HCC cells acquire resistance to lenvatinib by activating the EGFR–STAT3–ABCB1 pathway, identifying combined treatment with erlotinib as a strategy to overcome acquired resistance and improve the clinical benefit of lenvatinib.
Abstract
Lenvatinib is an inhibitor of multiple receptor tyrosine kinases that was recently authorized for first-line treatment of hepatocellular carcinoma (HCC). However, the clinical benefits derived from lenvatinib are limited, highlighting the urgent need to understand mechanisms of resistance. We report here that HCC cells develop resistance to lenvatinib by activating EGFR and stimulating the EGFR–STAT3–ABCB1 axis. Lenvatinib resistance was accompanied by aberrant cholesterol metabolism and lipid raft activation. ABCB1 was activated by EGFR in a lipid raft–dependent manner, which significantly enhanced the exocytosis of lenvatinib to mediate resistance. Furthermore, clinical specimens of HCC showed a correlation between the activation of the EGFR–STAT3–ABCB1 pathway and lenvatinib response. Erlotinib, an EGFR inhibitor that has also been shown to inhibit ABCB1, suppressed lenvatinib exocytosis, and combined treatment with lenvatinib and erlotinib demonstrated a significant synergistic effect on HCC both in vitro and in vivo. Taken together, these findings characterize a mechanism of resistance to a first-line treatment for HCC and offer a practical means to circumvent resistance and treat the disease.
Significance:
HCC cells acquire resistance to lenvatinib by activating the EGFR–STAT3–ABCB1 pathway, identifying combined treatment with erlotinib as a strategy to overcome acquired resistance and improve the clinical benefit of lenvatinib.
Introduction
Hepatocellular carcinoma (HCC) is one of the most common malignancies with high probabilities of metastatic recurrence, treatment failure, and a poor prognosis (1). Clinically, most HCCs are already in advanced stages with limited treatment options when the initial diagnosis is made. Recently, much progress in molecular targeted therapy and immunotherapy has provided new hope for patients with advanced HCCs. Among of them, sorafenib, lenvatinib, and atezolizumab/bevacizumab combination have been approved as the first-line therapy (2). Lenvatinib is the second authorized first-line medication for advanced HCC after sorafenib. It is an orally administered multitarget tyrosine kinase receptor inhibitor that inhibits the VEGFR 1/2/3, FGFR 1/2/3/4, platelet-derived growth factor receptor α and the proto-oncogenes KIT and RET (3, 4). However, the relatively rapid emergence of resistance to lenvatinib treatment limits the overall therapeutic benefit, highlighting the urgent need to investigate the molecular mechanisms and identify new therapeutic strategies to overcome the drug resistance.
Lipid rafts are a 10 to 200 nm, heterogeneous, high fluidity regions of the cell membrane that are rich in sterols, sphingolipids, and cholesterol (5), and can stably form a platform for the proteins and lipids that participate in a wide range of biological activities (6). A variety of receptor tyrosine kinases (RTK) and functional proteins, such as EGFR and ATP-binding cassette (ABC) transporter family members, are concentrated in lipid rafts. The recruited proteins in lipid rafts vary on the basis of the stimulus (7, 8).
EGFR, also known as ERBB1 or HER1, belongs to the subfamily of RTKs. The homologous ligands of EGFR include EGF, TGFα, and amphiregulin (9, 10), which induce EGFR homodimerization or heterodimerization and lead to the phosphorylation of intracellular tyrosine residues that activate downstream pathways (such as the RAS–RAF–MEK–ERK, JAK–STAT, and PI3K–AKT pathways) and regulate a variety of biological processes, such as cell proliferation, antiapoptosis, metastasis, and metabolism. The disorder of EGFR promotes the occurrence and development of various tumors (11–13).
Excessive excretion has been regarded as a remarkable mechanism mediating drug resistance. Multidrug resistance transporter proteins are known for their contribution to resistance through the cellular efflux of drugs (14). ATP-binding cassette transporter B1 (ABCB1), also known as P-glycoprotein (Pgp), is a member of the ABC protein family. It is a membrane transporter (14, 15) that can transport a variety of substrates (250–1,250 Da) including phospholipids, sterols, cholic acids, peptides, metabolites, and drugs against the concentration gradient by consuming ATP (16). Lenvatinib is the specific transport substrate for ABCB1 and cannot be excreted by OAT1, OAT3, OATP1B1, OATP1B3, OCT1, OCT2, MATE1, MATE2-K, or the bile salt export pump (4).
In the current study, we aimed to explore the mechanism of acquired resistance to lenvatinib and identify a novel combination treatment to overcome lenvatinib resistance and increase the treatment response in HCC.
Materials and Methods
Patients, follow-up, and clinical specimens
Nine patients who received curative resection for HCC at the authors’ institutes from October 2019 to December 2020 were enrolled in this study. All patients who underwent surgical resection were confirmed to have HCC and provided written informed consent in accordance with ethical approval by the Ethics Committee of Huashan Hospital, Fudan University (Shanghai, China). The clinical samples were fixed in formalin solution within half an hour of tissue isolation or stored directly in liquid nitrogen. All patients were pathologically diagnosed with HCC without extrahepatic or lymphoid metastasis and in the IA–IIIA tumor–node–metastasis stage. None of them received any preoperative cancer treatment except for lenvatinib. The patients had complete clinical cases and follow-up data, including personal information, clinical diagnosis, medication records, laboratory results, and imaging reports. Routine blood, liver and kidney function, and tumor marker assessments were performed every month. Liver ultrasonography was performed every 2 months. CT or MRI scans were performed every 6 months or when recurrence was suspected. All these examinations were performed independently by doctors without knowledge of the study.
Cell culture
Human liver cancer cell lines (HuH7 and PLC/PRF/5), HEK293T cells, and the mouse liver cancer cell line Hep1–6 were purchased in 2018 from the Shanghai Cell Bank of the Chinese Academy of Sciences (Shanghai, China). All human cancer cells and mouse liver cancer cells were cultured in DMEM with 10% FBS, glutamine and 1% penicillin–streptomycin (Gibco) at 37°C and 5% CO2. To establish lenvatinib-resistant (LR) cell lines, HuH7 and PLC/PRF/5 cells were cultured with increasing doses of lenvatinib (Selleck) starting from 3 μmol/L and 20 μmol/L, respectively. The cell culture media was replaced every 48 hours until the cells spread across 90% of the culture dish and then passaged. After the passaged cells were replated, the concentration was increased by 0.5 μmol/L until HuH7 cells proliferated quickly at a concentration of 30 μmol/L lenvatinib and PLC/PRF/5 cells proliferated quickly at 60 μmol/L lenvatinib. This process took at least 6 months. The resistant cell strains were named HuH7 LR and PLC/PRF/5 LR. The identities of all cell lines were confirmed via short tandem repeat profiling and the Mycoplasma test results were negative. Cell stocks were conducted within five passages, and all experiments were completed within eight passages.
LR mouse model
Hep1–6 cells were subcutaneously injected into the right posterior flanks of 4-week-old male C57BL/6 mice (5 × 106 cells/mouse), followed by treatment with lenvatinib (10 mg/kg/d) when the tumor reached a volume of approximately 100 mm3 in size. After 28 days of treatment, mice were sacrificed. The largest tumor was divided into 1-mm3 pieces and implanted subcutaneously in the next generation of 4-week-old male C57/BL6 mice. After 3 generations of continuous screening, the resistant Hep1-6 cells were stable.
Cell viability assay
Cells were seeded at a density of 3,000 cells per well on 96-well plates and incubated overnight. The next day, the cells were rinsed, and fresh medium was added with DMSO or the indicated treatment over a 7-point concentration range for 72 hours. The viability of cells was then measured by a Cell Counting Kit-8 (CCK-8) assay. IC50 values were determined by GraphPad Prism 8.0 using a 3-parameter dose–response model.
RNA isolation, reverse transcription, and qPCR
RNA was isolated from cell lines using TRIzol reagent (Invitrogen). RNA was quantified using a Nanodrop ND-1000 (Thermo Fisher Scientific). Complementary DNA synthesis was performed using the PrimeScript RT Reagent Kit (Takara) according to the manufacturer's directions.
Real-time PCR was performed using SYBR Green (Takara) and an ABI PRISM 7900 Sequence Detection System (Thermo Fisher). The results were normalized to Gapdh for mRNA measurement. Fold change was calculated by the 2−ΔΔCt method where ΔΔCt = ΔCt (Target-Reference) Treatment – ΔCt (Target-Reference) control. All the primers are listed in Supplementary Table S1. qPCR was conducted three times with three repetitions.
Western blotting
Total protein was extracted by lysing cells in RIPA buffer containing protease inhibitor. Protein samples were separated by SDS-PAGE and transferred onto polyvinylidene fluoride membranes. After blocking with 5% nonfat milk in TBS-T, membranes were incubated with primary antibody. The following antibodies were used: anti-STAT3 (#9139, CST) and anti–p-STAT3 (#4113, CST), anti-GAPDH (#2118, CST), anti-ABCB1 (#13978, CST), anti-EGFR (#4267, CST), anti–pEGFR (#3777, CST), anti-AKT (#4685, CST), anti–pAKT (#4060, CST), anti-ERK (#4695, CST), anti–pERK (#4370, CST), anti-CAV1 (66067-1-Ig, Proteintech), anti-FLOT1 (A6220, ABclonal), and anti—CCAAT/enhancer-binding protein delta (CEBPD; ab65081, Abcam). The detailed information about antibodies was shown in Supplementary Table S2. Protein bands were detected by image acquisition using an ImageQuant LAS 4000 (GE Healthcare Life Sciences).
IHC staining
For IHC, 5 μmol/L paraffin-embedded sections of patient/mouse tumors were baked at 60°C for 1 hour, deparaffinized in xylene, and rehydrated in a graded series of ethanol solutions. Antigens were unmasked by microwave heating the samples in 10 mmol/L sodium citrate buffer (pH 6.0) for 15 minutes (5 minutes, 3 times), and the reaction was quenched using hydrogen peroxide 3%. After washing with PBS, the samples were incubated with the following primary antibodies overnight at 4°C: anti–pEGFR (#3777, CST), anti-EGFR (#4267, CST), anti-ABCB1 (#13978, CST), anti-STAT3 (A19566, ABclonal), anti-Ki67 (ab15580, Abcam), and anti–cleaved caspase-3 (#9664, CST). 3,3′-diaminobenzidine was used as a detection system. Quantification analyses were performed using ImageJ software based on the percentage of positively stained cells and the staining intensity per field in representative sections.
Immunofluorescence
For immunofluorescence (IF), cells seeded in the wells of glass-bottomed dishes were fixed with 4% paraformaldehyde for 30 minutes after being rinsed twice with 1×PBS. The following primary antibodies were used: anti–pEGFR (#3777, CST), anti-CAV1 (66067-1-Ig, Proteintech), and anti-ABCB1 (#13342, CST). The secondary antibodies Alexa Fluor 488 [Alexa Fluor 488-Labeled Goat Anti-Mouse IgG (H+L), Beyotime Biotechnology] and Alexa Fluor 555 (Alexa Fluor 555-Labeled Donkey Anti-Rabbit IgG, Beyotime Biotechnology) were used. Nuclei were counterstained with 4’,6-diamidino-2-phenylindole (DAPI) before imaging. A Leica confocal microscope was used to capture the images. Quantification analyses were performed using ImageJ software based on the fluorescence intensity per cell in representative sections.
Vectors and cell transfections
An expression vector mediated by lentivirus for human EGFR or ABCB1 was constructed. The sequence was amplified from the cDNA library via specific primers listed in Supplementary Table S3. Then, the harvested DNA was inserted into pCDH-puro expression vector (System Biosciences). In addition, four short hairpin RNAs (shRNA) targeting the human genes EGFR, ABCB1, STAT3, CEBPD were synthesized using the primers listed in Supplementary Table S3. ABCB1 shRNA, EGFR shRNA, STAT3 shRNA, CEBPD shRNA, and nontarget shRNA control (pLKO.1 TRC, Mission RNAi) constructs were purchased from Sigma (SIGMA). pTSB-CMV-STAT3-Y705A was generated using pTSB-CMV-STAT3 WT as a template with the QuikChange Site-Directed Mutagenesis Kit (Stratagene). pCDH-EGFR-WT-Flag and pCDH-EGFR-Kinase-Death (K721A) were gifts from Dr. Mien-Chie Hung's Lab in MD Anderson Cancer Center (Houston, USA). All these constructs and oligonucleotides were transfected into HCC cells using Lipofectamine 2000 according to the product manual (Invitrogen).
Total cellular cholesterol determination and cholesterol staining
Cells were cultured in DMEM (Gibco) with 10% lipid depleted FBS (S181L, Biowest) for 24 hours. The total cellular cholesterol content was measured using a commercial Cholesterol Assay Kit (E1005; Applygen Technologies, Inc.). Cholesterol staining was performed using a Cholesterol Assay Kit (ab133116, Abcam) according to the manufacturer's instructions.
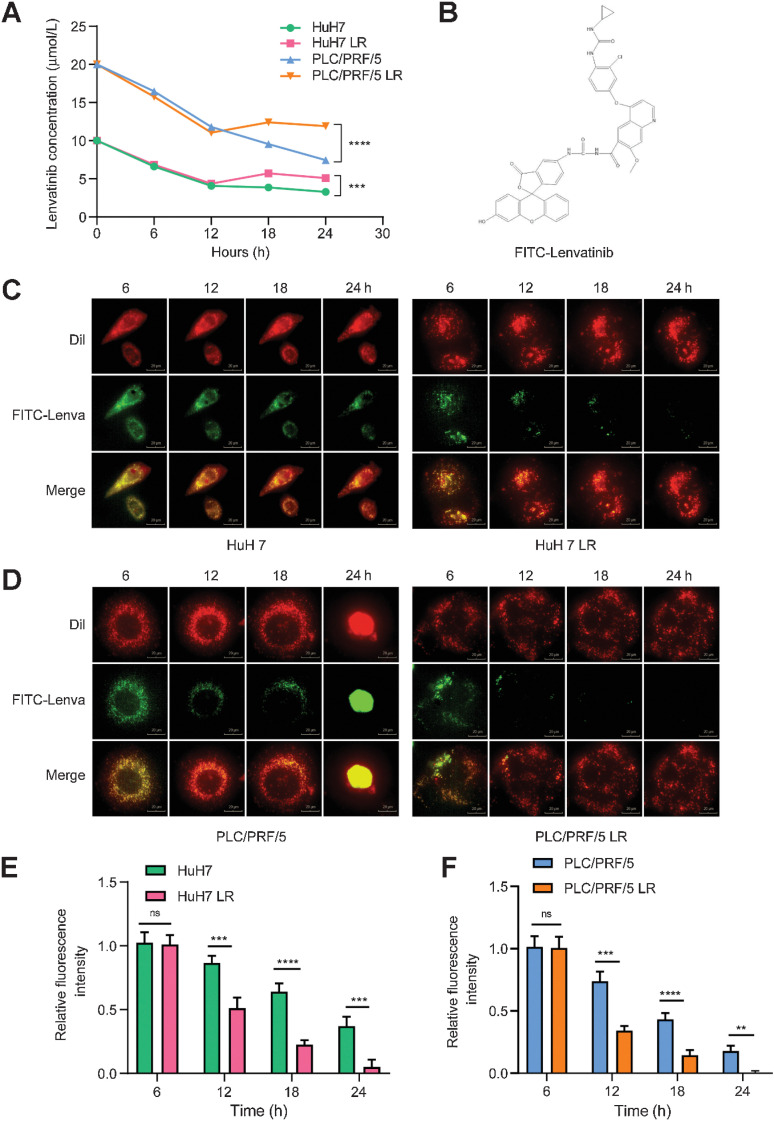
Lenvatinib efflux assay and concentration measurement
FITC-lenvatinib was purchased from Xi'an ruixi Biological Technology Co., Ltd. Cells were seeded in 24-well plates at 1 × 104 cells/well. Lenvatinib-sensitive (LS) and LR strains were stained with DiI dye (red) at working concentration of 10 μmol/L for 20 minutes. After rinsing with PBS, FITC-lenvatinib (green) at a concentration of 10 μmol/L was added to HuH7 and HuH7 LR cell media, and 20 μmol/L FITC-lenvatinib was added to PLC/PRF/5 and PLC/PRF/5 LR cell media. After 6 hours of treatment, the culture medium was replaced by fresh medium without FITC-lenvatinib and the cells were photographed every hour for the whole day to observe the changes of FITC-lenvatinib enrichment in the cells. Quantification analyses were performed using ImageJ software based on the fluorescence intensity per cell in representative sections. The concentration of lenvatinib in the culture supernatant was measured by Fuda Analytical Testing Group by LC-MS (Gas Temp: 200°C, N2 Flow:12 L/min, Sheath Gas Flow: 12 L/min, Sheath Gas Temp:350°C, VCap:300 V).
In vivo treatment studies
The animal research in this study was approved by the Shanghai Medical Experimental Animal Care Commission and performed according to the Guide for the Care and Use of Laboratory Animals. For LR strains, 5 × 106 cells were implanted subcutaneously into the right flanks of Balb/c nude mice. When tumors reached approximately 200 mm3, mice were randomized into four groups of 5 mice each and assigned to 5 days per week treatment with PBS, lenvatinib (10 mg/kg), erlotinib (30 mg/kg), and the drug combination with decreased doses (lenvatinib 3 mg/kg + erlotinib 10 mg/kg, 1/3 dose of each used alone) via oral gavage. Consistent with the procedures above, mice were treated with PBS, lenvatinib (Lenva, 10 mg/kg), Stattic (10 mg/kg), or their combination (lenvatinib 3 mg/kg + Stattic 3 mg/kg, 1/3 dose of each used alone). Tumor volume was determined using the modified ellipsoidal formula, tumor volume = ½ length × width2, based on digital calipers.
Protein profile
Tandem mass tag (TMT) proteome analysis was performed at Shanghai Applied Protein Technology Co. Ltd. (Shanghai, China).
Phospho-RTK array
A phospho-RTK array (R&D Systems) was performed according to the manufacturer's instructions.
Statistical analysis
Statistical analyses were performed using the Statistical Package for GraphPad Prism 8.0 software. Student t test and one- or two-way ANOVA were used for comparisons between groups. For all tests, significance was determined with a 95% confidence interval (ns, P > 0.05; *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001).
Data availability
All the data acquired and/or used in the study are available upon request from the corresponding author.
Results
Establishment of cell models with acquired resistance to lenvatinib
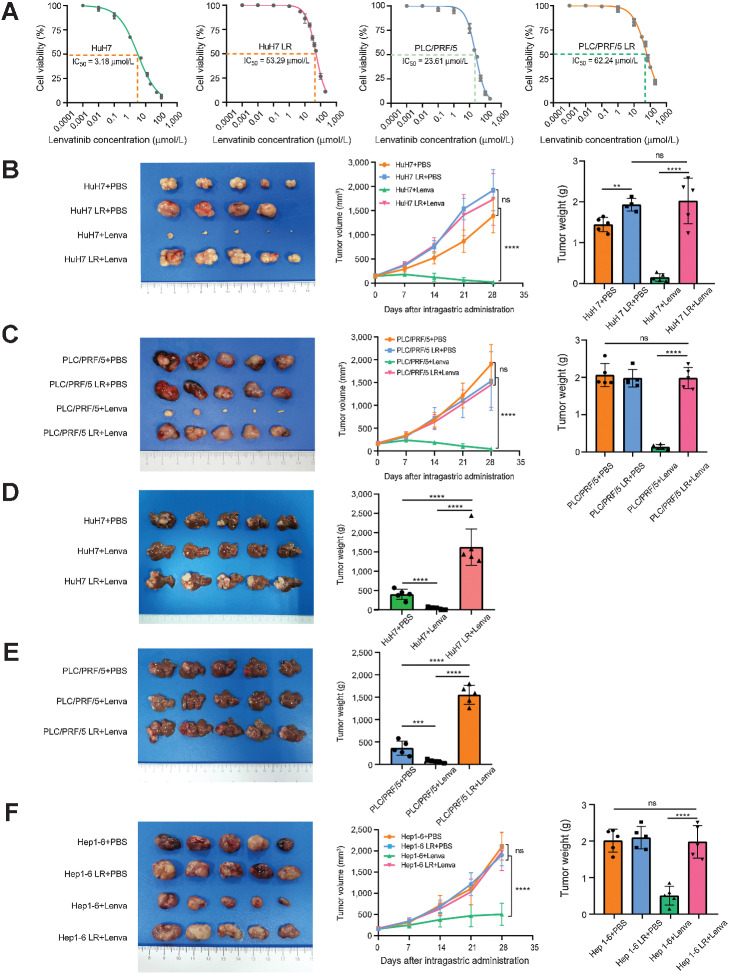
To obtain LR HCC cell models, we exposed HuH7 and PLC/PRF/5 cells to increasing concentrations of lenvatinib (starting from 3 μmol/L and 20 μmol/L, respectively) over 6 months and named them HuH7-LR and PLC/PRF/5-LR (LR strains). Compared with parent cells (LS strains), HuH7-LR and PLC/PRF/5-LR exhibited poor responses to lenvatinib as the IC50 values were 16.76 and 2.64 times greater than those of the LS strains, respectively (Fig. 1A; Supplementary Fig. S1A). After a month of abstinence from lenvatinib, IC50 values remained at 42.12 μmol/L and 52.41 μmol/L, respectively (Supplementary Fig. S1B). Moreover, LR strains were demonstrated considerably with elevated capabilities of resisting apoptosis, invasion, and metastasis (Supplementary Fig. S1C and S1D) but no obvious alteration in proliferation were observed (Supplementary Fig. S1E and S1F). To further validate their stabilities in vivo, we implanted the LR and LS strains subcutaneously or orthotopically into nude mice and tumor growth was monitored every 7 days (Supplementary Fig. S1G). As expected, treatment with lenvatinib failed to yield an antitumor effect in the LR groups as indicated by tumor volume and no significant reduction in tumor volume was observed in the LR group compared with the LS group after treatment with PBS (Fig. 1B–E). In addition, we established a LR mouse strain (Hep1-6 LR) by performing three cycles of lenvatinib treatment in C57BL/6 mice (Fig. 1F; Supplementary Fig. S1H). Taken together, these LR models could be very useful tools for elucidating the molecular mechanisms and processes of lenvatinib resistance, which were assessed in subsequent experiments.
Figure 1.
Establishment of LR cell models and mouse models. A, The IC50 value of HuH7, HuH7 LR, PLC/PRF/5, and PLC/PRF/5 LR cells via CCK-8. Each point on the dose–response curves represents three technical replicates. B and C, In subcutaneous xenograft mouse models, LS cells (HuH7 and PLC/PRF/5) and LR cells (HuH7 LR and PLC/PRF/5 LR) were implanted into the right flanks of BALB/c nude mice, followed by treatment with PBS or lenvatinib (10 mg/kg/d) when the tumor reached a volume of approximately 100 mm3 in size. Tumor growth was measured every 7 days. After 28 days of treatment, the mice were sacrificed. D and E, Tumor appearance, tumor growth, and tumor weight are shown. In in situ xenograft mouse models, the tumors derived from subcutaneous xenograft mouse models were divided into 1-mm3 sections and implanted under the liver capsule of 4-week-old male BALB/c nude mice, followed by treatment with PBS or lenvatinib (10 mg/kg/d) after 2 weeks. On the 28th day of treatment, the mice were sacrificed. Tumor appearance and tumor weight are shown. F, A 1-mm3 tumor block of Hep1-6 and Hep1-6 LR cells (3-generation continuous screening) was subcutaneously implanted into the right posterior flanks of each 4-week-old male C57BL/6 mouse, followed by treatment with lenvatinib (10 mg/kg/d) when the tumor reached a volume of approximately 100 mm3 in size. After 28 days of treatment, the mice were sacrificed. The tumor appearance, tumor growth, and tumor weight are shown. All the results are shown as mean ± SD (n = 5). One- or two-way ANOVA was used to analyze the data. **, P < 0.01; ***, P < 0.001; ****, P < 0.0001; ns, nonsignificant.
The EGFR–STAT3–ABCB1 pathway is activated in the LR HCC model
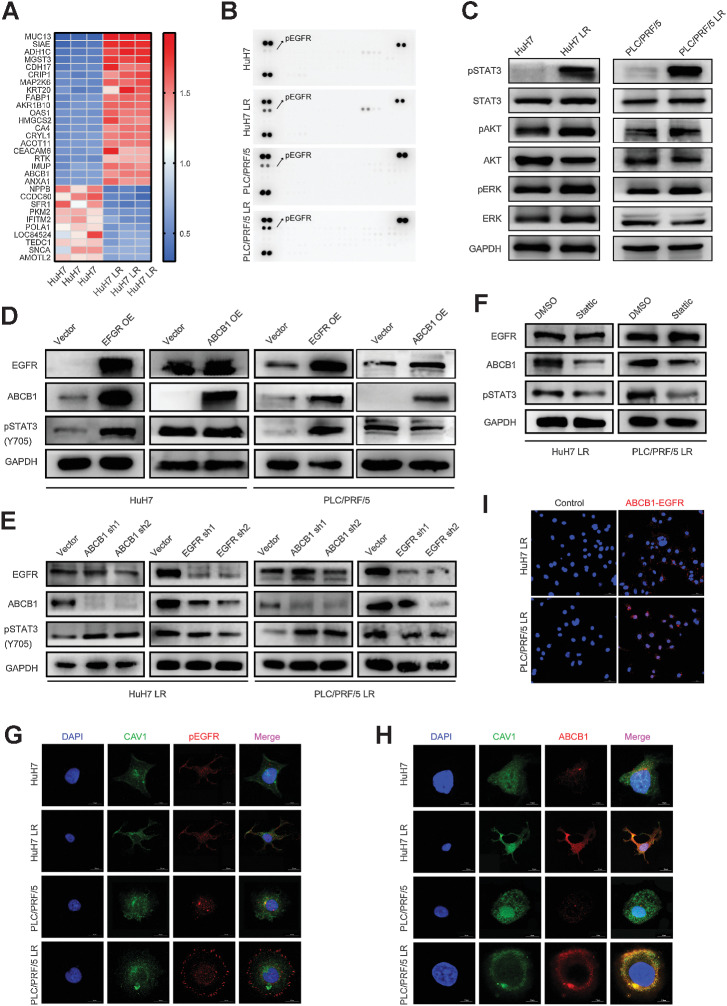
To investigate the mechanisms of acquired resistance to lenvatinib in HCC, we performed TMT proteome analyses in HuH7 and HuH7 LR cells. The results demonstrated that the expression levels of RTK family members and ABCB1 increased dramatically in HuH7 LR cells (Fig. 2A). Next, we examined the activation of RTKs using a human RTK phosphorylation array. Notably, the most dramatic alteration was observed in the pEGFR expression of LR strains compared with that of the control LS strains (Fig. 2B). These observations revealed that EGFR was significantly upregulated and activated in LR HCC cells.
Figure 2.
The EGFR–STAT3–ABCB1 axis is activated in LS strains. A, TMT proteome analysis was performed to analyze the proteins derived from HuH7 and HuH7 LR. The top 30 differently expressed proteins are shown in a heatmap, including 20 upregulated and 10 downregulated proteins. B, Phospho-RTK array of LS strains (HuH7 and PLC/PRF/5) versus LR strains (HuH7 LR and PLC/PRF/5 LR). The three spots at the top and bottom of the chips are the internal reference proteins. Positive spots indicated by arrows are pEGFR. C, Western blot of the protein expression of the EGFR downstream pathway. D, HuH7 and PLC/PRF/5 cells transfected with EGFR or ABCB1 overexpression vector were analyzed for the activation of EGFR, pSTAT3, and ABCB1. E, Western blot of the protein expression on EGFR, pSTAT3, and ABCB1 after knockdown of EGFR or ABCB1 expression in LR strains (HuH7 LR and PLC/PRF/5 LR). F, Western blot of the protein expression of EGFR, pSTAT3, and ABCB1 after LR strains (HuH7 LR and PLC/PRF/5 LR) were treated with or without Stattic (5 μmol/L) for 24 hours. G and H, CAV1 is a biomarker of lipid rafts. The cells were immunostained for EGFR or ABCB1 (red), CAV1 (green), and DNA (DAPI, blue). I, Representative fluorescence images of EGFR and ABCB1 anchored to lipid rafts are shown. Scale bars, 15 μm. A proximity ligation assay demonstrated that spatial colocalization existed between EGFR and ABCB1 (distance < 40 nm).
As previous studies have reported that there are three classical downstream signaling pathways of EGFR (RAS–RAF–MEK–ERK, JAK–STAT, and PI3K–AKT pathways; refs. 17, 18), we questioned which downstream signaling pathway was activated and mediated lenvatinib resistance. Western blot analysis demonstrated that pSTAT3 expression was remarkably altered (Fig. 2C). Because STAT3 has been implicated in regulating ABCB1 expression (19, 20), we overexpressed EGFR and ABCB1 in LS strains to validate the correlation among EGFR, STAT3, and ABCB1. The results showed that overexpression of EGFR significantly increased the levels of pSTAT3 and ABCB1. However, overexpression of ABCB1 did not produce an obvious effect on pSTAT3 or EGFR (Fig. 2D), thus indicating that ABCB1 locates in the downstream of both EGFR and STAT3. Moreover, we used shRNAs to knock down EGFR and ABCB1 in LR strains, and the results demonstrated that the depletion of EGFR in LR strains notably decreased the expression of pSTAT3 and ABCB1, whereas ABCB1 knockdown significantly increased the expression of pSTAT3 but not EGFR, indicating that STAT3 and ABCB1 might be bidirectionally regulated (Fig. 2E). In addition, compared with the LS strains transfected with a blank vector, the LS strains overexpressing ABCB1 and EGFR exhibited a significant IC50 shift and lenvatinib-resistant phenotype (Supplementary Fig. S2A–S2E), which suggested that EGFR and ABCB1 play critical roles in lenvatinib resistance.
A previous study has revealed that EGF binding leads to an increased phosphorylation of EGFR (21). Therefore, to further clarify the mechanism of EGFR feedback activation, we first measured EGF expression in the cell supernatants from LR strains (HuH7 LR and PLC/PRF/5 LR) as well as their parent LS strains (HuH7 and PLC/PRF/5) by ELISA, and found that EGF was obviously upregulated in the culture media of LR strains compared with that of LS strains (Supplementary Fig. S2F). After treated with human EGF (50 ng/mL) and/or erlotinib (10 μmol/L) for 1 hour, the increased EGF ligand in LS strains (HuH7 and PLC/PRF/5) was sufficient to trigger the activation of the EGFR–STAT3–ABCB1 pathway determined by Western blot (Supplementary Fig. S2G). Moreover, IC50 assays of HuH7 and PLC/PRF/5 cells treated with human EGF (50 ng/mL) and/or erlotinib (10 μmol/L) or with DMSO indicated that the supplementation of exogenous EGF ligand induced the resistance of HCC cells to lenvatinib (Supplementary Fig. S2H). In addition, to further define the role of EGFR in mediating the pathway activation and lenvatinib resistance in LR strains, we generated a constitutively active EGFR vector (EGFRWT) and a kinase-dead EGFR mutant (EGFRMUT) with inactive activity by mutating proton acceptor active residue lysine (K) at 721 to alanine (A) according to a previous study (22) and found that the suppression of the EGFR–STAT3–ABCB1 pathway could be rescued by the transfection of EGFRWT, but not EGFRMUT in EGFR-silenced LR strains (Supplementary Fig. S2I). These results indicated that EGFR activity is necessary for maintaining the resistance phenotype.
Next, we examined whether STAT3 activation affects pathway activation and lenavtinib response in HCC. First, we used shRNAs to knock down STAT3 expression in LR strains and found that it conferred the sensitivity of LR strains to lenvatinib as illustrated by decreased IC50 and LR colony formation (Supplementary Fig. S3A–S3C). Moreover, Western blot analyses revealed that knocking down STAT3 decreased the expression of ABCB1, which was rescued by ectopic expression of STAT3WT rather than STAT3Y705A mutant (Supplementary Fig. S3D). These suggest that STAT3 phosphorylation at Y705 mediates ABCB1 expression and affects the lenvatinib response. In addition, ABCB1 expression in LR strains was considerably suppressed by Stattic, a STAT3 inhibitor, but the EGFR expressison remained constant (Fig. 2F). This further supports that STAT3 is critical in regulating ABCB1 expression.
Furthermore, we investigated whether ABCB1 is a direct target of STAT3 in LR strains. The full-length ABCB1 promoter was cloned into a luciferase reporter plasmid and then the recombinant plasmid was cotransfected into 293T cells with blank or STAT3-containing plasmids. Unexpectedly, no obvious change in luciferase activity was observed (Supplementary Fig. S3E), indicating that STAT3 is not a direct transcription factor of ABCB1. Given that STAT3 was reported to regulate ABCB1 through CEBPD, which further binds to the ABCB1 promoter element and upregulates its transcription (19), we explored whether CEBPD expression was aberrated and induced ABCB1 expression in LR strains. As expected, CEBPD induction was observed in LR strains, and CEBPD knockdown using shRNAs attenuated the ABCB1 expression (Supplementary Fig. S3F and S3G). Taken together, these data indicate that ABCB1 is a downstream effector of STAT3 activation, and that feedback activation of the EGFR–STAT3–ABCB1 pathway triggers lenvatinib resistance in LR strains.
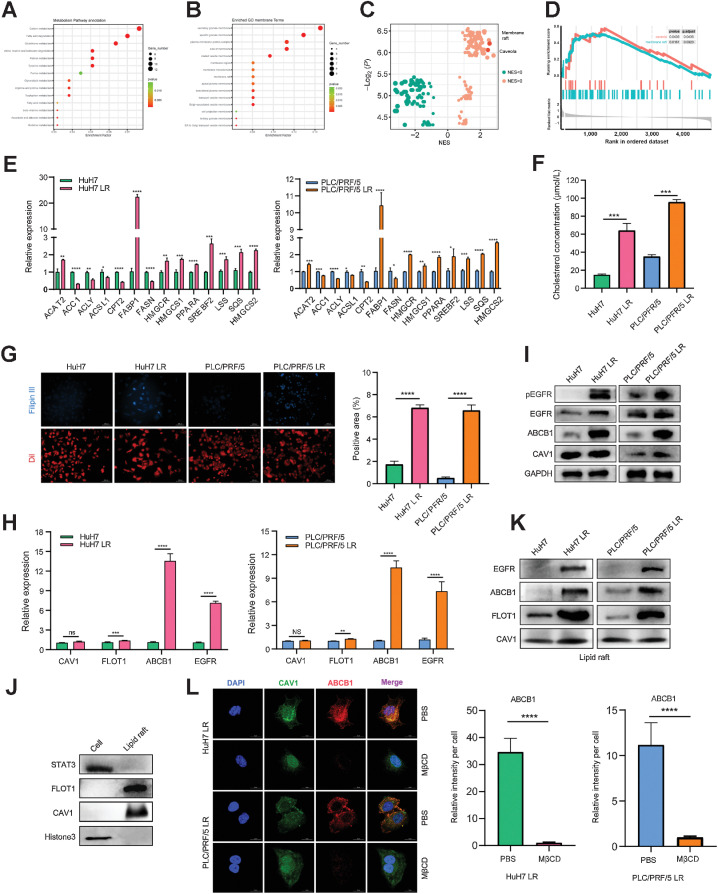
EGFR activates ABCB1 in a lipid raft–dependent manner
EGFR and ABCB1 are reported to be tightly connected to lipid rafts on the cell membrane (23, 24). In the current study, we demonstrated obvious colocalization between pEGFR, ABCB1 and the lipid raft marker CAV1 by IF (Fig. 2G and H). In addition, a spatial interaction between ABCB1 and EGFR was observed in the PLA assay (Fig. 2I). The proteomics data demonstrated a high level of lipid metabolism in LR strains and the processes of lipid raft synthesis and decomposition were extremely active (Fig. 3A–D). Given that cholesterol, one of the major components of lipid rafts, is critical for lipid raft stability and protein anchoring on lipid rafts, we used qRT-PCR to screen key enzymes of lipid metabolism to better understand the aberrant lipid metabolism pathways in the LR strains and identified significant changes in the key enzymes involved in cholesterol synthesis (Fig. 3E). Moreover, we quantified the total cholesterol in the LR and LS strains, and found that the total level of endogenous cholesterol increased markedly in the LR strains (Fig. 3F and G). We further determined the mRNA levels of lipid raft markers (CAV1 and FLOT1), ABCB1 and EGFR in HCC cells and found that, although there was no obvious difference in the overall levels of lipid raft markers between LR and LS strains, EGFR and ABCB1 were substantially upregulated in LR strains (Fig. 3H). Consistent results were confirmed at the protein level by Western Blot (Fig. 3I). Furthermore, we extracted lipid rafts from HCC cells and detected the protein levels of EGFR and ABCB1 by Western blotting. The results indicated that the samples extracted by the lipid raft kit were free of nucleoprotein and plasma protein contamination (Fig. 3J), and the expression levels of EGFR and ABCB1 in the lipid rafts of LR strains were significantly increased compared with those in the parent cells (Fig. 3K). These results were consistent with those observed in the above IF experiments (Fig. 2G and H). In addition, methyl-β-cyclodextrin (MβCD), which can efficiently destroy the lipid raft structure on the cell membrane, notably attenuated ABCB1 expression on the cell membrane, indicating that ABCB1 expression depends on the lipid raft structure (Fig. 3L). Taken together, these findings suggest that lenvatinib resistance is accompanied by aberrant cholesterol metabolism and lipid rafts activation. ABCB1 is activated by EGFR in a lipid raft–dependent manner.
Figure 3.
EGFR activates ABCB1 in a lipid raft–dependent manner. A and B, All differentially expressed genes were sorted in descending order by fold change and the normalized enrichment score (NES) was calculated for each gene set using the functional gene sets in MSigDB (literature vs. databases containing signaling pathways, physiologic function, and spatial structure, etc.). C and D, A total of 112 upregulated gene sets and 128 downregulated gene sets were obtained. NES > 1.0 or < 1.0, P < 0.05 was considered significant. (The NESs for lipid rafts and CAV1 were 2.2 and 2.3, respectively, suggesting functional enrichment in the LR group). E, qPCR results for the mRNA expression of the key enzymes in the lipid metabolic pathway in LS and LR strains. F, Total cellular cholesterol determination of LS and LR strains. G, Cholesterol staining in drug-resistant and drug-sensitive strains. Filipin III is a cholesterol-specific fluorescent dye (blue) and DiI is a membrane-structured fluorescent dye used for cells (red). H, qPCR results for the mRNA expression of CAV1, FLOT1, EGFR, and ABCB1 in LS and LR strains. I, Western blot of EGFR, pEGFR, CAV1, and ABCB1 expression in LS strains (HuH7 and PLC/PRF/5) versus LR strains (HuH7 LR and PLC/PRF/5 LR). J and K, Western blot of lipid raft markers (J) and protein levels (K) of EGFR and ABCB1 on lipid rafts. L, Representative IF images and quantitation for ABCB1 (red), CAV1 (green), and DNA (DAPI, blue) after treatment with PBS or MβCD. n = 5 independent experiences. Scale bars, 15 μm. All the results are shown as the mean ± SD. Two-tailed Student t test and one- or two-way ANOVA were used to analyze the data. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
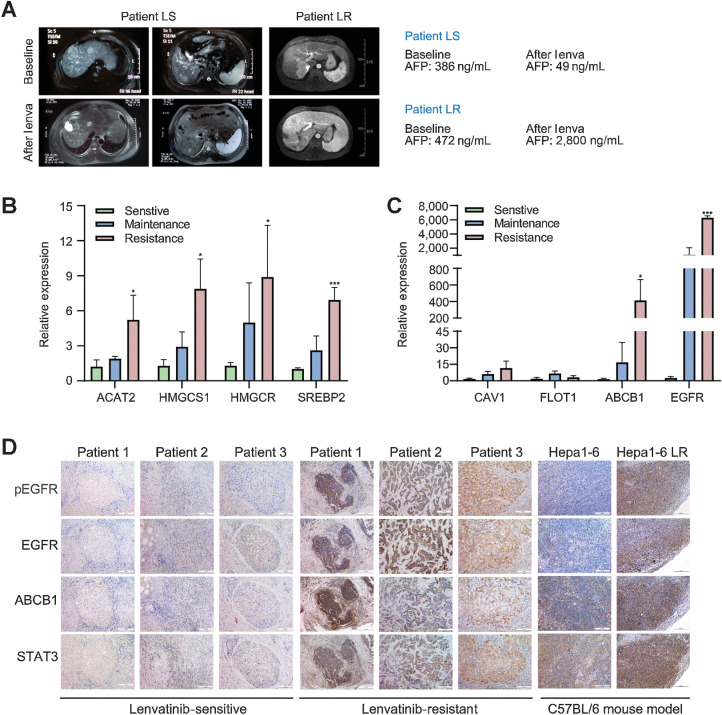
EGFR–STAT3–ABCB1 signaling activation is associated with the lenvatinib response in patients with HCC
To further validate the relevance between the activation of the EGFR–STAT3–ABCB1 pathway and lenvatinib response in HCC, we collected samples from 9 patients with HCC treated with lenvatinib, among whom 3 patients had a partial response (PR), 3 had stable disease (SD), and 3 had progression disease (PD) according to RESIST1.1 criteria (Fig. 4A). No prominent difference was observed in the mRNA levels of EGFR, ABCB1 and the key enzymes of cholesterol synthesis between the SD and PR groups, whereas their levels were increased significantly in the PD group compared with the PR and SD groups (P < 0.05). Similarly, the expression levels of lipid raft markers did not show significant differences among the three groups (Fig. 4B and C). In addition, IHC staining was performed in HCC tissues from clinical patients with acquired lenvatinib resistance as well as a subcutaneous implantation model (Hep1–6 LR) of mice to further validate the activation of the EGFR–STAT3–ABCB1 pathway in HCC resistance to lenvatinib (Fig. 4D). Collectively, these data suggest that the activation of the EGFR–STAT3–ABCB1 pathway is closely associated with the resistance of HCC to lenvatinib.
Figure 4.
Activation of the EGFR–STAT3–ABCB1 pathway is validated in LR patients and the Hep1–6 LR mouse model. The efficacy of lenvatinib treatment was evaluated according to RECIST1.1 criteria. In the preoperative efficacy evaluation, patients who reached PD were considered resistant to lenvatinib, and patients who reached PR or SD were considered sensitive to lenvatinib. A, Comparison of liver MRI images and AFP values before and after lenvatinib treatment between a patient in the sensitive group and a patient in the resistant group. B and C, Tissues from patients who reached PR, SD, and PD after lenvatinib treatment were used for validation at the mRNA level. D, IHC staining of liver cancer tissues from patients in the sensitive group and resistant group, and subcutaneous tumor tissues derived from mice that were implanted with the Hep1–6 and Hep1–6 LR strains. All the results are shown as the mean ± SD (n = 5). Two-way ANOVA was used to analyze the data. *, P < 0.05; ***, P < 0.001.
ABCB1 mediates lenvatinib resistance by enhancing its exocytosis from HCC cells
Because lenvatinib is a special substrate for Pgp (ABCB1), to further understand the role of ABCB1 in inducing lenvatinib resistance, we examined the concentration of lenvatinib in the culture medium of the LS and LR strains at different time points. We observed that the reduction in drug concentration in the supernatant of the LS and LR strains was consistent within the first 6 hours after treatment with lenvatinib. After that, different concentrations of lenvatinib were observed between the LS and LR strains, indicating the enhanced exocytosis of lenvatinib from the LR strains. After remaining approximately 6 hours, the lenvatinib concentration of both the LS and LR strains decreased again from the 12th hour after treatment to the endpoint of the experiments (Fig. 5A).
Figure 5.
Activation of the EGFR–STAT3–ABCB1 axis mediates lenvatinib resistance via drug exocytosis. A, Lenvatinib concentrations in cell culture media were determined. A concentration of 10 μmol/L lenvatinib was added to HuH7 and HuH7 LR cell media, and a concentration of 20 μmol/L of lenvatinib was added to PLC/PRF/5 and PLC/PRF/5 LR cell media. The concentration of lenvatinib in the cell supernatant was measured every 6 hours after drug addition. B, Molecular structure formula of FITC-lenvatinib (molecular weight 784.18 g/mol). C and D, Lenvatinib exocytosis assay for LS and LR strains. The LS and LR strains were stained with DiI dye (red) at a working concentration of 10 μmol/L for 20 minutes. After rinsing with PBS, FITC-lenvatinib (green) at a concentration of 10 μmol/L was added to HuH7 and HuH7 LR cell media, and 20 μmol/L FITC-lenvatinib was added to PLC/PRF/5 and PLC/PRF/5 LR cell media. After 6 hours of treatment, the culture medium was replaced by fresh medium without FITC-lenvatinib and photographed every hour to observe the change of FITC-lenvatinib enrichment in the cells. Scale bars, 20 μm. E and F, Histograms displaying the relative fluorescence intensity of FITC-lenvatinib (green). n = 5 independent experiments. All the results are shown as the mean ± SD. Two-way ANOVA was used to analyze the data. **, P < 0.01; ***, P < 0.001; ****, P < 0.0001; ns, nonsignificant.
To further validate these findings, we added a fluorescent FITC tag to lenvatinib (purity > 99.8%; Fig. 5B) and labeled the cells initially with Dil dye, which did not affect cell viability. Next, we added FITC-lenvatinib to the culture medium of the LS and LR strains and then changed the medium to remove the drug after 6 hours’ treatment. The cells were transferred to a high-connotation imaging analysis system (Living Cell Workstation) and photographed dynamically at 37°C and 5% CO2 for 24 hours. The imaging results were consistent with the solubility trend of lenvatinib. The intracellular fluorescence intensity of the LR strains significantly decreased over time. In contrast, the fluorescence quenching in LS strains was much slower than that in LR strains (Fig. 5C–F). These results indicated that the resistance to lenvatinib is at least in part due to the excretion of lenvatinib from HCC cells. Moreover, upregulation of EGFR and ABCB1 induced significant tolerance to lenvatinib in HCC cells (Supplementary Fig. S4A–S4E). Taken together, these data support that continuous exposure to lenvatinib induces the activation of the EGFR–STAT3–ABCB1 signaling pathway and thus enhances the exocytosis of lenvatinib from HCC cells.
Blocking EGFR phosphorylation and ABCB1 exocytosis has a synergistic effect with lenvatinib on HCC
We next investigated whether EGFR inhibitors could block the activation of the EGFR–STAT3–ABCB1 signaling pathway and thereby sensitize LR strains to lenvatinib. Erlotinib is a specific inhibitor targeting EGFR that can inhibit not only EGFR phosphorylation but also ABCB1-medicated exocytosis. Erlotinib alone did not show a striking effect on HCC cells, and no significant difference in IC50 values was observed between the LS and LR strains (Supplementary Fig. S5A–S5D).
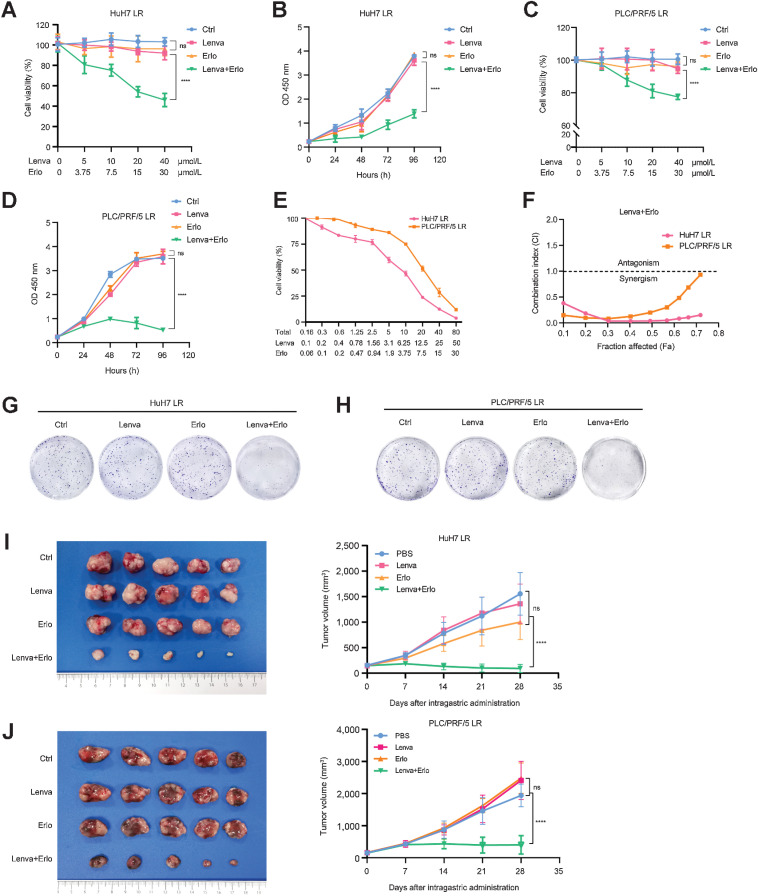
To evaluate the possible effect of the combination of lenvatinib and erlotinib in LR HCC, we treated the LR strains with the combination of erlotinib (10 μmol/L, 1/3 doses of IC50 to balance the efficacy with the toxic and side effects; ref. 25) and lenvatinib (a set of concentration gradients). The IC50 value of lenvatinib in HuH7 LR cells was significantly decreased from 53.29 μmol/L to 1.94 μmol/L, which was even lower than that in HuH7 cells (3.18 μmol/L; Supplementary Fig. S5E). Similar results were also found in PLC/PRF/5 LR cells with the IC50 value decreasing from 62.24 μmol/L to 20.42 μmol/L, which was lower than that in PLC/PRF/5 (23.61 μmol/L; Supplementary Fig. S5F). The potent effect on LR strains was also confirmed with fewer viable cells after the combination treatment (Supplementary Fig. S5G). Next, we used CCK-8 assays to determine the cell viabilities of HuH7 LR and PLC/PRF/5 LR strains, which were treated with the indicated concentration of erlotinib or lenvatinib alone, or their combination for 48 hours. The results showed that lenvatinib in combination with erlotinib demonstrated a more significant inhibition on the resistant cells compared with the lenvatinib or erlotinib alone (Fig. 6A–D). In addition, we used the Chou-Talalay algorithm to calculate their combination index (CI; ref. 26). The results showed that the CI < 1 in all the concentration gradients of lenvatinib, indicating that there is a significant synergistic effect between them (Fig. 6E and F). The long-term colony formation assays also revealed the synergistic effect in the combination group (Fig. 6G and H). Moreover, in a TUNEL apoptosis staining assay, LR strains treated with lenvatinib in combination erlotinib demonstrated much more apoptosis than that treated with lenvatinib or erlotinib alone (Supplementary Fig. S5H–S5K).
Figure 6.
Lenvatinib in combination with erlotinib has a synergistic effect on LR strains. A and B, HuH7 LR and PLC/PRF/5 LR strains were treated with the indicated concentration of erlotinib, lenvatinib alone, or their combination for 48 hours. Cells were then subjected to a CCK-8 assay to determine cell viability. C and D, HuH7 LR and PLC/PRF/5 LR strains were treated with the indicated concentration of lenvatinib, erlotinib alone, or their combination. Cells were then subjected to a CCK-8 assay to determine cell viability at the indicated time points. E and F, HuH7 LR and PLC/PRF/5 LR strains were treated with the indicated concentrations of lenvatinib and erlotinib for 48 hours. Cells were then subjected to a CCK-8 assay to determine cell viability, and the CI was determined. G and H, HuH7 LR and PLC/PRF/5 LR strains were treated with lenvatinib, erlotinib alone, or in combination. The remaining cells were stained after 14 days. I and J, Tumor appearance and tumor growth curves of subcutaneous implantation models of the HuH7 LR strain (I) and PLC/PRF/5 LR strain (J). When the tumor volume reached approximately 100 mm3, the mice were treated with PBS, lenvatinib (Lenva, 10 mg/kg), erlotinib (Erlo, 10 mg/kg), or their combination. All the results are shown as the mean ± SD. Two-way ANOVA was used to analyze the data. ****, P < 0.0001; ns, nonsignificant.
To further evaluate the synergistic effect of this combination treatment in vivo, we established subcutaneous LR models in nude mice using LR strains. Tumor-bearing mice were randomly divided into 4 groups with 5 mice in each group; the groups were treated with PBS, lenvatinib (10 mg/kg), erlotinib (30 mg/kg), and a combination with decreased doses (lenvatinib 3 mg/kg + erlotinib 10 mg/kg, 1/3 doses of each used alone). The results demonstrated that the combination therapy impaired tumor growth and decreased tumor burden more effectively than monotherapy (Fig. 6I and J), which indicated that the combination treatment exhibited a strong synergistic effect on tumor growth of LR strains in vivo. Similar results were further confirmed in the mouse model of lenvatinib resistance (Hep1–6 LR; Supplementary Fig. S5L). Moreover, the results indicated no obvious pathologic damage and no significant weight loss in the experimental and control groups, and no aberration was found in liver and kidney function (Supplementary Fig. S6A–S6D). In addition, we carried out IHC staining of Ki-67 and cleaved caspase-3 in the subcutaneous grafts of mice. The results indicated that the LR strains treated with their combination expressed higher cleaved caspase-3 and lower Ki-67 compared with that treated with the single agent alone (Supplementary Fig. S7A–S7C).
Given that STAT3 activation plays a remarkable role in mediating lenvatinib resistance, we further evaluated whether a STAT3 inhibitor had potent synergistic effect with lenvatinib. The results revealed that the combination of lenvatinib and Stattic (a pStat3 inhibitor) could significantly suppress the proliferation of LR strains, but Stattic alone had no obvious effect at these low concentrations (approximately less than 20 μmol/L; Supplementary Fig. S8A and S8B). Their synergistic effect was further confirmed in vivo (Supplementary Fig. S8C and S8D). In addition, we carried out IHC staining of Ki-67 and cleaved caspase-3 in the subcutaneous grafts of mice. Similar to the results of the combination of lenvatinib and erlotinib, LR strains treated with the lenvatinib in combination with Stattic expressed higher cleaved caspase-3 and lower Ki-67 compared with that treated with the single agent alone (Supplementary Fig. S8E and S8F).
Collectively, these results suggest that the combination of lenvatinib and erlotinib produces a strong synergistic effect on HCC with a decreased dose of lenvatinib and the treatment is safe with reliable outcomes.
Erlotinib blocks the EGFR–STAT3–ABCB1 pathway and inhibits the exocytosis of lenvatinib
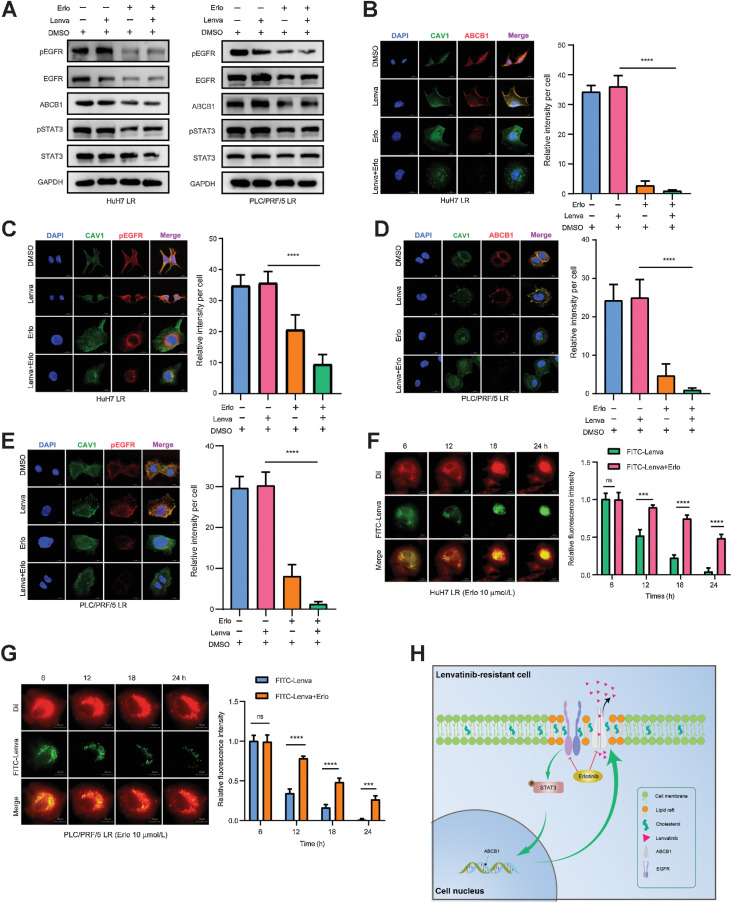
To elucidate the underlying mechanism of the synergistic effect induced by the combination of lenvatinib and erlotinib, we treated HuH7 LR cells and PLC/PRF/5 LR cells with drugs for 48 hours and detected alterations in the EGFR–STAT3–ABCB1 pathway using Western blotting. The results showed that erlotinib, rather than lenvatinib, significantly suppressed pEGFR and ABCB1expression, while their combination demonstrated a synergistic effect that achieved the same inhibition efficiency on pEGFR and ABCB1 levels with a 1/3 total dose (Fig. 7A). Consistent with Western blotting findings, the fluorescence intensities of ABCB1 and pEGFR on the cellular membrane of the combinational treatment group were significantly lower than those of the other groups. The fluorescence area was concentrated in the perinuclear cytoplasm of the treated cells (Fig. 7B–E), suggesting that the combination of lenvatinib with erlotinib induced significant downregulation and functional inhibition of pEGFR and ABCB1in the cellular membrane. Similar effects on pEGFR and ABCB1were further confirmed in xenograft models (Supplementary Fig. S7A–S7C). Moreover, we found that erlotinib significantly delayed the exocytosis of FITC-lenvatinib with a fluorescence retention time lasting until the end of 24 hours’ observation, during which, numerous apoptotic cells were observed (Fig. 7F and G). Taken together, these findings indicated that the combination with erlotinib could block the EGFR–STAT3–ABCB1 pathway and inhibit the exocytosis of lenvatinib, thus indicating a promising way to reverse lenvatinib resistance.
Figure 7.
Lenvatinib combined with erlotinib effectively blocks the EGFR–STAT3–ABCB1 pathway. A, HuH7 LR strains were treated with DMSO, without lenvatinib (Lenva, 20 μmol/L), erlotinib (Erlo, 30 μmol/L), or the combination (Lenva, 3 μmol/L + Erlo, 10 μmol/L) for 48 hours. PLC/PRF/5 LR strains were treated with DMSO, with/without lenvatinib (Lenva, 30 μmol/L), erlotinib (Erlo, 30 μmol/L), or the combination (Lenva, 10 μmol/L + Erlo, 10 μmol/L) for 48 hours. Western blotting was conducted to observe changes in the EGFR–STAT3–ABCB1 pathway. B–E, HuH7 LR (B and C) and PLC/PRF/5 LR (D and E) cells were treated with DMSO, lenvatinib, erlotinib, or the combination as the Western blotting experiment (A), fixed and stained with ABCB1, CAV1, and pEGFR antibodies. The fluorescence signals were analyzed by using confocal microscopy. Representative images and the quantitation of relative fluorescence intensity are shown. n = 5 independent experiments. Scale bars, 15 μm. F and G, LR strains were stained with DiI dye (red) at a working concentration of 10 μmol/L for 20 minutes. After rinsing with PBS, FITC-lenvatinib (green) at a concentration of 10 μmol/L was added to HuH7 and HuH7 LR cell media, and 20 μmol/L FITC-lenvatinib was added to PLC/PRF/5 and PLC/PRF/5 LR cell media. After 6 hours of treatment, the culture medium was replaced with fresh medium containing 10 μmol/L erlotinib and photographed every hour to observe the changes in FITC-lenvatinib enrichment in the cells. Representative images and the quantitation of relative fluorescence intensity are shown. n = 5 independent experiments. Scale bars, 20 μm. H, Working model of the combination of lenvatinib and erlotinib eliminating lenvatinib resistance induced by EGFR–STAT3–ABCB1 axis activation in HCC. All the results are shown as the mean ± SD. One- or two-way ANOVA was used to analyze the data. ***, P < 0.001; ****, P < 0.0001; ns, nonsignificant.
Discussion
Drug resistance has become one of the major problems for cancer therapy (27). As with sorafenib, the newly approved drug lenvatinib also faces the issue of drug resistance. According to incomplete statistics, over 60% of patients with HCC develop tolerance to lenvatinib within 1 year, and only a fraction of patients obtain long-term benefits. Therefore, extensive efforts are ongoing to understand the underlying mechanisms of response/resistance. According to the efficacy of HCC treatment, drug resistance can be classified into primary (resistance at the initial treatment) and acquired resistance (sensitive at the beginning but resistant later during the treatment process). On the basis of previous studies, the acquired resistance of HCC can be due to restricted drug uptake and increased exocytosis; abnormal drug metabolic pathways; bypass activation of treatment targets; enhanced DNA repair; imbalance of survival and apoptosis and tumor microenvironment factors (24, 28–36). For patients with HCC, sorafenib was the only molecularly targeted drug until a few years ago and its mechanism of acquired resistance has been explored for over a decade with little clarity. Therefore, the acquired resistance mechanism for newly approved drug lenvatinib needs to be unveiled, and it is critical to understand the underlying mechanisms of drug resistance, explore promising ways to overcome that and further improve the treatment response to benefit patients with HCC.
Through analysis of proteomics and RTK phosphorylation microarrays in models with acquired lenvatinib resistance, our study demonstrated that HCC cells developed resistance to lenvatinib by activating EGFR and stimulating the downstream EGFR–STAT3–ABCB1 axis, which was accompanied by aberrant cholesterol metabolism and lipid raft activation. ABCB1 was activated by EGFR in a lipid raft–dependent manner, which significantly enhanced the exocytosis of lenvatinib to mediate resistance (Fig. 7H). The combination of lenvatinib and erlotinib, an EGFR inhibitor for non–small cell lung cancer (NSCLC), yielded a significant synergistic effect to improve HCC patients’ clinical benefit with lenvatinib treatment. Coincidentally, a recent study also suggested that the combination of lenvatinib and an EGFR inhibitor (gefitinib) might be particularly beneficial for patients with high initial EGFR expresseion (37). Their results demonstrated that primary resistance to lenvatinib in patients with HCC is due to the inhibition of FGFR, which leads to feedback activation of the EGFR–PAK2–ERK5 signaling axis. Thus, both studies reinforced the idea that a combination of lenvatinib and EGFR inhibitors would further benefit patients with HCC by combating drug resistance (both primary and acquired).
For future studies, further exploration of aberrant cholesterol metabolism or the enhanced exocytosis from HCC cells will bring more clarity to our understanding of acquired drug resistance, which may provide us with better targets. Of note, ABCB1 is involved in acquired lenvatinib resistance in HCC. Although specialized targeted inhibitors of the key factor ABCB1 have been developed for the 3rd generation (Tariquidar), they have not been clinically authorized for therapy due to severe side effects. Similarly, the evaluation of Stattic, a STAT3 inhibitor, revealed significant cytotoxicity, weaking the possibility of its application in combination treatment. Erlotinib, a drug launched in 2004, has undergone 17 years of exploration and development, and in May 2020, the FDA approved erlotinib in combination with ramucirumab as a first-line treatment option for patients with advanced NSCLC with EGFR exon 19 deletion or exon 21 L858R mutation (38). This was the first approval of a combination of two RTK inhibitors in lung cancer therapy and provided the possibility for the combination of targeted drugs. Erlotinib and lenvatinib are administered orally and are well tolerated by patients without the use of invasive procedures, such as subcutaneous or intravenous injections (39). Moreover, this combination regimen offers a new treatment option for patients with immune dysfunction, those on immunosuppressive drugs after liver transplantation and those who cannot tolerate immunotherapy. Thus, EGFR inhibitors may be the best option for the treatment of HCC with acquired lenvatinib resistance.
In summary, our research has shed light on acquired lenvatinib resistance among patients with HCC. Combination treatment with lenvatinib and erlotinib can suppress the EGFR–STAT3–ABCB1 pathway, thus overcoming exocytosis-mediated resistance to lenvatinib.
Supplementary Material
Acknowledgments
This work was supported by the National Natural Science Foundation of China (81930074, 91959203) and the Program of Shanghai Academic Research Leader (20XD1400900).
The authors acknowledge the Dr. Mien-Chie Hung's Laboratory in MD Anderson Cancer Center (Houston, TX) and the members of the Huashan Hospital Central Laboratory, especially Xiao Xu for excellent technical assistance.
The publication costs of this article were defrayed in part by the payment of publication fees. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Footnotes
Note: Supplementary data for this article are available at Cancer Research Online (http://cancerres.aacrjournals.org/).
Authors' Disclosures
No disclosures were reported.
Authors' Contributions
B. Hu: Conceptualization, data curation, writing–original draft. T. Zou: Data curation, supervision, writing–review and editing. W. Qin: Data curation, formal analysis. X. Shen: Data curation, formal analysis. Y. Su: Writing–review and editing. J. Li: Resources. Y. Chen: Writing–original draft. Z. Zhang: Investigation. H. Sun: Resources. Y. Zheng: Methodology. C.Q. Wang: Investigation. Z. Wang: Resources. T.E. Li: Investigation. S. Wang: Investigation. L. Zhu: Visualization. X. Wang: Investigation. Y. Fu: Investigation. X. Ren: Investigation. Q. Dong: Conceptualization, funding acquisition, writing–review and editing. L.X. Qin: Conceptualization, funding acquisition.
References
- 1. Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2021;71:209–49. [DOI] [PubMed] [Google Scholar]
- 2. Llovet JM, Montal R, Sia D, Finn RS. Molecular therapies and precision medicine for hepatocellular carcinoma. Nat Rev Clin Oncol 2018;15:599–616. [DOI] [PubMed] [Google Scholar]
- 3. Kudo M. Lenvatinib may drastically change the treatment landscape of hepatocellular carcinoma. Liver Cancer 2018;7:1–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Al-Salama ZT, Syed YY, Scott LJ. Lenvatinib: a review in hepatocellular carcinoma. Drugs 2019;79:665–74. [DOI] [PubMed] [Google Scholar]
- 5. Pike LJ. Rafts defined: a report on the Keystone Symposium on Lipid Rafts and Cell Function. J Lipid Res 2006;47:1597–8. [DOI] [PubMed] [Google Scholar]
- 6. Lingwood D, Simons K. Lipid rafts as a membrane-organizing principle. Science 2010;327:46–50. [DOI] [PubMed] [Google Scholar]
- 7. Sezgin E, Levental I, Mayor S, Eggeling C. The mystery of membrane organization: composition, regulation, and roles of lipid rafts. Nat Rev Mol Cell Biol 2017;18:361–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lajoie P, Goetz JG, Dennis JW, Nabi IR. Lattices, rafts, and scaffolds: domain regulation of receptor signaling at the plasma membrane. J Cell Biol 2009;185:381–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Yarden Y, Sliwkowski MX. Untangling the ErbB signaling network. Nat Rev Mol Cell Biol 2001;2:127–37. [DOI] [PubMed] [Google Scholar]
- 10. Gschwind A, Fischer OM, Ullrich A. The discovery of receptor tyrosine kinases: targets for cancer therapy. Nat Rev Cancer 2004;4:361–70. [DOI] [PubMed] [Google Scholar]
- 11. Schlessinger J, Lemmon MA. Nuclear signaling by receptor tyrosine kinases: the first robin of spring. Cell 2006;127:45–8. [DOI] [PubMed] [Google Scholar]
- 12. Scaltriti M, Baselga J. The epidermal growth factor receptor pathway: a model for targeted therapy. Clin Cancer Res 2006;12:5268–72. [DOI] [PubMed] [Google Scholar]
- 13. Hynes NE, Lane HA. ERBB receptors and cancer: the complexity of targeted inhibitors. Nat Rev Cancer 2005;5:341–54. [DOI] [PubMed] [Google Scholar]
- 14. Gerlach JH, Kartner N, Bell DR, Ling V. Multidrug resistance. Cancer Surv 1986;5:25–46. [PubMed] [Google Scholar]
- 15. Sauna ZE, Smith MM, Müller M, Kerr KM, Ambudkar SV. The mechanism of action of multidrug resistance–linked P-glycoprotein. J Bioenerg Biomembr 2001;33:481–91. [DOI] [PubMed] [Google Scholar]
- 16. Robey RW, Pluchino KM, Hall MD, Fojo AT, Bates SE, Gottesman MM. Revisiting the role of ABC transporters in multidrug-resistant cancer. Nat Rev Cancer 2018;18:452–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Yarden Y, Shilo BZ. SnapShot: EGFR signaling pathway. Cell 2007;131:1018. [DOI] [PubMed] [Google Scholar]
- 18. Sabbah DA, Hajjo R, Sweidan K. Review on epidermal growth factor receptor (EGFR) structure, signaling pathways, interactions, and recent updates of EGFR inhibitors. Curr Top Med Chem 2020;20:815–34. [DOI] [PubMed] [Google Scholar]
- 19. Wang WJ, Li CF, Chu YY, Wang YH, Hour TC, Yen CJ, et al. Inhibition of the EGFR/STAT3/CEBPD axis reverses cisplatin cross-resistance with paclitaxel in the urothelial carcinoma of the urinary bladder. Clin Cancer Res 2017;23:503–13. [DOI] [PubMed] [Google Scholar]
- 20. Zhao L, Bin S, He HL, Yang JM, Pu YC, Gao CH, et al. Sodium butyrate increases P-gp expression in lung cancer by upregulation of STAT3 and mRNA stabilization of ABCB1. Anticancer Drugs 2018;29:227–33. [DOI] [PubMed] [Google Scholar]
- 21. Endres NF, Das R, Smith AW, Arkhipov A, Kovacs E, Huang Y, et al. Conformational coupling across the plasma membrane in activation of the EGF receptor. Cell 2013;152:543–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Danielsen AJ, Maihle NJ. Ligand-independent oncogenic transformation by the EGF receptor requires kinase domain catalytic activity. Exp Cell Res 2002;275:9–16. [DOI] [PubMed] [Google Scholar]
- 23. Klappe K, Hummel I, Hoekstra D, Kok JW. Lipid dependence of ABC transporter localization and function. Chem Phys Lipids 2009;161:57–64. [DOI] [PubMed] [Google Scholar]
- 24. Mineo C, James GL, Smart EJ, Anderson RG. Localization of epidermal growth factor-stimulated Ras/Raf-1 interaction to caveolae membrane. J Biol Chem 1996;271:11930–5. [DOI] [PubMed] [Google Scholar]
- 25. Dong Q, Du Y, Li H, Liu C, Wei Y, Chen MK, et al. EGFR and c-MET cooperate to enhance resistance to PARP inhibitors in hepatocellular carcinoma. Cancer Res 2019;79:819–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Chou TC. Drug combination studies and their synergy quantification using the Chou-Talalay method. Cancer Res 2010;70:440–6. [DOI] [PubMed] [Google Scholar]
- 27. Vasan N, Baselga J, Hyman DM. A view on drug resistance in cancer. Nature 2019;575:299–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Gao B, Yang FM, Yu ZT, Li R, Xie F, Chen J, et al. Relationship between the expression of MDR1 in hepatocellular cancer and its biological behaviors. Int J Clin Exp Pathol 2015;8:6995–7001. [PMC free article] [PubMed] [Google Scholar]
- 29. Ba HL, Mbatchi L, Gattacceca F, Evrard A, Lacarelle B, Blanchet B, et al. Pharmacogenetics and pharmacokinetics modeling of unexpected and extremely severe toxicities after sorafenib intake. Pharmacogenomics 2020;21:173–9. [DOI] [PubMed] [Google Scholar]
- 30. Caruso S, Calatayud AL, Pilet J, La Bella T, Rekik S, Imbeaud S, et al. Analysis of liver cancer cell lines identifies agents with likely efficacy against hepatocellular carcinoma and markers of response. Gastroenterology 2019;157:760–76. [DOI] [PubMed] [Google Scholar]
- 31. Ding X, He M, Chan AWH, Song QX, Sze SC, Chen H, et al. Genomic and epigenomic features of primary and recurrent hepatocellular carcinomas. Gastroenterology 2019;157:1630–45. [DOI] [PubMed] [Google Scholar]
- 32. Zhou SL, Zhou ZJ, Hu ZQ, Huang XW, Wang Z, Chen EB, et al. Tumor-associated neutrophils recruit macrophages and T-regulatory cells to promote progression of hepatocellular carcinoma and resistance to sorafenib. Gastroenterology 2016;150:1646–58. [DOI] [PubMed] [Google Scholar]
- 33. Xiang Q, Chen W, Ren M, Wang J, Zhang H, Deng DY, et al. Cabozantinib suppresses tumor growth and metastasis in hepatocellular carcinoma by a dual blockade of VEGFR2 and MET. Clin Cancer Res 2014;20:2959–70. [DOI] [PubMed] [Google Scholar]
- 34. Marin JJG, Briz O, Herraez E, Lozano E, Asensio M, Di Giacomo S, et al. Molecular bases of the poor response of liver cancer to chemotherapy. Clin Res Hepatol Gastroenterol 2018;42:182–92. [DOI] [PubMed] [Google Scholar]
- 35. Liu F, Dong X, Lv H, Xiu P, Li T, Wang F, et al. Targeting hypoxia-inducible factor-2α enhances sorafenib antitumor activity via β-catenin/C-Myc–dependent pathways in hepatocellular carcinoma. Oncol Lett 2015;10:778–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Zhu Y, Yang J, Xu D, Gao XM, Zhang Z, Hsu JL, et al. Disruption of tumor-associated macrophage trafficking by the osteopontin-induced colony-stimulating factor-1 signaling sensitizes hepatocellular carcinoma to anti–PD-L1 blockade. Gut 2019;68:1653–66. [DOI] [PubMed] [Google Scholar]
- 37. Jin H, Shi Y, Lv Y, Yuan S, Ramirez CFA, Lieftink C, et al. EGFR activation limits the response of liver cancer to lenvatinib. Nature 2021;595:730–4. [DOI] [PubMed] [Google Scholar]
- 38. Nakagawa K, Garon EB, Seto T, Nishio M, Ponce Aix S, Paz-Ares L, et al. Ramucirumab plus erlotinib in patients with untreated, EGFR-mutated, advanced non–small cell lung cancer (RELAY): a randomized, double-blind, placebo-controlled, phase III trial. Lancet Oncol 2019;20:1655–69. [DOI] [PubMed] [Google Scholar]
- 39. Minna JD, Dowell J. Erlotinib hydrochloride. Nat Rev Drug Discov 2005;Suppl:S14–5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All the data acquired and/or used in the study are available upon request from the corresponding author.