Abstract
Background
Histological grade is an important factor in the overall prognosis of patients with invasive breast cancer. Therefore, the non-invasive assessment of histological grade in breast cancer patients is an increasing concern for clinicians. We aimed to establish an MRI-based radiomics model for preoperative prediction of invasive breast cancer histological grade.
Methods
We enrolled 901 patients with invasive breast cancer and pre-operative MRI. Patients were randomly divided into the training cohort (n=630) and validation cohort (n=271) with a ratio of 7:3. A radiomics signature was constructed by extracting radiomics features from MRI images and was developed according to multivariate logistic regression analysis. The diagnostic performance of the radiomics model was assessed using receiver operating characteristic (ROC) curve analysis.
Results
Of the 901 patients, 618 (68.6%) were histological grade 1 or 2 while 283 (31.4%) were histological grade 3. The area under the ROC curve (AUC) of the radiomics model for the prediction of the histological grade was 0.761 (95% CI 0.728–0.794) in the training cohort and 0.722 (95% CI 0.664–0.777) in the validation cohort. The decision curve analysis (DCA) demonstrated the radiomics model’s clinical application value.
Conclusion
This is a preliminary study suggesting that the development of an MRI-based radiomics model can predict the histological grade of invasive breast cancer.
Keywords: breast cancer, histologic grade, magnetic resonance imaging, radiomics, signature
Introduction
Breast cancer is the most prevalent malignancy among women worldwide, accounting for almost 25% of all cancer cases in women. It is also the leading cause of cancer-related deaths in women worldwide.1 Studies have shown that tumor histological grade is an independent prognostic factor in specific subgroups of breast cancer patients, including estrogen receptor positivity,2 and lymph node metastasis.3 Invasive ductal carcinoma (IDC), the most common histological type of breast cancer, accounts for approximately 80% of all breast cancers. Therefore, accurate identification of tumor's histological grade in the IDC can provide a useful guide to the prognosis.
Magnetic resonance imaging (MRI) has been widely used in the study of breast cancer. Studies have shown that radiological analysis of MRI may help predict breast cancer subtypes,4–6 neoadjuvant chemotherapy responses,7,8 Ki-67 expression level,9–11 pathological stage,12 histological grade,11,13,14 tumor malignancy,15 and breast cancer recurrence.16,17 The eighth edition of the cancer staging system of the American Joint Committee on Cancer (AJCC), combined histological grade with anatomical staging to determine the clinical prognostic stage, where histological grade 3 luminal carcinoma is one stage higher than its anatomical stage.14,18 Therefore, we wish to distinguish histological grade 3 tumors from grade 1 or 2 tumors. However, to our knowledge, we used the largest sample size with effective predictors to develop and validate an MRI-based radiomics model for preoperative prediction of invasive breast cancer histological grade.
Materials and Methods
Patients
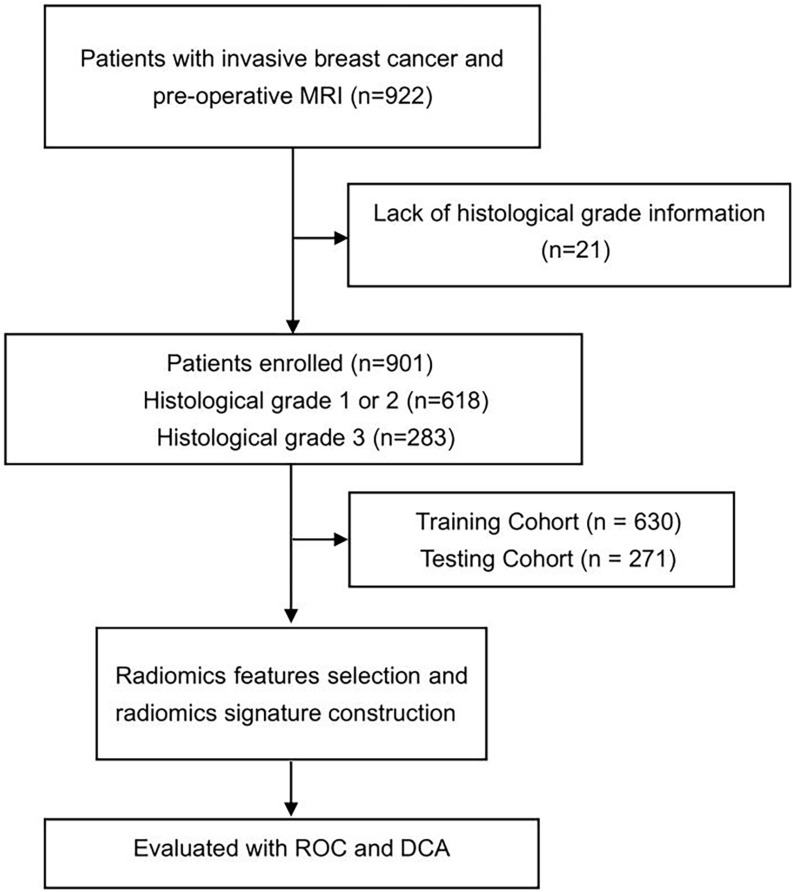
Patients were collected from The Cancer Imaging Archive (TCIA; http://www.cancerimagingarchive.net/) datasets.19, 922 patients whose pre-operative MRI acquired from 1 January 2000 to 23 March 2014 with post-operative pathology confirmed invasive breast cancer. Patients with prior breast surgery, history of breast cancer, or neoadjuvant therapy before the MRI acquisition were excluded. 901 patients were eventually included in this study, due to the lack of histological grade information for 21 patients. Patients were randomly divided into the training cohort (n=630) and validation cohort (n=271) with a ratio of 7:3. The study flow diagram is shown in Figure 1.
Figure 1.
The flow diagram of the study.
Imaging Acquisition, Segmentation, and Radiomics Feature Extraction
For the patients included in this study, cross-sectional breast MRI images were acquired in the prone position using a 1.5T or 3T scanner. Detailed scanner-related information and MR acquisition parameters are shown in the Tables S1 and S2, respectively. The following sequences were acquired by MRI: a non-fat-saturated T1-weighted sequence, a fat-saturated gradient-echo T1-weighted enhancement sequence, and four post-enhancement T1-weighted sequences typically acquired after intravenous contrast injection was used with a dose of 0.2 mL/kg body weight via peripheral veins.
A fuzzy C-means automatic segmentation20 was used to obtain the tumor mask. The radiomics feature of breast cancer was automatically extracted from the N4-corrected21 T1-non-fat saturated (T1-NFS) images and first post-contrast sequences. A comprehensive set of 529 radiomics features were collected from TCIA datasets19 that have been shown to be effective predictors and could quantify characteristics of the breast, tumor, and fibroglandular tissue. The process of image segmentation and radiomics feature extraction can be available in the TCIA datasets publication.4
Construction of the Radiomics Model
To avoid overfitting the radiomics signature, features were further selected in three steps before the radiomics signature was constructed. First, features with ICC >0.75 within the training cohort were retained. Second, statistically, significant features were screened out using the univariate logistic analysis in the training cohort. Third, the most valuable features were selected using the least absolute shrinkage and selection operator (LASSO). A radiomics score (Rad-score) was calculated by using a formula based on the radiomics features.
The Rad-score was used to establish a radiomics signature through multivariate logistic regression. The diagnostic performance of the radiomics signature model in predicting histological grade was evaluated based on the area under the receiver operator characteristic (ROC) curve (AUC) in both the training cohort and testing cohort. To evaluate the clinical effectiveness of the radiomics signature, a decision curve analysis (DCA) was performed by calculating the net benefit of a threshold probability range across the training and testing cohorts.
Statistical Analysis
Statistical tests were performed using SPSS 25.0 software (IBM) and R programming language (ver. 3.5.1, http://www.r-project.org). We chose 26 features from the training cohort and tested them on the testing cohort to investigate the prognostic usefulness of imaging features.
Results
Clinical Features of the Patients
Nine hundred and one patients were enrolled in the study as shown in Figure 1. The clinical features of invasive breast cancer in the training and testing cohorts are shown in Table 1. Of the 901 patients, Histologic grade 1 was observed in 164 patients, grade 2 in 454 patients, and grade 3 in 283 patients, and 618 (68.6%) were histological grade 1 or 2 while 283 (31.4%) were histological grade 3.
Table 1.
Clinical and Tumor Features in the Training and Validation Cohorts
| Characteristics | Training Cohort (N = 618) | Validation Cohort (N = 283) |
|---|---|---|
| Median age (years) | 53.28 | 49.86 |
| Race | ||
| White | 465 | 173 |
| Black | 108 | 93 |
| Others* | 36 | 8 |
| Not available | 9 | 9 |
| Menopausal status | ||
| Pre | 250 | 147 |
| Post | 356 | 132 |
| Not available | 12 | 4 |
| Tumor staging (size) | ||
| T1 | 302 | 95 |
| T2 | 246 | 145 |
| T3 | 59 | 31 |
| T4 | 8 | 12 |
| Not available | 3 | 0 |
| ER status | ||
| Positive | 536 | 137 |
| Negative | 82 | 146 |
| PR status | ||
| Positive | 486 | 103 |
| Negative | 132 | 180 |
| HER2 status | ||
| Positive | 89 | 66 |
| Negative | 529 | 217 |
Abbreviations: ER, estrogen receptor; PR, progesterone receptor; HER2, human epidermal growth factor; *Includes Asian, Native, Hispanic.
Construction of the Radiomics Model
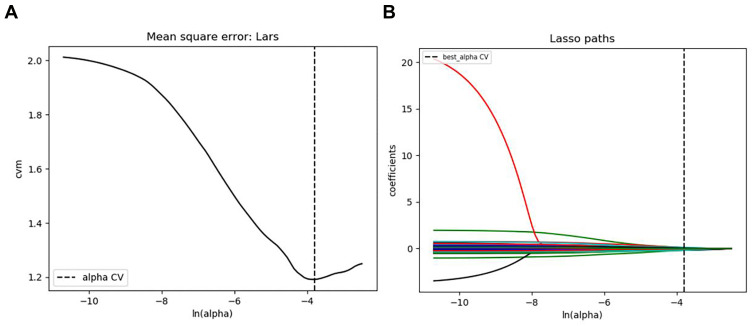
A total of 529 radiomics features were extracted from the MRI images. After univariate correlation analysis, 143 radiomics features showed significant differences in predicting histological grade. These features were sequentially imported into LASSO (Figure 2) to obtain the most valuable features, resulting in 26 useful features. Finally, the radiomics signature was established by using the 26 features (Table 2).
Figure 2.
The LASSO regression model was used to select radiomics features. (A) LASSO coefficient profiles of the 26 radiomics features. A coefficient profile plot was generated versus the selected log (λ) value using ten-fold cross-validation, where optimal λ resulted in 8 features with nonzero coefficients. (B) The 26 radiomics features’ LASSO coefficient profiles. The log (λ) sequence was used to create a coefficient profile plot. Using 10-fold cross-validation, the dotted vertical line was drawn at the value chosen.
Table 2.
Radiomics Features Selection from the CTE in the Training Cohort
| Radiomics Features | Coefficient |
|---|---|
| Volume_cu_mm_Tumor | −0.2952 |
| Inf_mea_of_corr2_Tumor | −0.3053 |
| Max_Enhancement_from_char_curv | −0.2096 |
| BreastDensity_T1 | −0.0853 |
| Grouping_based_variance_of_washout_slope_3D_tumor_Group_1 | 0.9801 |
| Grouping_based_variance_of_washout_slope_2D_tumorSlice_Group_2 | −0.1994 |
| Grouping_based_proportion_of_tumor_voxels_2D_tumorSlice_Group_1 | −0.2213 |
| Grouping_based_proportion_of_tumor_voxels_2D_tumorSlice_Group_2 | 0.0376 |
| Grouping_based_variance_of_peak_enhancement_slope_3D_tissue_PostCon_Group_3 | 0.0850 |
| Grouping_based_variance_of_washin_slope_3D_tissue_PostCon_Group_2 | 0.1022 |
| Grouping_based_variance_of_washout_slope_3D_tissue_PostCon_Group_1 | 0.0532 |
| Mean_norm_DHOG_max_timepoint_binsize_6_with_filling_Tumor | 0.1349 |
| SER_Washout_tumor_vol_cu_mm | −0.1489 |
| SER_map_Correlation2_tumor | −0.0687 |
| SER_map_Sum_of_Squares_variance_tumor | −0.1204 |
| PE_map_Cluster_Prominence_tumor | 0.1228 |
| WashinRate_map_Correlation2_tumor | 0.3493 |
| WashinRate_map_Max_Probability_tumor | 0.1759 |
| WashinRate_map_information_measure_correlation1_tumor | −0.0017 |
| WashinRate_map_skewness_tumor | −0.3948 |
| SER_map_skewness_tissue_T1 | 0.2037 |
| Peak_SER_tissue_PostCon | 0.2229 |
| SER_map_Cluster_Shade_tissue_PostCon | −0.3002 |
| PE_map_kurtosis_tissue_PostCon | −0.2192 |
| WashinRate_map_Autocorrelation_tissue_PostCon | 0.1409 |
| WashinRate_map_Cluster_Shade_tissue_PostCon | −0.0817 |
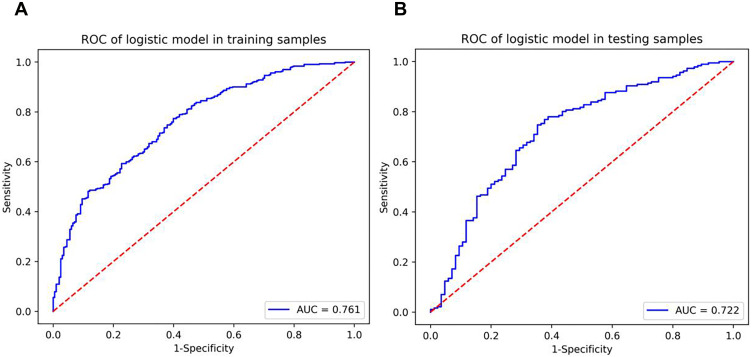
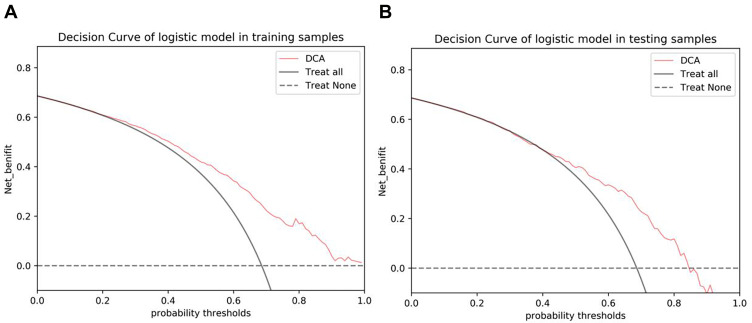
The radiomics model based on the training cohort and testing cohort is shown in Figure 3. Table 3 summarizes the discriminatory efficacies of the radiomics model. The AUC was 0.761 (95% CI 0.728–0.794) in the training cohort and 0.722 (95% CI 0.664–0.777) in the validation cohort for predicting histological grade. The DCA indicated that in most training and testing cohorts within reasonable threshold probabilities, the clinical radiomics nomogram added a greater overall net benefit in predicting histological grade in invasive breast cancer. The DCA for the radiomics model in the training and testing cohorts is shown in Figure 4.
Figure 3.
The receiver operator characteristic curves of the radiomics model. (A) The radiomics model of ROC curves in the training cohort. (B) The radiomics model of ROC curves in the validation cohort.
Table 3.
Performance of the Radiomics Model in the Training and Validation Cohorts
| Training Cohort (n=630) | Validation Cohort (n=271) | |
|---|---|---|
| Radiomics model | AUC (95% CI) SEN SPE ACC | AUC (95% CI) SEN SPE ACC |
| 0.761 (0.728–0.794) 0.771 0.601 0.717 | 0.722 (0.664–0.777) 0.763 0.624 0.720 |
Abbreviations: AUC, area under the curve; SEN, sensitivity; SPE, specificity; ACC, accuracy; 95% CI, 95% confidence interval.
Figure 4.
The decision curve analysis (DCA) of the radiomics model. (A) The radiomics model of DCA in the training cohort. (B) The radiomics model of DCA in the validation cohort.
Discussion
In the present study, we assessed the development and validation of a radiomics model to predict histological grade in invasive breast cancer. According to our findings, the radiomics signature could predict histological grade in both training and testing cohorts. At the same time, DCA demonstrated the radiomics model’s clinical application value.
Studies have shown that histological grade is a key factor in the overall prognosis of patients.22–25 Grading of breast tumors requires histological evaluation of tissue samples obtained by guided biopsy or surgical excision. Breast cancers are graded according to the Scarff-Bloom-Richardson grading system.26 This grading system relies on the frequency of cell mitosis (cytokinesis rate), Tubule formation (the percentage of tumors containing tubular structures), and the grade of nuclear pleomorphism.27
Saha et al4 analyzed a total of 922 individuals with invasive breast cancer and pre-operative MRI to predict the molecular, genomic, and proliferation characteristics. A computer algorithm was used to extract 529 features of the tumor and surrounding tissue from the MRI images. Those features could be found in the published literature as well as those developed in their research.
Our patients and radiomics features were collected from this study. Song et al14 used multiparametric MRI to create machine learning-based prediction models that performed well in predicting Ki-67 and histologic grade in patients with luminal breast tumors. Scholars28 reported that a total of 205 patients with contrast-enhanced spectral mammography examination invasive breast cancer were retrospectively enrolled and the radiomics model is a non-invasive predictive method that showed good application prospects in predicting histological grade. It is reported that the use of a combination of dynamic contrast-enhanced magnetic resonance imaging and T2-weighted imaging radiomic features could be used to predict histological grade in ductal breast carcinoma.29 Although these studies had good diagnostic efficacy, the number of cases is relatively small. We enrolled 901 patients in this study and developed an MRI radiomics model with effective predictors to predict histological grade in patients with invasive breast cancer.
A total of 529 radiomics features extracted from the MRI images were enrolled in our study. After univariate correlation analysis, 143 radiomics features showed significant differences in predicting histological grade. These features were sequentially imported into LASSO to obtain the most valuable features, resulting in 26 useful features. Finally, the radiomics signature was established by using the 26 features.
Our study had several limitations. First, our 901 breast cancer patients were acquired from different machines. The variability of imaging acquisition parameters is a source of noise and future studies should focus on uniform imaging parameters for scanning. Second, this was a retrospective study. The multicenter, large-sample, and prospective cases need to be included in future studies.
We developed an MRI-based radiomics model to predict histological grade in invasive breast cancer. This radiomics signature can provide a preliminary method to predict histological grade of invasive breast cancer.
Acknowledgments
We would like to thank the participants who contributed to our study.
Funding Statement
There is no funding to report.
Ethical Approval and Consent to Participate
The study protocol was approved by the institutional review board of the First Affiliated Hospital of Wannan Medical College. Written informed consent was not required due to the database is publicly available.19 The personal information of patients was strictly protected. The study was carried out in accordance with the tenets of the Declaration of Helsinki 1964 and its later amendments.
Disclosure
The authors declare that they have no conflicts of interest in relation to this work.
References
- 1.Sung H, Ferlay J, Siegel RL, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71(3):209–249. doi: 10.3322/caac.21660 [DOI] [PubMed] [Google Scholar]
- 2.Ehinger A, Malmström P, Bendahl PO, et al. Histological grade provides significant prognostic information in addition to breast cancer subtypes defined according to St Gallen 2013. Acta Oncol. 2017;56(1):68–74. doi: 10.1080/0284186x.2016.1237778 [DOI] [PubMed] [Google Scholar]
- 3.Rakha EA, El-Sayed ME, Lee AH, et al. Prognostic significance of Nottingham histologic grade in invasive breast carcinoma. J Clin Oncol. 2008;26(19):3153–3158. doi: 10.1200/jco.2007.15.5986 [DOI] [PubMed] [Google Scholar]
- 4.Saha A, Harowicz MR, Grimm LJ, et al. A machine learning approach to radiogenomics of breast cancer: a study of 922 subjects and 529 DCE-MRI features. Br J Cancer. 2018;119(4):508–516. doi: 10.1038/s41416-018-0185-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li H, Zhu Y, Burnside ES, et al. Quantitative MRI radiomics in the prediction of molecular classifications of breast cancer subtypes in the TCGA/TCIA data set. NPJ Breast Cancer. 2016;2:16012. doi: 10.1038/npjbcancer.2016.12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yuen S, Monzawa S, Yanai S, et al. The association between MRI findings and breast cancer subtypes: focused on the combination patterns on diffusion-weighted and T2-weighted images. Breast Cancer. 2020;27(5):1029–1037. doi: 10.1007/s12282-020-01105-z [DOI] [PubMed] [Google Scholar]
- 7.Liu Z, Li Z, Qu J, et al. Radiomics of multiparametric MRI for pretreatment prediction of pathologic complete response to neoadjuvant chemotherapy in breast cancer: a multicenter study. Clin Cancer Res. 2019;25(12):3538–3547. doi: 10.1158/1078-0432.Ccr-18-3190 [DOI] [PubMed] [Google Scholar]
- 8.Kim R, Chang JM, Lee HB, et al. Predicting axillary response to neoadjuvant chemotherapy: breast MRI and US in patients with node-positive breast cancer. Radiology. 2019;293(1):49–57. doi: 10.1148/radiol.2019190014 [DOI] [PubMed] [Google Scholar]
- 9.Lee JH, Yoon YC, Seo SW, Choi YL, Kim HS. Soft tissue sarcoma: DWI and DCE-MRI parameters correlate with Ki-67 labeling index. Eur Radiol. 2020;30(2):914–924. doi: 10.1007/s00330-019-06445-9 [DOI] [PubMed] [Google Scholar]
- 10.Li C, Song L, Yin J. Intratumoral and peritumoral radiomics based on functional parametric maps from breast DCE-MRI for prediction of HER-2 and Ki-67 status. J Magn Reson Imaging. 2021;54(3):703–714. doi: 10.1002/jmri.27651 [DOI] [PubMed] [Google Scholar]
- 11.Fan M, Yuan W, Zhao W, et al. Joint prediction of breast cancer histological grade and Ki-67 Expression level based on DCE-MRI and DWI radiomics. IEEE J Biomed Health Inform. 2020;24(6):1632–1642. doi: 10.1109/jbhi.2019.2956351 [DOI] [PubMed] [Google Scholar]
- 12.Burnside ES, Drukker K, Li H, et al. Using computer-extracted image phenotypes from tumors on breast magnetic resonance imaging to predict breast cancer pathologic stage. Cancer. 2016;122(5):748–757. doi: 10.1002/cncr.29791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xi G, He J, Kang D, et al. Nomogram model combining macro and micro tumor-associated collagen signatures obtained from multiphoton images to predict the histologic grade in breast cancer. Biomed Opt Express. 2021;12(10):6558–6570. doi: 10.1364/boe.433281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Song SE, Cho KR, Cho Y, et al. Machine learning with multiparametric breast MRI for prediction of Ki-67 and histologic grade in early-stage luminal breast cancer. Eur Radiol. 2022;32(2):853–863. doi: 10.1007/s00330-021-08127-x [DOI] [PubMed] [Google Scholar]
- 15.Vidić I, Egnell L, Jerome NP, et al. Support vector machine for breast cancer classification using diffusion-weighted MRI histogram features: preliminary study. J Magn Reson Imaging. 2018;47(5):1205–1216. doi: 10.1002/jmri.25873 [DOI] [PubMed] [Google Scholar]
- 16.Mazurowski MA, Saha A, Harowicz MR, Cain EH, Marks JR, Marcom PK. Association of distant recurrence-free survival with algorithmically extracted MRI characteristics in breast cancer. J Magn Reson Imaging. 2019;49(7):e231–e240. doi: 10.1002/jmri.26648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim JY, Kim JJ, Hwangbo L, et al. Kinetic heterogeneity of breast cancer determined using computer-aided diagnosis of preoperative MRI scans: relationship to distant metastasis-free survival. Radiology. 2020;295(3):517–526. doi: 10.1148/radiol.2020192039 [DOI] [PubMed] [Google Scholar]
- 18.Amin MB, Greene FL, Edge SB, et al. The eighth edition AJCC cancer staging manual: continuing to build a bridge from a population-based to a more ”personalized” approach to cancer staging. CA Cancer J Clin. 2017;67(2):93–99. doi: 10.3322/caac.21388 [DOI] [PubMed] [Google Scholar]
- 19.Clark K, Vendt B, Smith K, et al. The Cancer Imaging Archive (TCIA): maintaining and operating a public information repository. J Digit Imaging. 2013;26(6):1045–1057. doi: 10.1007/s10278-013-9622-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Page DL. Prognosis and breast cancer. Recognition of lethal and favorable prognostic types. Am J Surg Pathol. 1991;15(4):334–349. doi: 10.1097/00000478-199104000-00002 [DOI] [PubMed] [Google Scholar]
- 21.Tustison NJ, Avants BB, Cook PA, et al. N4ITK: improved N3 bias correction. IEEE Trans Med Imaging. 2010;29(6):1310–1320. doi: 10.1109/tmi.2010.2046908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Abubakar M, Chang-Claude J, Ali HR, et al. Etiology of hormone receptor positive breast cancer differs by levels of histologic grade and proliferation. Int J Cancer. 2018;143(4):746–757. doi: 10.1002/ijc.31352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang Y, Zhou Y, Mao F, Yao R, Sun Q. Ki-67 index, progesterone receptor expression, histologic grade and tumor size in predicting breast cancer recurrence risk: a consecutive cohort study. Cancer Commun. 2020;40(4):181–193. doi: 10.1002/cac2.12024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Singh K, He X, Kalife ET, Ehdaivand S, Wang Y, Sung CJ. Relationship of histologic grade and histologic subtype with oncotype Dx recurrence score; retrospective review of 863 breast cancer oncotype Dx results. Breast Cancer Res Treat. 2018;168(1):29–34. doi: 10.1007/s10549-017-4619-4 [DOI] [PubMed] [Google Scholar]
- 25.Escott CE, Zaenger D, Switchencko JM, et al. The influence of histologic grade on outcomes of elderly women with early stage breast cancer treated with breast conserving surgery with or without radiotherapy. Clin Breast Cancer. 2020;20(6):e701–e710. doi: 10.1016/j.clbc.2020.05.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Grajo JR, Barr RG. Strain elastography for prediction of breast cancer tumor grades. J Ultrasound Med. 2014;33(1):129–134. doi: 10.7863/ultra.33.1.129 [DOI] [PubMed] [Google Scholar]
- 27.Elston CW, Ellis IO. Pathological prognostic factors in breast cancer. I. The value of histological grade in breast cancer: experience from a large study with long-term follow-up. Histopathology. 1991;19(5):403–410. doi: 10.1111/j.1365-2559.1991.tb00229.x [DOI] [PubMed] [Google Scholar]
- 28.Mao N, Jiao Z, Duan S, Xu C, Xie H. Preoperative prediction of histologic grade in invasive breast cancer by using contrast-enhanced spectral mammography-based radiomics. J Xray Sci Technol. 2021;29(5):763–772. doi: 10.3233/xst-210886 [DOI] [PubMed] [Google Scholar]
- 29.Fan M, Liu Z, Xie S, et al. Integration of dynamic contrast-enhanced magnetic resonance imaging and T2-weighted imaging radiomic features by a canonical correlation analysis-based feature fusion method to predict histological grade in ductal breast carcinoma. Phys Med Biol. 2019;64(21):215001. doi: 10.1088/1361-6560/ab3fd3 [DOI] [PubMed] [Google Scholar]