Abstract
Brucellae have been reported to be phylogenetically related to bacteria of the family Rhizobiaceae. In the present study, we used a panel of monoclonal antibodies (MAbs) to Brucella outer membrane proteins (OMPs) to determine the presence of common OMP epitopes in some representative bacteria of this family, i.e., Ochrobactrum anthropi, Phyllobacterium rubiacearum, Rhizobium leguminosarum, and Agrobacterium tumefaciens, and also in bacteria reported to serologically cross-react with brucella, i.e., Yersinia enterocolitica O:9, Escherichia coli O:157, and Salmonella urbana. In particular, most MAbs to the Brucella outer membrane lipoproteins Omp10, Omp16, and Omp19 cross-reacted with O. anthropi and P. rubiacearum, which are actually the closest relatives of brucellae. Some of them also cross-reacted, but to a lower extent, with R. leguminosarum and A. tumefaciens. The putative Omp16 and Omp19 homologs in these bacteria showed the same apparent molecular masses as their Brucella counterparts. None of the antilipoprotein MAbs cross-reacted with Y. enterocolitica O:9, E. coli O:157, or S. urbana.
Brucellae are gram-negative, facultative, intracellular bacteria that can infect humans and many species of animals. Six species are recognized within the genus Brucella: B. abortus, B. melitensis, B. suis, B. ovis, B. canis, and B. neotomae (7). These classifications are based mainly on their differences in pathogenicity and host preference (7). The Brucella species constitute a very homogeneous group, as shown by their antigenic relatedness and by DNA-DNA hybridization studies (>90% DNA homology for all species) (8, 9, 25). On the basis of the 16S rRNA sequence, brucellae have been shown to belong to the family Rhizobiaceae (27). This family includes plant and animal pathogens, such as Agrobacterium, Bartonella, and Brucella, that are characteristically associated pericellularly or intracellularly with eukaryotic cells; plant endosymbionts, such as Rhizobium and Phyllobacterium; soil inhabitants, such as Mycoplana; and isolates from soil and from human clinical specimens, such as Ochrobactrum (14, 18, 19). Among all these bacteria, Ochrobactrum anthropi is the closest known relative of brucellae (14, 24, 27). This bacterium has gained interest in the past few years because of its isolation from immunocompromised hosts (1, 11–13). Recent reports have also described immunological cross-reactions between Brucella spp. and O. anthropi (23, 24). The antigens containing common epitopes were described as rough lipopolysaccharide and soluble and membrane proteins of unknown nature (23, 24). Since O. anthropi constitutes a heterogeneous group of bacteria on the basis of classical phenotypical characterization and DNA-DNA hybridization studies, further subdivision of the genus into two species, O. anthropi and O. intermedium, has recently been proposed (24). The latter, new species name has been suggested because of a closer genetic and antigenic relationship with brucellae than with O. anthropi (24). Additionally, brucellae also share epitopes, mainly on the smooth lipopolysaccharide (S-LPS), with bacteria reported earlier to serologically cross-react with Brucella, of which the most important is Yersinia enterocolitica O:9 (7).
The Brucella outer membrane contains three major proteins with molecular masses ranging from 25 to 27, 31 to 34, and 36 to 38 kDa (2, 6). The largest protein has been identified and characterized as a porin (10, 17). The genes coding for these proteins have been cloned and sequenced, and the current names for these outer membrane proteins (OMPs) are Omp25, Omp31, and Omp2b, respectively (4, 5, 17). The other OMPs identified so far by use of monoclonal antibodies (MAbs) are less abundant (minor) proteins with molecular masses of 10, 16.5, 19, and 89 kDa (2). Gene cloning, the predicted amino acid sequences, and the presence of particular protein motifs have identified the 10-, 16.5-, and 19-kDa OMPs as outer membrane lipoproteins (21, 22). The current names for these OMPs are Omp10, Omp16, and Omp19, respectively (21, 22). Omp16 actually belongs to the peptidoglycan-associated lipoprotein family of proteins found in many gram-negative bacteria (22). Homologs of Omp10 and Omp19 have not yet been reported for other bacteria. All of these proteins have been found as immunogenic proteins in infected cattle, sheep, and goats (3, 15, 16, 21, 28).
In the present study, we used MAbs to analyze the occurrence of epitopes common to Brucella OMPs in phylogenetically related bacteria of the family Rhizobiaceae and reported S-LPS-cross-reacting bacteria as well. The importance of the epitopes recognized by the MAbs in the antibody responses of Brucella-infected cattle and sheep has been previously shown by competitive enzyme-linked immunosorbent assay (ELISA) with these MAbs (3, 28). The occurrence of common epitopes could explain some of the serologic protein cross-reactivities reported between Brucella and Ochrobactrum (23, 24). In addition, the present study also led to the identification of new homologous proteins within the family Rhizobiaceae.
The strains studied that belong to the family Rhizobiaceae were O. anthropi 3301 (proposed as a reference strain for O. intermedium), O. anthropi 3331, Phyllobacterium rubiacearum Pr1, Rhizobium leguminosarum R11, and Agrobacterium tumefaciens At1 (26). The S-LPS-cross-reacting bacteria were Y. enterocolitica O:9 strain Ye8, Escherichia coli O:157 strain Ec2, and Salmonella urbana Su1 (26). B. abortus 544 (biovar 1) was used as a reference. Strains were grown on tryptic soy agar (Gibco BRL) supplemented with 0.1% (wt/vol) yeast extract (Difco) at 37°C. R. leguminosarum was cultured in tryptone-yeast medium at 30°C (20). MAbs used were those of previous studies (2, 3, 6, 21, 22, 26, 28), and they were used as hybridoma culture supernatants (twofold diluted in ELISA and immunoblotting).
The occurrence of cross-reacting epitopes was first screened by ELISA, performed as described previously (2, 5, 28). Microtiter plates were coated with bacterial suspensions in phosphate-buffered saline at an absorbance (600 nm) of 1.0. To improve accessibility of OMPs, bacteria were sonicated prior to coating (5). MAbs were used at a dilution of 1/2. Positive control MAbs were 3D6, specific for peptidoglycan (6), and A53/09G03/D02, specific for DnaK, previously shown to cross-react with O. anthropi and P. rubiacearum (26).
In particular, most MAbs to the outer membrane lipoproteins Omp10, Omp16, and Omp19 cross-reacted in ELISA with both O. anthropi 3301 and 3331 and P. rubiacearum (Table 1). Fewer MAbs against the three OMPs reacted with R. leguminosarum, and only one MAb, against Omp16, reacted weakly with A. tumefaciens. None of these MAbs reacted with the S-LPS-cross-reacting bacteria Y. enterocolitica O:9, E. coli O:157, and S. urbana. The MAb bindings observed correlated with the genetic closeness to brucellae. However, there was no significant difference in MAb bindings between O. anthropi 3301 (proposed as O. intermedium) and O. anthropi 3331.
TABLE 1.
Binding to MAbs to Brucella and related bacteria in ELISAa
| Specificity | MAb | Absorbance of MAb binding at
dilution of 1/2
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| B. abortus | O. anthropi 3301 | O. anthropi 3331 | P. rubiacearum | R. leguminosarum | A. tumefaciens | Y. enterocolitica O:9 | E. coli O:157 | S. urbana | ||
| Omp10 | A68/07G11/C10 | 2.320 | —a | — | — | — | — | — | — | — |
| A68/08E07/B11 | 2.303 | 1.060 | 1.008 | 0.992 | 1.067 | — | — | — | — | |
| Omp16 | A68/04G01/C06 | 2.243 | 2.591 | 2.349 | 2.860 | 2.860 | — | — | — | — |
| A76/08C03/G03 | 2.312 | 2.604 | 2.233 | — | 0.536 | 0.869 | — | — | — | |
| Omp19 | A68/25H10/A05 | 2.119 | 1.325 | 1.192 | 1.040 | — | — | — | — | — |
| A76/05C10/A08 | 2.100 | 1.358 | 1.254 | 0.987 | — | — | — | — | — | |
| A76/10D03/H02 | 2.260 | 2.179 | 2.227 | 1.452 | 0.667 | — | — | — | — | |
| A76/02A06/H10 | 1.516 | — | — | — | — | — | — | — | — | |
| DnaK | A53/09G03/D02 | 2.338 | 0.913 | 2.128 | 2.674 | — | — | — | — | — |
| PGb | 3D6 | 2.295 | 2.554 | 1.693 | 1.874 | 2.860 | 2.108 | 2.860 | 2.860 | 2.860 |
—, nonsigificant binding (absorbance below 0.5).
PG, peptidoglycan.
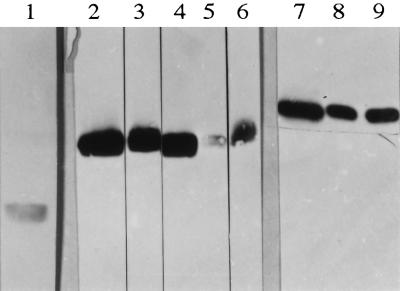
In immunoblotting after sodium dodecyl sulfate-polyacrylamide gel electrophoresis, performed as described previously (2, 28), the anti-Omp16 MAbs reacted strongly with O. anthropi 3301 and 3331, P. rubiacearum, and R. leguminosarum and weakly with A. tumefaciens, thus confirming the ELISA results (Fig. 1). The anti-Omp19 MAbs reacted strongly only with O. anthropi and P. rubiacearum, which is also in accordance with the ELISA results. The putative Omp16 and Omp19 homologs detected by the MAbs in these bacteria showed the same apparent molecular masses as their Brucella counterparts. The anti-Omp10 MAbs gave no positive reactions in immunoblotting and reacted only weakly with B. abortus, which was used as the control (Fig. 1).
FIG. 1.
Reactivity in immunoblotting of anti-Omp10 (lane 1), anti-Omp16 (lanes 2 to 6), and anti-Omp19 MAbs (lanes 7 to 9) after sodium dodecyl sulfate-polyacrylamide gel electrophoresis of B. abortus 544 (lanes 1, 2, and 7), O. anthropi (strains 3301 and 3331 gave the same result) (lanes 3 and 8), P. rubiacearum (lanes 4 and 9), A. tumefaciens (lane 5), and R. leguminosarum (lane 6).
In conclusion, the present study showed the presence of epitopes cross-reactive with Brucella outer membrane lipoproteins on genetically related bacteria, of which the most important is O. anthropi. Of particular interest are the lipoproteins Omp10 and Omp19, not yet reported for other bacteria. Thus, these proteins could constitute a new family of OMPs specifically encountered in Rhizobiaceae. As suggested by Velasco et al. (23), the immunoresponse of Brucella-infected hosts to protein antigens may not necessarily be specific for brucellae, and the presence of O. anthropi or related bacteria may explain previously described reactivities to OMPs in healthy animals (16). The outer membrane lipoproteins Omp10, Omp16, and Omp19 are the first identified among these OMPs.
Acknowledgments
We thank J. M. Verger and M. Grayon for supplying the strains. We also thank S. Baucheron for technical support.
REFERENCES
- 1.Alnor D, Frimondt-Moller N, Espersen F, Frederiksen W. Infections with the unusual human pathogens Agrobacterium species and Ochrobactrum anthropi. Clin Infect Dis. 1994;18:914–920. doi: 10.1093/clinids/18.6.914. [DOI] [PubMed] [Google Scholar]
- 2.Cloeckaert A, de Wergifosse P, Dubray G, Limet J N. Identification of seven surface-exposed Brucella outer membrane proteins by use of monoclonal antibodies: immunogold labeling for electron microscopy and enzyme-linked immunosorbent assay. Infect Immun. 1990;58:3980–3987. doi: 10.1128/iai.58.12.3980-3987.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cloeckaert A, Kerkhofs P, Limet J N. Antibody response to Brucella outer membrane proteins in bovine brucellosis: immunoblot analysis and competitive enzyme-linked immunosorbent assay using monoclonal antibodies. J Clin Microbiol. 1992;30:3168–3174. doi: 10.1128/jcm.30.12.3168-3174.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cloeckaert A, Verger J M, Grayon M, Vizcaino N. Molecular and immunological characterization of the major outer membrane proteins of Brucella. FEMS Microbiol Lett. 1996;145:1–8. doi: 10.1111/j.1574-6968.1996.tb08547.x. [DOI] [PubMed] [Google Scholar]
- 5.Cloeckaert A, Verger J-M, Grayon M, Zygmunt M S, Grépinet O. Nucleotide sequence and expression of the gene encoding the major 25-kilodalton outer membrane protein of Brucella ovis: evidence for antigenic shift, compared with other Brucella species, due to a deletion in the gene. Infect Immun. 1996;64:2047–2055. doi: 10.1128/iai.64.6.2047-2055.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cloeckaert A, Zygmunt M S, de Wergifosse P, Dubray G, Limet J N. Demonstration of peptidoglycan-associated Brucella outer membrane proteins by use of monoclonal antibodies. J Gen Microbiol. 1992;138:1543–1550. doi: 10.1099/00221287-138-7-1543. [DOI] [PubMed] [Google Scholar]
- 7.Corbel M J, Brinley-Morgan W J. Genus Brucella Meyer and Shaw 1920, 173AL. In: Krieg N R, Holt J G, editors. Bergey’s manual of systematic bacteriology. Vol. 1. Baltimore, Md: Williams & Wilkins; 1984. pp. 377–388. [Google Scholar]
- 8.Diaz R, Jones L M, Wilson J B. Antigenic relationship of Brucella ovis and Brucella melitensis. J Bacteriol. 1967;93:1262–1268. doi: 10.1128/jb.93.4.1262-1268.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Diaz R, Jones L M, Wilson J B. Antigenic relationship of the gram-negative organism causing canine abortion to smooth and rough brucellae. J Bacteriol. 1968;95:618–624. doi: 10.1128/jb.95.2.618-624.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Douglas J T, Rosenberg E Y, Nikaido H, Verstreate D R, Winter A J. Porins of Brucella species. Infect Immun. 1984;44:16–21. doi: 10.1128/iai.44.1.16-21.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ezzedine H, Mourad M, Van Ossel C, Logghe C, Squifflet J P, Renault F, Wauters G, Gigi J, Wilmotte L, Haxhe J J. An outbreak of Ochrobactrum anthropi bacteraemia in five organ transplant patients. J Hosp Infect. 1994;27:35–42. doi: 10.1016/0195-6701(94)90066-3. [DOI] [PubMed] [Google Scholar]
- 12.Gransden W R, Eykyn S J. Seven cases of bacteremia due to Ochrobactrum anthropi. Clin Infect Dis. 1992;15:1068–1069. doi: 10.1093/clind/15.6.1068. [DOI] [PubMed] [Google Scholar]
- 13.Haditsch M, Binder L, Tschurtschenthaler G, Watschinger R, Zauner G, Mittermayer H. Bacteremia caused by Ochrobactrum anthropi in an immunocompromised child. Infection. 1994;22:291–292. doi: 10.1007/BF01739922. [DOI] [PubMed] [Google Scholar]
- 14.Holmes B, Popoff M, Kiredjian M, Kersters K. Ochrobactrum anthropi gen. nov., sp. nov. from human clinical specimens and previously known as group Vd. Int J Syst Bacteriol. 1988;38:406–416. [Google Scholar]
- 15.Kovach M E, Elzer P H, Robertson G T, Chirhart-Gilleland R L, Christensen M A, Peterson K M, Roop R M., II Cloning and nucleotide sequence analysis of a Brucella abortus gene encoding an 18 kDa immunoreactive protein. Microb Pathog. 1997;22:241–246. doi: 10.1006/mpat.1996.0108. [DOI] [PubMed] [Google Scholar]
- 16.Letesson J J, Tibor A, van Eynde G, Wansard V, Weynants V, Denoel P, Saman E. Humoral immune responses of Brucella-infected cattle, sheep, and goats to eight purified recombinant Brucella proteins in an indirect enzyme-linked immunosorbent assay. Clin Diagn Lab Immunol. 1997;4:556–564. doi: 10.1128/cdli.4.5.556-564.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Marquis H, Ficht T A. The omp2 gene locus of Brucella abortus encodes two homologous outer membrane proteins with properties characteristic of bacterial porins. Infect Immun. 1993;61:3785–3790. doi: 10.1128/iai.61.9.3785-3790.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Moreno E. Evolution of Brucella. In: Plommet M, editor. Advances in brucellosis research. Wageningen, The Netherlands: Pudoc Scientific Publishers; 1992. pp. 198–218. [Google Scholar]
- 19.Moreno E, Stackebrandt E, Dorsch M, Wolters J, Busch M, Mayer H. Brucella abortus 16S rRNA and lipid A reveal a phylogenetic relationship with members of the alpha-2 subdivision of the class Proteobacteria. J Bacteriol. 1990;172:3569–3576. doi: 10.1128/jb.172.7.3569-3576.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Priefer U B. Genes involved in lipopolysaccharide production and symbiosis are clustered on the chromosome of Rhizobium leguminosarum biovar viciae VF39. J Bacteriol. 1989;171:6161–6168. doi: 10.1128/jb.171.11.6161-6168.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tibor A, Saman E, de Wergifosse P, Cloeckaert A, Limet J N, Letesson J-J. Molecular characterization, occurrence, and immunogenicity in infected sheep and cattle of two minor outer membrane proteins of Brucella abortus. Infect Immun. 1996;64:100–107. doi: 10.1128/iai.64.1.100-107.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tibor A, Weynants V, Denoel P, Lichtfouse B, De Bolle X, Saman E, Limet J N, Letesson J-J. Molecular cloning, nucleotide sequence, and occurrence of a 16.5-kilodalton outer membrane protein of Brucella abortus with similarity to PAL lipoproteins. Infect Immun. 1994;62:3633–3639. doi: 10.1128/iai.62.9.3633-3639.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Velasco J, Díaz R, Grilló M J, Barberán M, Marín C, Blasco J M, Moriyón I. Antibody and delayed-type hypersensitivity responses to Ochrobactrum anthropi cytosolic and outer membrane antigens in infections by smooth and rough Brucella spp. Clin Diagn Lab Immunol. 1997;4:279–284. doi: 10.1128/cdli.4.3.279-284.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Velasco J, Romero C, Lopez-Goni I, Leiva J, Diaz R, Moriyon I. Evaluation of the relatedness of Brucella spp. and Ochrobactrum anthropi and description of Ochrobactrum intermedium sp. nov., a new species with a closer relationship to Brucella spp. Int J Syst Bacteriol. 1998;48:759–768. doi: 10.1099/00207713-48-3-759. [DOI] [PubMed] [Google Scholar]
- 25.Verger J-M, Grimont F, Grimont P A D, Grayon M. Brucella, a monospecific genus as shown by deoxyribonucleic acid hybridization. Int J Syst Bacteriol. 1985;35:292–295. [Google Scholar]
- 26.Vizcaino N, Zygmunt M S, Verger J M, Grayon M, Cloeckaert A. Localization and characterization of a specific linear epitope of the Brucella DnaK protein. FEMS Microbiol Lett. 1997;154:117–122. doi: 10.1111/j.1574-6968.1997.tb12632.x. [DOI] [PubMed] [Google Scholar]
- 27.Yanagi M, Yamasato K. Phylogenetic analysis of the family Rhizobiaceae and related bacteria by sequencing of 16S rRNA gene using PCR and DNA sequencer. FEMS Microbiol Lett. 1993;107:115–120. doi: 10.1111/j.1574-6968.1993.tb06014.x. [DOI] [PubMed] [Google Scholar]
- 28.Zygmunt M S, Cloeckaert A, Dubray G. Brucella melitensis cell envelope protein and lipopolysaccharide epitopes involved in humoral immune responses of naturally and experimentally infected sheep. J Clin Microbiol. 1994;32:2514–2522. doi: 10.1128/jcm.32.10.2514-2522.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]