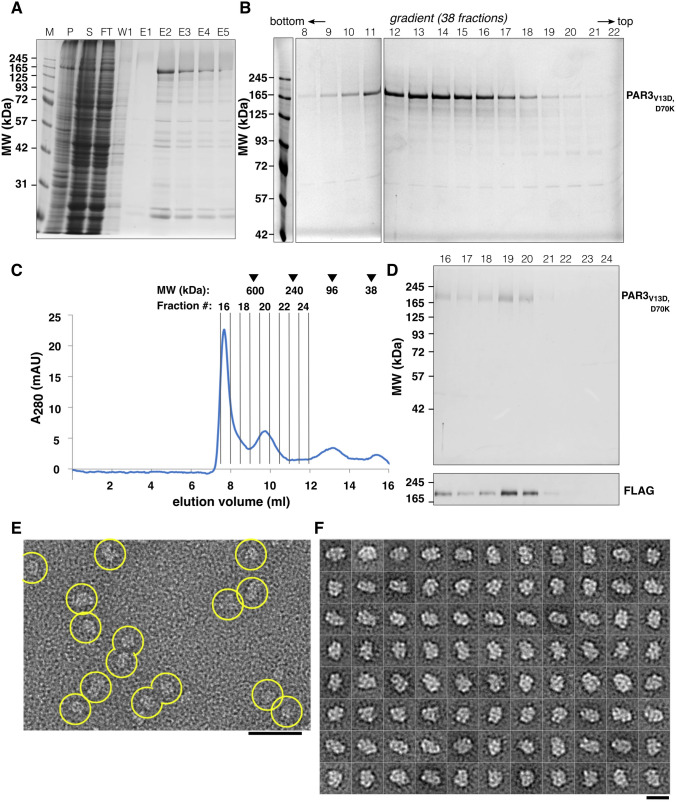
Initial expression tests of wild-type PAR3 alone indicated that wild-type PAR3 was not stable after elution (data not shown); thus, we investigated a mutant form of PAR3. To this end, we expressed a form of PAR3 mutated at two positions (V13D and D70K) that was reported earlier [29]. Expression tests with PAR3V13D,D70K alone (i.e., not in complex with PAR6–aPKC) showed that PAR3V13D,D70K was stable, and protein degradation could be minimized (Fig. 3A,B), which provided the ability to purify PAR3V13D,D70K in amounts sufficient for EM analysis. In SEC, PAR3V13D,D70K peaked in fraction 19–20 at an elution volume corresponding to a Stokes radius of ~ 6.1 nm, separate from void (Fig. 3C,D and Table 1). Another UV peak (fraction 16) visible nearby the void volume, however, contained smaller amounts of PAR3 as evinced by western blotting (Fig. 3D). PAR3V13D,D70K was subsequently run on a 5–20% glycerol gradient (Fig. 3B), where it peaked around fraction 12–13 out of 38 fractions corresponding to the ~ 7S region (approximate MW 133–182 kDa, compare Table 1). These results are consistent with a monomeric protein given the theoretical molecular weight of 141 kDa. EM images of PAR3V13D,D70K showed monodisperse, moderately elongated single particles (Fig. 3E), and the 2D class averages of PAR3V13D,D70K confirmed a well-defined structure with compact shape and maximum dimensions of ~ 20 nm (Fig. 3F).
Fig. 3.
PAR3V13D,D70K is stable in solution and forms elongated particles. A Affinity selection of N-terminal 3 × FLAG-tagged PAR3V13D,D70K visualized by Coomassie-stained SDS-PAGE (M, marker; P, pellet; S, supernatant; FT, flow through; W1, wash fraction 1; E1-E5, elution fraction 1–5). B 5–20% glycerol gradient fractionation of PAR3V13D,D70K as visualized by Coomassie-stained SDS-PAGE. Shown are fractions 8–22 out of 38 fractions in total. PAR3V13D,D70K forms a defined peak on the gradient. C In SEC, PAR3V13D,D70K peaks in fractions 19 and 20. Position of calibration proteins (in kDa) and fractions used for SDS-PAGE analysis (D) are indicated at the top. D SEC fractions as indicated in C are separated by SDS-PAGE and visualized by Coomassie staining (top). The presence of PAR3V13D,D70K was confirmed by anti-FLAG western blot (bottom). E Single particles observed by negative stain EM. F Representative 2D class averages of PAR3V13D,D70K showing particles up to 20 nm in diameter. The scale bars correspond to 50 nm E and 20 nm F, respectively
Discussion
Herein, we aimed at characterizing the non-polymerized building blocks as smallest units of the PAR complex that is formed by the PAR proteins PAR3, aPKC, and PAR6. To avoid polymerization of PAR3, we took advantage of two point mutations in the NTD of PAR3, V13D, and D70K. These mutations had been previously reported to prevent the PAR3 NTD from self-association [28], and were described to abolish interactions in the isolated NTD fragment of rat Par3 by preventing lateral packing into a helix [28, 29].
The sedimentation and SEC data we present herein are in favor of single copies of the largest subunit, PAR3, and of aPKC and PAR6 in the aPKC–PAR6 complex. For the aPKC–PAR6 complex, the presence of a single aPKC protein within the aPKC–PAR6 complex has independently been validated by the double tagging assay. In the aPKC–PAR6 heterodimer, aPKC and PAR6 interact via the PB1 domain present in both proteins [17]. In the crystal structure, the PB1 domains of aPKC and PAR6 form an asymmetric heterodimer, with one copy of the aPKC and PAR6, each [17]. Together with our pulldown data, these data support a 1:1 stoichiometry of aPKC and PAR6 as smallest unit.
It has been shown that PAR3 binds essentially two copies of PAR6 via its PDZ1 and PDZ3 domains, albeit at different affinities [18]. Both dissociation constants were however reported in the micromolar range [18]. Whether or not the local enrichment of the PAR proteins at the plasma membrane is sufficient to facilitate recruitment of two PAR6 copies (or two aPKC–PAR6 heterodimers) to the same PAR3 proteins under in vivo conditions at the cell membrane, will thus need further investigation.
By EM, we did not observe formation of specific multimers of the PAR3 protein, indicating that higher order assemblies that may have formed despite the engineering were not sufficient for visualization by EM. Further research will be required to investigate how the basic building blocks characterized here enrich into higher order PAR complex assemblies inside the cell. Especially, how the rather large PAR3 protein assembles into higher order complexes and whether or not PAR3 will adopt a helical assembly in vivo remains to be investigated. Future high-resolution cryo-EM reconstructions of the PAR complex based on these data are required to address these questions. Likewise, how the occurrence of alternative splicing of PAR3 shown here contributes to tissue-specific variants of the PAR complex will require more investigation. Overall, our current studies provide projection structures of the PAR components PAR3 and aPKC–PAR6 as a step toward a detailed structural and functional understanding of these components in the establishment and maintenance of cellular polarity.
Materials and Methods
Amplification of mRNA from Human Neural Cells
Full-length human PAR6, PAR3, and aPKC subtype iota (PKCɩ) were amplified with appropriate primers (Supplementary Table S1) and cDNA synthesized using mRNA derived from human neural cells as described previously [30]. The Maxima H minus first strand cDNA synthesis kit (ThermoFisher Scientific, Waltham, MA, U.S.A.) was used for cDNA synthesis. The vector pUC57 (ThermoFisher Scientific) was used to insert the DNA fragments using suitable restriction enzymes.
Plasmid Construction
PAR3 was subcloned into the vector pGS-BacA-21122 [34], a derivative of pACEBac1, which introduced a 3 × FLAG to the N-terminus of the expressed protein. PAR3 was studied as wild-type protein and as an engineered PAR3V13D,D70K with two amino acid changes, V13D and D70K, in the NTD. We introduced a mutation causing a kinase-dead mutant in mouse [35], PKCιK283R, into the human gene upon sequence alignment of the human PKCι (GenBank accession code: NM_002740.5) and mouse PKCι (GenBank accession code: BC021630.1) [35]. The QuickChange Lightning Site-Directed Mutagenesis kit (Agilent Technologies, Santa Clara, CA, U.S.A.) was used for site-directed mutagenesis. By Cre recombination, composite bacmids containing PAR3, PKCι, and PAR6 were created from acceptor and donor plasmids as described previously [36] for multi-protein expression in insect cells using the Multibac system [37, 38].
To study the dimeric aPKC–PAR6 complex, the coding sequence of aPKC was ligated into the acceptor vector pGS-BacA-21122 [34], and PAR6 was ligated into the donor vector pIDC. Furthermore, the 3 × FLAG tag from the aPKC plasmid was replaced by an HA tag to investigate aPKC self-oligomerization. The plasmids coding for PKCι and PAR6 were combined by Cre-LoxP reactions as outlined [36]. The plasmid constructs created in this study are listed in Supplementary Table S2. The sequence of relevant plasmid elements was confirmed by Sanger sequencing (Eurofins, Ebersberg, Germany or Macrogen Europe, Amsterdam, The Netherlands).
Protein Expression and Purification
The bacmid and virus preparations for PAR protein expression were performed as described previously [39]. High Five (BTI-TN-5B1-4) or Sf9 cells (both cell lines purchased from ThermoFisher Scientific) were infected with baculovirus carrying aPKC–PAR6 and PAR3V13D,D70K, respectively, and grown for 72 h. The cells were resuspended in lysis buffer (20 mM HEPES, pH 7.6; 10% glycerol; 400 mM NaCl; 1 mM EDTA; 1 mM PMFS (Sigma-Aldrich, St. Louis, MO, U.S.A.) supplemented with protease inhibitor (Complete ULTRA tablets, EDTA-free; Roche, Mannheim, Germany)). The cells were pelleted, and the supernatant was mixed with equilibrated anti-FLAG resin (100 µl; M2 anti-FLAG affinity gel, Sigma) and incubated at 4 °C for 3 h. After incubation, the sample was centrifuged at 700 × g for 10 min to remove the supernatant. The resin was washed three times with washing buffer (20 mM HEPES, pH 7.6; 5% glycerol; 400 mM NaCl). The resin was then transferred into a filter column (M105035F; MoBiTec, Göttingen, Germany) and centrifuged at 100 × g for 3 s. The resin was incubated with 100 µl FLAG-elution buffer (20 mM HEPES–NaOH, pH 7.6; 400 mM NaCl; 1 mM EDTA; 1 mM PMFS; 1 × protease inhibitor; 125 µg/ml 3 × FLAG peptide) for 30 min prior to elution as fraction E1. Fraction E2 was eluted as described above with 400 µl FLAG-elution buffer. A volume of 100 µl elution buffer (20 mM HEPES–NaOH pH 7.6, 400 mM NaCl, 1 mM EDTA, 1 mM PMFS, protease inhibitor) was then added to collect fraction E3. This step was repeated three times to elute the remaining proteins as fractions E4, E5, and E6. BCA assays were used to quantify protein amounts.
Size Exclusion Chromatography
The purified proteins were subjected to SEC on a Superdex 200 column (GE Healthcare, Little Chalfont, U.K.) for the dimeric aPKC–PAR6 and PAR3V13D,D70K in 20 mM HEPES (pH 7.6) supplemented with 400 mM NaCl. The Gel Filtration HMW Calibration Kit (GE Healthcare) was used for calibration of the SECs. For calibration of the elution volume as a function of Stoke’s radius Rs, Rs values reported in [32] were used.
Gradient Ultracentrifugation
The purified proteins were run in a 5–20% glycerol gradient for 17 h at 4 °C (20 mM HEPES, pH 7.6; 400 mM NaCl) at 352,996 × g for the dimeric aPKC–PAR6 complex or 274,824 × g for PAR3V13D,D70K. The gradients were fractionated into 38 fractions with 5 drops per fraction by fractionation from the bottom of the gradient using a P-1 peristaltic pump (GE Healthcare) as described previously [40]. For estimation of the apparent sedimentation coefficients (S), commercial standards were used (Sigma). As glycerol gradient peaks typically span over multiple fractions, an apparent S value range is given for all particles. Proteins were visualized by Coomassie staining and verified by western blotting.
Western Blot Analysis
The protein samples were added to SDS loading dye, heated to 95 °C, separated by SDS-PAGE and transferred to a nitrocellulose membrane (ThermoFisher Scientific). The antibodies anti-FLAG M2 (F1804; Sigma-Aldrich, 1:1000), anti-HA (Santa Cruz, sc-805, 1:200), anti-PKC (sc-216, Santa Cruz, 1:500), and anti-PAR6 (sc-33898, Santa Cruz, 1:500) were used as primary antibodies, anti-mouse IgG-Peroxidase (Sigma-Aldrich, 1:10,000), anti-rabbit IgG-Peroxidase (Sigma-Aldrich, 1:5000), and anti-goat IgG-Peroxidase (Sigma-Aldrich, 1:5000) for PAR3, aPKC, and PAR6, respectively, as secondary antibodies. The membranes were developed using SuperSignal West Pico or Femto Chemiluminescent Substrate (ThermoFisher Scientific). The detection was done by an ImageQuant LAS4010 system (GE Healthcare). The images were analyzed by ImageQuant TL toolbox version 8.1 following the company’s instructions and quantified by ImageStudio Lite (LI-COR, Lincoln, NE, U.S.A.).
EM Image Acquisition
A volume of glutaraldehyde (Sigma-Aldrich) corresponding to a final concentration of 0.075% was added to the protein samples followed by incubation overnight at 4 °C before grid preparation. Negative staining samples were prepared using the sandwich carbon method with home-made carbon film and uranyl formate or uranyl acetate (2%) [41]. The images were taken in a Tecnai T12 electron microscope (FEI, Eindhoven, The Netherlands) with a Multiscan 794 CCD camera (Gatan, Pleasanton, U.S.A.) operated at 120 kV at a nominal magnification of 52,000 × , which corresponded to an apparent magnification of 63,160x. The pixel size on the specimen level was 3.8 Å/pixel.
EM Image Processing
The particles on the images were selected manually. Determination of defocus and astigmatism of the EM images was done by fitting contrast transfer function (CTF) curves to the power spectra of the images [42]. The particle images were extracted, corrected for CTF-effects, and merged. The data set characteristics are summarized in Table S3. The classification and averaging of particles followed standard methods [43] and were performed in the statistical framework R [44] with 3–10 rounds of particle alignment followed by principal component analysis and unbiased classification using hierarchical ascendant and k-means classification (Table S3).
PKC Kinase Activity
The activity of purified dimeric aPKC–PAR6 complex was tested using the PKC Kinase Activity Assay Kit (Abcam, Cambridge, U.K.) with a protein concentration dilution series of 5 ng, 10 ng, and 20 ng of the purified protein complex following the manufacturer’s instructions, and the standard error of the mean (SEM) was used for visualization (n = 3 replicates). As positive control, 36 ng of control protein (Abcam) was used. A multimode plate reader (EnSpire, PerkinElmer, Waltham, MA, U.S.A.) was used to measure the reaction signal.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
We thank S. N. Stubbe and G. Ayoubi for excellent technical assistance.
Author Contributions
B.S. and M.M.G. designed and directed the project. L.T.M.L. carried out most of the experiments with support from S.D. and B.S. L.T.M.L, M.M.G., and B.S. analyzed and interpreted the data. L.T.M.L., J.R.N., M.M.G., and B.S. drafted the manuscript. M.M.G and B.S. revised the manuscript.
Funding
Open Access funding enabled and organized by Projekt DEAL. This work has been supported by the Danish Council for Independent Research and the Fabrikant Einar Willumsens Mindelegat to BS. MMG acknowledges support by the Lundbeck Foundation’s Fellowship program and the Sapere Aude Program of the Danish Council for Independent Research. JRN is supported by “Henny og Sophie Clausen og møbelarkitekt Aksel Clausens Fond.” The Center for Stochastic Geometry and Advanced Bioimaging is supported by the Villum Foundation.
Data Availability
Additional data are given in the Supporting Material.
Code Availability
Not applicable.
Declarations
Conflict of interest
The authors declare no conflicts of interest or competing interests related to the manuscript. No human participants or animals were involved in the study. All authors approved the final manuscript.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Dotti CG, Sullivan CA, Banker GA. The establishment of polarity by hippocampal neurons in culture. Journal of Neuroscience. 1988;8:1454–1468. doi: 10.1523/JNEUROSCI.08-04-01454.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Krummel MF, Macara I. Maintenance and modulation of T cell polarity. Nature Immunology. 2006;7:1143–1149. doi: 10.1038/ni1404. [DOI] [PubMed] [Google Scholar]
- 3.Wallenfang MR, Seydoux G. Polarization of the anterior-posterior axis of C. elegans is a microtubule-directed process. Nature. 2000;408:89–92. doi: 10.1038/35040562. [DOI] [PubMed] [Google Scholar]
- 4.Thompson, B. J. (2021). Par-3 family proteins in cell polarity & adhesion. FEBS J. [DOI] [PMC free article] [PubMed]
- 5.Harris TJC. Protein clustering for cell polarity: Par-3 as a paradigm. F1000Res. 2017;6:1620. doi: 10.12688/f1000research.11976.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Holly RW, Jones K, Prehoda KE. A conserved PDZ-binding motif in aPKC interacts with par-3 and mediates cortical polarity. Current Biology. 2020;30(893–898):e895. doi: 10.1016/j.cub.2019.12.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Martin E, Girardello R, Dittmar G, Ludwig A. New insights into the organization and regulation of the apical polarity network in mammalian epithelial cells. FEBS Journal. 2021;288:7073–7095. doi: 10.1111/febs.15710. [DOI] [PubMed] [Google Scholar]
- 8.Kemphues KJ, Priess JR, Morton DG, Cheng NS. Identification of genes required for cytoplasmic localization in early C. elegans embryos. Cell. 1988;52:311–320. doi: 10.1016/S0092-8674(88)80024-2. [DOI] [PubMed] [Google Scholar]
- 9.Joberty G, Petersen C, Gao L, Macara IG. The cell-polarity protein Par6 links Par3 and atypical protein kinase C to Cdc42. Nature Cell Biology. 2000;2:531–539. doi: 10.1038/35019573. [DOI] [PubMed] [Google Scholar]
- 10.Lin D, Edwards AS, Fawcett JP, Mbamalu G, Scott JD, et al. A mammalian PAR-3-PAR-6 complex implicated in Cdc42/Rac1 and aPKC signalling and cell polarity. Nature Cell Biology. 2000;2:540–547. doi: 10.1038/35019582. [DOI] [PubMed] [Google Scholar]
- 11.Rodriguez-Boulan E, Macara IG. Organization and execution of the epithelial polarity programme. Nature Reviews Molecular Cell Biology. 2014;15:225–242. doi: 10.1038/nrm3775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shi SH, Jan LY, Jan YN. Hippocampal neuronal polarity specified by spatially localized mPar3/mPar6 and PI 3-kinase activity. Cell. 2003;112:63–75. doi: 10.1016/S0092-8674(02)01249-7. [DOI] [PubMed] [Google Scholar]
- 13.Tahirovic S, Bradke F. Neuronal polarity. Cold Spring Harbor Perspectives in Biology. 2009;1:a001644. doi: 10.1101/cshperspect.a001644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu Z, Yang Y, Gu A, Xu J, Mao Y, et al. Par complex cluster formation mediated by phase separation. Nature Communications. 2020;11:2266. doi: 10.1038/s41467-020-16135-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Atashrazm F, Ellis S. The polarity protein PARD3 and cancer. Oncogene. 2021;40:4245–4262. doi: 10.1038/s41388-021-01813-6. [DOI] [PubMed] [Google Scholar]
- 16.Suzuki A, Yamanaka T, Hirose T, Manabe N, Mizuno K, et al. Atypical protein kinase C is involved in the evolutionarily conserved par protein complex and plays a critical role in establishing epithelia-specific junctional structures. Journal of Cell Biology. 2001;152:1183–1196. doi: 10.1083/jcb.152.6.1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hirano Y, Yoshinaga S, Takeya R, Suzuki NN, Horiuchi M, et al. Structure of a cell polarity regulator, a complex between atypical PKC and Par6 PB1 domains. Journal of Biological Chemistry. 2005;280:9653–9661. doi: 10.1074/jbc.M409823200. [DOI] [PubMed] [Google Scholar]
- 18.Renschler FA, Bruekner SR, Salomon PL, Mukherjee A, Kullmann L, et al. Structural basis for the interaction between the cell polarity proteins Par3 and Par6. Sci Signal. 2018 doi: 10.1126/scisignal.aam9899. [DOI] [PubMed] [Google Scholar]
- 19.Itoh M, Sasaki H, Furuse M, Ozaki H, Kita T, et al. Junctional adhesion molecule (JAM) binds to PAR-3: A possible mechanism for the recruitment of PAR-3 to tight junctions. Journal of Cell Biology. 2001;154:491–497. doi: 10.1083/jcb.200103047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ebnet K, Suzuki A, Horikoshi Y, Hirose T, Brickwedde MZ, M. K.,, et al. The cell polarity protein ASIP/PAR-3 directly associates with junctional adhesion molecule (JAM) EMBO Journal. 2001;20:3738–3748. doi: 10.1093/emboj/20.14.3738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chan JR, Jolicoeur C, Yamauchi J, Elliott J, Fawcett JP, et al. The polarity protein Par-3 directly interacts with p75NTR to regulate myelination. Science. 2006;314:832–836. doi: 10.1126/science.1134069. [DOI] [PubMed] [Google Scholar]
- 22.Chen S, Chen J, Shi H, Wei M, Castaneda-Castellanos DR, et al. Regulation of microtubule stability and organization by mammalian Par3 in specifying neuronal polarity. Developmental Cell. 2013;24:26–40. doi: 10.1016/j.devcel.2012.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Arimura N, Kaibuchi K. Neuronal polarity: From extracellular signals to intracellular mechanisms. Nature Reviews Neuroscience. 2007;8:194–205. doi: 10.1038/nrn2056. [DOI] [PubMed] [Google Scholar]
- 24.Nagai-Tamai Y, Mizuno K, Hirose T, Suzuki A, Ohno S. Regulated protein-protein interaction between aPKC and PAR-3 plays an essential role in the polarization of epithelial cells. Genes to Cells. 2002;7:1161–1171. doi: 10.1046/j.1365-2443.2002.00590.x. [DOI] [PubMed] [Google Scholar]
- 25.Mizuno K, Suzuki A, Hirose T, Kitamura K, Kutsuzawa K, et al. Self-association of PAR-3-mediated by the conserved N-terminal domain contributes to the development of epithelial tight junctions. Journal of Biological Chemistry. 2003;278:31240–31250. doi: 10.1074/jbc.M303593200. [DOI] [PubMed] [Google Scholar]
- 26.Nance J, Zallen JA. Elaborating polarity: PAR proteins and the cytoskeleton. Development. 2011;138:799–809. doi: 10.1242/dev.053538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McCaffrey LM, Macara IG. Signaling pathways in cell polarity. Cold Spring Harbor Perspectives in Biology. 2012;4:a009654. doi: 10.1101/cshperspect.a009654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Feng W, Wu H, Chan LN, Zhang M. The Par-3 NTD adopts a PB1-like structure required for Par-3 oligomerization and membrane localization. EMBO Journal. 2007;26:2786–2796. doi: 10.1038/sj.emboj.7601702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang Y, Wang W, Chen J, Zhang K, Gao F, et al. Structural insights into the intrinsic self-assembly of Par-3 N-terminal domain. Structure. 2013;21:997–1006. doi: 10.1016/j.str.2013.04.004. [DOI] [PubMed] [Google Scholar]
- 30.Lin L, Yuan J, Sander B, Golas MM. In vitro differentiation of human neural progenitor cells into striatal GABAergic neurons. Stem Cells Translational Medicine. 2015;4:775–788. doi: 10.5966/sctm.2014-0083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Golas MM, Sander B, Bessonov S, Grote M, Wolf E, et al. 3D cryo-EM structure of an active step I spliceosome and localization of its catalytic core. Molecular Cell. 2010;40:927–938. doi: 10.1016/j.molcel.2010.11.023. [DOI] [PubMed] [Google Scholar]
- 32.Erickson HP. Size and shape of protein molecules at the nanometer level determined by sedimentation, gel filtration, and electron microscopy. Biol Proced Online. 2009;11:32–51. doi: 10.1007/s12575-009-9008-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Siegel LM, Monty KJ. Determination of molecular weights and frictional ratios of proteins in impure systems by use of gel filtration and density gradient centrifugation. Application to crude preparations of sulfite and hydroxylamine reductases. Biochimica et Biophysica Acta. 1966;112:346–362. doi: 10.1016/0926-6585(66)90333-5. [DOI] [PubMed] [Google Scholar]
- 34.Golas MM, Jayaprakash S, Le LTM, Zhao Z, Heras Huertas V, et al. Modulating the expression strength of the baculovirus/insect cell expression system: a toolbox applied to the human tumor suppressor SMARCB1/SNF5. Molecular Biotechnology. 2018;60:820–832. doi: 10.1007/s12033-018-0107-2. [DOI] [PubMed] [Google Scholar]
- 35.Wang C, Shang Y, Yu J, Zhang M. Substrate recognition mechanism of atypical protein kinase Cs revealed by the structure of PKCiota in complex with a substrate peptide from Par-3. Structure. 2012;20:791–801. doi: 10.1016/j.str.2012.02.022. [DOI] [PubMed] [Google Scholar]
- 36.Inui K, Zhao Z, Yuan J, Jayaprakash S, Le LTM, et al. Stepwise assembly of functional C-terminal REST/NRSF transcriptional repressor complexes as a drug target. Protein Science. 2017;26:997–1011. doi: 10.1002/pro.3142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Berger I, Fitzgerald DJ, Richmond TJ. Baculovirus expression system for heterologous multiprotein complexes. Nature Biotechnology. 2004;22:1583–1587. doi: 10.1038/nbt1036. [DOI] [PubMed] [Google Scholar]
- 38.Fitzgerald DJ, Berger P, Schaffitzel C, Yamada K, Richmond TJ, et al. Protein complex expression by using multigene baculoviral vectors. Nature Methods. 2006;3:1021–1032. doi: 10.1038/nmeth983. [DOI] [PubMed] [Google Scholar]
- 39.Le LTM, Nyengaard JR, Golas MM, Sander B. Vectors for expression of signal peptide-dependent proteins in baculovirus/insect cell systems and their application to expression and purification of the high-affinity immunoglobulin gamma Fc receptor I in complex with Its Gamma Chain. Molecular Biotechnology. 2018;60:31–40. doi: 10.1007/s12033-017-0041-8. [DOI] [PubMed] [Google Scholar]
- 40.Lin L, Voronovsky A, Raabe M, Urlaub H, Sander B, et al. Dual tagging as an approach to isolate endogenous chromatin remodeling complexes from Saccharomyces cerevisiae. Biochm Biophys Acta. 2015;1854:198–208. doi: 10.1016/j.bbapap.2014.11.009. [DOI] [PubMed] [Google Scholar]
- 41.Golas MM, Sander B, Will CL, Luhrmann R, Stark H. Molecular architecture of the multiprotein splicing factor SF3b. Science. 2003;300:980–984. doi: 10.1126/science.1084155. [DOI] [PubMed] [Google Scholar]
- 42.Sander B, Golas MM, Stark H. Automatic CTF correction for single particles based upon multivariate statistical analysis of individual power spectra. Journal of Structural Biology. 2003;142:392–401. doi: 10.1016/S1047-8477(03)00072-8. [DOI] [PubMed] [Google Scholar]
- 43.van Heel M, Frank J. Use of multivariate statistics in analysing the images of biological macromolecules. Ultramicroscopy. 1981;6:187–194. doi: 10.1016/0304-3991(81)90059-0. [DOI] [PubMed] [Google Scholar]
- 44.R-Core-Team (2021) R: A language and environment for statistical computing, R Foundation for Statistical Computing, Vienna, Austria.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Additional data are given in the Supporting Material.
Not applicable.