Abstract
Two reference monoclonal antibodies against the meningococcal P1.15 subtype PorA, MN3C5C and 2-1-P1.15, showed only partial concordant recognition of meningococcal isolates. Cyanogen bromide cleavage of P1.19,15 PorA, peptide mapping, and sequencing of porA regions demonstrated that 2-1-P1.15 was specific for subtype P1.19, and henceforth it is to be redesignated as 2-1-P1.19.
The PorA, or class 1 outer membrane porin protein, of Neisseria meningitidis serves as the subtyping antigen for characterization of meningococci (13). Monomeric PorA is a transmembrane protein with eight outer loops, of which the first and the fourth loop contain the hypervariable regions, called variable region 1 (VR1) and VR2, respectively (6, 18, 26). These two regions define the dual PorA subtype, designated P1.x,y, where “P1.” stands for the class 1 protein and “x” and “y” stand for numbers denoting the VR1 and VR2 domains, respectively. The two subtype regions of PorA are generally determined by an enzyme-linked immunosorbent assay (ELISA) (1) or a blotting assay (31, 33) with reference monoclonal antibodies (MAbs) directed against epitopes in VR1 or VR2; consequently, each PorA can bind two different subtype-specific MAbs. In addition, variations in VR1 and VR2 are analyzed by sequencing of porA genes (4, 8, 17–20, 23–26).
In a previous characterization of meningococcal isolates, the two reference MAbs against the common P1.15 subtype, MN3C5C (1) and 2-1-P1.15, did not show identical binding patterns (29). The epitope for MN3C5C has previously been mapped to a 3-amino-acid sequence in VR2 (19), but that for 2-1-P1.15 has not been reported. Because those MAbs have been used for serological characterization of several large strain collections (1, 3, 9, 27), the aim of our study was to elucidate the reason for their different specificities.
(Parts of this work were presented at the Tenth International Pathogenic Conference, Baltimore, Md., 8 to 13 September 1996 [28]).
For this purpose, whole-cell suspensions of 707 strains, isolated between 1987 and 1995 from patients with meningococcal disease in Norway, were screened on dot blots with a panel of serotype- and subtype-specific MAbs as described elsewhere (31). Strains that were positive with MN3C5C and 2-1-P1.15 on dot blots were also immunoblotted with those MAbs following sodium dodecyl sulfate (SDS) gel electrophoresis of boiled cell suspensions (31). PorA bands on the blots, as well as the corresponding PorA bands in SDS gels, stained with Coomassie brilliant blue, were scanned by densitometry (30). The rationale behind this analysis was that PorA epitope variants might be revealed by their weaker antibody binding after antigen denaturation. Isolates were also characterized by multilocus enzyme electrophoresis from the combination of alleles at 14 enzyme loci (7). Distinctive multilocus genotypes were designated as electrophoretic types (ETs).
For DNA sequencing of the porA gene, chromosomal DNA was isolated from a loopful of N. meningitidis cells, suspended in 400 μl of TE buffer (10 mM Tris-HCl–1 mM EDTA [pH 8.0]), essentially as described previously (10), except for a 2-h lysozyme treatment. One microliter of DNA, diluted 1:5, was amplified in a PCR assay (total volume, 50 μl) with the primer pair 5′-AAACTTACCGCCCTCGTA-3′ and 5′-TTAGAATTTGTGGCGCAAACCGAC-3′ (8). Sequencing of PCR products was performed as reported previously (8) or by automated sequencing using an ABI Prism 377 and the Big Dye Terminator Cycle Sequencing Kit (Perkin-Elmer Applied Biosystems).
The epitope for MAb 2-1-P1.15 was localized by reacting the MAb in an ELISA (22) with synthetic 25- to 29-mer peptides corresponding to loops 1 (VR1), 4 (VR2), and 5 of the subtype P1.19,15 PorA from reference strain H355 (18, 25). The peptides were used in the oxidized state and bound directly to the plate. Detailed epitope mapping was performed by the Geysen method with pins derivatized to allow cleavage of the completed peptides from the pins (15). Twenty-three overlapping decapeptides (each shifted along the sequence by 1 amino acid) that spanned all of VR1 from P1.19,15 PorA were prepared. A 4-amino-acid spacer (SGSG) was added N-terminally to each decapeptide, and the completed peptides were biotinylated at the N terminus before cleavage from the pins (15). The SGSG spacer served to raise the reactive peptides from the surface of the ELISA plate and allow for mobility and conformational freedom of the potentially reactive sequences. The biotinylated peptides were bound to ELISA plates previously coated with streptavidin (50 μl of 50 μg ml−1, dried overnight at 37°C). After three washes, peptides diluted to 50 μg ml−1 in phosphate-buffered saline were added, and the plates were incubated for 2 h at room temperature. The MAb was diluted 1:1,000 and allowed to react with the peptides overnight at room temperature. Alkaline phosphatase-labelled anti-mouse immunoglobulin G (1 μg ml−1) was used as the second antibody and incubated for 2 h at room temperature. The assay was completed and read as described previously (22).
Dot blot analysis showed that 25 of the 707 patient strains expressed PorA’s that reacted with both reference MAbs, MN3C5C and 2-1-P1.15, whereas 12 strains bound 2-1-P1.15 but not MN3C5C (Table 1). Five of the latter strains also expressed epitopes for the P1.1, P1.2, or P1.14 subtype-specific MAbs. All but 1 of the 37 strains belonged to serogroup B, and all strains expressed a class 3 PorB protein except for one nontypeable strain that appeared to lack the PorB in SDS gels. From multilocus enzyme electrophoresis, the 37 isolates that were 2-1-P1.15 positive belonged to three distinct clone complexes. Eleven strains were members of the ET-5 complex; these included the four serotype 15 strains and seven of the serotype 4 strains (Table 1). The other five serotype 4 isolates belonged to a clonal group (J1) previously identified in Russia (9). The remaining 21 strains, which expressed serotype 8, 17, or 19 or were nontypeable, were somewhat more heterogeneous in their genotypes. Thus, strains of various unrelated genotypes all carried the epitope for MAb 2-1-P1.15.
TABLE 1.
Reaction on dot blots of the two subtype reference MAbs 2-1-P1.15 and MN3C5C with meningococci isolated from patients in Norwaya
| Serological typing characteristicsb | No. of strains | Dot blot reactions with:
|
|
|---|---|---|---|
| 2-1-P1.15 | MN3C5C | ||
| NG:4:P1.15 | 1 | + | + |
| B:4:P1.15 | 9 | + | + |
| B:8,19:P1.15 | 5 | + | + |
| B:15:P1.15 | 1 | + | + |
| B:17:P1.15 | 1 | + | + |
| B:19:P1.15 | 5 | + | + |
| B:NT:P1.15 | 3 | + | + |
| B:1:P1.2,15 | 1 | + | − |
| B:4:P1.15 | 1 | + | − |
| B:4:P1.14,15 | 1 | + | − |
| B:8,19:P1.15 | 6 | + | − |
| B:15:P1.1,15 | 3 | + | − |
| Total | 37 | 37c | 25c |
MN3C5C was a gift from J. T. Poolman, then at the National Institute of Public Health and the Environment, Bilthoven, The Netherlands. Both this (21) and MAb 2-1-P1.15 (raised at the Walter Reed Army Institute of Research) were produced with reference strain H355 (B:15:P1.19,15) as the antigen.
Include serogroup, serotype, and subtype, separated by colons (13). NG, nongroupable. NT, nontypeable.
Total number of strains reacting positively with the MAb.
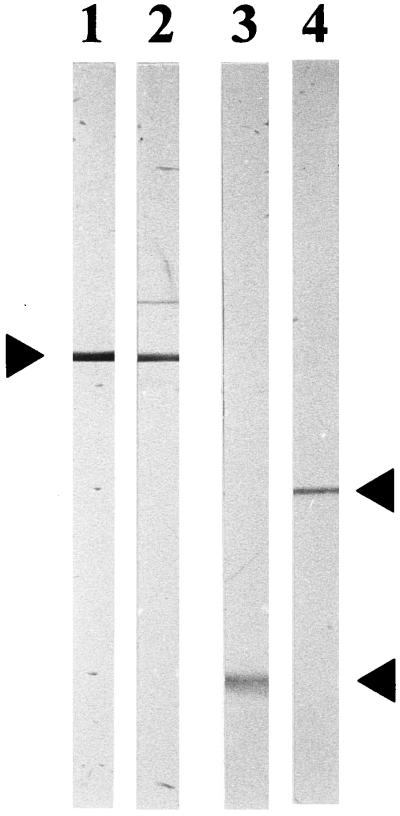
To determine whether the epitope for MAb 2-1-P1.15 was directed to VR1 of PorA or to VR2 as reported for MN3C5C (19), the two P1.15 reference MAbs were reacted on immunoblots with cyanogen bromide (CNBr)-treated PorA from the Cuban vaccine strain 385/83. This strain expresses the same P1.19,15 subtype PorA as reference strain H355 (14, 18). CNBr splits PorA at the single methionine located between VR1 and VR2 (18), resulting in VR1 with subtype P1.19 residing in loop 1 and VR2 with subtype P1.15 in loop 4 on two different fragments. Deoxycholate-extracted outer membrane vesicles (32) were separated in SDS gels, and the transparent PorA band was cut out from the gel after staining with imidazole-SDS and ZnSO4 (12). The gel piece was digested with CNBr in formic acid for 48 h at room temperature (16). Following drying, the digest was separated in SDS gels with 15% acrylamide, electrotransferred to 0.2-μm-pore-size nitrocellulose filters, and incubated with the MAbs. Figure 1 shows that MAb 2-1-P1.15 reacted with the VR1-containing fragment with a molecular mass of 16 kDa after CNBr cleavage, whereas MN3C5C bound to the 25-kDa fragment containing VR2. Thus, the epitope for 2-1-P1.15 was located on the part of PorA that comprised subtype P1.19, demonstrating that the specificity of this MAb was clearly distinct from that of MN3C5C.
FIG. 1.
Reaction of P1.19,15 PorA with the two reference MAbs MN3C5C and 2-1-P1.15 before and after cleavage with CNBr. The immunoblot shows untreated (lanes 1 and 2) and CNBr-treated (lanes 3 and 4) PorA incubated with MAb 2-1-P1.15 (lanes 1 and 3) or MAb MN3C5C (lane 2 and 4). Antibody binding was detected with peroxidase-conjugated antibodies. Arrows, from top to bottom, indicate the mobilities of PorA and its split products containing VR2 (25 kDa) and VR1 (16 kDa).
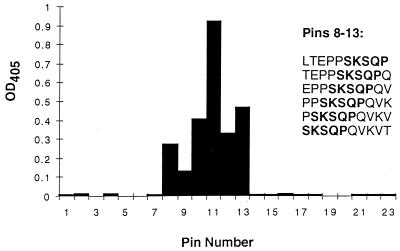
In support of this, ELISA analyses with 25- to 29-mer synthetic peptides, which corresponded to loop 1 (P1.19 region), loop 4 (P1.15 region), and loop 5 of the P1.19,15 PorA, resulted in a positive reaction of MAb 2-1-P1.15 with the loop 1 peptide only (ELISA titer of 1:12,800). More-detailed mapping with overlapping 10-mer peptides from the P1.19 VR1 region showed that 2-1-P1.15 gave a positive reaction only with peptides containing the sequence SKSQP (Fig. 2). The peptide giving the strongest reaction was PPSKSQPQVK. These results therefore confirmed that MAb 2-1-P1.15 was indeed a P1.19-specific reagent.
FIG. 2.
Epitope map for MAb 2-1-P1.15. Overlapping decapeptides from P1.19,15 PorA were synthesized on pins and extended by N-terminal addition of a SGSG spacer and biotin. The peptides were cleaved from the pins, applied to streptavidin-coated ELISA plates, and reacted with MAb 2-1-P1.15. The epitope common to all peptides reacting with the MAb was SKSQP.
The porA gene regions, encoding VR1 and VR2, were sequenced for 18 strains. These strains included some that reacted distinctly with the two reference MAbs on immunoblots, as well as those that showed less binding despite normal expression of denatured PorA protein in SDS gels. From the densitometric analysis described above, 9 of the 25 MN3C5C-positive strains bound MN3C5C weakly after immunoblotting, whereas 3 of the 2-1-P1.15-positive strains showed reduced binding or no binding, suggesting PorA epitope variants. The majority of the 2-1-P1.15-positive strains had a PPSKSQPQ sequence in VR1 (Table 2). This PorA sequence has previously been designated subtype P1.19 (11, 25). The three strains described above, which might express variants of the epitope for MAb 2-1-P1.15, did indeed express different modifications of the P1.19 sequence. These we have designated P1.19d (PRSKSQPQ; strain 19/92), P1.19e (PPSNSQPQ; strain 111/95), and P1.19f (PLSKSQPQ; strain 2/91), respectively, consistent with the proposal to designate minor subtype variants with lowercase letters (11, 25).
TABLE 2.
Deduced amino acid sequences in VR1 and VR2 of PorA from meningococcal isolates reacting with the two reference MAbs 2-1-P1.15 and MN3C5C
| Strain | Dot blot reaction
with:
|
Amino acid
sequence
|
Subtypeb | ||
|---|---|---|---|---|---|
| 2-1-P1.15 | MN3C5C | VR1 | VR2 | ||
| 52/93 | + | + | PPSKSQPQVKVTKA | HYTRQNNADVFVP | P1.19,15 |
| 9/93 | + | − | PPSKSQPQVKVTKA | YVDENKMVHA | P1.19,14dc |
| 36/93 | + | − | PPSKSQPQVKVTKA | YVAVENGVAKKVA | P1.19,1 |
| 19/92a | + | − | PRSKSQPQVKVTKA | HFVQQTPQSQPTLVP | P1.19d,c2c |
| 104/88 | + | − | PPSKSQPQVKVTKA | HFVQDKKGQPPTLVP | P1.19,10b |
| 111/95a | + | + | PPSNSQPQVKVTKA | HYTRQNNADVFVP | P1.19e,c15 |
| 35/92a | + | + | PPSKSQPQVKVTKA | HYTRQNNTDVFVP | P1.19,15a |
| 2/91a | + | − | PLSKSQPQVKVTKA | HYTRPNNTDVFVP | P1.19f,c15dc |
PorA variant with reduced binding of MN3C5C and/or 2-1-P1.15 on immunoblot as shown by densitometric analysis.
Subtype designations according to previous reports (11, 18, 20, 22, 25). In addition to the strains shown here, eight more strains with P1.19,15a PorA and two more with P1.19,15 PorA were sequenced.
New subtype variant described in this study.
All strains that bound MN3C5C on dot blots expressed either HYTRQNNA or HYTRQNNT in VR2 (Table 2). These two sequences have previously been designated P1.15 (11, 18) and P1.15a (20, 25), respectively. However, the HYTRQNNA sequence in reference strain H355 has also been labelled P1.15b (20). Since this designation was also used for another sequence (11), the assignment of the different P1.15 variants needs to be sorted out. The P1.15a sequence was found in all nine strains that reacted weakly with MN3C5C on immunoblots, supporting the usefulness of this method in picking epitope variants. Interestingly, while MN3C5C was produced with strain H355, expressing the P1.15 subtype (21), it was the P1.15a variant sequence that was used to map its epitope as NNT (19). Strain 2/91 failed to bind MN3C5C on dot blots despite the NNT sequence in VR2. The presence of a proline instead of a glutamine in front of that sequence probably blocked the binding of MN3C5C. This P1.15 variant was named P1.15d.
The two isolates which reacted with the subtype-specific MAbs P1.1 (strain 36/93) and P1.2 (strain 19/92) in addition to 2-1-P1.15 (Table 2) had VR2 sequences compatible with these reactions (18, 26). This was also true for strain 9/93, which bound the P1.14 subtype-specific MAb and expressed a variant of the VR2 in reference strain S3446 (B:14:P1.22a,14a) (11, 17, 22), here designated P1.14d. Thus, the sequencing studies confirmed the specificities of the different reference MAbs and resulted in detection of one new P1.15 variant, three P1.19 variants, and one P1.14 variant. No PCR product with porB-specific primers was obtained for the single nontypeable P1.19,15a isolate lacking a PorB protein in SDS gels (see above).
In conclusion, our results show that 2-1-P1.15 is not a VR2-specific reference MAb for the meningococcal P1.15 subtype, as previously believed, but is directed to subtype P1.19 located in VR1. No P1.19 reference MAb has yet been defined (2, 5), so we propose that MAb 2-1-P1.15 henceforth should be redesignated as 2-1-P1.19. Five percent (37 of 707) of the isolates in our strain collection bound this MAb. The coexpression of the epitopes for 2-1-P1.19 and MN3C5C on the majority of the clinical isolates (25 of 37; 68%) in this study and another study (29) probably explains why the different specificities of the two reference MAbs have not been noted previously.
Nucleotide sequence accession numbers.
The EMBL Nucleotide Sequence Database accession numbers for the sequences reported in Table 2 are AJ012189 and AJ012722 to AJ012736.
Acknowledgments
We are grateful to E. Holten and L. O. Frøholm for supplying the isolates from Norway and to J. T. Poolman, C. T. Sacchi, and C. E. Frasch for the generous gift of the MAbs. Torill Alvestad, Karin Bolstad, Tone Haslerud, and Berit Nyland provided excellent technical assistance.
This work was supported by grant C11/181/23 from the World Health Organization to D. A. Caugant.
REFERENCES
- 1.Abdillahi H, Poolman J T. Neisseria meningitidis group B serosubtyping using monoclonal antibodies in whole-cell ELISA. Microb Pathog. 1988;4:27–32. doi: 10.1016/0882-4010(88)90045-9. [DOI] [PubMed] [Google Scholar]
- 2.Ahrin F F, Moreau F, Coulton J W, Mills E L. Subtyping of Neisseria meningitidis strains isolated in Quebec, Canada: correlation between deduced amino acid sequences and serosubtyping techniques. Can J Microbiol. 1997;43:234–238. doi: 10.1139/m97-032. [DOI] [PubMed] [Google Scholar]
- 3.Ashton F E, Mancino L, Ryan A J, Poolman J T, Abdillahi H, Zollinger W D. Serotypes and subtypes of Neisseria meningitidis serogroup B strains associated with meningococcal disease in Canada, 1977–1989. Can J Microbiol. 1991;37:613–617. doi: 10.1139/m91-104. [DOI] [PubMed] [Google Scholar]
- 4.Barlow A K, Heckels J E, Clarke I N. The class 1 outer membrane protein of Neisseria meningitidis: gene sequence and structural and immunological similarities to gonococcal porins. Mol Microbiol. 1989;3:131–139. doi: 10.1111/j.1365-2958.1989.tb01802.x. [DOI] [PubMed] [Google Scholar]
- 5.Brooks J L, Fallon R J, Heckels J E. Sequence variation in class 1 outer membrane protein in Neisseria meningitidis isolated from patients with meningococcal infection and close household contacts. FEMS Microbiol Lett. 1995;128:145–150. doi: 10.1111/j.1574-6968.1995.tb07514.x. [DOI] [PubMed] [Google Scholar]
- 6.Butcher S J, Omar P J, Sarvas M, Runeberg-Nyman K. Sequence comparisons of class 1 genes from Neisseria meningitidis and a folding model of the class 1 protein. In: Achtman M, Kohl P, Marchal C, Morelli G, Seiler A, Thiesen B, editors. Neisseriae 1990. Berlin, Germany: Walter de Gruyter; 1990. pp. 193–198. [Google Scholar]
- 7.Caugant D A, Bol P, Høiby E A, Zanen H C, Frøholm L O. Clones of serogroup B Neisseria meningitidis causing systemic disease in The Netherlands, 1958–1986. J Infect Dis. 1990;162:867–874. doi: 10.1093/infdis/162.4.867. [DOI] [PubMed] [Google Scholar]
- 8.Caugant D A, Høiby E A, Frøholm L O, Brandtzaeg P. Polymerase chain reaction for case ascertainment of meningococcal meningitis: application to the cerebrospinal fluids collected in the course of the Norwegian meningococcal serogroup B protection trial. Scand J Infect Dis. 1996;28:149–153. doi: 10.3109/00365549609049066. [DOI] [PubMed] [Google Scholar]
- 9.Caugant D A, Mocca L F, Frasch C E, Frøholm L O, Zollinger W D, Selander R K. Genetic structure of Neisseria meningitidis populations in relation to serogroup, serotype, and outer membrane protein pattern. J Bacteriol. 1987;169:2781–2792. doi: 10.1128/jb.169.6.2781-2792.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Caugant D A, Sandven P, Eng J, Jeque J T, Tønjum T. Detection of rifampin resistance among isolates of Mycobacterium tuberculosis from Mozambique. Microb Drug Resist. 1995;1:321–326. doi: 10.1089/mdr.1995.1.321. [DOI] [PubMed] [Google Scholar]
- 11.Feavers I M, Fox A J, Gray S, Jones D M, Maiden M C J. Antigenic diversity of meningococcal outer membrane protein PorA has implications for epidemiological analysis and vaccine design. Clin Diagn Lab Immunol. 1996;3:444–450. doi: 10.1128/cdli.3.4.444-450.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fernandez-Patron C, Calero M, Collazo R P, Garcia J R, Madrazo J, Musacchio A, Soriano F, Estrada R, Frank R, Castellanos-Serra L R, Mendez E. Protein reverse staining: high-efficiency microanalysis of unmodified proteins detected on electrophoresis gels. Anal Biochem. 1995;224:203–211. doi: 10.1006/abio.1995.1031. [DOI] [PubMed] [Google Scholar]
- 13.Frasch C E, Zollinger W D, Poolman J T. Serotype antigens of Neisseria meningitidis and a proposed scheme for designation of serotypes. Rev Infect Dis. 1985;7:504–510. doi: 10.1093/clinids/7.4.504. [DOI] [PubMed] [Google Scholar]
- 14.Guillén G, Alvarez A, Lemos G, Paredes T, Silva R, Martín A. Comparison of DNA sequence of nine different genes for the class 1 outer membrane protein from Neisseria meningitidis. Biotecnol Apl. 1993;2:108–113. [Google Scholar]
- 15.Loomis-Price L D, Levi M, Burnett P R, van Hamont J E, Shafer R A, Wahren B, Birx D L. Linear epitope mapping of humoral responses induced by vaccination with recombinant HIV-1 envelope protein GP-160. J Ind Microbiol Biotechnol. 1997;19:58–65. doi: 10.1038/sj.jim.2900410. [DOI] [PubMed] [Google Scholar]
- 16.Mahboud A, Richard C, Delacourte A, Han K K. Application of chemical cleavage procedures to the peptide mapping of neurofilament triplet protein bands in sodium dodecyl sulfate-polyacrylamide gel electrophoresis. Anal Biochem. 1986;154:171–182. doi: 10.1016/0003-2697(86)90511-7. [DOI] [PubMed] [Google Scholar]
- 17.Maiden M C J, Bygraves J A, McCarvil J, Feavers I M. Identification of meningococcal serosubtypes by polymerase chain reaction. J Clin Microbiol. 1992;30:2835–2841. doi: 10.1128/jcm.30.11.2835-2841.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Maiden M C J, Suker J, McKenna A J, Bygraves J A, Feavers I M. Comparison of the class 1 outer membrane proteins of eight serological reference strains of Neisseria meningitidis. Mol Microbiol. 1991;5:727–736. doi: 10.1111/j.1365-2958.1991.tb00743.x. [DOI] [PubMed] [Google Scholar]
- 19.McGuinness B, Barlow A K, Clarke I N, Farley J E, Anilionis A, Poolman J T, Heckels J E. Deduced amino acid sequences of class 1 protein (PorA) from three strains of Neisseria meningitidis. J Exp Med. 1990;171:1871–1882. doi: 10.1084/jem.171.6.1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McGuinness B T, Lambden P R, Heckels J E. Class 1 outer membrane protein of Neisseria meningitidis: epitope analysis of the antigenic diversity between strains, implications for subtype definition and molecular epidemiology. Mol Microbiol. 1993;7:505–514. doi: 10.1111/j.1365-2958.1993.tb01141.x. [DOI] [PubMed] [Google Scholar]
- 21.Poolman, J. T. 1998. Personal communication.
- 22.Saunders N B, Brandt B L, Warren R L, Hansen B D, Zollinger W D. Immunological and molecular characterization of three variant subtype P1.14 strains of Neisseria meningitidis. Infect Immun. 1998;66:3218–3222. doi: 10.1128/iai.66.7.3218-3222.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Saunders N B, Shoemaker D R, Brandt B L, Zollinger W D. Confirmation of suspicious cases of meningococcal meningitis by PCR and enzyme-linked immunosorbent assay. J Clin Microbiol. 1997;35:3215–3219. doi: 10.1128/jcm.35.12.3215-3219.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Saunders N B, Zollinger W D, Rao V B. A rapid and sensitive PCR strategy for amplification and sequencing of porA from a single colony-forming unit of Neisseria meningitidis. Gene. 1993;137:153–162. doi: 10.1016/0378-1119(93)90001-j. [DOI] [PubMed] [Google Scholar]
- 25.Suker J, Feavers I M, Achtman M, Morelli G, Wang J F, Maiden M C J. The porA gene in serogroup A meningocooci: evolutionary stability and mechanism of genetic variation. Mol Microbiol. 1994;12:253–265. doi: 10.1111/j.1365-2958.1994.tb01014.x. [DOI] [PubMed] [Google Scholar]
- 26.van der Ley P, Heckels J E, Virji M, Hoogerhout P, Poolman J T. Topology of outer membrane porins in pathogenic Neisseria spp. Infect Immun. 1991;59:2963–2971. doi: 10.1128/iai.59.9.2963-2971.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wedege E, Caugant D A, Frøholm L O, Høiby E A, Rosenqvist E. Characterization of meningococcal isolates from Norwegian patients in the years 1987 to 1991. In: Conde-Glez C J, Morse S, Rice P, Sparling F, Calderón E, editors. Pathobiology and immunology of Neisseriaceae. Cuernavaca, Morelos, Mexico: Instituto Nacional de Salud Publica; 1994. pp. 318–324. [Google Scholar]
- 28.Wedege, E., D. A. Caugant, and W. D. Zollinger. P1.19 specificity of a previous P1.15 reference monoclonal antibody demonstrated by blotting methods, porA sequencing and peptide mapping, p. 538–539. In W. D. Zollinger, C. E. Frasch, and C. D. Deal (ed.), Abstracts of the Tenth International Pathogenic Conference, Baltimore, Md.
- 29.Wedege E, Frøholm L O. Comparison of monoclonal antibodies for serotyping and subtyping of Neisseria meningitidis. In: Achtman M, Kohl P, Marchal C, Morelli G, Seiler A, Thiesen B, editors. Neisseriae 1990. Berlin, Germany: Walter de Gruyter; 1991. pp. 147–151. [Google Scholar]
- 30.Wedege E, Høiby E A, Rosenqvist E, Bjune G. Immune responses against major outer membrane antigens of Neisseria meningitidis in vaccinees and controls who contracted meningococcal disease during the Norwegian serogroup B protection trial. Infect Immun. 1998;66:3223–3231. doi: 10.1128/iai.66.7.3223-3231.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wedege E, Høiby E A, Rosenqvist E, Frøholm L O. Serotyping and subtyping of Neisseria meningitidis isolates by co-agglutination, dot-blotting and ELISA. J Med Microbiol. 1990;31:195–201. doi: 10.1099/00222615-31-3-195. [DOI] [PubMed] [Google Scholar]
- 32.Zollinger W D, Mandrell R E, Altieri P, Berman S, Lowenthal J, Artenstein M S. Safety and immunogenicity of a Neisseria meningitidis type 2 protein vaccine in animals and humans. J Infect Dis. 1978;137:728–739. doi: 10.1093/infdis/137.6.728. [DOI] [PubMed] [Google Scholar]
- 33.Zollinger W D, Moran E E, Connelly H, Mandrell R E, Brandt B. Monoclonal antibodies to serotype 2 and serotype 15 outer membrane proteins of Neisseria meningitidis and their use in serotyping. Infect Immun. 1984;46:260–266. doi: 10.1128/iai.46.1.260-266.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]