Inhibition of mDia2/MIRO1-mediated mitochondrial positioning in CAFs induces mitochondrial dysfunction and suppresses tumor growth, revealing a promising therapeutic strategy to target tumor–stroma cross-talk.
Abstract
Cancer-associated fibroblasts (CAF) are key regulators of tumorigenesis. Further insights into the tumor-promoting mechanisms of action of CAFs could help improve cancer diagnosis and treatment. Here we show that the formin mDia2 regulates the positioning and function of mitochondria in dermal fibroblasts, thereby promoting a protumorigenic CAF phenotype. Mechanistically, mDia2 stabilized the mitochondrial trafficking protein MIRO1. Loss of mDia2 or MIRO1 in fibroblasts or CAFs reduced the presence of mitochondria and ATP levels near the plasma membrane and at CAF-tumor cell contact sites, caused metabolic alterations characteristic of mitochondrial dysfunction, and suppressed the secretion of protumorigenic proteins. In mouse models of squamous carcinogenesis, genetic or pharmacologic inhibition of mDia2, MIRO1, or their common upstream regulator activin A inhibited tumor formation. Consistently, co-upregulation of mDia2 and MIRO1 in the stroma of various human cancers negatively correlated with survival. This work unveils a key role of mitochondria in the protumorigenic CAF phenotype and identifies an activin A–mDia2–MIRO1 signaling axis in CAFs with diagnostic and therapeutic potential.
Significance:
Inhibition of mDia2/MIRO1-mediated mitochondrial positioning in CAFs induces mitochondrial dysfunction and suppresses tumor growth, revealing a promising therapeutic strategy to target tumor–stroma cross-talk.
Graphical Abstract
Introduction
Cancer-associated fibroblasts (CAF) are major components of the tumor microenvironment (TME), which, together with mutations and epigenetic alterations in the tumor cells, plays a crucial role in determining malignancy. CAFs most often promote tumor growth via paracrine control of cancer cells (1). This involves increased expression of cytokines, growth factors, and extracellular matrix (ECM) proteins and possibly alterations in protein secretion pathways. Recent studies identified cytoskeletal abnormalities in CAFs (2), including perturbations of the actomyosin cytoskeleton, which affect actin-based protrusions, cell contractility, and motility (3, 4). A key regulator of the actin cytoskeleton is the formin mDia2 (DIAPH3; refs. 5, 6), which is upregulated in CAFs and promotes the CAF phenotype by enhancing the formation of filopodia and cell migration, as well as the expression of CAF marker genes (7, 8). In dermal fibroblasts and skin CAFs, mDia2 is a downstream target of the protumorigenic cytokine activin A, a member of the transforming growth factor beta (TGFβ) superfamily that is highly expressed by skin cancer cells and also by cells in the TME (9). Activin A induced the expression of genes that encode profibrotic and protumorigenic proteins in dermal fibroblasts, which in turn stimulate cancer cell proliferation and migration (7, 10). The activin A–induced increase in mDia2 levels prevents p53 nuclear accumulation, thereby promoting the expression of CAF marker genes and protumorigenic activities of fibroblasts (7). This paracrine axis is highly relevant to human cancer as overexpression of both activin A and mDia2 correlates with poor prognosis in several malignancies (7).
Here we discovered that mitochondrial positioning and function play a key role in regulating the skin CAF phenotype and cancer progression, and depend on mDia2 and its ability to stabilize the atypical Rho-GTPase MIRO1 (RHOT1). Targeting the activin A–mDia2–MIRO1 signaling axis and the resulting mitochondrial/metabolic alterations in CAFs is therefore a promising strategy for the treatment of different human cancers.
Materials and Methods
Animals
NOD/SCID (NOD.CB17-Prkdcscid/NCrCrl) mice were housed under specific pathogen-free conditions and received food and water ad libitum. Mouse maintenance and all animal experiments were approved by the veterinary authorities of Zurich, Switzerland (Kantonales Veterinäramt Zurich).
Human skin cancer samples and CAFs
Normal human skin and skin cancer samples were obtained anonymously from the Department of Dermatology, University Hospital of Zurich (in the context of the Biobank project), approved by the local and cantonal Research Ethics Committees. Normal skin was from healthy adult volunteers or from the edges of skin tumors (SCC), which had been diagnosed by experienced pathologists. Written informed consent was obtained from all subjects, and the experiments conformed to the principles set out in the WMA Declaration of Helsinki and the Department of Health and Human Services Belmont Report.
CAFs were directly isolated from skin SCC biopsies alongside their paired normal skin fibroblasts as described previously, with some modifications (11). Briefly, skin samples were digested with trypsin, followed by digestion with collagenase. Cells were cultivated with J2 feeder cells. Fibroblasts/CAFs were separated from SCC cells upon short incubation with trypsin and then cultivated without feeder cells in DMEM, 10% FBS, and 1% penicillin/streptomycin (P/S). The CAF phenotype was confirmed by the expression of skin CAF markers (12), and fibroblast cultures isolated from SCCs that did not have CAF properties were excluded.
For fibroblast/CAF isolation from mouse ear tumors, we adapted a previously published protocol (13). Briefly, the tumor was minced into small pieces and incubated with 2.5 mL of a collagenase type II solution (500 U/ml; Worthington Biochemical Corporation) for 1 hour at 37°C with manual agitation every 15 minutes. The cell suspension was poured through a 100-μm cell strainer, and the content was centrifuged at 1,200 rpm for 5 minutes. The resulting cell pellet was resuspended in 8 mL of DMEM/10% FBS/P/S and plated in two 6-cm dishes. The medium was changed the following day, and cells were passaged prior to confluency.
Cell lines
HaCaT keratinocytes and SCC13 cells were obtained from Prof. Petra Boukamp, Leibniz Institute Düsseldorf, Germany. Authentication of the cell lines was performed by the “Deutsche Sammlung von Mikroorganismen und Zellkulturen” (DSMZ) GmbH, Braunschweig, Germany. Absence of Mycoplasma was confirmed by PCR using the PCR Mycoplasma Test Kit I/C (PromoKine) on a monthly basis.
siRNA-mediated knockdown
Knockdown of MIRO1 and mDia2 in primary human fibroblasts was performed using previously characterized siRNAs (14–16). Cells were transfected with MIRO1, mDia2, or control siRNAs using Lipofectamine RNAiMAX Transfection Reagent (Thermo Fisher Scientific) according to the manufacturer's instructions and incubated for 24–48 hours prior to analysis. To visualize/confirm the knockdown efficiency in vivo, the siRNAs were prelabeled with fluorescein using the Label IT siRNA Tracker (MIR 7216, MIRUS Bio LCC).
Generation of human fibroblasts with shRNA-mediated knockdown of mDia2
Primary human dermal fibroblasts (HDF) were infected with lentiviruses pLKO.1-shmDia2 TRCN0000150903, which target mouse mDia2 and human DIAPH3. The construct had been previously characterized for specificity, and results had been confirmed with another shRNA (17, 18).
Measurement of ATP and LDH levels
ATP levels were measured using the CellTiter-Glo assay according to the manufacturer's instructions (Promega). Briefly, cells were plated in triplicate in opaque 96-well plates. After 24 hours of incubation, they were incubated for 10 minutes with CellTiter-Glo reagent, and luminescence was measured using a 96-well plate reader. LDH assays were performed as described previously (19).
Analysis of cytokine concentrations
The concentrations of several cytokines in the supernatant of fibroblasts were measured by LEGENDplex human inflammation panel 1 (#740808, BioLegend) according to the manufacturer's protocol. All samples were analyzed in technical duplicates, and averages of biological replicates were used for statistical analysis. Flow cytometry was performed using a BD LSRFortessa, and data were analyzed using FlowJo v10 (Tristar).
Seahorse XF cell Mito Stress Test
Fibroblasts were plated in XF96 Seahorse plates at 100,000 cells per well in DMEM/10% FCS/1% PS, and the medium was subsequently switched to the Seahorse XF base medium (103335-100, Agilent Technologies) supplemented with 10 mmol/L glucose or galactose, 1 mmol/L sodium pyruvate and 2 mmol/L glutamine (assay concentration according to the manufacturer of the kit: https://www.agilent.com/cs/library/usermanuals/public/XF_Cell_Mito_Stress_Test_Kit_User_Guide.pdf) and further incubated in a CO2-free incubator for 1 hour. Oligomycin, FCCP (carbonyl cyanide-p-trifluoromethoxyphenylhydrazone), and antimycin A/rotenone were prepared in the XF assay medium with final concentrations of 1 μmol/L, 1.5 μmol/L, and 1/0.1 μmol/L, respectively, and provided by the Seahorse XF Cell Mito Stress Test Kit (#103015-100; Agilent Technologies). The compounds were serially injected to measure OCR of cells in the XF96 plate.
Transfection of human fibroblasts
Wild-type or mutant Flag-tagged mDia2 cDNAs (17, 20) were subcloned into the retroviral pMX vector (Cell Biolabs). pGEM-MIRO1 and cyto-Ruby3-iATPSnFR1.0 (ATP biosensors; ref. 21) plasmids were purchased from Sino Biological Inc. (#HG15898-G) or Addgene (#102551), respectively. Plasmid vectors and Lipofectamine 2000 (#11668030, Invitrogen) were incubated with Opti-MEM I Reduced Serum Medium (#31985062, Thermo Fisher Scientific) at room temperature for 20 minutes. The mixture was then added to primary human fibroblasts. After 6 hours of incubation, the medium was replaced by a normal culture medium containing 10% FBS. Cells were allowed to recover for 24–48 hours prior to analysis.
Endoplasmic reticulum and mitochondrial staining
Fibroblasts were seeded on glass coverslips in 24-well plates with complete medium and incubated at 37°C. They were stained with ER-Tracker Blue-White DPX (Thermo Fisher Scientific, E12353), MitoTracker Green (Thermo Fisher Scientific, M7514), CellROX-Orange (Thermo Fisher Scientific, C10443), MitoSOX-Red (Thermo Fisher Scientific, M36008), or MitoTracker Red CMXRos (Thermo Fisher Scientific, M7512) diluted 1:2,000 in complete medium for 45 minutes at 37°C. Subsequently, cells were washed and fixed with 3.7% formaldehyde in complete medium for 15 minutes at 37°C. After washing, they were mounted with Mowiol (Sigma-Aldrich). Images were acquired with an Axio Imager.A1 microscope (Carl Zeiss AG).
RNA isolation and qRT-PCR
RNA isolation and quantitative qRT-PCR were performed as described (13) using the primers listed in Supplementary Table S1. Values obtained for the first control were set to 1.
Histology, immunostaining, and image analysis
Histologic analysis and immunostainings were performed as described (13) using the antibodies listed in Supplementary Table S2. Immunofluorescence images were analyzed in Fiji (22) and normalized to cell number for in vitro experiments with at least 9 microscopic fields of view for each condition analyzed. The curvature of ECM fibers in vivo was analyzed with the ImageJ quantification tool TWOMBLI (23). Mitochondrial networks were analyzed using MiNA (24). Mitochondrial and ATP distribution were determined by measurement of MitoTracker or ATP biosensor fluorescence intensity relative to the highest intensity value. We calculated the average for every 10% incremental distance from the perinuclear region to the plasma membrane using ImageJ (Fiji) plot-profile tool. The relative distance is indicated with values from 1 to 10; at least 100 to 200 intensity profiles were measured. All images were processed in an identical way by adjusting brightfield/contrast and subtracting background signal to identify cell edge/contour using a wide-field microscope.
Proximity ligation assay
Proximity ligation assays (PLA) were performed with mDia2 (sc-293288, Santa Cruz), MIRO1 (NBP1-59021, Novus Biologicals), and TRAK1 (H00022906-M01A, Thermo Fisher Scientific) antibodies (1:500) and MitoTracker Green as described for fluorescence microscopy, followed by ligation of probes specific for each antibody using a PLA Kit (DUO92101, Sigma-Aldrich). Samples treated without primary antibodies were used as negative controls.
Preparation of protein lysates and Western blot analysis
Preparation of protein lysates and Western blot were performed as previously described (13). Some lysates were collected from cells pretreated with the proteasome inhibitor epoxomicin (10 μmol/L; S2619, Selleckchem), the mDia2 inhibitor SMIFH2 (25 μmol/L; S4826, Sigma-Aldrich) or recombinant activin A (20 ng/mL; #120-14, PeproTech).
Antibodies against INHBA (sc166503, Santa Cruz; 1:500 diluted), mDia2 (recognizing both the murine and human proteins; 1:5,000 diluted; refs. 17, 18), αSMA (A2547, Sigma‐Aldrich; 1:500 diluted), HSP60 (ab59457, Abcam; 1:500 diluted), FN1 (ab2413, Abcam; 1:500 diluted), COL4A1 (#10710, Progen; 1:500 diluted), ELN (ab21610, Abcam; 1:500 diluted), MIRO1 (NBP1-59021, Novus Biologicals; 1:500 diluted), POSTN (ab14041, Abcam; 1:500 diluted), calnexin (NB300-518, Novus Biologicals; 1:500 diluted), GM130 (MABT1363, Sigma‐Aldrich; 1:500 diluted), GAPDH (5G4, HyTest, Turku; 1:10,000 diluted), α‐tubulin (T5168, Sigma‐Aldrich; 1:10,000 diluted), and vinculin (V4505, Sigma‐Aldrich; 1:2,000 diluted) were used. Secondary antibodies were anti‐rabbit or anti‐mouse IgG (W4011 and W4021, Promega; 1:8,000 diluted) conjugated with horseradish peroxidase, and chemiluminescence was determined using the WesternBright ECL Detection System (Advansta). Bands were visualized using Fusion Solo 6S (Witec AG), and intensity was quantified with ImageJ software (NIH). Loading controls of different molecular weights were used depending on the protein that was analyzed in the experiment.
Chromatin immunoprecipitation
HDFs (approximately 5 × 108 cells) at 80% to 90% confluency, which had been pretreated with activin A (20 ng/mL) for 6 hours, were collected, resuspended in DMEM, and subjected to chromatin immunoprecipitation (ChIP) as previously described (25). The purified DNA fragments were used for PCR amplification using the primers listed in Supplementary Table S3.
Migration assays
For scratch assays, cells were grown to 100% confluency and treated with 2 μg/mL mitomycin C (Sigma-Aldrich) for 2 hours to inhibit cell proliferation. One or several scratches were made into the cell layer using a sterile 200 μL pipette tip. Dead cells and debris were washed off with prewarmed PBS. The same area was photographed directly after scratching and at different time points thereafter.
To determine the capacity of the matrix deposited by fibroblasts to stimulate SCC cell migration, SCC13 cells were seeded on culture inserts (#80209, Ibidi) that were placed on top of the matrix that had been deposited by fibroblasts. The insert was then removed for migration assays.
Chemotactic transwell migration was assessed as described (7). Briefly, SCC13 cells were seeded on the insert of the transwell plates in DMEM/1% FBS and let to migrate for 24 hours toward CM from fibroblasts.
Isolation of fibroblast secretomes and matrisomes
Cells were plated at 70% to 90% confluency in DMEM/10% FBS/P/S. On the following day, they were pretreated with 2 μg/mL mitomycin C and cultured in starvation medium (DMEM/1% FBS/P/S) with 1 μg/mL doxycycline (DOX) for an additional 3 days. Conditioned media and ECM were prepared as previously described (13). The total secreted protein concentration was determined using the Bradford method (26).
Skin tumorigenesis assays
Tumorigenesis assays in mouse ear skin were performed as described (12). Briefly, 3 μL cell suspension (2 × 105 cancer cells in Hanks's buffer) was injected intradermally into the ear of NOD/SCID mice. For tumor cell/fibroblast coinjection experiments, we injected 105 cancer cells and an equal number of fibroblasts. Tumor formation was monitored over 2 to 5 weeks, followed by sacrifice of the animals and analysis of the isolated tumors.
For MIRO1-Reducer treatment experiments, SCC13 cells and primary human CAFs were coinjected intradermally, and tumor formation was observed during 2 weeks. Tumors were then treated with MIRO1-Reducer (2.5 μmol/L; #7091, Tocris) or vehicle (DMSO) every 3 to 4 days by direct injection into the tumor. Tumors were harvested 3 days after the last injection.
Spheroid formation and growth assay
Spheroid assays were performed and reported according to experimental parameters proposed by The MISpheroID Consortium (27). Twenty microliters of conditioned media from primary human skin fibroblasts (preconditioned medium: DMEM/1% FBS/1% P/S) and containing 2,000 SCC13 cells were placed on the lids of 6-cm culture plates using a hanging-drop method (28). To prevent dehydration, 5 mL PBS was added to the bottom. Cells were incubated at 37°C and 5% CO2. Spheroids were incubated for 3 days and analyzed for an increase in the spheroid area.
Bioinformatic analysis
Gene expression data for MIRO1 were acquired from publicly available gene expression profiling data of human cancers (GSE45001, GSE9014, and GSE40595) and analyzed using the GEO2R analysis tool in the GEO database (29) and Gene Expression Profiling Interactive Analysis 2.0 (GEPIA2; ref. 30). Single-cell RNA sequencing (scRNA-seq) data from melanomas were investigated through the use of publicly available scRNA data sets (https://singlecell.broadinstitute.org/single_cell). The correlation between MIRO1 expression and expression of other genes in pan-cancer was investigated using Tumor Immune Estimation Resource 2 (TIMER2; http://timer.cistrome.org/; ref. 31). The effect of MIRO1 knockdown in genome-wide knockdown screens was analyzed using DepMap (DepMap.org). Survival graphs were generated based on the PROGgene gene expression–based survival analysis web application by using median gene expression value as a dividing point, and statistical analysis of patient survival data was performed as described (32).
Ex vivo tumor explant cultures
Ear skin tumors were cut into fragments of approximately 0.5 cm2 and cultured at the air–liquid interface in DMEM/10% FBS supplemented with 5 μg/mL insulin (I5500), 0.1 nmol/L cholera toxin (C8052), 10 ng/mL epidermal growth factor (E4127), 50 IU/ml P/S and 0.4 μg/mL hydrocortisone (#386698; all from Sigma-Aldrich). After overnight incubation, they were treated with follistatin (50 ng/mL) (#120-13, PeproTech) for 6 hours and embedded in paraffin.
Mitochondrial extraction
Mitochondrial extraction was carried out using the Mitochondria Isolation Kit for Cultured Cells (#89874, Thermo Fisher Scientific). In brief, 20,000,000 primary human fibroblasts were trypsinized, suspended in 1 mL of complete DMEM, and centrifuged at 850 × g for 2 minutes. The supernatant was discarded, and 800 μL Reagent A (with 1× cOmplete EDTA-free cocktail proteinase inhibitor; Sigma-Aldrich) was added to the pellet. After vortexing at medium speed for 5 seconds, the samples were incubated on ice for 2 minutes. Ten microliters of Reagent B was then added and vortexed for 5 seconds. The cells were then incubated on ice for 5 minutes, vortexing every minute. 800 μL of Reagent C (with 1× cOmplete EDTA-free cocktail proteinase inhibitor) was then added, and the cells were inverted several times. Samples were centrifuged at 700 × g for 10 minutes at 4°C. The pellet contained the total cell lysate and was stored at −20°C. The supernatant was collected and centrifuged again at 3,000 g for 15 minutes. The resultant supernatant (cytosolic fraction) and the pellet (mitochondrial fraction) were stored at −20°C.
Statistical analysis
Statistical analysis was performed using the PRISM software, version 9 for Mac OS X or Windows (GraphPad Software Inc). For comparison of two groups, an unpaired Student t test was performed; for comparison of more than two groups, one-way or two-way ANOVA and Bonferroni multiple comparisons test were used. Nonsignificant (ns), P > 0.05; *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
Data availability
The data generated in this study are available within the article and its Supplementary Data files.
Results
mDia2 promotes protein secretion and ECM deposition by primary human fibroblasts
The key role of mDia2 in fibroblast-CAF reprogramming (7, 8) and the pivotal contribution of CAF-derived soluble factors and ECM proteins to cancer progression (33) prompted us to assess the involvement of mDia2 in the production of a protumorigenic matrisome and secretome. Knockdown of mDia2 in primary HDF using shRNA (sh-mDia2; ref. 17) indeed reduced the overall amounts of secreted proteins compared with cells transduced with lentiviruses carrying an empty vector (sh-EV cells; Fig. 1A). The levels of secreted fibronectin 1 (FN1) and collagen IV (COL4), and of activin A (encoded by the INHBA gene; ref. 9), which follow the classic endoplasmic reticulum (ER)-Golgi secretion pathway (34), were markedly reduced by mDia2 knockdown, whereas their intracellular levels were not affected (Fig. 1B–E). This suggested that mDia2 regulates protein secretion. Indeed, the ECM deposited by the sh-mDia2 cells was strongly depleted of FN1, COL1A1, and elastin (ELN), and the conditioned medium (CM) of sh-mDia2 cells contained significantly lower amounts of IFNα2, IL1β, and IL6, compared with control cells (Fig. 1F and G). Importantly, it failed to promote anchorage-independent spheroid growth of SCC13 cells (Fig. 1H). mDia2 knockdown HDFs also showed fewer stress fibers and reduced migratory abilities (Fig. 1I–K). These phenotypes, as well as the reduced FN1 secretion, were rescued by reexpression of wild-type mDia2, but not of an actin polymerization-deficient mDia2 mutant (Fig. 1J–L; refs. 17, 35). Neither wild-type nor mutant mDia2 expression affected the intracellular FN1 levels (Fig. 1M).
Figure 1.
mDia2 promotes protein secretion from HDFs. A, Quantification of total proteins secreted by sh-EV and sh-mDia2 HDFs. n = 3. B, Representative Western blots of CM of sh-EV and -mDia2 fibroblasts for fibronectin 1 (FN1), collagen (COL)4A1, and INHBA under reducing conditions. Ponceau S staining of the membrane served as loading control. C and D, Representative immunofluorescence images of sh-EV and sh-mDia2 fibroblasts stained for INHBA, FN1, and ELN (C), and quantification of intracellular fluorescence intensity (D). n = 3. E, Western blot for FN1, COL4A1, elastin (ELN), and vinculin (VIN, loading control) using total lysates of sh-EV or sh-mDia2 fibroblasts. n = 3. F, Representative FN1, COL1A1, and ELN immunofluorescence images and quantification of the stained area in the decellularized matrix from sh-EV and sh-mDia2 fibroblasts cultured in glucose-containing medium. n = 3. G, IFNα2, IL1β, and IL6 concentrations (ng/mL) in supernatants from sh-EV and sh-mDia2 fibroblasts. n = 3. H, Quantification of the SCC13 tumor spheroid area in hanging drop including CM from sh-mDia2 and sh-EV fibroblasts and representative images (left). The area of the spheroid formed in one control sample was set to 1. n = 10–11. I, Western blots of total lysate from sh-EV or sh-mDia2 fibroblasts transfected with expression vectors encoding wild-type (WT) mDia2, an actin-polymerization–deficient mDia2 mutant (IA) or empty vector (pMX) for mDia2 and GAPDH. J and K, Percentage of total cells with actin stress fibers (J) and scratch closure (K) of sh-EV and sh-mDia2 fibroblasts transfected with expression vectors encoding WT or IA mDia2 or pMX. n = 3 (J) and 6 (K). L and M, sh-EV or -mDia2 fibroblasts were transfected with expression vectors encoding WT or IA mDia2 or pMX. Quantification of FN1 immunofluorescence staining of the decellularized ECM (L), and representative Western blot of total cell lysate and data quantification (M). n = 3. Graphs show mean ± SEM. ns, P > 0.05; *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001 [one-way ANOVA with Bonferroni post hoc test (D, E, J, and K–M), or unpaired Student t test (A and F–H)]. Scale bars, 50 μm (C), 100 μm (F).
mDia2 regulates mitochondrial distribution and function in fibroblasts
To unravel how mDia2 regulates protein secretion, we analyzed key components of the secretory pathway. mDia2 knockdown HDFs showed the expected size increase (7, 35), but they neither exhibited obvious morphologic abnormalities of the ER, the Golgi apparatus, or the microtubule network nor significant differences in the levels of the ER or Golgi markers calnexin and GM130, respectively (Supplementary Fig. S1A–S1D).
Given that efficient protein secretion requires a high cellular metabolic activity and intact mitochondria (36), and that the actin cytoskeleton contributes to the structural integrity and dynamics of mitochondria (37), we stained mitochondria in mDia2 knockdown HDFs with MitoTracker. mDia2-depleted cells showed perinuclear clustering of mitochondria, whereas mitochondria in control fibroblasts were evenly distributed from perinuclear to peripheral regions (Fig. 2A). These results were verified with two published siRNAs (16) and with HDFs from two different donors (Supplementary Fig. S1E–S1I). There was also a small increase in mitochondrial mean branch length and network branch in sh-mDia2 fibroblasts, indicative of elongated and hyperfused mitochondria. Furthermore, the localization of mDia2 in primary human foreskin and breast skin fibroblasts was suggestive of an association with the mitochondrial network (Supplementary Fig. S2A and S2B). Costaining with MitoTracker supported this association in human fibroblasts, whereas the colocalization of mDia2 and mitochondria was less obvious in different epithelial cell lines (Supplementary Fig. S2C). Western blot analysis of subcellular fractions of HDFs confirmed the known localization of mDia2 in the cytoplasm of fibroblasts (7), but also showed its association with mitochondria. The mitochondria-binding protein MIRO1 and the mitochondrial matrix and housekeeping protein HSP60 served as positive controls (Fig. 2B). The partial colocalization of MIRO1 and mDia2 in skin fibroblasts bolsters this conclusion (Supplementary Fig. S2D). Proper mitochondrial distribution in mDia2 knockdown fibroblasts was restored upon reexpression of wild-type mDia2, but not of the IA mutant (Fig. 2C and D). Moreover, wild-type mDia2 partially rescued mitochondrial topology (Supplementary Fig. S2E).
Figure 2.
mDia2 regulates mitochondrial function in HDFs. A, Representative images of sh-EV and sh-mDia2 HDFs stained with MitoTracker (green), rhodamine-coupled phalloidin (red), and Hoechst (blue). The graph shows the fluorescence intensity of MitoTracker relative to the highest intensity value. The relative distance is indicated with values from 1 to 10 (e.g., on the top). N = 10. B, Representative Western blots for mDia2, MIRO1, and HSP60 using cytoplasmic (Ct) and mitochondrial (Mt) fractions, and total lysate (TL) of HDFs. C and D, sh-mDia2 fibroblasts were transfected with pMX‐WT or pMX‐IA expression vectors or pMX, and stained with MitoTracker (C). sh-EV fibroblasts were used for comparison. Bar graph shows the fluorescence intensity distribution of MitoTracker relative to the highest intensity value from the perinuclear region to the plasma membrane (D) as described in A. n = 6. E–G, Representative images of sh-EV and sh-mDia2 HDFs stained with CellROX (E) or MitoSOX (red; F), counterstained with Hoechst (blue). Bar graphs show quantification of staining intensity. Cells were analyzed for LDH release (G). n = 3–4. H, Relative intracellular ATP levels in sh-EV or sh-mDia2 HDFs transfected with pMX-WT, pMX-IA, or pMX. n = 3. I, Relative levels of mtDNA in sh-EV and sh-mDia2 HDFs determined by qPCR. n = 3. J, Western blot of total lysate of sh-EV or sh-mDia2 fibroblasts for HSP60 and GAPDH, and quantification of the HSP60/GAPDH ratio. n = 3. K, Quantification of FN1- and COL1A1-stained area in the decellularized matrix from sh-EV and sh-mDia2 fibroblasts cultured in galactose-containing medium. n = 3. L, Quantification of data from Seahorse analysis from sh-EV and sh-mDia2 HDFs cultured in galactose-containing medium. n = 6. Graphs show mean ± SEM. ns, P > 0.05; *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001 [one-way ANOVA with Bonferroni post hoc test (H), two-way ANOVA with Bonferroni post hoc test (A and D), or unpaired Student t test (E–G and I–L)]. Scale bars, 25 μm (A and C), 100 μm (E and F).
The altered position and topology of mitochondria in the mDia2 knockdown fibroblasts were accompanied by a strong increase in intracellular reactive oxygen species (ROS) and mitochondrial superoxide levels (Fig. 2E and F). However, this did not affect cell viability as shown by the measurement of lactate dehydrogenase (LDH) in the supernatant (Fig. 2G). Total intracellular ATP levels were only mildly affected (Fig. 2H). Reexpression of wild-type mDia2, but not of the IA mutant, rescued and even further increased intracellular ATP levels compared with control cells (Fig. 2H). Levels of mitochondrial DNA and HSP60 were only slightly reduced in the mDia2 knockdown cells (Fig. 2I and J), suggesting that the increased ROS levels are not a consequence of an increase in the number of mitochondria.
Replacing glucose in the cell culture medium by galactose allows unmasking mitochondrial dysfunctions, particularly defects in oxidative phosphorylation (OXPHOS), because (i) cells rely on OXPHOS for efficient ATP production and (ii) oxidation of galactose to pyruvate via glycolysis does not yield net ATP (38). When cultured in the presence of galactose, the mDia2 knockdown fibroblasts displayed an even more dramatic reduction in the secretion of FN1 and COL1A1 (Fig. 2K vs. Fig. 1F). This correlated with impairments in the mitochondrial oxygen respiration rate (OCR) and reduced ATP-linked mitochondrial respiration in mDia2 knockdown versus control fibroblasts (Fig. 2L). Only a slight reduction in the basal respiration rate was observed in medium with glucose, suggesting partial compensation by glycolysis (Supplementary Fig. S2F). The basal OCR/ECAR (extracellular acidification rate) ratio of mDia2 knockdown fibroblasts was significantly higher compared with control cells, indicating their relatively higher reliance on mitochondrial OXPHOS (Supplementary Fig. S2G). These results identify mitochondrial dysfunction in mDia2 knockdown fibroblasts.
mDia2 stabilizes the mitochondrial trafficking protein MIRO1
Our results raise the possibility that mDia2 affects proteins involved in mitochondrial trafficking. Among them, Rho-GTPase MIRO1/RHOT1 (39) was an attractive candidate. Indeed, MIRO1 protein, but not mRNA levels, was strongly reduced in the mDia2 knockdown fibroblasts (Fig. 3A and B). Knockdown of mDia2 in fibroblasts also reduced MIRO1 protein levels in the mitochondrial fraction (Fig. 3C). MIRO1 colocalized with mitochondria and clustered in the perinuclear region of sh-mDia2 fibroblasts (Fig. 3D).
Figure 3.
mDia2 binds and stabilizes MIRO1 in primary human fibroblasts. A, Representative Western blot of total lysate of sh-EV or sh-mDia2 fibroblasts for mDia2, MIRO1, GAPDH, and α-tubulin. B, qRT-PCR analysis for MIRO1 using RNA from sh-EV and sh-mDia2 HDFs. n = 3. C, Representative Western blot of mitochondrial fractions from sh-EV and sh-mDia2 fibroblasts probed for mDia2, MIRO1, and HSP60, and quantification of mitochondrial mDia2/HSP60 or MIRO1/HSP60 ratios. n = 3. D, Representative images of sh-EV and sh-mDia2 HDFs immunostained for MIRO1 (red), counterstained with MitoTracker (green), and Hoechst (blue). Dotted lines outline the cell border. E, Western blot of total lysate from sh-EV or sh-mDia2 fibroblasts transfected with pMX‐WT, pMX-IA, or pMX‐control for MIRO1 and GAPDH, and quantification of the MIRO1/GAPDH ratio. n = 3. F, Representative Western blots for MIRO1 and vinculin using total lysates of sh-EV and sh-mDia2 fibroblasts treated for 2 hours with epoxomicin (Epox; 10 μmol/L) and quantification of the MIRO1/VINCULIN ratio. n = 3 biological replicates. G, Representative images of sh-mDia2 fibroblasts treated with epoxomicin (10 μmol/L) or vehicle and stained with MitoTracker (green) and rhodamine-coupled phalloidin (red). Bar graphs show the fluorescence intensity distribution of MitoTracker relative to the highest intensity value from the perinuclear region to the plasma membrane and mean network branch quantified using the MiNA toolset. n = 22–29. H and I, Representative images showing PLA signals (red) in HDFs stained with mDia2 and MIRO1 antibodies and in the negative control (-Ctrl) without primary antibodies (H), counterstained with Hoechst (blue), either alone (H) or together with MitoTracker (green; I). Graphs show mean ± SEM. ns, P > 0.05; *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001 [one-way ANOVA with Bonferroni post hoc test (E–G for network analysis), two-way ANOVA with Bonferroni post hoc test (G for MitoTracker intensity), or unpaired Student t test (B and C)]. Scale bars, 50 μm.
Reexpression of wild-type, but not IA mDia2, partially restored MIRO1 protein levels in the mDia2 knockdown fibroblasts (Fig. 3E). Consistent with the findings that mDia2 increases proteasome activity (20) and that ubiquitinated MIRO1 is targeted for proteasomal degradation (40), the proteasome inhibitor epoxomicin partially rescued MIRO1 protein levels in the mDia2 knockdown fibroblasts and normalized their mitochondrial distribution and topology (Fig. 3F and G). The stabilization of MIRO1 by mDia2 is likely the consequence of their physical interaction as supported by PLA (Fig. 3H). PLA combined with MitoTracker staining showed that the mDia2–MIRO1 complex localizes to mitochondria (Fig. 3I).
mDia2 and MIRO1 levels in human cancer stroma are predictors of poor prognosis
We next tested if MIRO1 is abnormally expressed and functionally relevant in human cancer tissue. We first made use of the Cancer Dependency Map (Depmap), a genome-wide loss-of-function screening database (41), to determine the effect of MIRO1 knockdown on cancer features. Dependency score analysis showed that MIRO1 plays an integral role in the proliferation and survival of various cancer cells (Supplementary Fig. S3A). Analysis of data from The Cancer Genome Atlas (TCGA) revealed a consistent upregulation of MIRO1 in the stroma of liver, breast, and ovarian cancers in comparison with the stroma of healthy tissue (Fig. 4A), which was not observed in bulk tumor tissue (Supplementary Fig. S3B). scRNA-seq showed remarkable coexpression of DIAPH3 and MIRO1 in melanoma CAFs (Fig. 4B). Strong expression of MIRO1 positively correlates with that of mDia2 and INHBA, but not of TGFB1, in different cancers (Fig. 4C). Kaplan–Meier survival analysis showed a strong association of high coexpression of mDia2 and MIRO1 with decreased survival of patients with liver or lung cancer (Fig. 4D). Notably, numbers of MIRO1-positive cells were much higher in the stroma of epithelial skin cancers than in healthy skin (Fig. 4E). Costaining of SCC sections for MIRO1 and the pan-fibroblast marker PDGFR-α indicated that many of the MIRO1-positive cells are indeed fibroblasts. Coexpression of both proteins was less pronounced in normal skin (Supplementary Fig. S3C). These findings point to a protumorigenic effect of MIRO1 in CAFs of different human cancers, although an additional protumorigenic function of this protein in cancer cells or other cells of the TME cannot be excluded.
Figure 4.
Overexpression of mDia2 and MIRO1 in CAFs promotes tumorigenesis. A, Expression of MIRO1 in liver, breast, and ovarian cancer stroma (CS) vs. stroma of respective normal tissues (NS) based on data sets GSE45001 (N = 10 per group), GSE9014 (N = 6 and 53), and GSE40595 (N = 8 and 36). B, Single-cell expression profiles of mDia2/DIAPH3 and MIRO1 in immune cells, endothelial cells, and CAFs of melanomas based on data from Single-Cell Portal (Broad Institute). C, Correlation analysis between MIRO1 and mDia2, INHBA, and TGFB1 gene expression across different cancers using TIMER2. UVM, uveal melanoma; LUAD, lung adenocarcinoma; PAAD, pancreatic ductal adenocarcinoma; SKCM, skin cancer cutaneous melanoma; BRCA-Lum, luminal breast cancer; LIHC, liver hepatocellular carcinoma. Red or blue indicates a statistically significant positive/negative correlation (Spearman, P < 0.05), respectively, and gray denotes a nonsignificant result. D, Kaplan–Meier survival curves based on TCGA and GTEx data. High expression of mDia2/DIAPH3 and MIRO1 in patients with liver or lung cancer correlates with poor survival. E, Representative sections from normal human skin, cSCC, and BCC stained for MIRO1. Scale bars, 100 μm. T, tumor cells. F, qRT-PCR analysis for MIRO1 using RNA from CAFs or NFs from two SCC patients. n = 3. G, Representative Western blot of total lysates of NFs and CAFs from patient No. 1 for MIRO1, CAF markers, and GAPDH. H, Quantification of SCC13 tumor spheroid area in hanging drop including CM from NF and CAFs. n = 10–11. I, Mitochondrial distribution in NFs or CAFs from patient No.1. n = 15–19. J, Representative images and bar graph showing fluorescence intensity distribution of ATP biosensor (cyto-Ruby3-iATPSnFR1.0) relative to the highest intensity value from the perinuclear region to the plasma membrane of NFs and CAFs (patient No. 1). n = 19. K, Mitochondrial branch length of NFs or CAFs (patient No.1). n = 15–19. L, Representative immunofluorescence images of decellularized matrices obtained from NFs and CAFs (patient No. 1) stained for FN1 and COL1A1 and quantification of the stained area. n = 3. M, Quantification of SCC13 tumor spheroid area in hanging drop including CM from NFs transfected with an expression vector encoding human MIRO1 (pGEM‐MIRO1 “+”) or empty vector (pGEM‐EV “−”). The spheroid area in one control sample was set to 1. n = 10–12. N, SCC13 cells plated on either decellularized matrix from NFs transfected with pGEM‐MIRO1 or pGEM‐EV. After 3 days, the number of Ki67-positive SCC13 cells was quantified. n = 3. O, Representative FN1 and COL1A1 immunofluorescence stainings and quantification of the stained area in the decellularized matrix derived from NFs transfected with pGEM‐MIRO1 or pGEM‐EV. n = 3. P and Q, sh-EV and sh-mDia2 fibroblasts were transfected with pGEM‐MIRO1 (“+”) or pGEM‐EV (“−”). Expression of MIRO1 was analyzed by immunofluorescence 24 hours after transfection (P). n = 3. Bar graph shows the fluorescence intensity distribution of MitoTracker relative to the highest intensity value from the perinuclear region to the plasma membrane (Q). n = 6–8. Bar graphs show mean ± SEM. ns, P > 0.05; *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001 [one-way ANOVA with Bonferroni post hoc test (P), two-way ANOVA with Bonferroni post hoc test (I, J, and Q), unpaired Student t test (A, F, H, and K–O)]. Scale bars, 50 μm.
To assess the functional relevance of the high MIRO1 levels in CAFs for cutaneous SCC pathogenesis, we used low-passage primary CAFs and matched normal HDFs (NF) from human SCC lesions and nontumorigenic skin, respectively. The cultured CAFs showed a significantly higher expression of MIRO1, mDia2, and other CAF markers, such as INHBA and FN1, compared with NFs, and they strongly promoted anchorage-independent growth of SCC13 cells in spheroids (Fig. 4F–H). The increased expression of mDia2 and MIRO1 in CAFs versus NFs did not further enhance peripheral mitochondrial positioning (Fig. 4I). However, using a fluorescent ATP sensor, we found that ATP was clearly detectable at the cell periphery of CAFs, but not of NFs from two different donors, whereas strong perinuclear fluorescence was observed in both CAFs and NFs (Fig. 4J; Supplementary Fig. S3D). Thus, ATP levels near the plasma membrane, which are required to supply energy for protein secretion (42), are higher in CAFs compared with NFs. Mitochondria were also more fragmented in CAFs, resulting in decreased mean branch length (Fig. 4K). MIRO1, in complex with TRAK1 and microtubule motor proteins, regulates anterograde mitochondrial movement in mammalian cells, the increase of which promotes cancer cell invasiveness and viability (43). Using PLA, we found a higher MIRO1–TRAK1 interaction in CAFs versus NFs (Supplementary Fig. S3E). These findings strongly suggest that mitochondrial trafficking and function are altered in CAFs, thereby promoting protein secretion. Indeed, deposition of FN1 and COL1A1 was strongly increased in the same CAFs (Fig. 4L). Overexpression of MIRO1 in NFs recapitulated the protumorigenic secretion-dependent effects of CAFs. In particular, isolated CM and decellularized ECM from NFs overexpressing MIRO1 promoted the spheroid formation and proliferation of the weakly malignant SCC13 cell line (Fig. 4M and N). This is in line with their increased deposition of the protumorigenic ECM proteins FN1 and COL1A1 (Fig. 4O). Treatment of MIRO1-overexpressing fibroblasts with FCCP to disrupt the mitochondrial membrane potential and consequent ATP production reduced the deposition of FN1 and COL1A1 (Supplementary Fig. S3F and S3G). This further supports the functional link between mitochondrial activity and the secretion of protumorigenic proteins by fibroblasts with high MIRO1 levels. Finally, overexpression of wild-type MIRO1 in mDia2 knockdown fibroblasts rescued their mitochondrial distribution defect (Fig. 4P and Q).
MIRO1 is a target of activin A/Smad2/3 signaling in fibroblasts
The co-upregulation of INHBA, mDia2, and MIRO1 in CAFs raised the question as to whether MIRO1 expression is also controlled by activin A. Indeed, treatment of HDFs with activin A increased the expression of MIRO1 (Fig. 5A). We identified a conserved SMAD binding element (SBE) within the 2-kb region upstream of the MIRO1 gene transcription start site (TSS), and activin A promoted the binding of SMAD2/3 to this SBE (Fig. 5B). In vivo, MIRO1 was strongly expressed in the stroma of xenograft tumors that formed upon injection of SCC13 cells overexpressing INHBA (7) into the ear skin of immunodeficient NOD/SCID mice (Fig. 5C). Vice versa, overexpression of the secreted activin antagonist follistatin in SCC13 cells (7) reduced the expression of MIRO1 in the stroma, and a similar effect was observed when biopsies from tumors generated by wild-type SCC13 cells were treated ex vivo with follistatin (Fig. 5D and E). Characterization of HDFs with DOX-inducible expression of follistatin (Fb FST) showed that both the mRNA and protein levels of MIRO1 and mDia2 were significantly downregulated (Fig. 5F–I). Furthermore, Fb FST exhibited changes in mitochondrial distribution (Fig. 5J), resembling those observed in mDia2-depleted fibroblasts. These abnormalities as well as the reduced efficacy of CM of Fb FST to promote anchorage-independent growth of SCC13 cells in spheroids were rescued by overexpression of MIRO1 in these cells (Fig. 5K–M). These results demonstrate that MIRO1 is a direct and functionally relevant target of activin A.
Figure 5.
MIRO1 is a target of activin A–SMAD2/3 signaling. A, HDFs were treated with 20 ng/mL activin A or vehicle for 6 hours and analyzed for MIRO1 expression by qRT-PCR. n = 3. B, ChIP using lysates from activin A– or vehicle-treated fibroblasts and a SMAD2/3 antibody, and amplification of the bound DNA with MIRO1 primers (top). Preimmune serum (IgG) was used as a negative control. Alignment of the sequences within the 2-kb region upstream of the MIRO1 gene TSS of different species that harbor a SMAD2/3 binding element (SBE; red and italics; bottom). C and D, Representative images of sections from ear skin tumors formed by SCC13 cells with DOX-inducible lentiviral overexpression of INHBA (SCC13 Act; C) or FST (SCC13 FST; D) or transduced with EV, immunostained for MIRO1 (green) or αSMA (red), and counterstained with Hoechst (blue). Graphs show the percentage of MIRO1- relative to αSMA–positive area (percentage of MIRO1+ area in stroma). n = 3 tumors per group. Tumors were collected 5 or 12 weeks after the initial injection of SCC13 Act or -FST. This accounts for the higher MIRO1 positivity of EV in D compared with C. E, Tumors formed by SCC13 Act cells were treated ex vivo for 6 hours with 50 ng/mL follistatin, immunostained for MIRO1 (green) or αSMA (red), and counterstained with Hoechst (blue). Graph shows the percentage of MIRO1- relative to αSMA–positive area. n = 4. F and G, qRT-PCR for FST (F) and MIRO1 (G) using RNA collected 24 hours after DOX treatment from clonally expanded HDFs transduced with empty vector (Fb-EV) or with DOX-inducible expression of FST (Fb FST1, clone 1; Fb FST2, clone 2). n = 3. H, Western blots for MIRO1 and GAPDH using total lysates of Fb FST (clone 1) and Fb-EV. I, qRT-PCR for DIAPH3 using RNA collected 24 hours after DOX treatment from HDFs transduced with Fb-EV or Fb FST1. n = 3. J, Representative images of Fb-EV or -FST1 stained with MitoTracker (green) and Hoechst (blue). K and L, Fb FST1 and control fibroblasts were transfected with pGEM‐MIRO1 (“+”) or pGEM‐EV (“−”). Expression of MIRO1 was analyzed by immunofluorescence 24 hours after transfection (K). n = 3. Graph shows fluorescence intensity distribution of MitoTracker relative to the highest intensity value from the perinuclear region to the plasma membrane (L). n = 8. M, Quantification of SCC13 spheroid area in hanging drops including CM from Fb FST1 and their controls transfected with pGEM‐MIRO1 or pGEM‐EV. The area of the spheroid in one control sample was set to 1. n = 8. Bar graphs show mean ± SEM. ns, P > 0.05; *, P < 0.05; **, P < 0.01; ***, P < 0.001 [one-way ANOVA with Bonferroni post hoc test (F, G, K, and M), two-way ANOVA with Bonferroni post hoc test (L), or unpaired Student t test (A, C–E, and I)]. Scale bars, 50 μm (C–E), 25 μm (J).
MIRO1 is required for efficient protein secretion by CAFs
Given the crucial role of mDia2 in the conversion of murine dermal fibroblasts into protumorigenic CAFs (7), we determined if knockdown of MIRO1 had a similar effect as knockdown of mDia2. MIRO1 silencing in patient-derived CAFs (Fig. 6A and B; Supplementary Fig. S4A) had no effect on the expression of major skin CAF marker genes. Accordingly, nuclear p53 levels in si-MIRO1 cells were similar to those in control cells (Fig. 6C). In addition, MIRO1 silencing did not affect the expression of other small GTPases (RHOA, CDC42, RALA, and RHOT2; Fig. 6D; refs. 12, 44). However, it caused the expected perinuclear mitochondrial clustering, reduced the deposition of COL1A1 and also enhanced the levels of mitochondrial superoxide (Fig. 6E–G). When we cultured si-MIRO1 cells in a medium containing high glucose, total cellular ATP levels were not reduced by MIRO1 knockdown (Supplementary Fig. S4B), as previously shown with other cells (45, 46). However, the distribution of intracellular ATP was affected, with ATP being reduced at the cell periphery (Supplementary Fig. S4C). When cells were cultured in a medium containing galactose instead of glucose, MIRO1 knockdown caused a significant reduction of total cellular ATP levels and an even more dramatic increase in mitochondrial superoxide (Supplementary Fig. S4D and S4E vs. Fig. 6G). This correlated with lower proliferation and migration rates of si-MIRO1 CAFs in medium with galactose (Supplementary Fig. S4F and S4G). Survival of MIRO1 knockdown cells was reduced in medium with galactose, but not in medium with glucose (Fig. 6H). Basal and maximal OCR as well as ATP-linked OCR were significantly lower in si-MIRO1 versus control CAFs cultured in the presence of galactose, but not in the presence of glucose (Fig. 6I). These results suggest that MIRO1 knockdown cells fail to increase their oxygen consumption capacity via OXPHOS when cultured in galactose medium and further points to their mitochondrial dysfunction.
Figure 6.
Knockdown of MIRO1 causes metabolic dysfunction and impaired protein secretion in primary human skin CAFs. A and B, CAFs from patient No. 1 were transfected with MIRO1 (si-MIRO1) or control (si-Ctrl) siRNAs. The knockdown was confirmed by qRT-PCR (A) and immunofluorescence staining (B). C, Quantification of nuclear p53 in CAFs transfected with Ctrl or MIRO1 siRNAs. n = 3. D, qRT-PCR for CAF marker genes and genes encoding different small GTPases using RNA from si-MIRO1 or si-Ctrl CAFs. n = 3. E, Representative images of si-MIRO1 or si-Ctrl CAFs (patient No.1) stained for MIRO1 (red), counterstained with MitoTracker (green) and Hoechst (blue). F, Quantification of COL1A1 -stained area in the decellularized matrix from si-Ctrl or si-MIRO1 CAFs. n = 3. G, Relative levels of mitochondrial superoxide based on MitoSOX fluorescence intensity in CAFs (patient No.1) transfected with si-Ctrl or si-MIRO1. n = 3. H, CAFs (patient No. 1) transfected with si-Ctrl or si-MIRO1 were analyzed for LDH release. n = 3. I, Seahorse assay for the OCR in CAFs (patient No. 1) transfected with si-Ctrl or si-MIRO1 and quantification of basal respiration, maximal respiratory capacity, and ATP production in glucose (top) or galactose (bottom) medium. n = 5–6. J, Quantification of total proteins secreted by si-MIRO1 or si-Ctrl CAFs (patient No. 1) in glucose medium. n = 6. K and L, Representative images of immunofluorescence stainings for FN1 and COL1A1 (K) and quantification of the stained area in the decellularized matrix (L) from si-Ctrl or si-MIRO1 CAFs (patient No. 1) in glucose medium. n = 3. M, Quantification of intracellular fluorescence in si-Ctrl or si-MIRO1 CAFs (patient No. 1) immunostained for INHBA or ACTA2. n = 3. Bar graphs show mean ± SEM. ns, P > 0.05; *, P < 0.05; **, P < 0.01 [two-way ANOVA with Bonferroni post hoc test (D), unpaired Student t test (other graphs)]. Scale bars, 50 μm (E) and 100 μm (B and K). Data shown in K and L were reproduced with CAFs from a second patient.
Overall protein secretion was already reduced in the MIRO1 knockdown CAFs when cultured in glucose medium, and they deposited much less FN1 and COL1A1 (Fig. 6J–L). However, expression of the CAF markers INHBA and ACTA2 was comparable at the RNA (Fig. 6D) and protein levels (Fig. 6M). Similar results were obtained using a second siRNA (Supplementary Fig. S4H and S4I). HDFs with DOX-inducible overexpression of INHBA (Fb Act), which acquire a CAF phenotype (7), confirmed these findings. In these cells, the knockdown of MIRO1 significantly reduced the secretion of mature activin A and resulted in the concomitant intracellular accumulation of the INHBA precursor, whereas INHBA mRNA levels were not affected (Supplementary Fig. S5A–S5E). Taken together, we observed similar changes in protein secretion and mitochondrial positioning in fibroblasts with either MIRO1 or mDia2 knockdown.
mDia2 and MIRO1 position mitochondria at CAF–tumor cell contact sites to support the protumorigenic activity of CAFs
We next analyzed cocultures of SCC13 cells with either sh-EV or sh-mDia2 fibroblasts and found that mitochondria in control fibroblasts were much closer to SCC13 cells (Fig. 7A and B). Pharmacologic inhibition of formins in SCC patient–derived CAFs by 25 μmol/L SMIFH2, a dose that is optimal for mDia2 inhibition (18), reduced the levels of mDia2 and also those of MIRO1 (Fig. 7C). This correlated with a reduction in F-actin staining and perinuclear mitochondrial clustering in SMIFH2-treated CAFs (Fig. 7D). Importantly, SMIFH2-treated HDFs displayed reduced secretion of FN1 and COL1A1, but no significant changes in their intracellular levels (Supplementary Fig. S6A and S6B). These data support the relevance of mDia2-dependent regulation of MIRO1 and mitochondria in NFs and patient-derived CAFs, which may affect cancer cells by regulating protein secretion.
Figure 7.
Depletion of MIRO1 in CAFs suppresses skin tumorigenesis. A and B, sh-EV or -mDia2 HDFs cocultured for 7 days with SCC13 cells immunostained for E-cadherin (red) and counterstained with MitoTracker (green) and Hoechst (blue; A). The mitochondrial distribution relative to the highest intensity value from the perinuclear region of fibroblasts to the interface with SCC13 cells was quantified (B). Relative distances range from 1 to 10. n = 4–6. C, Representative Western blots for MIRO1, mDia2, and GAPDH using total lysates of NFs and CAFs (patient No. 1) cultured for 24 hours with SMIFH2 (25 μmol/L) or vehicle. The dotted line indicates cutting of the membrane; all samples were loaded on the same gel. D, Representative images of NFs and CAFs cultured with SMIFH2 (25 μmol/L) or vehicle and stained with rhodamine‐coupled phalloidin (red), MitoTracker (green), and Hoechst (blue). E, Two-dimensional cocultures of SCC13 cells with si-Ctrl or si-MIRO1 CAFs at day 7. SCC13 cells were identified by E-cadherin (red) staining and counterstained with MitoTracker (green) and Hoechst (blue). The mitochondrial distribution relative to the highest intensity value from the perinuclear region of fibroblasts to the interface with SCC13 cells was quantified. Relative distances range from 1 to 10. n = 12–14. F, Chemotactic transwell migration of SCC13 cells using CM from si-Ctrl or si-MIRO1 CAFs. n = 3. G, Quantification of the SCC13 spheroid area in a single hanging drop including CM from si-Ctrl or si-MIRO1 CAFs. The spheroid area in one si-Ctrl sample was set to 1. N = 6. H, Tumor volume at different time points 2 weeks after injection (n = 4) and representative photos of 2-week-old tumors formed upon intradermal coinjection of SCC13 cells with si-MIRO1 or si-Ctrl CAFs. Inset, area where tumor cells invade into the cartilage. Asterisk, cartilage; arrow, site of invasion. I, Representative images of COL1A1 immunofluorescence and Herovici staining of tumors formed by SCC13 cells transfected with si-MIRO1 or si-Ctrl CAFs at day 15. J, Quantification of the percentage of the COL1A1-positive area and curvature of fibers per field of view. n = 3–4. K, Increase in SCC13 (coinjected with CAFs) tumor volume (mm3) upon injection of the tumors with MIRO1-Reducer (2.5 μmol/L per injection) or vehicle (DMSO) once every 3 to 4 days for 18 days. Red arrows, treatment time points. n = 5. Bar graphs show mean ± SEM. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001 [two-way ANOVA with Bonferroni post hoc test (B, E, H, and K), unpaired Student t test (F, G, and J)]. Scale bars, 25 μm (A, D, and E) and 50 μm (I).
To test the relevance of MIRO1 in CAFs for squamous carcinogenesis, we cocultured SCC13 cells with si-MIRO1 CAFs. These cells exhibited a similar defective mitochondrial distribution as sh-mDia2 fibroblasts (Fig. 7E). Consistently, incubation of SCC13 cells with CM collected from si-MIRO1 or control patient CAFs showed that the CM from the si-MIRO1 CAFs was significantly less efficient in promoting migration and anchorage-independent growth of the tumor cells in chemotactic transwell and spheroid formation assays (Fig. 7F and G). When we plated SCC13 cells on the decellularized matrix deposited by either control or si-MIRO1 CAFs, the colony-forming and migratory capacities of the cancer cells were significantly lower on the matrix deposited by si-MIRO1 CAFs (Supplementary Fig. S7A and S7B).
In vivo, intradermal coinjection of SCC13 cells (Fig. 7H) or of highly malignant A431 carcinoma cells (Supplementary Fig. S7C) and control primary human SCC CAFs into the ear of NOD/SCID mice resulted in rapid and efficient tumor formation. The tumors showed invasive growth, which was much less pronounced when si-MIRO CAFs were used (Fig. 7H, inset). Reduced expression of MIRO1 in the stroma of the few palpable tumors that developed under these conditions and in fibroblasts isolated from resected tumors was still visible at the assay endpoint (Supplementary Fig. S7D–S7G). This correlated with the significantly reduced deposition of collagen and increased collagen curvature (Fig. 7I–J), a feature of nonmalignant matrix remodeling (47). Tumor cell proliferation was also reduced, whereas there was no difference in the area covered by blood or lymphatic vessels (Supplementary Fig. S7H and S7I). These data clearly demonstrate a protumorigenic effect of MIRO1 in human fibroblasts. Finally, we assessed the therapeutic potential of MIRO1 inhibition in vivo. Palpable ear skin xenograft tumors formed by SCC13 cells and primary human CAFs were injected with “MIRO1-Reducer” (48) every 3 to 4 days over 18 days to induce degradation of MIRO1. Remarkably, this treatment strongly suppressed tumor growth (Fig. 7K) and correlated with reduced MIRO1 protein levels in the fibroblasts isolated from the treated tumors (Supplementary Fig. S7J).
Taken together, these results demonstrate that MIRO1 is required for the efficient secretion of proteins from CAFs, which promotes cancer cell proliferation and malignant SCC growth. Furthermore, they show that mDia2 acts through MIRO1 and mitochondria to control protumorigenic protein secretion and matrix deposition in vivo. Finally, they identify mDia2 and MIRO1 as important stromal targets for cancer therapy.
Discussion
We discovered a key role of mDia2 in the regulation of protein secretion by HDFs and CAFs, which involves stabilization of MIRO1 as well as appropriate positioning of mitochondria and ATP production at the cell periphery. Although recent studies have highlighted the importance of cellular metabolism for matrix protein synthesis in fibroblasts (49) and of metabolic reprogramming in the acquisition of a CAF phenotype (50), our work reveals that proper mitochondrial distribution and activity strongly affect the secretion of protumorigenic proteins by these cells. Knockdown of mDia2 impaired this process and led to intracellular accumulation of otherwise secreted proteins. Our biochemical and microscopy data demonstrate that mDia2 is associated with mitochondria in skin fibroblasts. These findings are consistent with actin assembly being implicated in reorganizing and shuffling mitochondria during mitosis and mitophagy (37, 51). Some of the molecular players involved in these actin-based processes are known (52), but neither mDia2’s association with mitochondria nor its role in the regulation of mitochondrial distribution and function had been described. Furthermore, we discovered that mDia2 interacts with MIRO1 and that MIRO1 is essential to sustain the protumorigenic activities of skin CAFs by allowing appropriate mitochondrial distribution and metabolic functions. First, we show that not only mDia2 but also MIRO1 is overexpressed in the stroma of various cancers, and that their combined overexpression correlates with reduced survival of patients with liver, lung, or pancreatic cancer. Second, knockdown of either mDia2 or MIRO1 strongly reduced the protumorigenic effects of patient-derived skin cancer CAFs in vitro and in vivo. The reduction in mDia2 and/or MIRO1 expression was associated with inefficient OXPHOS, reduced ATP levels—in particular at the cell periphery—and significantly reduced deposition of matrix proteins by CAFs. This is not the result of a transcriptional effect, as MIRO1 silencing did not alter the expression of several CAF marker genes. Rather, impaired mitochondrial distribution and function in these cells provide a likely explanation for the observed secretory deficiency. This is consistent with the important role of MIRO1 in the release of neurotransmitters and insulin from presynaptic nerve terminals or pancreatic β-cells, respectively (53, 54), and the positioning of mitochondria near secretory sites, thereby supplying ATP to support exocytosis (55). The coexpression of mDia2 and MIRO1 is at least in part controlled by activin A-Smad2/3 signaling. Two independent mechanisms allow mDia2 to regulate MIRO1 expression: the first mechanism involves a positive feedback loop whereby high mDia2 levels promote INHBA/activin A expression in fibroblasts and its secretion, which in turn increases mDia2 and MIRO1 transcription in these cells (ref. 7 and this study). The second relies on the ability of mDia2 to stabilize MIRO1 in fibroblasts, possibly by preventing its proteasomal degradation (this study). The mDia2–MIRO1 interaction may contribute to this effect, as mDia2 binds to ubiquitinated proteins and reduces proteasomal activity (20).
Given that optimal MIRO1 expression is key to the maintenance of a protumorigenic CAF phenotype, follistatin, neutralizing activin antibodies, or soluble activin receptor antagonists (56), which allow concomitant suppression of both mDia2 and MIRO1 activities, could be exploited to prevent the fibroblast-to-CAF reprogramming during tumor progression and may also reduce the protumorigenic activities of already existing CAFs. Suppression of mitochondrial activity, e.g., through cancer drugs that target mitochondrial metabolism in the tumor cells, is a promising approach for cancer treatment (57). Our results strongly suggest that perturbing mitochondrial positioning and function in CAFs may be therapeutically equally important. Therefore, we advocate further exploration of the potential of activin antagonists, the rational design of mDia2-specific formin inhibitors, and the development of next-generation MIRO1 inhibitors for cancer treatment.
Supplementary Material
Acknowledgments
The authors thank Drs. Pino Bordignon and Gian-Paolo Dotto, University of Lausanne, for help with the ear tumorigenesis assays, Dr. Petra Boukamp, Leibniz Institute for Environmental Medicine, Düsseldorf, for SCC13 and HaCaT cells, Dr. Irini Vgenopoulou, ETH Zurich, for valuable information on Seahorse analysis, and Drs. Jürg Hafner and Gaetana Restivo, University of Zurich, for SCC biopsies or help with CAF isolation. This work was supported by grants from Cancer Research Switzerland (KFS-4510-08-2018 to S. Werner), the Swiss National Science Foundation (grants 31003A_169204 and 31003B-189364 to S. Werner), the Wilhelm-Sander Foundation (grants 2014-059.3 to S. Werner and 2019-075.1 to Hans-Dietmar Beer for the position of M. Di Filippo), University Medicine Zurich (Project SKINTEGRITY to S. Werner), and the ETH Zurich (Open ETH Project SKINTEGRITY.CH and core funding to S. Werner and M. Kopf).
The publication costs of this article were defrayed in part by the payment of publication fees. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Footnotes
Note: Supplementary data for this article are available at Cancer Research Online (http://cancerres.aacrjournals.org/).
Authors' Disclosures
No disclosures were reported.
Authors' Contributions
M. Cangkrama: Conceptualization, data curation, formal analysis, validation, investigation, visualization, methodology, writing–original draft, writing–review and editing. H. Liu: Validation, investigation, visualization, methodology. J. Whipman: Validation, investigation, visualization, methodology. M. Zubair: Validation, investigation, visualization, methodology. M. Matsushita: Validation, investigation, visualization, methodology. M. Di Filippo: Methodology. M. Kopf: Resources, validation. M. Innocenti: Conceptualization, resources, supervision, writing–review and editing. S. Werner: Conceptualization, resources, data curation, supervision, funding acquisition, writing–original draft, project administration, writing–review and editing.
References
- 1. Sanz-Moreno V, Gaggioli C, Yeo M, Albrengues J, Wallberg F, Viros A, et al. ROCK and JAK1 signaling cooperate to control actomyosin contractility in tumor cells and stroma. Cancer Cell 2011;20:229–45. [DOI] [PubMed] [Google Scholar]
- 2. Baghban R, Roshangar L, Jahanban-Esfahlan R, Seidi K, Ebrahimi-Kalan A, Jaymand M, et al. Tumor microenvironment complexity and therapeutic implications at a glance. Cell Commun Signal 2020;18:59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Sahai E, Astsaturov I, Cukierman E, DeNardo DG, Egeblad M, Evans RM, et al. A framework for advancing our understanding of cancer-associated fibroblasts. Nat Rev Cancer 2020;20:174–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Melchionna R, Spada S, Di Modugno F, D'Andrea D, Di Carlo A, Panetta M, et al. The actin modulator hMENA regulates GAS6-AXL axis and pro-tumor cancer/stromal cell cooperation. EMBO Rep 2020;21:e50078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Innocenti M. New insights into the formation and the function of lamellipodia and ruffles in mesenchymal cell migration. Cell Adh Migr 2018;12:401–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Isogai T, Innocenti M. New nuclear and perinuclear functions of formins. Biochem Soc Trans 2016;44:1701–8. [DOI] [PubMed] [Google Scholar]
- 7. Cangkrama M, Wietecha M, Mathis N, Okumura R, Ferrarese L, Al-Nuaimi D, et al. A paracrine activin A–mDia2 axis promotes squamous carcinogenesis via fibroblast reprogramming. EMBO Mol Med 2020;12:e11466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Nishida Y, Nagatsuma AK, Kojima M, Gotohda N, Ochiai A. Novel stromal biomarker screening in pancreatic cancer patients using the in vitro cancer-stromal interaction model. BMC Gastroenterol 2020;20:411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Cangkrama M, Wietecha M, Werner S. Wound repair, scar formation, and cancer: converging on activin. Trends Mol Med 2020;26:1107–17. [DOI] [PubMed] [Google Scholar]
- 10. Wietecha MS, Pensalfini M, Cangkrama M, Muller B, Jin J, Brinckmann J, et al. Activin-mediated alterations of the fibroblast transcriptome and matrisome control the biomechanical properties of skin wounds. Nat Commun 2020;11:2604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Purdie KJ, Pourreyron C, South AP. Isolation and culture of squamous cell carcinoma lines. Methods Mol Biol 2011;731:151–9. [DOI] [PubMed] [Google Scholar]
- 12. Procopio MG, Laszlo C, Al Labban D, Kim DE, Bordignon P, Jo SH, et al. Combined CSL and p53 downregulation promotes cancer-associated fibroblast activation. Nat Cell Biol 2015;17:1193–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hiebert P, Wietecha MS, Cangkrama M, Haertel E, Mavrogonatou E, Stumpe M, et al. Nrf2-mediated fibroblast reprogramming drives cellular senescence by targeting the matrisome. Dev Cell 2018;46:145–61. [DOI] [PubMed] [Google Scholar]
- 14. Okumoto K, Ono T, Toyama R, Shimomura A, Nagata A, Fujiki Y. New splicing variants of mitochondrial Rho GTPase-1 (Miro1) transport peroxisomes. J Cell Biol 2018;217:619–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Li Q, Yao L, Wei Y, Geng S, He C, Jiang H. Role of RHOT1 on migration and proliferation of pancreatic cancer. Am J Cancer Res 2015;5:1460–70. [PMC free article] [PubMed] [Google Scholar]
- 16. Li W, Sancho A, Chung WL, Vinik Y, Groll J, Zick Y, et al. Differential cellular responses to adhesive interactions with galectin-8- and fibronectin-coated substrates. J Cell Sci 2021;134:jcs252221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Isogai T, van der Kammen R, Goerdayal SS, Heck AJ, Altelaar AF, Innocenti M. Proteomic analyses uncover a new function and mode of action for mouse homolog of Diaphanous 2 (mDia2). Mol Cell Proteomics 2015;14:1064–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Isogai T, van der Kammen R, Innocenti M. SMIFH2 has effects on Formins and p53 that perturb the cell cytoskeleton. Sci Rep 2015;5:9802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Seltmann K, Meyer M, Sulcova J, Kockmann T, Wehkamp U, Weidinger S, et al. Humidity-regulated CLCA2 protects the epidermis from hyperosmotic stress. Sci Transl Med 2018;10:eaao4650. [DOI] [PubMed] [Google Scholar]
- 20. Isogai T, van der Kammen R, Bleijerveld OB, Goerdayal SS, Argenzio E, Altelaar AF, et al. Quantitative proteomics illuminates a functional interaction between mDia2 and the proteasome. J Proteome Res 2016;15:4624–37. [DOI] [PubMed] [Google Scholar]
- 21. Lobas MA, Tao R, Nagai J, Kronschlager MT, Borden PM, Marvin JS, et al. A genetically encoded single-wavelength sensor for imaging cytosolic and cell surface ATP. Nat Commun 2019;10:711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, et al. Fiji: an open-source platform for biological-image analysis. Nat Methods 2012;9:676–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wershof E, Park D, Barry DJ, Jenkins RP, Rullan A, Wilkins A, et al. A FIJI macro for quantifying pattern in extracellular matrix. Life Sci Alliance 2021;4:e202000880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Valente AJ, Maddalena LA, Robb EL, Moradi F, Stuart JA. A simple ImageJ macro tool for analyzing mitochondrial network morphology in mammalian cell culture. Acta Histochem 2017;119:315–26. [DOI] [PubMed] [Google Scholar]
- 25. Wilanowski T, Caddy J, Ting SB, Hislop NR, Cerruti L, Auden A, et al. Perturbed desmosomal cadherin expression in grainy head-like 1-null mice. EMBO J 2008;27:886–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Dowell JA, Johnson JA, Li L. Identification of astrocyte secreted proteins with a combination of shotgun proteomics and bioinformatics. J Proteome Res 2009;8:4135–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Peirsman A, Blondeel E, Ahmed T, Anckaert J, Audenaert D, Boterberg T, et al. MISpheroID: a knowledgebase and transparency tool for minimum information in spheroid identity. Nat Methods 2021;18:1294–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Foty R. A simple hanging drop cell culture protocol for generation of 3D spheroids. J Vis Exp 2011:2720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Barrett T, Wilhite SE, Ledoux P, Evangelista C, Kim IF, Tomashevsky M, et al. NCBI GEO: archive for functional genomics data sets—update. Nucleic Acids Res 2013;41:D991–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Tang Z, Kang B, Li C, Chen T, Zhang Z. GEPIA2: an enhanced web server for large-scale expression profiling and interactive analysis. Nucleic Acids Res 2019;47:W556–W60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Li T, Fu J, Zeng Z, Cohen D, Li J, Chen Q, et al. TIMER2.0 for analysis of tumor-infiltrating immune cells. Nucleic Acids Res 2020;48:W509–W14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Goswami CP, Nakshatri H. PROGgene: gene expression based survival analysis web application for multiple cancers. J Clin Bioinforma 2013;3:22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Cox TR. The matrix in cancer. Nat Rev Cancer 2021;21:217–38. [DOI] [PubMed] [Google Scholar]
- 34. Lodberg A. Principles of the activin receptor signaling pathway and its inhibition. Cytokine Growth Factor Rev 2021;60:1–17. [DOI] [PubMed] [Google Scholar]
- 35. Beli P, Mascheroni D, Xu D, Innocenti M. WAVE and Arp2/3 jointly inhibit filopodium formation by entering into a complex with mDia2. Nat Cell Biol 2008;10:849–57. [DOI] [PubMed] [Google Scholar]
- 36. Martinez J, Marmisolle I, Tarallo D, Quijano C. Mitochondrial bioenergetics and dynamics in secretion processes. Front Endocrinol 2020;11:319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Moore AS, Coscia SM, Simpson CL, Ortega FE, Wait EC, Heddleston JM, et al. Actin cables and comet tails organize mitochondrial networks in mitosis. Nature 2021;591:659–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Rossignol R, Gilkerson R, Aggeler R, Yamagata K, Remington SJ, Capaldi RA. Energy substrate modulates mitochondrial structure and oxidative capacity in cancer cells. Cancer Res 2004;64:985–93. [DOI] [PubMed] [Google Scholar]
- 39. Lopez-Domenech G, Covill-Cooke C, Ivankovic D, Halff EF, Sheehan DF, Norkett R, et al. Miro proteins coordinate microtubule- and actin-dependent mitochondrial transport and distribution. EMBO J 2018;37:321–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Wang X, Winter D, Ashrafi G, Schlehe J, Wong YL, Selkoe D, et al. PINK1 and Parkin target Miro for phosphorylation and degradation to arrest mitochondrial motility. Cell 2011;147:893–906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Tsherniak A, Vazquez F, Montgomery PG, Weir BA, Kryukov G, Cowley GS, et al. Defining a cancer dependency map. Cell 2017;170:564–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Verstreken P, Ly CV, Venken KJ, Koh TW, Zhou Y, Bellen HJ. Synaptic mitochondria are critical for mobilization of reserve pool vesicles at Drosophila neuromuscular junctions. Neuron 2005;47:365–78. [DOI] [PubMed] [Google Scholar]
- 43. Onodera Y, Nam JM, Horikawa M, Shirato H, Sabe H. Arf6-driven cell invasion is intrinsically linked to TRAK1-mediated mitochondrial anterograde trafficking to avoid oxidative catastrophe. Nat Commun 2018;9:2682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Langert KA, Pervan CL, Stubbs EB Jr. Novel role of Cdc42 and RalA GTPases in TNF-alpha mediated secretion of CCL2. Small GTPases 2014;5:e29260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Ding L, Lei Y, Han Y, Li Y, Ji X, Liu L. Vimar is a novel regulator of mitochondrial fission through Miro. PLoS Genet 2016;12:e1006359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Schuler MH, Lewandowska A, Caprio GD, Skillern W, Upadhyayula S, Kirchhausen T, et al. Miro1-mediated mitochondrial positioning shapes intracellular energy gradients required for cell migration. Mol Biol Cell 2017;28:2159–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Liang Y, Lv Z, Huang G, Qin J, Li H, Nong F, et al. Prognostic significance of abnormal matrix collagen remodeling in colorectal cancer based on histologic and bioinformatics analysis. Oncol Rep 2020;44:1671–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Hsieh CH, Li L, Vanhauwaert R, Nguyen KT, Davis MD, Bu G, et al. Miro1 marks Parkinson's disease subset and Miro1 reducer rescues neuron loss in Parkinson's models. Cell Metab 2019;30:1131–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Schworer S, Berisa M, Violante S, Qin W, Zhu J, Hendrickson RC, et al. Proline biosynthesis is a vent for TGFbeta-induced mitochondrial redox stress. EMBO J 2020;39:e103334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Avagliano A, Granato G, Ruocco MR, Romano V, Belviso I, Carfora A, et al. Metabolic reprogramming of cancer associated fibroblasts: the slavery of stromal fibroblasts. Biomed Res Int 2018;2018:6075403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Kruppa AJ, Buss F. Actin cages isolate damaged mitochondria during mitophagy. Autophagy 2018;14:1644–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Majstrowicz K, Honnert U, Nikolaus P, Schwarz V, Oeding SJ, Hemkemeyer SA, et al. Coordination of mitochondrial and cellular dynamics by the actin-based motor Myo19. J Cell Sci 2021;134:jcs255844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Gao J, Sang M, Zhang X, Zheng T, Pan J, Dai M, et al. Miro1-mediated mitochondrial dysfunction under high nutrient stress is linked to NOD-like receptor 3 (NLRP3)-dependent inflammatory responses in rat pancreatic beta cells. Free Radic Biol Med 2015;89:322–32. [DOI] [PubMed] [Google Scholar]
- 54. Stephen TL, Higgs NF, Sheehan DF, Al Awabdh S, Lopez-Domenech G, Arancibia-Carcamo IL, et al. Miro1 regulates activity-driven positioning of mitochondria within astrocytic processes apposed to synapses to regulate intracellular calcium signaling. J Neurosci 2015;35:15996–6011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Villanueva J, Viniegra S, Gimenez-Molina Y, Garcia-Martinez V, Exposito-Romero G, del Mar Frances M, et al. The position of mitochondria and ER in relation to that of the secretory sites in chromaffin cells. J Cell Sci 2014;127:5105–14. [DOI] [PubMed] [Google Scholar]
- 56. Inman GJ, Nicolas FJ, Callahan JF, Harling JD, Gaster LM, Reith AD, et al. SB-431542 is a potent and specific inhibitor of transforming growth factor-beta superfamily type I activin receptor-like kinase (ALK) receptors ALK4, ALK5, and ALK7. Mol Pharmacol 2002;62:65–74. [DOI] [PubMed] [Google Scholar]
- 57. Porporato PE, Filigheddu N, Pedro JMB, Kroemer G, Galluzzi L. Mitochondrial metabolism and cancer. Cell Res 2018;28:265–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data generated in this study are available within the article and its Supplementary Data files.



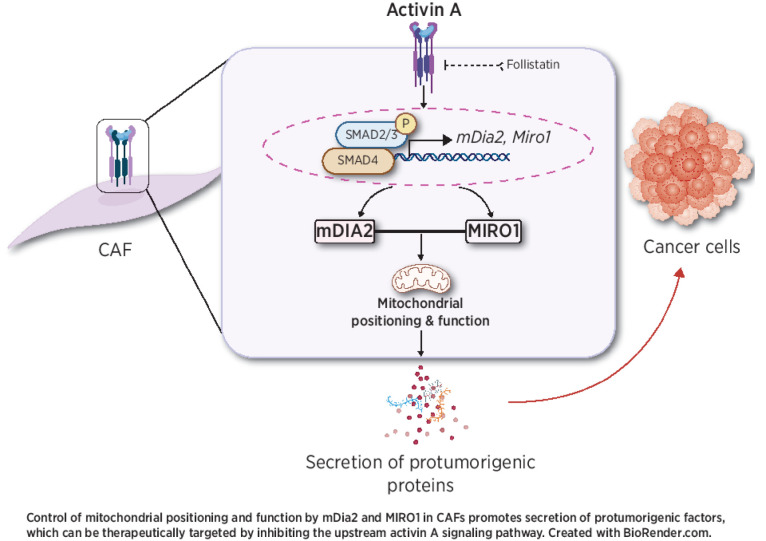
![Figure 1. mDia2 promotes protein secretion from HDFs. A, Quantification of total proteins secreted by sh-EV and sh-mDia2 HDFs. n = 3. B, Representative Western blots of conditioned media (CM) of sh-EV and -mDia2 fibroblasts for fibronectin 1 (FN1), collagen (COL)4A1, and INHBA under reducing conditions. Ponceau S staining of the membrane served as loading control. C and D, Representative immunofluorescence images of sh-EV and sh-mDia2 fibroblasts stained for INHBA, FN1, and ELN (C), and quantification of intracellular fluorescence intensity (D). n = 3. E, Western blot for FN1, COL4, elastin (ELN), and vinculin (VIN, loading control) using total lysates of sh-EV or sh-mDia2 fibroblasts. n = 3. F, Representative FN1, COL1A1, and ELN immunofluorescence images and quantification of the stained area in the decellularized matrix from sh-EV and sh-mDia2 fibroblasts cultured in glucose-containing medium. n = 3. G, IFNα2, IL1β, and IL6 concentrations (ng/mL) in supernatants from sh-EV and sh-mDia2 fibroblasts. n = 3. H, Quantification of the SCC13 tumor spheroid area in hanging drop including CM from sh-mDia2 and sh-EV fibroblasts and representative images (left). The area of the spheroid formed in one control sample was set to 1. n = 10–11. I, Western blots of total lysate from sh-EV or sh-mDia2 fibroblasts transfected with expression vectors encoding wild-type mDia2 (WT), an actin-polymerization–deficient mDia2 mutant (IA) or empty vector (pMX) for mDia2 and GAPDH. J and K, Percentage of total cells with actin stress fibers (J) and scratch closure (K) of sh-EV and sh-mDia2 fibroblasts transfected with expression vectors encoding WT or IA mDia2 or pMX. n = 3 (J) and 6 (K). L and M, sh-EV or -mDia2 fibroblasts were transfected with expression vectors encoding WT or IA mDia2 or pMX. Quantification of FN1 immunofluorescence staining of the decellularized ECM (L), and representative Western blot of total cell lysate and data quantification (M). n = 3. Graphs show mean ± SEM. ns P > 0.05; *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001 [one-way ANOVA with Bonferroni post hoc test (D, E, J, K–M), or unpaired Student t test (A, F–H)]. Scale bars: 50 μm (C), 100 μm (F).](https://cdn.ncbi.nlm.nih.gov/pmc/blobs/c90f/9574377/670d92a6422d/3701fig1.jpg)
![Figure 2. mDia2 regulates mitochondrial function in HDFs. A, Representative images of sh-EV and sh-mDia2 HDFs stained with MitoTracker (green), rhodamine-coupled phalloidin (red), and Hoechst (blue). The graph shows the fluorescence intensity of MitoTracker relative to the highest intensity value. The relative distance is indicated with values from 1 to 10 (e.g., on the top). N = 10. B, Representative Western blots for mDia2, MIRO1, and HSP60 using cytoplasmic (Ct) and mitochondrial (Mt) fractions, and total lysate (TL) of HDFs. C and D, sh-mDia2 fibroblasts were transfected with pMX‐WT or pMX‐IA expression vectors or pMX, and stained with MitoTracker (C). sh-EV fibroblasts were used for comparison. Bar graph shows the fluorescence intensity distribution of MitoTracker relative to the highest intensity value from the perinuclear region to the plasma membrane (D) as described in A. n = 6. E–G, Representative images of sh-EV and sh-mDia2 HDFs stained with CellROX (E) or MitoSOX (F) (red), counterstained with Hoechst (blue). Bar graphs show quantification of staining intensity. Cells were analyzed for LDH release (G). n = 3–4. H, Relative intracellular ATP levels in sh-EV or sh-mDia2 HDFs transfected with pMX-WT, pMX-IA, or pMX. n = 3. I, Relative levels of mtDNA in sh-EV and sh-mDia2 HDFs determined by qPCR. n = 3. J, Western blot of total lysate of sh-EV or sh-mDia2 fibroblasts for HSP60 and GAPDH, and quantification of the HSP60/GAPDH ratio. n = 3. K, Quantification of FN1- and COL I-stained area in the decellularized matrix from sh-EV and sh-mDia2 fibroblasts cultured in galactose-containing medium. n = 3. L, Quantification of data from Seahorse analysis from sh-EV and sh-mDia2 HDFs cultured in galactose-containing medium. n = 6. Graphs show mean ± SEM. ns P > 0.05; *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001 [one-way ANOVA with Bonferroni post hoc test (H), two-way ANOVA with Bonferroni post hoc test (A, D), or unpaired Student t test (E–G, I–L)]. Scale bars: 25 μm (A, C), 100 μm (E, F).](https://cdn.ncbi.nlm.nih.gov/pmc/blobs/c90f/9574377/45cd8dae2eeb/3701fig2.jpg)
![Figure 3. mDia2 binds and stabilizes MIRO1 in primary human fibroblasts. A, Representative Western blot of total lysate of sh-EV or sh-mDia2 fibroblasts for mDia2, MIRO1, GAPDH, and α-tubulin. B, qRT-PCR analysis for MIRO1 using RNA from sh-EV and sh-mDia2 HDFs. n = 3. C, Representative Western blot of mitochondrial fractions from sh-EV and sh-mDia2 fibroblasts probed for mDia2, MIRO1, and HSP60, and quantification of mitochondrial mDia2/HSP60 or MIRO1/HSP60 ratios. n = 3. D, Representative images of sh-EV and sh-mDia2 HDFs immunostained for MIRO1 (red), counterstained with MitoTracker (green), and Hoechst (blue). Dotted lines outline the cell border. E, Western blot of total lysate from sh-EV or sh-mDia2 fibroblasts transfected with pMX‐WT, pMX-IA, or pMX‐control for MIRO1 and GAPDH, and quantification of the MIRO1/GAPDH ratio. n = 3. F, Representative Western blots for MIRO1 and vinculin using total lysates of sh-EV and sh-mDia2 fibroblasts treated for 2 hours with epoxomicin (Epox; 10 μmol/L), and quantification of the MIRO1/VINCULIN ratio. n = 3 biological replicates. G, Representative images of sh-mDia2 fibroblasts treated with Epox (10 μmol/L) or vehicle and stained with MitoTracker (green) and rhodamine-coupled phalloidin (red). Bar graphs show the fluorescence intensity distribution of MitoTracker relative to the highest intensity value from the perinuclear region to the plasma membrane and mean network branch quantified using the MiNA toolset. n = 22–29. H and I, Representative images showing PLA signals (red) in HDFs stained with mDia2 and MIRO1 antibodies and in the negative control (-Ctrl) without primary antibodies (H), counterstained with Hoechst (blue), either alone (H) or together with MitoTracker (green) (I). Graphs show mean ± SEM. ns, P > 0.05; *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001 [one-way ANOVA with Bonferroni post hoc test (E–G for network analysis), two-way ANOVA with Bonferroni post hoc test (G for MitoTracker intensity), or unpaired Student t test (B, C)]. Scale bars: 50 μm.](https://cdn.ncbi.nlm.nih.gov/pmc/blobs/c90f/9574377/3a152f627570/3701fig3.jpg)
![Figure 4. Overexpression of mDia2 and MIRO1 in CAFs promotes tumorigenesis. A, Expression of MIRO1 in liver, breast, and ovarian cancer stroma (CS) vs. stroma of respective normal tissues (NS) based on data sets GSE45001 (N = 10 per group), GSE9014 (N = 6 and 53), and GSE40595 (N = 8 and 36). B, Single-cell expression profiles of mDia2/DIAPH3 and MIRO1 in immune cells, endothelial cells, and CAFs of melanomas based on data from Single-Cell Portal (Broad Institute). C, Correlation analysis between MIRO1 and mDia2, INHBA, and TGFB1 gene expression across different cancers using TIMER2. UVM: uveal melanoma; LUAD: lung adenocarcinoma; PAAD: pancreatic ductal adenocarcinoma; SKCM: skin cancer cutaneous melanoma, BRCA-Lum: luminal breast cancer; LIHC: liver hepatocellular carcinoma. The red or blue color indicates a statistically significant positive/negative correlation (Spearman, P < 0.05), respectively, and gray denotes a nonsignificant result. D, Kaplan–Meier survival curves based on TCGA and GTEx data. High expression of mDia2/DIAPH3 and MIRO1 in patients with liver or lung cancer correlates with poor survival. E, Representative sections from normal human skin, cSCC, and BCC stained for MIRO1. Scale bars: 100 μm. T: tumor cells. F, qRT-PCR analysis for MIRO1 using RNA from CAFs or NFs from two SCC patients. n = 3. G, Representative Western blot of total lysates of NFs and CAFs from patient No. 1 for MIRO1, CAF markers, and GAPDH. H, Quantification of SCC13 tumor spheroid area in hanging drop including CM from NF and CAFs. n = 10–11. I, Mitochondrial distribution in NFs or CAFs from patient No.1. n = 15–19. J, Representative images and bar graph showing fluorescence intensity distribution of ATP biosensor (cyto-Ruby3-iATPSnFR1.0) relative to the highest intensity value from the perinuclear region to the plasma membrane of NFs and CAFs (patient No. 1). n = 19. K, Mitochondrial branch length of NFs or CAFs (patient No.1). n = 15–19. L, Representative immunofluorescence images of decellularized matrices obtained from NFs and CAFs (patient No. 1) stained for FN1 and COL1A1 and quantification of the stained area. n = 3. M, Quantification of SCC13 tumor spheroid area in hanging drop including CM from NFs transfected with an expression vector encoding human MIRO1 (pGEM‐MIRO1 “+”) or empty vector (pGEM‐EV “–”). The spheroid area in one control sample was set to 1. n = 10–12. N, SCC13 cells plated on either decellularized matrix from NFs transfected with pGEM‐MIRO1 or pGEM‐EV. After 3 days, the number of Ki67-positive SCC13 cells was quantified. n = 3. O, Representative FN1 and COL1A1 immunofluorescence stainings and quantification of the stained area in the decellularized matrix derived from NFs transfected with pGEM‐MIRO1 or pGEM‐EV. n = 3. P and Q, sh-EV and sh-mDia2 fibroblasts were transfected with pGEM‐MIRO1 (“+”) or pGEM‐EV (“–”). Expression of MIRO1 was analyzed by immunofluorescence 24 hours after transfection (P). n = 3. Bar graph shows the fluorescence intensity distribution of MitoTracker relative to the highest intensity value from the perinuclear region to the plasma membrane (Q). n = 6–8. Bar graphs show mean ± SEM. ns P > 0.05; *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001 [one-way ANOVA with Bonferroni post hoc test (P), two-way ANOVA with Bonferroni post hoc test (I, J, Q), unpaired Student t test (A, F, H, K–O)]. Scale bars: 50 μm.](https://cdn.ncbi.nlm.nih.gov/pmc/blobs/c90f/9574377/fcd7be390ad1/3701fig4.jpg)
![Figure 5. MIRO1 is a target of activin A–SMAD2/3 signaling. A, HDFs were treated with 20 ng/mL activin A or vehicle for 6 hours and analyzed for MIRO1 expression by qRT-PCR. n = 3. B, ChIP using lysates from activin A– or vehicle-treated fibroblasts and an SMAD2/3 antibody, and amplification of the bound DNA with MIRO1 primers (top). Preimmune serum (IgG) was used as a negative control. Alignment of the sequences within the 2-kb region upstream of the MIRO1 gene TSS of different species that harbor a SMAD2/3 binding element (SBE; in red and italics, bottom). C and D, Representative images of sections from ear skin tumors formed by SCC13 cells with doxycycline (DOX)-inducible lentiviral overexpression of INHBA (SCC13 Act; C) or FST (SCC13 FST; D) or transduced with EV, immunostained for MIRO1 (green) or α-SMA (red), and counterstained with Hoechst (blue). Graphs show the percentage of MIRO1- relative to α-SMA–positive area (percentage of MIRO1+ area in stroma). n = 3 tumors per group. Tumors were collected 5 or 12 weeks after the initial injection of SCC13 Act or -FST. This accounts for the higher MIRO1 positivity of EV in D compared with C. E, Tumors formed by SCC13 Act cells were treated ex vivo for 6 hours with 50 ng/mL follistatin, immunostained for MIRO1 (green) or α-SMA (red), and counterstained with Hoechst (blue). Graph shows the percentage of MIRO1- relative to α-SMA–positive area. n = 4. F and G, qRT-PCR for FST (F) and MIRO1 (G) using RNA collected 24 hours after DOX treatment from clonally expanded HDFs transduced with empty vector (Fb-EV) or with DOX-inducible expression of FST (Fb FST1: clone 1, Fb FST2: clone 2). n = 3. H, Western blots for MIRO1 and GAPDH using total lysates of Fb FST (clone 1) and Fb-EV. I, qRT-PCR for DIAPH3 using RNA collected 24 hours after DOX treatment from HDFs transduced with Fb-EV or Fb FST1. n = 3. J, Representative images of Fb-EV or -FST1 stained with MitoTracker (green) and Hoechst (blue). K and L, Fb FST1 and control fibroblasts were transfected with pGEM‐MIRO1 (“+”) or pGEM‐EV (“–”). Expression of MIRO1 was analyzed by immunofluorescence 24 hours after transfection (K). n = 3. Graph shows fluorescence intensity distribution of MitoTracker relative to the highest intensity value from the perinuclear region to the plasma membrane (L). n = 8. M, Quantification of SCC13 spheroid area in hanging drops including CM from Fb FST1 and their controls transfected with pGEM‐MIRO1 or pGEM‐EV. The area of the spheroid in one control sample was set to 1. n = 8. Bar graphs show mean ± SEM. ns P > 0.05; *, P < 0.05; **, P < 0.01; ***, P < 0.001 [one-way ANOVA with Bonferroni post hoc test (F, G, K, M), two-way ANOVA with Bonferroni post hoc test (L), or unpaired Student t test (A, C–E, I)]. Scale bars: 50 μm (C–E), 25 μm (J).](https://cdn.ncbi.nlm.nih.gov/pmc/blobs/c90f/9574377/c2adc40c9f81/3701fig5.jpg)
![Figure 6. Knockdown of MIRO1 causes metabolic dysfunction and impaired protein secretion in primary human skin CAFs. A and B, CAFs from patient No. 1 were transfected with MIRO1 (si-MIRO1) or control (si-Ctrl) siRNAs. The knockdown was confirmed by qRT-PCR (A) and immunofluorescence staining (B). C, Quantification of nuclear p53 in CAFs transfected with Ctrl or MIRO1 siRNAs. n = 3. D, qRT-PCR for CAF marker genes and genes encoding different small GTPases using RNA from si-MIRO1 or si-Ctrl CAFs. n = 3. E, Representative images of si-MIRO1 or si-Ctrl CAFs (patient No.1) stained for MIRO1 (red), counterstained with MitoTracker (green) and Hoechst (blue). F, Quantification of COL1-stained area in the decellularized matrix from si-Ctrl or si-MIRO1 CAFs. n = 3. G, Relative levels of mitochondrial superoxide based on MitoSOX fluorescence intensity in CAFs (patient No.1) transfected with si-Ctrl or si-MIRO1. n = 3. H, CAFs (patient No. 1) transfected with si-Ctrl or si-MIRO1 were analyzed for LDH release. n = 3. I, Seahorse assay for the OCR in CAFs (patient No. 1) transfected with si-Ctrl or si-MIRO1 and quantification of basal respiration, maximal respiratory capacity, and ATP production in glucose (top) or galactose (bottom) medium. n = 5–6. J, Quantification of total proteins secreted by si-MIRO1 or si-Ctrl CAFs (patient No. 1) in glucose medium. n = 6. K and L, Representative images of immunofluorescence stainings for FN1 and COL1A (I) and quantification of the stained area in the decellularized matrix (J) from si-Ctrl or si-MIRO1 CAFs (patient No. 1) in glucose medium. n = 3. M, Quantification of intracellular fluorescence in si-Ctrl or si-MIRO1 CAFs (patient No. 1) immunostained for INHBA or ACTA2. n = 3. Bar graphs show mean ± SEM. ns P > 0.05; *, P < 0.05; **, P < 0.01 [two-way ANOVA with Bonferroni post hoc test (D), unpaired Student t test (other graphs)]. Scale bars: 50 μm (E) and 100 μm (B, K). Data shown in K and L were reproduced with CAFs from a second patient.](https://cdn.ncbi.nlm.nih.gov/pmc/blobs/c90f/9574377/ad528a5bff89/3701fig6.jpg)
![Figure 7. Depletion of MIRO1 in CAFs suppresses skin tumorigenesis. A and B, sh-EV or -mDia2 HDFs cocultured for 7 days with SCC13 cells immunostained for E-cadherin (red) and counterstained with MitoTracker (green) and Hoechst (blue) (A). The mitochondrial distribution relative to the highest intensity value from the perinuclear region of fibroblasts to the interface with SCC13 cells was quantified (B). Relative distances range from 1 to 10. n = 4–6. C, Representative Western blots for MIRO1, mDia2, and GAPDH using total lysates of NFs and CAFs (patient No. 1) cultured with SMIFH2 (25 μmol/L) or vehicle. The dotted line indicates cutting of the membrane; all samples were loaded on the same gel. D, Representative images of NFs and CAFs cultured with SMIFH2 (25 μmol/L) or vehicle and stained with rhodamine‐coupled phalloidin (red), MitoTracker (green), and Hoechst (blue). E, Two-dimensional cocultures of SCC13 cells with si-Ctrl or si-MIRO1 CAFs at day 7. SCC13 cells were identified by E-cadherin (red) staining and counterstained with MitoTracker (green) and Hoechst (blue). The mitochondrial distribution relative to the highest intensity value from the perinuclear region of fibroblasts to the interface with SCC13 cells was quantified. Relative distances range from 1 to 10. n = 12–14. F, Chemotactic transwell migration of SCC13 cells using CM from si-Ctrl or si-MIRO1 CAFs. n = 3. G, Quantification of the SCC13 spheroid area in a single hanging drop including CM from si-Ctrl or si-MIRO1 CAFs. The spheroid area in one si-Ctrl sample was set to 1. N = 6. H, Tumor volume at different time points 2 weeks after injection (n = 4) and representative photos of 2-week-old tumors formed upon intradermal coinjection of SCC13 cells with si-MIRO1 or si-Ctrl CAFs. An inset panel indicates the area where tumor cells invade into the cartilage. Asterisk indicates cartilage; arrow indicates site of invasion. I, Representative images of COL1A1 immunofluorescence and Herovici staining of tumors formed by SCC13 cells transfected with si-MIRO1 or si-Ctrl CAFs at day 15. J, Quantification of the percentage of the COL1A1-positive area and curvature of fibers per field of view. n = 3–4. K, Increase in SCC13 (coinjected with CAFs) tumor volume (mm3) upon injection of the tumors with MIRO1-Reducer (2.5 μmol/L per injection) or vehicle (DMSO) once every 3–4 days for 18 days. Red arrows indicate the treatment time points. n = 5. Bar graphs show mean ± SEM. ns P > 0.05; *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001 [two-way ANOVA with Bonferroni post hoc test (B, E, H, K), unpaired Student t test (F, G, J)]. Scale bars: 25 μm (A, D, E) and 50 μm (I).](https://cdn.ncbi.nlm.nih.gov/pmc/blobs/c90f/9574377/919ce5371ba9/3701fig7.jpg)