Abstract
EGR1, a short-lived transcription factor, regulates several biological processes, including cell proliferation and tumor progression. Whether and how EGR1 is regulated by Cullin-RING ligases (CRLs) remains elusive. Here, we report that MLN4924, a small molecule inhibitor of neddylation, causes EGR1 accumulation by inactivating SCFFBXW7 (CRL1), which is a new E3 ligase for EGR1. Specifically, FBXW7 binds to EGR1 via its consensus binding motif/degron, whereas cancer-derived FBXW7 mutants showed a much reduced EGR1 binding. SiRNA-mediated FBXW7 knockdown caused EGR1 accumulation, whereas FBXW7 overexpression reduced EGR1 levels. Likewise, FBXW7 knockdown significantly extended EGR1 protein half-life, while FBXW7 overexpression promotes polyubiquitylation of wild-type EGR1, but not EGR1-S2A mutant with the binding site abrogated. GSK3β kinase is required for the FBXW7-EGR1 binding, and for enhanced EGR1 degradation by wild type FBXW7, but not by FBXW7 mutants. Likewise, GSK3β knockdown or treatment with GSK3β inhibitor significantly increased the EGR1 levels and extended EGR1 protein half-life, while reducing EGR1 polyubiquitylation. Hypoxia exposure reduces the EGR1 levels via enhancing the FBXW7-EGR1 binding, and FBXW7-induced EGR1 polyubiquitylation. Biologically, EGR1 knockdown suppressed cancer cell growth, whereas growth stimulation by FBXW7 knockdown is partially rescued by EGR1 knockdown. Thus, EGR1 is a new substrate of the GSK3β-FBXW7 axis, and the FBXW7-EGR1 axis coordinately regulates growth of cancer cells.
Keywords: EGR1, FBXW7, Cullin-RING ligase (CRL), Ubiquitylation, GSK3β
Abbreviations: CPD, Cdc4 phosphodegron; CHX, Cycloheximide; CRL, Cullin-RING ligase; Cul, Cullin; DMEM, Dulbecco’s Modified Eagle Medium; DMSO, Dimethyl sulfoxide; EGR1, Early growth response-1; FBXW7, F-box and WD repeat domain containing 7; GSK3β, Glycogen synthase kinase-3β; HIF1α, Hypoxia Inducible Factor-1α; IB, Immunoblotting; IP, Immunoprecipitation; NAE, NEDD8-activating enzyme; SCF, SKP1-Cullin1-F-box protein; Ub, Ubiquitin; UPS, Ubiquitin-proteasome system; WT, Wild type
Introduction
Early growth response 1 (EGR1) is a transcription factor and a member of the zinc finger EGR family [1]. EGR1 contains a highly conserved DNA-binding domain that binds to a consensus motif GCG (G/T) GGGCG [2] to transactivate the expression of down-stream target genes, including P53, PTEN, TGFβ1, fibronectin [3,4], BAX [5], NAG1 [6], Cyclin D1 [7], CDKL1 [8], Leptin [9], MMP1 [10], HIF1α [11] and GGPPS (geranylgeranyl diphosphate synthase) [12]. On the other hand, EGR1 itself is stress-responsive protein and subjected to upregulation by growth factors, inflammation and radiation among the others [13]. For example, EGR1 was transcriptionally upregulated by the MAPK-ERK pathway [14], [15], [16], and the JNK-c-Jun pathway [5], but downregulated by the loss of E-cadherin, leading to Pten inhibition and PI3K/AKT activation [17].
EGR1 was reported to regulate a variety of cellular processes, including proliferation, differentiation, angiogenesis, apoptosis, tumor invasion and metastasis [13]. In general, EGR1 acts as a tumor suppressor, since loss of the EGR1 often occurs in human cancer [3,18], and EGR1 upregulate few typical tumor suppressor proteins such as PTEN and p53 [3,4]. On the other hand, EGR1 was found to act as an oncogenic protein in prostate cancer [19]. Thus, the function of EGR1 in human cancer could be context dependent. While EGR1 is mainly regulated at the transcriptional levels [1], EGR1 was also found to be regulated at the posttranslational levels, such as by phosphorylation [20], [21], [22], [23], acetylation [24], sumoylation [22], and possibly by ubiquitylation [25,26]. Whether and how EGR1 is subjected to ubiquitylation and degradation by SCF E3 ligase is totally unknown.
SCF (SKP1-Cullin1-F-box protein) ubiquitin ligase, also known as Cullin-RING ligase-1 (CRL1), is a member of the largest E3 ligases, which regulate many biological processes by promoting ubiquitylation and degradation of a variety of key signaling proteins [27,28]. SCF is a multiple component complex, consisting of an adaptor protein SKP1; a scaffold protein Cullin-1 (Cul-1); a RING protein, RBX1 or RBX2, which binds to ubiquitin-loaded E2, required for core enzymatic activity; and a F-box protein that is substrate-recognizing subunit, and bind to a phosphorylated substrate [29]. In mammalian cells, there are 69 F-box proteins [30]. Among them, FBXW7 (F-box and WD repeat domain-containing 7) is one of the best studied. FBXW7 is characterized as a typical tumor suppressor, given the fact that a) FBXW7 promotes ubiquitylation and degradation of many oncogenic proteins such as c-MYC, NOTCH1, mTOR, MCL-1, c-JUN, and Cyclin-E [31,32]; b) FBXW7 is frequently mutated, deleted or downregulated in human cancer, which is also associated with poor prognosis of cancer patients [33]; and c) Studies with genetically engineered mouse models showed that Fbxw7+/− mice have increased susceptibility to radiation-induced tumorigenesis with Fbxw7 acting as a p53-dependent, haploinsufficient tumor suppressor gene [34], and conditional deletion of Fbxw7 in the T cell lineage caused thymic hyperplasia and eventually developed thymic lymphoma due to c-Myc accumulation [35]. However, under certain conditions, FBXW7 also promotes ubiquitylation and degradation of some tumor suppressors. The examples include Nf1 during mouse embryogenesis [36], p53 upon DNA damage [37]. Furthermore, a recent study showed that in B-cell lymphoma, FBXW7 acts as an oncogenic protein by promoting ubiquitylation and degradation of tumor suppressor KMT2D [38]. Thus, FBXW7 has diverse activity in a manner dependent of cellular context.
In this study, we report that transcription factor EGR1 is a new substrate of SCFFBXW7. Specifically, FBXW7 binds to EGR1 via its consensus binding motif to promote EGR1 polyubiquitylation, thus shortening EGR1 protein half-life, whereas FBXW7 knockdown caused EGR1 accumulation by extending its protein half-life. GSK3β kinase is required for FBXW7-mediated EGR1 ubiquitylation and degradation. Furthermore, short term hypoxia exposure reduces EGR1 protein levels via enhancing FBXW7-EGR1 binding and FBXW7-mediated EGR1 polyubiquitylation. Finally, EGR1 knockdown inhibits growth of cancer cells and abrogated growth stimulation by FBXW7 knockdown. Thus, the GSK3β-FBXW7 axis controls the levels of EGR1, whereas the FBXW7-EGR1 axis coordinately regulates cancer cell growth.
Materials and methods
Cell culture
HEK293 cells, lung cancer cell lines H1299 and A549, mouse MEF cells, were maintained in Dulbecco's Modified Eagle's Medium (DMEM) containing 10% (v/v) fetal bovine serum (FBS) and 1% penicillin-streptomycin. Lung cancer cell lines H23, H125, H358, H460, H520, H1792, H2170, human prostate cancer cell lines Du145, human colorectal cancer DLD1 cells were maintained in RPMI 1640 Medium with 10% FBS and 1% penicillin-streptomycin. Cells were incubated at 37°C in a humidified incubator with 5% CO2.
Antibodies, chemical reagents, plasmids, and siRNA oligoes
Antibodies were used in this work: EGR1 (Cell Signaling Technology, #4153), c-MYC (Cell Signaling Technology, #5605), Cul-1 (Proteintech, 12895-1-AP), Cul-2 (Abcam, ab166917), Cul-3 (Cell Signaling Technology, #2759), Cul-4A (Cell Signaling Technology, #2699), Cul-4B (Proteintech, 12916-1-AP), Cul-5 (Abcam, ab184177), FBXW7 (Bethyl, A301-720A), Notch1 (Cell Signaling Technology, #4147), FLAG (Sigma-Aldrich, F1804), HA (Sigma-Aldrich, A2095), β-Actin (Santa Cruz, sc-47778), GSK3α/β (Cell Signaling Technology, #5676), p-GSK3β(Ser9) (Cell Signaling Technology, #5558), p-β-catenin(Ser33/37/Thr41) (Cell Signaling Technology, #9561), HIF1α (Novus, NB100-479).
Cycloheximide (CHX) and metformin were purchased from Sigma-Aldrich. MG132, GSK3i-IX and Cell Counting Kit-8 (CCK8) were purchased from MedChemExpress (MCE).
The plasmid constructs expressing FLAG-tagged FBXW4, FBXW5, FBXW7, FBXW8, FBXL5, β-TrCP1, FBXO4, and HA-tagged FBXW7 were used and described previously [39]. GSK3β, FBXW7 R479Q and R505C constructs were provided by Dr. Wenyi Wei. EGR1 plasmid was provided by Dr. Eugene Chen. EGR1 mutants were generated using the QuikChange Site-Directed mutagenesis kit (Agilent Technologies, #200522) by PCR, according to the manufacturer's instructions.
The sequence of siRNA oligos used in this study are as follows:
siCUL-1: 5’-GCUCUACACUCAUGUUUAU-3’;
siCUL-2: 5’-GCCCUUACGUCAGUUGUAAAUUACA-3’;
siCUL-3: 5’-GAGUGUAUGAGUUCCUAUU-3’;
siCUL-4A: 5’-GAACUUCCGAGACAGACCU-3’;
siCUL-4B: 5’-CACCGUCUCUAGCUUUGCUAA-3’;
siCUL-5: 5’-CUGGAGGACUUGAUACCGGAA-3’;
siFBXW7-1: 5’-ACAGGACAGUGUUUACAAA-3’;
siFBXW7-2: 5’-UGAUACAUCAAUCCGUGUUUG-3’;
siGSK3β-1: 5’-CAUGAAAGUUAGCAGAGACAA-3’;
siGSK3β-2: 5’-AGCAAAUCAGAGAAAUGAAC-3’;
siEGR1-1: 5’-CAUCUCUCUGAACAACGAGAA-3’;
siEGR1-2: 5’-GCCAAGCAAACCAAUGGUGAU-3’.
Transfection and lentiviral infection
Cells were transfected with various plasmids or siRNA oligonucleotides using Lipofectamine 2000, according to the manufacturer's instructions, followed by various indicated assays 48h or 72h post transfection. For lentivirus-based shRNAs targeting, cells were infected with the lentivirus for 48 h along with 8 μg/mL polybrene for stable cell lines, followed by various indicated assays.
Immunoprecipitation and immunoblotting
Cells transfected with plasmids or siRNAs were harvested and lysed for immunoprecipitation (IP) and immunoblotting (IB). For direct IB, cells were lysed in lysis buffer (50 mM Tris pH 7.5, 0.15 M NaCl, 1% NP-40, 0.1% SDS, 0.5% sodium deoxycholate, 50 mM NaF, 1 mM EDTA, 1 mM DTT, 1 mM Na3VO4) containing protease inhibitors and phosphatase inhibitors (Roche). The protein concentrations of the cell lysates were measured and same amounts of cell lysates were loaded on SDS-PAGE polyacrylamide gels, transferred to a PVDF membrane after electrophoresis. After blocking with 5% (w/v) milk, the membrane was stained with the indicated antibodies.
For immunoprecipitation, cells after various treatments (and in some cases, pretreat cells with 20 μM MG132 for 6h before harvesting) were harvested and cell lysates were incubated with FLAG-beads in a rotating incubator overnight at 4°C, or the appropriate antibody followed by 3-4 h incubation with Protein G Sepharose beads (GE Healthcare) at 4°C. Then the immunoprecipitates were washed five times with lysis buffer before subjected to IB.
Quantitative RT-PCR
To detect mRNA expression, total RNA was extracted using Trizol reagents (#15596018, Invitrogen) and transcribed into cDNA using PrimeScript™ RT reagent kit (RR037A, TaKaRa), followed by the instructions of TB Green Premix Ex TaqTM (RR420A, TaKaRa). The β-Actin was used as a loading control. The ratio of the relative expression of target genes was calculated by using 2−ΔΔCT method. Each assay was repeated for three times. The primers for EGR1 and β-Actin were purchased from Tsingke Biotechnology, and the sequences of primers are as follows:
EGR1: Forward: 5’- CACCTGACCGCAGAGTCTTTT-3’,
EGR1: Reverse: 5’-CGGCCAGTATAGGTGATGGG-3’,
β-Actin: Forward: 5’-TCACCCACACTGTGCCCATCTAC-3’,
β-Actin: Reverse: 5’- GGAACCGCTCATTGCCAATG-3’.
Half-life analysis
After transfection with siRNAs, plasmids or treated with small molecule inhibitors, cells were treated with 100 μg/ml cycloheximide (CHX, Sigma-Aldrich) for indicated time periods. Cells were then harvested and lysed for IB.
The in vivo ubiquitylation assay
HEK293 cells were transfected with the mentioned plasmids or siRNA oligos for 48 hrs, and treated with MG132 (20 μM) for 6 hrs before harvesting. Cells were lysed in a lysis buffer (6 M guanidinium-HCl, 10 mM Tris-HCl pH 8.0, 0.1 M Na2HPO4/NaH2PO4, 5mM Imidazole, 10 mM β-mercaptoethanol) and sonicated. Cell lysates were then incubated with Ni-NTA agarose beads (Qiagen) for 4-5 h at room temperature (RT). Beads were washed once with each of buffer A (8 M urea, 0.1M Na2HPO4/NaH2PO4, 10 mM Tris-HCl pH 8.0 and 10mM β-mercaptoethanol), buffer B (8 M urea, 10 mM Tris-HCl pH 6.3, 0.1 M Na2HPO4/NaH2PO4 and 10 mM β-mercaptoethanol) with 0.2% Triton X-100 and buffer B with 0.1% Triton X-100. Proteins were eluted from beads with elution buffer (200 mM imidazole, 0.15 M Tris-HCl pH 6.8, 30% glycerol, 0.72 M β-mecaptoethanol, 5% SDS) and then were analyzed by IB.
Hypoxia exposure
Cells in normal culture condition were transfected with indicated plasmids for ∼48 hours to reach ∼80% confluence prior to hypoxia exposure. Hypoxia exposure was performed at 37°C in a cell incubator filled with 1% O2, 5% CO2 and N2 for 1 or 4 hours, respectively. Cells were then harvested for various assays.
CCK8 assay
Cells were transfected with indicated siRNAs for 48 hrs, then seeded onto 96-well plates (2 × 103 per well) in triplicate, cultured in medium containing 2.5% FBS for indicated time periods, followed by the CCK8 assay, according to the manufacturer's instructions (MedChemExpress). The OD value for each well was read at wavelength 450 nm on a microplate reader.
Statistical analysis
The two-tailed Student's t-test or one-way ANOVA were performed with the data from three independent replicates. P < 0.05 was considered statistically significant.
Results
EGR1 is a putative substrate of SCF/CRL1 via FBXW7 binding
Cullin-RING ligases (CRLs) is the largest family of E3 ubiquitin ligases, responsible for ∼20% of proteins doomed to be degraded by the UPS in eukaryotic cells [40]. CRLs are actively involved in ubiquitylation and degradation of many short-lived proteins, particularly transcription factors, such as c-MYC [41], Notch1 [42], and c-JUN [43,44]. To determine whether transcription factor EGR1 is also subjected to CRL regulation, we first treated three lines of lung cancer cells with MLN4924, a small-molecule inhibitor of NEDD8-activating enzyme (NAE), leading to inhibition of cullin neddylation and inactivation of all CRL family members [45]. Interestingly, we found that EGR1, which is generally expressed at low levels in lung cancer cell lines [46], is accumulated, along with c-MYC, serving as a positive control, after MLN4924 treatment in all three lines tested (Fig. 1A), suggesting that EGR1 is likely a substrate of one of CRLs.
Fig. 1.
EGR1 is a putative substrate of SCF/CRL1 via FBXW7 binding. A. EGR1 accumulated after MLN4924 treatment. H23, H520, H1299 cells were treated with DMSO or MLN4924(1μM) for 24 hours, followed by IB with indicated antibodies (Abs). SE: short exposure, LE: long exposure. B. EGR1 accumulated after Cul-1 knockdown. H520 cells were transfected with siRNA oligos of Cullins as indicated, followed by IB with indicated Abs. C. Ectopically expressed FBXW7 binds to EGR1. HEK293 cells were transfected with indicated plasmids, followed by IP with FLAG beads and IB with indicated Abs. F is short for FLAG. D & E. Reduced EGR1 binding by cancer-derived FBXW7 mutants. HEK293 cells (D) were transfected with indicated plasmids, followed by IP with FLAG beads and IB with indicated Abs. H520 cells (E) were transfected with plasmids as indicated, followed by IP with HA Ab and IB with indicated Abs. F. Exogenous FBXW7 binds to wild type EGR1, but not its EGR1-S2A mutant on phosphorylation sites. Endogenous FBXW7 was first knockdown in HEK293 cells, FLAG-FBXW7 and wild type EGR1/mutant EGR1 were then transfected, followed by IP with FLAG beads and IB with indicated Abs. WCE: whole cell extract.
To determine the involving CRLs, we knocked down six cullins individually in H520 and HEK293 cells, and found that EGR1 is accumulated upon cullin-1 knockdown in both cell lines, along with the knockdown of Cul-2 and Cul-4A, but in a cell line dependent manner (Fig. 1B & S1A). We then focused on SCF/CRL1 and determined which F-box protein, a substrate-recognizing subunit of SCF, is responsible for EGR1 degradation. A total of seven plasmids encoding FLAG-tagged F-Box proteins were transfected into HEK293 cells, followed by FLAG-beads immunoprecipitation and immunoblotting for EGR1. The FBXW7 was the only one that pulled down endogenous EGR1 (Fig. 1C). The FBXW7-EGR1 binding was further confirmed in H1299 lung cancer cells (Fig. S1B), but was significantly reduced when examined using two human cancer-derived FBXW7 mutants, FBXW7-R479Q or FBXW7-R505C, with mutations located in the WD40-repeat substrate-binding domain [47] (Figure 1D & 1E).
It is well-known that FBXW7 binding to various phosphorylated substrates through their Cdc4 phosphodegron (CPD, L/I/PT/SPXXS/T) for subsequent polyubiquitylation via K48 linkage and proteasomal degradation [48,49]. We searched EGR1 amino acid sequence and indeed identified a typical CPD motif (175PSPAAS180), which is completely evolutionarily conserved in mammals (Fig. S1C). Since phosphorylation of two serine/threonine residues on CPD motif are known to be required for FBXW7 binding and subsequent ubiquitylation, we constructed a phosphorylation-dead mutant EGR1-S2A, carrying double S->A mutations (176S->A and 180S->A) on its CPD motif (Fig. S1C). Indeed, this EGR1-S2A mutant almost completely lost FBXW7 binding (Fig. 1F), confirming the requirement of these two serine residues in mediating the FBXW7-EGR1 binding. Finally, we determined potential correlation of protein levels between FBXW7 and EGR1 in 8 lung cancer cell lines, and found that although EGR1 levels are low in most lung cancer lines, a general tendency of inverse correlation between two proteins can be observed (Fig. S1D). Taken together, these results highly suggest that EGR1 is a new substrate of SCF/CRL1 E3, mediated by FBXW7 binding in a CPD motif dependent manner.
FBXW7 negatively regulates EGR1 protein levels
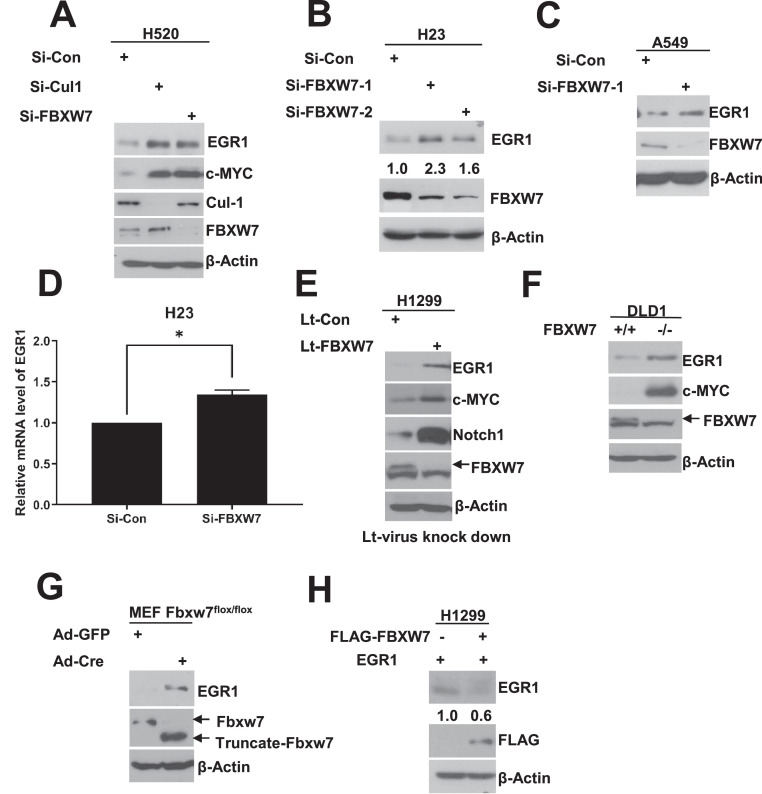
To further confirm that EGR1 is indeed a new substrate of SCFFBXW7, we next determined whether manipulation of Cullin-1 or FBXW7 would affect cellular levels of EGR1. Indeed, in three lung cancer cell lines, siRNA-based knockdown of either Cullin-1 (Fig. 2A) or FBXW7 (Fig. 2A-2C) caused accumulation of EGR1 protein levels, whereas FBXW7 knockdown had minor effect on EGR1 mRNA levels (Fig. 2D, S2A & S2B). Furthermore, the lentivirus-mediated FBXW7 knockdown also caused EGR1 accumulation in lung cancer cells (Fig. 2E), so did in DLD1 colon cancer cells with FBXW7 deletion (Fig. 2F), as well as in mouse embryonic fibroblast upon Ad-Cre infection to delete mouse Fbxw7 (Fig. 2G). Finally, we ectopically expressed FBXW7 and found that it reduced the EGR1 protein level in lung cancer cells (Fig. 2H). Collectively, it is a general phenomenon that EGR1 is indeed subjected to negative regulation by FBXW7.
Fig. 2.
FBXW7 negatively regulates EGR1 protein levels. A. EGR1 accumulated after Cul-1 or FBXW7 knockdown in H520 cells. H520 cells were transfected with indicated siRNA oligos, followed by IB with anti-EGR1 and c-MYC. B&C. FBXW7 knockdown increases the endogenous protein levels of EGR1. H23 cells (B) or A549 cells (C) were transfected as indicated and then subjected to IB with indicated Abs. D. H23 cells were transfected with siRNA targeting FBXW7 for 48 hours and then subjected to qRT-PCR. Shown is mean ± SEM (n=3). *p < 0.05. E. EGR1 accumulated after FBXW7 knockdown. H1299 cells were infected with Lt-FBXW7 or Lt-Con, followed by IB with indicated Abs. F. EGR1 was accumulated in DLD1 FBXW7-/- cells, in contrast to DLD1 FBXW7+/+ cells. G. EGR1 accumulated in Fbxw7-KO MEF cells. Fbxw7fl/fl MEF cells were infected with adenovirus encoding Cre-recombinase (Ad-Cre) or Ad-GFP control for 72 hours, then followed by IB with indicated Abs. H. EGR1 reduced after FBXW7 overexpression. H1299 cells were transfected with plasmids encoding both FBXW7 and EGR1, followed by IB with indicated Abs.
FBXW7 shortens EGR1 protein half-life and promotes EGR1 polyubiquitylation
Having established that FBXW7 negatively regulates EGR1 protein levels, we next determined whether FBXW7 controls EGR1 protein half-life. Three different lung cancer cell lines, along with HEK293 cells, were transfected with either siCon/lenti-Con (as a negative control) or siFBXW7/Lenti-FBXW7 (to knockdown FBXW7), followed by cyclohexamide (CHX) treatment to block protein synthesis for various time points. In all cases, FBXW7 knockdown or depletion remarkably extended the protein half-life of EGR1, an observation also seen in c-MYC, a known substrate of FBXW7, serving as a positive control (Fig. 3A-3D). Thus, FBXW7 indeed affects the stability of EGR1.
Fig. 3.
FBXW7 shortens EGR1 protein half-life and promotes EGR1 polyubiquitylation. A-D. FBXW7 knockdown extends the half-life of EGR1. H23 cells (A), H520 cells (B), and HEK293 cells (C) were transfected with Si-FBXW7 or Si-Con for 72 hrs, whereas H1299 cells (D) were infected with Lt-FBXW7 or Lt-Con. Cells were then treated with cyclohexamide (CHX) for indicated time periods, followed by IB with indicated Abs. The band density was qualified with ImageJ and the decay curves were generated and shown on the right. E. FBXW7 promotes polyubiquitylation of EGR1. HEK293 cells were first transfected with Si-FBXW7 for 24 hours and then transfected indicated plasmid for 48 hours,Cells were treated with 20μM MG132 for 6 h before harvesting, followed by His tag pull-down and then IB with indicated Abs. F. Endogenous FBXW7 promotes polyubiquitylation of EGR1-WT, but not EGR1-S2A. Wild type EGR1 or EGR1-S2A mutants were transfected into HEK293 cells, along with or without His-Ub as indicated, Cells were treated with 20μM MG132 for 6 h before harvesting, followed by His tag pull-down and IB with indicated Abs.
We further determined whether FBXW7 would promote the polyubiquitylation of EGR1 for enhanced degradation, using a classic in vivo ubiquitylation assay. We found that ectopically expressed FBXW7 indeed promoted the polyubiquitylation of ectopically expressed EGR1 in HEK293 cells (Fig. 3E), whereas endogenous FBXW7 only promoted polyubiquitylation of the wild-type EGR1, but not the mutant EGR1-S2A, which failed to bind to FBXW7 (Fig. 3F). Taken together, we concluded that FBXW7 acts as an E3 ubiquitin ligase that promotes ubiquitylation of EGR1 in a binding site dependent manner.
GSK3β is required for EGR1 ubiquitylation and degradation
It is well-established that it is prerequisite that a FBXW7 substrate has to be phosphorylated on the serine/threonine residues on the CPD motif prior to FBXW7 binding [50]. Given glycogen synthase kinase 3β (GSK3β) is a known kinase responsible for phosphorylating most substrates of FBXW7, including c-MYC [41], c-JUN [43,44] and MCL1 [51], we determined possible involvement of GSK3β in EGR1 degradation. Indeed, we found that the FBXW7-EGR1 binding was significantly inhibited by GSK3β inhibitor GSK3i-IX (Fig. 4A). Co-overexpression of both GSK3β and FBXW7 caused a dose-dependent reduction of endogenous EGR1 (Fig. 4B), which can be fully rescued by the proteasome inhibitor MG132 (Fig. 4B & 4C), indicating the involvement of protein degradation. Furthermore, GSK3β enhanced degradation of endogenous EGR1 by wild-type FBXW7, but not by two FBXW7 mutants (Fig. 4D). Likewise, GSK3β inhibitor treatment significantly increased the protein levels of EGR1 (Fig. 4E & 4F). Note that we used p-β-cateninSer33/37/Thr41, a well-known target of GSK3β [52,53] as a control for drug effectiveness. GSK3i-IX treatment significantly inhibited the level of p-β-cateninSer33/37/Thr41, indicating GSK3i-IX is effective to inhibit GSK3β activity (Fig. 4F). Consistently, the siRNA-based GSK3β knockdown caused accumulation of EGR1 protein (Fig. 4G) and extended EGR1 protein half-life (Fig. 4H, Fig. S3A), so did the GSK3β inhibitor in a few lung cancer cell lines (Fig. 4I & 4J, Fig. S3B&3C). Finally, we found that either GSK3β inhibitor treatment or GSK3β knockdown significantly inhibited polyubiquitylation of EGR1 (Fig. 4K). Taken together, our results showed that GSK3β facilitates the FBXW7-EGR1 binding and subsequent ubiquitylation and degradation of EGR1 by FBXW7, and EGR1 is a new substrate of the GSK3β-FBXW7 axis.
Fig. 4.
GSK3β is required for EGR1 ubiquitylation and degradation. A. The binding between FBXW7 and EGR1 depends on GSK3β activity. H520 cells were treated with GSK3 inhibitor GSK3i-IX or DMSO for 6 hours, followed by IP with anti-FBXW7 and IB with indicated Abs. B. GSK3β and FBXW7 co-overexpression reduces endogenous EGR1 levels in a dose-dependent manner and can be rescued by MG132. H23 cells were transfected as indicated for 48 hours, then treated with MG132/DMSO for 6 hours, followed by IB. C. Reduction of exogenous EGR1 after GSK3β and FBXW7 co-overexpression is rescued by MG132. H1299 cells were transfected as indicated, then treated with MG132/DMSO for 6 hours, followed by IB. D. GSK3β enhanced EGR1 degradation by wild-type FBXW7, but not by FBXW7 mutants. HEK293 cells were transfected with indicated plasmids, followed by IB with indicated Abs. E&F. EGR1 accumulated after inhibition of GSK3β activity. Du145 cells (E) or H23 cells (F) were treated with GSK3i-IX or DMSO for 6 hours, followed by IB with indicated Abs. G. EGR1 accumulated after GSK3β knockdown. H23 cells were treated with indicated siRNAs for 48 hours, followed by IB. H-J. Inactivation of GSK3β extends half-life of EGR1. H23 cells were transfected with Si-GSK3β or Si-Con for 72 hours (H), or H23 cells (I) or H520 cells (J) were pretreated with GSK3i-IX for 2 hours, then treated them with CHX for indicated time periods, followed by IB with indicated Abs. K. GSK3β is responsible for EGR1 phosphorylation-dependent ubiquitylation. HEK293 were transfected as indicated, then treated with MG132 and GSK3i-IX/DMSO for 6 hours, followed by His tag pull-down and then IB with indicated Abs.
The stress conditions that regulate the EGR1 levels
We next determined under what stressed condition that the GSK3β-FBXW7 axis would regulate EGR1 levels. It has been established that dephosphorylation of GSK3β at serine 9 increases its activity, which is then commonly used as an activity marker for GSK3β [54,55]. A previous study showed that the combination of low glucose and metformin treatment would activate GSK3β (as indicated by dephosphorylation of GSK3βSer9) through the PP2A-GSK3β-MCL-1 axis [55]. We therefore used the same stressed condition and found that in H23 lung cancer cells with high level of endogenous EGR1, combination of low glucose and metformin treatment indeed activated GSK3β (as evidence by lack of GSK3βSer9 detection), and caused complete elimination of EGR1, which is completely rescued by MG132 treatment (Fig. 5A).
Fig. 5.
The stress conditions that regulate the EGR1 levels. A. Combination of low glucose and metformin treatment reduces EGR1 levels, which is rescued by MG132. H23 cells were cultured in normal glucose medium (11.1mM) or low glucose medium (1.11mM), and replenished every 6 hours in the absence or presence of metformin (5mM)[55], then treated with MG132/DMSO for 6 hours. Cell lysates were prepared and subjected to IB with indicated Abs. t-GSK3β: total GSK3β. B & C. EGR1 levels fluctuate upon hypoxia in H23 and H1299 cells. H23 cells (B) or H1299 cells (C) were exposed to 1% O2 for indicated periods, cell lysates were prepared and subjected to IB with indicated Abs. D & E. Hypoxia enhances the binding between FBXW7 and EGR1. H23 cells (D) or H1299 cells (E) were transfected with indicated plasmids, then with or without exposure to 1% O2 for 4 hours, followed by FLAG IP and IB. WCE: whole cell extract. F. Hypoxia promotes FBXW7-mediated EGR1 polyubiquitylation. H1299 cells were transfected as indicated, then treated with MG132 for 6 hours and subjected to 1% O2 for 1 or 4 hours, respectively, followed by in vivo polyubiquitylation assay.
Furthermore, we found that short exposure of both H23 and H1299 cells to hypoxia (1% O2) reduced EGR1 levels (Fig. 5B & 5C). Although it had minimal, if any, effect on the levels of FBXW7 or GSK3β, hypoxia increased FBXW7-EGR1 binding (Fig. 5D & 5E), and FBXW7-mediated EGR1 polyubiquitylation (Fig. 5F). Collectively, EGR1 levels are subjected to regulation by a stressed condition that activates GSK3β or by hypoxia.
The FBXW7-EGR1 axis coordinately regulates growth of cancer cells
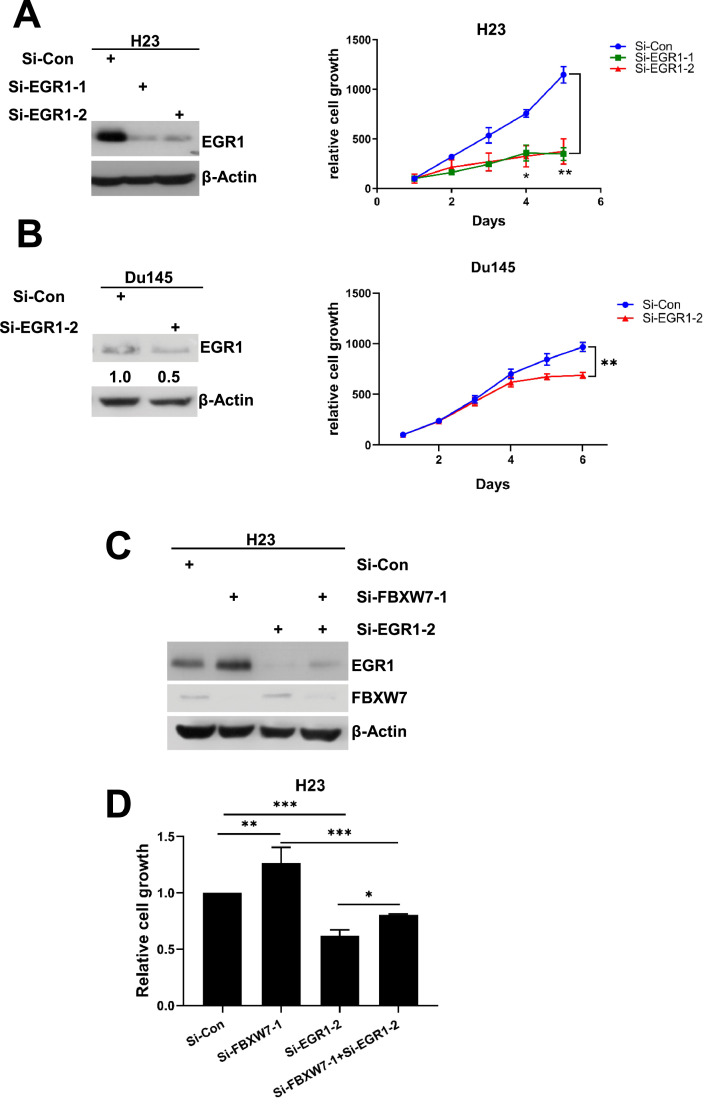
Finally, we determined biological implication of FBXW7 regulation of EGR1. While EGR1 was reported to have tumor suppressive activity in general [13], it has oncogenic activity in prostate cancer [56]. To this end, we silenced EGR1 in both H23 lung cancer cells and Du145 prostate cancer cells and found that EGR1 knockdown significantly inhibited growth of both cancer cell lines (Fig. 6A & 6B). We further determined possible interaction between FBXW7 and EGR1 in H23 lung cancer cells and found that FBXW7 knockdown promoted cell growth, as expected, which can be largely rescued by simultaneous knockdown of EGR1 (Fig. 6C & 6D), indicating FBXW7 regulation of EGR1 has the biological implication, in which both proteins coordinately regulate the growth of cancer cells.
Fig. 6.
The FBXW7-EGR1 axis coordinately regulates growth of cancer cells. A and B. EGR1 knockdown inhibits growth of cancer cells. H23 (A) and Du145 (B) cells were transfected with indicated siRNA oligos targeting EGR1 for 48 hours, one portion of cells were subjected to IB (left panels), whereas the other portion of cells were plated in 96-well plate and cultured for indicated periods of time, then subjected to CCK8 assay for cell growth (right panels). C and D. FBXW7 knockdown promotes cell growth which is partially rescued by EGR1 knockdown. H23 cells were transfected with indicated siRNA oligos for 48 hours. One portion of cells was then subjected to IB with indicated Abs (C), the other portion was then plated in 96-well plate and grew for 6 days, followed by CCK8 assay for cell growth (D). Shown is mean ± SEM (n=3). *p < 0.05, ⁎⁎p < 0.01, ⁎⁎⁎p < 0.001.
Discussion
EGR1 is an important transcription factor that regulates many key biological processes, including proliferation, differentiation, angiogenesis, apoptosis, invasion and metastasis [13]. Given its biological significance, it is not surprised that EGR1 is subjected to fine regulation at the multiple levels, particularly at the transcriptional level [4,10,11]. However, it is quite surprising that very little is known as to how EGR1, a short-lived important transcription factor, is regulated by the UPS. Although two E3 ligases MDM2 and RNF114 were implicated in regulation of EGR1 turnover in leukemia and gastric cancer cells, respectively, both studies did not provide detailed characterization to convincingly show that EGR1 is indeed a substrate of these two E3 ligases [25,26].
In this study, we reported that EGR1 is a new substrate of SCFFBXW7 E3 ligase. We first observed that MLN4924, a general small molecule inhibitor of entire CRL E3s by blocking neddylation of all Cullins, caused accumulation of EGR1. Although our subsequent study was focused on SCF/CRL1-FBXW7, it is possible that other CRLs, such as CRL2 and CRL4A might also be involved in regulation of EGR1 turnover, since siRNA-based knockdown of cullin-2 and cullin-4A also caused EGR1 accumulation to some extent in a cell line dependent manner.
Among seven tested F-box proteins that act as a substrate-recognizing subunit of SCF/CRL1, FBXW7 is the only one that binds to EGR1 in FBXW7 binding motif dependent manner. Following this lead, we further characterized EGR1 as a bona fide substrate of the FBXW7-GSK3β axis, as evidenced by a) FBXW7 knockdown/ knockout caused accumulation of EGR1; b) FBXW7 knockdown extended EGR1 protein half-life in various cancer cell lines; c) FBXW7 overexpression promoted EGR1 polyubiquitylation; and d) GSK3β inactivation reduced the EGR1-FBXW7 binding, EGR1 polyubiquitylation, and extended EGR1 protein half-life. Thus, EGR1 becomes a new member of FBXW7 substrate family, subjecting to FBXW7 negative regulation.
We next addressed under what stressed conditions, that regulate FBXW7-GSK3β activity, would affect EGR1 turnover. We first used one previously reported condition in which GSK3β is activated when cells were exposed to low glucose in combination with metformin. Under such a condition, EGR1 levels were indeed remarkably reduced, which can be fully rescued by proteasome inhibitor MG132, indicating that GSK3β activation targets EGR1 for degradation. We further found that short-term hypoxia exposure decreased EGR1 levels via increasing its FBXW7 binding, leading to enhanced polyubiquitylation. Thus, EGR1 levels are indeed subjected to regulation by GSK3β-FBXW7 axis under stressed conditions.
What is the biological significance of EGR1 degradation by FBXW7, since many previous reports showed that EGR1 has tumor suppressive activity in various types of human cancer [18,57]. Furthermore, it was reported that EGR1 expression was very low in most NSCLC and low EGR1 expression was negatively correlated with patient survival [46]. On the other hand, it was reported that in many prostate cancer tissues, EGR1 activity was elevated due to EGR1 overexpression, which promotes the androgen-independent growth of prostate carcinoma cells in in vitro cultured cells and in vivo nude mice model [58]. EGR1 was also reported to regulate angiogenic and osteoclastogenic factors and promote bone and brain metastasis of Du145 prostate cancer cells [56], thus acting as an oncogenic protein. In this study, we found that in both H23 lung cancer cells with high EGR1 expression and Du145 prostate cancer cells, EGR1 knockdown significantly inhibited the growth of cancer cells, indicating that EGR1 acts as an oncogenic protein under these cellular contexts. Furthermore, growth stimulation induced by FBXW7 knockdown in H23 cells can be largely rescued by simultaneous EGR1 knockdown, indicating in H23 lung cancer cells, the FBXW7-EGR1 axis did coordinately regulates cancer cell growth. It will be of great interest in defining what is the determinant for EGR1 to act as an oncogenic or tumor suppressive protein, and under what pathological conditions, EGR1 is subjected to negative regulation of FBXW7 with biological implications.
Data availability
All data are contained within the article.
Author's contributions
L.Y, J.Z. and Y.S. designed the experiments, analyzed and interpreted the data. L.Y. and J.Z. performed experiments and drafted the manuscript. Y.S. supervised the study, revised and finalized the manuscript. All authors have reviewed and approved the manuscript.
Funding
This work is supported in part by the National Key R&D Program of China (2016YFA0501800 to YS); and Zhejiang Provincial Natural Science Foundation of China (LD22H300003 to YS).
Declaration of Competing Interest
The authors declare that they have no conflicts of interest with the contents of this article.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.neo.2022.100839.
**********Appendix. Supplementary materials
References
- 1.Sukhatme V.P., et al. A zinc finger-encoding gene coregulated with c-fos during growth and differentiation, and after cellular depolarization. Cell. 1988;53:37–43. doi: 10.1016/0092-8674(88)90485-0. [DOI] [PubMed] [Google Scholar]
- 2.Christy B., Nathans D. DNA binding site of the growth factor-inducible protein Zif268. Proc Natl Acad Sci U S A. 1989;86:8737–8741. doi: 10.1073/pnas.86.22.8737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Virolle T., et al. The Egr-1 transcription factor directly activates PTEN during irradiation-induced signalling. Nat Cell Biol. 2001;3:1124–1128. doi: 10.1038/ncb1201-1124. [DOI] [PubMed] [Google Scholar]
- 4.Baron V., Adamson E.D., Calogero A., Ragona G., Mercola D. The transcription factor Egr1 is a direct regulator of multiple tumor suppressors including TGFbeta1, PTEN, p53, and fibronectin. Cancer Gene Ther. 2006;13:115–124. doi: 10.1038/sj.cgt.7700896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang C., et al. EGR-1/Bax pathway plays a role in vitamin E δ-tocotrienol-induced apoptosis in pancreatic cancer cells. J Nutr Biochem. 2015;26:797–807. doi: 10.1016/j.jnutbio.2015.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shin D.Y., et al. Induction of apoptosis by pectenotoxin-2 is mediated with the induction of DR4/DR5, Egr-1 and NAG-1, activation of caspases and modulation of the Bcl-2 family in p53-deficient Hep3B hepatocellular carcinoma cells. Oncol Rep. 2008;19:517–526. [PubMed] [Google Scholar]
- 7.Xiao D., Chinnappan D., Pestell R., Albanese C., Weber H.C. Bombesin regulates cyclin D1 expression through the early growth response protein Egr-1 in prostate cancer cells. Cancer Res. 2005;65:9934–9942. doi: 10.1158/0008-5472.CAN-05-1830. [DOI] [PubMed] [Google Scholar]
- 8.Shao S., et al. Egr‑1 inhibits colon cancer cell proliferation, migration and invasion via regulating CDKL1 at the transcriptional level. Oncol Rep. 2021;46 doi: 10.3892/or.2021.8120. [DOI] [PubMed] [Google Scholar]
- 9.Kim J., et al. Leptin is a direct transcriptional target of EGR1 in human breast cancer cells. Mol Biol Rep. 2019;46:317–324. doi: 10.1007/s11033-018-4474-3. [DOI] [PubMed] [Google Scholar]
- 10.Yeo H., et al. Transcription factor EGR-1 transactivates the MMP1 gene promoter in response to TNFα in HaCaT keratinocytes. BMB Rep. 2020;53:323–328. doi: 10.5483/BMBRep.2020.53.6.290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sperandio S., et al. The transcription factor Egr1 regulates the HIF-1alpha gene during hypoxia. Mol Carcinog. 2009;48:38–44. doi: 10.1002/mc.20454. [DOI] [PubMed] [Google Scholar]
- 12.Shen N., et al. An early response transcription factor, Egr-1, enhances insulin resistance in type 2 diabetes with chronic hyperinsulinism. J Biol Chem. 2011;286:14508–14515. doi: 10.1074/jbc.M110.190165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang B., et al. The role of the transcription factor EGR1 in cancer. Front Oncol. 2021;11 doi: 10.3389/fonc.2021.642547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schwachtgen J.L., Houston P., Campbell C., Sukhatme V., Braddock M. Fluid shear stress activation of egr-1 transcription in cultured human endothelial and epithelial cells is mediated via the extracellular signal-related kinase 1/2 mitogen-activated protein kinase pathway. J Clin Invest. 1998;101:2540–2549. doi: 10.1172/JCI1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim J.-H., Jeong I.-Y., Lim Y., Lee Y.H., Shin S.Y. Estrogen receptor beta stimulates Egr-1 transcription via MEK1/Erk/Elk-1 cascade in C6 glioma cells. BMB Rep. 2011;44:452–457. doi: 10.5483/BMBRep.2011.44.7.452. [DOI] [PubMed] [Google Scholar]
- 16.Kuo P.-L., Chen Y.-H., Chen T.-C., Shen K.-H., Hsu Y.-L. CXCL5/ENA78 increased cell migration and epithelial-to-mesenchymal transition of hormone-independent prostate cancer by early growth response-1/snail signaling pathway. J Cell Physiol. 2011;226:1224–1231. doi: 10.1002/jcp.22445. [DOI] [PubMed] [Google Scholar]
- 17.Lau M.T., Klausen C., Leung P.C.K. E-cadherin inhibits tumor cell growth by suppressing PI3K/Akt signaling via β-catenin-Egr1-mediated PTEN expression. Oncogene. 2011;30:2753–2766. doi: 10.1038/onc.2011.6. [DOI] [PubMed] [Google Scholar]
- 18.Joslin J.M., et al. Haploinsufficiency of EGR1, a candidate gene in the del(5q), leads to the development of myeloid disorders. Blood. 2007;110:719–726. doi: 10.1182/blood-2007-01-068809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gitenay D., Baron V.T. Is EGR1 a potential target for prostate cancer therapy? Future Oncol. 2009;5 doi: 10.2217/fon.09.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Srivastava S., et al. Estrogen blocks M-CSF gene expression and osteoclast formation by regulating phosphorylation of Egr-1 and its interaction with Sp-1. J Clin Invest. 1998;102:1850–1859. doi: 10.1172/JCI4561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cao X., Mahendran R., Guy G.R., Tan Y.H. Protein phosphatase inhibitors induce the sustained expression of the Egr-1 gene and the hyperphosphorylation of its gene product. J Biol Chem. 1992;267:12991–12997. [PubMed] [Google Scholar]
- 22.Yu J., et al. PTEN regulation by Akt-EGR1-ARF-PTEN axis. EMBO J. 2009;28:21–33. doi: 10.1038/emboj.2008.238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huang R.P., Adamson E.D. The phosphorylated forms of the transcription factor, Egr-1, bind to DNA more efficiently than non-phosphorylated. Biochem Biophys Res Commun. 1994;200:1271–1276. doi: 10.1006/bbrc.1994.1588. [DOI] [PubMed] [Google Scholar]
- 24.Yu J., de Belle I., Liang H., Adamson E.D. Coactivating factors p300 and CBP are transcriptionally crossregulated by Egr1 in prostate cells, leading to divergent responses. Mol Cell. 2004;15:83–94. doi: 10.1016/j.molcel.2004.06.030. [DOI] [PubMed] [Google Scholar]
- 25.Jing Y., et al. Mutant NPM1-regulated lncRNA HOTAIRM1 promotes leukemia cell autophagy and proliferation by targeting EGR1 and ULK3. J Exp Clin Cancer Res. 2021;40:312. doi: 10.1186/s13046-021-02122-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Feng Z., et al. RNF114 silencing inhibits the proliferation and metastasis of gastric cancer. J Cancer. 2022;13:565–578. doi: 10.7150/jca.62033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Deshaies R.J., Joazeiro C.A.P. RING domain E3 ubiquitin ligases. Annu Rev Biochem. 2009;78:399–434. doi: 10.1146/annurev.biochem.78.101807.093809. [DOI] [PubMed] [Google Scholar]
- 28.Nakayama K.I., Nakayama K. Ubiquitin ligases: cell-cycle control and cancer. Nat Rev Cancer. 2006;6:369–381. doi: 10.1038/nrc1881. [DOI] [PubMed] [Google Scholar]
- 29.Cardozo T., Pagano M. The SCF ubiquitin ligase: insights into a molecular machine. Nat Rev Mol Cell Biol. 2004;5:739–751. doi: 10.1038/nrm1471. [DOI] [PubMed] [Google Scholar]
- 30.Jin J., et al. Systematic analysis and nomenclature of mammalian F-box proteins. Genes Dev. 2004;18:2573–2580. doi: 10.1101/gad.1255304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Davis R.J., Welcker M., Clurman B.E. Tumor suppression by the Fbw7 ubiquitin ligase: mechanisms and opportunities. Cancer Cell. 2014;26:455–464. doi: 10.1016/j.ccell.2014.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang Z., Liu P., Inuzuka H., Wei W. Roles of F-box proteins in cancer. Nat Rev Cancer. 2014;14:233–247. doi: 10.1038/nrc3700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yeh C.-H., Bellon M., Nicot C. FBXW7: a critical tumor suppressor of human cancers. Mol Cancer. 2018;17:115. doi: 10.1186/s12943-018-0857-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mao J.H., et al. Fbxw7/Cdc4 is a p53-dependent, haploinsufficient tumour suppressor gene. Nature. 2004;432:775–779. doi: 10.1038/nature03155. [DOI] [PubMed] [Google Scholar]
- 35.Onoyama I., et al. Conditional inactivation of Fbxw7 impairs cell-cycle exit during T cell differentiation and results in lymphomatogenesis. J Exp Med. 2007;204:2875–2888. doi: 10.1084/jem.20062299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tan M., et al. SAG/RBX2/ROC2 E3 ubiquitin ligase is essential for vascular and neural development by targeting NF1 for degradation. Dev Cell. 2011;21:1062–1076. doi: 10.1016/j.devcel.2011.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cui D., et al. FBXW7 confers radiation survival by targeting p53 for degradation. Cell Rep. 2020;30 doi: 10.1016/j.celrep.2019.12.032. [DOI] [PubMed] [Google Scholar]
- 38.Saffie R., et al. FBXW7 triggers degradation of KMT2D to favor growth of diffuse large B-cell lymphoma cells. Cancer Res. 2020;80:2498–2511. doi: 10.1158/0008-5472.CAN-19-2247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yang F., et al. FBXW2 suppresses migration and invasion of lung cancer cells via promoting β-catenin ubiquitylation and degradation. Nat Commun. 2019;10:1382. doi: 10.1038/s41467-019-09289-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Berndsen C.E., Wolberger C. New insights into ubiquitin E3 ligase mechanism. Nat Struct Mol Biol. 2014;21:301–307. doi: 10.1038/nsmb.2780. [DOI] [PubMed] [Google Scholar]
- 41.Welcker M., et al. The Fbw7 tumor suppressor regulates glycogen synthase kinase 3 phosphorylation-dependent c-Myc protein degradation. Proc Natl Acad Sci U S A. 2004;101:9085–9090. doi: 10.1073/pnas.0402770101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mo J.-S., et al. Integrin-linked kinase controls Notch1 signaling by down-regulation of protein stability through Fbw7 ubiquitin ligase. Mol Cell Biol. 2007;27:5565–5574. doi: 10.1128/MCB.02372-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wei W., Jin J., Schlisio S., Harper J.W., Kaelin W.G. The v-Jun point mutation allows c-Jun to escape GSK3-dependent recognition and destruction by the Fbw7 ubiquitin ligase. Cancer Cell. 2005;8:25–33. doi: 10.1016/j.ccr.2005.06.005. [DOI] [PubMed] [Google Scholar]
- 44.Gu Q., Bowden G.T., Normolle D., Sun Y. SAG/ROC2 E3 ligase regulates skin carcinogenesis by stage-dependent targeting of c-Jun/AP1 and IkappaB-alpha/NF-kappaB. J Cell Biol. 2007;178:1009–1023. doi: 10.1083/jcb.200612067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Soucy T.A., et al. An inhibitor of NEDD8-activating enzyme as a new approach to treat cancer. Nature. 2009;458:732–736. doi: 10.1038/nature07884. [DOI] [PubMed] [Google Scholar]
- 46.Ferraro B., Bepler G., Sharma S., Cantor A., Haura E.B. EGR1 predicts PTEN and survival in patients with non-small-cell lung cancer. J Clin Oncol. 2005;23:1921–1926. doi: 10.1200/JCO.2005.08.127. [DOI] [PubMed] [Google Scholar]
- 47.Fan J., et al. Clinical significance of FBXW7 loss of function in human cancers. Mol Cancer. 2022;21:87. doi: 10.1186/s12943-022-01548-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nash P., et al. Multisite phosphorylation of a CDK inhibitor sets a threshold for the onset of DNA replication. Nature. 2001;414:514–521. doi: 10.1038/35107009. [DOI] [PubMed] [Google Scholar]
- 49.Finley D. Recognition and processing of ubiquitin-protein conjugates by the proteasome. Annu Rev Biochem. 2009;78:477–513. doi: 10.1146/annurev.biochem.78.081507.101607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Welcker M., Clurman B.E. FBW7 ubiquitin ligase: a tumour suppressor at the crossroads of cell division, growth and differentiation. Nat Rev Cancer. 2008;8:83–93. doi: 10.1038/nrc2290. [DOI] [PubMed] [Google Scholar]
- 51.Inuzuka H., et al. SCF(FBW7) regulates cellular apoptosis by targeting MCL1 for ubiquitylation and destruction. Nature. 2011;471:104–109. doi: 10.1038/nature09732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hart M., et al. The F-box protein beta-TrCP associates with phosphorylated beta-catenin and regulates its activity in the cell. Curr Biol. 1999;9:207–210. doi: 10.1016/s0960-9822(99)80091-8. [DOI] [PubMed] [Google Scholar]
- 53.Liu C., et al. beta-Trcp couples beta-catenin phosphorylation-degradation and regulates Xenopus axis formation. Proc Natl Acad Sci U S A. 1999;96:6273–6278. doi: 10.1073/pnas.96.11.6273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cohen P., Frame S. The renaissance of GSK3. Nat Rev Mol Cell Biol. 2001;2:769–776. doi: 10.1038/35096075. [DOI] [PubMed] [Google Scholar]
- 55.Elgendy M., et al. Combination of hypoglycemia and metformin impairs tumor metabolic plasticity and growth by modulating the PP2A-GSK3β-MCL-1 axis. Cancer Cell. 2019;35 doi: 10.1016/j.ccell.2019.03.007. [DOI] [PubMed] [Google Scholar]
- 56.Li L., et al. EGR1 regulates angiogenic and osteoclastogenic factors in prostate cancer and promotes metastasis. Oncogene. 2019;38:6241–6255. doi: 10.1038/s41388-019-0873-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Liu J., et al. Concurrent down-regulation of Egr-1 and gelsolin in the majority of human breast cancer cells. Cancer Genomics Proteom. 2007;4:377–385. [PubMed] [Google Scholar]
- 58.Yang S.-Z., Eltoum I.A., Abdulkadir S.A. Enhanced EGR1 activity promotes the growth of prostate cancer cells in an androgen-depleted environment. J Cell Biochem. 2006;97:1292–1299. doi: 10.1002/jcb.20736. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are contained within the article.