Abstract
The orientation of cell polarity depends on the position of the centrosome, the main microtubule‐organizing center (MTOC). Microtubules (MTs) transmit pushing forces to the MTOC as they grow against the cell periphery. How the actin network regulates these forces remains unclear. Here, in a cell‐free assay, we used purified proteins to reconstitute the interaction of a microtubule aster with actin networks of various architectures in cell‐sized microwells. In the absence of actin filaments, MTOC positioning was highly sensitive to variations in microtubule length. The presence of a bulk actin network limited microtubule displacement, and MTOCs were held in place. In contrast, the assembly of a branched actin network along the well edges centered the MTOCs by maintaining an isotropic balance of pushing forces. An anisotropic peripheral actin network caused the MTOC to decenter by focusing the pushing forces. Overall, our results show that actin networks can limit the sensitivity of MTOC positioning to microtubule length and enforce robust MTOC centering or decentering depending on the isotropy of its architecture.
Keywords: actin network, centrosome positioning, microtubule, MTOC, synthetic cell
Subject Categories: Cell Adhesion, Polarity & Cytoskeleton; Cell Cycle
In vitro reconstitution in a cell‐free system reveals how pushing forces in microtubules growing against the actin network contribute to centrosome positioning.

Introduction
The network of microtubules (MTs) supports the construction of cell body plan and directs its symmetry axes (Bornens, 2008; Vignaud et al, 2012; Meiring et al, 2020). The asymmetric organization of MTs directs the preferential orientation of vesicle transport and the position of key sensory organelles and thereby orients the main functions of polarized cells (Harris et al, 2009; Vladar et al, 2012; Bornens, 2018; Meiring et al, 2020). The centrosome is the main microtubule‐organizing center (MTOC), so its position is a key determinant of cell polarity (Bornens, 2008; Tang & Marshall, 2012). The positioning of centrosome, either at the cell center or at the cell periphery, in contact with the plasma membrane, is important for ciliogenesis, immune reactions, cell division, epithelial‐to‐mesenchymal transition, or neuronal development (Elric & Etienne‐Manneville, 2014; Stinchcombe & Griffiths, 2014; Burute et al, 2017; Pitaval et al, 2017; Shao et al, 2020).
Centrosome position is mainly controlled by combinations of pushing and pulling forces produced in the MT network (Burakov et al, 2003; Zhu et al, 2010; Pavin et al, 2012; Ma et al, 2014; Letort et al, 2016; Jimenez et al, 2021). Homogeneous distribution of minus‐end directed molecular motors, pulling on MTs in the cytoplasm or at the cell cortex, can enforce centrosome centering (Koonce et al, 1999; Kimura & Kimura, 2011; Wu et al, 2011; Laan et al, 2012). Heterogeneous distribution of motors, due to local accumulations, can locally increase pulling forces and enforce MTOC decentering up to the contact with the plasma membrane (Dujardin et al, 2003; Yi et al, 2013). Regulation of centrosome positioning by such pulling forces is robust: centering, in the case of homogeneous distribution of motors, or peripheral positioning, in the case of heterogeneous distribution, are both poorly sensitive to variations of MT length (Letort et al, 2016). On the opposite, production of pushing forces is much more sensitive to variations of MT length and hence appears as a less reliable positioning mechanism. MT‐based pushing is ineffective if MTs are too short to reach the spatial boundaries. Longer MTs may allow centering if their length corresponds precisely to the length of its confining region. However, MTs longer than this critical length will induce an abrupt transition from centering to decentering (Holy et al, 1997; Faivre‐Moskalenko & Dogterom, 2002; Pinot et al, 2009; Laan et al, 2012).
Actin networks are involved in centrosome positioning and have been proposed to modulate the pushing forces produced by MTs, but the underlying mechanisms are obscure (Chevrier et al, 2002; Burakov et al, 2003; Brito et al, 2005; Hale et al, 2011; Pelletier et al, 2020; Jimenez et al, 2021). There are numerous examples of physical interactions between MTs and actin filaments (Jiang et al, 2012; López et al, 2014; Colin et al, 2018; Farhadi et al, 2018; Dogterom & Koenderink, 2019; Inoue et al, 2019). Acting as obstacles, capturing sites, or stabilizing sheaths, actin architectures may regulate and organize the spatial distribution of forces in the MT network, by either amplifying or buffering local asymmetries. However, how specific actin architectures, such as dense cortical networks, bundles of linear filaments, or cytoplasmic mesh, specifically impact forces production and propagation along MTs, remains poorly understood.
Here, we show that the presence and architecture of actin networks affect the spatial distribution of MTs and the displacements of MTOC either toward or away from the geometrical center of the well. Our results revealed how various actin networks have a distinct and specific impact on the distribution of pushing forces and confer some robustness to the mechanism of MTOC positioning by making it less sensitive to variations of MT length.
Results
aMTOC positioning in microwells
It is difficult to directly assess the mechanical effects of actin networks on the production of pushing forces by MTs in living cells because of the biochemical and structural complexities of cell interior, and the presence of motors exerting pulling forces on MTs. Therefore, to reconstitute the interaction of an aster of dynamic MTs with actin networks, we designed an in vitro reconstitution assay using purified proteins and microfabrication techniques.
We investigated how an astral array of MTs self‐organizes in a cell‐sized compartment. 3D microwell can be used to impose physical barrier to MT growth (Laan et al, 2012; Inoue et al, 2020). As compared to lipid droplets (Pinot et al, 2009; Juniper et al, 2018), microwells offer the possibility to control the size and shape of the container (Colin et al, 2020). Actual boundaries are made of lipids in living systems. Therefore, we started by setting up the coating of microwell with a lipid bilayer (Zieske & Schwille, 2014) and further closed the upper surface of the container with a layer of mineral oil (Fig 1A and B, Appendix Fig S1A and B). Lipids were properly diffusing in the bottom plane and the vertical edges of the microwell (Appendix Fig S1C). However, there was a problem of tubulin precipitation, as reported previously (Weis et al, 2010; Baumann & Surrey, 2014). After 30–60 min of incubation of tubulin in the TicTac or the BRB80 buffer, the tubulin precipitated (Appendix Fig S2A) and microtubule dynamics stopped. A screening of the biochemical conditions (Appendix Fig S2B–F) suggested that at lower temperature (22°C) and with high concentration of BSA and GTP, tubulin precipitation could be delayed, up to 2 h (Fig 1C). As purified centrosomes generate a quite variable number of MTs (Inoue et al, 2019), we chose to work with artificial MTOCs (aMTOCs) made of short stabilized pieces of MTs grafted on a polystyrene bead (Fig 1D). They efficiently generated 15–20 dynamic MTs per aMTOC (Fig EV1A–D).
Figure 1. aMTOC positioning in microwells.
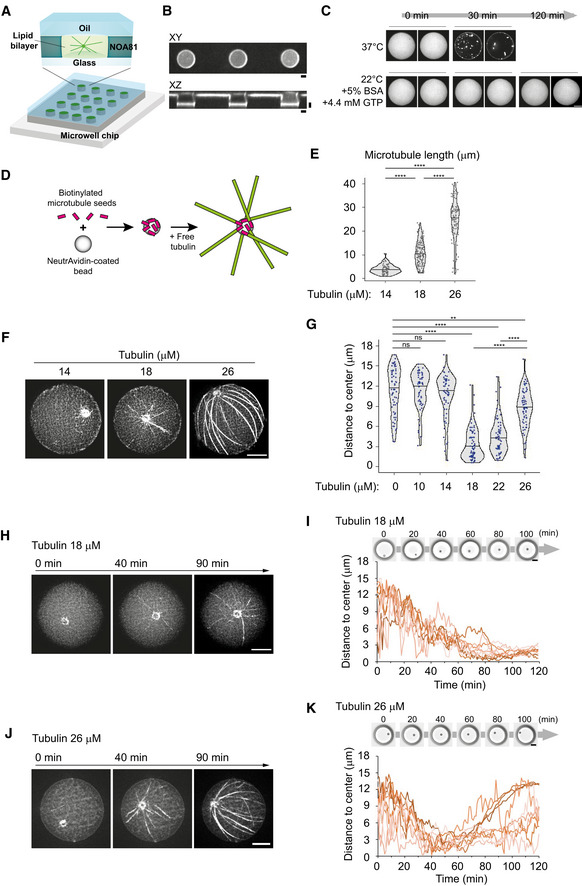
- Scheme of microwells (see also Appendix Fig S1A and B).
- Images of fluorescence‐labeled lipid. XY and XZ views of microwells were shown.
- A screening of biochemical conditions to slow down tubulin precipitation (see also Appendix Fig S2 and Materials and Methods).
- Scheme of preparation of an artificial MTOC (aMTOC). Biotinylated MT seeds were attached on NeutrAvidin‐coated beads. By the addition of free tubulin, MT polymerization occurs from the beads (see also Materials and Methods).
- MT length at the indicated tubulin concentrations in microwells. (Tubulin 14 μM, n = 71; 18 μM, n = 134; 26 μM, n = 97 MTs (8 wells, respectively)) ****P < 0.0001 (Kruskal–Wallis test with Dunn's multiple comparison test).
- Representative images of MT asters with various tubulin concentrations. Images were taken at 2 h after sample preparation.
- Distance from aMTOC to center of the well at the indicated tubulin concentrations (2 h after sample preparation). (Tubulin 0 μM, n = 62; 10 μM, n = 59; 14 μM, n = 60; 18 μM, n = 68; 22 μM, n = 60; 26 μM, n = 65 wells) **P < 0.01, ****P < 0.0001, ns (not significant) > 0.1 (Kruskal–Wallis test with Dunn's multiple comparison test).
- Time‐lapse imaging of MT aster formation at 18 μM of tubulin.
- aMTOC position over time at 18 μM of tubulin. Bright‐field images were taken at 1 min intervals. Positions of 10 individual aMTOCs were shown with different colors.
- Time‐lapse imaging of MT aster formation at 26 μM of tubulin.
- aMTOC position over time at 26 μM of tubulin. Bright‐field images were taken at 1 min intervals. Positions of 10 individual aMTOCs were shown with different colors.
Data information: Scale bar = 10 μm. Violin plots were shown with the median (horizontal line).
Figure EV1. Characterization of an artificial MTOC.
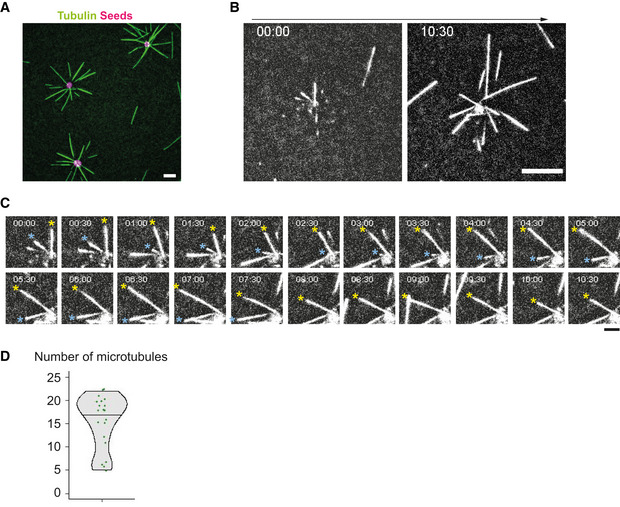
- Observation of MT aster formation using TIRF microscope. Scale bar = 10 μm.
- Time‐lapse imaging of the MT aster formation using TIRF. Scale bar = 10 μm.
- Magnified images of (B). Time interval 30 s. Time bar indicates (min:sec). MTs showing dynamic instability were indicated with asterisks. Scale bar = 5 μm.
- Number of MTs emanating from the aMTOCs in microwells. Tubulin 18 μM. n = 20 wells. Violin plots were shown with the median (horizontal line).
We first analyzed the sensitivity of aMTOC positioning to the ratio of MT length over container length by varying tubulin concentrations in microwells of controlled size. As tubulin concentration has been increased from 14 to 26 μM, the average length of MTs varied from 4 to 25 μm (Fig 1E and F). The radius of the microwell was close to 19 μm. Below 18 μM, MTs were shorter than 10 μm and aMTOC adopted a random position (Figs 1G and EV2A, and B). At 18 μM of tubulin, MTs were longer and could reach the microwell boundaries (Figs 1E and F, and EV2C, and Movie EV1). In about 60 min (Fig 1H and I, and Movie EV1), most aMTOC reached the center of the microwell and remained there (Figs 1G and EV2B). Centering was also efficient at 22 μM of tubulin (Fig 1G). At 26 μM of tubulin, most MTs were longer than 20 μm (Fig 1E). As they grew, they first ensured a proper centering but after an hour, MT elongation and slippage along microwell edges broke the network symmetry and MTs pushed aMTOC away from the center (Figs 1J and K, and EV2D, and Movie EV2). Taken together, in our experimental system, aMTOC positioning appeared highly sensitive to the tubulin concentration and the MT length. Above a critical concentration of 22 μM, MT elongation and reorientation could bias the distribution of pushing forces and promote aMTOC decentering, as previously described in water‐in‐oil droplets (Pinot et al, 2009).
Figure EV2. Characterization of aMTOC positioning in microwells.
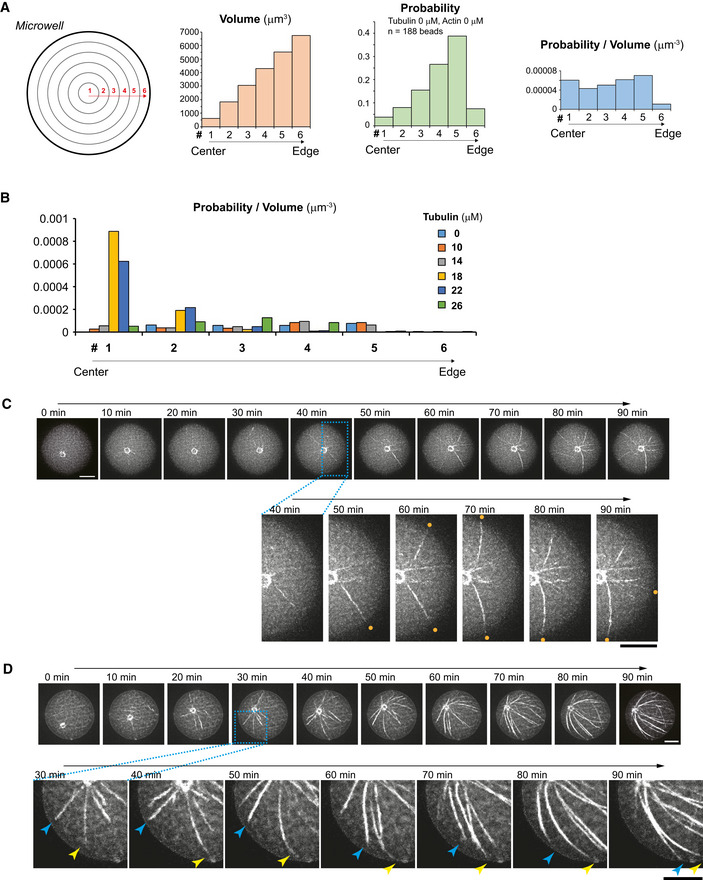
- Consideration of the probability of aMTOC distribution. When the volumes of six segments (equal intervals from well center) are considered as shown in the left scheme, the volume per segment increases from well center toward well edge (left histogram), suggesting that the probability of the aMTOC distribution increases from well center toward the edge. Middle histogram shows experimental probability of the aMTOC position in the absence of free tubulin and actin (n = 188 wells). The probability tended to increase from well center toward the edge. However, the distribution near the edge was restricted, because of the size of aMTOCs (bead 1.5 μm radius + MT seeds). Right histogram indicates probability per volume, suggesting almost random distribution of the aMTOCs in microwells. Volume per segment was calculated based on the approximate size of microwells (37.5 μm in diameter and 20 μm in height).
- Distribution of aMTOC in microwells at the indicated tubulin concentrations. Probability per volume was calculated as shown in (A). Data shown in Fig 1G were used.
- Time‐lapse imaging of MT aster positioning at 18 μM of tubulin shown in Fig 1H. In magnified images, the orange dots indicate the MTs hitting the well edge.
- Time‐lapse imaging of MT aster positioning at 26 μM of tubulin shown in Fig 1J. In magnified images, the blue and yellow arrow heads indicate the MTs slipping along the well edge, respectively.
Data information: Scale bar = 10 μm.
Previous works based on numerical simulations suggested that the friction along the boundaries of the container might prevent the symmetry break by enforcing a vortex‐like structure in the network that preserves MTOC centering (Letort et al, 2016). In cells, the actin filaments form distinct cortical and cytoplasmic networks that might restrict MT lateral translocation and aster displacement (Field & Lénárt, 2011; Blanchoin et al, 2014). Therefore, we tested how these various architectures might impact either centering or decentering mechanisms.
Assembly of various actin architectures in microwells
Rather than working with preassembled and stabilized actin filaments, we chose to grow actin filaments in the microwells in order to control their position and architecture by controlling the mechanism of actin network assembly. Unbranched actin network could be formed in the bulk of the microwell simply by spontaneously assembling 4 μM of actin monomers (Fig 2A and B, and Appendix Fig S3A). Alternatively, to limit the cytoplasmic pool, and favor the assembly of a dense cortical layer, actin filaments were nucleated near the lipid layer by a Nucleation Promoting Factor (NPF) attached to the lipid in the presence of the Arp2/3 complex and actin monomers in the solution (Fig 2C and D, and Appendix Fig S3A,B).
Figure 2. Assembly of various actin architectures in microwells.
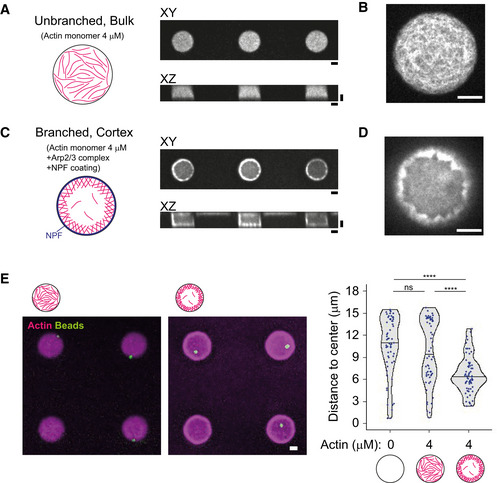
-
A, BUnbranched, bulk actin network. XY and XZ views were shown. Higher magnification image (XY view) was shown in (B).
-
C, DCortical branched actin network. XY and XZ views were shown. NPF (streptavidin‐tagged WA) was coated on the lipid‐biotin. Higher magnification image (XY view) was shown in (D).
-
EaMTOC position in the absence of free tubulin. Left, representative images showing actin and the aMTOCs in microwells. Right, measurement of distance from aMTOC to center of the well (2 h after sample preparation). Violin plots were shown with the median (horizontal line). (Actin 0 μM, n = 60, Actin 4 μM Bulk n = 60, Actin 4 μM Cortex n = 63 wells) ****P < 0.0001, ns (not significant) > 0.1 (Kruskal–Wallis test with Dunn's multiple comparison test).
Data information: Scale bar = 10 μm.
We first tested the impact of these two actin architectures on aMTOC positioning independently of MTs (Fig 2E). Unbranched actin filaments in the bulk had no visible impact on bead position as compared to similar conditions without them (Fig 2E and Appendix Fig S3C). Position of the beads was a bit centered in the presence of cortical actin, as the thickness of the cortical layer restricted the available space for beads (Fig 2E and Appendix Fig S3C and D).
These results showed that actin filaments in the bulk and branched cortical network can be reconstituted in 3D microwells and have a distinct impact on aMTOC position independently of MTs. The two networks may also have specific effects on MT slippage or deformation and as such distinct impacts on the force distribution in the MT network and, therefore, on the positioning of the MTOC.
Bulk actin network impairs aMTOC displacement and aster self‐centering
aMTOC displacements depend on the production of pushing forces by MT polymerization against container boundaries. However, the morphology of aster confers them a large effective cross‐section that limits their displacements by viscous drag. We first tested whether the density of actin network in the bulk could impact the production of pushing forces against effective boundaries, reorganize the spatial distribution of MTs, and thus affect the inner balance of force production by MT polymerization.
To test whether bulk actin network could impair the centering process, we worked in conditions where MT length was comparable to the microwell radius (i.e., at 18 μM tubulin). aMTOC position was not dramatically affected by 1 μM of actin but appeared decentered with 4 μM of actin (Figs 3A and B‐left, and EV3A). Indeed, time‐lapse imaging revealed that aMTOC remained stuck at their initial position in the presence of 4 μM of actin (Fig 3C). The path length of the MTOC was severely limited in the presence of 4 μM of actin, suggesting that the motion of the MTOC was restricted in the presence of actin filaments (Fig EV3B). Importantly, at this concentration, unbranched actin network had no impact on MT elongation (Fig 3D). In addition, higher concentrations of tubulin, 26 μM, although capable of promoting MT elongation, could not overcome aster immobilization by the bulk actin meshwork (Figs 3B‐right and EV3C). This suggested that the defective centering was due to friction resisting aster displacement rather than steric effects blocking MT polymerization (Colin et al, 2018; Inoue et al, 2019). Indeed, implementing physical hindrance to aster displacement by taking into account steric effect of actin filaments along MT aster in numerical simulations was sufficient to account for the immobilization of MTOC by linear bulk actin filaments (Figs 3E–G and EV3D–F, and Movie EV3). Therefore, we conclude that the presence of a dense network of actin filaments in the bulk can affect MT aster centering by resisting aster translocation rather than impairing MT elongation.
Figure 3. Bulk actin network impairs aMTOC displacement and aster self‐centering.
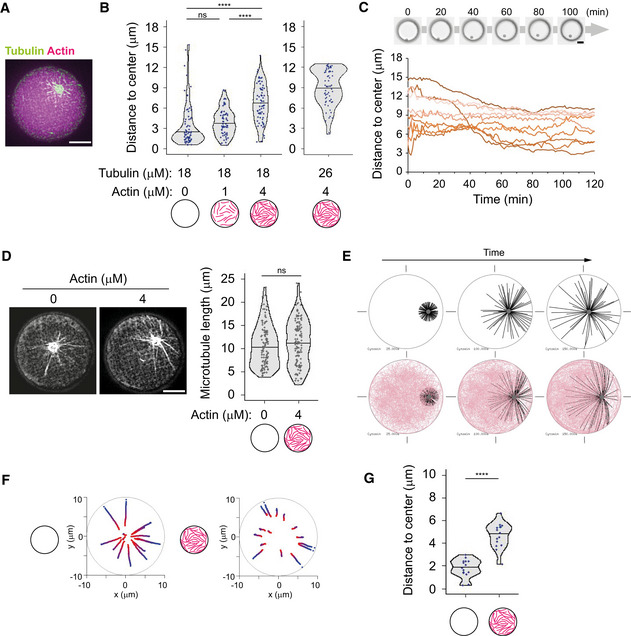
- Representative image of MT aster in the presence of bulk actin network. Tubulin 18 μM and actin 4 μM.
- Distance from aMTOC to well center (2 h after sample preparation). Left: Tubulin 18 μM. (Actin 0 μM, n = 68; 1 μM, n = 65; 4 μM, n = 71 wells). Right: Tubulin 26 μM Actin 4 μM (n = 66 wells). ****P < 0.0001, ns (not significant) > 0.1 (Kruskal–Wallis test with Dunn's multiple comparison test).
- aMTOC position over time. Tubulin 18 μM and Actin 4 μM. Bright‐field images were taken at 1‐min intervals. Positions of 10 individual aMTOC were shown with different colors.
- Measurement of MT length in the absence or presence of bulk actin network. Tubulin 18 μM. Images were taken 2 h after sample preparation. (Actin 0 μM, n = 109; 4 μM, n = 118 MTs (from 8 wells, respectively)). ns (not significant) > 0.1 (Mann–Whitney U test).
- Simulations in the absence (top) or presence (bottom) of actin filaments. Different time points (From left, 25, 100, and 150 s) were shown. MTOC—gray, MT—black, actin—pink.
- Trajectories of MTOCs from blue (0 s) to red (150 s). Fifteen simulations per condition. The initial position (0 s) was randomly chosen.
- Final position of MTOC (at 150 s). Fifteen simulations per condition. ****P < 0.0001 (Mann–Whitney U test).
Data information: Violin plots were shown with the median (horizontal line). Scale bar = 10 μm in (A), (C), and (D).
Figure EV3. aMTOC positioning in the presence of bulk actin network.
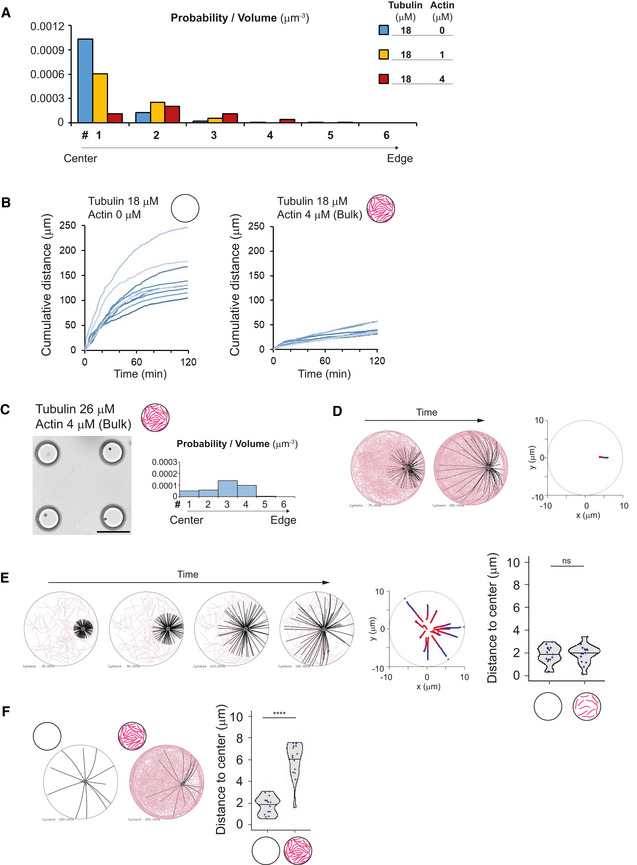
- The path length (cumulative distance) of the aMTOC during the time‐lapse imaging was measured for the indicated conditions. The data of individual aMTOC were shown with different colors (10 aMTOCs per condition). Images were taken at 1‐min intervals.
- Simulation in the presence of bulk actin network. Even with longer MT formation (compared with Fig 3E), the MTOC centering was not occurred. Different time points (75 and 250 s) were shown. Right graph shows trajectories from blue (0 s) to red (250 s). MTOC—gray, MT—black, actin—pink.
- Simulations in the presence of lower density of actin. Different time points (25, 50, 100, and 150 s) were shown. Middle graph shows trajectories from blue (0 s) to red (150 s). Right graph shows the final position of MTOC (at 150 s). Data of the absence of actin (left) are same as shown in Fig 3G. ns (not significant) > 0.1 (Mann–Whitney U test). Fifteen simulations per condition.
- Simulations in the presence of smaller number of MTs. The images represent the time point at 150 s. Right graph shows the final position of MTOC (at 150 s). ****P < 0.0001 (Mann–Whitney U test). Fifteen simulations per condition.
Data information: Violin plots were shown with the median (horizontal line).
These data also suggested that restricting the actin network to the periphery might specifically limit MT slippage without impairing aster translocation.
Cortical branched actin meshwork favors aster centering
To test whether cortical actin could counteract MT slippage and MTOC decentering, experiments were performed in the presence of long MTs (i.e., at 26 μM tubulin) (Fig 4A). As described above, the cortical network clustered aMTOC in a smaller volume and thus induced a partial centering (Figs 2E‐right and 4B‐left). Interestingly, this centering was significantly improved by the growth of long MTs (Figs 4B‐left and EV4A). MT length was not significantly changed even in the absence or presence of cortical actin, suggesting that this effect is not due to interference of actin with MT elongation (Fig 4C). Instead, MTs appeared longer than the radius of the microwell and the network adopted a vortex‐like structure (Fig 4D). Time‐lapse imaging showed that in the absence of cortical actin, MTs were pivoting around the aMTOC, whereas they maintained their orientation and grew along the edge in the presence of cortical actin (Fig 4E–H, Fig EV4B and D, and Movie EV4) The path length of the MTOC was shortened in the presence of cortical actin, suggesting that the presence of cortical actin stabilized the position of the MTOC by restricting MT slippage (Fig EV4E). MT pivoting in the absence of cortical actin appeared associated with aMTOC decentering, whereas MT sneaking into the cortical actin was associated with aMTOC stable centering and maintenance at the center (Fig 4I and J). Numerical simulations in which friction was restricted to the cell periphery displayed similar aMTOC displacements and final positions (Figs 4K–M and EV4F, and G and Movie EV5), demonstrating that local steric interactions between cortical actin and MTs are indeed sufficient to prevent MTOC decentering. From these results, we concluded that cortical actin network can counteract the effect of MT elongation and aster decentering by restricting MT slippage and thus maintaining a regular distribution of force application sites along the cortex and around the aMTOC. In addition, even at lower concentrations of tubulin (18 μM), MT asters displayed a proper centering mechanism in the presence of cortical actin (Figs 4B‐right and EV4H). This showed that cortical actin can enforce a robust centering that is less sensitive to MT length. Importantly, cortical actin network of low density could not enforce MTOC centering, suggesting that resistance to MT slippage depends on actin network density (Fig EV4I and J).
Figure 4. Cortical branched actin meshwork favors aster centering.
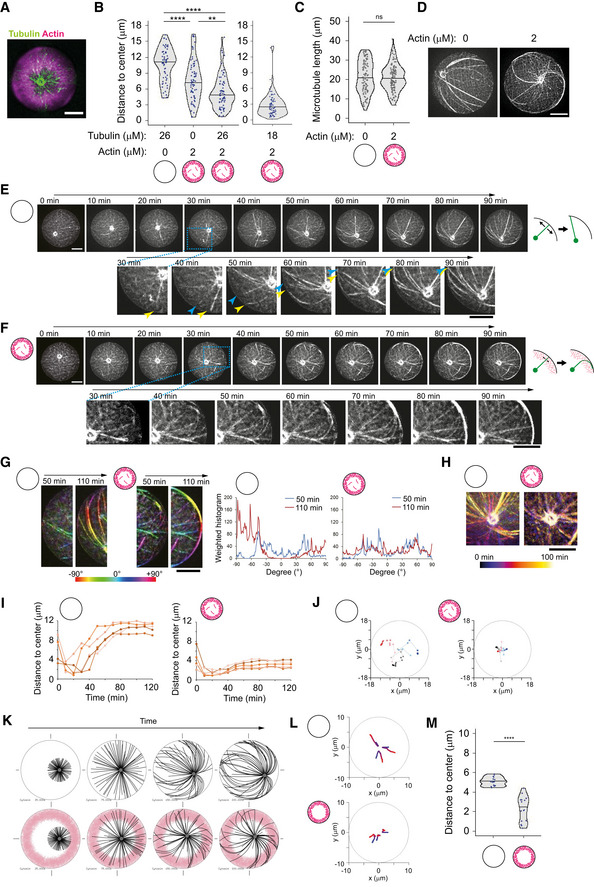
-
ARepresentative image of MT aster with cortical actin. Partial maximum projection was shown. Tubulin 26 μM, actin 2 μM, and Arp2/3 complex were added into NPF (WA)‐coated microwells.
-
BDistance from aMTOC to well center (2 h after sample preparation). Left: Tubulin 26 μM Actin 0 μM, n = 61, Tubulin 0 μM Actin 2 μM, n = 73, Tubulin 26 μM Actin 2 μM, n = 70 wells. Tubulin 18 μM Actin 2 μM, n = 76 wells. **P < 0.01, ****P < 0.0001, ns (not significant) > 0.1 (Kruskal–Wallis test with Dunn's multiple comparison test).
-
CMeasurement of MT length in the absence or presence of cortical actin. Tubulin 26 μM. Images were taken 2 h after sample preparation. Actin 0 μM, n = 96, 2 μM cortex n = 104 MTs (from 6 wells, respectively). ns (not significant) > 0.1 (Mann–Whitney U test).
-
DRepresentative images of MT organization in the absence or presence of cortical actin. Tubulin 26 μM.
-
E, FTime‐lapse imaging of MT aster positioning at 26 μM in the absence (E) or presence (F) of cortical actin. Final actin structure was shown in (A). Magnified images were also shown. In magnified images in (E), the blue and yellow arrowheads indicate the MTs slipping along the well edge, respectively. Right schemes indicate how MTs behave along cell boundary. In the absence of actin, MTs slipped and reoriented along well boundary as they grew. In contrast, MT reorientation was restricted in the presence of actin, although MTs can grow through actin network and along well boundary.
-
GOrientation of MTs near the well edge in the absence or presence of cortical actin. Orientation of MTs was shown with different colors. Right graph indicates the measurement of the MT orientation using Orientation J. The different time points were shown with different colors. Another example is also shown in Fig EV4D. In the absence of cortical actin, the MT orientation dynamically changed over time, whereas in the presence of cortical actin, the MT orientation was not significantly changes.
-
HMT motion around the aMTOC shown in (E) and (F). Temporal‐color coded images were shown. The position of aMTOC was centered at each time point in the image.
-
IaMTOC position over time in the absence (left) or presence (right) of cortical actin. Five representative data per condition were shown.
-
JRepresentative trajectories of aMTOCs in microwells from light colors (0 min) to dark colors (120 min). Time‐lapse imaging was performed for 2 h at 10‐min intervals. Three trajectories per condition were shown with different colors.
-
KSimulations in the absence or presence of actin. Different time points (From left, 25, 75, 150, and 200 s) were shown. MTOC—gray, MT—black, actin—pink.
-
LRepresentative trajectories of MTOC from blue (0 s) to red (200 s). The initial position (0 s) was randomly chosen within 4 μm from the cell center. Three simulations per condition.
-
MFinal position of MTOC (at 200 s). Fifteen simulations per condition. ****P < 0.0001 (Mann–Whitney U test).
Data information: Violin plots were shown with the median (horizontal line). Scale bar = 10 μm in (A) and (D)–(H).
Figure EV4. Characterization of aMTOC positioning and MT behaviors in the absence or presence of cortical actin network.
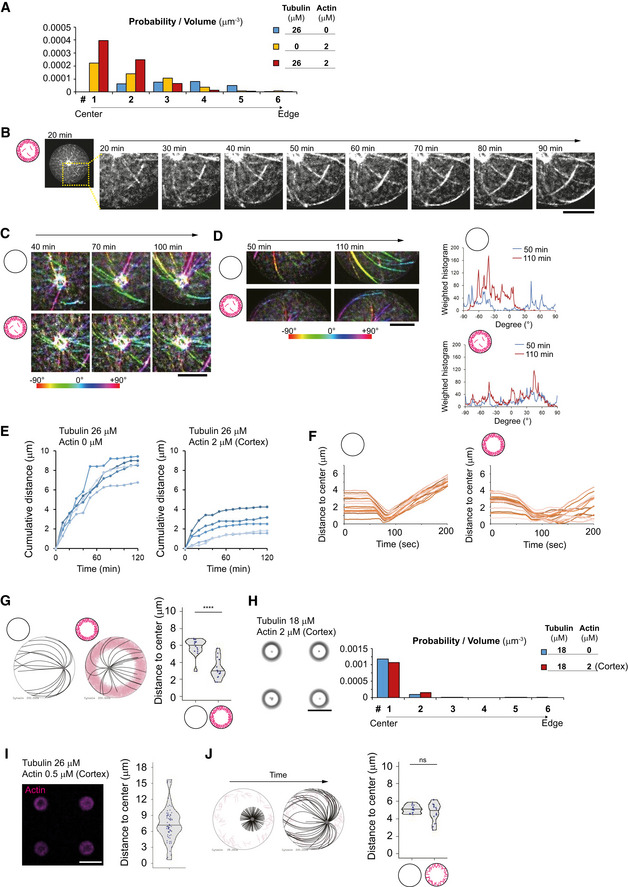
- Time‐lapse imaging of MT aster positioning at 26 μM of tubulin in the presence of cortical actin shown in Fig 4F. Magnified images were shown. Scale bar = 10 μm.
- Orientation of MTs around the MTOCs in the absence or presence of cortical actin. The samples were same as shown in Fig 4E and F. Orientation of MTs was shown with different colors. Scale bar = 10 μm.
- Other representatives showing orientation of MTs near the well edge in the absence or presence of cortical actin. Orientation of MTs was shown with different colors. Right graph indicates the measurement of the MT orientation using Orientation J. The different time points were shown with different colors. Scale bar = 10 μm.
- The path length (cumulative distance) of the aMTOC during the time‐lapse imaging was measured for the indicated conditions. The data of individual aMTOC were shown with different colors (10 aMTOCs per condition). Images were taken at 10‐min intervals.
- Simulations of MTOC position over time in the absence (left) or presence (right) of cortical actin. Fifteen simulations per condition were shown with different colors.
- Simulations in the presence of smaller number of MTs. The images represent the time point at 200 s. Right graph shows the final position of MTOC (at 200 s). ****P < 0.0001 (Mann–Whitney U test). Fifteen simulations per condition.
- aMTOC position in the presence of 26 μM tubulin and with less dense cortical actin assembled at a lower concentration of actin (0.5 μM). Right panel shows measurement of distance from aMTOC to center of the well (2 h after sample preparation). n = 71 wells. Scale bar = 50 μm.
- Simulations in the presence of lower density of cortical actin. The time points at 200 s were shown. Right graph shows the final position of MTOC (at 200 s). Data of the absence of actin (left) are same as shown in Fig 4M. ns (not significant) > 0.1 (Mann–Whitney U test). Fifteen simulations per condition.
Data information: Violin plots were shown with the median (horizontal line).
These results also suggested that a heterogeneous pattern of cortical friction might create an asymmetry in the angular distributions of MTs and an alignment of pushing forces leading to aMTOC decentering.
Asymmetric cortical actin meshwork induces aster decentering
We reasoned that with lower actin filament density, which would be cross‐linked to each other, we could enforce the asymmetry of the actin network growing from the walls of the microwells (Ierushalmi et al, 2020). Indeed, we found that with 0.5 μM instead of 2 μM of actin and 100 nM of α‐actinin, the cortical network grew from all edges but formed an asymmetric cortex (Figs 5A and EV5A, and B). In cells, the inner region of the cytoplasm that is almost devoid of actin filament was defined as the actin inner zone (Fig EV5B) (Jimenez et al, 2021). The growth of MTs from the asters did not seem to have an impact on the asymmetric architecture of the actin network (Fig 5B). To analyze aster positioning with respect to this asymmetry, images were reoriented in order to align horizontally the center of the actin inner zone and the center of the microwell (Figs 5C and D, and EV5C, and D). To test the potential guiding effect of asymmetric cortical actin networks, we worked in decentering conditions, that is, 26 μM tubulin, in which aMTOCs are randomly distributed in the microwell in the absence of actin (Figs 5E and EV5E, and F). By its thickness, the cortical actin network constrained asters positioning and limits aMTOC dispersion even in the absence of MTs (Figs 5F and EV5E, and F). However, as anticipated, the reorientation of the microwells with respect to the asymmetry of the actin network revealed that as MTs grew from the asters they shifted the aMTOCs toward the center of the actin inner zone (Figs 5G–I and EV5E–G and Movie EV6). Interestingly, in conditions imposing shorter MTs and an efficient centering of the aster, that is, 18 μM of tubulin, the asters in the presence of the asymmetric actin network appeared decentered toward the center of the actin inner zone as well (Figs 5J and EV5G). Numerical simulations confirmed that a heterogeneous friction pattern due to variable thickness in the cortical actin network along microwell boundary was sufficient to push the MTOC away from the thicker actin layer and thus promote aster decentering (Figs 5K–M and EV5H, and I, and Movie EV7). Overall, these results showed that the cortical actin network architecture can direct the position of MTOCs, either at the center or away from it, depending on its heterogeneity. It controls the force balance at the MTOC by modulating the pattern of friction resisting MT slippage and thus directing the localization of the sites of application of pushing forces.
Figure 5. Asymmetric cortical actin meshwork induces aster decentering.
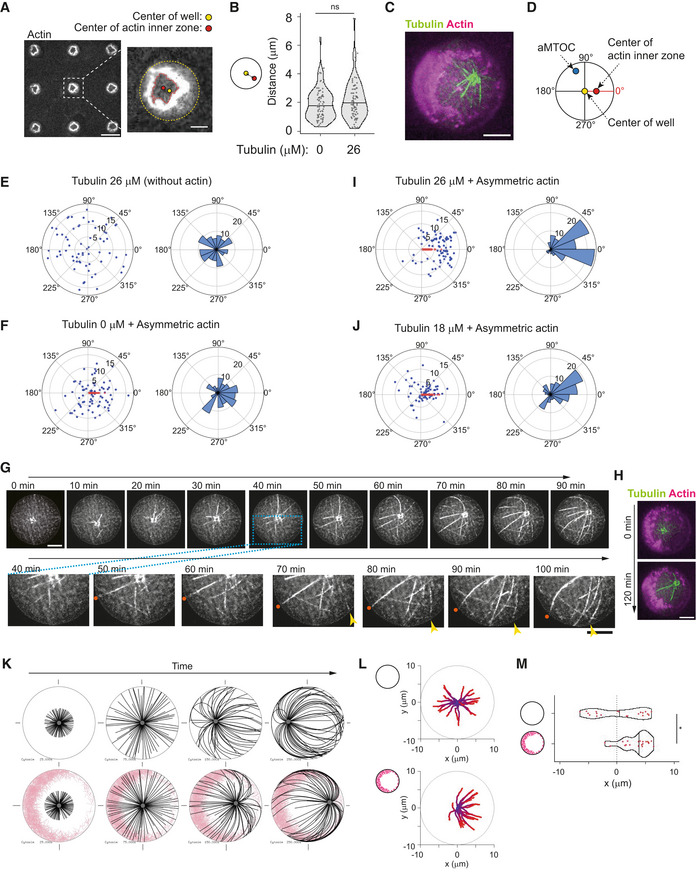
-
ARepresentative image of asymmetric actin cortex. Actin 0.5 μM, α‐actinin 100 nM, and Arp2/3 complex were added into NPF (WA)‐coated microwells. In the right image, the center of the well and the center of the actin inner zone were indicated. A single slice of the image was shown. Scale bar = left 50 μm, right 10 μm.
-
BDistance between well center and center of the actin inner zone. (Tubulin 0 μM, n = 74; 26 μM, n = 79 wells) ns (not significant) > 0.1 (Mann–Whitney U test).
-
CRepresentative image of MT aster with asymmetric actin. Tubulin 26 μM. Partial maximum projection was shown.
-
DScheme of angle measurements. Angles from the well center to the aMTOC and the center of the actin inner zone were measured. In (F), (I), and (J), the wells were reoriented based on each XY coordinates in order to align the angles from well center to center of the actin inner zone at 0° (see also Fig EV5C and D).
-
E, FDistributions of aMTOCs. Left, aMTOC positions (μm) relative to well center. Right, Angular distributions (%) of aMTOCs from well center. (Tubulin 26 μM Actin 0 μM, n = 65, Tubulin 0 μM Actin 0.5 μM, n = 74) Blue and red dots indicate the position of aMTOC and the center of the actin inner zone after alignment, respectively.
-
GTime‐lapse imaging of MT aster in the presence of asymmetric actin. Tubulin 26 μM. In magnified images, the arrow head indicates the MT slipping along the well edge and the orange dot indicates the MT hitting the well edge.
-
HActin network structure of (H) at the initial (0 min) and final time point (120 min). Partial maximum projection was shown.
-
I, JDistributions of aMTOCs. Left, aMTOC positions (μm) relative to well center. Right, angular distributions (%) of aMTOCs from well center. (Tubulin 26 μM Actin 0.5 μM, n = 79 wells, Tubulin 18 μM Actin 0.5 μM, n = 68 wells) Blue and red dots indicate the position of aMTOC and the center of the actin inner zone after alignment, respectively.
-
KSimulations in the absence (top) or presence (bottom) of asymmetric actin (see also Fig EV5H). Initial position was set to the cell center. Time point: 25, 75, 150, and 250 s. MTOC—gray, MT—black, actin—pink.
-
LTrajectories of MTOCs from blue (0 s) to red (250 s). Twenty simulations. Initial position: cell center.
-
MFinal position of MTOC along X‐axis (at 250 s). 0 indicates the center along X‐axis in cells. Twenty simulations per condition. *P < 0.1 (Mann–Whitney U test).
Data information: Violin plots were shown with the median (horizontal line). Scale bar = 10 μm in (C), (G), and (H).
Figure EV5. aMTOC positioning in the presence of asymmetric actin network.
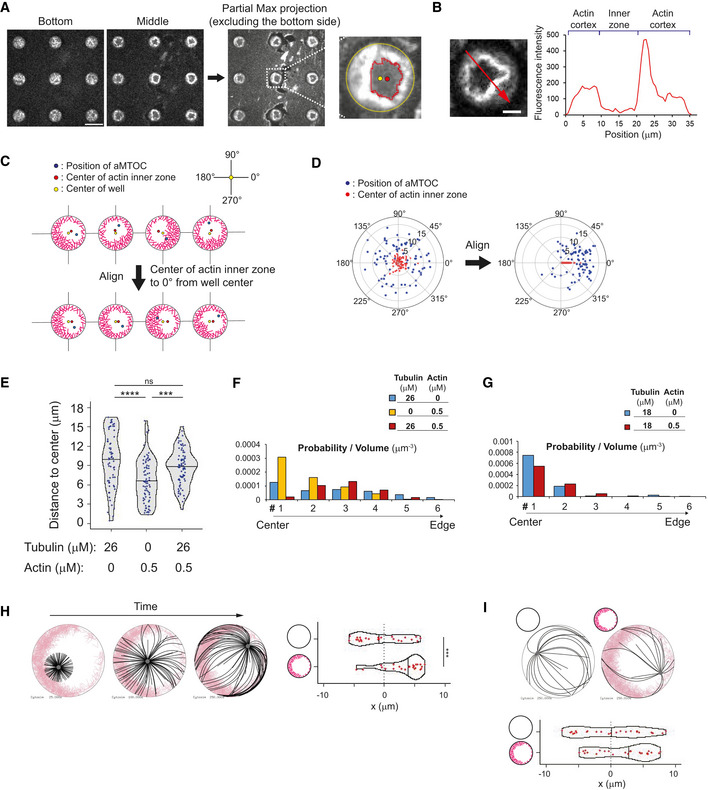
-
ADetection of actin inner zone. Actin 0.5 μM and a‐actinin 100 nM with Arp2/3 complex and NPF (WA) coating. The bottom and top were excluded from the maximum projection to visualize the vertical edges of actin. Using the partial max projection images and defining a threshold of actin intensity, the region of actin inner zone was detected and the center (centroid) was measured. Scale bar = 50 μm.
-
BMeasurement of the fluorescence intensity of actin. The red arrow in the image indicates the region used for the line scan analysis. Right graph indicates the result of the line scan analysis. Background signals outside the wells were subtracted. The less actin region inside the cortex was defined as the actin inner zone. Scale bar = 10 μm.
-
C, DTo analyze the aMTOC position with respect to the asymmetry of cortical actin, the angles from the well center to the center of the actin inner zone were aligned at the same degree (0°). It means that wells were reoriented in order to align the angles from the well center to the center of the actin inner zone.
-
EDistance from aMTOC to well center (2 h after sample preparation). Data shown in Fig 5E and F, and I were used. (Tubulin 26 μM Actin 0 μM, n = 65, Tubulin 0 μM Actin 0.5 μM, n = 74, Tubulin 26 μM Actin 0.5 μM, n = 79 wells) ***P < 0.001, ****P < 0.0001, ns (not significant) > 0.1 (Kruskal–Wallis test with Dunn's multiple comparison test). Violin plots were shown with the median (horizontal line).
- F, G
-
HSimulations in the presence of asymmetric actin when the initial position was randomly chosen within 4 μm from cell center. Twenty‐five simulations per condition. Different time points (From left, 25, 100, and 250 s) were shown. Even if the initial position is off‐centered, the MTOC tended to migrate toward the thinner side of the actin network. ***P < 0.001 (Mann–Whitney U test). Violin plots were shown with the median (vertical line).
-
ISimulations in the presence of smaller number of MTs. The images represent the time point at 250 s. Lower graph shows the final position of MTOC (at 250 s). Twenty‐five simulations per condition. Violin plots were shown with the median (vertical line).
Discussion
Our results suggest that actin networks make aster positioning both more robust and more versatile. Indeed, in the absence of actin filaments, asters displayed abrupt transitions from centering to decentering depending on MTs length. By contrast, bulk actin filaments resisted asters displacements and MTOC were held in place, independently of the presence of MTs. Moreover, cortical actin networks specifically favored aster centering over a broad range of MT lengths. The presence of asymmetric actin resulted in decentering of the aster. From these observations, we propose that actin networks can modulate the sensitivity of MT aster positioning to variation of MT length.
It has been reported that the presence of cytoplasmic actin network can affect MT organization (Dahlgaard et al, 2007; Field & Lénárt, 2011). The immobilization of MTOC by bulk actin filaments in our system is reminiscent of the regulation of MT aster in Xenopus extract and sea urchin embryo (Colin et al, 2018; Xie et al, 2022). Bulk actin network might resist the MTOC displacement by passive friction along the large cross‐section area formed by radiating MTs. On the contrary, the robust polarization of MT network and decentering of MTOC by asymmetric organization of the cortical actin is reminiscent of various mechanisms of mutual polarization of the actin and microtubule networks, such as ciliogenesis and immune synapse formation (Stinchcombe & Griffiths, 2014; Ritter et al, 2015; Pitaval et al, 2017) or MTOC positioning at the rear of migrating leukocytes (Kopf & Kiermaier, 2021; Mastrogiovanni et al, 2021). In these conditions, as in our reconstitution system, the actin network might control MTOC decentering by concentrating the distribution of the MT‐based pushing forces in dense cortical regions. So, our reconstitution assay based on a minimal set of components might properly account for the actual mechanism regulating MT aster positioning by pushing forces in living cells.
However, the conditions of our reconstitution assay did not include the key role played by molecular motors that move the MTOC by pulling on microtubules (Yi et al, 2013) and by regulating their dynamics (Hooikaas et al, 2020) during cell polarization. Furthermore, our experimental conditions and the use of short pieces of MTs attached to a bead did not offer us the possibility to control MT pivoting around the aMTOC, a property that could have promoted symmetry break and amplified MTOC decentering (Baumgärtner & Tolić, 2014; Letort et al, 2016; Fong et al, 2021). In addition, we studied aMTOC positioning in response to the production of pushing forces in cylindrical and rigid microwells. Softer materials would be necessary to study whether the forces produced by growing microtubules (Bornens et al, 1989; Fygenson et al, 1997) could deform the container, force microtubule relocalization, destabilize the central position, break network symmetry, and promote MTOC decentering.
Building a synthetic cell from scratch is a powerful strategy to improve our understanding of cell biology and pave the way toward new living materials (Salehi‐Reyhani et al, 2017). Here, we established a way to combine dynamic MTs and actin filaments in cell‐sized confinement. Our findings and techniques could be used as a step toward the reconstitution of the polarization process in synthetic cells.
Materials and Methods
Protein expression and purification
Tubulin was purified from fresh bovine brain by three cycles of temperature‐dependent assembly/disassembly in Brinkley Buffer 80 (BRB80: 80 mM Pipes pH 6.8, 1 mM EGTA and 1 mM MgCl2) (Shelanski, 1973). MAP‐free tubulin was purified by cation‐exchange chromatography (EMD SO, 650M, Merck) in 50 mM Pipes, pH 6.8, supplemented with 0.2 mM MgC12, and 1 mM EGTA. Fluorescently labeled tubulin (ATTO‐488‐ or ATTO‐647‐labeled) and biotinylated tubulin were prepared by following the previously published method (Hyman et al, 1991). Actin was purified from rabbit skeletal‐muscle acetone powder. Monomeric Ca‐ATP‐actin was purified by gel‐filtration chromatography on Sephacryl S‐300 at 4°C in G buffer (2 mM Tris–HCl, pH 8.0, 0.2 mM ATP, 0.1 mM CaCl2, 1 mM NaN3, and 0.5 mM dithiothreitol (DTT)). Actin was labeled on lysines with Alexa‐568. The Arp2/3 complex, recombinant GST‐α‐actinin 4 and GST‐WA (a truncated version of human WASP) were purified in accordance with previous methods (Ennomani et al, 2016; Boujemaa‐Paterski et al, 2017).
Snap‐Streptavidin‐WA (pETplasmid) was expressed in Rosettas 2 (DE3) pLysS (Merck, 71403). Culture was grown in TB medium supplemented with 30 μg/ml kanamycin and 34 μg/ml chloramphenicol, then 0.5 mM isopropyl β‐D‐1‐ thiogalactopyranoside (IPTG) was added, and protein was expressed overnight at 16°C. Pelleted cells were resuspended in Lysis buffer (20 mM Tris pH8, 500 mM NaCl, 1 mM EDTA, 15 mM Imidazole, 0,1% TritonX100, 5% Glycerol, 1 mM DTT). Following sonication and centrifugation, the clarified extract was loaded on a Ni Sepharose high performance column (GE Healthcare Life Sciences, ref 17526802). Resin was washed with Wash buffer (20 mM Tris pH8, 500 mM NaCl, 1 mM EDTA, 30 mM Imidazole, 1 mM DTT). Protein was eluted with Elution buffer (20 mM Tris pH8, 500 mM NaCl, 1 mM EDTA, 300 mM Imidazole, 1 mM DTT). Purified protein was dialyzed overnight at 4°C with storage buffer (20 mM Tris pH8, 150 mM NaCl, 1 mM EDTA, 1 mM DTT), concentrated with Amicon 3KD (Merck, ref UFC900324). Aliquots were flash‐frozen in liquid nitrogen and stored at −80°C.
Preparation of an artificial MTOC
To prepare microtubule seeds, the mixture containing 3 μM of fluorescent‐labeled tubulin, 7 μM of biotinylated tubulin and 0.5 mM GMPCPP (Jena Bioscience, NU‐405S) in BRB80 buffer was incubated at 37°C for 40 min. After the incubation, 10 μM of Taxol was added and the mixture was incubated at room temperature for 10 min. The microtubule seeds were then pelleted by centrifugation at 20,238 g for 10 min and were resuspended in the BRB80 supplemented with 0.5 mM GMPCPP and 10 μM Taxol. The seeds were flash‐frozen and stored in liquid nitrogen.
To prepare the Neutravidin‐coated beads, the polystyrene beads containing surface primary amino groups (PolySciences, 17145‐5, Diameter 3 μm) were incubated with 10 mM of Sulfo‐NHS‐LC–LC‐Biotin (ThermoFisher, 21338) at room temperature for 40 min to modify their surface with biotin. The beads were washed with phosphate‐buffered saline (PBS) and then with HKEM buffer (10 mM HEPES (pH 7.5), 50 mM KCl, 5 mM MgCl2, 1 mM EGTA) supplemented with 0.1% bovine serum albumin (BSA). The beads were incubated with 1 mg/ml of Neutravidin (ThermoFisher, 31000) at 15°C for 30 min or at 4°C for 2 h. After washing the beads with HKEM buffer supplemented with 0.1% BSA, the beads were resuspended in 200 μl of HKEM buffer supplemented with 0.1% BSA. The beads solution was then mixed with 10 μl of the microtubule seeds. The mixture was incubated under rotation at room temperature overnight. Before mixing the aMTOCs (microtubule seeds + beads) with the reaction mixture containing free tubulin, the solution containing aMTOCs was washed with HKEM supplemented with 0.1% BSA to remove excess seeds and Taxol.
Preparation of small unilamellar vesicles (SUV)
L‐α‐phosphatidylcholine (EggPC) (Avanti, 840051C), 1,2‐distearoyl‐sn‐glycero‐3 phosphoethanolamine‐N‐[biotinyl(polyethylene glycol)‐2000] (DSPE‐PEG(2000)‐Biotin) (Avanti, 880129C) and ATTO 647N‐labeled DOPE (ATTO‐TEC, AD 647N‐161 dehydrated) were used. Lipids were mixed in glass tubes as follows: Type 1 (99% EggPC (10 mg/ml) and 1% DOPE‐ATTO390 (1 mg/ml)), Type 2 (98.75% EggPC (10 mg/ml) and 0.25% DSPE‐PEG‐Biotin (10 mg/ml) and 1% DOPE‐ATTO390 (1 mg/ml)), Type 3 (99.5% EggPC (10 mg/ml), 0.25% DSPE‐PEG‐Biotin (10 mg/ml) and 0.25% DOPE‐ATTO647N (1 mg/ml)). The mixture was dried with nitrogen gas. The dried lipids were incubated in a vacuum overnight. After that, the lipids were hydrated in the SUV buffer (10 mM Tris (pH 7.4), 150 mM NaCl, 2 mM CaCl2). The mixture was sonicated on ice. The mixture was then centrifuged for 10 min at 20,238 g to remove large structures. The supernatants were collected and stored at 4°C. The final concentration of lipids was adjusted to 0.5 mg/ml. The Type 2 SUV was used to bind snap‐streptavidin‐WA onto the lipid layer. The Type 3 SUV was used to visualize lipids on microwells. In other experiments, the Type 1 SUV was used.
Construction of microwells
The master mold (approximately 20 μm of thickness) was fabricated through photolithography using SU8 3025 (MicroChem) and then vapor silanized with Trichloro (1H,1H,2H,2H‐perfluoro‐octyl) silane (Sigma, 448931). To make 1st PDMS, the mixture of prepolymer and curing agent (Dow, SYLGARD 184 silicone elastomer kit) was poured onto the master mold. It was baked at 70°C for 2 h. The 1st PDMS was then vapor silanized with Trichloro (1H,1H,2H,2H‐perfluoro‐octyl) silane. The 2nd PDMS was made from the silanized 1st PDMS as a template. The 2nd PDMS was cut into small pieces and used as PDMS stamps.
Glasses were cleaned by successive chemical treatments: 30 min in acetone with sonication, 15 min in ethanol (96%), washing ultrapure water, 2 h in HellmanexIII (2% in water, Hellma), and washing in ultrapure water. The glasses were then dried. The slide glasses were oxidized in plasma cleaner (diener) for 2 min at 80% power and then incubated overnight in a solution of 1 mg/ml of mPEG‐Silane (30 kDa, PSB‐2014, Creative PEG works), 96% ethanol, and 0.1%(v/v) HCl. The slide glasses were then dried and stored at 4°C.
To make microwell chip on a cover glass, the PDMS stamp was placed on the cleaned cover glass (20 mm × 20 mm, No. 1), facing the pillar surface of the stamp onto the glass. NOA81(Norland Products) drop was put at the side of the PDMS stamp to fill the space between the PDMS pillars with NOA81. The NOA81 was cured with UV light (UVKUB2, 100%, 12 min). The PDMS stamp and excess NOA81 were then removed.
Sample preparation
To make a reaction chamber, a cover glass with microwell chip was first oxidized in plasma cleaner (diener) for 2 min at power of 80%. The cover glass with microwell chip was attached onto the silane‐PEG‐coated slide glass with two double‐sided tapes (70 μm thickness), facing the side of microwell chip to the slide glass. The SUV solution (0.5 mg/ml) was introduced into the chamber and incubated for 10 min to make a supported lipid bilayer on the surface of microwell chip. It was washed with the SUV buffer to remove excess SUV and then washed with HKEM buffer supplemented with 0.1% BSA. Unless otherwise noted, microtubule and actin assembly were induced by diluting tubulin dimers (20% labeled) and/or actin monomers (10% labeled) in the reaction mixture containing the TicTac buffer (10 mM Hepes, 16 mM Pipes (pH 6.8), 50 mM KCl, 1 mM EGTA, 5 mM MgCl2) supplemented with 5% BSA, 4.4 mM GTP, 2.7 mM ATP, 20 mM DTT, 20 μg/ml catalase, 3 mg/ml glucose, 100 μg/ml glucose oxidase. Microtubule aster formation was induced by adding microtubule seeds‐coated beads (aMTOCs) into the mixture.
To induce branched actin assembly from the edge of microwells, the SUV solution containing DSPE‐PEG‐Biotin (Type 2 SUV) was used for lipid coating. Before introducing the reaction mixture into the chamber, HKEM buffer containing 200 nM of snap‐streptavidin‐WA (used as an NPF) and 0.1% BSA was loaded into the chamber and incubated for 5 min. The excess WA was then removed by perfusing HKEM buffer supplemented with 0.1% BSA. 80 nM of the Arp2/3 complex was added in the reaction mixture. To make asymmetric cortical actin structures, 100 nM of GST‐α‐actinin 4 was also added.
The reaction mixture was introduced into the chamber immediately after the preparation of the mixture. Mineral oil (Paragon Scientific, RTM13) was then loaded into the chamber in order to close the wells. Unless otherwise noted, the chamber was incubated at room temperature (22–23°C) in order to prevent tubulin precipitation. The final position of the aMTOCs was analyzed at 2 h after sample preparation. Experiments were repeated to confirm the reproducibility.
For initial screening of biochemical conditions (in Fig 1C and Appendix Fig S2), TicTac buffer supplemented with 0.1% BSA, 1 mM GTP, 2.7 mM ATP, 20 mM DTT, 20 μg/ml catalase, 3 mg/ml glucose, 100 μg/ml glucose oxidase was used as a control buffer solution. BSA (Sigma, A7030), Polyethylene Glycol (PEG) 3 k (Sigma, P3640), PEG 20 k (Sigma, 95172), Glycerol (CARLO ERBA), Ficoll400 (Sigma, F9378) and Dextran 40 k (Sigma, 31389) were added in the solution at the indicated concentrations. The samples were incubated at the indicated temperatures.
Microscopy
Microtubule asters and actin filaments in microwells were visualized using a confocal spinning‐disk system (EclipseTi‐E Nikon inverted microscope equipped with a CSUX1‐A1 Yokogawa confocal head, an Evolve EMCCD camera (Photometrics), a Nikon CFI Plan‐Apo ×60 NA1.4 oil immersion objective, a Nikon CFI S‐Fluor ×100 NA1.30 oil immersion objective and a Nikon ×20 NA0.75 dry objective). Time‐lapse imaging was performed using Metamorph software (Universal Imaging). For time‐lapse imaging of microtubule asters in microwells, images were taken every 5 or 10 min to avoid photo‐damages of microtubules. Photo‐bleaching experiments of lipids were also performed on this system using an iLas2 device.
To measure the position of aMTOC in microwells, the images were acquired using an upright Axioimager M2 Zeiss microscope equipped with an EC Plan‐Neofluar dry ×20 NA0.5 dry objective and CoolSNAP EZ camera (Photometrics) or using the confocal spinning‐disk system described above.
To visualize individual microtubules and actin filaments, an objective‐based total internal reflection fluorescence (TIRF) microscopy instrument composed of a Nikon Eclipse Ti, an azimuthal iLas2 TIRF illuminator (Roper Scientific), a ×60 NA1.49 TIRF objective lens, a ×100 NA1.49 TIRF objective lens and an Evolve EMCCD camera (Photometrics) was used. This system was also used to visualize tubulin precipitation in microwells. Excitation was achieved using 491, 561, and 642 nm lasers.
Image processing and measurements
To visualize microtubule asters in microwells, the images were processed to improve the signal‐to‐noise ratio. Background subtraction was performed using Fiji (NIH). To further improve the signal‐to‐noise ratio, deconvolution was also performed for some of the images (Figs 1F and 3D, and 4D) using DeconvolutionLab2 in Fiji before background subtraction. Background subtraction was not performed to show free tubulin signals in Fig 1C and Appendix Fig S2A–E. Maximum projection was performed to show microtubule asters using Fiji. To visualize actin networks along the vertical edges, maximum projection was performed excluding the bottom and the top images of microwells (partial maximum projection). Microtubule length was manually measured using a 3D distance measurement tool in Fiji. Kymograph and temporal‐color coded images were generated using plugins in Fiji. The orientation of MTs was measured and visualized using OrientationJ in Fiji. The orientation was evaluated for every pixel, and the histogram was weighted by the coherency parameter.
Measurement of the center (centroid) of wells and the center of aMTOCs was performed using Fiji with bright‐field images. Microwells containing only a single aMTOC were analyzed. Distance from the aMTOC to center of the well was measured from each XY coordinates. The actin inner zone (AIZ) was determined by setting thresholds of the fluorescence signals of actin. Then, the center of the AIZ was measured. The angles of the aMTOC and the center of the AIZ relative to the well center were calculated from each XY coordinate. To analyze aMTOC positioning relative to the center of the AIZ, wells were reoriented based on each XY coordinates in order to align the angles from the well center to center of the AIZ (see also Fig EV5C and D).
Numerical simulations
Simulations were performed using Cytosim software (Nedelec & Foethke, 2007). The motion of elastic filaments and solids surrounded by a viscous fluid was calculated using Langevin dynamics (Nedelec & Foethke, 2007). The main parameters used in the simulation were presented in Appendix Table S1.
In the simulations, repulsive steric effects between actin filaments and microtubule aster were considered. As a limitation in the simulation, steric repulsion also occurs between actin filaments and between microtubules at the same steric force. Attractive steric forces between filaments were not included in the simulations. The steric parameters were adapted from the range in previous studies testing steric interactions between microtubules or actin filaments using Cytosim (Letort et al, 2015; Rickman et al, 2019). Because of excessive computational costs, it was difficult to perform simulations of centrosome positioning with dense actin filaments. Therefore, to reduce computational costs, the effective diameter of microtubules and actin filaments was set to 100 nm (Letort et al, 2015; Rickman et al, 2019) and the cell size was set to 10 μm in radius. In addition, the total time simulated was set to 150 to 250 s. The simulations were performed in the two‐dimensional mode.
Bulk actin network was made by adding actin filaments in the cell without fixation of their position. To make actin meshwork near the cell periphery, the actin nucleation factors and branching factors were positioned near the cell periphery (within 7–9.2 μm from the cell center). The position of actin nucleation factors was fixed, so that the position of one of the ends of actin filaments was fixed at the initial position. When the actin branching factor binds to an existing actin filament, it nucleates a new actin filament from the existing filament. Asymmetric actin network was made by asymmetrically localizing the actin nucleation factors and the actin branching factors.
Statistics
Statistical tests were performed using R statistical software. Statistical test, sample sizes, and P‐values are described in each figure legend.
Author contributions
Shohei Yamamoto: Conceptualization; data curation; formal analysis; investigation; visualization; methodology; writing – original draft; writing – review and editing. Jérémie Gaillard: Data curation; validation; project administration. Benoit Vianay: Data curation; project administration. Christophe Guerin: Data curation. Magali Orhant‐Prioux: Data curation. Laurent Blanchoin: Conceptualization; funding acquisition; investigation; methodology; project administration; writing – review and editing. Manuel Théry: Conceptualization; funding acquisition; investigation; methodology; writing – original draft; project administration; writing – review and editing.
Disclosure and competing interests statement
The authors declare that they have no conflict of interest.
Supporting information
Appendix S1
Expanded View Figures PDF
Movie EV1
Movie EV2
Movie EV3
Movie EV4
Movie EV5
Movie EV6
Movie EV7
PDF+
Acknowledgements
We thank Laura Schaedel for helping in the design and manufacturing of microwells and Gaelle Letort for advices and discussions about Cytosim. This work was supported by the European Research Council (Consolidator Grant 771599 (ICEBERG) to MT and Advanced Grant 741773 (AAA) to LB). S.Y. was supported by fellowships from the EMBO (ALTF 652‐2019), Astellas Foundation for research on metabolic disorders and Mochida memorial foundation for medical and pharmaceutical research. This work was also supported by the MuLife imaging facility, which is funded by GRAL, a program from the Chemistry Biology Health Graduate School of University Grenoble Alpes (ANR‐17‐EURE‐0003).
The EMBO Journal (2022) 41: e111631.
Contributor Information
Laurent Blanchoin, Email: laurent.blanchoin@cea.fr.
Manuel Théry, Email: manuel.thery@cea.fr.
Data availability
This study includes no data deposited in external repositories. The data that support the findings of this study are available from the corresponding author upon request.
References
- Baumann H, Surrey T (2014) Motor‐mediated cortical versus astral microtubule organization in lipid‐monolayered droplets. J Biol Chem 289: 22524–22535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumgärtner S, Tolić IM (2014) Astral microtubule pivoting promotes their search for cortical anchor sites during mitosis in budding yeast. PLoS One 9: e93781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanchoin L, Boujemaa‐Paterski R, Sykes C, Plastino J (2014) Actin dynamics, architecture, and mechanics in cell motility. Physiol Rev 94: 235–263 [DOI] [PubMed] [Google Scholar]
- Bornens M (2008) Organelle positioning and cell polarity. Nat Rev Mol Cell Biol 9: 874–886 [DOI] [PubMed] [Google Scholar]
- Bornens M (2018) Cell polarity: having and making sense of direction—on the evolutionary significance of the primary cilium/centrosome organ in Metazoa. Open Biol 8: 180052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bornens M, Paintrand M, Celati C (1989) The cortical microfilament system of lymphoblasts displays a periodic oscillatory activity in the absence of microtubules: implications for cell polarity. J Cell Biol 109: 1071–1083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boujemaa‐Paterski R, Suarez C, Klar T, Zhu J, Guérin C, Mogilner A, Théry M, Blanchoin L (2017) Network heterogeneity regulates steering in actin‐based motility. Nat Commun 8: 1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brito DA, Strauss J, Magidson V, Tikhonenko I, Khodjakov A, Koonce MP (2005) Pushing forces drive the comet‐like motility of microtubule arrays in Dictyostelium. Mol Biol Cell 16: 3334–3340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burakov A, Nadezhdina E, Slepchenko B, Rodionov V (2003) Centrosome positioning in interphase cells. J Cell Biol 162: 963–969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burute M, Prioux M, Blin G, Truchet S, Letort G, Tseng Q, Bessy T, Lowell S, Young J, Filhol O et al (2017) Polarity reversal by centrosome repositioning primes cell scattering during epithelial‐to‐mesenchymal transition. Dev Cell 40: 168–184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chevrier V, Piel M, Collomb N, Saoudi Y, Frank R, Paintrand M, Narumiya S, Bornens M, Job D (2002) The Rho‐associated protein kinase p160ROCK is required for centrosome positioning. J Cell Biol 157: 807–817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colin A, Singaravelu P, Théry M, Blanchoin L, Gueroui Z (2018) Actin‐network architecture regulates microtubule dynamics. Curr Biol 28: 2647–2656.e4 [DOI] [PubMed] [Google Scholar]
- Colin L, Chevallier A, Tsugawa S, Gacon F, Godin C, Viasnoff V, Saunders TE, Hamant O (2020) Cortical tension overrides geometrical cues to orient microtubules in confined protoplasts. Proc Natl Acad Sci USA 117: 32731–32738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahlgaard K, Raposo AASF, Niccoli T, St Johnston D (2007) Capu and spire assemble a cytoplasmic actin mesh that maintains microtubule organization in the Drosophila oocyte. Dev Cell 13: 539–553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dogterom M, Koenderink GH (2019) Actin–microtubule crosstalk in cell biology. Nat Rev Mol Cell Biol 20: 38–54 [DOI] [PubMed] [Google Scholar]
- Dujardin DL, Barnhart LE, Stehman SA, Gomes ER, Gundersen GG, Vallee RB (2003) A role for cytoplasmic dynein and LIS1 in directed cell movement. J Cell Biol 163: 1205–1211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elric J, Etienne‐Manneville S (2014) Centrosome positioning in polarized cells: common themes and variations. Exp Cell Res 328: 240–248 [DOI] [PubMed] [Google Scholar]
- Ennomani H, Letort G, Guérin C, Martiel JL, Cao W, Nédélec F, De La Cruz EM, Théry M, Blanchoin L (2016) Architecture and connectivity govern actin network contractility. Curr Biol 26: 616–626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faivre‐Moskalenko C, Dogterom M (2002) Dynamics of microtubule asters in microfabricated chambers: the role of catastrophes. Proc Natl Acad Sci USA 99: 16788–16793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farhadi L, Fermino Do Rosario C, Debold EP, Baskaran A, Ross JL (2018) Active self‐organization of actin‐microtubule composite self‐propelled rods. Front Phys 6: 75 [Google Scholar]
- Field CM, Lénárt P (2011) Bulk cytoplasmic actin and its functions in meiosis and mitosis. Curr Biol 21: R825–R830 [DOI] [PubMed] [Google Scholar]
- Fong KK, Davis TN, Asbury CL (2021) Microtubule pivoting enables mitotic spindle assembly in S. cerevisiae . J Cell Biol 220: e202007193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fygenson DK, Marko JF, Libchaber A (1997) Mechanics of microtubule‐based membrane extension. Phys Rev Lett 79: 4497–4500 [Google Scholar]
- Hale CM, Chen WC, Khatau SB, Daniels BR, Lee JSH, Wirtz D (2011) SMRT analysis of MTOC and nuclear positioning reveals the role of EB1 and LIC1 in single‐cell polarization. J Cell Sci 124: 4267–4285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris TJC, Sawyer JK, Peifer M (2009) How the cytoskeleton helps build the embryonic body plan: models of morphogenesis from Drosophila . Curr Top Dev Biol 89: 55–85 [DOI] [PubMed] [Google Scholar]
- Holy TE, Dogterom M, Yurke B, Leibler S (1997) Assembly and positioning of microtubule asters in microfabricated chambers. Proc Natl Acad Sci USA 94: 6228–6231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooikaas PJ, Damstra HG, Gros OJ, van Riel WE, Martin M, Smits YT, van Loosdregt J, Kapitein LC, Berger F, Akhmanova A (2020) Kinesin‐4 KIF21B limits microtubule growth to allow rapid centrosome polarization in T cells. Elife 9: e62876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyman A, Drechsel D, Kellogg D, Salser S, Sawin K, Steffen P, Wordeman L, Mitchison T (1991) Preparation of modified tubulins. Methods Enzymol 196: 478–485 [DOI] [PubMed] [Google Scholar]
- Ierushalmi N, Malik‐Garbi M, Manhart A, Shah EA, Goode BL, Mogilner A, Keren K (2020) Centering and symmetry breaking in confined contracting actomyosin networks. Elife 9: e55368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue D, Obino D, Pineau J, Farina F, Gaillard J, Guerin C, Blanchoin L, Lennon‐Duménil A, Théry M (2019) Actin filaments regulate microtubule growth at the centrosome. EMBO J 38: e99630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue D, Kabir AMR, Tokuraku K, Sada K, Kakugo A (2020) Mechanical stimulation‐induced orientation of gliding microtubules in confined microwells. Adv Mater Interfaces 7: 1902013 [Google Scholar]
- Jiang K, Toedt G, Montenegro Gouveia S, Davey NE, Hua S, Van Der Vaart B, Grigoriev I, Larsen J, Pedersen LB, Bezstarosti K et al (2012) A proteome‐wide screen for mammalian SxIP motif‐containing microtubule plus‐end tracking proteins. Curr Biol 22: 1800–1807 [DOI] [PubMed] [Google Scholar]
- Jimenez AJ, Schaeffer A, De Pascalis C, Letort G, Vianay B, Bornens M, Piel M, Blanchoin L, Théry M (2021) Acto‐myosin network geometry defines centrosome position. Curr Biol 31: 1206–1220.e5 [DOI] [PubMed] [Google Scholar]
- Juniper MPN, Weiss M, Platzman I, Spatz JP, Surrey T (2018) Spherical network contraction forms microtubule asters in confinement. Soft Matter 14: 901–909 [DOI] [PubMed] [Google Scholar]
- Kimura K, Kimura A (2011) Intracellular organelles mediate cytoplasmic pulling force for centrosome centration in the Caenorhabditis elegans early embryo. Proc Natl Acad Sci USA 108: 137–142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonce MP, Köhler J, Neujahr R, Schwartz J‐M, Tikhonenko I, Gerisch G (1999) Dynein motor regulation stabilizes interphase microtubule arrays and determines centrosome position. EMBO J 18: 6786–6792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopf A, Kiermaier E (2021) Dynamic microtubule arrays in leukocytes and their role in cell migration and immune synapse formation. Front Cell Dev Biol 9: 158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laan L, Pavin N, Husson J, Romet‐Lemonne G, Van Duijn M, López MP, Vale RD, Jülicher F, Reck‐Peterson SL, Dogterom M (2012) Cortical dynein controls microtubule dynamics to generate pulling forces that position microtubule asters. Cell 148: 502–514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letort G, Politi AZ, Ennomani H, Théry M, Nedelec F, Blanchoin L (2015) Geometrical and mechanical properties control actin filament organization. PLoS Comput Biol 11: e1004245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letort G, Nedelec F, Blanchoin L, Théry M (2016) Centrosome centering and decentering by microtubule network rearrangement. Mol Biol Cell 27: 2833–2843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- López MP, Huber F, Grigoriev I, Steinmetz MO, Akhmanova A, Koenderink GH, Dogterom M (2014) Actin‐microtubule coordination at growing microtubule ends. Nat Commun 5: 4778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma R, Laan L, Dogterom M, Pavin N, Jülicher F (2014) General theory for the mechanics of confined microtubule asters. New J Phys 16: 013018 [Google Scholar]
- Mastrogiovanni M, Di Bartolo V, Alcover A (2021) Cell polarity regulators, multifunctional organizers of lymphocyte activation and function. Biomed J 45: 299–309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meiring JCM, Shneyer BI, Akhmanova A (2020) Generation and regulation of microtubule network asymmetry to drive cell polarity. Curr Opin Cell Biol 62: 86–95 [DOI] [PubMed] [Google Scholar]
- Nedelec F, Foethke D (2007) Collective Langevin dynamics of flexible cytoskeletal fibers. New J Phys 9: 427 [Google Scholar]
- Pavin N, Laan L, Ma R, Dogterom M, Jülicher F (2012) Positioning of microtubule organizing centers by cortical pushing and pulling forces. New J Phys 14: 105025 [Google Scholar]
- Pelletier JF, Field CM, Fürthauer S, Sonnett M, Mitchison TJ (2020) Co‐movement of astral microtubules, organelles and F‐actin by dynein and actomyosin forces in frog egg cytoplasm. Elife 9: e60047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinot M, Chesnel F, Kubiak JZ, Arnal I, Nedelec FJ, Gueroui Z (2009) Effects of confinement on the self‐organization of microtubules and motors. Curr Biol 19: 954–960 [DOI] [PubMed] [Google Scholar]
- Pitaval A, Senger F, Letort G, Gidrol X, Guyon L, Sillibourne J, Théry M (2017) Microtubule stabilization drives 3D centrosome migration to initiate primary ciliogenesis. J Cell Biol 216: 3713–3728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rickman J, Nédélec F, Surrey T (2019) Effects of spatial dimensionality and steric interactions on microtubule‐motor self‐organization. Phys Biol 16: 046004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritter ATT, Asano Y, Stinchcombe JC, Dieckmann NMGMG, Chen B‐CC, Gawden‐Bone C, van Engelenburg S, Legant W, Gao L, Davidson MWW et al (2015) Actin depletion initiates events leading to granule secretion at the immunological synapse. Immunity 42: 864–876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salehi‐Reyhani A, Ces O, Elani Y (2017) Artificial cell mimics as simplified models for the study of cell biology. Exp Biol Med 242: 1309–1317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao W, Yang J, He M, Yu XY, Lee CH, Yang Z, Joyner AL, Anderson KV, Zhang J, Tsou MFB et al (2020) Centrosome anchoring regulates progenitor properties and cortical formation. Nature 580: 106–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shelanski ML (1973) Chemistry of the filaments and tubules of brain. J Histochem Cytochem 21: 529–539 [DOI] [PubMed] [Google Scholar]
- Stinchcombe JC, Griffiths GM (2014) Communication, the centrosome and the immunological synapse. Philos Trans R Soc Lond B Biol Sci 369: 20130463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang N, Marshall WF (2012) Centrosome positioning in vertebrate development. J Cell Sci 125: 4951–4961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vignaud T, Blanchoin L, Théry M (2012) Directed cytoskeleton self‐organization. Trends Cell Biol 22: 671–682 [DOI] [PubMed] [Google Scholar]
- Vladar EK, Bayly RD, Sangoram AM, Scott MP, Axelrod JD (2012) Microtubules enable the planar cell polarity of Airway Cilia. Curr Biol 22: 2203–2212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weis F, Moullintraffort L, Heichette C, Chrétien D, Garnier C (2010) The 90‐kDa heat shock protein Hsp90 protects tubulin against thermal denaturation. J Biol Chem 285: 9525–9534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Misra G, Russell RJ, Ladd AJC, Lele TP, Dickinson RB (2011) Effects of dynein on microtubule mechanics and centrosome positioning. Mol Biol Cell 22: 4834–4841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie J, Najafi J, Le Borgne R, Verbavatz JM, Durieu C, Sallé J, Minc N (2022) Contribution of cytoplasm viscoelastic properties to mitotic spindle positioning. Proc Natl Acad Sci USA 119: e2115593119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi J, Wu X, Chung AH, Chen JK, Kapoor TM, Hammer JA (2013) Centrosome repositioning in T cells is biphasic and driven by microtubule end‐on capture‐shrinkage. J Cell Biol 202: 779–792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Burakov A, Rodionov V, Mogilner A (2010) Finding the cell center by a balance of dynein and myosin pulling and microtubule pushing: a computational study. Mol Biol Cell 21: 4418–4427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zieske K, Schwille P (2014) Reconstitution of self‐organizing protein gradients as spatial cues in cell‐free systems. Elife 3: e03949 [DOI] [PMC free article] [PubMed] [Google Scholar]


