Abstract
Background
We established the first prospective cohort to understand how infection with dengue virus is influenced by vector-specific determinants such as humoral immunity to Aedes aegypti salivary proteins.
Methods
Children aged 2–9 years were enrolled in the PAGODAS (Pediatric Assessment Group of Dengue and Aedes Saliva) cohort with informed consent by their guardians. Children were followed semi-annually for antibodies to dengue and to proteins in Ae. aegypti salivary gland homogenate using enzyme-linked immunosorbent assays and dengue-specific neutralization titers. Children presented with fever at any time for dengue testing.
Results
From 13 July to 30 August 2018, we enrolled 771 children. At baseline, 22% (173/770) had evidence of neutralizing antibodies to 1 or more dengue serotypes. By April 2020, 51 children had symptomatic dengue while 148 dengue-naive children had inapparent dengue defined by neutralization assays. In a multivariate model, individuals with higher antibodies to Ae. aegypti salivary proteins were 1.5 times more likely to have dengue infection (hazard ratio [HR], 1.47 [95% confidence interval {CI}, 1.05–2.06]; P = .02), particularly individuals with inapparent dengue (HR, 1.64 [95% CI, 1.12–2.41]; P = .01).
Conclusions
High levels of seropositivity to Ae. aegypti salivary proteins are associated with future development of dengue infection, primarily inapparent, in dengue-naive Cambodian children.
Clinical Trials Registration
Keywords: Aedes aegypti, Cambodia, dengue, mosquito saliva, pediatric cohort
Results from a longitudinal pediatric cohort in Cambodia demonstrated that children with high levels of antibodies to Aedes aegyptimosquito saliva were more likely to get dengue, but were also less likely to have clinical symptoms.
Mosquito-borne diseases are emerging and reemerging, continuing to cause global morbidity and mortality [1, 2]. The understanding of both transmission and pathogenesis is inextricably linked to the mosquito vector whose saliva serves as a driver for pathogen establishment in the host. Since the 1940s, scientists have observed the immunological effects of vector saliva on the human host [3]. Over the past few decades, research linked insect vector saliva to enhanced transmission and virulence of pathogens, from various arboviruses to Plasmodium and Leishmania parasites, in various animal models [4–7]. Only recently were these results translated into the clinic with a first-in-human mosquito saliva peptide vaccine, but questions remain on how the host immune response to vector saliva may help—or hinder—in the context of disease outcomes [8].
Dengue virus (DENV) is arguably the most widespread, and as a consequence best-studied, mosquito-borne virus [2]. The reach of its vectors, Aedes albopictus and Aedes aegypti, will include nearly a billion new inhabitants by 2050 [9]. Prior studies have evaluated Ae. aegypti mosquito saliva immunity as a biomarker of mosquito exposure in humans, but the relationship to disease development has yet to be established [10–12]. In retrospective and cross-sectional studies, there are no appreciable difference in immunoglobulin G (IgG) levels for specific peptide markers or total anti-saliva antibodies between DENV-positive and DENV-negative individuals at point-of-care [13]. This may be attributed to an unknown baseline immune status and the complicated nature of DENV infection, whose symptoms and severity are driven by antibody-dependent enhancement if a person has prior exposure to DENV or another flavivirus [14]. DENV infection is often clinically inapparent, and thereby harder to detect or confirm via conventional tests, especially in populations living in areas with multiple circulating flaviviruses [2, 15].
In 2018, we launched the longitudinal PAGODAS (Pediatric Assessment Group of Dengue and Aedes Saliva) cohort study in periurban Cambodia to lay the epidemiological groundwork for investigations into the relationship between human immune response to Ae. aegypti mosquito saliva and dengue [16]. We hypothesize that a baseline level of saliva-specific Ae. aegypti antibodies is associated with future DENV infection. We fit statistical and geospatial models to DENV infection histories defined by neutralization titers to better understand how Ae. aegypti saliva antibodies are associated with disease outcomes in a real-world scenario, an at-risk dengue-naive subset of the population in a hyperendemic area for DENV transmission. These data will be useful for public health interventions that harness anti-mosquito saliva antibodies as targets for vector-based vaccine strategies or as epidemiological tools to guide vector control efforts in areas at high risk for mosquito-borne diseases.
METHODS
Study Population
PAGODAS is an ongoing prospective cohort study of approximately 771 children aged 2–9 years in Chbar Mon, Kampong Speu, Cambodia [16]. Chbar Mon is a periurban area with rural, transitional, and denser urban environments. All children live within 5 km from Kampong Speu District Referral Hospital with a catchment area encompassing approximately 50000 inhabitants. Enrollment occurred in 2018 with follow-up semi-annual sampling during rainy and dry seasons. The cohort is stratified for equal numbers of sex and age in 2-year blocks. All households were georeferenced using house visits with hand-held GPS devices. Demographic data including age, sex, school, and behavioral risk factors (eg, insecticide use) were collected via mobile devices using standardized forms in REDCap databases [17].
Study Oversight
The study protocol was approved by the institutional review boards at the US National Institutes of Health and the National Ethics Committee on Human Research in Cambodia. The guardians of all pediatric participants provided signed informed consent to participate in the study.
Clinical and Laboratory Procedures
Sera were collected every 6 months to measure dengue and Ae. aegypti saliva-specific immune responses. At baseline, participants were defined as dengue-naive, seropositive to 1 serotype (monotypic immunity) or seropositive to >1 serotype (multitypic immunity) based on screening PanBio Dengue Indirect IgG enzyme-linked immunosorbent assay (ELISA) and confirmed by evidence of DENV1–4 neutralizing antibody titers >1:40. For those positive on screening DENV ELISA at baseline or at follow-up scheduled visits, neutralizing antibodies to DENV were determined by plaque reduction neutralization tests (PRNTs) in Vero cells as previously described [18]. If an individual was positive on ELISA but negative for the presence of DENV-specific neutralizing antibodies, then serostatus was defined by following repeat neutralization titers. The reciprocal of the lowest calculated dilution which reduced the virus by 50% (PRNT50) was reported as the neutralizing titer. Seropositivity to a particular serotype was defined as PRNT50 >1:40 given that Cambodia is hyperendemic for DENV.
Participants received thermometers and were instructed to report to hospital at the first sign of fever, defined as axillary temperature of 37.5ºC, to undergo a DENV rapid test (SD Bioline DengueDuo NS1 Ag/immunoglobulin M [IgM]/IgG). If positive, viremia was confirmed via a multiplex reverse-transcription polymerase chain reaction (RT-PCR) for DENV, Zika virus (ZIKV), and chikungunya virus (Fast Track Diagnostics ZDC Multiplex kit). Study staff collected blood for PCR and complete blood count. All DENV-positive cases were admitted to the hospital per national guidelines.
Clinical and laboratory data were also collected via REDCap [17]. Clinically apparent dengue cases are defined as those presenting with fever or other symptoms and confirmed to have DENV1–4 via RT-PCR. Clinically inapparent dengue cases are defined as those who do not present with PCR-confirmed DENV during a sick visit, but seroconverted PRNT50 titers from <1:40 to >1:40 for at least 1 DENV serotype.
Quantification of total IgG to Ae. aegypti whole salivary gland homogenate (SGH), also herein referred to as salivary protein, was performed via ELISA in 96-well Immulon plates in duplicate via antibody binding to whole Ae. aegypti SGH 2 µL/ug concentration diluted in carbonate-bicarbonate buffer, reported as arbitrary ELISA units [19]. Each plate included sera (1:200 dilution) from all time points, a negative control (nonreactive human sera to SGH), an internal control (a pool of positive sera to SGH with optical density [OD] set to 0.25 at 450nm), and blank wells. The controls were chosen accordingly from a stored bank of human sera, well-characterized by reactivity to the saliva of various vectors, at the Laboratory of Malaria and Vector Research. Plate-to-plate normalization was done by multiplication of all OD values by a correcting factor to achieve the preset internal control OD 0.25 at 450nm. Secondary antibodies (goat anti-human IgG [1:10 000; Millipore-Sigma A1543], goat anti-human IgM, and goat anti- human immunoglobulin A [IgA] [1:3000, Southern Biotech]) were used for isotypes, and mouse anti-human IgG1–4 (1:1000, Southern Biotech) were used for subclasses. Aedes aegypti IgG SGH antibody levels are classified as high/low using a cut point of 0.17 with log-transformed OD values, and all values below limit of detection at 0.13 were imputed as 0.13 for the hazard ratio (HR) calculations.
Statistical Analysis
The cohort endpoints are (1) seropositivity to DENV1–4 per ELISA with confirmatory PRNT50 assays presented as proportions by each season for each year; (2) seropositivity to Ae. aegypti SGH presented as proportions for each season each year of the study; and (3) HRs of infection comparing at-risk subjects with seroconversion to Ae. aegypti salivary protein to those without seroconversion at that time. A Cox proportional hazards model was used to model the time to any DENV infection, comparing those with high vs low immunoreactivity to Ae. aegypti saliva proteins, and controlling for many covariates including time-varying covariates and competing risks via Lunn–McNeil method. Missing visits were assumed to be completely at random and to have no events. This may bias the time-to-event to be later, but bias is equally applied to those with and without saliva protein seropositivity at baseline, having minimal effect on the effect estimates. The difference between groups in continuous secondary outcomes were tested by nonparametric Wilcoxon rank-sum test, or a 2-sample t test for inferences on differences in means. To test for differences among >2 groups, either a Kruskal–Wallis rank test (for nonparametric inferences) or an F-test (for inferences on the means) was used to test for overall differences. Multiple comparisons were adjusted using Tukey test.
A smoothed surface was generated from baseline seroreactivity to Ae. aegypti saliva proteins among DENV-naive participants using a quartic kernel density function, weighted by raw OD readings, with a radius around each point of 100 m (based on hypothesized Aedes mosquito ranges) [20]. An independent grid with 100 m × 100 m cells was generated, then overlaid on the smoothed surface, while DENV infections were overlaid on top of the grid using QGIS version 3.4.9. We calculated anti–Ae. aegypti SGH mean intensity for each cell using the Zonal Statistics function and tabulated DENV count per cell using a spatial join function. We used a generalized additive negative binomial regression model to investigate associations between antibodies to Ae. aegypti salivary proteins and the DENV count within each cell. We included an interaction term (smoothed spline function for each cell coordinates from their centroids to control for unmeasured space-dependent variables) and an offset term for the count of participants living within each cell.
All analyses were conducted using R, R cran version 4.0.3, and the mgcv package.
RESULTS
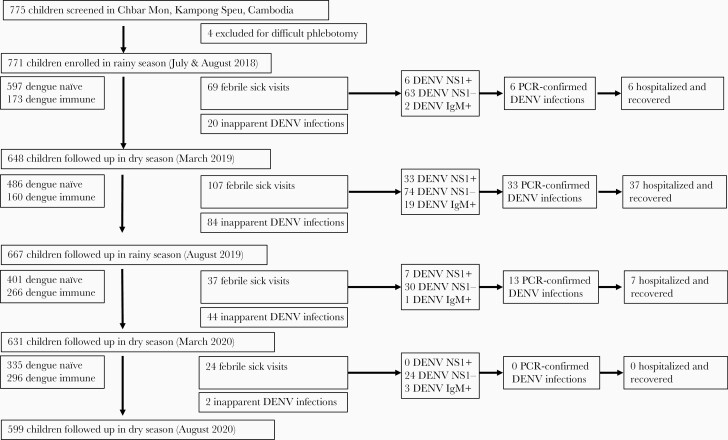
In July and August 2018 (rainy season 1), 771 children were enrolled into the cohort and contributed data to baseline seroprevalence of DENV and Ae. aegypti salivary proteins (Figure 1). We followed the participants in March 2019 (dry season 1; 648 children), August 2019 (rainy season 2; 667 children), March 2020 (dry season 2; 631 children), August 2020 (rainy season 3; 599 children) (Figure 1). The study population characteristics are shown in Table 1. Male-to-female ratio and age stratifications in 2-year blocks were equally distributed in the cohort (Table 1).
Figure 1.
Study flowchart. Seven hundred seventy-five children were screened for enrollment into the cohort; 771 children were enrolled and followed up every 6 months (± 1 month) for pan–dengue virus (DENV) and Aedes aegypti salivary gland homogenate enzyme-linked immunosorbent assay (ELISA) screening for seroprevalence. In between visits, children had semi-active surveillance for dengue by being encouraged to visit the hospital for febrile episodes. Children with symptomatic polymerase chain reaction–confirmed dengue were hospitalized per Cambodian national guidelines. Clinically inapparent dengue infections were any children who seroconverted via pan-DENV ELISA screen then confirmed via DENV1–4 50% plaque reduction neutralization test titers >1:40, but never had a clinically apparent or symptomatic DENV episode. Abbreviations: DENV, dengue virus; IgM, immunoglobulin M; PCR, polymerase chain reaction.
Table 1.
Cohort Demographics by Dengue Seropositivity Status at Baseline
| Characteristic | Total | Naive | Monotypic | Multitypic |
|---|---|---|---|---|
| No. | 771 | 598 | 115 | 58 |
| Age, y, median (IQR) | 6.0 (4.0–8.0) | 5.0(3.0–7.0) | 8.0(6.0–9.0) | 8.0(5.25–9.0) |
| Age at baseline, y | ||||
| 2–3 | 178 (23.1) | 164 (27.4) | 9 (7.8) | 5 (8.6) |
| 4–5 | 206 (26.7) | 183 (30.6) | 13 (11.3) | 10 (17.2) |
| 6–7 | 189 (24.5) | 147 (24.6) | 30 (26.1) | 12 (20.7) |
| 8–9 | 198 (25.7) | 104 (17.4) | 63 (54.8) | 31 (53.5) |
| Female sex | 394 (51.1) | 299(50.0) | 65(56.5) | 30(51.7) |
| Subject in school | 409 (53.0) | 275(46.0) | 92(80.0) | 42(72.4) |
| Housing type | ||||
| Floatingvillage/boat | 1 (0.1) | 0(0.0) | 1(0.9) | 0(0.0) |
| House | 769 (99.7) | 597(99.8) | 114(99.1) | 58(100.0) |
| Temporaryhousing | 1 (0.1) | 1(0.2) | 0(0.0) | 0(0.0) |
| Socioeconomic class | ||||
| Very poor | 1 (0.1) | 1(0.2) | 0(0.0) | 0(0.0) |
| Lower | 170 (22.0) | 134(22.4) | 25(21.7) | 11(19.0) |
| Middle | 599 (77.7) | 462(77.3) | 90(78.3) | 47(81.0) |
| Upper | 1 (0.1) | 1(0.2) | 0(0.0) | 0(0.0) |
| No. of domestic water containers in the home | ||||
| 1 | 196 (25.4) | 143(23.9) | 37(32.2) | 16(27.6) |
| 2 | 146 (18.9) | 120(20.1) | 16(13.9) | 10(17.2) |
| 3 | 127 (16.5) | 91(15.2) | 20(17.4) | 16(27.6) |
| 4 | 94 (12.2) | 81(13.5) | 11(9.6) | 2(3.4) |
| 5 | 89 (11.6) | 65(10.9) | 15(13.0) | 9(15.5) |
| ≥6 | 119 (15.5) | 98(16.4) | 16(13.9) | 5(8.6) |
| No. of toilets in the home | ||||
| 0 | 24 (3.1) | 20(3.3) | 3(2.6) | 1(1.7) |
| 1 | 537 (69.6) | 421(70.4) | 74(64.3) | 42(72.4) |
| 2 | 158 (20.5) | 120(20.1) | 28(24.3) | 10(17.2) |
| ≥3 | 52 (6.7) | 37(6.2) | 10(8.7) | 5(8.6) |
| Insecticide spray | ||||
| No | 351 (45.5) | 276(46.2) | 51(44.3) | 24(41.4) |
| Yes | 420 (54.5) | 322(53.8) | 64(55.7) | 34(58.6) |
| Larvicide applied to water | ||||
| No | 632 (82.0) | 494(82.6) | 94(81.7) | 44(75.9) |
| Yes | 139 (18.0) | 104(17.4) | 21(18.3) | 14(24.1) |
| How often mosquito coils burned | ||||
| Never | 264 (34.2) | 219(36.6) | 33(28.7) | 12(20.7) |
| Rarely(about1 time/mo) | 36 (4.7) | 23(3.8) | 9(7.8) | 4(6.9) |
| Sometimes(about1 time/wk) | 174 (22.6) | 138(23.1) | 22(19.1) | 14(24.1) |
| Often(about3 times/wk) | 8 (1.0) | 5(0.8) | 0(0.0) | 3(5.2) |
| Daily | 289 (37.5) | 213(35.6) | 51(44.3) | 25(43.1) |
Data are presented as No. (%) unless otherwise stated.
Abbreviation: IQR, interquartile range.
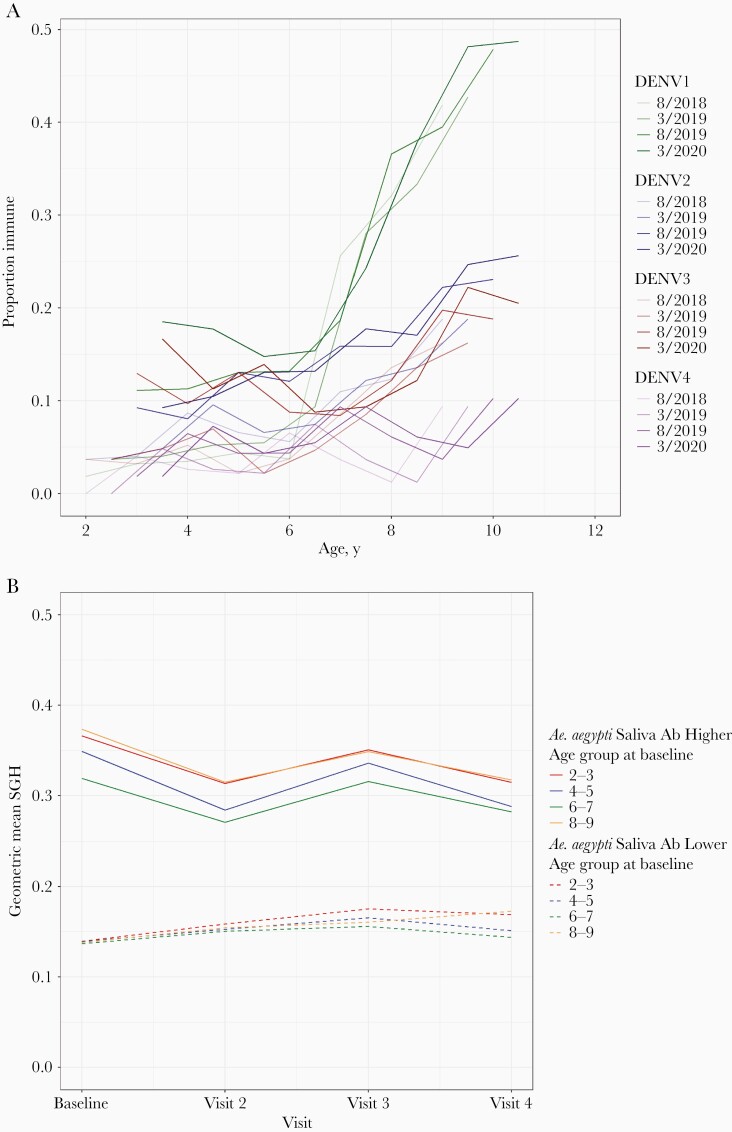
Overall dengue seroprevalence was estimated via indirect IgG ELISA screening and confirmation of positive IgG individuals by PRNT50 assay >1:40 for each of the 4 dengue serotypes. At baseline enrollment in 2018, DENV seroprevalence by indirect IgG ELISA was 37% (95% confidence interval [CI], 33.7%–0.7%; 286/770; 1 participant’s baseline sample was compromised). Confirmatory DENV neutralization assays decreased the seroprevalence to 22.5% (95% CI, 19.6%–25.6%; 173/770) with 15% (95% CI, 12.5%–17.7%; 115/770) as monotypic and 7.5% (95% CI, 5.8%–9.6%; 58/770) as multitypic, yielding an 18.9% false positivity rate (95% CI, 15.9%–22.3%) by ELISA alone (113 false positives and 597 true negatives; Supplementary Figure 1). Over the 24 months of follow-up, the most notable increase was with DENV1 seroprevalence, the result of epidemic dengue fever in 2019. Serotype-specific seroprevalence by DENV neutralization assays (PRNT50) stratified by age and time is detailed in Figure 2A; Supplementary Table 2, with DENV-1 and DENV-2 being most common and all multitypic seropositivity increasing with age (Figure 2A; Supplementary Figure 1). The median age of dengue-naive children to contract symptomatic dengue was 6.33 years vs 5.44 years for clinically inapparent dengue (P = .009).
Figure 2.
Seroprevalence to dengue and Aedes aegypti salivary proteins. Seroprevalence to dengue and Aedes aegypti saliva over the first 24 months of study follow-up with sampling every 6 months alternating rainy and dry seasons (eg, baseline and visit 3 are rainy seasons). A, Serotype-specific dengue seroprevalence confirmed by neutralization assays, by age. B, Binary antibody response to Aedes aegypti salivary proteins (higher/lower). Abbreviations: DENV, dengue virus; SGH, salivary gland homogenate.
Over 24 months of study follow-up, 237 children presented with fever in between scheduled visits (Figure 1). Of these, 52 episodes of apparent dengue were confirmed by PCR (in 51 children, 1 of whom had 2 infections). Most (n = 37) occurred from March to August 2019 when Cambodia’s worst-recorded dengue outbreak occurred. Surprisingly, the majority of the apparent dengue cases were dengue-naive (40/52 [78%]) (Supplementary Table 1). There were no cases of dengue shock syndrome, but 1 child developed dengue with severe anemia (hemoglobin 4.3mg/dL) requiring blood transfusion and otherwise recovered. PCR-confirmed dengue cases had a significant leukopenia and thrombocytopenia compared to dengue-negative febrile individuals (median white blood cell count, 4.9 vs 9.4, P < .001; median platelets, 221000/µL vs 285000/µL, P = .029; Supplementary Table 1). All children with apparent dengue fully recovered. A total of 148 clinically inapparent dengue cases (148/202 [73.3%] of dengue cases) were detected via neutralization assays performed at follow-up visits until March 2020 (Figure 1). Curtailed activities due to COVID-19 had a moderate impact on follow-up during 2020, but dengue infections remained at an all-time low nationally and at study site with only 2 additional seroconversions by August 2020.
Total IgG antibody levels to Ae. aegypti salivary proteins were significantly different between rainy and dry seasons, but highly correlated at an individual level (geometric mean arbitrary ELISA units: 0.263 in wet seasons vs 0.244 in dry seasons; P < .001) (Figure 2B; Supplementary Tables 3 and 4). We randomly selected 135 subjects at baseline to evaluate IgG subclasses and IgA and IgM isotypes for seroreactivity to Ae. aegypti salivary proteins. The predominant subclass was IgG4 with few individuals displaying IgG1-specific seroreactivity (Supplementary Figure 2A). We also detected production of IgM and IgA isotypes specific to Ae. aegypti salivary proteins in this cohort (Supplementary Figure 2B).
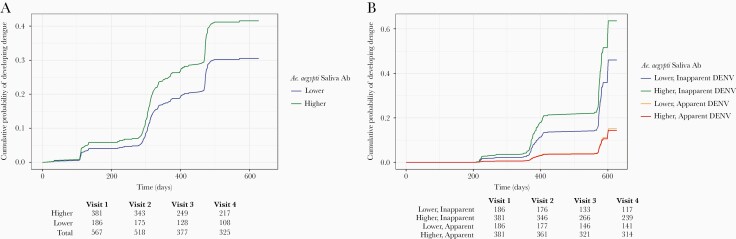
In the dengue-naive population at baseline (n = 597), higher levels of Ae. aegypti saliva antibodies increased the risk of DENV infection. Participants with higher levels of Ae. aegypti saliva antibodies had 1.47 times the risk (95% CI, 1.05–2.05) of developing DENV infection after adjusting for sex, age, insecticide use, larvicide use, school attendance, and mosquito coil use in addition to time-varying temperature and rainfall (Table 2; Figure 3A; Supplementary Tables 5 and 6). We observed that insecticide use, defined colloquially as spraying the interior of one’s house with commercial over-the-counter insect spray, typically intended for roaches or ants, was associated with lower DENV infection (HR, 0.66 [95% CI, .48–.91]). Higher temperature, time-lagged by the previous visit, was associated with increased risk of DENV (HR, 1.12 [95% CI, 1.00–1.25] per increase in °C) (Table 2). The DENV-naive population showed an increased risk of inapparent DENV with higher Ae. aegypti saliva antibodies (HR, 1.64 [95% CI, 1.12–2.41]; P = .013); however, the risk for apparent DENV was near 1 (HR, 0.95 [95% CI, .49–1.83]; P = .87) with no significant difference between subgroups (Supplementary Table 7).
Table 2.
Risk Factors for Dengue Seroconversion
| Risk Factor | Hazard Ratio | (95% CI) | P Value |
|---|---|---|---|
| Age (per year) | 1.0896 | (.9866–1.2035) | .0905 |
| Sex | |||
| Male | 1.0495 | (.7816–1.4092) | .7480 |
| Female | Reference | ||
| Educational status | |||
| Not in school | 0.8586 | (.5568–1.3240) | .4902 |
| In school | Reference | ||
| Socioeconomic class | |||
| Upper/middle | 1.0011 | (.6811–1.4714) | .9957 |
| Lower/very poor | Reference | ||
| No. of domestic water containers in the home | 0.9999 | (.9413–1.0621) | .9961 |
| No. of toilets in the home | 1.1479 | (.9608–1.3716) | .1287 |
| Use of insecticide spray | |||
| Uses insecticide spray | 0.6644 | (.4846–.9109) | .0111 |
| Does not use insecticide spray | Reference | ||
| Use of larvicide | |||
| Applies larvicide to water | 1.3025 | (.8878–1.9107) | .1765 |
| Does not apply larvicide | Reference | ||
| How often mosquito coils burned | |||
| Never | 1.2735 | (.9142–1.7740) | .1528 |
| Sometimes/rarely (1–3 times/wk) | 0.7176 | (.4731–1.0886) | .1186 |
| Daily/often | Reference | ||
| Aedes aegypti salivary protein antibodies | |||
| High | 1.4723 | (1.0533–2.0578) | .0236 |
| Low | Reference | ||
| Average rainfall | 0.9985 | (.9946–1.0024) | .4437 |
| Average temperature | 1.1207 | (1.0027–1.2527) | .0448 |
Abbreviation: CI, confidence interval.
Figure 3.
Hazard risk of Aedes aegypti salivary protein antibody levels as predictor of dengue infection in a dengue-naive population at baseline. Cox proportional hazards model showing the effects of higher (green) vs lower (blue) Ae. aegypti mosquito salivary protein antibody levels on the risk of dengue infection (A); and the effect of Ae. aegypti salivary protein antibody levels on the risk of inapparent dengue (green and blue) vs apparent, or symptomatic, dengue (red and orange) (B). Steep increases every 6 months are artifact of sampling scheme for inapparent dengue cases compared to apparent cases that were diagnosed on a rolling basis. Abbreviations: Ab, antibody; DENV, dengue virus.
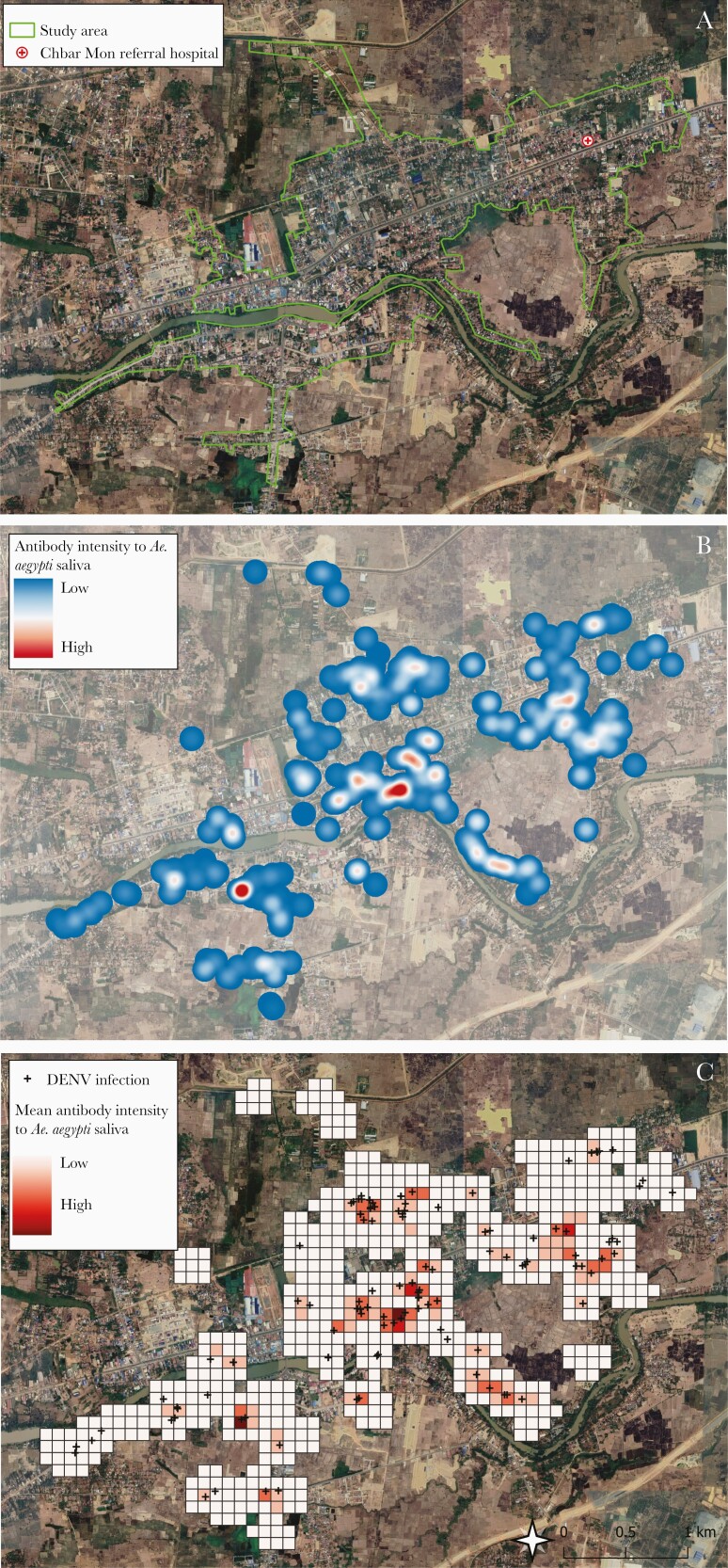
Results from the grid cell analysis indicated that areas with high immunoreactivity to Ae. aegypti salivary proteins overlapped with apparent and inapparent DENV cases (Figure 4). Bivariable analysis demonstrated a dose-response between increasing DENV cases and Ae. aegypti saliva reactivity (Supplementary Figure 3). The negative binomial regression analysis indicated that a 1-unit increase in mean Ae. aegypti saliva antibody level was associated with a 29% increase in DENV infection incidence in that 100-m grid cell (incidence rate ratio, 1.29 [95% CI, 1.02–1.63]).
Figure 4.
Map of study area and visual description of geospatial model of antibody levels to Aedes aegpyti salivary proteins on predicting dengue risk. A, Aerial view of study site (designated by green lines) with hospital (red cross). B, Smoothed surface depicting intensity of participants’ seroreactivity to Ae. aegypti salivary proteins, generated from optical density values. C, Grid cells (100 m × 100 m) and mean Ae. aegypti salivary protein antibody values at the grid cell level (higher values are darker red), with both apparent and inapparent dengue virus infections overlaid (black crosses). Abbreviation: DENV, dengue virus.
DISCUSSION
Here, we use detailed DENV infection histories to show that higher levels of antibodies to Ae. aegypti saliva are associated with DENV infection, particularly inapparent dengue infection. While it is intuitive that infection is associated with vector contact, our cohort is the first study designed to show an empirical association between infection status and the accumulation of measurable antibody responses to mosquito saliva proteins. These data lay the future groundwork for (1) consideration of saliva-specific antibodies as yet another factor in the complex clinical picture of arboviral immunopathology; (2) development of Ae. aegypti mosquito saliva–based biomarkers that can prospectively monitor the risk of DENV disease prior to infection; and (3) mapping human antibody specificity to salivary Ae. aegypti proteins to understand its association with the inapparent dengue outcome in a pediatric population.
Prior studies contributed to understanding anti-saliva antibodies at point-of-care in individuals, often with secondary DENV infections, in endemic areas [10–12]. These data logically follow the assumption that risk of disease increases as an individual is increasingly exposed to a vector, but saliva-specific antibodies have not yet predicted disease a priori. Indeed, dengue-viremic individuals present with increased antibody titers to Ae. aegypti SGH or extract, specifically IgG4 and some IgG1 as also seen in our study, when compared to uninfected individuals [11, 12]. A retrospective assessment of secondary DENV infections revealed increased seroreactivity to aegyptin, a 30-kDa Ae. aegypti mosquito saliva protein that binds collagen, in Thai children without increased vascular permeability vs those with hemorrhagic features [11]. The Ae. aegypti N-terminus 34kDa salivary peptide, a short-term indicator of exposure before and after vector control implementation [21], did not retrospectively distinguish between individuals who did or did not develop DENV infection in Laos, but would be potentially valuable in a prospective cohort design like here [13]. Previous field studies of Ae. aegypti saliva protein immunity may have been limited in drawing conclusions because of retrospective study design, convenience sampling at point-of-care, unknown baseline DENV seropositivity status, or lack of statistical power [10, 11, 21, 22]. These issues highlight the importance of (1) using longitudinal analysis of dengue-naive populations given the complicated interplay of flavivirus immunity driving disease presentation; and (2) performing neutralization assays to accurately define baseline infection histories in flavivirus-endemic areas [2, 11–13]. Indeed, in our study, indirect IgG ELISA misclassified nearly one-fifth of the DENV-naive population as DENV-seropositive at baseline compared to “gold standard” neutralization assays that are costly and time-consuming. This discrepancy highlights the critical need for more accurate and easily deployable tools to confirm dengue serostatus, a prerequisite to receive the only licensed dengue vaccine and a logistical barrier to widespread use [23].
DENV can be a life-threatening disease, for which there is no easily accessible antiviral treatment or vaccine for prevention, making the ability to predict disease of great clinical importance [2, 15]. Despite widespread mosquito sampling, entomological indicators poorly predict dengue transmission [24]. Application of mosquito saliva biomarkers to predict disease “hot spots” would afford the opportunity to intervene in the interim with vector control in high-risk areas to prevent infection until vaccines are widely available. Like others, we speculate that a mosquito saliva-based biomarker, ideally field-optimized via a finger prick, would be high-throughput and cost-saving in assessing the effectiveness of vector interventions or targeted rationing of vector control resources [21]. Our geospatial model supports additional evidence for the future use of saliva-based biomarkers to predict disease, showing strong overlap between preexisting immunity to Ae. aegypti salivary proteins and subsequent DENV infections.
While we lack a comprehensive and mechanistic understanding of the factors driving symptomatic and inapparent DENV infection, a clear link exists between severe disease and antibody-dependent enhancement as a result of heterologous DENV infection [25]. However, a variety of host and pathogen factors also contribute to clinical manifestations: (1) infecting DENV serotype or genotype; (2) preinfection flavivirus-specific neutralization antibody levels; (3) antibody boosting by exposure to circulating DENV; and (4) onset of DENV-specific T-cell IFN-γ responses [25–27]. The resulting pathology of arboviral infection is likely multifactorial across the pathogen-host-vector triad, although the vector is often overlooked [7, 15, 28]. The question is how vector-specific determinants of disease, such as higher preexisting levels of Ae. aegypti saliva antibodies, may play a role in an individual’s clinical course of infection. Our results suggest an association between increased Ae. aegypti saliva antibodies and inapparent DENV infection rather than symptomatic disease, though this may be attributed to the larger number of inapparent DENV cases. A large body of evidence in animal models demonstrates that increased mosquito saliva–specific antibody levels (generated via mosquito bites or needle inoculation of saliva) impact clinical outcomes such as level of viremia, level of dissemination to distant organs, and time to death [29–31]. Particularly, co-inoculation of Ae. aegypti salivary protein “aegyptin” with DENV altered the immune responses at the bite site in the hours immediately following infection in mosquito saliva–naive animals [5]. Yet, how the resulting anti-saliva antibodies may alter the microenvironment is not clear since salivary proteins have diverse functions.
On the whole, salivary proteins appear to promote infection and disease [28]. Hence, passive immunization with Ae. aegypti salivary proteins (NeST1, AgBR1, and anti-LTRIN) prevented ZIKV replication and improved clinical outcomes in mice via immunomodulation of the inflammatory milieu [32–34]. Similarly, peripheral blood mononuclear cells from human participants vaccinated with synthetic Anopheles gambiae saliva peptides had higher IFN-γ levels than those who received placebo [8]. The initial findings here are the first to show an association of increased humoral immunity to Ae. aegypti saliva and DENV clinical outcomes, but these findings are limited to a DENV-naive pediatric population. We chose a DENV-naive population since dengue-experienced individuals possess a preexisting, serotype-specific immune response that can impact outcomes. Unexpectedly, the symptomatic DENV burden was predominantly in DENV-naive children in our cohort. Yet, as expected, the majority of cases were inapparent, underlining the utility of a salivary biomarker that tracks risk in this difficult-to-surveil population as seen here. Another consideration is that mosquito saliva–specific antibodies do not wane to zero in Southeast Asia lowlands given the regional abundance of Ae. aegypti mosquitos year-round, hence the detectable antibodies in the dry season here. Studies in regions with demarcated seasons are better scenarios in which to study the waning of saliva-specific antibody responses and their role in infection dynamics [10].
The next challenging steps include identifying specific proteins or peptides within the Ae. aegypti salivary repertoire that confer differential risk of disease [11, 29]. Immunogenic salivary proteins may not possess a clear function, or even an advantageous effect like the viral inhibitory properties of D7 that make it a poor vaccine target [29]. Conversely, less immunogenic proteins such as those discovered via yeast display may have critical disease-promoting properties that can be targeted in a vector-based vaccine [32, 33]. Further understanding of mosquito saliva immunity in endemic populations yields clinical promise in 2 critical areas: (1) the utility of using mosquito saliva antibodies as an epidemiological tool to predict risk of disease and guide vector control interventions; and (2) the potential to identify targets for Aedes species saliva-based vaccine approaches for disease prophylaxis [8, 21].
Supplementary Data
Supplementary materials are available at The Journal of Infectious Diseases online. Supplementary materials consist of data provided by the author that are published to benefit the reader. The posted materials are not copyedited. The contents of all supplementary data are the sole responsibility of the authors. Questions or messages regarding errors should be addressed to the author.
Notes
Acknowledgments. We thank the participating children and their parents in Chbar Mon, Kampong Speu. We also acknowledge the clinical staff of Kampong Speu District Referral Hospital for their excellent patient care and collaboration in this project. We thank Jesus Valenzuela for his critical review of the manuscript. We thank Steve Whitehead, Anna Durbin, and Emerito Amaro-Carambot for their expertise and technical assistance in transfer of their dengue neutralization assay protocols to Cambodia. The clinical protocol is available at https://clinicaltrials.gov/ct2/show/NCT03534245 and published elsewhere [16]. Dengue sequences are available in the sequence read archive at the National Center for Biotechnology Information.
Disclaimer. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the US government.
Financial support. This work was supported by the Division of Intramural Research at the National Institute of Allergy and Infectious Diseases and in part with federal funds from the National Cancer Institute (contract number 75N910D00024, task order number 75N91019F00130 to A. M.).
Presented in part: 68th Annual Meeting of the American Society of Tropical Medicine and Hygiene, National Harbor, Maryland, November 2019.
Contributor Information
Jessica E Manning, Laboratory of Malaria and Vector Research, National Institute of Allergy and Infectious Diseases, National Institutes of Health, Bethesda, Maryland, USA; International Center of Excellence in Research, National Institute of Allergy and Infectious Diseases, National Institutes of Health, Phnom Penh, Cambodia.
Sophana Chea, International Center of Excellence in Research, National Institute of Allergy and Infectious Diseases, National Institutes of Health, Phnom Penh, Cambodia; National Center for Parasitology, Entomology, and Malaria Control, Ministry of Health, Phnom Penh, Cambodia.
Daniel M Parker, University of California, Irvine, Irvine, California, USA.
Jennifer A Bohl, Laboratory of Malaria and Vector Research, National Institute of Allergy and Infectious Diseases, National Institutes of Health, Bethesda, Maryland, USA.
Sreyngim Lay, International Center of Excellence in Research, National Institute of Allergy and Infectious Diseases, National Institutes of Health, Phnom Penh, Cambodia; National Center for Parasitology, Entomology, and Malaria Control, Ministry of Health, Phnom Penh, Cambodia.
Allyson Mateja, Clinical Monitoring Research Program Directorate, Frederick National Laboratory for Cancer Research, Frederick, Maryland, USA.
Somnang Man, International Center of Excellence in Research, National Institute of Allergy and Infectious Diseases, National Institutes of Health, Phnom Penh, Cambodia; National Center for Parasitology, Entomology, and Malaria Control, Ministry of Health, Phnom Penh, Cambodia.
Sreynik Nhek, International Center of Excellence in Research, National Institute of Allergy and Infectious Diseases, National Institutes of Health, Phnom Penh, Cambodia; National Center for Parasitology, Entomology, and Malaria Control, Ministry of Health, Phnom Penh, Cambodia.
Aiyana Ponce, Laboratory of Malaria and Vector Research, National Institute of Allergy and Infectious Diseases, National Institutes of Health, Bethesda, Maryland, USA.
Sokunthea Sreng, International Center of Excellence in Research, National Institute of Allergy and Infectious Diseases, National Institutes of Health, Phnom Penh, Cambodia; National Center for Parasitology, Entomology, and Malaria Control, Ministry of Health, Phnom Penh, Cambodia.
Dara Kong, International Center of Excellence in Research, National Institute of Allergy and Infectious Diseases, National Institutes of Health, Phnom Penh, Cambodia; National Center for Parasitology, Entomology, and Malaria Control, Ministry of Health, Phnom Penh, Cambodia.
Soun Kimsan, National Center for Parasitology, Entomology, and Malaria Control, Ministry of Health, Phnom Penh, Cambodia; National Dengue Control Program, Ministry of Health, Phnom Penh, Cambodia.
Claudio Meneses, Laboratory of Malaria and Vector Research, National Institute of Allergy and Infectious Diseases, National Institutes of Health, Bethesda, Maryland, USA.
Michael P Fay, Biostatistics Branch, National Institute of Allergy and Infectious Diseases, National Institutes of Health, Bethesda, Maryland, USA.
Seila Suon, International Center of Excellence in Research, National Institute of Allergy and Infectious Diseases, National Institutes of Health, Phnom Penh, Cambodia; National Center for Parasitology, Entomology, and Malaria Control, Ministry of Health, Phnom Penh, Cambodia.
Rekol Huy, National Center for Parasitology, Entomology, and Malaria Control, Ministry of Health, Phnom Penh, Cambodia.
Chanthap Lon, Laboratory of Malaria and Vector Research, National Institute of Allergy and Infectious Diseases, National Institutes of Health, Bethesda, Maryland, USA; International Center of Excellence in Research, National Institute of Allergy and Infectious Diseases, National Institutes of Health, Phnom Penh, Cambodia.
Rithea Leang, National Center for Parasitology, Entomology, and Malaria Control, Ministry of Health, Phnom Penh, Cambodia; National Dengue Control Program, Ministry of Health, Phnom Penh, Cambodia.
Fabiano Oliveira, Laboratory of Malaria and Vector Research, National Institute of Allergy and Infectious Diseases, National Institutes of Health, Bethesda, Maryland, USA.
REFERENCES
- 1. Fauci AS, Morens DM.. Zika virus in the Americas—yet another arbovirus threat. N Engl J Med 2016; 374:601–4. [DOI] [PubMed] [Google Scholar]
- 2. Bhatt S, Gething PW, Brady OJ, et al. The global distribution and burden of dengue. Nature 2013; 496:504–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Mellanby K. Man’s reaction to mosquito bites. Nature 1946; 158:554. [DOI] [PubMed] [Google Scholar]
- 4. Kamhawi S, Belkaid Y, Modi G, Rowton E, Sacks D.. Protection against cutaneous leishmaniasis resulting from bites of uninfected sand flies. Science 2000; 290:1351–4. [DOI] [PubMed] [Google Scholar]
- 5. McCracken MK, Christofferson RC, Grasperge BJ, Calvo E, Chisenhall DM, Mores CN.. Aedes aegypti salivary protein “aegyptin” co-inoculation modulates dengue virus infection in the vertebrate host. Virology 2014; 468–470:133–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Pingen M, Bryden SR, Pondeville E, et al. Host inflammatory response to mosquito bites enhances the severity of arbovirus infection. Immunity 2016; 44:1455–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Dudley DM, Newman CM, Lalli J, et al. Infection via mosquito bite alters Zika virus tissue tropism and replication kinetics in rhesus macaques. Nat Commun 2017; 8:2096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Manning JE, Oliveira F, Coutinho-Abreu IV, et al. Safety and immunogenicity of a mosquito saliva peptide-based vaccine: a randomised, placebo-controlled, double-blind, phase 1 trial. Lancet 2020; 395:1998–2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ryan SJ, Carlson CJ, Mordecai EA, Johnson LR.. Global expansion and redistribution of Aedes-borne virus transmission risk with climate change. PLoS Negl Trop Dis 2019; 13:e0007213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Londono-Renteria B, Cardenas JC, Cardenas LD, et al. Use of anti-Aedes aegypti salivary extract antibody concentration to correlate risk of vector exposure and dengue transmission risk in Colombia. PLoS One 2013; 8:e81211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Machain-Williams C, Mammen MP Jr, Zeidner NS, et al. Association of human immune response to Aedes aegypti salivary proteins with dengue disease severity. Parasite Immunol 2012; 34:15–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Cardenas JC, Drame PM, Luque-Burgos KA, et al. IgG1 and IgG4 antibodies against Aedes aegypti salivary proteins and risk for dengue infections. PLoS One 2019; 14:e0208455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ndille EE, Dubot-Pérès A, Doucoure S, et al. Human IgG antibody response to Aedes aegypti Nterm-34kDa salivary peptide as an indicator to identify areas at high risk for dengue transmission: a retrospective study in urban settings of Vientiane city, Lao PDR. Trop Med Int Health 2014; 19:576–80. [DOI] [PubMed] [Google Scholar]
- 14. Katzelnick LC, Narvaez C, Arguello S, et al. Zika virus infection enhances future risk of severe dengue disease. Science 2020; 369:1123–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Christofferson RC, Parker DM, Overgaard HJ, et al. Current vector research challenges in the greater Mekong subregion for dengue, malaria, and other vector-borne diseases: a report from a multisectoral workshop March 2019. PLoS Negl Trop Dis 2020; 14:e0008302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Manning JE, Oliveira F, Parker DM, et al. The PAGODAS protocol: pediatric assessment group of dengue and Aedes saliva protocol to investigate vector-borne determinants of Aedes-transmitted arboviral infections in Cambodia. Parasit Vectors 2018; 11:664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Harris PA, Taylor R, Thielke R, Payne J, Gonzalez N, Conde JG.. Research electronic data capture (REDCap)—a metadata-driven methodology and workflow process for providing translational research informatics support. J Biomed Info 2009; 42:377–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Durbin AP, Karron RA, Sun W, et al. Attenuation and immunogenicity in humans of a live dengue virus type-4 vaccine candidate with a 30 nucleotide deletion in its 3’-untranslated region. Am J Trop Med Hyg 2001; 65:405–13. [DOI] [PubMed] [Google Scholar]
- 19. Oliveira F, Rowton E, Aslan H, et al. A sand fly salivary protein vaccine shows efficacy against vector-transmitted cutaneous leishmaniasis in nonhuman primates. Sci Transl Med 2015; 7:290ra90. [DOI] [PubMed] [Google Scholar]
- 20. McDonald PT. Population characteristics of domestic Aedes aegypti (Diptera: culicidae) in villages on the Kenya coast I. Adult survivorship and population size. J Med Entomol 1977; 14:42–8. [DOI] [PubMed] [Google Scholar]
- 21. Elanga Ndille E, Doucoure S, Poinsignon A, et al. Human IgG antibody response to aedes nterm-34kDa salivary peptide, an epidemiological tool to assess vector control in chikungunya and dengue transmission area. PLoS Negl Trop Dis 2016; 10:e0005109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Londono-Renteria BL, Shakeri H, Rozo-Lopez P, et al. Serosurvey of human antibodies recognizing Aedes aegypti D7 salivary proteins in Colombia. Front Public Health 2018; 6:111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lopez AL, Adams C, Ylade M, et al. Determining dengue virus serostatus by indirect IgG ELISA compared with focus reduction neutralisation test in children in Cebu, Philippines: a prospective population-based study. Lancet Glob Health 2021; 9:e44–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Bowman LR, Runge-Ranzinger S, McCall PJ.. Assessing the relationship between vector indices and dengue transmission: a systematic review of the evidence. PLoS Negl Trop Dis 2014; 8:e2848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Katzelnick LC, Gresh L, Halloran ME, et al. Antibody-dependent enhancement of severe dengue disease in humans. Science 2017; 358:929–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Alexander LW, Ben-Shachar R, Katzelnick LC, et al. Boosting can explain patterns of fluctuations of ratios of inapparent to symptomatic dengue virus infections. Proc Natl Acad Sci U S A 2021; 118:e2013941118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wijeratne DT, Fernando S, Gomes L, et al. Quantification of dengue virus specific T cell responses and correlation with viral load and clinical disease severity in acute dengue infection. PLoS Negl Trop Dis 2018; 12:e0006540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Conway MJ, Colpitts TM, Fikrig E.. Role of the vector in arbovirus transmission. Annu Rev Virol 2014; 1:71–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Reagan KL, Machain-Williams C, Wang T, Blair CD.. Immunization of mice with recombinant mosquito salivary protein D7 enhances mortality from subsequent West Nile virus infection via mosquito bite. PLoS Negl Trop Dis 2012; 6:e1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Machain-Williams C, Reagan K, Wang T, Zeidner NS, Blair CD.. Immunization with Culex tarsalis mosquito salivary gland extract modulates West Nile virus infection and disease in mice. Viral Immunol 2013; 26:84–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Donovan MJ, Messmore AS, Scrafford DA, Sacks DL, Kamhawi S, McDowell MA.. Uninfected mosquito bites confer protection against infection with malaria parasites. Infect Immun 2007; 75:2523–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Wang Y, Marin-Lopez A, Jiang J, Ledizet M, Fikrig E.. Vaccination with Aedes aegypti AgBR1 delays lethal mosquito-borne Zika virus infection in mice. Vaccines 2020; 8:145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Hastings AK, Uraki R, Gaitsch H, et al. Aedes aegypti NeSt1 protein enhances Zika virus pathogenesis by activating neutrophils. J Virol 2019; 93:e00395–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Jin L, Guo X, Shen C, et al. Salivary factor LTRIN from Aedes aegypti facilitates the transmission of Zika virus by interfering with the lymphotoxin-β receptor. Nat Immunol 2018; 19:342–53. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.