Abstract
Rhamnose-rich cell wall polysaccharides (Rha-CWPSs) have emerged as crucial cell wall components of numerous Gram-positive, ovoid-shaped bacteria—including streptococci, enterococci, and lactococci—of which many are of clinical or biotechnological importance. Rha-CWPS are composed of a conserved polyrhamnose backbone with side-chain substituents of variable size and structure. Because these substituents contain phosphate groups, Rha-CWPS can also be classified as polyanionic glycopolymers, similar to wall teichoic acids, of which they appear to be functional homologs. Recent advances have highlighted the critical role of these side-chain substituents in bacterial cell growth and division, as well as in specific interactions between bacteria and infecting bacteriophages or eukaryotic hosts. Here, we review the current state of knowledge on the structure and biosynthesis of Rha-CWPS in several ovoid-shaped bacterial species. We emphasize the role played by multicomponent transmembrane glycosylation systems in the addition of side-chain substituents of various sizes as extracytoplasmic modifications of the polyrhamnose backbone. We provide an overview of the contribution of Rha-CWPS to cell wall architecture and biogenesis and discuss current hypotheses regarding their importance in the cell division process. Finally, we sum up the critical roles that Rha-CWPS can play as bacteriophage receptors or in escaping host defenses, roles that are mediated mainly through their side-chain substituents. From an applied perspective, increased knowledge of Rha-CWPS can lead to advancements in strategies for preventing phage infection of lactococci and streptococci in food fermentation and for combating pathogenic streptococci and enterococci.
Keywords: polysaccharide, rhamnose, rhamnan, rhamnopolysaccharide, cell wall, gram-positive bacteria, ovoid-shaped, teichoic acid, multicomponent glycosylation system, GT-C fold glycosyltransferase, bacteriophage, antibiotic development
Abbreviations: AFM, atomic force microscopy; CPS, capsular polysaccharides; CWPS, cell wall polysaccharides; EPA, enterococcal polysaccharide antigen; GAC, group A carbohydrates; GAS, group A Streptococcus; GBC, group B carbohydrates; GCC, group C carbohydrates; GCS, group C Streptococcus; GroP, glycerol-phosphate; hGIIA, human Group IIA phospholipase A2; HR-MAS NMR, high-resolution magic angle spinning nuclear magnetic resonance; LCP, LytR-CpsA-Psr; LTA, lipoteichoic acid; RBP, receptor-binding protein; RGP, rhamnose-glucose polysaccharide; Rha-CWPS, rhamnose-rich cell wall polysaccharide; SCC, serotype c carbohydrate; TEM, transmission electron microscopy; WTA, wall teichoic acid
Bacteria living in complex ecological niches interact with a wide range of other organisms, such as other bacterial members of the local microbiota, bacteriophage predators, and mammalian host cells. Many of these relationships are mediated by glycan structures covering the bacterial surface, which can interface with the outside world via glycan–protein or glycan–glycan interactions (1, 2). There are several types of glycans, which exhibit a high level of structural diversity. In Gram-negative bacteria, outer membranes are typically rich in lipopolysaccharides, which are formed of a conserved lipid and variable O-antigen polysaccharide chain (3). In both Gram-positive and Gram-negative bacteria, capsular polysaccharides (CPS) form a thick outer layer around bacterial cells, while exopolysaccharides are released in the surrounding medium (4). Finally, in Gram-positive bacteria, wall teichoic acids (WTAs) or cell wall polysaccharides (CWPSs) are covalently anchored onto peptidoglycan and partly embedded inside the peptidoglycan layer (5).
The protective Gram-positive cell wall comprises a thick peptidoglycan sacculus, which maintains a bacterium’s shape and resistance to internal turgor pressure and protects it against external environmental threats (6). The peptidoglycan meshwork is decorated with proteins and other glycopolymers. WTA are commonly found in this setting and can represent up to half the mass of the cell wall like in Bacillus subtilis (7). WTA are anionic glycopolymers, of which the best-characterized examples are made of alditol-phosphate monomers (glycerol- or ribitol-phosphate), possibly including other monosaccharides in their backbone chain and side-chain substituents (mainly sugars and D-alanine). A wide range of functions has been attributed to WTA, including regulation of cell wall growth and morphology, biofilm formation, interactions with bacteriophages, resistance to host cationic antimicrobial peptides, and pathogenicity (7, 8, 9). However, certain Gram-positive bacteria, mainly ovoid-shaped cocci belonging to the genera Streptococcus, Enterococcus, or Lactococcus, synthesize WTA in small amounts or not at all, and most often lack WTA biosynthesis genes. Strikingly, these bacteria instead produce a large amount of CWPS (up to 50% of cell wall mass) that are characterized by a high content of L-rhamnose (Rha), a 6-deoxyhexose commonly found in bacteria but not in humans (10). Different appellations are encountered in the literature for these glycopolymers, such as rhamnopolysaccharide, rhamnose polysaccharide, or, when Glc is present in the backbone chain, rhamnose-glucose polysaccharide (RGP). In this review, we will refer to them as rhamnose-rich CWPS (Rha-CWPS).
Rha-CWPS were initially studied as group-specific polysaccharides (or C-substances) that enabled the serotype classification of streptococci (groups A to G, further extended to V) referred to as Lancefield classification (11). These groups encompass pathogenic species responsible for mild or severe infections in human and animal as well as nonpathogenic species that are part of the commensal human microbiota or widely used in food fermentation. Rha-CWPS share Rha as their major monosaccharide component but differ in their global composition (12). They were found to have a common polyrhamnose core in group A Streptococcus (GAS) (Streptococcus pyogenes), group C Streptococcus (GCS) (including certain strains of Streptococcus dysgalactiae), and Streptococcus mutans, with variation in monosaccharide or disaccharide substituents among species (Fig. 1) (10). In group B (Streptococcus agalactiae), a more complex Rha-CWPS, with multiantenna branching structures, was characterized (13). Because of these affiliations, these Rha-CWPS are also called group A carbohydrates (GAC), group B carbohydrates (GBC), or group C carbohydrates (GCC). It is important to note that the Lancefield serological classification is not able to distinguish streptococci at the species level as defined nowadays on the basis of genomic sequences; this means that strains of different genetic species may synthesize identical Rha-CWPS, whereas within other species, different strains express different types of Rha-CWPS. In more recent years, Rha-CWPS have also been structurally characterized in nonpathogenic food lactic acid bacteria such as Lactococcus lactis (formerly Streptococcus lactis, group N Streptococcus) (14) and Streptococcus thermophilus (15, 16). In these bacteria, Rha-CWPS have received significant attention for their role as receptors for numerous infecting bacteriophages that threaten milk fermentation in the food industry (17, 18, 19). Finally, in pathogenic Enterococcus faecalis (formerly Streptococcus faecalis, group D Streptococcus), Rha-CWPS (also named enterococcal polysaccharide antigen, EPA) was found to influence virulence and colonizing potential in the host gut (20, 21, 22). In all three of these latter species, Rha-CWPS are complex heteropolysaccharides made of a polyrhamnose chain substituted with variable oligosaccharide or polysaccharide chains (Fig. 1).
Figure 1.
Overview of the main Rha-CWPS structures available in streptococci, lactococci, and enterococci.A, schematic representation of their rhamnan backbone. Symbols ∗ and # indicate the branching sites of side-chain substituents represented in panel B. For Streptococcus mutans, the letters refer to the serotypes. B, schematic representation of side-chain substituents. When several substituents are attached on the same rhamnan, they are labeled with the symbols ∗ or # corresponding to the branching sites in Panel A. For polymeric side-chains, the black star (★) indicates the reducing end, which is linked to rhamnan in the final structure; except for GBS, their attachment site is unknown. Phosphoglycerol (GroP) modification of sugars is partial (around 25% on GlcNAc in Streptococcus pyogenes, 70% on GalNAc in Lactococcus lactis IL1403, undetermined for others). GroP could be present on other side chains and lost during extraction and purification. Rha-CWPS, Rhamnose-rich cell wall polysaccharide.
Rha-CWPS are not present in all streptococci. For example, they are absent in Streptococcus pneumoniae, whose cell wall instead contains WTA and lipoteichoic acids (LTAs) (23) and is covered with a capsule, the structure of which is highly variable among strains and allows the definition of at least 90 serotypes (24). However, a Rha-CWPS was also recently characterized in a more distantly related bacterial species, Ruminococcus gnavus, an anaerobic ovoid-shaped Gram-positive bacterium that is a component of the human gut microbiota (25). This finding suggests that the occurrence of Rha-CWPS could be broader than previously anticipated.
The importance and distinctiveness of Rha-CWPS in Gram-positive bacteria were previously presented in a comprehensive review (10). Since then, however, significant progress has been achieved. Novel structures have been identified, subtle but essential modifications of known structures have been discovered, and significant advances have been made regarding their biosynthesis and roles in bacterial physiology and interactions with the environment. Here, we present an update on what is currently known regarding the structure, biosynthesis, and roles of Rha-CWPS, focusing on a few well-characterized ovoid-shaped bacterial species. We describe the complex and diversified arsenal of enzymatic activities dedicated to side-chain substituent biosynthesis, including multicomponent transmembrane glycosylation systems. These systems involve GT-C–fold glycosyltransferases using polyprenyl-monophosphate–linked sugar donors. We discuss emerging data that reveal the importance of Rha-CWPS in bacterial cell wall architecture and cell division. Finally, we summarize their roles, with a particular emphasis on the influence of side-chain substituents, as bacteriophage receptors and in host interactions.
Diversity of Rha-CWPS chemical structures
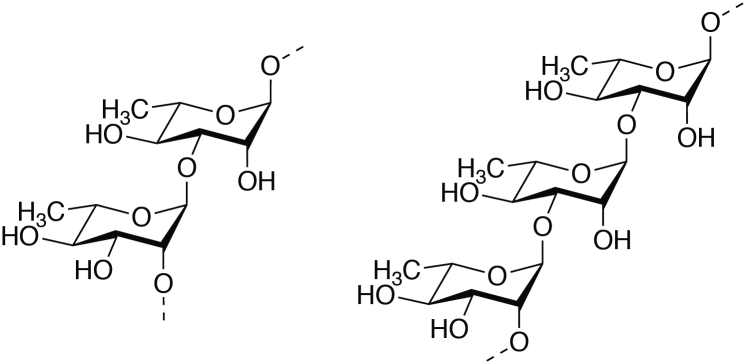
Among different bacterial species, Rha-CWPS share several common structural features, upon which an astonishing variety of chemical refinements are constructed by sophisticated glycopolymer modification systems. The polyrhamnose core is relatively conserved, while the side-chain substituents are characterized by extensive diversity in both size and structure among and even within species. The constituent repeating units of the rhamnose-rich backbone differ with respect to the number of L-Rha residues (between 2 and 5) and the glycosidic linkages between them (Fig. 1A). The most conserved subunit structure is made of two L-Rha residues with alternating α-1,2- and α-1-3-L-Rha (Fig. 2). In certain cases, the repeating unit contains one glucose (Glc), thus accounting for the name RGP. For the sake of simplicity in this text, we will refer to this linear Rha-rich backbone as rhamnan in all cases. The rhamnan polymer is synthesized with a linker unit made of GlcNAc, which may be N-deacetylated to glucosamine (GlcNH2) in certain cases (26). In addition, a cap structure made of sugar has been suggested by experimental data (14, 27) but it is not known at present whether this is a conserved feature of Rha-CWPS.
Figure 2.
Structures of repeating units of rhamnan backbone chains. (Left) Disaccharide subunit (→2-α-L-Rha-(1→3)-α-L-Rha-) identified in several streptococci (GAS, Streptococcus mutans, GCS, GGS) and Lactococcus lactis IL1403. (Right) Trisaccharide subunit (→2-α-L-Rha-(1→2)-α-L-Rha-(1→)3-α-L-Rha-) identified in L. lactis MG1363 and 3107. GAS, group A Streptococcus; GCS, group C Streptococcus; GGS, group G Streptococcus.
Two classes of Rha-CWPS can be distinguished based on their side-chain substituents: the first group has only simple monosaccharide or disaccharide substituents, while the second possesses complex oligosaccharide or polysaccharide chain substituents (Fig. 1B). In S. pyogenes (GAS), GlcNAc monosaccharide is grafted onto the polyrhamnose backbone in all serotypes and is the immunodominant epitope, allowing serological discrimination of GAS (28). Interestingly, the GlcNAc substituents were recently found to be themselves (partly) modified by a glycerol-phosphate (GroP) group (29). It is likely that this modification had not been detected earlier because of the acidic conditions used for extracting Rha-CWPS from the cell wall, which lead to GroP loss. In S. mutans, Glc is the common substituent in three different serotypes, each with a different chemical bond between Glc and Rha: α-1,2- (serotype c), β-1,2- (serotype e), and α-1,3 (serotype f) (30). In addition, GroP was recently shown to be attached to the serotype c carbohydrate and possibly to others as well (31). In GCS, disaccharide substituents comprise two GalNAc units, with a certain proportion of subunits devoid of substituents (32), whereas in group G Streptococcus, they are composed of Rha-GalNAc (33) (Fig. 1B).
In the second class of Rha-CWPS, substituents are far more complex. In S. agalactiae (GBS), a multiantenna polysaccharide was structurally characterized several decades ago as GBC common to all GBS strains (13). The tri-Rha terminal motifs of the phospho-octasaccharide subunits constitute the major antigenic determinant of GBS serological classification. More recently, two elements were highlighted in this complex structure: a stem oligosaccharide made of five Rha residues and a substituting multiantenna structure made of repeated octasaccharide that contains one phosphate (10) (Fig. 1B). The gene cluster involved in GBC biosynthesis is part of the S. agalactiae core genome (34), in agreement with the conserved GBC structure among strains. In contrast, different strains of L. lactis synthesize complex Rha-CWPS that exhibit high structural diversity (35) (Fig. 1B). This structural diversity parallels the genetic variations found within the large gene cluster responsible for their biosynthesis (36). To date, more than 10 chemical structures have been established by 2D-NMR spectroscopy studies in different strains. The variable part of the structures determines their sensitivity to bacteriophages, which suggests the influence of a coevolutionary arms race between phage receptor-binding proteins and their saccharidic ligands at the bacterial surface (17, 19). In general, Rha-CWPS in L. lactis appear to be composed of a rhamnan chain (14) that is covalently linked to a variable oligosaccharide or polysaccharide component exposed at the bacterial surface (Fig. 1B) (37, 38). In several strains, the rhamnan chain is made of identical tri-Rha subunits, while in L. lactis IL1403, it is a disaccharide subunit (27) (Fig. 2) and, in strain UC509.9, a tetrasaccharide with three Rha and one Glc (39) (Fig. 1A). The rhamnan chain may also be substituted with lateral monosaccharide Glc; this was found in very low amounts in strains MG1363 and 3107 (40) but was present on each tri-Rha subunit in strain SK11 (Fig. 1) (36). In most described structures, the polysaccharide substituents are made of pentasaccharide or hexasaccharide subunits containing a phosphate group (Fig. 1B) (37, 38, 41). The oligosaccharide substituents consist of a trisaccharide in strain IL1403 (27) or a hexasaccharide in strain UC509.9 (39), with notable nonstoichiometric GroP substituents in both strains. In S. thermophilus, the structures of two Rha-CWPS (called RGP in the original studies) established by NMR revealed an identical backbone structure, with subunits made of two L-Rha and one Glc, together with lateral oligosaccharide side chains containing similar sugars but different structures (15, 16) (Fig. 1). Finally, in strain V583 of E. faecalis, a complex structure was described for EPA (another type of Rha-CWPS), with a rhamnan backbone covalently bound to teichoic acid–like chains (42) (Fig. 1). Since the epa chromosomal locus exhibits variability among strains, structural diversity is likely in the decorating chains (43).
A conserved ABC transporter–dependent pathway for rhamnan backbone biosynthesis
Biosynthesis of rhamnose-containing polysaccharides requires the sugar-nucleotide dTDP-Rha as a precursor. dTDP-L-Rha is synthesized from Glc-1-phosphate through a four-step enzymatic process involving the enzymes RmlA, B, C, and D (44) (Fig. 3). In L. lactis, all four rmlABCD genes are found in the gene cluster encoding Rha-CWPS biosynthesis (14), while in S. pyogenes and S. agalactiae, only certain rml genes are present in that locus, with others clustered in another part of the genome (45, 46). Because inactivation of one of the rml genes generally leads to severe growth and morphology defects, and even death, the Rml enzymes have been proposed as possible drug targets (47).
Figure 3.
General scheme proposed for rhamnan biosynthesis involving an ABC transporter–dependent pathway. The gene names refer to Lactococcus lactis, but this scheme is generally conserved for all rhamnan chains. Synthesis is initiated at the cytoplasmic side of the membrane, by the transfer of GlcNAc-P onto C55-P by a phosphoglycosyltransferase (TagO). The enzymes RmlA, B, C, and D are involved in synthesis of the rhamnose precursor dTDP-L-Rha. The rhamnosyltransferase RgpA transfers the first rhamnosyl residue onto the C55-PP-GlcNAc lipid intermediate acceptor and the chain is further elongated by two other rhamnosyltransferases, RgpB and RgpF. Chain polymerization is possibly terminated by the addition of a sugar (GlcNAc residue in L. lactis) by RgpE. The ABC transporter (comprising RgpC and RgpD) catalyzes the transfer of rhamnan chains to the outer side of the cytoplasmic membrane. Following export, rhamnan is most probably anchored covalently to peptidoglycan acceptor sites via a phosphodiester linkage to MurNAc (or GlcNAc) residues of the glycan chains, by an LCP transferase (LcpA in L. lactis). On the right side is represented the GT-C–fold glycosyltransferase (WpsJ) involved in side-chain addition onto rhamnan (detailed in Fig. 4). LCP, LytR-CpsA-Psr.
Commonly, besides rml genes, the chromosomal loci involved in rhamnan biosynthesis (most often named rgp) encode several glycosyltransferases, including three predicted rhamnosyltransferases as well as two genes encoding an ABC transporter. Pioneer work on S. mutans proposed a biosynthesis scheme based on an ABC transporter–dependent pathway (48), which appears to be conserved in S. pyogenes (28) as well as in L. lactis (14) and E. faecalis (42). In this pathway, the polysaccharide chain is assembled on a lipid carrier in the cytoplasm and transported across the inner membrane by an ABC transporter. Biosynthesis is initiated on the cytoplasmic side of the cell membrane on a lipid acceptor, presumably undecaprenyl-phosphate (C55-P), by phosphoglycosyltransferase-catalyzed transfer of a sugar-phosphate from the corresponding nucleotide sugar precursor donor (49). In S. mutans, the first enzyme of the biosynthesis pathway was identified (outside the rgp locus) as RgpG, a phosphoglycosyltransferase from the TagO/WecA family that has undecaprenyl-phosphate N-acetylglucosaminyl 1-P transferase activity (50). Thus, GlcNAc-P is the first sugar added to the lipid precursor, giving rise to C55-PP-GlcNAc (Fig. 3). In the final structure linked to peptidoglycan, the GlcNAc residue located at the polymer reducing end constitutes a linkage unit. In most other species, including S. pyogenes, S. agalactiae, and L. lactis, homologs of RgpG (named gacO, gbsO, and tagO, respectively) are also located outside the rgp locus (14, 28, 51), whereas, in E. faecalis, the homolog ef2198 is located at the 5′ end of the epa locus (42). The next step is catalyzed by the rhamnosyltransferase RgpA, which transfers the first rhamnosyl residue from the dTDP-L-Rha precursor onto the GlcNAc-primed lipid intermediate acceptor (52). Two other glycosyltransferases (RgpB and RgpF) elongate the chain, catalyzing the transfer of L-Rha into positions 3 and 2, respectively, of the previous L-Rha. Notably, in L. lactis MG1363, rhamnan is composed of trisaccharide repeating subunits with two 2-linked and one 3-linked α-L-Rha (-2-α-L-Rha-2-α-L-Rha-3-α-L-Rha-) (Fig. 2). The biosynthesis of such a structure requires that one of the two enzymes, RgpB or RgpF, sequentially adds two α-1,2-linked L-Rha, while the other adds only one α-1,3-linked L-Rha; the way in which this process is regulated is not clear at present. In S. thermophilus, the Rha-rich linear core chain is composed of a trisaccharide repeating unit that contains two 2-linked Rha and one 6-linked Glc (-6-α-Glc-2-α-Rha-2-α-Rha-) (Fig. 1). The conserved part of the biosynthesis locus encodes three glycosyltransferases, two with high sequence identity with L. lactis RgpA and RgpB and the third with significantly lower sequence identity with L. lactis RgpF (15). These observations suggest that S. thermophilus RgpF could be the glycosyltransferase that adds Glc in the rhamnan subunits.
Mass spectrometry analysis of rhamnan from L. lactis has detected chains with an average length of 33 Rha residues in strain MG1363 (14) and up to 24 Rha residues in strain IL1403 (27). In addition, mass spectrometry data from L. lactis MG1363 suggest that a terminal cap sugar (GlcNAc or Glc) is present at the nonreducing end of the rhamnan polysaccharide chains. Similarly, in L. lactis IL1403, a putative terminal Glc was detected by NMR. The RgpE glycosyltransferase present in the locus was proposed to be involved in chain termination (35), thereby regulating chain length by a mechanism similar to that previously proposed for other polysaccharides synthesized by an ABC-transporter pathway (3). However, no experimental evidence is available regarding this hypothesis; in L. lactis, rgpE appears to be essential and could not be inactivated (35). Proteins sharing moderate sequence identity with RgpE are encoded in the rgp cluster in S. pyogenes (annotated GacF), S. mutans, and S. thermophilus but their function is also not elucidated. RgpC and RgpD encode a permease and an ATP-binding protein, respectively, and together constitute the ABC transporter responsible for exporting the elongated chain outside the cytoplasmic membrane. In the final step, the rhamnan chain is anchored covalently onto peptidoglycan, most probably by a transferase in the LytR-CpsA-Psr (LCP) family (53), which catalyzes the transfer of the phospho-chain onto MurNAc residues of the peptidoglycan chains (14) (Fig. 3), as described below in more detail.
Certain lactobacilli, but not all of them, synthesize CWPS with a high Rha content (54, 55), but these are not considered part of the Rha-CWPS family because of poor sequence homology at the level of their biosynthesis gene clusters. Instead of being synthesized by an ABC transporter–dependent pathway, like rhamnan (48), lactobacilli CWPS are synthesized by a Wzx/Wzy-dependant pathway (56). In the Wzx/Wzy pathway, repeat units are synthesized on C55-PP at the cytoplasmic side of the membrane, transported to the inner side by a Wzx-type flippase, and polymerized by a Wzy polymerase that transfers the growing CWPS chain from its lipid carrier to the nonreducing end of a newly flipped C55-P-repeat unit (4, 57).
Attachment of side-chain substituents onto the rhamnan backbone: the main role of multicomponent glycosylation systems
As mentioned above, the rhamnan backbone of all Rha-CWPS is substituted with glycan moieties of various sizes and structures, which may be specific to one bacterial species or highly variable among strains within a species. The addition of side-chain substituents to the rhamnan backbone is accomplished through the activity of multicomponent transmembrane glycosylation systems. These systems, which are conserved in both Gram-negative and Gram-positive bacteria, are composed of three or four proteins that perform extracytoplasmic glycosylation of glycopolymers such as lipopolysaccharides, WTA, LTA, and CWPS (58, 59, 60). They involve two types of glycosyltransferases belonging to GT-A or GT-C superfamilies that are defined on the basis of structural features (61). GT-A–fold enzymes are characterized by a Rossman domain with several α-helices flanking a β-sheet core on both sides. They contain a DXD motif and a divalent cation at their catalytic site and exclusively use nucleotide-sugar donors as substrates (62). GT-C–fold glycosyltransferases are integral membrane proteins with a modified DXD signature and exclusively utilize lipid-linked sugar donors (63).
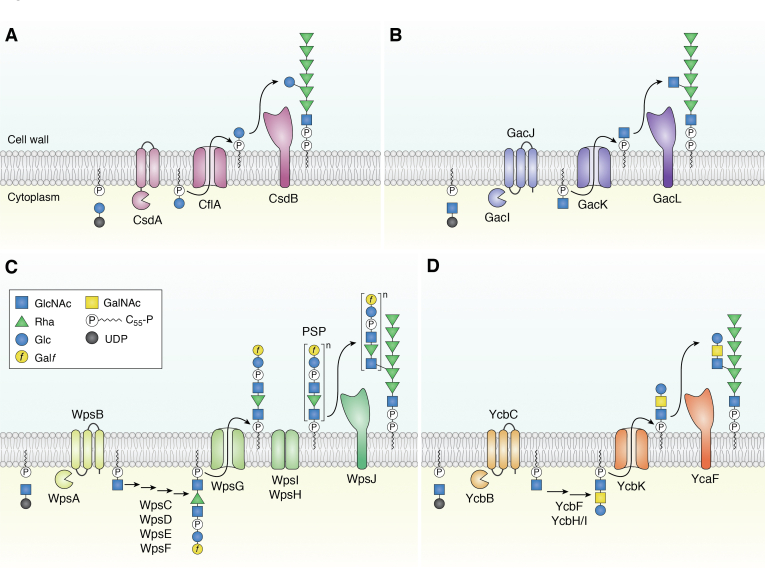
The three-component glycosylation system includes: (i) a membrane-associated GT-A–fold glycosyltransferase, which transfers the modifying sugar from the UDP-sugar donor to a lipid carrier, C55-P, to form a C55-P-sugar intermediate at the inner side of the cytoplasmic membrane; (ii) a flippase of the GtrA family with four transmembrane segments, which flips the C55-P-sugar through the membrane; and (iii) an integral membrane glycosyltransferase with a GT-C fold, which transfers the sugar from the lipid carrier to the final glycopolymer at the outer side of the cytoplasmic membrane (Fig. 4A) (58). More recently, a four-component glycosylation system has also been described, featuring a GT-A–fold glycosyltransferase and a small membrane protein activator to catalyze C55-P-sugar formation, a flippase with 12 transmembrane segments belonging to the Wzx rather than the GtrA family, and finally, a GT-C fold glycosyltransferase (Fig. 4B) (60). Unlike the GT-A–fold components, the GT-C fold glycosyltransferases in these systems do not exhibit sequence conservation, which reflects their specificity toward their diverse glycan acceptors. They are instead identified as integral membrane proteins (with 8–13 transmembrane helices) that share predicted structural similarity (as detected by HHpred software (64)) with other membrane-associated GT-C–fold glycosyltransferases that have an established 3D-structure, including 4-amino-4-deoxy-L-arabinose (L-Ara4N) transferase (ArnT) and archaeal oligosaccharyltransferase (AlgB).
Figure 4.
Schematic representation of side-chain substituent synthesis and addition to rhamnan chains by multi-component glycosylation systems.A, three-component glycosylation system. The model depicts extracytoplasmic addition of Glc to rhamnan in Lactococcus lactis. This system includes (i) a membrane-linked GT-A fold glycosyltransferase (CsdA), which transfers the modifying sugar (Glc) from the UDP-sugar donor to the lipid carrier, C55-P, to form a C55-P-sugar intermediate at the inner side of the cytoplasmic membrane; (ii) a flippase of the GtrA family which flips the C55-P-sugar through the membrane (CflA); and (iii) an integral membrane glycosyltransferase with a GT-C fold (CsdB) which transfers the sugar from the lipid carrier to the final glycopolymer (rhamnan) at the outer side of the cytoplasmic membrane. Such a system is also used to add Glc onto PSP subunits in L. lactis. B, four-component glycosylation system. The scheme depicts the extracytoplasmic addition of GlcNAc onto rhamnan in Streptococcus pyogenes. This system uses two proteins instead of one to generate C55-P-sugar (GlcNAc in this example): a glycosyltransferase with a GT-A fold (GacI) and a small membrane protein activator (GacJ) to catalyze the process. The remaining two components are a flippase (GacK) belonging to the Wzx family and a GT-C–fold glycosyltransferase (GacL), which transfers the sugar from the lipid carrier to the final glycopolymer (rhamnan) at the outer side of the cytoplasmic membrane. In S. pyogenes, GlcNAc is partly modified by GroP (see Fig. 1B), added extracellularly by GacH (not shown on the scheme). C, complex modification system derived from the four-component glycosylation system allowing to synthesize polymeric PSP substituents as described in L. lactis MG1363 (type C strain). Synthesis starts by the transfer of GlcNAc to C55-P by WpsA/WpsB (GacI/GacJ homologs). The repeat unit is then synthesized on the C55-P-GlcNAc lipid precursor at the intracellular face of the cytoplasmic membrane by the successive addition of (phospho)monosaccharides catalyzed by various glycosyltransferases (WpsC,D,E,F). The linear pentasaccharide PSP repeat unit is flipped to the outside of the cytoplasmic membrane by a flippase of the Wzx family (WpsG). It is polymerized by WpsI, with WpsH as a copolymerase and/or chain-length regulator. Finally, the GT-C–fold glycosyltransferase WpsJ transfers the PSP onto rhamnan. It is worth noting that, in L. lactis MG1363, the final PSP (with an hexasaccharide repeat unit) also has a side-chain Glc (not shown on the scheme, see Fig. 1B), presumably added outside the cytoplasmic membrane by a distinct three-component glycosylation mechanism (see Fig. 3A). D, complex modification system derived from the four-component glycosylation system allowing to synthesize oligosaccharide substituents as described in L. lactis IL1403 (type B strain). In type A and type B strains, the side-chain substituents are oligosaccharides. The mechanism proposed for synthesis and anchoring is similar to the one described for polymeric PSP in panel C, without the proteins involved in the polymerization of subunits (WpsI and H). In IL1403, GalNAc is partly modified by GroP (see Fig. 1B), presumably added intracellularly (not shown on the scheme). GAC, group A carbohydrates; GroP, glycerol-phosphate.
Monosaccharide substituents
In L. lactis, a three-component glycosylation system—comprising the two glycosyltransferases CsdA (GT-A fold) and CsdB (GT-C fold) encoded by two adjacent genes, and a flippase (CflA) encoded in another chromosomal locus—was shown to be involved in the addition of side-chain Glc to the rhamnan backbone (Fig. 4A) (40). In WT L. lactis NZ9000 grown in laboratory conditions, the amount of Glc grafted onto rhamnan is very low. However, in a mutant strain that overexpressed the two csdAB genes, nearly all [-2-α-Rha-2-α-Rha-3-α-Rha-] subunits were found to possess a side-chain Glc connected to the 3-Rha. The typical level of rhamnan glucosylation appears to vary between strains, since NMR analysis of L. lactis SK11 revealed that each rhamnan subunit structure contained a side-chain Glc (36). The environmental or cellular signals that could possibly modulate rhamnan glucosylation have not been identified.
In S. pyogenes, elucidation of the mechanism of side-chain GlcNAc addition to the polyrhamnose backbone of GAC revealed that this process relies on a four-component glycosylation system (Fig. 4B) (60). In the first step, a GT-A–fold glycosyltransferase, GacI, transfers GlcNAc from UDP-GlcNAc to C55-P; this activity is strongly enhanced by a second small membrane protein (GacJ) with three transmembrane helices. The next step is performed by a flippase (GacK) belonging to the Wzx family and in the final step, a polytopic GT-C glycosyltransferase, GacL, catalyzes the transfer of GlcNAc onto rhamnan.
Homologs of GacI and GacJ are present in E. faecalis and numerous other bacteria, suggesting that similar modes of GlcNAc grafting onto rhamnan can occur in other bacteria (60). In E. faecalis V583, the Rha-CWPS (EPA) has a rhamnan backbone made of five Rha, with side-chain monosaccharides that include Glc and GlcNAc (Fig. 1A). The proteins involved in the addition were not identified experimentally, but it is possible that the GacI/J homologs EF2190 and EF2189, which are present in the epa locus of E. faecalis, could be involved in GlcNAc side-chain linkage (42). Glc side chains might then be added by an as-yet-unidentified three-component glycosylation system.
Complex oligosaccharides or polysaccharide substituents
In L. lactis, the structural diversity of Rha-CWPS is strikingly high, mostly as a result of extensive variability in complex side-chain substituents (Fig. 1B). In addition to the Glc monosaccharide found on rhamnan (most often in low amounts as described above), the rhamnan chain is also decorated with oligosaccharide or polysaccharide substituents. CWPS biosynthesis is encoded in a large genomic cluster of around 20 kb, with a more-conserved 5′-positioned region encoding rhamnan synthesis and a more-variable 3′-located region that was shown to encode the biosynthesis of oligosaccharides or polysaccharide substituents. Bioinformatic analysis of this gene cluster has led to the identification of four genotypes of L. lactis strains, that is, types A, B, C, and D (36). In C-type strains, for which most structures are known, polysaccharide substituents (called polysaccharide pellicle or PSP) are composed of repeating pentasaccharide or hexasaccharide subunits that contain phosphate for most of them (Fig. 1B) (36, 37, 38, 41). (Fig. 1B). They have been shown to be covalently attached to the rhamnan backbone (14), although the exact nature of the chemical linkage remains to be established. In A- and B-type strains, Rha-CWPS structures consist of a rhamnose-rich chain or rhamnan, with short oligosaccharide substituents made of six or three residues (27, 39) (Fig. 1B).
Based on experimental data and bioinformatic analysis, recent work has proposed a comprehensive biosynthesis scheme for the complete heteropolysaccharide, comprising both rhamnan and PSP in L. lactis (35). In the proposed model, a complex multicomponent glycosylation system—which appears to be an evolutionary refinement of the four-component system—allows complex PSP chains to be synthesized and attached to the rhamnan backbone as glycosidic substituents (Fig. 4C). PSP synthesis is initiated by the WpsA and WpsB proteins (homologs of GacI/GacJ), which transfer GlcNAc from UDP-GlcNAc to C55-P, generating a monophosphate intermediate, C55-P-GlcNAc. In the case of L. lactis MG1363, the PSP subunit is then further elongated by four glycosyltransferases, WpsC/D/E/F, in the cytoplasm. The flippase WpsG, belonging to the Wzx family, flips the C55-P-oligosaccharide outside the cytoplasmic membrane, and a polytopic GT-C–fold glycosyltransferase, WpsJ, transfers the PSP (or a first PSP subunit) from the lipid intermediate onto rhamnan. In C-type strains, the polymerization of oligosaccharide subunits to generate PSP involves WpsI, a GT-C fold glycosyltransferase that is able to use C55-P-oligosaccharide as a donor. This process is thought to be regulated by WpsH, which is predicted to have two transmembrane helices and one large extracellular loop, similar to the Wzz proteins that have been proposed to act as copolymerases regulating (lipo)polysaccharide length in the Wzx/Wzy pathway (57). Remarkably, WpsA/B, WpsG flippase, and GT-C–fold WpsJ are all conserved components encoded in the cwps gene cluster in all L. lactis strains. Therefore, the proposed model for synthesis and attachment of side-chain substituents to rhamnan, involving a multicomponent glycosylation system, is relevant for all L. lactis strains with PSP substituents (e.g., type C strains) or oligosaccharide substituents (e.g., type A and B strains) (Fig. 4D). Notably, WpsI and H are absent in type A and B strains, which is consistent with their proposed function in PSP polymerization (35, 36).
In S. thermophilus, two different structures of Rha-CWPS (named RGP) have been recently described, characterized by trisaccharide or tetrasaccharide side chains with a GlcNAc at their reducing end (15, 16) (Fig. 1). Homologs of genes encoding WpsA, B, H, and J are present in the rgp biosynthesis locus. Thus, the process of anchoring the side-chain oligosaccharide onto the rhamnan chain likely follows a similar scheme as that proposed for L. lactis (Fig. 4D). Interestingly, in pathogenic S. agalactiae, homologs of WpsA, WpsB, WpsH, and WpsI are encoded in the biosynthesis locus of GBC (46), which suggests some degree of conservation between the complex scheme employed for L. lactis Rha-CWPS biosynthesis and that used for GBC.
Teichoic acid–like structures as substituents
The complete structure of the Rha-CWPS (EPA) of E. faecalis V583 was recently established (42). It is composed of a rhamnan backbone with partial substitution of monosaccharides including Glc and GlcNAc and in addition, highly complex decorations of teichoic-like structures (Fig. 1). These structures consist of phosphopolysaccharide chains that are reminiscent of the phosphate-containing PSP chains of L. lactis type C strains. However, they are characterized by the presence of ribitol, a typical constituent of WTA (7), in their subunit (indeed, these chains were previously isolated as WTA (65)). A high degree of genetic diversity has been identified in the locus encoding EPA biosynthesis, suggesting that these decorations may be likewise diverse in structure. Notably, a similarly variable genetic locus has been identified in Enterococcus faecium, indicating that this bacterial species also synthesizes complex Rha-rich CWPS with variable substituents (43).
Interestingly, NMR experiments detected poly-GroP chains associated with Rha-CWPS isolated from R. gnavus, which may thus potentially represent another case of a teichoic acid substituent of Rha-CWPS (25). In the type D strain L. lactis 184, the PSP substituent of rhamnan was found to contain one ribitol-P in its subunit and alanine substituents and could also be classified as a teichoic acid–like structure (36). Despite this difference, strain 184 still possesses all of the features described above for rhamnan modification by a complex four-component mechanism, including WpsA/B, WpsG flippase, and WpsJ GT-C–fold glycosyltransferase (35). In contrast, the way in which teichoic acid chains are assembled and transferred onto rhamnan chains in E. faecalis is less clear. The current model for this biosynthesis pathway (42) hypothesizes that the teichoic acid subunits are synthesized as a C55-PP-subunit inside the cytoplasm (similarly to WTA), flipped outside the cytoplasmic membrane by a Wzx flippase, and polymerized by a Wzy-like polymerase, at which point the chains might subsequently be attached extracellularly to rhamnan by a protein in the LCP family.
Secondary modifications of glycosidic substituent groups
Another layer of complexity in the structures of certain Rha-CWPS arises from chemical modifications of the glycosidic substituents of rhamnan chains. These modifications include the addition of side-chain monosaccharides or GroP groups.
NMR analysis of several type C strains of L. lactis, such as MG1363 or SMQ388, has revealed side-chain Glc branched onto the oligosaccharide subunit of PSP (Fig. 1B) (37, 41). Although the Glc side-chain is present in every subunit, it is not synthesized in the cytoplasm as part of the subunit but was shown to be grafted onto it on the outer side of the cytoplasmic membrane by a three-component system (40) (Fig. 4A). The two glycosyltransferases involved, CsdC and CsdD (GT-A and GT-C fold, respectively), appear to work in concert with the putative flippase CflA. Remarkably, the CflA protein is also involved in the addition of Glc to rhamnan by CsdAB in the same strain, as well as the addition of Gal on teichoic acid chains by CsdEF (40). The fact that the CflA protein is shared by three different three-component glycosylation systems in L. lactis suggests the existence of possible cross-talk between the three systems dedicated to the glycosylation of cell wall glycopolymers.
Another type of secondary modification described in S. pyogenes rhamnan (GAC) consists of the addition of a GroP group to the C6 hydroxyl group of GlcNAc substituents (29). This decoration, found on approximately 25% of GlcNAc, provides negatively charged groups that transform GAC into a polyanionic cell wall glycopolymer. GroP transfer is catalyzed by the enzyme GacH, which is a membrane protein in the same family as LtaS (involved in synthesizing the poly(GroP) backbone chains of LTA (66)). As for LtaS, the most likely donor substrate for GacH is phosphatidylglycerol, and GroP addition is thought to take place extracellularly. Since GacH is conserved in numerous streptococci, GroP substituents could be a widely distributed modification of Rha-CWPS in these bacteria, as confirmed in particular in S. mutans (29).
In type A and B strains of L. lactis, NMR analysis of Rha-CWPS has also detected nonstoichiometric GroP substituents located on branching oligosaccharide side chains of rhamnan (Fig. 1B) (27, 39). However, no GacH homologs are present in these L. lactis genomes, and the genes involved in GroP addition have not yet been identified. One putative candidate has been reported from the cwps cluster of the type B strain IL1403: the gene ycbG, which encodes a protein with a LicD domain (36) that is associated with phosphocholine or phosphosugar transferase activity (23) and is predicted by HH-Pred software to exhibit structural similarity with fukutin, a eukaryotic ribitol-P transferase (67). Moreover, the same cwps locus also encodes a putative glycerol-3-phosphate-cytidylyltransferase (TagD1) involved in CDP-Gro synthesis, which could provide a donor substrate to YcbG. Since YcbG is devoid of a predicted signal peptide, GroP modifications would most likely be added intracellularly. Homologs of these two proteins are also present in strain UC509.9, in which Rha-CWPS also bear oligosaccharide side chains partly substituted with GroP (Fig. 1B).
As described above, GroP groups provide negative charges to components of Rha-CWPS. Other structures contain chain substituents with oligosaccharide subunits linked by phosphodiester bonds, like PSP in L. lactis or teichoic acid–like structures in E. faecalis (Fig. 1). Thus, as a whole, Rha-CWPS appear to be polyanionic cell wall glycopolymers, similar to WTA. In a few cases, no phosphate group has been identified in the Rha-CWPS structures, but there is a possibility that it could be lost during acidic extraction procedures, as was reported for GAC in S. pyogenes (26).
Covalent attachment of Rha-CWPS to peptidoglycan by LCP transferases
LCP proteins were first identified as the enzymes involved in the ligation of WTA onto peptidoglycan (68). They are phosphotransferases that catalyze the transfer of the reducing end of the glycopolymer chain to peptidoglycan from a lipid carrier–bound intermediate (C55-PP-glycopolymer); this is accomplished via the formation of a phosphodiester bond with the C6 hydroxyl group of MurNAc (or GlcNAc) residues of the glycan chains of peptidoglycan (53, 69, 70). LCP proteins, initially characterized in B. subtilis, are widely distributed in Gram-positive bacteria. The typical structure has an N-terminal transmembrane domain for anchoring and a C-terminal catalytic domain. Several paralogs are often present within a single species and their respective roles remain to be assigned. Nevertheless, it is clear that their function encompasses anchoring of WTA as well as other CWPS (53).
The extraction of Rha-CWPS from the cell wall generally requires harsh acid treatments or the digestion of peptidoglycan by mutanolysin, indicating that, like WTA, they are also covalently bound to peptidoglycan. The involvement of LCP proteins in the anchoring of rhamnan to peptidoglycan was investigated in L. lactis (14). Two lcp paralogs, named lcpA and lcpB, are present in L. lactis but neither is in the close vicinity of the cwps gene cluster. Experimental data pointed toward the involvement of LcpA but not LcpB in anchoring rhamnan onto peptidoglycan: the lcpB gene could be deleted whereas lcpA could not. In a mutant strain with attenuated expression of lcpA, alterations in the cell wall structure were noted, along with the presence of free rhamnan in the cell wall (revealed by high-resolution magic angle spinning nuclear magnetic resonance (HR-MAS NMR) spectroscopy). These data strongly suggested that LcpA, but not LcpB, participates in the anchoring of rhamnan to peptidoglycan. Similarly, in S. mutans, the two LCP proteins, BrpA and Psr, were found to play a role in Rha-CWPS attachment to the cell envelope. Their inactivation significantly reduced quantities of cell wall–associated Rha-CWPS and led to concurrent extracellular accumulation in the culture supernatant (71).
Generally speaking, Rha-CWPS are most likely covalently linked via a GlcNAc-phosphodiester bond to the C6 hydroxyl group of MurNAc. Until very recently, though, the chemical structure of the linkage unit had not been experimentally established. Work in S. pyogenes revealed that GAC is linked to peptidoglycan through glucosamine-1-phosphate rather than the expected GlcNAc-P, which explains why GAC is not released from cell walls by mild acid treatment (26). This GlcN residue is formed by enzymatic N-deacylation of the GlcNAc residue located at the reducing end of the rhamnan chain after its synthesis, and this modification is catalyzed by a de-N-acetylase named PplD. Homologs of this enzyme are present in many streptococci and lactococci, in which they probably perform the same function. In L. lactis, extraction of rhamnan requires harsh acid treatment, which suggests similar de-N-acetylation of GlcNAc in the linkage unit. Importantly, by introducing positive charges (located on GlcN) inside the cell wall, this modification has functional implications, one of them being an increased resistance to host cationic antimicrobial peptides.
In S. agalactiae, biochemical evidence was obtained that Rha-CWPS (GBC) is covalently bound to MurNAc, whereas capsular polysaccharide is attached via a phosphodiester bond to GlcNAc (72). The presence of a phosphodiester bond between GBC and peptidoglycan is likely if an LCP protein is involved in its attachment to peptidoglycan.
Interestingly, the C6 hydroxyl group of MurNAc, on which Rha-CWPS are covalently bound, is also the site of O-acetyl group addition, catalyzed by O-acetyltransferase, on the peptidoglycan chain. Peptidoglycan O-acetylation has an important role in bacterial physiology and can, in particular, modify peptidoglycan sensitivity to cleavage by endogenous peptidoglycan hydrolases (73, 74). Since both peptidoglycan modifications likely take place at the septum during cell division, competition between Rha-CWPS and O-acetyl groups for attachment could constitute a possible regulatory mechanism of cell wall biogenesis, which warrants further investigation.
Localization of Rha-CWPS in the cell wall
The initial characterization of Rha-CWPS in streptococci as carbohydrate antigens that enable serotype identification of streptococci suggests that they are accessible to antibodies at the bacterial surface. Nowadays, the spatial organization of the glycopolymer constituents in the cell wall can be assessed at the nanoscale with powerful biophysical techniques (75). The application of both transmission electron microscopy (TEM) and atomic force microscopy (AFM) in L. lactis revealed that the variable PSP side-chains of Rha-CWPS form a layer outside the bacterial cell (37). With TEM, PSP was visualized as a dark layer outside the cell, which disappeared in a mutant devoid of PSP. Topographic imaging of the surface of live bacteria by AFM revealed that the surface of WT bacteria was smooth, while in the absence of PSP, the surface appeared rough. The surface localization of PSP was also supported by an analysis of intact bacterial cells by HR-MAS NMR, a technique that makes it possible to directly analyze surface-exposed and/or flexible components of intact bacteria (76, 77). HR-MAS NMR was applied to WT L. lactis and revealed the presence of surface-exposed flexible PSP, but not rhamnan (14). Instead, in a mutant devoid of PSP, rhamnan was detected at the surface, indicating that it became exposed in the absence of PSP and/or became more flexible inside the cell wall. These results demonstrated that the rhamnan core is trapped inside the peptidoglycan layer, while the PSP substituents are exposed at the bacterial surface (Fig. 5). This spatial organization is consistent with PSP’s role as a receptor that is recognized by bacteriophage receptor-binding protein (RBP) and required for phage adsorption at the bacterial surface (38). When AFM was conducted with tips functionalized (with LysM domain, specifically) to recognize GlcNAc of glycan chains in peptidoglycan, it revealed that peptidoglycan was accessible at the surface only in mutant L. lactis devoid of PSP pellicle (Fig. 5). Moreover, the results of this study indicated that peptidoglycan may be organized as cable-like structures around the main axis of the cell (78). Since these experiments were performed before the discovery of rhamnan, it would be interesting to investigate whether rhamnan participates in the formation of these cable-like structures.
Figure 5.
Schematic view of the localization of Rha-CWPS in the cell wall of various Gram-positive bacteria. The peptidoglycan network is represented in gray. Rhamnan polymers are shown as chains of green circles and polymeric side-chain substituents (PSP or teichoic acid (TA)-like) as chains of orange circles. When present, capsular polysaccharides (CPSs) are shown as chains of purple circles. LTA are shown as chains of brown hexagons. The position of the different polysaccharide chains relatively to the peptidoglycan network is deduced from the results of the various biophysical approaches applied on the different bacterial species, as described in the text. Rha-CWPS, Rhamnose-rich cell wall polysaccharide; LTA, lipoteichoic acid.
Similar HR-MAS NMR studies on E. faecalis cells revealed that the teichoic acid decorations of Rha-CWPS are exposed at the bacterial surface, together with the CPS produced by this bacterium (Fig. 5). Like in L. lactis, the rhamnan chain was embedded in the cell wall, reducing its mobility and surface exposure (42).
In GBS, TEM and AFM analyses indicated that peptidoglycan is covered by a thin layer of Rha-CWPS (GBC) that is overlaid by a thicker layer of capsular polysaccharide (79). In addition, AFM molecular imaging with specific lectin probes showed that removal of CPS enables the detection of peptidoglycan at the bacterial surface, indicating that GBC does not prevent peptidoglycan detection and thus that it is most probably partly embedded in the peptidoglycan layer (Fig. 5).
Finally, in S. thermophilus, the short oligosaccharide substituents of the Rha-rich backbone were shown to be involved in phage adsorption (15). They were detected by a fluorescence microscopy experiment on whole bacterial cells in which the RBP of an infecting phage was fused with GFP, thus confirming that oligosaccharide side chains are exposed at the bacterial surface.
Role in bacterial cell morphogenesis and division
In Gram-positive bacteria, the peptidoglycan sacculus is considered the major structural component of the cell wall as it plays a primary role in maintaining cell shape and integrity during growth and division. Peptidoglycan biosynthesis is a complex process requiring numerous enzymes and regulators to be assembled in protein complexes which are finely regulated in time and space (80, 81). However, it now appears that other cell wall glycopolymers, such as WTA (82, 83) and Rha-CWPS, are also essential for cell wall homeostasis and cell division.
In L. lactis, efforts to inactivate the tagO gene, which initiates rhamnan chain biosynthesis, have not been successful, suggesting that rhamnan is an essential component of the cell wall. When tagO expression was attenuated, dramatic alterations were observed in cell shape and septum positioning, which provides evidence for the importance of Rha-CWPS in cell wall biogenesis (14). Remarkably, the impairment of PSP (side-chain substituents) synthesis also resulted in prominent alterations to cell shape and division, leading to the chaining and sedimentation of L. lactis mutants (35, 37). This phenotype was observed in particular, in a NZ9000-wpsA mutant in which the initial PSP biosynthesis step—the transfer of GlcNAc-P to C55-P—was compromised. With the inactivation of wpsA, C55-P is not sequestered in PSP biosynthetic intermediates, and thus this mutation should not impair peptidoglycan synthesis. Therefore, these results suggest that PSP, the variable component of L. lactis Rha-CWPS, is required for the proper functioning of the cell division machinery in L. lactis (35). In GBS and GAS, complete loss of Rha-CWPS production was obtained by inactivating the gbsO or gacO genes. The resulting mutants exhibited severe growth defects, morphological alterations, and cell division abnormalities (28, 51). In the gbcO mutant strain, reduced peptidoglycan cross-linking was observed, indicating functional alterations in the machinery of peptidoglycan synthesis. In addition, mislocation of the peptidoglycan hydrolase PcsB, which plays a significant role in cell division and separation, was detected (51). A recent study elegantly illuminated the function of Rha-CWPS in S. mutans and more specifically, that of its decorations (31). The S. mutans serotype c carbohydrate (SCC) has a rhamnan backbone with side-chain Glc that is decorated by GroP chemical groups. It was demonstrated that this modification is required first to ensure proper localization of the peptidoglycan hydrolase AtlA, which is involved in daughter cell separation and in addition, to control the localization of MapZ, a protein previously shown to guide FtsZ-ring positioning at the cell equator during cell division (84, 85). AtlA was shown to bind to SCC rhamnan through its Bsp domains, but this binding is prevented by GroP modifications, thus suggesting that immature SCC recruits AtlA. In the absence of GroP modifications on SCC, AtlA was mislocated, resulting in bacterial autolysis. To date, the mechanism by which MapZ recognizes immature SCC is unknown. This study highlights the critical role of Rha-CWPS modifications in cell division in S. mutans.
Role of Rha-CWPS as bacteriophage receptors
Bacteriophages—viruses that infect bacteria—have evolved to recognize, adhere to, infect, and replicate in their bacterial host, which typically results in host death by cell lysis. In the context of increasing antibiotic resistance, phages that target bacterial pathogens are considered promising new tools for therapy (86). In contrast, phages that infect lactic acid bacteria in the course of food fermentation have serious detrimental effects, particularly in milk fermentation, leading to poor quality products and economic losses (18, 87). In all cases, deciphering the molecular basis of the host-phage recognition mechanisms is of critical importance. An early and essential step for the success of the phage infection cycle is the adsorption of phage particles to the host surface, which occurs through specific recognition of a component—either a protein or glycopolymer—of the bacterial outer layer. Because of their exposure on the cell surface, Rha-CWPS are exploited as phage receptors in numerous species.
Early studies examined the involvement of Rha-CWPS in bacteriophage adsorption at the surface of many streptococci, with a specific focus on the side-chain substituents of the rhamnan backbone (10). For example, in S. mutans, the α-1,2-linked Glc side chain of serotype c is recognized by specific phages for adsorption (88). Similarly, monosaccharide substituents of WTA are also crucial determinants of phage adsorption in Listeria monocytogenes (89) or Staphylococcus aureus (90).
In L. lactis, the isolation of bacteriophage-insensitive mutants and the identification of the mutations responsible pointed toward CWPS as receptors for phages in the 936 family (now termed skunaviruses), at a time when the L. lactis Rha-CWPS structure was not yet characterized at the molecular level (91). At present, hundreds of phages are known to infect lactic acid bacteria, particularly L. lactis, making this system one of the most advanced models of phage–host interactions that involve a carbohydrate receptor (17). The variable part of Rha-CWPS, the PSP or oligosaccharide component, has been shown to serve as the ligand of numerous siphophage RBPs (19, 92). Moreover, differences in the structure of lactococcal PSP act as the primary factor determining hosts’ bacteriophage sensitivity and explain, at least partially, the narrow host range of lactococcal phages (38). A study utilizing surface plasmon resonance was able to characterize the molecular-level interactions between phage p2 RBP and its cognate saccharidic receptor (93). The authors reported that the relatively low affinity (compared to that observed for a protein receptor) measured between p2 RBP and its saccharidic receptor is most likely compensated by the high avidity effect provided by the multiple receptor-binding sites of the baseplate. Moreover, the interaction between strain SMQ388 and phage 1358 was investigated by determining the crystal structure of the phage RBP in complex with a chemically synthesized trisaccharide derived from the host PSP (94). Further modeling of the complete hexasaccharide subunit of SMQ388 PSP showed it docked into the RBP crevice. In addition to RBP, the distal tail (Dit) protein of siphophages contains other carbohydrate-binding modules that have an auxiliary role in infection. They recognize the same receptor as RBP and may help orient the viral particles for DNA injection (95). Glucosylation of both rhamnan and PSP was shown to increase resistance to phage predation, particularly in the presence of high salt concentrations (conditions encountered during cheese manufacturing) (40). Recent work performed on S. thermophilus also showed that the variable short oligosaccharide side chains of Rha-CWPS constitute the molecular moiety recognized by phages for adsorption (15). Overall, evidence from L. lactis and S. thermophilus points to the variable components of complex Rha-CWPS acting as phage receptors, with their structural variability serving as the primary determinant of sensitivity to phages. The progress achieved in deciphering the molecular-level interactions between bacteriophages and their bacterial hosts has opened the door for new strategies for selecting dairy starter strains that are resistant to phage attack (36).
In E. faecalis, an inhabitant of the human gastrointestinal tract and a cause of nosocomial infections, phage therapy could provide a means of curing infections by multidrug-resistant enterococci (96). Several studies have shown that the enterococcal Rha-CWPS, EPA, is the receptor of some lytic phages, and in particular, that EPA decorations play a role in phage adsorption (97, 98). Exposure to specific phages, both in vitro and in vivo in the mouse gut, results in the emergence of resistant mutants with mutations located in the variable epa genes, which could possibly hamper phage therapy. However, these mutants had reduced colonization potential and/or reduced sensitivity to cell wall–targeting antibiotics (beta-lactams or vancomycin), possibly because of fitness defects. Thus, the most promising strategy to target enterococci may be the use of phage therapy in combination with antibiotics (99, 100).
Role of Rha-CWPS in bacteria–host interactions
Rha-CWPS, both as a whole and through their side-chain modifications, appear to affect the virulence of pathogenic streptococci and enterococci. In an in vivo infection model, the deletion of rgpF in S. mutans—leading to the interruption of Rha-CWPS synthesis—resulted in attenuated virulence in Galleria mellonella (101). In this case, though, the absence of Rha-CWPS resulted in fitness defects in bacterial cells, which could contribute to the observed phenotype. In S. pyogenes, the absence of GlcNAc side chains on GAC led to attenuated virulence in animal models, which was associated with increased sensitivity to neutrophils and serum killing as well as with a higher susceptibility to the human antimicrobial peptide cathelicidin LL-37 (28, 102). In contrast, S. pyogenes mutants devoid of gacI (involved in GlcNAc side-chain addition onto rhamnan) and gacH (involved in GroP addition to GlcNAc) exhibited significantly increased resistance to human Group IIA phospholipase A2 (hGIIA). hGIIA is a secreted cationic antimicrobial protein of the human immune system that is particularly active against Gram-positive pathogens, which it kills by degrading phospholipids of the bacterial membrane. The resistance of gacI and gacH mutants most probably arises from the reduction in negative charges (the phosphate groups of GroP bound to GlcNAc) at the bacterial surface or inside the cell wall, which are required for hGIIA binding and diffusion through the cell wall to reach the membrane (29, 103). A gacO mutant of S. agalactiae, which did not produce GBC at all, was also resistant to hGIIA, possibly for the same reason: the absence of the negatively charged GBC that contains a significant amount of phosphate (103). Interestingly, inactivation of the de-N-acetylase PplD in S. pyogenes, which deacetylates the linkage GlcNAc, had the inverse effect on bacterial sensitivity to cationic antimicrobial peptides. By producing positively charged GlcN, PplD contributes to the neutralization of the negative charges displayed by GroP, thus increasing protection against cationic antimicrobial peptides (26).
In E. faecalis, the Rha-CWPS EPA was also found to contribute to pathogenesis, specifically through its side-chain decorations, which were characterized in strain V583 as teichoic-like chains (42). Mutations in the variable section of the epa gene cluster, which is involved in the biosynthesis of EPA decorations, were found to impair intestinal colonization in a mouse model (20). Likewise, the decorations were also found to be essential for virulence in a zebrafish infection model (104). The presence of EPA has been associated with increased resistance to phagocytosis (21) and its decorations appear to be involved in phagocyte evasion (104). A similar result was found in L. lactis: increased phagocytosis by macrophages was observed for mutants lacking PSP chains made of phosphohexasaccharide subunits (37). Specifically, EPA decorations, which were formerly considered to be WTA, were reported to confer resistance to complement-mediated phagocytosis by neutrophils via the mannose-binding lectin pathway (65). As expected from their structure, the absence of EPA decoration results in a lower negative cell surface charge but was unexpectedly found to increase resistance to lysozyme (104).
In the cell wall of R. gnavus, a prevalent gut microbe associated with the inflammatory bowel disorder Crohn’s disease, Rha-CWPS was shown to have a proinflammatory character, inducing TLR-4–dependent secretion of pro-inflammatory cytokines (TNFα) by dendritic cells (25). In this case, the potential role of teichoic acid–like substituents in these immunomodulatory effects remains to be investigated.
Overall, Rha-CWPS and their negatively charged side-chain modifications, either formed of small moieties or long chains, play a critical role in host colonization and the interactions of bacteria with different effectors of the host immune system. Because of their role in pathogenesis, different steps of the biosynthesis pathway could constitute novel targets for the design of antimicrobial compounds.
Since they are conserved between strains of the same species or serotype, Lancefield group carbohydrates have been proposed as potential antigens for vaccine design, especially to fight S. pyogenes infections (105). In animal models, protective antibodies directed against GAS were generated as a result of exposure to the poly-rhamnose backbone conjugated to a carrier protein (28). These antibodies also promoted opsonophagocytosis by neutrophils in vitro. However, for vaccine design, detailed knowledge of the structure of the immunizing antigen is of prime importance since, for example, antibodies induced against GlcNAc side-chain substituents of GAC could cross-react with human tissue antigens, which would strongly limit vaccine development (26, 106).
Other roles of Rha-CWPS in bacterial adaptation to the environment
Rha-CWPS also contribute to protection from stress, resistance to antimicrobials, and adaptation to a biofilm versus planktonic lifestyle. Increased expression of the Rml enzymes involved in dTDP-L-Rha biosynthesis was observed in E. faecalis biofilms compared to planktonic cultures, suggesting that EPA serves as a major component of the extracellular matrix. EPA was also shown to protect bacterial cells against high salt concentration (107). In S. mutans, Rha-CWPS were found to protect cells against various hostile conditions, including saline, high temperature, acid, and oxidative stresses (101). In addition, disruption of RGP biosynthesis led to enhanced sensitivity of bacterial cells to β-lactam antibiotics, which target peptidoglycan transpeptidation (108). Similarly, loss of EPA by E. faecalis is associated with increased susceptibility to several cell-wall–targeting antibiotics (99, 100). In S. thermophilus, exposure to bacitracin resulted in the increased expression of rmlC, which is involved in dTDP-Rha biosynthesis through a response involving a two-component system, which suggests a role for Rha-CWPS in protection against this antibiotic (109).
Conclusions and outlook
Rha-CWPS are highly abundant cell wall glycopolymers that are covalently attached to the peptidoglycan sacculus in many Gram-positive, ovoid-shaped cocci in the genera Streptococcus, Enterococcus, or Lactococcus, which lack typical WTA. The wealth of knowledge accumulated in recent years definitively demonstrates that Rha-CWPS are polyanionic glycopolymers that fulfill essential functions similar to those of WTA in bacterial physiology, phage binding, and interactions with the host. Current research has provided new clues for fighting phage attacks during food fermentation or using phage therapy against multidrug-resistant pathogens and has raised the possibility of predicting the sensitivity of a specific strain to a given phage. In particular, Rha-CWPS have been revealed as essential glycopolymers for cell wall homeostasis and bacterial fitness in pathogenic streptococci or enterococci, and therefore their biosynthesis pathway represents an attractive target for the development of novel antimicrobials.
The side-chain modifications attached to the rhamnan backbone play critical roles in numerous functions of Rha-CWPS, and the complex arsenal of enzymes involved in their biosynthesis has been deciphered in several cocci. More studies are needed to biochemically confirm or elucidate all steps of side-chain substituent biosynthesis, particularly for complex substituents. In addition, the spatiotemporal control of the different molecular events—including rhamnan anchoring to peptidoglycan by LCP proteins and the attachment of side-chain substituents to rhamnan by GT-C–fold glycosyltransferases—remains to be investigated.
Finally, the question of how Rha-CWPS biosynthesis is coordinated with that of peptidoglycan remains challenging. One approach for addressing this topic could be the use of metabolic labeling coupled to high-resolution microscopy techniques to visualize the localization and dynamics of the synthesis of Rha-CWPS relative to that of peptidoglycan (110, 111). Results obtained in S. mutans indicate a direct interaction between Rha-CWPS and certain proteins essential in cell division and point out a regulatory role of the negatively charged side-chain substituents of Rha-CWPS (31). Whether and how a similar role might be carried out by the complex and variable decorations of the rhamnan chain present in lactococci and enterococci warrants further investigation. More generally, the possible interactions between the proteins of the cell division machinery and the enzymes of the Rha-CWPS biosynthesis pathway—or the Rha-CWPS themselves—represent an exciting area of future study. This will provide new insights into the cell division mechanisms in ovoid-shaped Gram-positive bacteria and pave the way for future identification of targets for novel antimicrobials for mitigating the rise of multiple antibiotic resistance.
Conflict of interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this article.
Acknowledgments
We are grateful to all past and present collaborators mentioned in this review. We thank L. Higgins for English reading of the article and Luciana Giono for artistic work. We apologize to those researchers whose work could not be cited because of limited space.
Author contribution
M.-P. C.-C. conceptualization; H. G. and S. K. investigation; H. G. and S. K. writing–original draft; M.-P. C.-C. writing–review and editing.
Funding and additional information
The work of M.-P. C.-C. and S. K. was supported by funds from INRAE and Université Paris-Saclay (POC in labs project). H. G. received a PhD fellowship from the Ministère de l’Enseignement Supérieur, de la Recherche et de l’Innovation.
Edited by Chris Whitfield
References
- 1.Mostowy R.J., Holt K.E. Diversity-generating machines: genetics of bacterial sugar-coating. Trends Microbiol. 2018;26:1008–1021. doi: 10.1016/j.tim.2018.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Porter N.T., Martens E.C. The critical roles of polysaccharides in gut microbial ecology and physiology. Annu. Rev. Microbiol. 2017;71:349–369. doi: 10.1146/annurev-micro-102215-095316. [DOI] [PubMed] [Google Scholar]
- 3.Whitfield C., Williams D.M., Kelly S.D. Lipopolysaccharide O-antigens-bacterial glycans made to measure. J. Biol. Chem. 2020;295:10593–10609. doi: 10.1074/jbc.REV120.009402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Whitfield C., Wear S.S., Sande C. Assembly of bacterial capsular polysaccharides and exopolysaccharides. Annu. Rev. Microbiol. 2020;74:521–543. doi: 10.1146/annurev-micro-011420-075607. [DOI] [PubMed] [Google Scholar]
- 5.Weidenmaier C., Peschel A. Teichoic acids and related cell-wall glycopolymers in Gram-positive physiology and host interactions. Nat. Rev. Microbiol. 2008;6:276–287. doi: 10.1038/nrmicro1861. [DOI] [PubMed] [Google Scholar]
- 6.Martinez B., Rodriguez A., Kulakauskas S., Chapot-Chartier M.P. Cell wall homeostasis in lactic acid bacteria: Threats and defences. FEMS Microbiol. Rev. 2020;44:538–564. doi: 10.1093/femsre/fuaa021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brown S., Santa Maria J.P., Jr., Walker S. Wall teichoic acids of gram-positive bacteria. Annu. Rev. Microbiol. 2013;67:313–336. doi: 10.1146/annurev-micro-092412-155620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sumrall E.T., Keller A.P., Shen Y., Loessner M.J. Structure and function of Listeria teichoic acids and their implications. Mol. Microbiol. 2020;113:627–637. doi: 10.1111/mmi.14472. [DOI] [PubMed] [Google Scholar]
- 9.van Dalen R., Peschel A., van Sorge N.M. Wall teichoic acid in Staphylococcus aureus host interaction. Trends Microbiol. 2020;28:985–998. doi: 10.1016/j.tim.2020.05.017. [DOI] [PubMed] [Google Scholar]
- 10.Mistou M.Y., Sutcliffe I.C., van Sorge N.M. Bacterial glycobiology: Rhamnose-containing cell wall polysaccharides in gram-positive bacteria. FEMS Microbiol. Rev. 2016;40:464–479. doi: 10.1093/femsre/fuw006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lancefield R.C. A serological differentiation of human and other groups of hemolytic streptococci. J. Exp. Med. 1933;57:571–595. doi: 10.1084/jem.57.4.571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pritchard D.G., Coligan J.E., Speed S.E., Gray B.M. Carbohydrate fingerprints of streptococcal cells. J. Clin. Microbiol. 1981;13:89–92. doi: 10.1128/jcm.13.1.89-92.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Michon F., Brisson J.R., Dell A., Kasper D.L., Jennings H.J. Multiantennary group-specific polysaccharide of group B Streptococcus. Biochemistry. 1988;27:5341–5351. doi: 10.1021/bi00414a059. [DOI] [PubMed] [Google Scholar]
- 14.Sadovskaya I., Vinogradov E., Courtin P., Armalyte J., Meyrand M., Giaouris E., et al. Another brick in the wall: A rhamnan polysaccharide trapped inside peptidoglycan of Lactococcus lactis. MBio. 2017;8:e01303–e01317. doi: 10.1128/mBio.01303-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lavelle K., Sadovskaya I., Vinogradov E., Kelleher P., Lugli G.A., Ventura M., et al. Brussowvirus SW13 requires a cell surface-associated polysaccharide to recognise its Streptococcus thermophilus host. Appl. Environ. Microbiol. 2021;88 doi: 10.1128/AEM.01723-21. e01723-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McDonnell B., Hanemaaijer L., Bottacini F., Kelleher P., Lavelle K., Sadovskaya I., et al. A cell wall-associated polysaccharide is required for bacteriophage adsorption to the Streptococcus thermophilus cell surface. Mol. Microbiol. 2020;114:31–45. doi: 10.1111/mmi.14494. [DOI] [PubMed] [Google Scholar]
- 17.Mahony J., Cambillau C., van Sinderen D. Host recognition by lactic acid bacterial phages. FEMS Microbiol. Rev. 2017;41:S16–S26. doi: 10.1093/femsre/fux019. [DOI] [PubMed] [Google Scholar]
- 18.Romero D.A., Magill D., Millen A., Horvath P., Fremaux C. Dairy lactococcal and streptococcal phage-host interactions: An industrial perspective in an evolving phage landscape. FEMS Microbiol. Rev. 2020;44:909–932. doi: 10.1093/femsre/fuaa048. [DOI] [PubMed] [Google Scholar]
- 19.Lavelle K., Sinderen D.V., Mahony J. Cell wall polysaccharides of Gram positive ovococcoid bacteria and their role as bacteriophage receptors. Comput. Struct. Biotechnol. J. 2021;19:4018–4031. doi: 10.1016/j.csbj.2021.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rigottier-Gois L., Madec C., Navickas A., Matos R.C., Akary-Lepage E., Mistou M.Y., et al. The surface rhamnopolysaccharide epa of Enterococcus faecalis is a key determinant of intestinal colonization. J. Infect Dis. 2015;211:62–71. doi: 10.1093/infdis/jiu402. [DOI] [PubMed] [Google Scholar]
- 21.Teng F., Singh K.V., Bourgogne A., Zeng J., Murray B.E. Further characterization of the epa gene cluster and Epa polysaccharides of. Enterococcus Faecalis. Infect Immun. 2009;77:3759–3767. doi: 10.1128/IAI.00149-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ramos Y., Sansone S., Morales D.K. Sugarcoating it: enterococcal polysaccharides as key modulators of host-pathogen interactions. PLoS Pathog. 2021;17 doi: 10.1371/journal.ppat.1009822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vollmer W., Massidda O., Tomasz A. The cell wall of Streptococcus pneumoniae. Microbiol. Spectr. 2019;7 doi: 10.1128/microbiolspec.gpp3-0018-2018. GPP3-0018-2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yother J. Capsules of Streptococcus pneumoniae and other bacteria: Paradigms for polysaccharide biosynthesis and regulation. Annu. Rev. Microbiol. 2011;65:563–581. doi: 10.1146/annurev.micro.62.081307.162944. [DOI] [PubMed] [Google Scholar]
- 25.Henke M.T., Kenny D.J., Cassilly C.D., Vlamakis H., Xavier R.J., Clardy J. Ruminococcus gnavus, a member of the human gut microbiome associated with Crohn's disease, produces an inflammatory polysaccharide. Proc. Natl. Acad. Sci. U. S. A. 2019;116:12672–12677. doi: 10.1073/pnas.1904099116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rush J.S., Parajuli P., Ruda A., Li J., Pohane A.A., Zamakhaeva S., et al. PplD is a de-N-acetylase of the cell wall linkage unit of streptococcal rhamnopolysaccharides. Nat. Commun. 2022;13:590. doi: 10.1038/s41467-022-28257-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vinogradov E., Sadovskaya I., Courtin P., Kulakauskas S., Grard T., Mahony J., et al. Determination of the cell wall polysaccharide and teichoic acid structures from Lactococcus lactis IL1403. Carbohydr. Res. 2018;462:39–44. doi: 10.1016/j.carres.2018.04.002. [DOI] [PubMed] [Google Scholar]
- 28.van Sorge N.M., Cole J.N., Kuipers K., Henningham A., Aziz R.K., Kasirer-Friede A., et al. The classical lancefield antigen of group A Streptococcus is a virulence determinant with implications for vaccine design. Cell Host Microbe. 2014;15:729–740. doi: 10.1016/j.chom.2014.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Edgar R.J., van Hensbergen V.P., Ruda A., Turner A.G., Deng P., Le Breton Y., et al. Discovery of glycerol phosphate modification on streptococcal rhamnose polysaccharides. Nat. Chem. Biol. 2019;15:463–471. doi: 10.1038/s41589-019-0251-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nakano K., Ooshima T. Serotype classification of Streptococcus mutans and its detection outside the oral cavity. Future Microbiol. 2009;4:891–902. doi: 10.2217/fmb.09.64. [DOI] [PubMed] [Google Scholar]
- 31.Zamakhaeva S., Chaton C.T., Rush J.S., Ajay Castro S., Kenner C.W., Yarawsky A.E., et al. Modification of cell wall polysaccharide guides cell division in. Streptococcus Mutans. Nat. Chem. Biol. 2021;17:878–887. doi: 10.1038/s41589-021-00803-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Neiwert O., Holst O., Duda K.A. Structural investigation of rhamnose-rich polysaccharides from Streptococcus dysgalactiae bovine mastitis isolate. Carbohydr. Res. 2014;389:192–195. doi: 10.1016/j.carres.2013.12.018. [DOI] [PubMed] [Google Scholar]
- 33.Pritchard D.G., Rener B.P., Furner R.L., Huang D.H., Krishna N.R. Structure of the group G streptococcal polysaccharide. Carbohydr. Res. 1988;173:255–262. doi: 10.1016/s0008-6215(00)90821-8. [DOI] [PubMed] [Google Scholar]
- 34.Tettelin H., Masignani V., Cieslewicz M.J., Donati C., Medini D., Ward N.L., et al. Genome analysis of multiple pathogenic isolates of Streptococcus agalactiae: Implications for the microbial "pan-genome". Proc. Natl. Acad. Sci. U. S. A. 2005;102:13950–13955. doi: 10.1073/pnas.0506758102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Theodorou I., Courtin P., Palussiere S., Kulakauskas S., Bidnenko E., Pechoux C., et al. A dual-chain assembly pathway generates the high structural diversity of cell-wall polysaccharides in. Lactococcus Lactis. J. Biol. Chem. 2019;294:17612–17625. doi: 10.1074/jbc.RA119.009957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mahony J., Frantzen C., Vinogradov E., Sadovskaya I., Theodorou I., Kelleher P., et al. The CWPS Rubik's cube: linking diversity of cell wall polysaccharide structures with the encoded biosynthetic machinery of selected Lactococcus lactis strains. Mol. Microbiol. 2020;114:582–596. doi: 10.1111/mmi.14561. [DOI] [PubMed] [Google Scholar]
- 37.Chapot-Chartier M.P., Vinogradov E., Sadovskaya I., Andre G., Mistou M.Y., Trieu-Cuot P., et al. The cell surface of Lactococcus lactis is covered by a protective polysaccharide pellicle. J. Biol. Chem. 2010;285:10464–10471. doi: 10.1074/jbc.M109.082958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ainsworth S., Sadovskaya I., Vinogradov E., Courtin P., Guerardel Y., Mahony J., et al. Differences in lactococcal cell wall polysaccharide structure are major determining factors in bacteriophage sensitivity. MBio. 2014;5 doi: 10.1128/mBio.00880-14. e00880-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vinogradov E., Sadovskaya I., Grard T., Murphy J., Mahony J., Chapot-Chartier M.P., et al. Structural studies of the cell wall polysaccharide from Lactococcus lactis UC509.9. Carbohydr. Res. 2018;461:25–31. doi: 10.1016/j.carres.2018.03.011. [DOI] [PubMed] [Google Scholar]
- 40.Theodorou I., Courtin P., Sadovskaya I., Palussiere S., Fenaille F., Mahony J., et al. Three distinct glycosylation pathways are involved in the decoration of Lactococcus lactis cell wall glycopolymers. J. Biol. Chem. 2020;295:5519–5532. doi: 10.1074/jbc.RA119.010844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Farenc C., Spinelli S., Vinogradov E., Tremblay D., Blangy S., Sadovskaya I., et al. Molecular insights on the recognition of a Lactococcus lactis cell wall pellicle by the phage 1358 receptor binding protein. J. Virol. 2014;88:7005–7015. doi: 10.1128/JVI.00739-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Guerardel Y., Sadovskaya I., Maes E., Furlan S., Chapot-Chartier M.P., Mesnage S., et al. Complete structure of the enterococcal polysaccharide antigen (EPA) of vancomycin-resistant Enterococcus faecalis V583 reveals that EPA decorations are teichoic acids covalently linked to a rhamnopolysaccharide backbone. mBio. 2020;11 doi: 10.1128/mBio.00277-20. e00277-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Palmer K.L., Godfrey P., Griggs A., Kos V.N., Zucker J., Desjardins C., et al. Comparative genomics of enterococci: Variation in Enterococcus faecalis, clade structure in E. faecium, and defining characteristics of E. gallinarum and E. casseliflavus. mBio. 2012;3 doi: 10.1128/mBio.00318-11. e00318-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Giraud M.F., Naismith J.H. The rhamnose pathway. Curr. Opin. Struct. Biol. 2000;10:687–696. doi: 10.1016/s0959-440x(00)00145-7. [DOI] [PubMed] [Google Scholar]
- 45.van der Beek S.L., Le Breton Y., Ferenbach A.T., Chapman R.N., van Aalten D.M., Navratilova I., et al. GacA is essential for Group A Streptococcus and defines a new class of monomeric dTDP-4-dehydrorhamnose reductases (RmlD) Mol. Microbiol. 2015;98:946–962. doi: 10.1111/mmi.13169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sutcliffe I.C., Black G.W., Harrington D.J. Bioinformatic insights into the biosynthesis of the Group B carbohydrate in Streptococcus agalactiae. Microbiology. 2008;154:1354–1363. doi: 10.1099/mic.0.2007/014522-0. [DOI] [PubMed] [Google Scholar]
- 47.van der Beek S.L., Zorzoli A., Canak E., Chapman R.N., Lucas K., Meyer B.H., et al. Streptococcal dTDP-L-rhamnose biosynthesis enzymes: Functional characterization and lead compound identification. Mol. Microbiol. 2019;111:951–964. doi: 10.1111/mmi.14197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shibata Y., Yamashita Y., Ozaki K., Nakano Y., Koga T. Expression and characterization of streptococcal rgp genes required for rhamnan synthesis in Escherichia coli. Infect Immun. 2002;70:2891–2898. doi: 10.1128/IAI.70.6.2891-2898.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lukose V., Walvoort M.T.C., Imperiali B. Bacterial phosphoglycosyl transferases: Initiators of glycan biosynthesis at the membrane interface. Glycobiology. 2017;27:820–833. doi: 10.1093/glycob/cwx064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yamashita Y., Shibata Y., Nakano Y., Tsuda H., Kido N., Ohta M., et al. A novel gene required for rhamnose-glucose polysaccharide synthesis in Streptococcus mutans. J. Bacteriol. 1999;181:6556–6559. doi: 10.1128/jb.181.20.6556-6559.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Caliot E., Dramsi S., Chapot-Chartier M.P., Courtin P., Kulakauskas S., Pechoux C., et al. Role of the group B antigen of Streptococcus agalactiae: A peptidoglycan-anchored polysaccharide involved in cell wall biogenesis. PLoS Pathog. 2012;8 doi: 10.1371/journal.ppat.1002756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zorzoli A., Meyer B.H., Adair E., Torgov V.I., Veselovsky V.V., Danilov L.L., et al. Group A, B, C, and G Streptococcus lancefield antigen biosynthesis is initiated by a conserved α-D-GlcNAc-β-1,4-L-rhamnosyltransferase. J. Biol. Chem. 2019;294:15237–15256. doi: 10.1074/jbc.RA119.009894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Stefanovic C., Hager F.F., Schaffer C. LytR-CpsA-psr glycopolymer transferases: essential bricks in gram-positive bacterial cell wall assembly. Int. J. Mol. Sci. 2021;22:908. doi: 10.3390/ijms22020908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Vinogradov E., Sadovskaya I., Grard T., Chapot-Chartier M.P. Structural studies of the rhamnose-rich cell wall polysaccharide of Lactobacillus casei BL23. Carbohydr. Res. 2016;435:156–161. doi: 10.1016/j.carres.2016.10.002. [DOI] [PubMed] [Google Scholar]
- 55.Garcia-Vello P., Sharma G., Speciale I., Molinaro A., Im S.H., De Castro C. Structural features and immunological perception of the cell surface glycans of Lactobacillus plantarum: A novel rhamnose-rich polysaccharide and teichoic acids. Carbohydr. Polym. 2020;233 doi: 10.1016/j.carbpol.2020.115857. [DOI] [PubMed] [Google Scholar]
- 56.Tytgat H.L., Lebeer S. The sweet tooth of bacteria: Common themes in bacterial glycoconjugates. Microbiol. Mol. Biol. Rev. 2014;78:372–417. doi: 10.1128/MMBR.00007-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Islam S.T., Lam J.S. Synthesis of bacterial polysaccharides via the Wzx/Wzy-dependent pathway. Can J. Microbiol. 2014;60:697–716. doi: 10.1139/cjm-2014-0595. [DOI] [PubMed] [Google Scholar]
- 58.Mann E., Whitfield C. A widespread three-component mechanism for the periplasmic modification of bacterial glycoconjugates. Can. J. Chem. 2016;94:883–893. [Google Scholar]
- 59.Rismondo J., Gillis A., Grundling A. Modifications of cell wall polymers in gram-positive bacteria by multi-component transmembrane glycosylation systems. Curr. Opin. Microbiol. 2021;60:24–33. doi: 10.1016/j.mib.2021.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rush J.S., Edgar R.J., Deng P., Chen J., Zhu H., van Sorge N.M., et al. The molecular mechanism of N-acetylglucosamine side-chain attachment to the Lancefield group A carbohydrate in Streptococcus pyogenes. J. Biol. Chem. 2017;292:19441–19457. doi: 10.1074/jbc.M117.815910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lairson L.L., Henrissat B., Davies G.J., Withers S.G. Glycosyltransferases: Structures, functions, and mechanisms. Annu. Rev. Biochem. 2008;77:521–555. doi: 10.1146/annurev.biochem.76.061005.092322. [DOI] [PubMed] [Google Scholar]
- 62.Liu J., Mushegian A. Three monophyletic superfamilies account for the majority of the known glycosyltransferases. Protein Sci. 2003;12:1418–1431. doi: 10.1110/ps.0302103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bai L., Li H. Protein N-glycosylation and O-mannosylation are catalyzed by two evolutionarily related GT-C glycosyltransferases. Curr. Opin. Struct. Biol. 2021;68:66–73. doi: 10.1016/j.sbi.2020.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zimmermann L., Stephens A., Nam S.Z., Rau D., Kubler J., Lozajic M., et al. A completely reimplemented MPI bioinformatics toolkit with a new HHpred server at its core. J. Mol. Biol. 2018;430:2237–2243. doi: 10.1016/j.jmb.2017.12.007. [DOI] [PubMed] [Google Scholar]
- 65.Geiss-Liebisch S., Rooijakkers S.H., Beczala A., Sanchez-Carballo P., Kruszynska K., Repp C., et al. Secondary cell wall polymers of Enterococcus faecalis are critical for resistance to complement activation via mannose-binding lectin. J. Biol. Chem. 2012;287:37769–37777. doi: 10.1074/jbc.M112.358283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Percy M.G., Grundling A. Lipoteichoic acid synthesis and function in gram-positive bacteria. Annu. Rev. Microbiol. 2014;68:81–100. doi: 10.1146/annurev-micro-091213-112949. [DOI] [PubMed] [Google Scholar]
- 67.Kuwabara N., Imae R., Manya H., Tanaka T., Mizuno M., Tsumoto H., et al. Crystal structures of fukutin-related protein (FKRP), a ribitol-phosphate transferase related to muscular dystrophy. Nat. Commun. 2020;11:303. doi: 10.1038/s41467-019-14220-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kawai Y., Marles-Wright J., Cleverley R.M., Emmins R., Ishikawa S., Kuwano M., et al. A widespread family of bacterial cell wall assembly proteins. EMBO J. 2011;30:4931–4941. doi: 10.1038/emboj.2011.358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Schaefer K., Matano L.M., Qiao Y., Kahne D., Walker S. In vitro reconstitution demonstrates the cell wall ligase activity of LCP proteins. Nat. Chem. Biol. 2017;13:396–401. doi: 10.1038/nchembio.2302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Li F.K.K., Rosell F.I., Gale R.T., Simorre J.P., Brown E.D., Strynadka N.C.J. Crystallographic analysis of Staphylococcus aureus LcpA, the primary wall teichoic acid ligase. J. Biol. Chem. 2020;295:2629–2639. doi: 10.1074/jbc.RA119.011469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.De A., Liao S., Bitoun J.P., Roth R., Beatty W.L., Wu H., et al. Deficiency of RgpG causes major defects in cell division and biofilm formation, and deficiency of LytR-CpsA-psr family proteins leads to accumulation of cell wall antigens in culture medium by Streptococcus mutans. Appl. Environ. Microbiol. 2017;83 doi: 10.1128/AEM.00928-17. e00928-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Deng L., Kasper D.L., Krick T.P., Wessels M.R. Characterization of the linkage between the type III capsular polysaccharide and the bacterial cell wall of group B Streptococcus. J. Biol. Chem. 2000;275:7497–7504. doi: 10.1074/jbc.275.11.7497. [DOI] [PubMed] [Google Scholar]
- 73.Bernard E., Rolain T., Courtin P., Guillot A., Langella P., Hols P., et al. Characterization of O-acetylation of N-acetylglucosamine: A novel structural variation of bacterial peptidoglycan. J. Biol. Chem. 2011;286:23950–23958. doi: 10.1074/jbc.M111.241414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sychantha D., Brott A.S., Jones C.S., Clarke A.J. Mechanistic pathways for peptidoglycan O-acetylation and de-O-acetylation. Front Microbiol. 2018;9:2332. doi: 10.3389/fmicb.2018.02332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Viljoen A., Foster S.J., Fantner G.E., Hobbs J.K., Dufrene Y.F. Scratching the surface: bacterial cell envelopes at the nanoscale. mBio. 2020;11 doi: 10.1128/mBio.03020-19. e03020-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Li W., Lee R.E., Lee R.E., Li J. Methods for acquisition and assignment of multidimensional high-resolution magic angle spinning NMR of whole cell bacteria. Anal Chem. 2005;77:5785–5792. doi: 10.1021/ac050906t. [DOI] [PubMed] [Google Scholar]
- 77.Sadovskaya I., Guerardel Y. Simple protocol to purify cell wall polysaccharide from gram-positive bacteria and assess its structural integrity. Methods Mol. Biol. 2019;1954:37–45. doi: 10.1007/978-1-4939-9154-9_4. [DOI] [PubMed] [Google Scholar]
- 78.Andre G., Kulakauskas S., Chapot-Chartier M.P., Navet B., Deghorain M., Bernard E., et al. Imaging the nanoscale organization of peptidoglycan in living Lactococcus lactis cells. Nat. Commun. 2010;1:27. doi: 10.1038/ncomms1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Beaussart A., Pechoux C., Trieu-Cuot P., Hols P., Mistou M.Y., Dufrene Y.F. Molecular mapping of the cell wall polysaccharides of the human pathogen Streptococcus agalactiae. Nanoscale. 2014;6:14820–14827. doi: 10.1039/c4nr05280c. [DOI] [PubMed] [Google Scholar]
- 80.Rohs P.D.A., Bernhardt T.G. Growth and division of the peptidoglycan matrix. Annu. Rev. Microbiol. 2021;75:315–336. doi: 10.1146/annurev-micro-020518-120056. [DOI] [PubMed] [Google Scholar]
- 81.Ducret A., Grangeasse C. Recent progress in our understanding of peptidoglycan assembly in Firmicutes. Curr. Opin. Microbiol. 2021;60:44–50. doi: 10.1016/j.mib.2021.01.011. [DOI] [PubMed] [Google Scholar]
- 82.Atilano M.L., Pereira P.M., Yates J., Reed P., Veiga H., Pinho M.G., et al. Teichoic acids are temporal and spatial regulators of peptidoglycan cross-linking in Staphylococcus aureus. Proc. Natl. Acad. Sci. U. S. A. 2010;107:18991–18996. doi: 10.1073/pnas.1004304107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Schirner K., Marles-Wright J., Lewis R.J., Errington J. Distinct and essential morphogenic functions for wall- and lipo-teichoic acids in Bacillus subtilis. EMBO J. 2009;28:830–842. doi: 10.1038/emboj.2009.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Fleurie A., Lesterlin C., Manuse S., Zhao C., Cluzel C., Lavergne J.P., et al. MapZ marks the division sites and positions FtsZ rings in Streptococcus pneumoniae. Nature. 2014;516:259–262. doi: 10.1038/nature13966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Holeckova N., Doubravova L., Massidda O., Molle V., Buriankova K., Benada O., et al. LocZ is a new cell division protein involved in proper septum placement in Streptococcus pneumoniae. mBio. 2014;6:e01700–e01714. doi: 10.1128/mBio.01700-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Shen Y., Loessner M.J. Beyond antibacterials - exploring bacteriophages as antivirulence agents. Curr. Opin. Biotechnol. 2021;68:166–173. doi: 10.1016/j.copbio.2020.11.004. [DOI] [PubMed] [Google Scholar]
- 87.Samson J.E., Moineau S. Bacteriophages in food fermentations: New frontiers in a continuous arms race. Annu. Rev. Food Sci. Technol. 2013;4:347–368. doi: 10.1146/annurev-food-030212-182541. [DOI] [PubMed] [Google Scholar]
- 88.Shibata Y., Yamashita Y., van der Ploeg J.R. The serotype-specific glucose side chain of rhamnose-glucose polysaccharides is essential for adsorption of bacteriophage M102 to Streptococcus mutans. FEMS Microbiol. Lett. 2009;294:68–73. doi: 10.1111/j.1574-6968.2009.01546.x. [DOI] [PubMed] [Google Scholar]
- 89.Sumrall E.T., Shen Y., Keller A.P., Rismondo J., Pavlou M., Eugster M.R., et al. Phage resistance at the cost of virulence: listeria monocytogenes serovar 4b requires galactosylated teichoic acids for InlB-mediated invasion. PLoS Pathog. 2019;15 doi: 10.1371/journal.ppat.1008032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Winstel V., Xia G., Peschel A. Pathways and roles of wall teichoic acid glycosylation in Staphylococcus aureus. Int. J. Med. Microbiol. 2014;304:215–221. doi: 10.1016/j.ijmm.2013.10.009. [DOI] [PubMed] [Google Scholar]
- 91.Dupont K., Janzen T., Vogensen F.K., Josephsen J., Stuer-Lauridsen B. Identification of Lactococcus lactis genes required for bacteriophage adsorption. Appl. Environ. Microbiol. 2004;70:5825–5832. doi: 10.1128/AEM.70.10.5825-5832.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Chapot-Chartier M.-P. Interactions of the cell-wall glycopolymers of lactic acid bacteria with their bacteriophages. Front. Microbiol. 2014;5:236. doi: 10.3389/fmicb.2014.00236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Bebeacua C., Tremblay D., Farenc C., Chapot-Chartier M.P., Sadovskaya I., van Heel M., et al. Structure, adsorption to host, and infection mechanism of virulent lactococcal phage p2. J. Virol. 2013;87:12302–12312. doi: 10.1128/JVI.02033-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.McCabe O., Spinelli S., Farenc C., Labbe M., Tremblay D., Blangy S., et al. The targeted recognition of Lactococcus lactis phages to their polysaccharide receptors. Mol. Microbiol. 2015;96:875–886. doi: 10.1111/mmi.12978. [DOI] [PubMed] [Google Scholar]
- 95.Hayes S., Vincentelli R., Mahony J., Nauta A., Ramond L., Lugli G.A., et al. Functional carbohydrate binding modules identified in evolved dits from siphophages infecting various gram-positive bacteria. Mol. Microbiol. 2018;110:777–795. doi: 10.1111/mmi.14124. [DOI] [PubMed] [Google Scholar]
- 96.Bolocan A.S., Upadrasta A., Bettio P.H.A., Clooney A.G., Draper L.A., Ross R.P., et al. Evaluation of phage therapy in the context of Enterococcus faecalis and its associated diseases. Viruses. 2019;11:366. doi: 10.3390/v11040366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Ho K., Huo W., Pas S., Dao R., Palmer K.L. Loss-of-function mutations in epaR confer resistance to ΦNPV1 infection in Enterococcus faecalis OG1RF. Antimicrob. Agents Chemother. 2018;62 doi: 10.1128/AAC.00758-18. e00758-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Lossouarn J., Briet A., Moncaut E., Furlan S., Bouteau A., Son O., et al. Enterococcus faecalis countermeasures defeat a virulent picovirinae bacteriophage. Viruses. 2019;11:48. doi: 10.3390/v11010048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Canfield G.S., Chatterjee A., Espinosa J., Mangalea M.R., Sheriff E.K., Keidan M., et al. Lytic bacteriophages facilitate antibiotic sensitization of Enterococcus faecium. Antimicrob. Agents Chemother. 2021;65 doi: 10.1128/AAC.00143-21. e00143-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Chatterjee A., Johnson C.N., Luong P., Hullahalli K., McBride S.W., Schubert A.M., et al. Bacteriophage resistance alters antibiotic-mediated intestinal expansion of enterococci. Infect Immun. 2019;87 doi: 10.1128/IAI.00085-19. e00085-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Kovacs C.J., Faustoferri R.C., Quivey R.G., Jr. RgpF is required for maintenance of stress tolerance and virulence in. Streptococcus Mutans. J. Bacteriol. 2017;199 doi: 10.1128/JB.00497-17. e00497-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Henningham A., Davies M.R., Uchiyama S., van Sorge N.M., Lund S., Chen K.T., et al. Virulence role of the GlcNAc side chain of the lancefield cell wall carbohydrate antigen in non-M1-serotype group A Streptococcus. mBio. 2018;9 doi: 10.1128/mBio.02294-17. e02294-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.van Hensbergen V.P., Movert E., de Maat V., Luchtenborg C., Le Breton Y., Lambeau G., et al. Streptococcal Lancefield polysaccharides are critical cell wall determinants for human Group IIA secreted phospholipase A2 to exert its bactericidal effects. PLoS Pathog. 2018;14 doi: 10.1371/journal.ppat.1007348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Smith R.E., Salamaga B., Szkuta P., Hajdamowicz N., Prajsnar T.K., Bulmer G.S., et al. Decoration of the enterococcal polysaccharide antigen EPA is essential for virulence, cell surface charge and interaction with effectors of the innate immune system. PLoS Pathog. 2019;15 doi: 10.1371/journal.ppat.1007730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Kabanova A., Margarit I., Berti F., Romano M.R., Grandi G., Bensi G., et al. Evaluation of a Group A Streptococcus synthetic oligosaccharide as vaccine candidate. Vaccine. 2010;29:104–114. doi: 10.1016/j.vaccine.2010.09.018. [DOI] [PubMed] [Google Scholar]
- 106.Gao N.J., Rodas Lima E., Nizet V. Immunobiology of the classical lancefield group A streptococcal carbohydrate antigen. Infect Immun. 2021;89 doi: 10.1128/IAI.00292-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Solheim M., La Rosa S.L., Mathisen T., Snipen L.G., Nes I.F., Brede D.A. Transcriptomic and functional analysis of NaCl-induced stress in Enterococcus faecalis. PLoS One. 2014;9 doi: 10.1371/journal.pone.0094571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Kovacs C.J., Faustoferri R.C., Bischer A.P., Quivey R.G., Jr. Streptococcus mutans requires mature rhamnose-glucose polysaccharides for proper pathophysiology, morphogenesis and cellular division. Mol. Microbiol. 2019;112:944–959. doi: 10.1111/mmi.14330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Thevenard B., Besset C., Choinard S., Fourcassie P., Boyaval P., Monnet V., et al. Response of Streptococcus thermophilus LMD-9 to bacitracin: Involvement of a BceRS/AB-like module and of the rhamnose-glucose polysaccharide synthesis pathway. Int. J. Food Microbiol. 2014;177:89–97. doi: 10.1016/j.ijfoodmicro.2014.02.011. [DOI] [PubMed] [Google Scholar]
- 110.Radkov A.D., Hsu Y.P., Booher G., VanNieuwenhze M.S. Imaging bacterial cell wall biosynthesis. Annu. Rev. Biochem. 2018;87:991–1014. doi: 10.1146/annurev-biochem-062917-012921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Banahene N., Kavunja H.W., Swarts B.M. Chemical reporters for bacterial glycans: development and applications. Chem. Rev. 2022;122:3336–3413. doi: 10.1021/acs.chemrev.1c00729. [DOI] [PMC free article] [PubMed] [Google Scholar]