Abstract
Background
Dengue virus (DENV) often circulates endemically. In such settings with high levels of transmission, it remains unclear whether there are risk factors that alter individual infection risk.
Methods
We tested blood taken from individuals living in multigenerational households in Kamphaeng Phet province, Thailand for DENV antibodies (N = 2364, mean age 31 years). Seropositivity ranged from 45.4% among those 1–5 years old to 99.5% for those >30 years. Using spatially explicit catalytic models, we estimated that 11.8% of the susceptible population gets infected annually.
Results
We found that 37.5% of the variance in seropositivity was explained by unmeasured household-level effects with only 4.2% explained by spatial differences between households. The serostatus of individuals from the same household remained significantly correlated even when separated by up to 15 years in age.
Conclusions
These findings show that despite highly endemic transmission, persistent differences in infection risk exist across households, the reasons for which remain unclear.
Keywords: Dengue virus, serology, force of infection, drivers of transmission
We explored the drivers of dengue infection in an endemic setting testing blood taken from multigenerational households and looking for antibodies against dengue virus. We showed that most of the variance in seropositivity was explained by unmeasured house-level effects.
Dengue virus (DENV) is an arbovirus transmitted by Aedes mosquitoes [1]. The 4 serotypes have circulated in Thailand for decades, with all provinces in the country reporting cases throughout the year [2]. Human immunity, climate, and the genotypes in circulation are known to shape annual infection risk; however, there are likely a myriad of other unidentified human, entomological, and environmental factors that lead to differences in risk of individuals, households, and communities living within the same region [3–5]. The need to identify modifiable risk factors of infection is especially important because we still do not have clearly effective vaccines or widely available vector control technologies.
Dengue virus transmission tends to occur over small spatial scales [6, 7], resulting in the local clustering of cases in both space and time [8, 9]. In addition, the diversity of circulating viruses has also been shown to be closely linked to the number of individuals living in the community [10]. However, it remains unclear the extent to which such strong local spatial structure ultimately leads to different experiences of infection across communities.
Understanding underlying heterogeneities in DENV risk is complicated. In particular, relying on passive surveillance systems can be problematic. Most infections are not severe enough to result in healthcare visits. Across infectious diseases, the subset of infections that are detected by surveillance systems may be strongly affected by heterogeneities in healthcare seeking habits [11]. Surveillance systems themselves also are not perfect and often rely on cases that present at large tertiary care healthcare facilities, which tend to be located in urban centers. Mosquito data may provide an alternative approach to indirectly capture spatial heterogeneity in the underlying level of infection. Attempts to characterize the impact of mosquito abundance on infection risk have largely relied on mosquito larval indices (eg, container, household, and Breteau indices), with no consistent relationship identified [12, 13]. Adult mosquito densities have been associated with dengue virus incidence in some studies, although the robustness of this metric across diverse environments remains unexplored [12, 14, 15].
A more direct approach to understand spatial heterogeneities in dengue risk is through studies that explore the serostatus in community residents [16, 17]. By identifying whether individuals from different communities have DENV-specific antibodies, we can identify risk factors linked to their serostatus. Furthermore, we can combine the age of individuals and their serostatus to inform mathematical models that reconstruct the risk of infection and assess how it differs across space [18–20]. In this study, we use baseline data from a large multigenerational cohort study that recruited individuals across a large province in Thailand, and we also conducted mosquito trapping experiments, to estimate the underlying probability of infection and explore the drivers of infection risk.
METHODS
Cohort Details
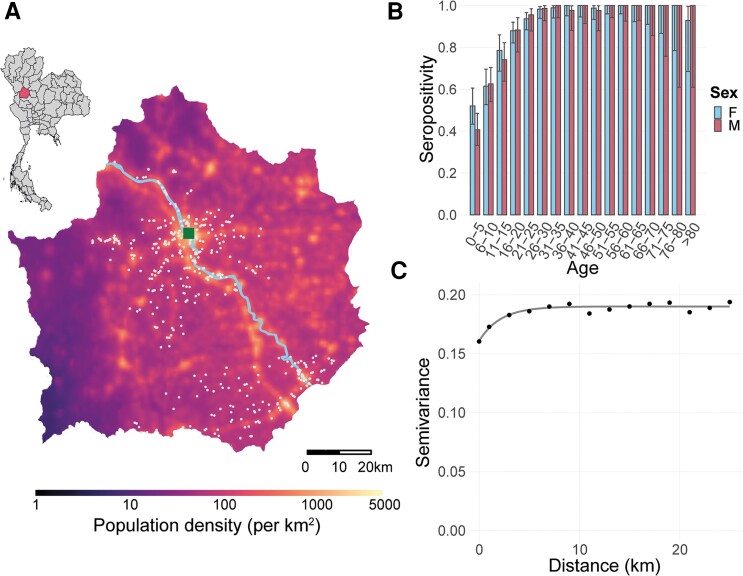
The details of the cohort study have been published elsewhere [21]. In brief, the cohort is located in Kamphaeng Phet province in Northern Thailand (Figure 1A). Between 2015 and 2021, expectant mothers and their households were screened for enrollment. A minimum of 4 multigenerational residents was required for enrollment of the household (the pregnant woman, her newborn, another child aged less than 15 years, and a grandparent of the newborn), but all household members were allowed the opportunity to participate. Blood was taken from all consenting household members at baseline as well as annually thereafter. In addition, household members were asked to complete questionnaires about individual and household characteristics. Approximately 23% of the population of the province of Kamphaeng Phet live within 1 km of one of our study households.
Figure 1.
Cohort details. (A) Map of study households in Kamphaeng Phet on a map of population density. Light blue line represents the river. Inset shows location of Kamphaeng Phet within Thailand. The white dots represent the location of the households. (B) Seropositivity by age and sex. (C) Semivariance in seropositivity across the province with a fitted exponential model.
Mosquito Data
In each year of the study, study teams randomly selected at least 100 households and used BG traps to collect mosquitoes indoors for an 8-hour period. Because households were randomly selected each year, some households were selected more than once and some households were never selected. All mosquitoes were counted and speciated [22]. In addition, potential breeding sites (including water containers, organic “containers” [eg, coconut shells], and plastic waste [eg, buckets, plastic bottles]) and larval abundance were enumerated. We calculated the container index for each household, defined as the proportion of water-holding containers that contain larvae or pupae.
Characterizing Urbanicity
We captured the urbanicity of a household using several different metrics. First, we obtained WorldPop estimates of the number of individuals living throughout the province. Second, we used satellite imagery to capture differences in land use and urban development across the study area. Finally, a summary statistic measuring whether an area surrounding a household was “developed” or not was created. More details on this designation can be found in the Supplementary Materials.
Determination of Serostatus
All serum samples were tested using hemagglutination inhibition assay to all 4 DENV serotypes [23]. We considered individuals to be seropositive (ie, had been infected with DENV in their lifetime) if they had a titer of 10 or greater to any serotype. Because Japanese encephalitis vaccination is routinely administered, which can result in cross-reacting dengue antibodies, we conducted sensitivity analyses in which we used a higher cutpoint of 20 to define seropositivity.
Capturing Spatial Dependence
To capture spatial dependence in serostatus, the number of mosquitoes, and the container index by household, we fit semivariograms to the status of each individual (or household for the mosquito measures) using an exponential model using the geoR package in R [24]. Semivariograms are a way of measuring the spatial autocorrelation between the labels (in this case serostatus, the number of mosquitoes and the container index) attached to points on a map. The extent of spatial dependence is the distance when the semivariogram becomes horizontal. When semivariograms give a completely horizontal line, there is no evidence of spatial dependence.
Estimating the Force of Infection
We constructed spatially explicit serocatalytic models using R-INLA. We fit the baseline (enrollment) serostatus of each individual using a binomial model with a cloglog link function with log(age) as an offset. The benefit of this approach is that it allowed us to directly estimate the force (or “hazard”) of infection [25]. We included a Matérn spatial correlation structure and a random intercept for each household. All individuals under 1 year in age were removed due to the potential presence of maternal antibodies. We initially fit a model using age only to estimate the overall force of infection. We then fit separate univariate models using the different individual-, household-, and community-level covariates of interest. In each model, log(age) was included as an offset to allow us to explore the impact of that covariate on the force of infection, taking into account the strong dependence on age. Because almost all individuals over 30 years in age were seropositive, we used only individuals under 30 years as the main model, but we also calculated a separate estimate for when we included all individuals. We also ran a multivariable model in which we included all terms, except in instances in which 2 terms were collinear (number of different types of containers as well as the different variables used to characterize urbanicity). We also only included a single measure of the number of containers and excluded the mosquito measures because they were only conducted on a subset of households.
To convert the estimated instantaneous force (or “hazard”) of infection to a measure of the proportion of the susceptible population infected each year, we used the following expression: 1 − exp( − λ), where λ is the estimated force of infection.
Estimating the Basic Reproductive Number
The basic reproductive number (R0) is the number of secondary infections generated by a primary infection as mediated by the vector in a completely susceptible population. We provide an average R0 estimate across all serotypes derived from our force of infection estimate and the national population age-structure. Our estimation assumes that individuals can be independently infected by the 4 serotypes and are subsequently immune to further infection by the same serotype [26]. The formulae can be found in the Supplementary Materials.
Spatial Prediction
We used the same spatially structured model structure to estimate the force of infection throughout the province using the log(age) as an offset and the Matérn spatial correlation structure. We could not include the covariates from our multivariable regression model in the spatial prediction because covariate values are not available outside our study households. We divided up the province in 1 km × 1 km cells. We then fit a serocatalytic model using the age of individuals only with a Matérn spatial covariance matrix. We then used the fitted model to predict the force of infection in each cell throughout the province.
We next estimated the number of infections occurring in each cell [16] (see Supplementary Materials for details). Because symptomatic infections tend to occur from primary and secondary infections, we present both the total number of primary/secondary infections as well as all infections. To assess the proportion of infections that are detected, we also extracted the average number of DENV cases reported to the national surveillance system from Kamphaeng Phet in the period 2003–2020.
Prediction Model Performance
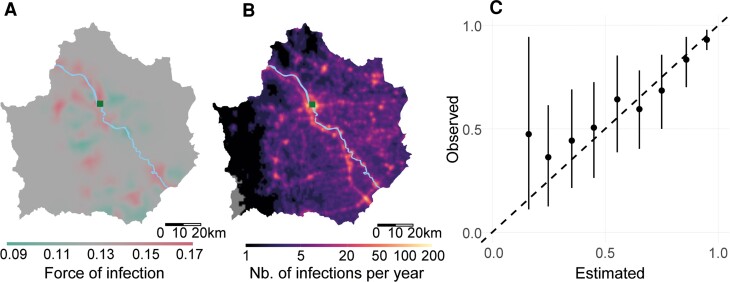
To assess the performance of the prediction model, we conducted additional analyses in which we predicted the serostatus for individuals in the model using 3 different methods. First, we fitted the model using data from all individuals and then predicted the serostatus on the same individuals. Second, we randomly removed 20% of individuals then fit the model using data from the remaining individuals. We then predicted the serostatus in the individuals not included in the model fitting process. Finally, we removed all individuals from clusters of households (representing 20% of all individuals in the dataset), where each cluster consisted of households within 10 km of randomly selected index households. We again fit the model using data from the remaining individuals and predicted the serostatus in the individuals not included in the model fitting process. For the second and third approaches, we created 500 such training and/or testing datasets. We then compared the predicted serostatus with that actually observed for those individuals. The presented spatial predictions (Figure 3) are when data from all individuals were included in the model fitting process.
Figure 3.
Spatial prediction. (A) Predicted force of infection across the province. (B) Predicted number of infections in the province. (C) Estimated versus predicted seropositivity in locations held out of the model fitting process.
Code Availability
All of the codes used in these analyses are available on a GitHub repository (pdgcam/kpp_drivers_dengue).
RESULTS
We recruited 2364 individuals from 649 households, representing 86.7% of the total number of people living in these households (Figure 1A). Individuals had a mean age of 31 years (range 0–100; mean age of males = 29 years, mean age of females = 32 years) and 58.9% were female (Table 1). The mean population density of households was 394 per km2 but ranged from 46 in the rural areas to 4540 in the urban Muang area. We found that 46.9% of households were located in developed areas based upon land use classification methods. We did not identify any spatial correlation in the number of mosquitoes or the container index trapped across households, including when we considered data from each year separately (Supplementary Figure S1 and S2).
Table 1.
Number of Study Subjects and Baseline Serostatus, by Individual-, Household-, and Community-Level Covariates
| Covariate | Total (N = 2376) | Seronegative (%) (N = 342) | Seropositive (%) (N = 2034) | |
|---|---|---|---|---|
| Individual Level | ||||
| Age | 1–5 | 291 | 159 (55) | 132 (45) |
| 6–10 | 253 | 96 (38) | 157 (62) | |
| 11–15 | 169 | 40 (24) | 129 (76) | |
| 16–20 | 226 | 27 (12) | 199 (88) | |
| 21–25 | 210 | 12 (6) | 198 (94) | |
| 26–30 | 189 | 3 (2) | 186 (98) | |
| 31–35 | 154 | 1 (1) | 153 (99) | |
| 36–40 | 118 | 1 (1) | 117 (99) | |
| 41–45 | 108 | 0 (0) | 108 (100) | |
| 46–50 | 125 | 2 (2) | 123 (98) | |
| 51–55 | 156 | 0 (0) | 156 (100) | |
| 56–60 | 141 | 0 (0) | 141 (100) | |
| 61–65 | 97 | 0 (0) | 97 (100) | |
| 66–70 | 62 | 0 (0) | 62 (100) | |
| 71–75 | 37 | 0 (0) | 37 (100) | |
| 76–80 | 20 | 0 (0) | 20 (100) | |
| >80 | 20 | 1 (5) | 19 (95) | |
| Sex | Female | 1399 | 163 (12) | 1236 (88) |
| Male | 977 | 179 (18) | 798 (82) | |
| Occupation | Employed | 654 | 11 (2) | 643 (98) |
| Farmer | 560 | 10 (2) | 550 (98) | |
| Student | 437 | 129 (30) | 308 (70) | |
| Unemployed | 725 | 192 (26) | 533 (74) | |
| Household Level | ||||
| No. of containers total | 0–10 | 324 | 43 (13) | 281 (87) |
| 11–20 | 568 | 84 (15) | 484 (85) | |
| 21–30 | 595 | 82 (14) | 513 (86) | |
| 31–40 | 295 | 40 (14) | 255 (86) | |
| 41–50 | 191 | 38 (20) | 153 (80) | |
| >50 | 400 | 54 (14) | 346 (87) | |
| No. of water containers - plastic | 0 | 783 | 110 (14) | 673 (86) |
| 1–10 | 969 | 128 (13) | 841 (87) | |
| 11–20 | 352 | 52 (15) | 300 (85) | |
| >20 | 272 | 52 (19) | 220 (81) | |
| House type | Poles | 413 | 54 (13) | 359 (87) |
| Single | 1875 | 273 (15) | 1602 (85) | |
| Shop-house style | 88 | 15 (17) | 73 (83) | |
| Garbage management | Burn | 1067 | 165 (15) | 902 (85) |
| Bury | 5 | 0 (0) | 5 (100) | |
| Pick-up | 1170 | 164 (14) | 1006 (86) | |
| Dump | 134 | 13 (10) | 121 (90) | |
| Highest household education | No school/primary | 461 | 73 (16) | 388 (84) |
| Secondary | 898 | 141 (16) | 757 (84) | |
| High school | 774 | 97 (13) | 677 (87) | |
| Higher | 243 | 31 (13) | 212 (87) | |
| Concrete household construction | Yes | 2067 | 303 (15) | 1764 (85) |
| No | 309 | 39 (13) | 270 (87) | |
| Type of roof | Not zinc | 622 | 102 (16) | 520 (84) |
| Zinc | 1754 | 240 (14) | 1514 (86) | |
| Water nearby | No | 739 | 109 (15) | 630 (85) |
| Yes | 1637 | 233 (14) | 1404 (86) | |
| Water supply | Other | 219 | 33 (15) | 186 (85) |
| Pipe | 2157 | 309 (14) | 1848 (86) | |
| Door screens | No | 2049 | 285 (14) | 1764 (86) |
| Yes | 327 | 57 (17) | 270 (83) | |
| Toilets outside | No | 1208 | 171 (14) | 1037 (86) |
| Yes | 1168 | 171 (15) | 997 (85) | |
| Community-level | ||||
| population density | Low | 1717 | 255 (15) | 1462 (85) |
| Med | 364 | 49 (13) | 315 (87) | |
| High | 295 | 38 (13) | 257 (87) | |
| Level of urbanicity | Low | 991 | 157 (16) | 834 (84) |
| Mid | 946 | 134 (14) | 812 (86) | |
| High | 439 | 51 (12) | 388 (88) | |
| Development index | Undeveloped | 1265 | 201 (16) | 1064 (84) |
| Developed | 1111 | 141 (13) | 970 (87) | |
Among participants who were >1 year old, 86% were seropositive (2029 of 2364). Female participants, who were slightly older on average, had a slightly higher seropositivity than males (88% vs 82%). We found a strong pattern of seropositivity by age, with 45.4% of 1- to 5-year-olds being seropositive (95% confidence interval [CI], 39.7–51.1) but 99.5% of over 30-year-olds being seropositive (95% CI, 98.9–99.8) (Figure 1B). We found there was spatial dependence in the serostatus of individuals up to a distance of 6.9 km (Figure 1C).
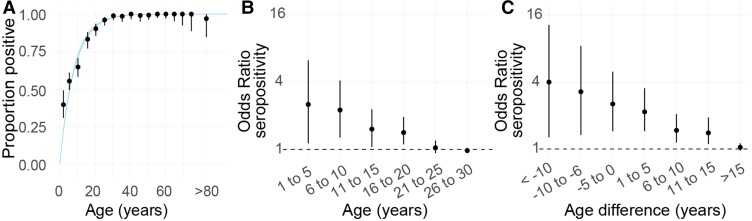
We initially fit a serocatalytic model using the ages of individuals only (Figure 2). We estimated an annual force of infection of 0.126 (95% CI, .116–.136), meaning that an average of 11.8% of the susceptible population gets infected with DENV each year. We estimated the R0 was 2.60 (95% CI, 2.44–2.76). The household-level random effect explained 37.5% of the variance in individual risk of being seropositive, and 4.2% of the variance could be explained through spatial differences outside the household. In models that included all age groups, the annual force of infection was 0.124 (95% CI, .115–.133), and R0 was 2.57 (95% CI, 2.43–2.71). In the base model, we used the serostatus and age of individuals from the moment of their enrollment (range, 2015–2018). We repeated the analysis using the age and serostatus from the blood draws from a single year (2018). We found that our results remained unchanged with an annual force of infection of 0.122 (95% CI, .113–.132).
Figure 2.
Serocatalytic model. (A) Proportion seropositive by 5-year age group (dots, with 95% confidence interval [CI]). The fit of a serocatalytic model with 95% CI is represented by the blue line. (B) Odds ratio of an individual being seropositive if a member in the household is seropositive by age of the individual with 95% CI. (C) Odds ratio of being seropositive if a member in the household is seropositive by age difference (age minus age of the household member) with 95% CI.
We found that overall, household members had 1.26 the odds (95% CI, 1.06–1.54) of being seropositive if an individual from that household was seropositive than if this individual was seronegative, with a consistent pattern across age groups (restricting the analysis to those under 30 years) (Figure 2B). This difference was maintained for pairs of household members who were up to 15 years difference in age (Figure 2C). By contrast, an individual that lived in a different household <1 km away from a seropositive individual had 1.01 the odds (95% CI, .95–1.09) of being seropositive than if they lived <1 km away from a seronegative individual (Supplementary Figure S3).
We used our regression approach to identify individual-, household-, and community-level risk factors associated with the force of infection (Table 2). Individuals from households that had door screens had 0.69 (95% CI, .50–.94; adjusted hazard ratio [aHR] 0.70; 95% CI, .49–1.01) the hazard of being seropositive compared with individuals from households that did not have door screens. The population density surrounding the household was associated with being seropositive, with each 1000 increase in population size associated with 1.10 the hazard of being seropositive (95% CI, .98–1.25; aHR 1.14; 95% CI, 1.00–1.31). Our more nuanced description of urban development also appeared to explain some differences in infection risk. Individuals who were categorized as living in a developed environment through this metric had 1.32 the hazard (95% CI, 1.05–1.66) of being seropositive than individuals who lived elsewhere. For the mosquito measures, the number of adult Aedes aegypti mosquitoes trapped in a household gave a hazard ratio of 1.01 (95% CI, 1.00–1.02) and the container index gave a hazard ratio of 1.67 (95% CI, .77–3.71). Models that used individuals of all ages or used a higher cutpoint to define seropositivity gave consistent results (Supplementary Tables S1–S2).
Table 2.
Results of Univariate and Multivariable Binomial Regression
| Covariate | Univariate | Multivariable | |
|---|---|---|---|
| HR (95% CI) | aHR (95% CI) | ||
| Individual Level | |||
| Sex | Female | REF | REF |
| Male | 0.90 (.74–1.10) | 0.88 (.72–1.08) | |
| Occupation | Employed | REF | REF |
| Farmer | 1.00 (.98–1.03) | 1.00 (.98–1.02) | |
| Student | 1.00 (.96–1.02) | 1.00 (.97–1.02) | |
| Unemployed | 1.00 (.98–1.03) | 1.00 (.98–1.02) | |
| Household Level | |||
| No. of water containers | 1.00 (1.00–1.01) | 1.02 (.94–1.10) | |
| No. of water containers - plastic | 1.00 (.99–1.01) | … | |
| No. of plastic bottles | 0.92 (.73–1.17) | … | |
| House Type | Poles | REF | REF |
| Single | 1.00 (.98–1.02) | 1.00 (.98–1.02) | |
| Townhouse | 1.00 (.98–1.02) | 1.00 (.98–1.02) | |
| Garbage management | Car collection | REF | REF |
| Burnt/buried/dumped | 1.00 (.79–1.26) | 1.03 (.79–1.33) | |
| Highest household education | No school/primary | 1.00 (.98–1.02) | 1.00 (.98–1.02) |
| Secondary | 1.00 (.98–1.02) | 1.00 (.98–1.02) | |
| High School | 1.00 (.98–1.02) | 1.00 (.98–1.02) | |
| Degree | REF | REF | |
| Concrete house | 1.12 (.87–1.43) | 1.07 (.75–1.54) | |
| Zinc roof | 1.27 (.99–1.65) | 1.22 (.91–1.65) | |
| Nearby source of water | 1.10 (.86–1.40) | 1.11 (.85–1.45) | |
| Water supply by pipe | 0.85 (.56–1.27) | 0.91 (.59–1.41) | |
| Door screens | 0.69 (.50–.94) | 0.70 (.49–1.01) | |
| Mean container index | 1.67 (.77–3.71) | … | |
| Adult aegypti mosquitoes captured | 1.01 (1.00–1.02) | … | |
| Toilets outside | 0.93 (.74–1.17) | 0.88 (.69–1.13) | |
| Community level | |||
| Population density | 1.10 (.98–1.25) | 1.14 (1.00–1.31) | |
| Level of urbanicity | 3.00 (1.09–8.40) | … | |
| Development index | 1.32 (1.05–1.66) | … | |
Abbreviations: aHR, adjusted hazard ratio; CI, confidence interval; HR, hazard ratio; REF, Reference category.
We used our framework to predict the force of infection throughout Kamphaeng Phet province. We found that the force of infection varied between 0.09 and 0.17, with communities along the river having a slightly increased force of infection than elsewhere, potentially a proxy for urbanization (Figure 3A). Using these estimates of the spatial heterogeneity in infection risk, the number of individuals living in the whole province, and the age structure of the population from the 2015 national census, we estimated the average number of infections each year in the province. We estimated there were an average of 27 517 primary and secondary infections and 54 001 total infections annually. We estimated that 9.5% of infections in the province occur within 5 km of the main city (Muang). The national surveillance system reported an average of 703 yearly infections for the province between 2013 and 2020. Assuming that all infections detected by the surveillance system are from primary and secondary infections, due to a lower rate of symptomatic disease in tertiary and/or quaternary infections, suggests that 2.6% of primary and/or secondary infections (and 1.3% of all infections) are detected by the surveillance system.
To assess the performance of our prediction model, we removed spatial clusters of households representing 20% of individuals and used the rest of the data to fit the model. We were able to accurately estimate the serostatus in the individuals excluded from the model-fitting process (Figure 3C, Supplementary Figure S4).
DISCUSSION
We have used the results of a large household study to explore underlying differences in risk for DENV infection. We found that DENV was highly endemic with 11.8% of the susceptible population being infected each year, with limited spatial variability in risk between households. We estimated that there are over 50 000 infections annually, only 1% of which are detected by the national surveillance system.
Our estimate of the mean force of infection of 0.13 is similar to that previously found for other provinces in Thailand, including estimates of 0.12 in Bangkok and 0.15 in Rayong province [20, 27]. We found limited heterogeneity in risk across the province, although households that were located in more urban environments had an elevated estimated risk, with a hazard ratio of 3.0 for urban settings compared with rural ones, consistent with studies elsewhere [16, 28]. By contrast, we found that the household appears to be a central determinant of infection risk, with one third of the overall variance in individual risk being explained by differences across households. This household effect appears to persist for many years, with seronegative individuals being more likely to live with other seronegative individuals, even when they are separated by up to 15 years in age. The correlation in serostatus disappeared when we considered individuals living in different households, even when those households were nearby. Virtually all individuals ultimately do become infected, but the household of residence appears to change the age of first infection by many years.
It remains unclear whether this key role played by the household is due to substantial within-household transmission or whether there are household-specific factors that can lead to long-term changes in risk. Among the potential household covariates we considered, the presence of door screens resulted in a 12% reduced hazard compared with households that had no screens. Overall, 13.1% of households had door screens installed. Door and window screens have previously been shown to be protective in studies in Puerto Rico, Australia, and Taiwan [29–32]. This modifiable household-level risk factor may represent an important opportunity to interrupt DENV transmission, worthy of further attention and research. Nevertheless, the use of door screens could only explain a fraction of the household effect on risk, and we did not identify other household-level factors linked to infection risk, despite considering many different facets of the household. More nuanced measures of household-level characteristics, including behaviors of household members, are needed.
We found that the number of adult mosquitoes trapped in the households was not linked to infection risk. The relationship between mosquito indices and infection risk has been highly variable across studies and regions [12]. The most consistent correlation has been between adult mosquito densities and DENV risk [14, 16], although this remains an imperfect proxy for actual exposure (ie, bite from an infected mosquito). In this study, we obtained mosquitoes from a subset of households (44.2%) with most households only providing data from a single time point. The large year-to-year differences in mosquito numbers may make levels from a single time point uninformative of lifetime risk. Longitudinal measures of mosquito number may be more predictive of underlying dengue risk [33]. Alternatively, the overall number of adult mosquitoes may not be linked to infection risk. In support of this latter hypothesis, cluster studies have found that only a small proportion (approximately 1%) of Aedes aegypti mosquitoes found around the households of confirmed cases have detectable virus [34]. In the context where only a small fraction of mosquitoes are involved in transmission, the overall number of mosquitoes may not be predictive of infection risk. This effective saturation of mosquitoes may be particularly relevant in Kamphaeng Phet where most households had both immature and adult mosquitoes at any household visit.
Our study has limitations. Our results come from a single snapshot of serostatus, and we have not considered time-varying changes in the force of infection or differences in the risk of infection by age. Nevertheless, even our simple model has been able to reconstruct the observed seropositivity by age. Our study design meant that we only recruited multigenerational households with an expectant mother. This may not be representative of all households in the region. The mean age of our cohort is also younger than the wider population. However, our modeling approach explicitly accounts for the age structure in the population, so this would not affect our estimates. We used a simple cutpoint to define seropositivity based on hemagglutination inhibition titers. Cross-reactivity with Japanese encephalitis vaccine may result in false positives; however, in a sensitivity analysis, we used a higher cutpoint that would be unlikely to result in false positives and obtained consistent results. Our estimate of R0 relied on assumptions of no cross-protection between serotypes and up to 4 infections over a lifetime. These assumptions may lead to underestimates in R0.
CONCLUSIONS
Kamphaeng Phet is likely typical of the dengue-endemic world, with high levels of year-on-year transmission at the population scale across the rural to urban gradient. Our findings show that despite the pervasiveness of the pathogen, individual households appear to have persistently different levels of risk to other neighboring households in the community, the reasons for which require further study.
Supplementary Data
Supplementary materials are available at The Journal of Infectious Diseases online (http://jid.oxfordjournals.org/). Supplementary materials consist of data provided by the author that are published to benefit the reader. The posted materials are not copyedited. The contents of all supplementary data are the sole responsibility of the authors. Questions or messages regarding errors should be addressed to the author.
Supplementary Material
Contributor Information
Gabriel Ribeiro dos Santos, Department of Genetics, University of Cambridge, United Kingdom.
Darunee Buddhari, Department of Virology, Armed Forces Research Institute of Medical Sciences, Bangkok, Thailand.
Sopon Iamsirithaworn, Department of Disease Control, Ministry of Public Health, Tiwanond, Nonthaburi, Thailand.
Direk Khampaen, Department of Disease Control, Ministry of Public Health, Tiwanond, Nonthaburi, Thailand.
Alongkot Ponlawat, Department of Entomology, Armed Forces Research Institute of Medical Sciences, Bangkok, Thailand.
Thanyalak Fansiri, Department of Entomology, Armed Forces Research Institute of Medical Sciences, Bangkok, Thailand.
Aaron Farmer, Department of Virology, Armed Forces Research Institute of Medical Sciences, Bangkok, Thailand.
Stefan Fernandez, Department of Virology, Armed Forces Research Institute of Medical Sciences, Bangkok, Thailand.
Stephen Thomas, Department of Medicine, SUNY Upstate Medical University, Syracuse, New York, USA; Department of Microbiology and Immunology, SUNY Upstate Medical University, Syracuse, New York, USA; Institute for Global Health and Translational Sciences, SUNY Upstate Medical University, Syracuse, New York, USA.
Isabel Rodriguez Barraquer, Department of Medicine, University of California, San Francisco, San Francisco, California, USA.
Anon Srikiatkhachorn, Department of Cell and Molecular Biology, Institute for Immunology and Informatics, University of Rhode Island, Providence, Rhone Island, USA; Faculty of Medicine, King Mongkut’s Institute of Technology Ladkrabang, Bangkok, Thailand.
Angkana T Huang, Department of Genetics, University of Cambridge, United Kingdom; Department of Virology, Armed Forces Research Institute of Medical Sciences, Bangkok, Thailand.
Derek A T Cummings, Department of Biology, University of Florida, Gainesville, USA; Emerging Pathogens Institute, University of Florida, Gainesville, USA.
Timothy Endy, Department of Medicine, SUNY Upstate Medical University, Syracuse, New York, USA; Department of Microbiology and Immunology, SUNY Upstate Medical University, Syracuse, New York, USA; Institute for Global Health and Translational Sciences, SUNY Upstate Medical University, Syracuse, New York, USA; Coalition for Epidemic Preparedness Innovations (CEPI), Washington, District of Columbia, USA.
Alan L Rothman, Department of Cell and Molecular Biology, Institute for Immunology and Informatics, University of Rhode Island, Providence, Rhone Island, USA.
Henrik Salje, Department of Genetics, University of Cambridge, United Kingdom; Department of Biology, University of Florida, Gainesville, USA.
Kathryn B Anderson, Department of Virology, Armed Forces Research Institute of Medical Sciences, Bangkok, Thailand; Department of Medicine, SUNY Upstate Medical University, Syracuse, New York, USA; Department of Microbiology and Immunology, SUNY Upstate Medical University, Syracuse, New York, USA; Institute for Global Health and Translational Sciences, SUNY Upstate Medical University, Syracuse, New York, USA.
Notes
Disclaimer . Material has been reviewed by the Walter Reed Army Institute of Research. There is no objection to its presentation and/or publication. The opinions or assertions contained herein are the private views of the author, and are not to be construed as official, or as reflecting true views of the Department of the Army or the Department of Defense. The investigators have adhered to the policies for protection of human subjects as prescribed in AR 70-25.
Financial support . This work was funded by the National Institutes of Health (Grant Number P01AI034533) and the European Research Council (Grant Number 804744).
References
- 1. Halstead SB. Dengue. Lancet 370:1644–52. [DOI] [PubMed] [Google Scholar]
- 2. Cummings DAT, Iamsirithaworn S, Lessler JT, et al. The impact of the demographic transition on dengue in Thailand: insights from a statistical analysis and mathematical modeling. PLoS Med 2009; 6:e1000139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Vu TTH, Holmes EC, Duong V, et al. Emergence of the Asian 1 genotype of dengue virus serotype 2 in Viet Nam: in vivo fitness advantage and lineage replacement in South-East Asia. PLoS Negl Trop Dis 2010; 4:e757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Salje H, Cummings DAT, Rodriguez-Barraquer I, et al. Reconstruction of antibody dynamics and infection histories to evaluate dengue risk. Nature 2018; 557:719–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. van Panhuis WG, Choisy M, Xiong X, et al. Region-wide synchrony and traveling waves of dengue across eight countries in Southeast Asia. Proc Natl Acad Sci U S A 2015; 112:13069–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Harrington LC, Scott TW, Lerdthusnee K, et al. Dispersal of the dengue vector Aedes aegypti within and between rural communities. Am J Trop Med Hyg 2005; 72:209–20. [PubMed] [Google Scholar]
- 7. Salje H, Wesolowski A, Brown TS, et al. Reconstructing unseen transmission events to infer dengue dynamics from viral sequences. Nat Commun 2021; 12:1810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bhoomiboonchoo P, Gibbons RV, Huang A, et al. The spatial dynamics of dengue virus in Kamphaeng Phet, Thailand. PLoS Negl Trop Dis 2014; 8:e3138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Anderson KB, Stewart-Ibarra AM, Buddhari D, et al. Key findings and comparisons from analogous case-cluster studies for dengue virus infection conducted in Machala, Ecuador, and Kamphaeng Phet, Thailand. Front Public Health 2020; 8:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Salje H, Lessler J, Maljkovic Berry I, et al. Dengue diversity across spatial and temporal scales: Local structure and the effect of host population size. Science 2017; 355:1302–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Nikolay B, Salje H, Sturm-Ramirez K, et al. Evaluating hospital-based surveillance for outbreak detection in Bangladesh: analysis of healthcare utilization data. PLoS Med 2017; 14:e1002218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Bowman LR, Runge-Ranzinger S, McCall PJ. Assessing the relationship between vector indices and dengue transmission: a systematic review of the evidence. PLoS Negl Trop Dis 2014; 8:e2848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Fustec B, Phanitchat T, Hoq MI, et al. Complex relationships between Aedes vectors, socio-economics and dengue transmission-Lessons learned from a case-control study in northeastern Thailand. PLoS Negl Trop Dis 2020; 14:e0008703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ong J, Aik J, Ng LC. Short report: Adult Aedes abundance and risk of dengue transmission. PLoS Negl Trop Dis 2021; 15:e0009475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Parra MCP, Fávaro EA, Dibo MR, et al. Using adult Aedes aegypti females to predict areas at risk for dengue transmission: a spatial case-control study. Acta Trop 2018; 182:43–53. [DOI] [PubMed] [Google Scholar]
- 16. Salje H, Paul KK, Paul R, et al. Nationally-representative serostudy of dengue in Bangladesh allows generalizable disease burden estimates. Elife 2019; 8:e42869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Bailly S, Rousset D, Fritzell C, et al. Spatial distribution and burden of emerging arboviruses in French Guiana. Viruses 2021; 13:1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Imai N, Dorigatti I, Cauchemez S, Ferguson NM. Estimating dengue transmission intensity from sero-prevalence surveys in multiple countries. PLoS Negl Trop Dis 2015; 9:e0003719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Salje H, Cauchemez S, Alera MT, et al. Reconstruction of 60 years of chikungunya epidemiology in the Philippines demonstrates episodic and focal transmission. J Infect Dis 2016; 213:604–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Rodríguez-Barraquer I, Buathong R, Iamsirithaworn S, et al. Revisiting Rayong: shifting seroprofiles of dengue in Thailand and their implications for transmission and control. Am J Epidemiol 2014; 179:353–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Anderson KB, Buddhari D, Srikiatkhachorn A, et al. An innovative, prospective, hybrid cohort-cluster study design to characterize dengue virus transmission in multigenerational households in Kamphaeng Phet, Thailand. Am J Epidemiol 2020; 189:648–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Fansiri T, Buddhari D, Pathawong N, et al. Entomological risk assessment for dengue virus transmission during 2016–2020 in Kamphaeng Phet, Thailand. Pathogens 2021; 10:1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Drăgănescu N, Girjabu E, Iacobescu V. Investigations on the presence of hemagglutination-inhibiting antibodies to dengue virus in apparently healthy subjects of several counties in Romania. Virologie 1975; 26:23–5. [PubMed] [Google Scholar]
- 24. Ribeiro JR, Diggle PJ. geoS: a geostatistical library for S-PLUS. Lancaster: Department of Mathmatics and Statistics, Lancaster University, 1999. [Google Scholar]
- 25. Bjørnstad ON. Epidemics: models and data using R. Cham: Springer; Nature Switzerland AG, 2018. [Google Scholar]
- 26. Ferguson NM, Donnelly CA, Anderson RM. Transmission dynamics and epidemiology of dengue: insights from age-stratified sero-prevalence surveys. Philos Trans R Soc Lond B Biol Sci 1999; 354:757–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hoang Quoc C, Henrik S, Isabel RB, et al. Synchrony of dengue incidence in Ho Chi Minh City and Bangkok. PLoS Negl Trop Dis 2016; 10:e0005188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Chew CH, Woon YL, Amin F, et al. Rural-urban comparisons of dengue seroprevalence in Malaysia. BMC Public Health 2016; 16:824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Waterman SH, Novak RJ, Sather GE, Bailey RE, Rios I, Gubler DJ. Dengue transmission in two Puerto Rican communities in 1982. Am J Trop Med Hyg 1985; 34:625–32. [DOI] [PubMed] [Google Scholar]
- 30. Ko YC, Chen MJ, Yeh SM. The predisposing and protective factors against dengue virus transmission by mosquito vector. Am J Epidemiol 1992; 136:214–20. [DOI] [PubMed] [Google Scholar]
- 31. Bowman LR, Donegan S, McCall PJ. Is dengue vector control deficient in effectiveness or evidence?: systematic review and meta-analysis. PLoS Negl Trop Dis 2016; 10:e0004551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. McBride WJ, Mullner H, Muller R, Labrooy J, Wronski I. Determinants of dengue 2 infection among residents of Charters Towers, Queensland, Australia. Am J Epidemiol 1998; 148:1111–6. [DOI] [PubMed] [Google Scholar]
- 33. Cromwell EA, Stoddard ST, Barker CM, et al. The relationship between entomological indicators of Aedes aegypti abundance and dengue virus infection. PLoS Negl Trop Dis 2017; 11:e0005429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Yoon I-K, Getis A, Aldstadt J, et al. Fine scale spatiotemporal clustering of dengue virus transmission in children and Aedes aegypti in rural Thai villages. PLoS Negl Trop Dis 2012; 6:e1730. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.