Abstract
The small multidrug resistance (SMR) family is composed of widespread microbial membrane proteins that fulfill different transport functions. Four functional SMR subtypes have been identified, which variously transport the small, charged metabolite guanidinium, bulky hydrophobic drugs and antiseptics, polyamines, and glycolipids across the membrane bilayer. The transporters possess a minimalist architecture, with ∼100-residue subunits that require assembly into homodimers or heterodimers for transport. In part because of their simple construction, the SMRs are a tractable system for biochemical and biophysical analysis. Studies of SMR transporters over the last 25 years have yielded deep insights for diverse fields, including membrane protein topology and evolution, mechanisms of membrane transport, and bacterial multidrug resistance. Here, we review recent advances in understanding the structures and functions of SMR transporters. New molecular structures of SMRs representing two of the four functional subtypes reveal the conserved structural features that have permitted the emergence of disparate substrate transport functions in the SMR family and illuminate structural similarities with a distantly related membrane transporter family, SLC35/DMT.
Keywords: multidrug transporter, membrane transport, structure-function, substrate specificity, evolution, quaternary ammonium cations, SLC35, guanidinium, polyamine, flippase
Abbreviations: EM, electron microscopy; GEBA, Genomic Encyclopedia of Bacteria and Archaea; TM, transmembrane
From atomistic descriptions of membrane transport mechanism to global spread of multidrug resistance over the last century, small multidrug resistance (SMR) proteins have provided broad insights along multiple research fronts since the family’s discovery in the mid 1990s (1, 2). With just four transmembrane helices and ∼100 residues, SMR proteins are among nature’s smallest membrane transport proteins, making them ideal systems for biochemical and biophysical investigation. These same properties impeded high resolution structural characterization for many years, however, since the proteins are almost entirely embedded in the membrane, too small for cryo-EM, and with little polar surface area to form crystal contacts. Recently, new high resolution crystal structures have been determined for two functionally distinct SMR subtypes (3, 4). These complement the body of mechanistic data that have been assembled over the years and provide an opportunity to consider the molecular underpinnings of functional diversity among SMR transporters.
In general, the SMRs transport positively charged solutes across the membrane coupled to the antiport of protons (Fig. 1A). The resting membrane potential and pH gradient of most bacteria implies that they typically function in the active efflux of substrates. Four major functional subtypes have been described within the SMR family, and according to our bioinformatic analysis described later in this review, at least 97% of bacterial SMR genes correspond to one of these four subtypes. The first, and likely primal, SMR subtype transports guanidinium ion, a small cationic byproduct of nitrogen metabolism, and is referred to as Gdx (guanidinium export) (5). These are also known by the name of the gene encoding them, sugE. The second subtype, which we refer to as Qac (quaternary ammonium cation), are promiscuous exporters of hydrophobic cationic compounds, including quaternary ammonium antiseptics like benzalkonium and cetyltrimethylammonium (also known as cetrimonium), and polyaromatic cationic biocides like methyl viologen (also known as paraquat), acriflavine, and ethidium (1, 6, 7). The promiscuous transport phenotype of the Qac subtype gave the SMR family its name (2), and this subtype includes the well-studied multidrug exporter from Escherichia coli, EmrE. Associated gene names for the Qac transporters include emrE, ebrA/ebrB, qacE, qacG, qacH, and others. The third subtype (gene name mdtI/mdtJ) has been implicated in the transport of small polyamine metabolites like spermidine and putrescine (8), and the fourth subtype (gene name arnE/arnF) acts as a glycolipid flippase (9). In this review, we will first describe unique topological considerations shared by all four SMR subtypes, then analyze the occurrence and distribution of the different SMR subtypes among bacterial genomes. We will review recent advances in our understanding of each SMR subtype, with particular emphasis on recent high resolution structures (Table 1), and finally, analyze structural homology between the SMRs and a distantly related family of transporters, SLC35 (also known as Drug/Metabolite Transport (DMT)).
Figure 1.
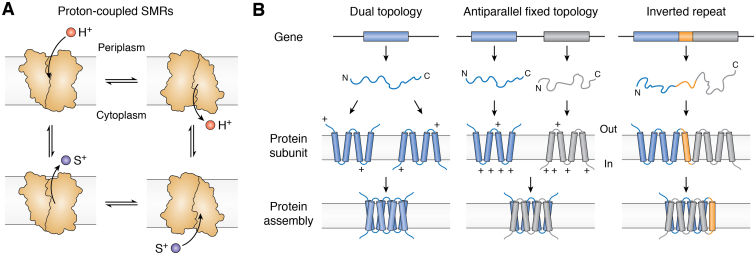
Transport scheme and transporter topologies.A, simplified scheme of the transport cycle for proton-coupled SMRs. B, cartoons showing some potential transporter topologies. SMR proteins are found as dual topology homodimers and fixed topology heterodimers (left and center). Dual topology proteins are characterized by a balanced distribution of positively charged residues (arginines and lysines, indicated by + symbols) on the extramembrane loops and termini, whereas subunits that assemble as antiparallel fixed dimers have oppositely biased positive charge distributions. For a protein with an even number of helices, evolution of inverted repeat topology (right) requires the insertion of a transmembrane helix (orange) to enforce antiparallel architecture of fused 4-TM subunits.
Table 1.
Structural data and models available for SMR transporters Gdx-Clo and EmrE
| Protein | Substrate | Method (max. resolution) | PDB | Reference |
|---|---|---|---|---|
| EmrE | Tetraphenylphosphonium (TPP+) | Electron microscopy with 2D crystals (7.5 Å) | Data: EMD-1087 Model: 2I68 |
Data (12) Model (58) |
| Gdx-Clo | Gdm+ | Crystallography (3.50 Å) | 6WK5 | (4) |
| Gdx-Clo | PhenylGdm+ | Crystallography (2.53 Å) | 6WK8 | (4) |
| Gdx-Clo | OctylGdm+ | Crystallography (2.32 Å) | 6WK9 | (4) |
| Gdx-Clo | None (pH 5.0) | Crystallography (2.32 Å) | 7SZT | (3) |
| aEmrE3 | None (pH 5.2) | Crystallography (2.85 Å) | 7MH6 | (3) |
| aEmrE3 | Methyl viologen | Crystallography (3.13 Å) | 7MGX | (3) |
| aEmrE3 | TPP+ | Crystallography (3.36 Å) | 7SV9 | (3) |
| aEmrE3 | Methyltriphenylphosphonium | Crystallography (3.22 Å) | 7SSU | (3) |
| aEmrE3 | Benzyltrimethylammonium | Crystallography (3.91 Å) | 7T00 | (3) |
| aEmrE3 | Harmane | Crystallography (3.91 Å) | 7SVX | (3) |
| bEmrE S64V | Tetra(4-fluorophenyl) phosphonium/pH 5.8 | NMR | 7JK8 | (80) |
| bEmrE S64V | Tetra(4-fluorophenyl) phosphonium/pH 8.0 | NMR | 7SFQ | (81) |
SMR family topology
In general, bacterial membrane proteins are inserted into the membrane according to the ‘positive inside rule’, in which the cytoplasmic face of the protein has an excess of positively charged residues arginine and lysine relative to the periplasmic face (10). The SMR proteins were among the first membrane proteins to be identified as possessing unusual “dual topology” architecture (11, 12). Dual topology proteins lack the typical biased charge distribution and are thus inserted into the membrane in both inward- and outward-facing orientations (Fig. 1B, left), where they can oligomerize with antiparallel subunits (13, 14). Experimental evidence suggests that individual EmrE subunits achieve their topology cotranslationally or via limited posttranslational annealing (13, 14, 15). The subunits can interact with each other during the immediate posttranslational protein folding stage (16, 17) but do not undergo major reorientations within the membrane after insertion to form the antiparallel homodimers (13).
In addition to dual topology homodimers, there are also numerous examples of SMR gene duplications that have given rise to co-expressed genes within a single operon that assemble as obligate heterodimers (8, 18, 19, 20) (Fig. 1B, middle). Sometimes called “paired SMRs” or PSMRs (7), SMRs with this arrangement are found among all four functional subtypes and likely evolved via multiple independent duplication events (5). In the great majority of these cases, the paired protomers exhibit opposite charge biases, which determine the orientation of each subunit in the membrane and enforces the antiparallel assembly (11).
Although dual topology and fixed antiparallel topology are only rarely observed among membrane proteins (11, 21, 22, 23), the assembly may be an evolutionary antecedent to an architecture that is extremely common among membrane transport proteins, the inverted repeat (Fig. 1B, right), in which a single protein possesses structurally homologous domains arranged antiparallel with respect to each other (24, 25, 26). Unlike other membrane protein families that include dual topology members (27, 28), no simple inverted repeat representatives have been detected among the SMRs, suggesting that internally fused SMR proteins might not be evolutionarily advantageous (29). Alternatively, the fusion of 4-transmembrane (TM) dual topology proteins might simply be an evolutionarily rare event, since this process requires addition of a transmembrane linker helix to connect the N and C termini of the two monomers (27) (Fig. 1B, right).
Distribution of SMR genes among bacterial genomes
To gauge the distribution of SMR genes across diverse microbes, we evaluated bacterial genomes from the Joint Genome Institute’s curated set of ∼1000 Genomic Encyclopedia of Bacteria and Archaea (GEBA) genomes (30). This set of genomes was selected to reduce sampling bias and maximize phylogenetic diversity in microbial sequences used for evolutionary studies. Available sequencing data tend to be biased toward pathogenic isolates (30), and because many SMRs play a role in multidrug resistance, they are particularly prone to horizontal gene transfer via plasmids and other transposable sequences (4, 31, 32, 33, 34). It is therefore particularly important to use a phylogenetically representative dataset to gain a balanced view of SMR distribution among bacteria. Although archaea do possess SMR transporters (35, 36), we excluded archaea from this analysis.
SMR genes were identified from GEBA genomes with HMMER3.3.2 (37) using a profile Hidden Markov Model (profile HMM) constructed for the SMR family (pfam 00893). Profile HMMs for each subtype (Gdx, Qac, polyamine transport, and lipid transport) were constructed from functionally annotated clusters in a sequence similarity network of reference SMR proteins (4), and SMR sequences were assigned to the subtype that corresponded to the lowest e-value calculated by HMMR. SMR sequences were annotated “other” if the e-value was >10−20. Consensus sequences for each SMR subtype are shown in Figure 2, A and B, and sequence information and annotations for individual SMR proteins from this set of genomes is available for download from the Deep Blue Data repository hosted by the University of Michigan with unique identifier doi.org/10.7302/0ynd-b343.
Figure 2.
Sequence conservation of the four major SMR transporter subtypes.A, alignment of representative proteins of the Gdx, Qac, and polyamine transport subtypes. Sequences from E. coli EmrE for Qac, Clostridales Gdx-Clo for Gdx, and the inward- and outward-facing polyamine transporter subunits from proteobacterium Photorhabdus australis. Sequences are numbered to correspond to the EmrE sequence and colored according to sequence conservation within that subtype using ConSurf (142). Highly conserved and mechanistically important residues including the central Glu, the Tyr switch, and the binding site Trp are indicated with asterisks. The TM3 GXG fulcrum motif is indicated by the horizontal line. B, representative sequence of lipid transporter from proteobacterium Microvirgula aerodenitrificans with sequence conservation analyzed and colored as in panel (A). Residues that align with the central Glu, the Tyr switch, the binding site Trp, and the GXG fulcrum are indicated.
Approximately 2/3 of the bacterial genomes from the GEBA set have at least one gene encoding an SMR protein, and ∼1/3 of the GEBA genomes encode two or more SMR genes (Fig. 3A). This count of genomes with multiple SMR genes reflects both paired SMR genes that encode heterodimers, as well as genomes with more than one SMR functional subtype. The majority of SMR genes have no other SMR gene within 100 base pairs, suggesting they are expressed independently. As expected for dual topology proteins (11), the Arg/Lys bias distribution for these genetic singletons is centered at 0, and only 3% encode protomers with an Arg/Lys bias greater than ±2 (Fig. 3B). SMR genes are also found as adjacent gene pairs. In our dataset, >95% of adjacent gene pairs encode subunits with opposite Arg/Lys biases. The Arg/Lys distributions are centered around +4 and −4 for inward- and outward-facing protomers, respectively, which assemble to form heterodimeric transporters (Fig. 3B). Approximately 10% of Qac transporters and ∼20% of Gdx transporters are encoded by such paired genes, as are all polyamine transporters (Fig. 3C). In the GEBA genome set, all the SMR lipid transporters are encoded by singleton genes, although functional pairs have been identified in some bacteria (9).
Figure 3.
Identification and annotation of SMR-coding genes from GEBA genomes.A, proportion of GEBA genomes that possess one or more SMR genes. B, left, positive charge (Arg/Lys) bias (cytoplasmic face) for SMR subunits encoded by genetic singletons (defined as no other SMR genes within 100 base pairs). Right, positive charge (Arg/Lys) bias (cytoplasmic face) for SMR subunits encoded by adjacent gene pairs (within 100 base pairs). The positive charge bias is given by (Arg+Lys+N-terminal amino group)termini, loop 2 – (Arg+Lys)loop1, loop3. C, transporter subtypes identified from GEBA genomes. Functional annotation is based on sequence comparison to functionally annotated gene clusters using HMMER (37) as described in the text. For this annotation, proteins encoded by singleton genes with unbiased positive charge distribution are annotated as homodimers, and proteins encoded by adjacent SMR genes with opposite charge biases are annotated as heterodimers. D, proportion of all GEBA genomes that encode Gdx, Qac, or both subtypes. E, frequency of genes encoding Gdx (light blue), Qac (magenta), or both subtypes (dark blue) for major bacterial phyla. Phylogenetic relationships and distances between clades from (143). F, proportion of all genomes and Proteobacterial genomes in the GEBA genome set that encode polyamine and lipid transport SMRs (yellow and orange slices, respectively). GEBA, Genomic Encyclopedia of Bacteria and Archaea.
Most bacterial phyla possess genes encoding SMRs. The most prevalent SMRs are Gdx, which are found in about ∼50% of all bacterial genomes, including 80% of Actinobacteria, half of Proteobacteria and Bacteroides, and ∼30% of Firmicutes (Fig. 3, D and E). Qac genes are also frequent, found in ∼25% of bacterial genomes overall, including ∼50% of Proteobacteria, ∼30% of Actinobacteria, and ∼25% of Firmicutes. Many species possess both Gdx and Qac transporters. The lipid and polyamine transporters are less common and found mainly in Proteobacteria, where they are found in <10% of species (Fig. 3F). Thus, the SMR transporters are widespread among bacteria and dual topology Gdx and Qac transporters are the predominant SMR variants.
Guanidinium exporters (Gdx)
Although they are the most common SMRs encoded in bacterial genomes, the Gdx transporters were also the last to be functionally annotated. The proteins were originally reported to play a role in activity of the chaperone GroEL and called SUG (Suppressor of GroEL mutations) (38). However, this phenotype was later shown to be artefactual (7, 35, 39). With high sequence similarity to the multidrug export Qac subtype and frequent association with horizontally transferred multidrug resistance gene arrays (4, 40, 41), early characterization efforts focused on resistance to antiseptics and biocides (39, 42, 43, 44). The subtype appeared to contribute to low levels of resistance to a narrow subset of drugs, but the activity was not robust, and the proteins from this subtype remained poorly characterized until their physiological role in export of guanidinium (Gdm+) was established (5, 45). These proteins were renamed Gdx (Guandinium export) rather than SUG to reflect their proper functional annotation (5).
Gdm+ has been recognized as a byproduct of bacterial metabolism since the late 1800s, when high concentrations of Gdm+ were found in spoiled meats (46). But the molecular players have only begun to emerge since 2017, beginning with the discovery of riboswitch-controlled operons dedicated to Gdm+ metabolism and transport (45). Four unrelated classes of Gdm+ riboswitches have been identified (45, 47, 48, 49, 50), along with three distinct enzymatic pathways for utilizing Gdm+ as a nitrogen source (51, 52, 53, 54), including as a sole nitrogen source by some bacteria (53, 55). The bacteria that do not consume Gdm+—about half of those with Gdm+ riboswitches—instead produce and export endogenous Gdm+, likely as a metabolic waste product (5, 45). The riboswitches bind Gdm+ with KD values between ∼60 to 200 μM (45, 47, 48, 49) to upregulate expression of the associated transporters and enzymes. These proteins have somewhat higher Km values for Gdm+, between ∼200 μM −1 mM (4, 45, 51, 53), suggesting that Gdm+ accumulation becomes toxic to cells and must be mitigated within this range.
Like other SMR transporters, the Gdx harness the bacteria’s proton motive force to drive transport, exporting Gdm+ with strict 2:1 H+:Gdm+ antiport stoichiometry (5, 56). To prevent export of useful guanidinylated metabolites, the Gdx are highly selective for Gdm+ over other physiological compounds with guanidinyl moieties, such as arginine, agmatine, and creatine (5). However, electrophysiological transport experiments show that the Gdx are not exquisitely selective for Gdm+ either—although Gdx proteins strictly exclude guanidinyl metabolites with polar substituents, like arginine, they transport guanidinyl compounds with single hydrophobic substitutions at WT-like levels (4).
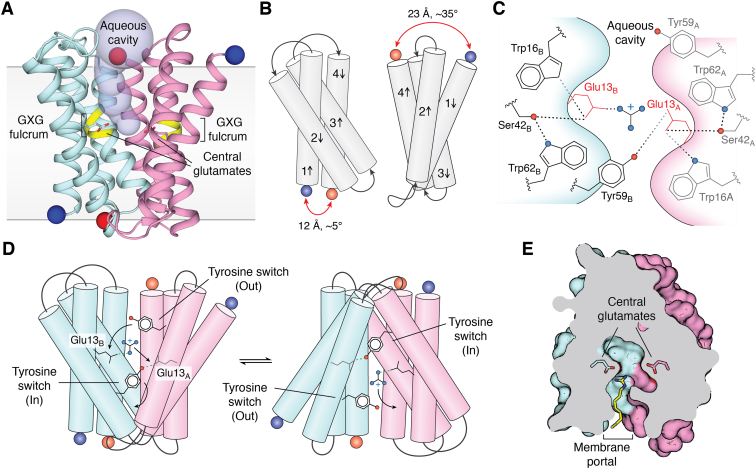
The Gdx subtype yielded the first high resolution crystal structures from the SMR family, of a protein from Clostridales oral taxon 876, referred to as Gdx-Clo (4). Many of the structural features observed for this homolog had been proposed for Qac protein EmrE based on prior biophysical and biochemical experiments, establishing common structural attributes of the SMR family. Gdx-Clo possesses the expected antiparallel topology, and the two subunits assemble as an asymmetric homodimer with an aqueous cavity opened to one side of the membrane (Fig. 4A). Dimerization is mediated primarily by TM helix 4. The extramembrane loops also form extensive hydrogen bonded cross-subunit interactions to seal the closed side of the transporter. Each subunit of the dimer is composed of two discrete lobes delineated by a conserved Gly-Ile-Gly motif that acts as a fulcrum in TM helix 3 (TM3). The subunits differ according to a ∼35° rotation between N- and C-terminal lobes that stems from a difference in the angle of the Gly-Ile-Gly kink (Fig. 4B). The outward facing to inward facing conformational transition involves a structural swap between the two subunits, each changing the degree of rotation between the N- and C-terminal lobes. As a result, the inward- and outward-open conformations are 2-fold symmetric to each other, and the structural swap opens an identical, symmetry-related aqueous cavity on the opposite side of the membrane (Fig. 4A). This elegant mechanism for the alternating access of the substrate-binding site—a prototype for the “rocker-switch” mechanism used by many other transporters—was first predicted and demonstrated for EmrE (57, 58), and the same TM3 fulcrum motif (Gly-hydrophobic-Gly or GXG) is conserved in the Gdx, Qac, and polyamine subtypes (Fig. 2).
Figure 4.
Structure and mechanism of Gdx.A, crystal structure of Gdx-Clo (PDB:6WK9). Subunits are colored in cyan and pink, with N and C termini rendered as blue and red spheres, respectively. Central glutamates (E13A and E13B in Gdx-Clo) are shown as sticks, the conserved TM3 GXG fulcrum is colored yellow, and the aqueous vestibule is shown as a light blue surface. The approximate membrane boundaries are shown. B, structural comparison of individual subunits. To highlight the structural difference, the different angles between helix 1 and 4 and the different distances between the N and C termini (blue and red spheres) are indicated. C, diagram showing the hydrogen bond network in the substrate-binding site. D, proposed tyrosine switch mechanism. Rotameric movements of the tyrosine switch are indicated by arrows and hydrogen bonds indicated by dashed green lines. In this view, one of the central glutamates is occluded from view in each panel (E13A in the left panel and E13B in the right panel). The occluded glutamate is shown in a lighter gray color. E, top-down view of Gdx-Clo in complex with octylGdm+ (yellow stick rendering) sliced at the midpoint of the membrane (PDB:6WK9). The central glutamates are shown as sticks. The alkyl tail of the substrate extends out of the binding site through the membrane portal. PDB, Protein Data Bank.
The substrate-binding site is located at the bottom of the aqueous cavity, with the positively charged Gdm+ situated between a pair of negatively charged glutamates, E13A and E13B, one contributed by each subunit. These “central glutamates” are conserved and essential in the Gdxs, the Qacs, and the polyamine transporters (5, 8, 59). In addition to binding positively charged substrate, the central glutamates are protonatable at physiological pH and carry protons across the membrane during the opposing leg of the antiport cycle (59, 60). This common binding site for the small molecule substrate and the two antiported protons favors alternating binding site occupancy by the substrates and sets the 2:1 H+:Gdm+ stoichiometry measured for Gdx (5).
In Gdx-Clo, the central glutamates are fixed in position by a polarized stack of alternating hydrogen bond donors and acceptors, including W16, S42, and W62 (4) (Fig. 4C). Mutation of any one of these residues to remove H-bonding capacity substantially impairs transport function (4). In the structures, the substrate Gdm+ is directly coordinated by E13B, whereas E13A is deflected away from the Gdm+ by a cross-subunit interaction with Y59B (4). Y59A, in contrast, points away from the substrate-binding pocket and into the aqueous vestibule. The divergent poses of Y59 presented a mechanistic proposal for conformational change by the SMR transporters (4, 61) whereby the rotameric switch of Y59A from the aqueous vestibule toward E13B displaces the substrate Gdm+ from its interaction with E13B. The Gdm+, in turn, engages with E13A, displacing Y59B, which undergoes the converse rotameric switch, away from the central glutamates. This “tyrosine switch” has been proposed to trigger the global conformational swap that opens an aqueous cavity to the other side of the membrane, where Y59B ultimately rests (Fig. 4D) (4). This tyrosine is almost perfectly conserved in all SMR subtypes and is mechanistically essential in all three SMR subtypes in which the effect of its mutation has been tested (3, 4, 8, 62), suggesting that the tyrosine switch is fundamental to transport by the SMRs.
In addition to the central glutamates and the tyrosine switch, the structures of Gdx-Clo revealed a third structural feature that is likely to be conserved among other SMR subtypes, the membrane portal (4). This portal is defined by TM2A and TM2B, which form one side of the binding pocket and splay apart on the open side of the transporter. The gap between these helices is lined by hydrophobic sidechains and could, in principle, permit substrate access between the aqueous substrate-binding site and the membrane interior (Fig. 4E). In EmrE, spectroscopic experiments lead to the suggestion that the hydrophobic residues lining this portal act as a gate that permits access for the lipophilic substrates to the binding site (61, 63). Similar lateral openings are well-described features of both lipid and drug transport proteins (64, 65, 66), permitting hydrophobic or amphipathic substrates to diffuse between, or have simultaneous access to, the membrane and the substrate-binding pocket.
The significance of the portal for the Gdx subtype is less readily apparent, however, since the physiological substrate Gdm+ is small and hydrophilic and would be expected to access the binding pocket directly from aqueous solution. Nonetheless, structures of Gdx-Clo with phenyl-bound and octylGdm+-bound showed that these non-natural substrates utilize the membrane portal to accommodate their hydrophobic substituents, while their guanidinyl headgroups bind between the central glutamates in the binding pocket, similar to Gdm+ (4). These structures rationalize prior observations that Gdx transport hydrophobic, singly substituted guanidiniums (4, 5). Moreover, the positioning of these non-natural substrates also suggested a mechanism to select against natural guanidinyl metabolites (4). Should a compound such as arginine or agmatine enter the binding site in the same orientation, its polar tail would likewise be positioned to extend from the binding pocket through the membrane portal. But the hydrophobic membrane interior would not favorably interact with the polar substituents, and thus, the membrane itself could contribute to selectivity against natural guanidinylated metabolites (4). The membrane portal might also explain the association of Gdx-encoding genes with multidrug resistance gene arrays in environmental reservoirs (4, 40, 41): hydrophobic guanidinyl compounds that enter the biosphere via municipal wastewater or farm runoff present microbes with a persistent low-grade toxic threat (67, 68, 69) that could be mitigated by a Gdx exporter. Examples of such common biocides include the agricultural antifungal dodine (decylGdm+) and pharmaceuticals like metformin, which is excreted into wastewater, where it is slow to degrade and accumulates to levels of environmental concern (70, 71). Likewise, this portal may explain prior observations that cationic detergents bind to Gdx homologs (39).
Drug and antiseptic exporters (Qac)
Transporters of the Qac subtype garnered early attention for their role in bacterial multidrug resistance. Frequently found in clinical and agricultural isolates (7, 72), this SMR subtype confers resistance to the quaternary ammonium compounds used as common hospital and household antiseptics. These antimicrobial agents were introduced in the 1930s, and evolutionary analysis suggests that it was around this time that the immediate ancestor of the clinically important vector for multidrug resistance, the class I integron, emerged (73). This ancestral class I integron likely consisted of an integron/integrase sequence to capture drug resistance genes, a transposable element to facilitate its spread among microbial populations, and a single resistance gene: an SMR transporter of the Qac subtype (73). Sequence analysis suggests that Qac SMRs have been dynamically associated with these and other drug resistance gene arrays over the last hundred years, gained and lost multiple times as these elements have spread among both pathogenic and environmental bacteria (74, 75). Today, Qac transporters remain adaptive to subinhibitory concentrations of quaternary ammonium antiseptics found in wastewater and surface runoff and remain among the most common genes isolated from human-adjacent environments (31, 32, 33, 34). By conferring this selective advantage against ubiquitous environmental biocides, the Qac SMRs coselect for other resistance genes in the cassettes that provide resistance against more potent clinical antibiotics (76), contributing to the continued spread of multidrug resistance.
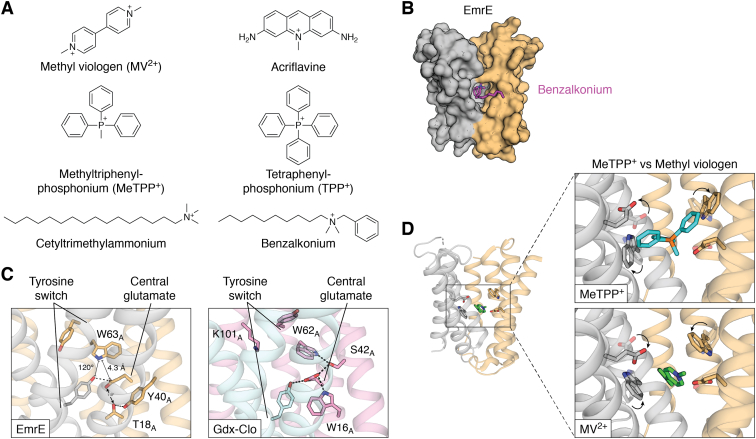
Meanwhile, the E. coli variant, EmrE, has become one of the best studied bacterial multidrug exporters over the last 25 years. EmrE was originally shown to transport a variety of polyaromatic, cationic antimicrobial compounds (1) (Fig. 5A). Early, low resolution electron microscopy (EM) of 2D crystals demonstrated the unusual antiparallel architecture and established an elementary understanding of the helical connectivity and protein fold (12, 58). Although high resolution structural information lagged, biochemical and biophysical studies provided a detailed molecular picture of the protein and its transport cycle. As a reference, we provide a summary of the scanning mutagenesis studies that have been performed for EmrE in Table 2. Although too extensive to discuss individually here (see reference (77) for an in-depth review of EmrE mutagenesis), these studies provided a functional grounding for the interpretation of electron paramagnetic resonance (EPR) distance measurements (63), models based on the low-resolution EM data with computationally predicted sidechains (61, 78, 79), models derived from NMR chemical shifts and substrate/protein distance restraints (80, 81), and ultimately, the high resolution crystal structures, as discussed later (3).
Figure 5.
Substrate binding to EmrE.A, examples of planar aromatic, quaternary ammonium/phosphonium, and alkylated substrates transported by EmrE. B, model of EmrE with benzalkonium bound (3). EmrE subunits in gray and tan surface rendering with benzalkonium shown as purple stick representation. The substrate’s alkyl tail extends from the binding site into the membrane via the conserved membrane portal. C, comparison of EmrE (PDB:7MH6) and Gdx-Clo (PDB:7SZT) substrate-binding sites with putative hydrogen bonds (distance <3.5 Å, angle 140–180°) represented as dashed lines. Distances/angles are shown for potential H-bond partners that possess nonoptimal geometry. D, EmrE substrate-binding site with different substrates bound. Top panel: bulky quaternary phosphonium substrate, methyltriphenylphosphonium (PDB:7SSU). Bottom panel: planar aromatic substrate, methyl viologen (PDB:7MGX). To aid comparison, sidechain positions in the presence of the alternative substrate is rendered as lightly colored, transparent sticks. Arrows indicate movements of the central glutamates and binding site tryptophan (W63) that accommodate the differently sized substrates. PDB, Protein Data Bank.
Table 2.
Selected scanning mutagenesis studies of EmrE
| Mutations tested | Substrates tested | Assay | Reference |
|---|---|---|---|
| Scan: all residues to Ala, Gly, and Val | Ethidium, acriflavine, methyl viologen | Bacterial resistance | (88) |
| Scan: all residues to Cys | Ethidium | Bacterial resistance | (85) |
| 33 variants of mechanistically important residues | 12 drugs including polyaromatics, quaternary ammonium cations, and antiseptics with alkyl tails | Bacterial resistance | (89) |
| 18 mutations in helix 1, loop 1, and helix 2 to Cys | TPP+ | Binding (purified, reconstituted protein) | (144) |
| 48 mutations throughout protein to Cys | Methyl viologen, acriflavine, ethidium | Bacterial resistance | (145) |
| All Glycines (12) mutated to Cys, Ala, Pro | Methyl viologen, acriflavine, ethidium | Bacterial resistance | (146) |
| All Tyrosines (5) mutated to various residues. 18 total mutants tested. | Methyl viologen, TPP+ | Transport and binding experiments | (62) |
| All Tryptophans (4) mutated to Cys. | TPP+, methyl viologen, acriflavine, ethidium | Transport, binding, and bacterial resistance experiments | (86) |
| All acidic and basic residues (10) mutated to various residues | Ethidium, acriflavine, methyl viologen | Transport and bacterial resistance experiments | (60) |
These EmrE crystal structures were determined with structurally diverse substrates bound, including planar polyaromatics and tetrahedral quaternary phosphoniums and ammoniums (Table 1). The electron density determined using X-ray crystallography shows high correspondence with previous low resolution EM data obtained for EmrE in lipid bilayers (12), implying that the crystal structures represent a native, low energy conformation. The molecular details of the crystal structures also agree with specific predictions from spectroscopic and mutagenic studies (63). Recent models from the NMR experiments (80, 81) exhibit notable structural differences with the crystal structures (3) and with the available computational and EM models (58, 61). These differences, and potential reasons for the differences, are discussed in depth in (3) and will only be briefly summarized here. In the NMR models, the subunits of the dimer are arranged more parallel with respect to each other, and the loops are unpacked, permitting aqueous access to the binding site from both sides of the membrane. The hydrogen bond network in the binding site is also rearranged with respect to the recent crystal structures. It is possible that the NMR models represent functional intermediate states, such as those suggested by prior EPR experiments (63), and that the differences between the crystallography or EM density (which largely agree with each other) and NMR models and are due to differences in the experimental conditions. However, it is also important to note that the NMR models are based on relatively few experimental measurements of distances between backbone atoms and bound substrate and that the models (including sidechain placement) are generated computationally, based on these distance restraints and backbone chemical shift measurements. Since the recent crystal structures are in the best agreement with the EM maps and are the only structural models of EmrE with experimental electron density that supports sidechain placement without further computational modeling, we will focus our analysis on these crystal structures (3). Note that previous EmrE crystal structures (82) (Protein Data Bank codes 3B5D, 3B62, 3B61) are low quality and incomplete (only Cα atoms are modeled) and experimental maps are unavailable in the Protein Data Bank. These prior structures are broadly considered inadequate for molecular inference (3, 61, 78, 79) and should not be used.
Both the recent structures and the long history of functional data show that EmrE has many major mechanistic features in common with Gdx-Clo, including the central glutamates in the binding pocket that contribute to alternate binding of drug and protons (59, 83, 84), the tyrosine switch engaged in a cross-subunit interaction (61, 62), the GXG fulcrum that kinks TM3 and defines the N- and C-terminal lobes of each subunit (58, 85), and the hydrophobic portal that permits access between the binding pocket and the hydrophobic interior of the inner lipid membrane for hydrophobic substrates and substrate substituents (61, 63). Based on information from the crystal structures of EmrE with benzyltrimethylammonium (the headgroup of the common household antiseptic benzalkonium) and Gdx-Clo with octylGdm+, a model for benzalkonium binding was constructed (3), illustrating how the membrane portals of both the Gdx and Qac subtypes can be exploited to bind substrates with extended alkyl substituents (Fig. 5B).
Given the structural similarities and high sequence conservation, why then can EmrE bind and transport a much more diverse range of substrates than Gdx-Clo? The crystal structures in complex with substrates suggest that, despite shared sequences, EmrE and Gdx-Clo also have important structural differences. Although many of the binding pocket residues that serve as hydrogen bond donors or acceptors are conserved in EmrE, they do not form as an extensive an inter-residue H-bond network as is observed in the binding site of Gdx-Clo. Peripheral binding site residues W63 and S42 are both present, but they do not H-bond with other EmrE sidechains (4). Many potential H-bond interactions have either poor geometry or longer interaction distances than ideal (Fig. 5C). As a result, the central glutamates and conserved binding site Trp (W63 in EmrE) are comparatively unconstrained and able to adopt different rotamers in the presence of different substrates. EmrE’s central glutamates move closer together or farther apart to accommodate flat planar substrates or bulky quaternary compounds (Fig. 5D). At the same time, the binding site Trp rotates over ∼80° to stack against aromatic rings of different substrates bound in different poses (3) (Fig. 5D). Unlike Gdx-Clo, in which the residues that contribute to the H-bond stack stabilizing the central glutamates cannot be altered (4), many of the analogous mutations to reduce H-bonding capacity are tolerated in EmrE (3). Notably, the binding site Trp, W63, which had been shown to be essential in all previous studies with aromatic substrates (85, 86, 87, 88), is not required for the transport of nonaromatic substrates by EmrE (3, 89).
These structural observations are in accord with NMR and computational studies that suggest that EmrE possesses an unusual degree of structural plasticity that might contribute to substrate polyspecificity (79, 90). However, it should be emphasized that the observed structural perturbations are limited to the sidechains. Larger conformational changes involving the backbone are not necessary to explain the binding of diverse substrates to EmrE, and such perturbations are not observed in any of the five drug-bound crystal structures (3). Likewise, the low pH, proton-bound crystal structures of EmrE (as well as Gdx-Clo) do not exhibit major structural differences relative to the substrate-bound structures, with only local changes in the position of the central glutamates (3). However, EPR studies have suggested that upon protonation, the conformational ensemble of EmrE becomes more heterogenous than in the presence of the drug TPP+ (63). Thus, the doubly protonated state might not exist in a single predominant stable conformation and the reported low pH crystal structure may capture only one species in this ensemble.
Although the crystal structures provided essential insight into the molecular basis for substrate binding by EmrE, the transport of these disparate substrates poses additional problems, requiring the choreography of substrate binding and dissociation, conformational exchange, and proton antiport. Spectroscopic techniques, including NMR and EPR, have been integral to fleshing out a dynamic picture of EmrE. NMR studies in lipid membranes and in bicelles have identified various mutations that slow or eliminate conformational change but preserve substrate binding, isolating residues involved in the first process and not the second (88, 91). NMR measurements have shown that the kinetic behavior of EmrE, including the rate of conformational exchange, differs depending on the substrate, demonstrating that different substrates have different affinities for the transition state of the conformational exchange, as they do in the ground state (92). Likewise, protonation of the central glutamates accelerates conformational transition (93), perhaps reflecting the same ground state destabilization that leads to heterogeneity in the conformational ensemble upon protonation (63). The reduction of free energy of conformational transitions upon substrate binding is a classical requirement for coupled substrate antiport (94).
However, emerging evidence also suggests that under certain conditions, EmrE violates tenets of classic transport mechanisms. Conformational exchange of the apo (proton- and drug-free) and single proton–bound transporter have been reported (83, 90, 95), as has simultaneous binding of proton with some drugs such that both can be carried across the membrane together (80, 96). The conformational exchange rate of EmrE with different substrates is not tightly correlated with the rate of substrate transport, hinting that different substrates, particularly high affinity substrates, might undergo futile cycles and remain bound as the transporter transits between inward and outward open states (92). Kinetic modeling (97) suggests scenarios in which the microscopic rate constants measured for each potential binding event and conformational transition in the transport cycle combine to reduce the stoichiometry noticeably from the 2:1 H+:substrate stoichiometry measured for Gdx (5) and many EmrE substrates (6). These studies suggest that, in some cases, the specific proton and substrate gradients and substrate-binding energy may even lead to cycles of substrate import (96, 97).
A limitation of such free exchange transport models is that they permit potential pathways for proton leak. If the inward-to-outward facing transition of the unoccupied or singly protonated transporter is not energetically prohibitive in living bacteria, such transport cycles would contribute to the dissipation of the proton motive force. For EmrE, different mechanisms have been proposed to explain the apparent absence of detrimental leak pathways in vivo and in vitro. In one proposal, the central glutamates are electrostatically independent so that the proton cannot “hop” from the glutamate with the lower pKa to that with the higher pKa, preventing proton release from the singly protonated state after the conformational swap (84). Alternatively, it has been suggested that the proton pathway is gated by a C-terminal histidine residue that is highly conserved among the Qac subtype, which occludes the binding pocket in the absence of drug, preventing proton leak (98). It should also be mentioned that the quaternary phosphonium substrates used in these transport experiments are not encountered by bacteria outside the laboratory. Thus, the transport properties for such anthropogenic chemicals have not been optimized by purifying selection in bacterial populations over evolutionary time. While it is possible that proton slippage and deviations from ideal stoichiometry are evolved properties of the transporter to handle diverse substrates (96), it is also possible that these mechanistic features reflect nonoptimized transport of non-native compounds and that transport of native substrates (whether the drug-like molecules produced by microbes in competitive niches or yet-unknown metabolites) is more parsimonious. Native substrates of the Qac transporters have yet to be identified, however, so this remains an open question.
Polyamine transporters
Polyamines, such as spermidine, putrescine, and cadaverine, play myriad roles in diverse bacteria (99, 100, 101, 102). These small, charged metabolites are synthesized or taken up from the environment by bacteria, where they can be used as synthetic precursors to siderophores (103, 104) or structural components of the cell wall (105, 106), contribute to oxidative stress resistance (107, 108), or interact with nucleic acids to modulate translation (102). Polyamines also serve as signals for the induction of virulence genes (109, 110, 111) and surface behaviors like biofilm formation and swarming (112, 113, 114, 115, 116). However, excessive accumulation of polyamines is toxic (117). A subset of transporters from the SMR family has been implicated in polyamine export in E. coli and Shigella (8, 118). These proteins are proposed to serve as a “safety valve” when intracellular polyamines accumulate to toxic levels (118). The transporters form heterodimers, and their genes are always found as pairs, annotated mdtI and mdtJ in E. coli (8). Native expression is low and is regulated by accumulation of polyamines and bile salts (8, 118). Although biochemical information is relatively limited for the polyamine transporters, mutagenesis coupled with growth assays has demonstrated that key mechanistic residues for Qac and Gdx function, including the central glutamates, the tyrosine switch, and the binding site tryptophans, are critical for function of the MdtIJ complex (8), implying that the polyamine transporters share mechanistic similarities with the more extensively characterized Gdx and Qac subtypes.
Lipid transport proteins
The most distantly related members of the SMR family are reported to act as glycolipid flippases. The SMR genes (annotated arnE and arnF in E. coli and Salmonella enterica) are found in larger biosynthetic operons that contribute to the chemical modification of lipid A in the outer membrane with 4-amino-4-deoxy-l-arabinose (L-Ara4N) (119). This synthetic pathway contributes to polymyxin resistance by reducing the electrostatic interactions of lipid A with the cationic polymyxin antibiotic (119). In S. enterica serovar Typhimurium, the role of this SMR subtype is to transport undecaprenyl phosphate aminoarabinose (9), a lipid that carries L-Ara4N from its site of synthesis in the cytoplasm to the periplasmic leaflet, on its way toward lipid A in the outer membrane (120). Deletion of the SMR transport proteins prevents localization of L-Ara4N to the outer membrane and thus prevents cells from acquiring polymyxin resistance via this route (9). There are indications that the S. enterica flippase has somewhat broader specificity, including genetic complementation of flippase deletion mutants in synthetic pathways that require transport of different glycolipids (121, 122).
The lipid SMRs are the only subtype that transports noncationic substrates, and these proteins often possess an asparagine in place of the central glutamate (Fig. 2). This replacement also suggests that lipid transport may not be proton coupled, since the central glutamates are also responsible for proton binding in proton-coupled SMRs. Because undecaprenyl phosphate aminoarabinose is synthesized in the cytoplasmic leaflet (120), transport of the lipid down its concentration gradient to the periplasmic leaflet via facilitated diffusion might be sufficient. However, this has not been established experimentally. A membrane portal similar to that observed in structures of EmrE and Gdx-Clo would be an obviously useful feature for lipid transport, permitting the substrate’s prenyl tail access to the membrane while the polar headgroup is ensconced within its protein-binding site, a familiar feature of other lipid flippases (123). Indeed, in this SMR subtype, the hydrophobic character of the TM2 residues is retained, and we therefore conjecture that the membrane portal is conserved as well. The tyrosine switch is also conserved in the lipid SMRs, despite their overall low sequence similarity with other SMR subtypes.
Structural relationship between the SMR and the SLC35/DMT folds
Although other dual topology transporter families have representatives with inverted repeat topology (27, 28), the SMR family does not possess such internally fused transporters with detectable sequence homology (29). However, structural analysis of transporters from the DMT superfamily, which possess the SLC35/DMT fold, suggests that the SMR fold might nonetheless have been preserved by evolution as an inverted repeat. Structures of transporters with the SLC35/DMT fold, including a bacterial aromatic amino acid exporter, a protozoan drug exporter, and eukaryotic organellar sugar/nucleotide transporters (124, 125, 126, 127, 128), possess striking structural homology to the bacterial SMRs (TM helix RMSD 2.6–3.8 Å) despite sharing no sequence similarity (Fig. 6A). DMT and SMR transporters have previously been proposed to be evolutionarily related (129, 130). However, the SLC35/DMT and SMR structures also bear two notable differences. First, each domain of the SLC35 inverted repeat is composed of five TM helices. In the 3D structure, this pair of inserted helices pack against the membrane portal defined by helices 2A and 2B of the SMR transporters, sealing the portal and eliminating access to the substrate-binding site from the membrane (4). The second major difference between the SMR and SLC35/DMT structures is in the helix connectivity. Whereas in the SMRs, each transport domain is composed of a single, independently folded monomer; in the SLC35/DMT proteins, the transport domains do not correspond simply to the N- and C-terminal halves of the protein. The first transport domain is composed of helices 1, 2, 8, 9, and 10 and the second is composed of helices 3, 4, 5, 6, and 7 (Fig. 6A).
Figure 6.
Structural relationship between the SMR and SLC35 folds.A, structural comparisons of Gdx-Clo (PDB:6WK9) (4) and SLC35 CMP-sialic acid transporter (PDB: 6I1R) (125). For the SMR fold, the individual subunits are colored in gray and blue and the membrane portal is labeled. For the SLC35/DMT fold, helices involved in a potential domain swap are colored in green (for the blue monomer) and yellow (for the gray monomer). The transmembrane (TM) helix insertions are indicated. The two transport domains are outlined by the dashed turquoise line. B, a potential pathway for divergent evolution of the SMR and SLC35/DMT folds. Topology is shown as a top-down cartoon, as in the left panels of (A). Panel 1: SMR topology with membrane portal indicated. Panel 2: insertion of a TM helix (helices I1 and I2) inverts helix 1 and 1∗ with respect to the other helices in the subunit, introducing clashes and disrupting helical packing (red x symbols). The original packing can be restored by swapping the positions of the yellow and green helices, indicated by arrows. Panel 3: Dual topology ancestor of the SLC35/DMT fold possesses a helix insertion that seals the membrane portal and a domain swap involving the yellow and green helices that preserves structural homology with the SMR fold. Panel 4: a duplication/fusion event links the C terminus of the first subunit (helix 5) with the N terminus of the second subunit (helix 6) and fixes this topology in the SLC35/DMT lineage. The transport domains are outlined by the turquoise dashed line, as in the SLC35 structure in panel (A). PDB, Protein Data Bank.
Although the possibility that the similarities between the SMR and SLC35/DMT folds arose via convergent evolution cannot be ruled out; structural correspondence between proteins with similar functions is usually considered evidence for evolutionary relatedness (131, 132). Moreover, we propose that the topological differences between the SMRs and SLC35/DMT folds can be plausibly explained by a divergent evolutionary pathway (Fig. 6B). This model posits an ancestral, dual topology transporter with the SMR fold and a TM helix insertion between TM helices 1 and 2. The introduction of this new TM helix would enforce a reorientation of TM helices 2, 3, and 4 relative to TM helix 1, disrupting the packing between TM helix 1 and the other TM helices in that monomer (Fig. 6B, panel 2). The 3D SMR fold could be preserved, however, by a domain swap during dimer assembly (Fig. 6B, panels 2 and 3) such that the now-inverted TM1 from the first monomer trades positions with TM1∗ in the opposite subunit and vice versa (Fig. 6B, panel 3). TM helix domain swaps have been observed in other membrane proteins (133, 134) and for engineered EmrE concatamers (29), and an analogous mechanism has been proposed for the evolution of the structurally similar, but topologically distinct, Pnu vitamin transporters and SWEET sugar exporters (135). Finally, a subsequent duplication/fusion of the 5-helix, domain-swapped ancestral dual topology transporter would give rise to the extant SLC35/DMT fold (Fig. 6B, panel 4). Duplication/fusion of 4-TM dual topology transporters are not unprecedented (27), but this process is more common for dual topology transporters with an odd number of transmembrane helices, since the N and C termini are on the same side of the membrane and connecting them does not require introduction of a transmembrane linker (28, 29). Although no sequence homology can be detected between the SLC35/DMT transporters and SMRs or even between symmetry-related helices of the SLC35/DMTs, lack of sequence homology is not uncommon in other families of evolutionary divergent transporters with shared folds (24, 136).
Conclusions and perspective
The SMRs have provided a tremendously productive system for studying membrane protein evolution, transport mechanism, and microbial multidrug resistance. After 25 years of such studies, the SMR transporters finally joined the high resolution structural era in 2021. These recent crystal structures representing two of the four known functional subtypes have provided a platform for analyzing prior functional studies and understanding the structural features that contribute to substrate binding and transport for each SMR subtype: Gdm+, drugs, polyamines, and glycolipids. Moreover, structural homology with SLC35/DMT transporters suggests that, contrary to the proposal that the SMRs are unusual in having not evolved fused, inverted repeat architecture (29), it is probable that the SMR fold has indeed been preserved through this evolutionary mechanism, albeit with a helical insertion and domain swap along the way. These recent advances in understanding the molecular architecture bolster ongoing efforts to develop antimicrobials that target SMR proteins, either by inhibiting transporter assembly in order to sensitize bacteria to transported compounds (78, 137, 138) or by hijacking the nominal exporters to import antimicrobial compounds instead (139).
In addition, the molecular framework described here opens the door for future integrative functional, structural, and computational studies to understand how the SMR scaffold has been tailored to transport diverse substrates as the family has evolved. Such lines of inquiry are urgently important as bacteria continue to evolve around us. The Qac and Gdx subtypes, in particular, have found new roles in human-impacted environments, conferring bacterial resistance to household antiseptics (89), “dead-end” metabolites from the degradation of metformin (140) and other pharmaceuticals that accumulate in the environment or the human microbiome, and other agricultural and industrial chemicals (4). Genes encoding SMR transporters are currently spreading among bacterial populations, encountering new physiological contexts and substrate transport demands, driving co-selection of co-localized antimicrobial resistance genes in environmental reservoirs (76), and influencing microbial population compositions in the human microbiome and human-impacted environments (32, 76, 141), as the SMR transporters contribute to the ongoing story of natural selection at the human–microbe interface.
Data availability
Sequences and annotation for SMR proteins identified in the bacterial GEBA genome set are available for download from the Deep Blue Data repository hosted by the University of Michigan with unique identifier doi.org/10.7302/0ynd-b343.
Conflict of interest
The authors declare that they have no competing interests with the contents of this article.
Acknowledgments
Author contributions
C. B. M. data curation; O. E. B., T. J. Y., and R. B. S. writing–original draft; O. E. B., T. J. Y., C. B. M., and R. B. S. writing–review & editing; O. E. B., T. J. Y., and R. B. S. visualization; R. B. S. project administration; R. B. S. funding acquisition.
Funding and additional information
This work was supported by National Institutes of Health grant R35 GM128768 to R. B. S. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Edited by Roger Colbran
Footnotes
Present address for Christian B. Macdonald: Department of Bioengineering and Therapeutic Sciences, University of California San Francisco, San Francisco, CA 94158, USA.
References
- 1.Yerushalmi H., Lebendiker M., Schuldiner S. EmrE, an Escherichia coli 12-kDa multidrug transporter, exchanges toxic cations and H+ and is soluble in organic solvents. J. Biol. Chem. 1995;270:6856–6863. doi: 10.1074/jbc.270.12.6856. [DOI] [PubMed] [Google Scholar]
- 2.Paulsen I.T., Skurray R.A., Tam R., Saier M.H., Jr., Turner R.J., Weiner J.H., et al. The SMR family: a novel family of multidrug efflux proteins involved with the efflux of lipophilic drugs. Mol. Microbiol. 1996;19:1167–1175. doi: 10.1111/j.1365-2958.1996.tb02462.x. [DOI] [PubMed] [Google Scholar]
- 3.Kermani A.A., Burata O.E., Koff B.B., Koide A., Koide S., Stockbridge R.B. Crystal structures of bacterial small multidrug resistance transporter EmrE in complex with structurally diverse substrates. Elife. 2022;11 doi: 10.7554/eLife.76766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kermani A.A., Macdonald C.B., Burata O.E., Ben Koff B., Koide A., Denbaum E., et al. The structural basis of promiscuity in small multidrug resistance transporters. Nat. Commun. 2020;11:6064. doi: 10.1038/s41467-020-19820-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kermani A.A., Macdonald C.B., Gundepudi R., Stockbridge R.B. Guanidinium export is the primal function of SMR family transporters. Proc. Natl. Acad. Sci. U. S. A. 2018;115:3060–3065. doi: 10.1073/pnas.1719187115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rotem D., Schuldiner S. EmrE, a multidrug transporter from Escherichia coli, transports monovalent and divalent substrates with the same stoichiometry. J. Biol. Chem. 2004;279:48787–48793. doi: 10.1074/jbc.M408187200. [DOI] [PubMed] [Google Scholar]
- 7.Bay D.C., Rommens K.L., Turner R.J. Small multidrug resistance proteins: a multidrug transporter family that continues to grow. Biochim. Biophys. Acta. 2008;1778:1814–1838. doi: 10.1016/j.bbamem.2007.08.015. [DOI] [PubMed] [Google Scholar]
- 8.Higashi K., Ishigure H., Demizu R., Uemura T., Nishino K., Yamaguchi A., et al. Identification of a spermidine excretion protein complex (MdtJI) in Escherichia coli. J. Bacteriol. 2008;190:872–878. doi: 10.1128/JB.01505-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yan A., Guan Z., Raetz C.R. An undecaprenyl phosphate-aminoarabinose flippase required for polymyxin resistance in Escherichia coli. J. Biol. Chem. 2007;282:36077–36089. doi: 10.1074/jbc.M706172200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.von Heijne G. Control of topology and mode of assembly of a polytopic membrane protein by positively charged residues. Nature. 1989;341:456–458. doi: 10.1038/341456a0. [DOI] [PubMed] [Google Scholar]
- 11.Rapp M., Granseth E., Seppala S., von Heijne G. Identification and evolution of dual-topology membrane proteins. Nat. Struct. Mol. Biol. 2006;13:112–116. doi: 10.1038/nsmb1057. [DOI] [PubMed] [Google Scholar]
- 12.Ubarretxena-Belandia I., Baldwin J.M., Schuldiner S., Tate C.G. Three-dimensional structure of the bacterial multidrug transporter EmrE shows it is an asymmetric homodimer. EMBO J. 2003;22:6175–6181. doi: 10.1093/emboj/cdg611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fluman N., Tobiasson V., von Heijne G. Stable membrane orientations of small dual-topology membrane proteins. Proc. Natl. Acad. Sci. U. S. A. 2017;114:7987–7992. doi: 10.1073/pnas.1706905114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Woodall N.B., Yin Y., Bowie J.U. Dual-topology insertion of a dual-topology membrane protein. Nat. Commun. 2015;6:8099. doi: 10.1038/ncomms9099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Van Lehn R.C., Zhang B., Miller T.F., 3rd. Regulation of multispanning membrane protein topology via post-translational annealing. Elife. 2015;4 doi: 10.7554/eLife.08697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nicolaus F., Metola A., Mermans D., Liljenstrom A., Krc A., Abdullahi S.M., et al. Residue-by-residue analysis of cotranslational membrane protein integration in vivo. Elife. 2021;10 doi: 10.7554/eLife.64302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mermans D., Nicolaus F., Fleisch K., von Heijne G. Cotranslational folding and assembly of the dimeric E. coli inner membrane protein EmrE. bioRxiv. 2022 doi: 10.1101/2022.04.02.486806. [preprint] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Masaoka Y., Ueno Y., Morita Y., Kuroda T., Mizushima T., Tsuchiya T. A two-component multidrug efflux pump, EbrAB, in Bacillus subtilis. J. Bacteriol. 2000;182:2307–2310. doi: 10.1128/jb.182.8.2307-2310.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jack D.L., Storms M.L., Tchieu J.H., Paulsen I.T., Saier M.H., Jr. A broad-specificity multidrug efflux pump requiring a pair of homologous SMR-type proteins. J. Bacteriol. 2000;182:2311–2313. doi: 10.1128/jb.182.8.2311-2313.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ganas P., Mihasan M., Igloi G.L., Brandsch R. A two-component small multidrug resistance pump functions as a metabolic valve during nicotine catabolism by Arthrobacter nicotinovorans. Microbiology (Reading) 2007;153:1546–1555. doi: 10.1099/mic.0.2006/004234-0. [DOI] [PubMed] [Google Scholar]
- 21.Stockbridge R.B., Robertson J.L., Kolmakova-Partensky L., Miller C. A family of fluoride-specific ion channels with dual-topology architecture. Elife. 2013;2 doi: 10.7554/eLife.01084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stockbridge R.B., Kolmakova-Partensky L., Shane T., Koide A., Koide S., Miller C., et al. Crystal structures of a double-barrelled fluoride ion channel. Nature. 2015;525:548–551. doi: 10.1038/nature14981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sanghai Z.A., Liu Q., Clarke O.B., Belcher-Dufrisne M., Wiriyasermkul P., Giese M.H., et al. Structure-based analysis of CysZ-mediated cellular uptake of sulfate. Elife. 2018;7 doi: 10.7554/eLife.27829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Forrest L.R. Structural symmetry in membrane proteins. Annu. Rev. Biophys. 2015;44:311–337. doi: 10.1146/annurev-biophys-051013-023008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Keller R., Ziegler C., Schneider D. When two turn into one: evolution of membrane transporters from half modules. Biol. Chem. 2014;395:1379–1388. doi: 10.1515/hsz-2014-0224. [DOI] [PubMed] [Google Scholar]
- 26.Sarti E., Aleksandrova A.A., Ganta S.K., Yavatkar A.S., Forrest L.R. EncoMPASS: an online database for analyzing structure and symmetry in membrane proteins. Nucl. Acids Res. 2019;47:D315–D321. doi: 10.1093/nar/gky952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Macdonald C.B., Stockbridge R.B. A topologically diverse family of fluoride channels. Curr. Opin. Struct. Biol. 2017;45:142–149. doi: 10.1016/j.sbi.2017.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lolkema J.S., Dobrowolski A., Slotboom D.J. Evolution of antiparallel two-domain membrane proteins: tracing multiple gene duplication events in the DUF606 family. J. Mol. Biol. 2008;378:596–606. doi: 10.1016/j.jmb.2008.03.005. [DOI] [PubMed] [Google Scholar]
- 29.Lloris-Garcera P., Seppala S., Slusky J.S., Rapp M., von Heijne G. Why have small multidrug resistance proteins not evolved into fused, internally duplicated structures? J. Mol. Biol. 2014;426:2246–2254. doi: 10.1016/j.jmb.2014.03.012. [DOI] [PubMed] [Google Scholar]
- 30.Mukherjee S., Seshadri R., Varghese N.J., Eloe-Fadrosh E.A., Meier-Kolthoff J.P., Goker M., et al. 1,003 reference genomes of bacterial and archaeal isolates expand coverage of the tree of life. Nat. Biotechnol. 2017;35:676–683. doi: 10.1038/nbt.3886. [DOI] [PubMed] [Google Scholar]
- 31.Gaze W.H., Abdouslam N., Hawkey P.M., Wellington E.M. Incidence of class 1 integrons in a quaternary ammonium compound-polluted environment. Antimicrob. Agents Chemother. 2005;49:1802–1807. doi: 10.1128/AAC.49.5.1802-1807.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhu Y.G., Zhao Y., Li B., Huang C.L., Zhang S.Y., Yu S., et al. Continental-scale pollution of estuaries with antibiotic resistance genes. Nat. Microbiol. 2017;2 doi: 10.1038/nmicrobiol.2016.270. [DOI] [PubMed] [Google Scholar]
- 33.An X.L., Chen Q.L., Zhu D., Zhu Y.G., Gillings M.R., Su J.Q. Impact of wastewater treatment on the prevalence of integrons and the genetic diversity of integron gene cassettes. Appl. Environ. Microbiol. 2018;84 doi: 10.1128/AEM.02766-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Domingues S., da Silva G.J., Nielsen K.M. Global dissemination patterns of common gene cassette arrays in class 1 integrons. Microbiology (Reading) 2015;161:1313–1337. doi: 10.1099/mic.0.000099. [DOI] [PubMed] [Google Scholar]
- 35.Bay D.C., Turner R.J. Diversity and evolution of the small multidrug resistance protein family. BMC Evol. Biol. 2009;9:140. doi: 10.1186/1471-2148-9-140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ninio S., Schuldiner S. Characterization of an archaeal multidrug transporter with a unique amino acid composition. J. Biol. Chem. 2003;278:12000–12005. doi: 10.1074/jbc.M213119200. [DOI] [PubMed] [Google Scholar]
- 37.Eddy S.R. Accelerated profile HMM searches. PLoS Comput. Biol. 2011;7 doi: 10.1371/journal.pcbi.1002195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Greener T., Govezensky D., Zamir A. A novel multicopy suppressor of a groEL mutation includes two nested open reading frames transcribed from different promoters. EMBO J. 1993;12:889–896. doi: 10.1002/j.1460-2075.1993.tb05729.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chung Y.J., Saier M.H., Jr. Overexpression of the Escherichia coli sugE gene confers resistance to a narrow range of quaternary ammonium compounds. J. Bacteriol. 2002;184:2543–2545. doi: 10.1128/JB.184.9.2543-2545.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jeong H., Nasir A. A preliminary list of horizontally transferred genes in prokaryotes determined by tree reconstruction and reconciliation. Front. Genet. 2017;8:112. doi: 10.3389/fgene.2017.00112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Slipski C.J., Jamieson T.R., Zhanel G.G., Bay D.C. Riboswitch-associated guanidinium-selective efflux pumps frequently transmitted on proteobacterial plasmids increase Escherichia coli biofilm tolerance to disinfectants. J. Bacteriol. 2020;202 doi: 10.1128/JB.00104-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bay D.C., Turner R.J. Spectroscopic analysis of the intrinsic chromophores within small multidrug resistance protein SugE. Biochim. Biophys. Acta. 2011;1808:2233–2244. doi: 10.1016/j.bbamem.2011.05.005. [DOI] [PubMed] [Google Scholar]
- 43.Sikora C.W., Turner R.J. SMR proteins SugE and EmrE bind ligand with similar affinity and stoichiometry. Biochem. Biophys. Res. Commun. 2005;335:105–111. doi: 10.1016/j.bbrc.2005.07.051. [DOI] [PubMed] [Google Scholar]
- 44.Nishino K., Yamaguchi A. Analysis of a complete library of putative drug transporter genes in Escherichia coli. J. Bacteriol. 2001;183:5803–5812. doi: 10.1128/JB.183.20.5803-5812.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nelson J.W., Atilho R.M., Sherlock M.E., Stockbridge R.B., Breaker R.R. Metabolism of free guanidine in bacteria is regulated by a widespread riboswitch class. Mol. Cell. 2017;65:220–230. doi: 10.1016/j.molcel.2016.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vaughan V.C., Novy F.G. Lea Brothers & Co; Philadelphia: 1891. Ptomaines, Leucomaines, and Bacterial Proteids: Or the Chemical Factors in the Causation of Disease. [Google Scholar]
- 47.Sherlock M.E., Breaker R.R. Biochemical validation of a third guanidine riboswitch class in bacteria. Biochemistry. 2017;56:359–363. doi: 10.1021/acs.biochem.6b01271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sherlock M.E., Malkowski S.N., Breaker R.R. Biochemical validation of a second guanidine riboswitch class in bacteria. Biochemistry. 2017;56:352–358. doi: 10.1021/acs.biochem.6b01270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lenkeit F., Eckert I., Hartig J.S., Weinberg Z. Discovery and characterization of a fourth class of guanidine riboswitches. Nucl. Acids Res. 2020;48:12889–12899. doi: 10.1093/nar/gkaa1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Salvail H., Balaji A., Yu D., Roth A., Breaker R.R. Biochemical validation of a fourth guanidine riboswitch class in bacteria. Biochemistry. 2020;59:4654–4662. doi: 10.1021/acs.biochem.0c00793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schneider N.O., Tassoulas L.J., Zeng D., Laseke A.J., Reiter N.J., Wackett L.P., et al. Solving the conundrum: widespread proteins annotated for urea metabolism in bacteria are carboxyguanidine deiminases mediating nitrogen assimilation from guanidine. Biochemistry. 2020;59:3258–3270. doi: 10.1021/acs.biochem.0c00537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang B., Xu Y., Wang X., Yuan J.S., Johnson C.H., Young J.D., et al. A guanidine-degrading enzyme controls genomic stability of ethylene-producing cyanobacteria. Nat. Commun. 2021;12:5150. doi: 10.1038/s41467-021-25369-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Funck D., Sinn M., Fleming J.R., Stanoppi M., Dietrich J., Lopez-Igual R., et al. Discovery of a Ni(2+)-dependent guanidine hydrolase in bacteria. Nature. 2022;603:515–521. doi: 10.1038/s41586-022-04490-x. [DOI] [PubMed] [Google Scholar]
- 54.Wang B.D., Dong T., Myrlie A., Gu L., Zhu H., Xiong W., et al. Photosynthetic production of the nitrogen-rich compound guanidine. Green. Chem. 2019;21:2928–2937. [Google Scholar]
- 55.Sinn M., Hauth F., Lenkeit F., Weinberg Z., Hartig J.S. Widespread bacterial utilization of guanidine as nitrogen source. Mol. Microbiol. 2021;116:200–210. doi: 10.1111/mmi.14702. [DOI] [PubMed] [Google Scholar]
- 56.Thomas N.E., Feng W., Henzler-Wildman K.A. A solid-supported membrane electrophysiology assay for efficient characterization of ion-coupled transport. J. Biol. Chem. 2021;297 doi: 10.1016/j.jbc.2021.101220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Morrison E.A., DeKoster G.T., Dutta S., Vafabakhsh R., Clarkson M.W., Bahl A., et al. Antiparallel EmrE exports drugs by exchanging between asymmetric structures. Nature. 2012;481:45–50. doi: 10.1038/nature10703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Fleishman S.J., Harrington S.E., Enosh A., Halperin D., Tate C.G., Ben-Tal N. Quasi-symmetry in the cryo-EM structure of EmrE provides the key to modeling its transmembrane domain. J. Mol. Biol. 2006;364:54–67. doi: 10.1016/j.jmb.2006.08.072. [DOI] [PubMed] [Google Scholar]
- 59.Muth T.R., Schuldiner S. A membrane-embedded glutamate is required for ligand binding to the multidrug transporter EmrE. EMBO J. 2000;19:234–240. doi: 10.1093/emboj/19.2.234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yerushalmi H., Schuldiner S. An essential glutamyl residue in EmrE, a multidrug antiporter from Escherichia coli. J. Biol. Chem. 2000;275:5264–5269. doi: 10.1074/jbc.275.8.5264. [DOI] [PubMed] [Google Scholar]
- 61.Vermaas J.V., Rempe S.B., Tajkhorshid E. Electrostatic lock in the transport cycle of the multidrug resistance transporter EmrE. Proc. Natl. Acad. Sci. U. S. A. 2018;115:E7502–E7511. doi: 10.1073/pnas.1722399115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rotem D., Steiner-Mordoch S., Schuldiner S. Identification of tyrosine residues critical for the function of an ion-coupled multidrug transporter. J. Biol. Chem. 2006;281:18715–18722. doi: 10.1074/jbc.M602088200. [DOI] [PubMed] [Google Scholar]
- 63.Dastvan R., Fischer A.W., Mishra S., Meiler J., McHaourab H.S. Protonation-dependent conformational dynamics of the multidrug transporter EmrE. Proc. Natl. Acad. Sci. U. S. A. 2016;113:1220–1225. doi: 10.1073/pnas.1520431113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Aller S.G., Yu J., Ward A., Weng Y., Chittaboina S., Zhuo R., et al. Structure of P-glycoprotein reveals a molecular basis for poly-specific drug binding. Science. 2009;323:1718–1722. doi: 10.1126/science.1168750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.He X., Szewczyk P., Karyakin A., Evin M., Hong W.X., Zhang Q., et al. Structure of a cation-bound multidrug and toxic compound extrusion transporter. Nature. 2010;467:991–994. doi: 10.1038/nature09408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Payandeh J., Scheuer T., Zheng N., Catterall W.A. The crystal structure of a voltage-gated sodium channel. Nature. 2011;475:353–358. doi: 10.1038/nature10238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Metabolic Pathways of Agrochemicals Part Two: Insecticides and Fungicides. Royal Society of Chemistry; Cambridge, UK: 1999. [Google Scholar]
- 68.Zahn D., Mucha P., Zilles V., Touffet A., Gallard H., Knepper T.P., et al. Identification of potentially mobile and persistent transformation products of REACH-registered chemicals and their occurrence in surface waters. Water Res. 2019;150:86–96. doi: 10.1016/j.watres.2018.11.042. [DOI] [PubMed] [Google Scholar]
- 69.Peter K.T., Tian Z., Wu C., Lin P., White S., Du B., et al. Using high-resolution mass spectrometry to identify organic contaminants linked to urban stormwater mortality syndrome in coho salmon. Environ. Sci. Technol. 2018;52:10317–10327. doi: 10.1021/acs.est.8b03287. [DOI] [PubMed] [Google Scholar]
- 70.Scheurer M., Michel A., Brauch H.J., Ruck W., Sacher F. Occurrence and fate of the antidiabetic drug metformin and its metabolite guanylurea in the environment and during drinking water treatment. Water Res. 2012;46:4790–4802. doi: 10.1016/j.watres.2012.06.019. [DOI] [PubMed] [Google Scholar]
- 71.Blair B.D., Crago J.P., Hedman C.J., Klaper R.D. Pharmaceuticals and personal care products found in the Great Lakes above concentrations of environmental concern. Chemosphere. 2013;93:2116–2123. doi: 10.1016/j.chemosphere.2013.07.057. [DOI] [PubMed] [Google Scholar]
- 72.Gillings M.R. Class 1 integrons as invasive species. Curr. Opin. Microbiol. 2017;38:10–15. doi: 10.1016/j.mib.2017.03.002. [DOI] [PubMed] [Google Scholar]
- 73.Gillings M., Boucher Y., Labbate M., Holmes A., Krishnan S., Holley M., et al. The evolution of class 1 integrons and the rise of antibiotic resistance. J. Bacteriol. 2008;190:5095–5100. doi: 10.1128/JB.00152-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Gillings M.R., Holley M.P., Stokes H.W. Evidence for dynamic exchange of qac gene cassettes between class 1 integrons and other integrons in freshwater biofilms. FEMS Microbiol. Lett. 2009;296:282–288. doi: 10.1111/j.1574-6968.2009.01646.x. [DOI] [PubMed] [Google Scholar]
- 75.Gillings M.R., Xuejun D., Hardwick S.A., Holley M.P., Stokes H.W. Gene cassettes encoding resistance to quaternary ammonium compounds: a role in the origin of clinical class 1 integrons? ISME J. 2009;3:209–215. doi: 10.1038/ismej.2008.98. [DOI] [PubMed] [Google Scholar]
- 76.Pal C., Bengtsson-Palme J., Kristiansson E., Larsson D.G. Co-occurrence of resistance genes to antibiotics, biocides and metals reveals novel insights into their co-selection potential. BMC Genomics. 2015;16:964. doi: 10.1186/s12864-015-2153-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Schuldiner S. EmrE, a model for studying evolution and mechanism of ion-coupled transporters. Biochim. Biophys. Acta. 2009;1794:748–762. doi: 10.1016/j.bbapap.2008.12.018. [DOI] [PubMed] [Google Scholar]
- 78.Ovchinnikov V., Stone T.A., Deber C.M., Karplus M. Structure of the EmrE multidrug transporter and its use for inhibitor peptide design. Proc. Natl. Acad. Sci. U. S. A. 2018;115:E7932–E7941. doi: 10.1073/pnas.1802177115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Jurasz J., Baginski M., Czub J., Wieczor M. Molecular mechanism of proton-coupled ligand translocation by the bacterial efflux pump EmrE. PLoS Comput. Biol. 2021;17 doi: 10.1371/journal.pcbi.1009454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Shcherbakov A.A., Hisao G., Mandala V.S., Thomas N.E., Soltani M., Salter E.A., et al. Structure and dynamics of the drug-bound bacterial transporter EmrE in lipid bilayers. Nat. Commun. 2021;12:172. doi: 10.1038/s41467-020-20468-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Shcherbakov A.A., Spreacker P.J., Dregni A.J., Henzler-Wildman K.A., Hong M. High-pH structure of EmrE reveals the mechanism of proton-coupled substrate transport. Nat. Commun. 2022;13:991. doi: 10.1038/s41467-022-28556-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Chen Y.J., Pornillos O., Lieu S., Ma C., Chen A.P., Chang G. X-ray structure of EmrE supports dual topology model. Proc. Natl. Acad. Sci. U. S. A. 2007;104:18999–19004. doi: 10.1073/pnas.0709387104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Morrison E.A., Robinson A.E., Liu Y., Henzler-Wildman K.A. Asymmetric protonation of EmrE. J. Gen. Physiol. 2015;146:445–461. doi: 10.1085/jgp.201511404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Li J., Sae Her A., Traaseth N.J. Asymmetric protonation of glutamate residues drives a preferred transport pathway in EmrE. Proc. Natl. Acad. Sci. U. S. A. 2021;118 doi: 10.1073/pnas.2110790118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Amadi S.T., Koteiche H.A., Mishra S., McHaourab H.S. Structure, dynamics, and substrate-induced conformational changes of the multidrug transporter EmrE in liposomes. J. Biol. Chem. 2010;285:26710–26718. doi: 10.1074/jbc.M110.132621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Elbaz Y., Tayer N., Steinfels E., Steiner-Mordoch S., Schuldiner S. Substrate-induced tryptophan fluorescence changes in EmrE, the smallest ion-coupled multidrug transporter. Biochemistry. 2005;44:7369–7377. doi: 10.1021/bi050356t. [DOI] [PubMed] [Google Scholar]
- 87.Brill S., Sade-Falk O., Elbaz-Alon Y., Schuldiner S. Specificity determinants in small multidrug transporters. J. Mol. Biol. 2015;427:468–477. doi: 10.1016/j.jmb.2014.11.015. [DOI] [PubMed] [Google Scholar]
- 88.Wu C., Wynne S.A., Thomas N.E., Uhlemann E.M., Tate C.G., Henzler-Wildman K.A. Identification of an alternating-access dynamics mutant of EmrE with impaired transport. J. Mol. Biol. 2019;431:2777–2789. doi: 10.1016/j.jmb.2019.05.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Saleh M., Bay D.C., Turner R.J. Few conserved amino acids in the small multidrug resistance transporter EmrE influence drug polyselectivity. Antimicrob. Agents Chemother. 2018;62 doi: 10.1128/AAC.00461-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Cho M.K., Gayen A., Banigan J.R., Leninger M., Traaseth N.J. Intrinsic conformational plasticity of native EmrE provides a pathway for multidrug resistance. J. Am. Chem. Soc. 2014;136:8072–8080. doi: 10.1021/ja503145x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Leninger M., Sae Her A., Traaseth N.J. Inducing conformational preference of the membrane protein transporter EmrE through conservative mutations. Elife. 2019;8 doi: 10.7554/eLife.48909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Morrison E.A., Henzler-Wildman K.A. Transported substrate determines exchange rate in the multidrug resistance transporter EmrE. J. Biol. Chem. 2014;289:6825–6836. doi: 10.1074/jbc.M113.535328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Gayen A., Leninger M., Traaseth N.J. Protonation of a glutamate residue modulates the dynamics of the drug transporter EmrE. Nat. Chem. Biol. 2016;12:141–145. doi: 10.1038/nchembio.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Jencks W.P. Utilization of binding energy and coupling rules for active transport and other coupled vectorial processes. Met. Enzymol. 1989;171:145–164. doi: 10.1016/s0076-6879(89)71010-7. [DOI] [PubMed] [Google Scholar]
- 95.Gayen A., Banigan J.R., Traaseth N.J. Ligand-induced conformational changes of the multidrug resistance transporter EmrE probed by oriented solid-state NMR spectroscopy. Angew. Chem. Int. Ed. Engl. 2013;52:10321–10324. doi: 10.1002/anie.201303091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Robinson A.E., Thomas N.E., Morrison E.A., Balthazor B.M., Henzler-Wildman K.A. New free-exchange model of EmrE transport. Proc. Natl. Acad. Sci. U. S. A. 2017;114:E10083–E10091. doi: 10.1073/pnas.1708671114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Hussey G.A., Thomas N.E., Henzler-Wildman K.A. Highly coupled transport can be achieved in free-exchange transport models. J. Gen. Physiol. 2020;152 doi: 10.1085/jgp.201912437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Thomas N.E., Wu C., Morrison E.A., Robinson A.E., Werner J.P., Henzler-Wildman K.A. The C terminus of the bacterial multidrug transporter EmrE couples drug binding to proton release. J. Biol. Chem. 2018;293:19137–19147. doi: 10.1074/jbc.RA118.005430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Michael A.J. Polyamine function in archaea and bacteria. J. Biol. Chem. 2018;293:18693–18701. doi: 10.1074/jbc.TM118.005670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Di Martino M.L., Campilongo R., Casalino M., Micheli G., Colonna B., Prosseda G. Polyamines: emerging players in bacteria-host interactions. Int. J. Med. Microbiol. 2013;303:484–491. doi: 10.1016/j.ijmm.2013.06.008. [DOI] [PubMed] [Google Scholar]
- 101.Wortham B.W., Patel C.N., Oliveira M.A. Polyamines in bacteria: pleiotropic effects yet specific mechanisms. Adv. Exp. Med. Biol. 2007;603:106–115. doi: 10.1007/978-0-387-72124-8_9. [DOI] [PubMed] [Google Scholar]
- 102.Shah P., Swiatlo E. A multifaceted role for polyamines in bacterial pathogens. Mol. Microbiol. 2008;68:4–16. doi: 10.1111/j.1365-2958.2008.06126.x. [DOI] [PubMed] [Google Scholar]
- 103.Codd R., Soe C.Z., Pakchung A.A.H., Sresutharsan A., Brown C.J.M., Tieu W. The chemical biology and coordination chemistry of putrebactin, avaroferrin, bisucaberin, and alcaligin. J. Biol. Inorg. Chem. 2018;23:969–982. doi: 10.1007/s00775-018-1585-1. [DOI] [PubMed] [Google Scholar]
- 104.Abergel R.J., Wilson M.K., Arceneaux J.E., Hoette T.M., Strong R.K., Byers B.R., et al. Anthrax pathogen evades the mammalian immune system through stealth siderophore production. Proc. Natl. Acad. Sci. U. S. A. 2006;103:18499–18503. doi: 10.1073/pnas.0607055103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Hirao T., Sato M., Shirahata A., Kamio Y. Covalent linkage of polyamines to peptidoglycan in Anaerovibrio lipolytica. J. Bacteriol. 2000;182:1154–1157. doi: 10.1128/jb.182.4.1154-1157.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Takatsuka Y., Kamio Y. Molecular dissection of the Selenomonas ruminantium cell envelope and lysine decarboxylase involved in the biosynthesis of a polyamine covalently linked to the cell wall peptidoglycan layer. Biosci. Biotechnol. Biochem. 2004;68:1–19. doi: 10.1271/bbb.68.1. [DOI] [PubMed] [Google Scholar]
- 107.Barbagallo M., Di Martino M.L., Marcocci L., Pietrangeli P., De Carolis E., Casalino M., et al. A new piece of the Shigella pathogenicity puzzle: spermidine accumulation by silencing of the speG gene [corrected] PLoS One. 2011;6 doi: 10.1371/journal.pone.0027226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Chattopadhyay M.K., Tabor C.W., Tabor H. Polyamines protect Escherichia coli cells from the toxic effect of oxygen. Proc. Natl. Acad. Sci. U. S. A. 2003;100:2261–2265. doi: 10.1073/pnas.2627990100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Chan Y.Y., Chua K.L. Growth-related changes in intracellular spermidine and its effect on efflux pump expression and quorum sensing in Burkholderia pseudomallei. Microbiology (Reading) 2010;156:1144–1154. doi: 10.1099/mic.0.032888-0. [DOI] [PubMed] [Google Scholar]
- 110.Ware D., Jiang Y., Lin W., Swiatlo E. Involvement of potD in Streptococcus pneumoniae polyamine transport and pathogenesis. Infect. Immun. 2006;74:352–361. doi: 10.1128/IAI.74.1.352-361.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Jelsbak L., Thomsen L.E., Wallrodt I., Jensen P.R., Olsen J.E. Polyamines are required for virulence in Salmonella enterica serovar Typhimurium. PLoS One. 2012;7 doi: 10.1371/journal.pone.0036149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Sturgill G., Rather P.N. Evidence that putrescine acts as an extracellular signal required for swarming in Proteus mirabilis. Mol. Microbiol. 2004;51:437–446. doi: 10.1046/j.1365-2958.2003.03835.x. [DOI] [PubMed] [Google Scholar]
- 113.Kurihara S., Suzuki H., Tsuboi Y., Benno Y. Dependence of swarming in Escherichia coli K-12 on spermidine and the spermidine importer. FEMS Microbiol. Lett. 2009;294:97–101. doi: 10.1111/j.1574-6968.2009.01552.x. [DOI] [PubMed] [Google Scholar]
- 114.Patel C.N., Wortham B.W., Lines J.L., Fetherston J.D., Perry R.D., Oliveira M.A. Polyamines are essential for the formation of plague biofilm. J. Bacteriol. 2006;188:2355–2363. doi: 10.1128/JB.188.7.2355-2363.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Karatan E., Duncan T.R., Watnick P.I. NspS, a predicted polyamine sensor, mediates activation of Vibrio cholerae biofilm formation by norspermidine. J. Bacteriol. 2005;187:7434–7443. doi: 10.1128/JB.187.21.7434-7443.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Burrell M., Hanfrey C.C., Murray E.J., Stanley-Wall N.R., Michael A.J. Evolution and multiplicity of arginine decarboxylases in polyamine biosynthesis and essential role in Bacillus subtilis biofilm formation. J. Biol. Chem. 2010;285:39224–39238. doi: 10.1074/jbc.M110.163154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Fukuchi J., Kashiwagi K., Yamagishi M., Ishihama A., Igarashi K. Decrease in cell viability due to the accumulation of spermidine in spermidine acetyltransferase-deficient mutant of Escherichia coli. J. Biol. Chem. 1995;270:18831–18835. doi: 10.1074/jbc.270.32.18831. [DOI] [PubMed] [Google Scholar]
- 118.Leuzzi A., Di Martino M.L., Campilongo R., Falconi M., Barbagallo M., Marcocci L., et al. Multifactor regulation of the MdtJI polyamine transporter in Shigella. PLoS One. 2015;10 doi: 10.1371/journal.pone.0136744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Gunn J.S., Lim K.B., Krueger J., Kim K., Guo L., Hackett M., et al. PmrA-PmrB-regulated genes necessary for 4-aminoarabinose lipid A modification and polymyxin resistance. Mol. Microbiol. 1998;27:1171–1182. doi: 10.1046/j.1365-2958.1998.00757.x. [DOI] [PubMed] [Google Scholar]
- 120.Trent M.S., Ribeiro A.A., Lin S., Cotter R.J., Raetz C.R. An inner membrane enzyme in Salmonella and Escherichia coli that transfers 4-amino-4-deoxy-L-arabinose to lipid A: induction on polymyxin-resistant mutants and role of a novel lipid-linked donor. J. Biol. Chem. 2001;276:43122–43131. doi: 10.1074/jbc.M106961200. [DOI] [PubMed] [Google Scholar]
- 121.Larrouy-Maumus G., Skovierova H., Dhouib R., Angala S.K., Zuberogoitia S., Pham H., et al. A small multidrug resistance-like transporter involved in the arabinosylation of arabinogalactan and lipoarabinomannan in mycobacteria. J. Biol. Chem. 2012;287:39933–39941. doi: 10.1074/jbc.M112.400986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Chin C.Y., Zhao J., Llewellyn A.C., Golovliov I., Sjostedt A., Zhou P., et al. Francisella FlmX broadly affects lipopolysaccharide modification and virulence. Cell Rep. 2021;35 doi: 10.1016/j.celrep.2021.109247. [DOI] [PubMed] [Google Scholar]
- 123.Kalienkova V., Clerico Mosina V., Paulino C. The groovy TMEM16 family: molecular mechanisms of lipid scrambling and ion conduction. J. Mol. Biol. 2021;433 doi: 10.1016/j.jmb.2021.166941. [DOI] [PubMed] [Google Scholar]
- 124.Parker J.L., Newstead S. Structural basis of nucleotide sugar transport across the Golgi membrane. Nature. 2017;551:521–524. doi: 10.1038/nature24464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Nji E., Gulati A., Qureshi A.A., Coincon M., Drew D. Structural basis for the delivery of activated sialic acid into Golgi for sialyation. Nat. Struct. Mol. Biol. 2019;26:415–423. doi: 10.1038/s41594-019-0225-y. [DOI] [PubMed] [Google Scholar]
- 126.Tsuchiya H., Doki S., Takemoto M., Ikuta T., Higuchi T., Fukui K., et al. Structural basis for amino acid export by DMT superfamily transporter YddG. Nature. 2016;534:417–420. doi: 10.1038/nature17991. [DOI] [PubMed] [Google Scholar]
- 127.Kim J., Tan Y.Z., Wicht K.J., Erramilli S.K., Dhingra S.K., Okombo J., et al. Structure and drug resistance of the Plasmodium falciparum transporter PfCRT. Nature. 2019;576:315–320. doi: 10.1038/s41586-019-1795-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Barland N., Rueff A.-S., Cebrero G., Hutter C.A.J., Seeger M.A., Veening J.-W., et al. Mechanistic basis of choline import involved in teichoic acids and lipopolysaccharide modification. Sci. Adv. 2022;8 doi: 10.1126/sciadv.abm1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Vastermark A., Almen M.S., Simmen M.W., Fredriksson R., Schioth H.B. Functional specialization in nucleotide sugar transporters occurred through differentiation of the gene cluster EamA (DUF6) before the radiation of Viridiplantae. BMC Evol. Biol. 2011;11:123. doi: 10.1186/1471-2148-11-123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Jack D.L., Yang N.M., Saier M.H., Jr. The drug/metabolite transporter superfamily. Eur. J. Biochem. 2001;268:3620–3639. doi: 10.1046/j.1432-1327.2001.02265.x. [DOI] [PubMed] [Google Scholar]
- 131.Theobald D.L., Wuttke D.S. Divergent evolution within protein superfolds inferred from profile-based phylogenetics. J. Mol. Biol. 2005;354:722–737. doi: 10.1016/j.jmb.2005.08.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Dietmann S., Holm L. Identification of homology in protein structure classification. Nat. Struct. Biol. 2001;8:953–957. doi: 10.1038/nsb1101-953. [DOI] [PubMed] [Google Scholar]
- 133.Brohawn S.G., Campbell E.B., MacKinnon R. Domain-swapped chain connectivity and gated membrane access in a Fab-mediated crystal of the human TRAAK K+ channel. Proc. Natl. Acad. Sci. U. S. A. 2013;110:2129–2134. doi: 10.1073/pnas.1218950110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Singh A.K., Saotome K., Sobolevsky A.I. Swapping of transmembrane domains in the epithelial calcium channel TRPV6. Sci. Rep. 2017;7 doi: 10.1038/s41598-017-10993-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Jaehme M., Guskov A., Slotboom D.J. The twisted relation between Pnu and SWEET transporters. Trends Biochem. Sci. 2015;40:183–188. doi: 10.1016/j.tibs.2015.02.002. [DOI] [PubMed] [Google Scholar]
- 136.Theobald D.L., Miller C. Membrane transport proteins: Surprises in structural sameness. Nat. Struct. Mol. Biol. 2010;17:2–3. doi: 10.1038/nsmb0110-2. [DOI] [PubMed] [Google Scholar]
- 137.Poulsen B.E., Deber C.M. Drug efflux by a small multidrug resistance protein is inhibited by a transmembrane peptide. Antimicrob. Agents Chemother. 2012;56:3911–3916. doi: 10.1128/AAC.00158-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Bellmann-Sickert K., Stone T.A., Poulsen B.E., Deber C.M. Efflux by small multidrug resistance proteins is inhibited by membrane-interactive helix-stapled peptides. J. Biol. Chem. 2015;290:1752–1759. doi: 10.1074/jbc.M114.616185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Spreacker P.J., Thomas N.E., Beeninga W.F., Brousseau M., Porter C.J., Hibbs K.M., et al. Activating alternative transport modes in a multidrug resistance efflux pump to confer chemical susceptibility. bioRxiv. 2022 doi: 10.1101/2021.12.04.471113. [preprint] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Tassoulas L.J., Robinson A., Martinez-Vaz B., Aukema K.G., Wackett L.P. Filling in the gaps in metformin biodegradation: a new enzyme and a metabolic pathway for guanylurea. Appl. Environ. Microbiol. 2021;87 doi: 10.1128/AEM.03003-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Maier L., Pruteanu M., Kuhn M., Zeller G., Telzerow A., Anderson E.E., et al. Extensive impact of non-antibiotic drugs on human gut bacteria. Nature. 2018;555:623–628. doi: 10.1038/nature25979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Ashkenazy H., Abadi S., Martz E., Chay O., Mayrose I., Pupko T., et al. ConSurf 2016: an improved methodology to estimate and visualize evolutionary conservation in macromolecules. Nucl. Acids Res. 2016;44:W344–350. doi: 10.1093/nar/gkw408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Coleman G.A., Davin A.A., Mahendrarajah T.A., Szantho L.L., Spang A., Hugenholtz P., et al. A rooted phylogeny resolves early bacterial evolution. Science. 2021;372 doi: 10.1126/science.abe0511. [DOI] [PubMed] [Google Scholar]
- 144.Gutman N., Steiner-Mordoch S., Schuldiner S. An amino acid cluster around the essential Glu-14 is part of the substrate- and proton-binding domain of EmrE, a multidrug transporter from Escherichia coli. J. Biol. Chem. 2003;278:16082–16087. doi: 10.1074/jbc.M213120200. [DOI] [PubMed] [Google Scholar]
- 145.Mordoch S.S., Granot D., Lebendiker M., Schuldiner S. Scanning cysteine accessibility of EmrE, an H+-coupled multidrug transporter from Escherichia coli, reveals a hydrophobic pathway for solutes. J. Biol. Chem. 1999;274:19480–19486. doi: 10.1074/jbc.274.27.19480. [DOI] [PubMed] [Google Scholar]
- 146.Elbaz Y., Salomon T., Schuldiner S. Identification of a glycine motif required for packing in EmrE, a multidrug transporter from Escherichia coli. J. Biol. Chem. 2008;283:12276–12283. doi: 10.1074/jbc.M710338200. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Sequences and annotation for SMR proteins identified in the bacterial GEBA genome set are available for download from the Deep Blue Data repository hosted by the University of Michigan with unique identifier doi.org/10.7302/0ynd-b343.