Abstract

Targeted protein degradation (TPD) strategies exploit bivalent small molecules to bridge substrate proteins to an E3 ubiquitin ligase to induce substrate degradation. Few E3s have been explored as degradation effectors due to a dearth of E3-binding small molecules. We show that genetically induced recruitment to the GID4 subunit of the CTLH E3 complex induces protein degradation. An NMR-based fragment screen followed by structure-guided analog elaboration identified two binders of GID4, 16 and 67, with Kd values of 110 and 17 μM in vitro. A parallel DNA-encoded library (DEL) screen identified five binders of GID4, the best of which, 88, had a Kd of 5.6 μM in vitro and an EC50 of 558 nM in cells with strong selectivity for GID4. X-ray co-structure determination revealed the basis for GID4–small molecule interactions. These results position GID4-CTLH as an E3 for TPD and provide candidate scaffolds for high-affinity moieties that bind GID4.
Introduction
Over the last decade, targeted protein degradation (TPD) has evolved into a therapeutic strategy with great potential, with at least 17 TPD therapeutics in clinical trials at the beginning of 2022.1,2 TPD approaches are based on two classes of small molecules, namely, molecular glues and hetero-bifunctional agents termed proteolysis targeting chimeras (PROTACs), both of which function by recruiting a target protein of interest to an E3 ubiquitin ligase. This juxtaposition causes target polyubiquitination and subsequent degradation by the 26S proteasome, thereby eliminating target protein function. Molecular glues enhance or introduce complementarity between an E3 ligase and a neo-substrate3 and, to date, have been discovered serendipitously due to difficulties in de novo design. In contrast, the bivalent chemical architecture of PROTACs is well suited to rational design. PROTACs are composed of a moiety that binds an E3 ligase joined by a flexible linker to a moiety that specifically binds the target protein.4 Because PROTACs do not require the E3 ligase and target to engage each other directly, ligand development against the E3 ligase and target can be explored through modular combinations of binding and linker moieties. PROTACs also present many opportunities to repurpose existing small molecule binders against a multitude of targets for induced degradation.
Most PROTACs reported or in development exploit ligands to a restricted set of E3 ligases, most notably cereblon (CRBN)5,6 and Von Hippel–Lindau (VHL),7 which are substrate recruitment factors for two different cullin-RING E3 ligase complexes. The use of CRBN and VHL binding moieties stems from the availability of structurally well-characterized, high-affinity small molecule ligands with physiochemical properties compatible with therapeutic development.8 Only a handful of other E3 ligases have been explored for PROTAC development, with an untapped pool of over 600 E3 ligases encoded by the human genome that can potentially be exploited for TPD. Expansion of the E3 ligase toolbox for PROTAC development would improve the potential for spatial and temporal control of degradation within cells and tissues and the potential to maximize target degradation efficiency in specific contexts.9 Currently, structural information is available for a number of E3 ligases, some of which have well-characterized modes of substrate recognition.8 One such example is glucose-induced degradation protein 4 homolog (GID4), which functions as a substrate receptor in the human CTLH (C-terminal to LisH) E3 ligase complex and the orthologous GID complex in yeast.10−13 As GID4 is detectably expressed in most tissue types,14 it is an attractive new candidate E3 for targeted protein degradation strategies. Additionally, consistent with other subunits of the CTLH complex, GID4 localizes to the cytosol and nucleus,15 thereby providing an opportunity for induced degradation of targets in both cellular compartments.
GID4 comprises an eight-strand β-barrel that forms a deep cavity for substrate binding at one opening (PDB: 6CDC).16 GID4 recognizes short linear peptide motifs, termed degrons, at the free amino-terminus of substrate proteins, which often commence with a hydrophobic residue such as Pro, Val, Ile, and Phe.16−20 Co-structures of GID4 bound to degron peptides16−19 reveal a substantial degree of plasticity within the ligand binding pocket that appears well suited for engagement of small molecule ligands. Indeed, unpublished co-structures of two small molecules in complex with GID4 have been reported by the Structural Genomics Consortium (PDB: 7SLZ and 7S12).21
Here, we show genetically that GID4 induces the degradation of a protein substrate when brought into close proximity to GID4. We undertook two campaigns to discover novel small molecule binders of GID4 using fragment-based NMR and DNA-encoded library (DEL) screening approaches. Determination of X-ray structures of these compounds in complex with GID4 revealed distinct binding modes with the GID4 substrate recognition pocket. Structure–activity relationship (SAR) series based on these hits produced further optimized binders with dissociation constant (Kd) values ranging from 5.6 to 110 μM. The highest affinity binder engaged GID4 in cells with an EC50 value of 558 nM and high selectivity as assessed by mass-spectrometry-based thermal proteome profiling. The compounds and binding modes identified in this study provide a structural framework to support the development of high-affinity small molecule binders to GID4, which in turn will allow the exploration of GID4 and the CTLH complex in TPD design strategies.
Results and Discussion
GID4 Recruitment Induces the Degradation of a Model Protein Substrate
To investigate the potential of the CTLH E3 ligase complex for TPD, we assessed whether direct recruitment of the model substrate EGFP to the substrate receptor subunit GID4 could induce EGFP degradation in living cells. We developed a genetic reporter system that recapitulates proximity-dependent degradation by TPD effectors. E3 substrate recognition subunits GID4, CRBN (positive control), VHL (positive control) or unrelated protein Renilla luciferase (RLuc) (negative control), were fused to an anti-GFP nanobody (vhhGFP) and transfected into an HEK293T cell line that stably expresses EGFP (Figure 1A). Binding of the vhhGFP-E3 effector fusion to EGFP is predicted to cause ubiquitination and degradation of EGFP, manifesting as a decrease in EGFP fluorescence signal measured by flow cytometry. As for the established degradation effectors CRBN and VHL, but not RLuc, the GID4-vhhGFP fusion decreased EGFP fluorescence (Figure 1). The low efficacy of GID4 compared to VHL and CRBN in this assay may reflect a higher intrinsic activity of the VHL and CRBN E3 complexes or assay parameters including effector-vhhGFP expression levels, catalytic access to lysine residues in EGFP, the geometry of substrate reporter fusions (N- versus C-terminal tagging), and/or efficiency of E3 effector fusion incorporation into multi-subunit E3 ligases.22,23 Nonetheless, this result demonstrated that GID4 can target ectopically recruited substrates for degradation and provided a rationale for our subsequent efforts to develop small molecule binders to GID4.
Figure 1.

Renilla luciferase (RLuc), VHL, CRBN, or GID4 effector proteins fused by a poly Gly-Ser (GS) linker to a nanobody that binds GFP (vhhGFP) were transfected into the reporter cell line expressing EGFP fused to ABI1 alongside DsRed to monitor transfection efficiency. Forty-eight hours post-transfection, EGFP fluorescence was measured by flow cytometry for DsRed positive cells. (A) EGFP intensity overlapping histograms for VHL, CRBN, and GID4 (solid line) versus reporter cells transfected with an unrelated plasmid control (dotted line). (B) Median EGFP signal was normalized to control transfected reporter cells (dashed line). Experiment was done in duplicate. Statistical significance was calculated with unpaired two-tailed t test assuming equal variance and corrected for multiple hypotheses with the false discovery rate (FDR) approach of Benjamini, Krieger, and Yekutieli (***, p < 0.001; **, p < 0.01; *, p < 0.05). Note that the ABI1 element of the EGFP reporter was not used for effector recruitment in this experiment.
NMR Fragment Screen for Binders to GID4
To initiate the development of heterobifunctional degraders that recruit neo-substrates to the CTLH E3 ligase complex, we set out to discover small molecule binders of the GID4 substrate recognition subunit. Given the favorably modest size of the substrate binding domain of GID4 (residues 124–289; 19.5 kDa), we investigated the use of NMR in a small molecule screening assay format. N15-labeled GID4124–289 (referred to as GID4 hereafter) displayed an excellent H1-N15-HSQC spectrum in the presence of 2.5% DMSO, with 152 resonance peaks detected out of 163 expected using a 10 min acquisition time (Figure S1A). We demonstrated the potential of NMR to detect low-affinity binders using a degron peptide of sequence PGLWKSC that binds GID4 with a dissociation constant (Kd) of 1.9 μM.16 This peptide elicited marked global changes to the GID4 HSQC spectrum, with evident chemical shift perturbations and line broadening for many peaks (Figure S1B).
We screened the commercially available 1000-compound Maybridge Ro3 fragment library in 200 pools of five fragments at 5 mM each (Figure 2A). Pools were ranked based on the degree of HSQC chemical shift perturbations relative to a DMSO control. The top eight pools were chosen for deconvolution of candidate binders within each pool in an additional 40 NMR experiments. Nine individual fragments (denoted 1 to 9, see Figure 2B for chemical structures) yielded HSQC perturbations similar to the original hit pools (Figure S1C). For one of the hit pools, 1 and 3 both elicited HSQC perturbations that, when combined, resembled those observed in the original pool. Fresh powders of compounds 1 through 8 were purchased from commercial sources (9 was not available for purchase), and all produced HSQC perturbations similar to those observed in the hit deconvolution experiments (data not shown).
Figure 2.

(A) Workflow of the NMR screening approach to identify GID4 binders. (B) Chemical structures of hit fragments 1 to 9. (C) DSF data for the degron peptide PGLWKS and hit fragments 1 to 9. ΔTm values were calculated by subtracting the Tm of GID4 from the Tm of GID4 with the ligand. Each bar represents the mean ± SEM of determinations in triplicate. (B, C) Fragments that yielded co-crystal structures are colored.
To validate the NMR screen hits and obtain an approximate rank order of binding affinity, we used differential scanning fluorimetry (DSF) to measure melting curve changes upon binding. GID4 on its own exhibited a melting temperature (Tm) of 37 °C, and the addition of the PGLWKS degron peptide resulted in a 9.4 °C increase of the Tm (Figure 2C). Of the eight compounds purchased, six stabilized GID4 by greater than 1 °C, with 1, 4, and 7 producing the largest effects (6.7, 4.5, and 3.2 °C increases, respectively) (Figure 2C).
X-ray Crystal Structures of NMR Fragment Hits Bound to GID4
To understand hit compound binding modes to GID4, we set up co-crystallization trials of GID4 with each of the eight hit fragments. Co-crystals and structures were obtained for 1 (PDB: 7U3E), 4 (PDB:7U3F ), and 7 (PDB: 7U3H) (Figure 3; see Table S1 for X-ray structure determination and refinement statistics). The substrate recognition domain of GID4 consists of an eight-strand β-barrel with an insert containing three α-helices (Figure S2A). One end of the β-barrel contains a deep pocket flanked by four loops denoted L1 to L4 that comprise the degron peptide binding pocket. The co-crystal structures of 1, 4, and 7 revealed that all three fragments bind to this same pocket on GID4 (Figure S2A).
Figure 3.
(Left) Co-crystal structures of GID4 in complex with (A) 7 (PDB: 7U3H), (B) 4 (PDB: 7U3F), and (C) 1 (PDB: 7U3E) with hydrogen bonds depicted as black dashed lines. (Right) Graphical representation showing the key interactions between GID4 and each hit fragment. Hydrophobic interactions are represented as eyelashes, while hydrogen bonds are represented by blue dashed lines, each with an associated distance in angstroms.
Although the three fragment hits are structurally distinct, their binding to GID4 is anchored by a common hydrogen bond between the Glu237 side chain of GID4 and a secondary amino group within the fragments (Figure 3, Figure S2B). Notably, Glu237 coordinates the amino terminus of degron peptides in previously reported crystal structures.16,17,19 Each of the three fragments contains a hydrophobic aromatic ring that engages a hydrophobic pocket in GID4 lined by Tyr273, Phe254, Leu171, Ala252, Leu164, Ile161, and Leu159. This same pocket accommodates bulky hydrophobic groups in degron peptides (e.g., Phe in the FDVSWFMG degron).17 Similar to degron peptide binding, the binding of each hit fragment elicits major conformational changes to the substrate binding pocket of GID4 relative to the apo structure (PDB: 6CCR) (Figure S2A,B). These conformational changes differ considerably for each hit fragment, highlighting the plasticity of the degron binding cavity of GID4. The specific binding interactions and conformational changes evoked by each compound relative to the apo structure are as follows:
Binding mode of fragment 7
This fragment comprises a secondary amine flanked by fluorophenyl and dihydrothiazole moieties (Figure 2B, Figure S2C). Within racemate 7, the (R)-enantiomer was selectively bound to GID4 (Figure 3A). Binding of 7 drives a conformational shift in loop L2 (∼6.1 Å) away from the ligand binding cavity of GID4 (Figure S2B). Within the hydrophobic pocket of GID4, the fluorophenyl group of 7 forms π–π interactions with Phe254, while the dihydrothiazole group forms hydrophobic contacts with Tyr273 (Figure 3A). Favorable polar interactions include hydrogen bonds between the Glu237 side chain of GID4 and each of the two nitrogen atoms in 7, and between the Tyr258 side chain and the imine nitrogen of 7. Carbon atoms in the phenyl 2,3-positions, as well as sulfur atom and methyl substituent, are all oriented to the solvent and represent sites for potential fragment elaboration (Figure S2C).
Binding mode of fragment 4
This fragment comprises a tetrahydrothienopyridine group bearing a para-methoxyphenyl substituent (Figure 2B, Figure S2C). While 4 was synthesized as a racemic mixture, GID4 bound only to the (R)-stereoisomer (Figure 3B). Unlike 7, the binding of 4 drives a major conformational change in the N-terminal segment of loop L3 (∼13.7 Å) away from the ligand binding cavity of GID4 (Figure S2B). The C-terminal segment of loop L3 adopts an alpha-helical structure in apo GID4 that is unraveled and translated (∼8.3 Å) toward the binding cavity upon binding to 4. Within the hydrophobic pocket of GID4, the methoxyphenyl group of 4 forms π–π interactions with Phe254, while the tetrahydrothienopyridine ring forms hydrophobic interactions with Tyr273 (Figure 3B). Favorable polar interactions include hydrogen bonds between the Glu237 and Tyr258 side chains of GID4 and the secondary amine of 4. The thiazole ring of 4 notably orients toward the solvent (Figure S2C).
Binding mode of fragment 1
This fragment is composed of a bridged bicyclic piperazine protected with a single tert-butoxycarbonyl group (Figure 2B, Figure S2C). The co-crystal structure of 1 bound to GID4 shows exclusive binding of the (1S,4S)-enantiomer as opposed to its (1R,4R) counterpart (Figure 3C). Binding of 1 elicits similar conformational changes in loop L3 as binding of 4 (Figure S2B). Fragment 1 lacks a constituent occupying the deep hydrophobic pocket of GID4. Instead, the side chain of Phe254 intramolecularly fills the hydrophobic cavity, forming hydrophobic interactions with the bridged piperazine of 1. Favorable polar interactions include hydrogen bonds between the Glu237 side chain of GID4 and the secondary amine of 1 and between the backbone amine of Ser253 and carbonyl group of 1. The t-butyl group of 1 is oriented toward the solvent (Figure S2C).
Structure-Guided Optimization of GID4 Binding Fragments
With the binding mode of each hit fragment to GID4 in hand, we set out to optimize binding affinity by exploring SAR for each fragment. We first reassessed the binding affinity of GID4 for 7, 4, and 1 using isothermal titration calorimetry (ITC). All three fragments displayed binding signals that were too weak to quantify accurately (data not shown). We commercially sourced or synthesized analogs of 7, 4, and 1 and explored SAR for each as follows:
Fragment 7 SAR
An online search of the Molport chemical database for commercial compounds with 50% structural similarity to 7 yielded 458 compounds. We selected 22 analogs, 10–31, for purchase (see Experimental Procedures for details) that vary in substitution pattern and scaffold relative to 7 (Table 1). Each of these analogs was first assessed for binding to GID4 by DSF. The three analogs, 15, 16, and 20, each improved the stability of GID4 by at least 1 °C relative to 7 (Table 1). To compare the relative binding affinities of 15, 16, and 20 for GID4, we carried out competition studies using a fluorescently labeled PGLWKS substrate peptide as a probe in fluorescence polarization (FP) assays (Kd = 4.0 μM, Figure S3). Analog 15 was the weakest binder of the three analogs (IC50 = 264.0 μM), while 16 and 20 displayed comparable binding affinities (IC50 values of 148.5 and 113.2 μM, respectively) (Table 1, Figure S4). Analogs 16 and 20 differ from 15 by the constituents on their benzene rings where the meta-fluoro group in 20 is replaced with a meta-hydroxyl group in 16 and is entirely absent in 20. Further analysis of rac-16 by ITC yielded a Kd of 110 μM (Figure S5A).
Table 1. Structure–Activity Relationships of 7.
ΔTm values were calculated from DSF experiments by subtracting the Tm of GID4 from the Tm of GID4 in the presence of the compound. Each value represents the mean ± SEM of determinations in triplicate.
IC50 values were calculated by averaging displacement curves from FP competition assays. Each value represents the mean ± SEM of determinations in triplicate.
We determined the co-crystal structure of 16 bound to GID4 (PDB: 7U3I, Figure 4, Figure S5B, Table S1). In contrast to the selective binding of (R)-7, the co-structure of 16 unveiled preference for the (S)-enantiomer. Because only one stereoisomer in the 16 racemate was observed to bind GID4, its binding affinity is likely underestimated (Kd < 110 μM). On the phenyl moiety, 16 is decorated with a meta-hydroxyl as opposed to the para-fluoro of 7. This difference allows for an additional hydrogen bond between the hydroxyl group of 16 and the side chain of Ser253 and preferential binding of the (S)-stereoisomer. Globally, the binding cavity on GID4 remains unchanged upon binding to 7 or 16.
Figure 4.

(Left) Chemical structure of 16. (Middle) Co-crystal structure of GID4 in complex with 16 (PDB: 7U3I) with hydrogen bonds depicted as black dashed lines. (Right) Graphical representation showing the key interactions between GID4 and 16. Hydrophobic interactions are represented as eyelashes, while hydrogen bonds are represented by blue dotted lines, each with an associated distance in angstroms.
Fragment 4 SAR
An online search of the Molport chemical database for analogs with 50% structural similarity to 4 yielded 1000 compounds bearing little resemblance to the parent. Nonetheless, in silico docking analysis using Glide24−26 predicted analogs 32–40 to have improved affinity to GID4 (see Experimental Procedures for details; Figure S6). The nine analogs were purchased and tested by DSF, but none enhanced the stability of GID4 by >1.5 °C, suggesting a lack of binding (Figure S6). We next synthesized a series of 30 analogs, 41–71, to explore subtler modification of 4 (Table 2; see Experimental Procedures for synthetic routes). Of these, 15 analogs stabilized GID4 by at least 1 °C better than 4 in DSF experiments (Table 2). Further analysis of these 15 analogs using the FP competition assay revealed 67 as the best binder to GID4 with an IC50 of 18.9 μM (Table 2, Figure S4). Quantification of the binding affinity of 67 by ITC yielded a Kd of 17 μM (Figure S7A).
Table 2. Structure–Activity Relationships of 4.
| cmpd | X | R1 | R2 | R3 | R4 | ΔTm (°C)a | IC50 (μM)b |
|---|---|---|---|---|---|---|---|
| 4 | S | 4-MeO-Ph | N/A | N/A | N/A | 4.5 ± 1.4 | |
| 41 | S | H | N/A | N/A | N/A | 1.9 ± 0.7 | |
| 42 | S | pyrid-3-yl | N/A | N/A | N/A | 2.5 ± 0.4 | |
| 43 | S | Ph | N/A | N/A | N/A | 7.9 ± 0.5 | ND (>250) |
| 44 | O | Ph | N/A | N/A | N/A | 8.7 ± 0.5 | 90.8 ± 17.4 |
| 45 | NH | Ph | N/A | N/A | N/A | 9.2 ± 0.4 | 108.3 ± 16.2 |
| 46 | S | 4-AcO-Ph | N/A | N/A | N/A | 3.3 ± 0.8 | |
| 47 | S | 4-CF3-Ph | N/A | N/A | N/A | 2.7 ± 0.8 | |
| 48 | S | 4-CN-Ph | N/A | N/A | N/A | 2.0 ± 0.8 | |
| 49 | S | 4-Br-Ph | N/A | N/A | N/A | 4.2 ± 0.7 | |
| 50 | S | 4-Cl-Ph | N/A | N/A | N/A | 5.1 ± 0.8 | |
| 51 | S | 4-F-Ph | N/A | N/A | N/A | 3.8 ± 1.7 | |
| 52 | S | 4-NH2-Ph | N/A | N/A | N/A | 7.1 ± 0.3 | ND (>250) |
| 53 | S | 4-(HOCH2)-Ph | N/A | N/A | N/A | 4.4 ± 0.6 | |
| 54 | S | 4-Me-Ph | N/A | N/A | N/A | 5.9 ± 0.7 | 55.0 ± 3.7 |
| 55 | S | 3-Cl-Ph | N/A | N/A | N/A | 7.3 ± 0.7 | 49.3 ± 17.8 |
| 56 | S | 3-OH-Ph | N/A | N/A | N/A | 6.1 ± 0.6 | ND (>250) |
| 57 | S | 3-NH2-Ph | N/A | N/A | N/A | 6.6 ± 0.4 | 235.3 ± 103.4 |
| 58 | S | 3-MeO-Ph | N/A | N/A | N/A | 7.6 ± 0.7 | ND (>250) |
| 59 | S | 3-CN-Ph | N/A | N/A | N/A | 3.9 ± 0.6 | |
| 60 | S | 3-NO2-Ph | N/A | N/A | N/A | 4.9 ± 0.6 | |
| 61 | S | 2-F-Ph | N/A | N/A | N/A | 5.8 ± 0.7 | 149.1 ± 84.6 |
| 62 | S | 2-Cl-Ph | N/A | N/A | N/A | 5.0 ± 0.8 | |
| 63 | S | 2-NH2-Ph | N/A | N/A | N/A | 2.8 ± 0.2 | |
| 64 | S | 2-NO2-Ph | N/A | N/A | N/A | 4.1 ± 0.5 | |
| 65 | S | 2-CF3-Ph | N/A | N/A | N/A | 5.3 ± 0.7 | |
| 66 | N/A | N/A | Br | H | Ph | 7.0 ± 0.8 | 64.8 ± 8.8 |
| 67 | N/A | N/A | NH2 | H | Ph | 8.5 ± 0.9 | 18.9 ± 3.5 |
| 68 | N/A | N/A | OMe | H | Ph | 4.4 ± 0.7 | |
| 69 | N/A | N/A | H | OMe | Ph | 6.4 ± 0.9 | ND (>250) |
| 70 | N/A | N/A | H | H | 3-MeO-Ph | 7.8 ± 0.7 | 35.5 ± 2.8 |
| 71 | N/A | N/A | H | H | 4-MeO-Ph | 6.7 ± 0.7 | 89.8 ± 28.6 |
ΔTm values were calculated from DSF experiments by subtracting the Tm of GID4 from the Tm of GID4 in the presence of compound. Each value represents the mean ± SEM of determinations in triplicate.
IC50 values were calculated by averaging displacement curves from FP competition assays. Each value represents the mean ± SEM of determinations in triplicate. ND = not determined.
Fragment 67 harbors a 5-amino-tetrahydroisoquinoline scaffold in lieu of the tetrahydrothienopyridine in 4 and lacks the para-positioned methoxy substituent. As observed for 4, co-structure analysis of 67 (PDB: 7U3G) revealed a similar overall positioning within GID4 and exclusive binding to the (R)-enantiomer in the racemic mixture, suggesting that its binding affinity is also underestimated (Kd < 17 μM) (Figure 5, Figure S7B). The protruding amine group of 67 introduces a hydrogen bond with the side chain of Gln282.
Figure 5.
(Left) Chemical structure of 67. (Middle) Co-crystal structure of GID4 in complex with 67 (PDB: 7U3G) with hydrogen bonds depicted as black dashed lines. (right) Graphical representation showing the key interactions between GID4 and 67. Hydrophobic interactions are represented as eyelashes, while hydrogen bonds are represented by blue dashed lines, each with an associated distance in angstroms.
Fragment 1 SAR
An online search of the Molport chemical database for analogs of 1 yielded 922 compounds, most of which displayed major structural differences from 1. We conducted in silico docking analysis with Glide24−26 to prioritize nine analogs, 72–80, for purchase, but none enhanced the stability of GID4 relative to 1 as assessed by DSF (Figure S8). This prompted us to synthesize a series of analogs, 81–87, focused on the Boc group (Table S2). The seven synthetic analogs were tested by DSF and displayed no evidence of improved binding to GID4. Variants of the t-butyl group introduced truncation or polarity that abrogated the stabilization of GID4, suggesting that the hydrophobic interactions made by this substituent are essential binding elements of 1 (Figure 3C, Figure S2C).
.
DNA-Encoded Library Screen against GID4
The identification of low-molecular-weight (<250 Da) binders to GID4 with Kd values ranging from 17 to 110 μM using an NMR fragment screening and SAR optimization strategy suggested that it may be possible to identify more complex chemical moieties that bind GID4 with higher affinity. DNA encoded library (DEL) technology affords a means to screen vast libraries generated by combinatorial chemistry, typically using binding selection assays with recombinant proteins.27−29 We therefore screened a 4.4 billion-compound DEL offered through the DELopen program (https://hits.wuxiapptec.com/delopen) against recombinant GID4. Binding selections were carried out with three protein samples immobilized on nickel chelate affinity resin: GID4 wild-type (WT) alone, GID4 WT in the presence of PGLWKS degron peptide (Kd = 4.0 μM by FP; Figure S9A), and the GID4Q282A mutant that is defective in binding to the PGLWKS peptide (Figure S9A; see experimental workflow in Figure 6A). Selection against nickel chelate affinity resin alone was used as a nonspecific binding control. For each of the four samples, three successive rounds of DEL selections were carried out to enrich for robust binders. The second round of selections was chosen for deep sequencing based on the total retained DNA content. Sequenced hits were filtered to remove commonly encountered false positives and nonspecific binders to the affinity resin. Enrichment scores for 1430 hit molecules were calculated based on DNA copy number, sub-library size, and number of sequencing reads.30 Almost all hits produced comparable enrichment scores in the GID4 WT and GID4 WT + PGLWKS degron peptide samples, which may reflect the inability of the low-affinity PGLWKS degron peptide to completely occlude the GID4 binding site. In contrast, many hits in the GID4 WT sample displayed approximately 2-fold reduced enrichment scores in the GID4Q282A mutant sample, suggesting that these hits might engage the degron binding cavity of GID4. We focused our downstream validation studies on this latter class of hits.
Figure 6.

(A) Workflow of the DEL screening approach to identify GID4 binders. (B) Chemical structures of degron peptide PGLW and hit DEL compounds 88 to 92 aligned by their peptide bonds. Moieties in the three building block positions are colored red for BB3, blue for BB2, and green for BB1. (C, top) Displacement curves from FP competition assays of GID4 bound to PGLWKS-FITC in the presence of the indicated ligand competitor. Quantified IC50 values together with ΔTm values determined by DSF. ΔTm values were calculated by subtracting the Tm of GID4 from the Tm of GID4 with the ligand. Each value represents the mean ± SEM of determinations performed in triplicate.
Across the 904 molecules enriched (>100-fold enrichment) in the GID4 WT sample, patterns of recurring building blocks were apparent in each of the three building block positions (denoted BB1, BB2, and BB3), suggesting the presence of structurally related families (Figure 6B). We categorized the top 200 hits into five families based on the identity of the building block in position BB3. We next selected a high-scoring representative molecule (denoted 88 through 92) from each of the families for resynthesis without the DNA tag (Figure 6B). All five hit molecules can be viewed as di-peptides with non-natural R groups in building block positions BB2 and BB1 and incorporating an N-terminal substituent in BB3. Ordered from N to C termini, the N-terminus of BB2 is attached to BB3 and the C-terminus of BB2 is attached to BB1 through a peptide bond. The C-terminus of BB1 (corresponding to the position of DNA tag attachment in the original screen) is blocked by amidation with methylamine.
Validation of DEL Screen Hits
We first assessed the binding of each of the five DEL-derived molecules to GID4 by DSF using resynthesized hits. Four out of five molecules displayed robust stabilizing effects on GID4 with the best stabilizer, 88, producing a ΔTm of 16.0 °C relative to apo-GID4 and 6.6 °C relative to the PGLWKS degron peptide (Figure 6C). Molecules 91, 89, and 90 produced ΔTm values of 8.8, 8.7, and 4.5 °C, respectively, relative to apo GID4, while 92 had a marginal stabilizing effect (∼0.1 °C). The degree of GID4 stabilization by each DEL-derived molecule correlated approximately with affinity in the FP competition assay (IC50 = 5.4, 4.8, and 4.7 μM, for 88, 91, and 89 and unquantifiable values for 90 and 92, respectively) (Figure 6C). Given the similar size, chemical structure, and potencies (i.e., IC50 values) of 88, 89, and 91, we chose 88 as a representative molecule for follow-up analysis by ITC, which revealed a Kd of 5.6 μM for GID4 (Figure S9B).
To determine whether the original apparent 2-fold decrease in enrichment observed for 88 (and other DEL screen hits) in the GID4Q282A versus the GID4 WT DEL screen samples correlates with a decrease in affinity of 88 for GID4Q282Ain vitro, we measured the binding affinities by ITC. Although the Q282A mutation abrogated GID4 binding to the PGLWKS degron peptide as assessed by FP (Figure S9A), to our surprise, the same mutation enhanced binding to 88 (Kd = 0.28 μM) relative to GID4 WT (Kd = 5.6 μM) (Figure S9B). We speculate that the discrepancy between binding in the DEL screen (i.e., enrichment score) and in the ITC experiment may be due to the absence of linker and DNA tag sequences in the resynthesized 88 sample.
To determine if 88 could engage full-length GID4 in the context of a complex cellular lysate, we conducted cellular thermal shift assay (CETSA) experiments. Because we could not identify an antibody that detects endogenous GID4, we first generated a HEK293 cell line with tetracycline-inducible expression of 3xFLAG epitope-tagged full-length GID4 (denoted FLAG-GID4). Incubation of tetracycline-induced lysates with 50 μM 88 revealed a Tm shift of 5.4 °C, while 92, used as a negative binding control, produced a minor Tm shift of 0.3 °C relative to lysates treated with DMSO (Tm = 56.4 °C) (Figure 7AB, Figure S10A). For comparison, our best NMR fragment-derived binder, 67, produced a Tm shift of 3.8 °C, while the fragment binder 16 and the PGLWKS degron peptide produced Tm shifts of <0.5 °C at a concentration of 50 μM (Figure 7AB, Figure S10A). Next, we examined the dose-dependent stabilization of GID4 by conducting isothermal dose–response fingerprinting cellular thermal shift assay (ITDRF-CETSA) experiments (Figure 7CD, Figure S10B).31,32 Here, FLAG-GID4 expressing cell lysates were incubated with increasing compound concentrations and subjected to heat treatment at 57 °C (the Tm of FLAG-GID4 in the absence of a small molecule binder).
Figure 7.
(A) Representative western blot analysis of CETSA experiments in the presence of DMSO or 50 μM 88, 92, 67, 16, or PGLWKS peptide. (B) Band intensities were quantified across duplicates by densitometry and plotted. (C) Representative western blot analysis of ITDRF-CETSA experiments in the presence of 88, 67, 16, or PGLWKS peptide. Input refers to lysate that has not been heat treated. (D) Band intensities were quantified by densitometry from duplicate blots and plotted. (A, C) Anti-FLAG antibody was used to detect FLAG-GID4 from FLAG-GID4 expressing HEK293 cell lysates, while anti-SOD1 antibody was used to detect SOD1 as a loading control. See Figure S10A for duplicate blots. (B, D) Each point represents the mean ± SEM of determinations performed in duplicate.
Titrations of 88 (Kd = 5.6 μM; Figure S9B) displayed saturable binding with an EC50 of 558 nM, while our best NMR fragment binder, 67 (Kd = 17 μM; Figure S7A), showed nonsaturable stabilization of GID4 (EC50 value could not be determined) (Figure 7CD). In contrast, the weaker binders, 16 and the PGLWKS degron peptide, showed no stabilizing effects on GID4 at all concentrations tested (Figure 7CD, Figure S10B). These results confirm the ability of a subset of small molecules developed through both NMR fragment and DEL screening approaches to bind GID4 in cell lysates.
We next employed live-cell CETSA to test whether our best binder, 88, could permeate the cell membrane and engage FLAG-GID4 in living cells. Western blot analysis of cells treated with 88 showed a 3.8 °C stabilization of FLAG-GID4 relative to cells treated with DMSO, indicating that 88 is cell-permeable and can engage FLAG-GID4 in cells (Figure 8, Figure S11). To investigate the binding specificity of 88, we conducted thermal proteome profiling33 of HEK293 lysates treated with DMSO or 88.31 Of the 5663 endogenous proteins detected by mass spectrometry in this experiment, GID4 exhibited the greatest Tm shift of 7.4 °C in the presence of 88, whereas 14 other proteins, representing possible off-target binders, produced smaller Tm shifts ranging from −3.7 to 4.6 °C (Figure 9, Table S3). Together, these results demonstrate the ability of 88 to engage endogenous GID4 protein in cells with considerable selectivity.
Figure 8.
Representative western blot analysis of the live cell CETSA experiment using FLAG-GID4 expressing HEK293 cells in the presence of DMSO or 50 μM 88. Anti-FLAG antibody was used to detect FLAG-GID4, while anti-SOD1 antibody was used to detect SOD1 as a loading control. Band intensities were quantified by densitometry, and melting curves were plotted on the right. Each point represents the mean ± SEM of determinations performed in duplicate (see duplicate blots in Figure S11).
Figure 9.
Thermal proteome profiling of HEK293 lysates treated with 20 μM 88. (A) Scatter plot of ΔTm values calculated from the two replicates of 88 versus vehicle treatment experiment. 88-induced ΔTm values for proteins that passed the significance criteria are shown in red (see Table S3 for the list of shifted proteins). We note that an overall temperature drift in the 88 versus vehicle treated samples led us to implement stricter statistical tests for identifying proteins that were affected by the compound. (B) Melting curves for GID4 from two replicates of cells treated with 88 or the vehicle.
X-ray Crystal Structures of DEL Molecules Bound to GID4
To delineate binding modes to GID4, we set up co-crystallization trials with each of the five DEL-derived molecules. Co-crystals and structures were obtained for the three strongest stabilizers of GID4, namely, 88 (PDB: 7U3J), 89 (PDB: 7U3K), and 91 (PDB: 7U3L) (Figure 10, Figure S12; see Table S1 for X-ray structure determination and refinement statistics). Consistent with their ability to compete with degron peptide binding (Figure 6C), all three DEL molecules bound to the degron binding pocket of GID4 (Figure S12A). We note that all three DEL molecules possess two stereocenters, and while 89 and 91 were synthesized as homochiral (R,S)-molecules, 88 was synthesized as a diastereoisomeric mixture (roughly 50% each of (S,S) and (R,S)). The crystal structure confirmed the expected (R,S) chirality of 89 and 91 and showed exclusive binding of the (S,S)-stereoisomer of 88 to GID4. This result implied that the apparent Kd of 88 (5.6 μM) determined by ITC is likely underestimated by a factor of 2, which is in agreement with a higher-than-expected N value of 1.5 (Figure S9B).
Figure 10.
(Left) Co-crystal structures of GID4 in complex with DEL compound (A) 88 (PDB: 7U3J), (B) 89 (PDB: 7U3K), and (C) 91 (PDB: 7U3L) with hydrogen bonds depicted as black dashed lines. (Right) Graphical representation showing the key interactions between GID4 and each hit DEL compound. Hydrophobic interactions are represented as eyelashes, while hydrogen bonds are represented by blue dashed lines, each with an associated distance in angstroms.
Superimposition of the PGLW degron peptide co-structure with the three DEL molecule co-structures revealed remarkable similarity in binding modes, including the near-perfect overlap of (i) the backbone atoms of Pro1 with the backbone atoms of BB3, (ii) the backbone atoms of Gly2 with the backbone atoms of BB2, and (iii) the amino group of Leu3 with the amino group of BB1 (Figure 6B). Similarities in hydrogen bonding patterns include hydrogen bonds between (i) the secondary amine of BB3 and the GID4 side chains of Glu237 and Tyr258, (ii) the backbone amine of BB2 and the main chain carbonyl of Ser253, and (iii) the backbone amine of BB1 and the side chain carbonyl of Gln282 (Figure 10). Three distinct cavities in GID4 mediate analogous recognition of the hydrophobic portions in each DEL building block and the hydrophobic side chains in the PGLW degron peptide, as follows.
The deepest penetrating cavity lined by Phe254, Ile249, Leu171, Thr173, Ile161, and Leu159 side chains engages Pro1 of the degron and BB3 of the DEL molecules. In 88 and 91, the BB3 hydrophobic moiety is a substituted phenyl ring, whereas in 89, the hydrophobic moiety is a butyl group. A second cavity engages Gly2 of the degron and BB2 of the DEL molecules. In each of the three DEL molecules, the moieties occupy different spatial positions due to the large rearrangement of GID4 loops L2 and L3 (Figure S12A,B). While the thiophene of BB2 in 88 and the propenylbenzyl of BB2 in 91 similarly interact with Tyr139 and Tyr273, the 4-tert-butylbenzyl moiety of BB2 in 89 interacts with Ile249, Leu164, and Ile161. A third cavity, lined by Trp280 and Tyr139, borders Leu3 of the degron and BB1 of the DEL molecules. All three DEL molecules have an aryl ring-containing group in the BB1 position that overlaps well with the Leu3 side chain of the degron.
Analysis of Embedded SAR in the DEL Screen Hit List
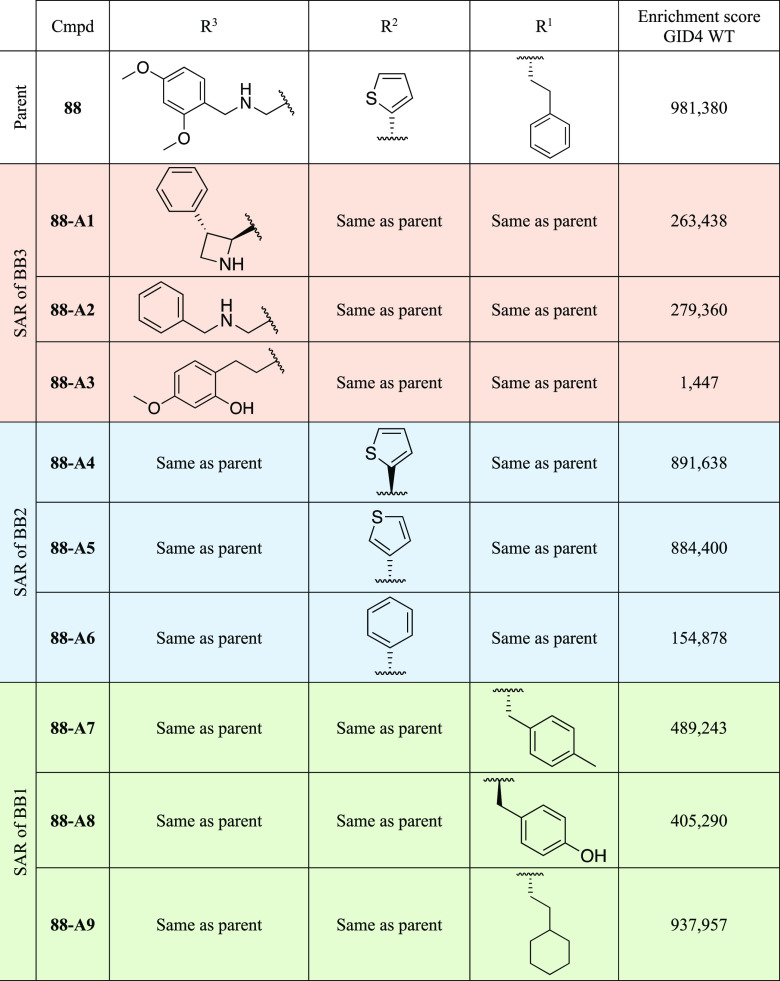
Patterns of recurring building blocks in the DEL screen hit list suggest rich SAR information that could guide future hit optimization. Using our best binder 88 as an exemplar, we searched for the top three most enriched building blocks in each of the BB3, BB2, and BB1 positions while maintaining the two other building block positions fixed (Table 3). Enrichment scores in the GID4 WT sample served as a rough proxy for relative binding affinity. The most enriched moieties in the BB3 position all contain an aromatic ring that can be rationalized by the hydrophobic nature of the pocket to which it binds. Analogs 88-A1 and 88-A2 but not 88-A3 have an amino group, and the absence of this group correlates with a dramatic loss of enrichment for 88-A3 (Table 3). This likely reflects a loss of binding affinity because all known GID4 binders characterized to date (whether degron peptides or NMR-derived fragments and DEL-derived molecules in this study) contain an amino group that hydrogen bonds with Glu237 of GID4. The top three most enriched moieties in the BB2 position (present in 88-A4 to 88-A6) all contain an aromatic moiety (either a thiophene moiety or a phenyl group), which is again consistent with the hydrophobic nature of the binding pocket that they engage. The top three most enriched moieties in the BB1 position (present in 88-A7 to 88-A9) contain phenyl or cyclohexane ring structures that correlate with the hydrophobic nature of the binding pocket.
Table 3. Structure–Activity Relationships of 88.

Comparisons to the Binding Mode of PFI-7
The Structural Genomics Consortium (SGC) has reported an unpublished small molecule binder of GID4 called PFI-7 (https://www.thesgc.org/chemical-probes/PFI-7). PFI-7 binds GID4 with a Kd of 80 nM as determined by surface plasmon resonance, an IC50 of 4.1 μM in an FP competition assay, and an EC50 of 600 nM in a NanoBRET cellular target engagement assay. Furthermore, the PFI-7 binding mode has been determined by X-ray crystallography (PDB: 7SLZ).21 These performance measurements are superior to the fragment-derived molecules 16 and 67 (Kd of 16 = 110 μM and Kd of 67 = 17 μM) but similar to the DEL-derived binder 88 (Kd = 5.6 μM, IC50 = 5.4 μM, and EC50 = 558 nM), whose potency is likely 2-fold underrepresented due to the presence of two enantiomers in the racemic mixture used in our determinations. We note that the cellular EC50 values observed for both 88 and PFI-7 are lower than expected considering their respective potencies in vitro (i.e., IC50 values in FP competition assays). We speculate that this difference may be due to enhanced binding of 88 and PFI-7 to full-length GID4 in cell lysates compared to the isolated ligand binding domain of GID4 used in FP competition experiments.
Superimposition of the PFI-7-GID4 complex structure (PDB: 7SLZ) with the small molecule co-structures reported here reveals overlaps between PFI-7 and the lead NMR-derived fragments 16 and 67 (Figure S13A) and between PFI-7 and the BB3 to BB2 portions of the DEL molecules 88, 89, and 91 (Figure S13B). Notably, all binding events involve a hydrogen bond between an amino group in the small molecules and the Glu237 side chain in GID4 and overlapping hydrophobic interactions. Compared to fragment molecules 67 and 16, PFI-7 is ∼50% larger in size and has more extensive contacts with GID4. Common binding features of 16, 67, and PFI-7 include overlaps between the benzene rings of the fragments and the benzimidazole moiety of PFI-7, which make similar hydrophobic contacts with GID4. Another common feature of 16 and PFI-7 includes a second hydrogen-bonding interaction with Glu237 (in addition to that shared by all GID4 binders) involving the exocyclic nitrogen of 16 and the benzimidazole moiety of PFI-7. In contrast to the lead fragments, PFI-7 contains a cyclohexane and 2-indole moiety with no functional overlap with 16 or 67. These groups form hydrophobic and hydrophilic interactions with GID4 that may be partially responsible for the added potency of PFI-7 over 67 and 16.
Compared to the DEL-derived molecules 88, 89, and 91, PFI-7 forms unique hydrogen bonds with GID4 that likely account for its somewhat more efficient binding mode on a per mass basis (i.e., comparable potency with ∼30% less mass). The 2-indole moiety of PFI-7 resides in the same cavity as the BB2 building block of 88, 89, and 91 but forms a unique hydrogen bond with Glu237 of GID4. This hydrogen bond is likely strengthened by the cyclohexane moiety of PFI-7 that appears to restrain the 2-indole group orientation. Moreover, the benzimidazole group of PFI-7, which overlaps with the BB3 building blocks of 88, 89, and 91, also forms hydrogen bonds with GID4. Unique contacts exploited by 88, 89, and 91 but not PFI-7 are mediated by the BB1 building block and amine linker (used for DNA attachment in the original screen; Figure 6B), which engage in both hydrogen bonding and hydrophobic interactions with GID4.
Prospects for Further GID4 Binder Optimization
The majority of PROTACs reported to date exploit either CRBN via thalidomide and its analogs34 or VHL via hydroxyproline derivatives.7 Moieties that bind VHL with comparable binding potencies (IC50 values ranging 2.5 to 16 μM) to those described here for GID4 (IC50 values ranging from 5.4 to 18.9 μM for 88 and 67) can promote protein degradation when incorporated in a PROTAC.23,35,36 These comparable affinities suggest that our current GID4 binders may also be capable of inducing target protein degradation when incorporated in a PROTAC. However, given that most reported PROTACs exploit higher-affinity binders such as thalidomide and VHL032, further optimization of GID4 binders may be warranted before development into PROTACs. Four discernable optimization strategies that may improve the affinity of our small molecule binders to GID4 are fragment merging,37 fragment extension,38 design-based strategies, and further SAR exploration of suboptimal elements. With respect to fragment merging, whereby favorable features of different fragments are combined into a single molecule, one particular possibility is noteworthy. Merging of the 3-aminophenol group within 16 with the 5-amino-tetrahydroisoquinoline motif within 67 would afford a fragment chimera in which both hydrogen bonding and favorable hydrophobic contacts were increased (Figure S13C). With respect to fragment extension, crystal structures of previously reported degron peptides and the DEL screen-derived binders described here uncover fragment extension opportunities such as at the aniline nitrogen in 67 (Figure S13D).
The addition of favorable hydrogen bonding groups may be a particularly useful design strategy to enhance the affinity of the DEL-derived binder, 88. Introduction of hydrogen bonding atoms to the aromatic rings of 88, similar to those observed in PFI-7, is one attractive option. In PFI-7, constraint is introduced through the turn-inducing cis-cyclohexane group that positions constituents in optimal positions for potent interactions with GID4. Within DEL hit 88, introducing constraints that preorganize high-affinity poses such as linking the proximal BB3 resorcinol and BB2 thiophene side changes through the introduction of a macrocyclic structure may increase potency. Finally, with respect to further rounds of SAR analysis, analogs of the BB1 building block position in DEL-derived compounds 88, 89, and 91 may prove fruitful as the hydrophobic R groups display relatively poor complementarity with GID4 and might benefit from SAR exploration. The implementation of the strategies outlined above provides potential points of departure to improve the affinity of the small molecule binders to GID4 presented here.
Conclusions
In this study, we successfully applied both NMR fragment and DEL screening approaches to identify new small molecule ligands for GID4. The NMR fragment screen yielded three lower-affinity binders with estimated Kd values in the mM range. Modest structure-guided optimization of these hits yielded two GID4 binders with Kd values of 110 and 17 μM. Attractive features of these optimized binders include their small size (<250 Da) and non-peptide-like structures. The DEL screening approach yielded molecules with better apparent binding affinity to GID4. In particular, our best binder, 88, which had a Kd of 5.6 μM in vitro, was able to bind GID4 in cells and exhibited binding selectivity in cell lysates. Attractive features of the DEL screening approach included the embedded SAR information in an extensive hit list, overall lower cost, and rapid turnaround time afforded by the WuXi AppTec DELopen program. In addition, by virtue of the DEL library design, a linker attachment site for future PROTAC design can be inferred from the DNA barcode attachment site. Improvement of the drug-like properties of DEL hits may be achieved in part through machine-learning-based extraction of more subtly embedded SAR information from DEL profiles.39 The small molecule binders to GID4 resulting from this study provide a point of departure for further optimization of E3 binding moieties that may allow the CTLH E3 ligase complex to be co-opted for PROTAC design.
Experimental Procedures
Cell Culture
Generation of stable HEK293 cells integrated with N-terminally tagged 3X-FLAG-GID4 was performed using HEK293 Flp-In T-REx cells. GID4 ORF with a stop codon was cloned into the Gateway entry pDONR223 and then transferred to the pDEST-pcDNA5-3X-FLAG vector for expression. When cells reached 70% confluence, recombinant protein expression was induced with 1 μg/mL tetracycline for 16 h. The monoclonal HEK293T reporter cell line used for the induced proximity degradation assay was generated expressing ABI1-EGFP-IRES-TagBFP (blasticidin, 6 μg/mL). Single cells were sorted and expanded, and a clone showing high EGFP and TagBFP expression was selected for subsequent experiments. Cells were grown in high-glucose DMEM supplemented with 10% FBS at 37 °C and 5% CO2.
Induced Proximity Degradation Assay
Degradation assays were performed in a 48-well cell culture format by transient transfection of the clonal EGFP reporter cell line stably co-expressing ABI-GFP with 15 ng of transfection control plasmid expressing triple FLAG-tagged DsRed and 200 ng of the effector protein fused to vhhGFP using Lipofectamine 2000 (Life Technologies). Forty-eight hours post-transfection, cells were washed in PBS, treated with a cell dissociation buffer (1 mM EDTA, 150 mM NaCl, 5 mM NaHCO3, 0.1% glucose, 10 mM KCl), and resuspended in a fluorescence activated cell sorting (FACS) sorting buffer (1× PBS, 5 mM EDTA, 25 mM HEPES pH 7.0, 1% BSA). Cells were spun down in a microcentrifuge at 1000 rpm for 5 min. Cell pellets were resuspended in the FACS sorting buffer and analyzed using BD LSR Fortessa X20 (BD Biosciences; University of Toronto Faculty of Medicine Flow Cytometry Facility).
Cloning, Protein Expression, and Purification
The pET28-MHL plasmid containing GID4124–289 expression was a gift from Jinrong Min (Structural Genomics Consortium, Toronto). The Q282A mutation in GID4124–289 was generated using site-directed mutagenesis. Wild type and the Q282A mutant of GID124–289 were expressed in Escherichia coli BL21 (DE3) in terrific broth media for use in differential scanning fluorimetry, fluorescence polarization competition, ITC assays, X-ray crystallography, and DEL screening and in minimal media containing 15N for NMR screening. The protein was purified as previously described;16 briefly, the protein was purified by nickel chelate affinity chromatography and size-exclusion chromatography, yielding >95% pure protein as assessed by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE). Protein identity was confirmed by mass spectrometry. PGLWKSC and PGLWKS peptides and PGLWKS C-terminally conjugated to FITC (PGLWKS-FITC) were synthesized by Bio Basic and confirmed to be >98% pure (Markham, Ontario, Canada).
NMR Spectroscopy
NMR-heteronuclear single quantum coherence spectroscopy (HSQC) spectra were recorded at 30 °C on a 600 MHz Bruker AVANCE III spectrometer. The 600 MHz spectrometer was equipped with a 1.7 mm TCI CryoProbe. NMR samples of 40 μL contained 200 μM of 15N-GID4 124–289 in 20 mM HEPES pH 7.0, 100 mM NaCl, 0.5 mM TCEP, and 10% D2O. As a positive control, an HSQC spectrum was collected in the presence of 1 mM of peptide PGLWKSC. For the fragment screen, HSQC spectra of 200 pools, each containing five different compounds at final concentrations of 1 mM in 2.5% DMSO, were collected. A control sample containing no compound in 2.5% DMSO was acquired every 40 tubes to monitor GID4 stability over time. Deconvolution of eight pools required 40 additional NMR experiments where a unique fragment at a final concentration of 5 mM in 2.5% DMSO was tested in the same condition as for the screen. Data processing was conducted using NMRviewJ and NMRpipe,40 and NMR spectra were analyzed using Analysis.41 (1H, 15N) Chemical shift perturbations were calculated as a weighted average Δδav = [(ΔδH)2 + (ΔδN × 0.15)2]1/2.
Crystallography
Crystallization trials were conducted with 4 mg/mL of GID4124–289 and 5 mM of DMSO-solubilized 1, 4, 7, 16, or 67, or 1.25 mM of 88, 89, or 91 incubated in 20 mM Tris pH 7.5, 100 mM NaCl, and 0.25 mM TCEP with a final DMSO concentration of 2.5%. Sparse-matrix crystallization conditions were initially screened in sitting drop format at a 1:1 protein/mother liquor ratio. Crystallization conditions were optimized and harvested from hanging drops unless specified otherwise. Each GID4124–289-compound complex crystallized in different conditions. Fragment 1 was co-crystallized in 17% PEG3350, 0.1 M Bis–Tris pH 6.5, and 0.2 M ammonium acetate. Fragment 4 was co-crystallized in 15% PEG5000 and 0.1 M Bis–Tris pH 6.5. Fragment 7 was co-crystallized in 14% PEG3350, 0.1 M Bis–Tris pH 6.0, and 0.2 M ammonium formate. Fragment 16 was co-crystallized in 16% PEG3350, 0.1 M Bis–Tris pH 6.0, and 0.2 M ammonium formate. Fragment 67 was co-crystallized in 25% PEG3350, 0.1 M Bis–Tris pH 6.0, and 0.2 M ammonium acetate. Compound 88 was co-crystallized in 18% PEG6000 and 0.1 M HEPES pH 6.5. Compound 89 was co-crystallized in sitting drop format in 20% PEG6000, 0.1 M HEPES pH 7.0, and 0.2 M calcium chloride hexahydrate. Compound 91 was co-crystallized in sitting drop format in 20% PEG3350 and 0.2 M sodium fluoride. For cryoprotection, crystals were soaked in a well solution containing 30% glycerol. Diffraction data were collected on beamline NE-CAT (APS, Chicago, IL) and processed with XDS.42 Molecular replacement was performed using Phaser43 with the structure of GID4116–300 (PDB: 6CCR) as a search model. Refinement was performed using PHENIX,44 and model building was done in Coot.45 X-ray data collection and refinement statistics are shown in Table S1.
Analog Search and In Silico Docking
Purchasable analogs of the fragment hits were searched using the Structure Search feature from the Molport website (https://www.molport.com/shop/index) based on 50% structural similarity for 7 and 4 and 90% for 1. The compounds from search results of 4 and 1 were prepared using Ligprep (Schrodinger, NY), while GID4 from each co-structure (PDB: 7U3E and 7U3F) was prepared using PrepWizard (Schrodinger, NY) for bond order assignment, assessment of correct protonation states, and optimization of hydrogen bond assignment at pH 7.3. Restrained minimization was conducted using the OPLS3e force field. Receptor grids were calculated using the co-crystallized fragment as the center for each respective complex. The respective analog list for each GID4-fragment complex was docked using Glide24−26 (Schrodinger, NY) with a hydrogen bond formed between the NH of the fragment and the OH of the Glu237 side chain. Docking results were visually inspected and ordered by free energy of binding (MM-GBSA scoring).
Differential Scanning Fluorimetry
A real-time PCR instrument (LightCycler 480 II, Roche) was used to monitor protein unfolding via SYPRO orange fluorescent dye (Invitrogen, Carlsbad, CA). Experiments were performed with 55 μM of GID4124–289 and 4 mM of ligand in 20 mM Tris, 100 mM NaCl, 0.25 μM TCEP pH 7.5, 5× SYPRO orange, and 2.5% DMSO in a final volume of 20 μL. Samples were subjected to a temperature range of 20 to 85 °C with a ramp rate of 0.01 °C/s. Melting temperature values were deduced by identifying the minima of the first derivative of each curve in GraphPad Prism, along with standard error calculation.
Fluorescence Polarization (FP) Binding and Competition Assays
Binding experiments were performed with 20 nM PGLWKS-FITC and increasing concentrations of wild-type or Q282A mutant GID4124–289 proteins in a buffer containing 20 mM Tris pH 7.5, 100 mM NaCl, 1 mM DTT, 0.01% Brij-35, and 0.1 mg/mL BSA. Competitive displacement assays were performed in the same buffer using 7 μM GID4124–289 in complex with 20 nM PGLWKS-FITC and increasing concentrations of competitor peptide/compound in 2.5% DMSO. Samples were incubated for 30 min in 384-well plates (Corning, 3573), and fluorescence intensities were measured using a BioTek Synergy Neo plate reader with excitation and absorbance at 485/528 nm, respectively. Fluorescence polarization was calculated with the Gen5 Data Analysis Software. Graphs and IC50 values were generated using a four-parameter dose–response function in GraphPad Prism.
Isothermal Titration Calorimetry
Isothermal titration calorimetry experiments were performed at 23 °C using a MicroCal iTC200 (GE Healthcare). GID4124–289 (50–200 μM) or GID4Q282A (50 μM) was loaded into the sample cell, and 0.75–1.8 mM of the compound was loaded into the syringe in a solution containing 20 mM Tris, 100 mM NaCl, 0.5 mM TCEP pH 7.5, and 2.5% DMSO. To account for heat of dilution, a reference set was collected by titrating the compound into the buffer. Injections were carried out by serial injection of the experimental compound; for compounds 16 and 67, one injection of 0.5 μL was followed by 38 injections of 1 μL, at 150 s intervals, with data from the first two injections excluded due to pre-equilibration mixing; for compound 88, one injection of 1 μL was followed by 18 injections of 2 μL and one injection of 1.9 μL, at 180 s intervals, with data from the first injection excluded. All experiments were completed in duplicate. Data collection, analysis, and plotting were performed using the Origin 7 software. Peak areas were integrated, subtracted from the reference set, and then fitted by nonlinear regression using the one-site model. Binding isotherms provided the equilibrium association or binding constant (Ka), the change in enthalpy (ΔH), and the stoichiometry of binding (N).
Cellular Thermal Shift Assay (CETSA) in Lysates
Assays were conducted as previously described.46 FLAG-GID4 expressing HEK293 cells were washed in ice-cold PBS, pelleted by centrifugation (400g, 2 min), and frozen at −80 °C. Cell pellets were resuspended in CETSA lysis buffer (1:10 w/v in PBS will full protease inhibitor cocktail (Sigma Aldrich P8340) at a final concentration of 1:500). After freeze-thawing three times, the lysate was clarified by centrifugation at 100,000g for 20 min at 4 °C. Protein concentration (measured with the Pierce BCA assay) was adjusted to 2000 μg/mL, and lysates were flash-frozen and stored at −80 °C. Lysates were incubated with 50 μM compound (in 1% DMSO) for 10 min at room temperature and divided into 10 × 50 μL aliquots, which were each exposed to a different temperature for 3 min using a thermocycler (Veriti 96-Well Fast Thermal Cycler). The samples were then centrifuged at 100,000g for 20 min at 4 °C, and the soluble fractions were analyzed by SDS-PAGE gel and western blot. Mouse anti-FLAG antibody (AbCam, 18230) and mouse anti-SOD1 antibody (used as a thermally stable loading control; AbCam, 51254) were used at 1:3000 in 5% milk and TBST. Secondary antibody (Abcam, 7068) was used in a 1:15,000 dilution. Band intensities were quantified using ImageJ, and melting curves were built following a sigmoidal model in GraphPad Prism where each value was normalized relative to that at the lowest experimental temperature, which was given a value of 1.
Isothermal Dose–Response Fingerprinting Cellular Thermal Shift Assay (ITDRF-CETSA)
Assays were performed using the lysis and incubation protocol described for CETSA, with incubation at room temperature with the indicated compound concentrations at final DMSO concentrations of 1% prior to incubation at the single partial denaturation temperature of 57 °C, centrifugation, and western blot analysis. Band intensities were quantified using the ImageJ software, subtracted from the vehicle control, and modeled using a four-parameter dose–response function in GraphPad Prism.46
Live-Cell CETSA
FLAG-GID4 expressing HEK293 cells were grown in three 15 cm plates to 80% confluence before 1 h treatment with 50 μM 88 or 1% DMSO as the vehicle control. Cells were harvested as described above for CETSA, resuspended in 500 μL of the CETSA lysis buffer, and divided into 10 × 50 μL aliquots. Each aliquot was exposed to a different temperature for 3 min. Lysates were spiked with 1.6% NP-40 in lysis buffer47 and then processed and analyzed as described for CETSA.
Thermal Proteome Profiling
HEK293 Flp-In T-REx cells were lysed following the lysis protocol for CETSA and treated for 10 min at room temperature with 20 μM 88 or 1% DMSO as the vehicle control. Lysates were divided into 8 × 100 μL aliquots and each subjected to a different temperature for 3 min. Heat-treated samples were allowed to equilibrate at room temperature for 5 min and centrifuged at 100,000g for 20 min. Twenty-five microliters from each sample was digested overnight at 37 °C as previously described.48 The digests were concentrated using a Labconco Centrivap Concentrator and resuspended in 50% acetonitrile and 0.1% formic acid. The concentration of digests was measured using the Pierce Colorimetric Peptide Assay to determine the volume required to achieve 10 μg of peptide in the 37 °C-treated sample. The same volume was taken from the remaining samples for TMT labeling as previously described.49 Mixed TMT-labeled peptides were desalted using a Pierce peptide desalting kit and fractionated using high-pH offline fractionation as previously described.50 The resulting 60 fractions were concatenated into 20 fractions by combining multiple early, middle, and late fractions (i.e., fractions N, N + 20, N + 40) as previously described,51 dried, and resuspended in 13 μL of 5% formic acid. A second replicate of samples was created from new cell pellets from different days. All samples were analyzed using an Eksigent nanoLC 425 (Sciex) and an Orbitrap Fusion Lumos Tribrid mass spectrometer (ThermoFisher) using a 60 min gradient. Five microliters of each sample was loaded onto a packed tip C18 column made in-house (15 cm × 75 μm; Reprosil-Pur 120 C18-AQ, 3 μm (Dr. Maisch)) at 400 nL/min using the autosampler. Peptides were separated at 200 nL/min over 60 min with a linear gradient starting at 2.5% acetonitrile in 0.1% formic acid and ending at 35% acetonitrile in 0.1% formic acid. Following the separation gradient, acetonitrile concentration was increased to 80% over 10 min and then returned to starting conditions for 27 min to recondition the column. The Orbitrap Fusion Lumos mass spectrometer was operated with the Xcalibur 4.3 software in data-dependent acquisition mode with HCD and a 3 s cycle time. MS1 scans were performed with 120k resolution for 400–2000 m/z and MS2 scans at 50k resolution, isolation width of 0.7 m/z, and 32% collision energy, and AGC was set to 2e5 with a 40 ms maximum scan time. Precursors with charge states of 2–5 were selected for MS/MS with a dynamic exclusion time of 12 s and acquired in profile mode. Data were processed using MaxQuant (v 1.6.2.10)52 with a 1% FDR (PSM and protein) digested by trypsin with a maximum of two missed cleavages allowing for protein N-terminal acetylation, methionine oxidation, and a fixed modification of cysteine carbamidomethyl. TMTpro isobaric labels were selected with a reporter mass tolerance of 0.003 Da. The minimum peptide length was set to 7 and the maximum to 25 with a search tolerance of 20 ppm for MS/MS. Uniprot UP000005640 (20,598 genes and 80,027 proteins) was used for FASTA search file. Decoys were made from reverse sequences. Quantified proteins were analyzed using the TPP package in R.53 Data were normalized using the nonmelting protein SOD1 (superoxide dismutase; Uniprot SODC_HUMAN). Because a small 1 °C difference was identified between the median melting temperature of DMSO-treated samples and 88-treated samples, stringent statistical cutoffs were used. To be significant, hits were first identified as proteins with an adjusted p value below 0.05. The Tm difference between both DMSO replicates in either replicate was less than the Tm difference between DMSO and 88. Moreover, the sign of the Tm shift was the same for both replicates. Finally, the minimum slope was set to 0.06. Melting curves and thermal shifts were calculated as previously described.33 Poor quality denaturation curves were excluded from analysis by excluding proteins with denaturation curves with plateaus above 0.3 or an R2 value below 0.8. In total, 5663 proteins were quantified between both replicates. Approximately 80% of total proteins passed the minimum curve quality thresholds. Raw data can be accessed in MassIVE using the accession MSV000089890 freely accessible for reviewers with password CTLHGID4.
Chemistry
All solvents and commercially available reagents were used as obtained. All reactions involving air- or moisture-sensitive compounds were performed under a nitrogen atmosphere using oven- or flame-dried glassware. NMR analysis was performed on 400 MHz Bruker Avance III NMR and 500 MHz Agilent DD2 NMR spectrometers. Spectra were measured at 298 K unless indicated otherwise and were referenced relative to the solvent chemical shift. NMR chemical shifts are expressed in ppm, and coupling constants (J) are expressed in Hz. Data were reported as follows: (s) singlet, (d) doublet, (t) triplet, (q) quartet, (m) multiplet, and (br) broad. Chromatography purification of 41–71 was performed using an Isolera One system and prepacked RediSep Rf Normal-phase Silica (60 Å mesh) Flash Cartridges or RediSep Rf C18 Reverse-phase (60 Å mesh) Flash Cartridges. Purification of 88–92 by silica gel chromatography was conducted using Biotage automated MPLC systems, and purification by reverse-phase preparative HPLC was conducted using Gilson or Shimadzu systems. Purity for all tested compounds was determined through high-performance liquid chromatography, and all compounds were >95% pure by HPLC analysis.
Synthesis of Thienopyridines
General Procedure A
4-Phenyl-4,5,6,7-tetrahydrothieno[3,2-c]pyridine (43)
2-(2-Thienyl)-ethylamine (100 mg, 0.79 mmol), benzaldehyde (135 mg, 1.27 mmol), triethylamine (0.05 mL), and EtOH (1 mL) were mixed at room temperature. The reaction mixture was stirred at room temperature for 15 h and then evaporated in vacuo to obtain the crude oil. The crude oil was added to TFA (3.8 mL) at once (exothermic reaction). The reaction mixture was stirred at room temperature for 24–72 h and then evaporated in vacuo. The crude oil was suspended in dichloromethane and washed with 2 N NaOH. The aqueous phase was extracted with dichloromethane (three times). The organic phases were combined, dried with MgSO4, evaporated in vacuo with silica gel, and purified with Hex/EtOAc to afford corresponding 102 mg (60%) of 43 as colorless prisms. 1H NMR (400 MHz, CDCl3) δ ppm 7.33–7.25 (m, 5H), 7.01 (d, J = 5.1 Hz, 1H), 6.45 (d, J = 5.0 Hz, 1H), 5.01 (s, 1H), 3.33–3.28 (m, 1H), 3.13–3.08 (m, 1H), 3.01–2.96 (m, 1H), 2.87–2.82 (m, 1H). 13C NMR (101 MHz, CDCl3) δ ppm 143.9, 136.9, 134.5, 128.4, 128.5, 127.5, 126.5, 121.8, 60.3, 42.6, 26.1. MS (ESI) 216.2 (M + H).
General Procedure B
4,5,6,7-Tetrahydrothieno[3,2-c]pyridine (41)
The procedure described for compound 43 (general procedure A) with 2-(2-thienyl)-ethylamine (150 mg, 0.84 mmol), formaldehyde (1 mL, 1.7 mmol), triethylamine (0.1 mL), EtOH (2 mL), and TFA (6 mL) afforded 41 (19 mg, 16% yield) as a yellow oil. 1H NMR (400 MHz, CDCl3) δ ppm 7.12 (d, J = 5.11 Hz, 1H), 6.78 (d, J = 5.11 Hz, 1H), 3.93–4.07 (m, 2H), 3.24 (t, J = 5.67 Hz, 2H), 2.89 (t, J = 5.67 Hz, 2H), 2.82 (br. s., 2H).13C NMR (101 MHz, CDCl3) δ ppm 134.2, 133.4, 125.0, 122.0, 45.9, 43.6, 26.0. MS (ESI) 140.3 (M + H).
4-(Thiophen-3-yl)-4,5,6,7-tetrahydrothieno[3,2-c]pyridine (42)
The procedure described for compound 43 (general procedure A) with 2-(2-thienyl)-ethylamine (100 mg, 0.79 mmol), thiophene-3-carbaldehyde (88 mg, 0.79 mmol), triethylamine (0.05 mL), EtOH (1 mL), and TFA (4 mL) afforded 42 (97 mg, 55% yield). 1H NMR (500 MHz, CDCl3) δ ppm 7.27–7.35 (m, 1H), 7.11 (d, J = 2.89 Hz, 1H), 7.06 (d, J = 5.11 Hz, 2H), 6.64 (d, J = 5.33 Hz, 1H), 5.19 (s, 1H), 3.29 (td, J = 5.33, 12.44 Hz, 1H), 3.14 (ddd, J = 5.00, 7.28, 12.39 Hz, 1H), 2.82–3.01 (m, 2H), 1.89 (br. s., 1H). 13C NMR (101 MHz, CDCl3) δ ppm 141.8, 136.6, 134.8, 127.4, 126.2, 125.9, 122.6, 121.8, 55.0, 42.0, 26.1 MS (ESI) 222.2 (M + H).
4-Phenyl-4,5,6,7-tetrahydro-1H-pyrrolo[3,2-c]pyridine (45)
A solution of 2-(1H-pyrrol-2-yl)ethanamine (100 mg, 0.9 mmol) in DCM (2 mL) was treated at room temperature with benzaldehyde (95 mg, 0.9 mmol). The solution was cooled, and glacial acetic acid (109 mg, 1.8 mmol) was slowly added. The reaction mixture was diluted with DCM. Saturated NaHCO3 was added, and the organic phase was separated. The aqueous phase was extracted with DCM (30 mL), evaporated with silica gel, and purified with DCM/ACN to give 45 (105 mg, 59%). 1H NMR (400 MHz, DMSO-d6) δ ppm 2.68 (dt, J = 15.28, 3.92 Hz, 1 H) 2.76–2.92 (m, 1H) 2.99–3.14 (m, 1H) 3.18–3.33 (m, 1H) 5.16 (s, 1H) 5.49 (t, J = 2.44 Hz, 1H) 6.59 (t, J = 2.67 Hz, 1H) 7.25–7.47 (m, 5H) 10.71 (br. s., 1H). 13C NMR (101 MHz, DMSO-d6) δ ppm 141.6, 128.6, 128.1, 127.7, 124.0, 116.4, 115.6, 104.9, 57.4, 41.4, 21.7 MS (ESI) 199.2 (M + H).
4-(4,5,6,7-Tetrahydrothieno[3,2-c]pyridin-4-yl)phenyl Acetate (46)
The procedure described for compound 43 (general procedure A) with 2-(2-thienyl)-ethylamine (100 mg, 0.79 mmol), 4-formylphenyl acetate (129 mg, 0.78 mmol), triethylamine (0.05 mL), EtOH (1 mL), and TFA (4 mL) afforded 46 (141 mg, 69% yield). 1H NMR (400 MHz, CDCl3) δ ppm 7.31 (d, J = 8.44 Hz, 2H), 6.97–7.11 (m, 2H), 6.60–6.66 (m, 1H), 6.44–6.54 (m, 1H), 5.11 (s, 1H), 3.31 (td, J = 4.92, 12.39 Hz, 1H), 2.81–3.20 (m, 3H), 2.31 (s, 3H), 1.88 (s, 1H).13C NMR (101 MHz, CDCl3) δ ppm 169.5, 149.6, 140.9, 136.2, 135.1, 129.6, 126.3, 122.0, 121.6, 59.3, 42.2, 25.8, 21.2. MS (ESI) 274.2 (M + H).
4-(4-(Trifluoromethyl)phenyl)-4,5,6,7-tetrahydrothieno[3,2-c]pyridine (47)
The procedure described for compound 43 (general procedure A) with 2-(2-thienyl)-ethylamine (100 mg, 0.79 mmol), 4-(trifluoromethyl)benzaldehyde (137 mg, 0.79 mmol), triethylamine (0.05 mL), EtOH (1 mL), and TFA (4 mL) afforded 47 (123 mg, 55% yield). 1H NMR (400 MHz, CDCl3) δ ppm 7.61 (m, J = 8.22 Hz, 2H), 7.45 (m, J = 8.00 Hz, 2H), 7.06 (d, J = 5.11 Hz, 1H), 6.46 (d, J = 5.11 Hz, 1H), 5.12 (s, 1H), 3.32 (dt, J = 12.22, 4.78 Hz, 1H), 3.22–3.10 (m, 1H), 3.08–2.96 (m, 1H), 2.94–2.84 (m, 1H). 19F NMR (377 MHz, CDCl3) δ ppm −62.44 (s, 3F). 13C NMR (101 MHz, CDCl3) δ ppm 135.8, 135.3, 129.9, 129.6, 128.7, 125.9, 122.2, 59.6, 42.4, 26.0. MS (ESI) 284.1 (M + H).
4-(4-Bromophenyl)-4,5,6,7-tetrahydrothieno[3,2-c]pyridine (49)
The procedure described for compound 43 (general procedure A) with 2-(2-thienyl)-ethylamine (100 mg, 0.79 mmol), 4-bromobenzaldehyde (146 mg, 0.94 mmol), triethylamine (0.05 mL), EtOH (1 mL), and TFA (4 mL) afforded 49 (101 mg, 52% yield). 1H NMR (400 MHz, CDCl3) δ ppm 7.44–7.53 (m, 2H), 7.15–7.26 (m, 2H), 7.04 (d, J = 5.11 Hz, 1H), 6.47 (d, J = 5.33 Hz, 1H), 5.01 (s, 1H), 3.31 (td, J = 4.78, 12.22 Hz, 1H), 3.08–3.20 (m, 1H), 2.93–3.07 (m, 1H), 2.82–2.91 (m, 1H), 1.89 (s, 1H). 13C NMR (101 MHz, DMSO-d6) δ ppm 142.7, 141.3, 137.6, 134.4, 127.8, 126.2, 126.2, 122.0, 62.7, 59.0, 42.0, 25.1. MS (ESI) 294.0 (M + H).
4-(4-Fluorophenyl)-4,5,6,7-tetrahydrothieno[3,2-c]pyridine (51)
The procedure described for compound 43 (general procedure A) with 2-(2-thienyl)-ethylamine (100 mg, 0.79 mmol), 4-fluorobenzaldehyde (98 mg, 0.79 mmol), triethylamine (0.05 mL), EtOH (1 mL), and TFA (4 mL) afforded 51 (125 mg, 67% yield) as a white solid. 1H NMR (400 MHz, CDCl3) δ ppm 7.22–7.33 (m, 2H), 6.98–7.08 (m, 3H), 6.47 (d, J = 5.11 Hz, 1H), 5.04 (s, 1H), 3.26–3.41 (m, 1H), 3.10–3.21 (m, 1H), 2.95–3.07 (m, 1H), 2.81–2.94 (m, 1H), 1.89 (br. s., 1H).19F NMR (377 MHz, CDCl3) δ ppm −115.11 (s, 1F).13C NMR (101 MHz, CDCl3) δ ppm 163.0, 161.0, 139.6, 136.8, 135.1, 130.3, 126.2, 121.9, 115.3, 59.4, 43.6, 26.7. MS (ESI) 234.1 (M + H).
4-(4,5,6,7-Tetrahydrothieno[3,2-c]pyridin-4-yl)aniline (52)
4-(4-Nitrophenyl)-4,5,6,7-tetrahydrothieno[3,2-c]pyridine (100 mg, 0.66 mmol) was dissolved in EtOH (1.5 mL). HCl (20%, 0.2 mL) and Fe (40 mg, 0.71 mmol) were added to the mixture at room temperature. The resulting reaction mixture was stirred at 95 °C overnight. The mixture was diluted with CHzClz (20 mL) and washed with saturated NaHCO3 aq (10 mL). The organic layer was washed with brine (10 mL), dried over MgSO4, filtered, concentrated, evaporated in vacuo with silica gel, and purified with DCM/MeOH to afford 52 (102 mg, 60%) as an off-white solid. 1H NMR (400 MHz, DMSO-d6) δ ppm 7.23 (d, J = 5.11 Hz, 1H), 6.88–7.00 (m, 2H), 6.49–6.59 (m, 2H), 6.42 (d, J = 5.11 Hz, 1H), 4.97 (s, 1H), 4.37 (br. s., 2H), 3.15–3.27 (m, 1H), 2.90–3.09 (m, 2H), 2.76–2.88 (m, 1H). 13C NMR (101 MHz, DMSO-d6) δ ppm 148.3, 136.5, 133.5, 129.2, 128.4, 126.2, 122.4, 113.5, 58.4, 41.3, 24.3. MS (ESI) 231.2 (M + H).
(4-(4,5,6,7-Tetrahydrothieno[3,2-c]pyridin-4-yl)phenyl)methanol (53)
The procedure described for compound 43 (general procedure A) with 2-(2-thienyl)-ethylamine (100 mg, 0.79 mmol), 4-(hydroxymethyl)benzaldehyde (107 mg, 0.94 mmol), triethylamine (0.05 mL), EtOH (1 mL), and TFA (4 mL) afforded 53 (101 mg, 52% yield). 1H NMR (400 MHz, DMSO-d6) δ ppm 7.21–7.31 (m, 4H), 7.17 (d, J = 5.11 Hz, 1H), 6.39 (d, J = 5.33 Hz, 1H), 5.14 (br. s., 1H), 4.90 (s, 1H), 4.49 (s, 2H), 3.15 (td, J = 4.36, 11.28 Hz, 1H), 2.68–2.99 (m, 4H). 13C NMR (101 MHz, DMSO-d6) δ ppm 142.7, 141.3, 137.6, 134.4, 127.8, 126.2, 126.2, 122.0, 62.7, 59.0, 42.0, 25.1. MS (ESI) 246.4 (M + H).
4-(p-Tolyl)-4,5,6,7-tetrahydrothieno[3,2-c]pyridine (54)
The procedure described for compound 43 (general procedure A) with 2-(2-thienyl)-ethylamine (100 mg, 0.79 mmol), 4-methylbenzaldehyde (96 mg, 0.8 mmol), triethylamine (0.05 mL), EtOH (1 mL), and TFA (4 mL) afforded 54 (51 mg, 28% yield). 1H NMR (400 MHz, CDCl3) δ ppm 7.14–7.25 (m, 4H), 7.06 (d, J = 5.11 Hz, 1H), 6.48 (d, J = 5.13 Hz, 1H), 4.98 (s, 1H), 3.19–3.10 (m, 1H), 2.98–3.07 (m, 1H), 2.89–2.96 (m, 1H), 2.36–2.42 (m, 1H). 13C NMR (101 MHz, CDCl3) δ ppm 136.1, 135.3, 131.9, 129.6, 127.5, 126.7, 125.0, 123.2, 63.6, 42.4, 26.4, 20.3. MS (ESI) 230.1 (M + H).
4-(3-Chlorophenyl)-4,5,6,7-tetrahydrothieno[3,2-c]pyridine (55)
The procedure described for compound 43 (general procedure A) with 2-(2-thienyl)-ethylamine (100 mg, 0.79 mmol), 4-chlorobenzaldehyde (110 mg, 0.79 mmol), triethylamine (0.05 mL), EtOH (1 mL), and TFA (4 mL) afforded 55 (181 mg, 91% yield). 1H NMR (400 MHz, CDCl3) δ ppm 7.27–7.29 (m, 1H), 7.24–7.26 (m, 2H), 7.15–7.19 (m, 1H), 7.03 (d, J = 5.26 Hz, 1H), 6.46 (d, J = 5.26 Hz, 1H), 5.00 (t, J = 1.77 Hz, 1H), 3.23–3.35 (m, 1H), 3.07–3.16 (m, 1H), 2.94–3.03 (m, 1H), 2.80–2.88 (m, 1H), 1.82 (br. s., 1H). 13C NMR (101 MHz, CDCl3) δ ppm 141.7, 136.4, 135.7, 133.7, 130.2, 128.7, 127.7, 126.1, 123.8, 122.0, 59.7, 41.5, 26.0. MS (ESI) 250.0 (M + H).
3-(4,5,6,7-Tetrahydrothieno[3,2-c]pyridin-4-yl)phenol (56)
58 (50 mg, 0.2 mmol) was dissolved in DCM (4 mL). BBr3 in DCM (1 M, 0.6 mL, 0.6 mmol) was added at 0 °C and was stirred overnight at room temperature. NH4OH (1 mL) was added to the reaction mixture until pH 7, and the mixture was diluted with EtOAc. The organic layer was separated and washed with saturated NaHCO3 (10 mL). The organic layer was washed with brine (10 mL), dried over MgSO4, filtered, evaporated in vacuo with silica gel, and purified with DCM/MeOH to afford 56 (21 mg, 45%) as a white solid. 1H NMR (400 MHz, , DMSO-d6) δ ppm 9.26 (br. s., 1H), 7.18 (d, J = 5.11 Hz, 1H), 7.08–7.14 (m, 1H), 6.73 (d, J = 7.56 Hz, 1H), 6.68–6.71 (m, 1H), 6.63–6.67 (m, 1H), 6.42 (d, J = 5.11 Hz, 1H), 4.83 (s, 1H), 3.81 (dd, J = 7.33, 8.67 Hz, 1H), 3.07–3.19 (m, 1H), 2.80–2.97 (m, 2H). 13C NMR (101 MHz, DMSO-d6) δ ppm 157.2, 145.6, 137.5, 134.1, 128.9, 126.2, 125.4, 121.8, 115.0, 114.0, 59.1, 41.9, 25.5. MS (ESI) 232.2 (M + H).
3-(4,5,6,7-Tetrahydrothieno[3,2-c]pyridin-4-yl)aniline (57)
60 (130 mg, 0.86 mmol) was dissolved in EtOH (2 mL). HCl (20%, 0.4 mL) and Fe (65 mg, 1.16 mmol) were added to the mixture at room temperature. The resulting reaction mixture was stirred at 95 °C overnight. The mixture was diluted with CHzClz (30 mL) and washed with saturated NaHCO3 aq (20 mL). The organic layer was washed with brine (20 mL), dried over MgSO4, filtered, concentrated, evaporated in vacuo with silica gel, and purified with DCM/MeOH to afford 57 (104 mg, 52%) as an off-white solid. 1H NMR (400 MHz, DMSO-d6) δ ppm 7.15 (d, J = 5.11 Hz, 1H), 6.96 (t, J = 7.78 Hz, 1H), 6.38–6.58 (m, 4H), 4.98 (br. s., 2H), 4.75 (s, 1H), 3.17 (td, J = 4.36, 11.28 Hz, 1H), 2.62–2.99 (m, 3H).13C NMR (101 MHz, DMSO-d6) δ ppm 148.5, 144.3, 137.9, 134.0, 128.4, 126.3, 121.6, 115.9, 113.6, 112.8, 59.6, 42.2, 25.6. MS (ESI) 231.2 (M + H).
4-(3-Methoxyphenyl)-4,5,6,7-tetrahydrothieno[3,2-c]pyridine (58)
The procedure described for compound 43 (general procedure A) with 2-(2-thienyl)-ethylamine (100 mg, 0.79 mmol), 3-methoxybenzaldehyde (107 mg, 0.79 mmol), triethylamine (0.05 mL), EtOH (1 mL), and TFA (4 mL) afforded 58 (85 mg, 44% yield). 1H NMR (400 MHz, CDCl3) δ ppm 7.23–7.32 (m, 1H), 7.04 (d, J = 5.33 Hz, 1H), 6.82–6.94 (m, 3H), 6.53 (d, J = 5.11 Hz, 1H), 5.04 (s, 1H), 3.81 (s, 3H), 3.35 (td, J = 4.78, 12.22 Hz, 1H), 3.09–3.19 (m, 1H), 3.01 (d, J = 7.78 Hz, 1H), 2.82–2.92 (m, 1H), 1.99 (br. s., 1H).13C NMR (101 MHz, CDCl3) δ ppm 159.8, 145.3, 136.6, 135.0, 129.4, 126.4, 121.8, 120.7, 114.0, 113.0, 60.1, 55.2, 42.5, 26.1. MS (ESI) 246.4 (M + H).
3-(4,5,6,7-Tetrahydrothieno[3,2-c]pyridin-4-yl)benzonitrile (59)
The procedure described for compound 43 (general procedure A) with 2-(2-thienyl)-ethylamine (100 mg, 0.79 mmol), 3-formylbenzonitrile (107 mg, 0.81 mmol), triethylamine (0.05 mL), EtOH (1 mL), and TFA (4 mL) afforded 59 (109 mg, 56% yield). 1H NMR (400 MHz, CDCl3) δ ppm 7.57–7.65 (m, 3H), 7.43–7.50 (m, 1H), 7.08 (d, J = 5.33 Hz, 1H), 6.45 (d, J = 5.11 Hz, 1H), 5.09 (s, 1H), 3.23–3.35 (m, 1H), 3.10–3.20 (m, 1H), 2.96–3.08 (m, 1H), 2.83–2.96 (m, 1H), 1.80 (br. s., 1H). 13C NMR (101 MHz, CDCl3) δ ppm 135.4, 132.9, 132.0, 131.3, 129.3, 125.8, 122.5, 118.8, 59.2, 42.3, 26.0. MS (ESI) 241.3 (M + H).
4-(3-Nitrophenyl)-4,5,6,7-tetrahydrothieno[3,2-c]pyridine (60)
The procedure described for compound 43 (general procedure A) with 2-(2-thienyl)-ethylamine (100 mg, 0.79 mmol), 3-nitrobenzaldehyde (119 mg, 0.79 mmol), triethylamine (0.05 mL), EtOH (1 mL), and TFA (4 mL) afforded 60 (126 mg, 61% yield). 1H NMR (400 MHz, CDCl3) δ ppm 8.10–8.27 (m, 2H), 7.68 (td, J = 1.17, 7.67 Hz, 1H), 7.47–7.57 (m, 1H), 7.08 (d, J = 5.11 Hz, 1H), 6.46 (d, J = 5.33 Hz, 1H), 5.17 (s, 1H), 3.25–3.38 (m, 1H), 3.11–3.24 (m, 1H), 2.97–3.10 (m, 1H), 2.82–2.94 (m, 1H), 1.98 (br. s., 1H). 13C NMR (101 MHz, CDCl3) δ ppm 146.1, 135.7, 135.4, 134.6, 129.4, 125.7, 123.3, 122.7, 122.5, 59.3, 42.5, 26.0. MS (ESI) 261.0 (M + H).
4-(2-Fluorophenyl)-4,5,6,7-tetrahydrothieno[3,2-c]pyridine (61)
The procedure described for compound 43 (general procedure A) with 2-(2-thienyl)-ethylamine (100 mg, 0.79 mmol), 2-fluorobenzaldehyde (98 mg, 0.79 mmol), triethylamine (0.05 mL), EtOH (1 mL), and TFA (4 mL) afforded 61 (111 mg, 60% yield). 1H NMR (400 MHz, CDCl3) δ ppm 7.24–7.35 (m, 1H), 7.03–7.18 (m, 4H), 6.54 (d, J = 5.11 Hz, 1H), 5.48 (s, 1H), 3.24–3.35 (m, 1H), 3.09–3.20 (m, 1H), 2.86–3.05 (m, 2H), 1.97 (br. s., 1H).19F NMR (377 MHz, CDCl3) δ ppm −119.86 (s, 1F). 13C NMR (101 MHz, CDCl3) δ ppm 151.7, 139.8, 135.5, 130.0, 129.1, 126.0, 124.0, 122.0, 115.5, 52.6, 42.0, 26.0. MS (ESI) 234.1 (M + H).
4-(2-Chlorophenyl)-4,5,6,7-tetrahydrothieno[3,2-c]pyridine (62)
The procedure described for compound 43 (general procedure A) with 2-(2-thienyl)-ethylamine (100 mg, 0.79 mmol), 2-chlorobenzaldehyde (110 mg, 0.79 mmol), triethylamine (0.05 mL), EtOH (1 mL), and TFA (4 mL) afforded 62 (158 mg, 81% yield). 1H NMR (400 MHz, CDCl3) δ ppm 7.44 (dd, J = 1.56, 7.56 Hz, 1H), 7.22 (dtd, J = 1.56, 7.42, 18.50 Hz, 2H), 7.06–7.12 (m, 2H), 6.54 (d, J = 5.11 Hz, 1H), 5.59 (s, 1H), 3.19–3.27 (m, 1H), 3.10–3.18 (m, 1H), 2.86–3.06 (m, 2H), 2.04 (br. s., 1H). 13C NMR (101 MHz, CDCl3) δ ppm 140.7, 135.7, 135.4, 133.7, 130.2, 129.7, 128.7, 126.8, 126.2, 122.1, 55.8, 41.6, 26.1. MS (ESI) 250.0 (M + H).
4-(2-Nitrophenyl)-4,5,6,7-tetrahydrothieno[3,2-c]pyridine (64)
The procedure described for compound 43 (general procedure A) with 2-(2-thienyl)-ethylamine (100 mg, 0.79 mmol), 2-nitrobenzaldehyde (107 mg, 0.94 mmol), triethylamine (0.05 mL), EtOH (1 mL), and TFA (4 mL) afforded 64 (123 mg, 60% yield). 1H NMR (400 MHz, CDCl3) δ ppm 7.87 (dd, J = 1.22, 8.11 Hz, 1H), 7.47–7.56 (m, 1H), 7.39–7.46 (m, 1H), 7.22–7.32 (m, 1H), 7.08 (d, J = 5.33 Hz, 1H), 6.49 (d, J = 5.33 Hz, 1H), 5.55 (s, 1H), 3.11–3.28 (m, 2H), 2.97–3.07 (m, 1H), 2.83–2.95 (m, 1H), 2.38 (br. s., 1H). 13C NMR (101 MHz, CDCl3) δ ppm 150.0, 137.9, 136.3, 134.9, 132.6, 131.2, 128.3, 126.0, 124.1, 122.4, 54.2, 41.8, 26.0. MS (ESI) 261.3 (M + H).
4-(2-(Trifluoromethyl)phenyl)-4,5,6,7-tetrahydrothieno[3,2-c]pyridine (65)
The procedure described for compound 43 (general procedure A) with 2-(2-thienyl)-ethylamine (100 mg, 0.79 mmol), 2-(trifluoromethyl)benzaldehyde (123 mg, 0.79 mmol), triethylamine (0.05 mL), EtOH (1 mL), and TFA (4 mL) afforded 65 (133 mg, 59% yield). 1H NMR (400 MHz, CDCl3) δ ppm 7.72 (d, J = 8.00 Hz, 1H), 7.46–7.55 (m, 1H), 7.33–7.44 (m, 2H), 7.03 (d, J = 5.11 Hz, 1H), 6.37 (d, J = 5.33 Hz, 1H), 5.50 (s, 1H), 3.33–3.45 (m, 1H), 3.04–3.25 (m, 2H), 2.85–2.97 (m, 1H), 2.14 (br. s., 1H). 19F NMR (377 MHz, CDCl3) δ ppm −57.59 (s, 3F). 13C NMR (101 MHz, CDCl3) δ ppm 142.7, 136.5, 135.3, 132.0, 131.0, 127.4, 126.2, 125.4, 122.2, 55.2, 43.3, 26.0. MS (ESI) 284.1 (M + H).
N-(2-Bromophenethyl)benzamide
To a solution of 2-(2-bromophenyl)ethanamine (1 g, 5 mmol) and TEA (1.5 mL) in DCM (15 mL) at 0 °C was added benzoyl chloride (700 mg, 5 mmol) dropwise. The mixture was stirred at 20 °C overnight. The mixture was diluted with DCM (20 mL) and washed with saturated NaHCO3 aq (20 mL). The organic layer was washed by 1 N HCl (20 mL) and brine (20 mL), dried over MgSO4, filtered, and concentrated in vacuo to give N-(2-bromophenethyl)benzamide (1.45 g, 95% yield) as an off-white solid. MS (ESI) 305.1 (M + H).
5-Bromo-1-phenyl-3,4-dihydroisoquinoline (66-int)
To a solution of N-(2-bromophenethyl)benzamide (470 mg, 1.54 mmol) in xylene (3 mL) was added P2O5 (354 mg, 2.49 mmol) in two portions while stirring. POCl3 (0.7 mL) was added under nitrogen, and the reaction mixture was stirred at 130 °C for 2 days. The mixture was concentrated in vacuo, and the residue was diluted with DCM (30 mL) and quenched with cold saturated NaHCO3 aq (20 mL). The organic layer was dried over MgSO4, filtered, concentrated in vacuo, and purified with DCM/MeOH to give 67-int (240 mg, 54%), which was used for the next step. 1H NMR (400 MHz, CDCl3) δ ppm 7.63–7.67 (m, 1H), 7.56–7.60 (m, 2H), 7.41–7.49 (m, 3H), 7.23–7.27 (m, 1H), 7.12–7.18 (m, 1H), 3.83–3.94 (m, 2H), 2.89–2.99 (m, 2H). 13C NMR (101 MHz, CDCl3) δ ppm 166.9, 143.5, 138.7, 138.4, 134.7, 132.1, 130.8, 130.0, 129.6, 128.8, 128.2, 127.7, 127.2, 123.5, 47.2, 25.8. MS (ESI) 287.2 (M + H).
5-Bromo-1-phenyl-1,2,3,4-tetrahydroisoquinoline (66)
To a solution of 66-int (200 mg, 0.69 mmol) in MeOH (2 mL) at 20 °C was added NaBH4 (100 mg, 2.9 mmol) portionwise. The mixture was stirred at 20 °C for 12 h. The mixture was concentrated in vacuo, and the residue was diluted with EtOAc (30 mL) and washed by 1 N HCl (30 mL). The aqueous layer was separated and basified with saturated N2CO3 aq (25 mL). The mixture was extracted with EtOAc (25 mL × 2). The combined organic layer was dried over MgSO4, filtered, concentrated in vacuo, and purified with DCM/ACN to give 96 (150 mg, 75%) as an off-white solid. 1H NMR (400 MHz, CDCl3) δ ppm 7.50–7.58 (m, 1H), 7.33–7.41 (m, 5H), 7.02 (t, J = 7.89 Hz, 1H), 6.73–6.82 (m, 1H), 5.40 (s, 1H), 3.22–3.39 (m, 1H), 2.97–3.21 (m, 3H). 13C NMR (101 MHz, CDCl3) δ ppm 138.3, 133.2, 131.8, 130.0, 129.1, 128.9, 127.6, 125.3, 60.0, 39.9, 27.7. MS (ESI) 289.4 (M + H).
1-Phenyl-1,2,3,4-tetrahydroisoquinolin-5-amine (67)
96 (100 mg, 0.34 mmol), Pd2(dba)3 (95 mg, 0.1 mmol), and Xantphos (65 mg, 0.1 mmol) were mixed in toluene (2 mL). Benzophenone imine (65 mg, 0.35 mmol) and K3PO4 were added under nitrogen. The reaction mixture was heated at 100 °C overnight. The reaction mixture was cooled, and tetrahydrofuran (3 mL) with HCl (3 N, 3 mL) was added. The reaction mixture was stirred at room temperature for 1 h, and NaOH was added until pH = 7.5. The resulting mixture was extracted with CH2Cl2 (three times). The organic phases were combined, dried with MgSO4, evaporated in vacuo with silica gel, and purified with DCM/MeOH to afford corresponding 10 mg (13%) of 67 as an off-white solid.
1H NMR (400 MHz, CDCl3) δ ppm 7.55–7.68 (m, 2H), 7.39–7.50 (m, 3H), 7.03–7.15 (m, 1H), 6.86 (dd, J = 0.89, 8.00 Hz, 1H), 6.68–6.79 (m, 1H), 3.84–3.97 (m, 2H), 3.74 (br. s., 2H), 2.55–2.70 (m, 2H). 13C NMR (101 MHz, CDCl3) δ ppm 144.8, 144.1, 139.0, 128.7, 127.3, 125.7, 120.5, 118.4, 112.8, 62.1, 42.3, 24.7. MS (ESI) 225.4 (M + H).
5-Methoxy-1-phenyl-1,2,3,4-tetrahydroisoquinoline (68)
The procedure described for compound 43 (general procedure A) with 2-(2-methoxyphenyl)ethanamine (100 mg, 0.66 mmol), benzaldehyde (100 mg, 0.94 mmol), triethylamine (0.05 mL), EtOH (1 mL), and TFA (6 mL) afforded 68 (51 mg, 32% yield). 1H NMR (400 MHz, CDCl3) δ ppm 7.25–7.38 (m, 5H), 7.06 (t, J = 8.00 Hz, 1H), 6.74 (d, J = 8.00 Hz, 1H), 6.42 (d, J = 7.78 Hz, 1H), 5.12 (s, 1H), 3.89 (s, 3H), 3.30 (td, J = 5.28, 12.11 Hz, 1H), 3.04–3.15 (m, 1H), 2.85 (t, J = 5.89 Hz, 2H), 1.92 (br. s., 1H). 13C NMR (101 MHz, CDCl3) δ ppm 157.1, 144.8, 139.7, 129.0, 128.4, 127.3, 125.8, 124.7, 120.3, 107.4, 61.9, 55.3, 41.7, 23.9. MS (ESI) 240.1 (M + H).
1-(4-Methoxyphenyl)-1,2,3,4-tetrahydroisoquinoline (71)
The procedure described for compound 43 (general procedure A) with 2-phenylethanamine (100 mg, 0.66 mmol), 4-methoxybenzaldehyde (100 mg, 0.94 mmol), triethylamine (0.05 mL), EtOH (1 mL), and TFA (6 mL) afforded 71 (71 mg, 45% yield). 1H NMR (400 MHz, CDCl3) δ ppm 7.21–7.24 (m, 2H), 7.18 (d, J = 4.00 Hz, 2H), 7.04–7.11 (m, 1H), 6.86–6.94 (m, 2H), 6.80 (d, J = 7.56 Hz, 1H), 5.10 (s, 1H), 3.84 (s, 3H), 3.27–3.37 (m, 1H), 3.03–3.17 (m, 2H), 2.80–2.92 (m, 1H), 1.90–2.02 (m, 1H). 13C NMR (101 MHz, CDCl3) δ ppm 158.9, 138.7, 137.2, 135.5, 130.0, 129.0, 128.1, 126.2, 125.6, 113.8, 61.5, 55.3, 42.0, 29.8. MS (ESI) 240.1 (M + H).
(S)-2-((S)-2-(2-((2,4-Dimethoxybenzyl)amino)acetamido)-2-(thiophen-2-yl)acetamido)-N-methyl-4-phenylbutanamide (88)
For detailed information about the chemical synthesis of 88, see the Supporting Information. The compound was obtained as a 58:42 mixture of epimers. 1H NMR (CD3OD, 400 MHz) δ 7.43 (dd, J = 16.4, 4.4 Hz, 1H), 7.23–7.26 (m, 1H), 7.09–7.22 (m, 5H), 7.00–7.08 (m, 1H), 6.96 (d, J = 7.2 Hz, 1H), 6.52 (t, J = 2.4 Hz, 1H), 6.44–6.50 (m, 1H), 5.74 (d, J = 10.4 Hz, 1H), 4.13–4.36 (m, 1H), 3.76–3.81 (m, 8H), 3.36–3.43 (m, 2H), 2.71(d, J = 20.8 Hz, 3H), 2.35–2.67(m, 2H), 2.04–2.21 (m, 1H), 1.80–2.03 (m, 1H); m/z [M + H] + = 539.3; HPLC (Shimadzu) tR = 1.94 min (minor epimer), 2.02 min (major epimer); SFC (Waters UPCC) tR = 2.41 min (major epimer), 2.67 (minor epimer).
(2R)-2-[[2-(Butylamino)acetyl]amino]-3-(4-tert-butylphenyl)-N-[(1S)-1-[(3-methoxyphenyl)methyl]-2-(methylamino)-2-oxo-ethyl]propenamide (89)
For detailed information about the chemical synthesis of 89, see the Supporting Information. 1H NMR (CD3OD, 400 MHz) δ 7.28 (d, J = 8.4 Hz, 2H), 7.16–7.21 (m, 1H), 6.98 (d, J = 8.0 Hz, 2H), 6.73–6.80 (m, 3H), 4.52–4.64 (m, 3H), 3.76 (s, 3H), 3.50 (s, 2H), 3.04 (dd, J = 14.0, 5.6 Hz, 1H), 2.71–2.85 (m, 4H), 2.70 (s, 3H), 1.47–1.58 (m, 2H), 1.32–1.40 (m, 2H), 1.27 (s, 9H), 0.94 (t, J = 7.4 Hz, 3H); 13C NMR (DMSO-d6, 126 MHz) δ 171.4, 170.5, 170.1, 163.8, 159.2, 148.3, 139.6, 134.0, 129.1, 128.9, 124.5, 121.6, 114.8, 111.9, 54.9, 54.2, 52.9, 51.4, 48.5, 37.8, 37.3, 34.0, 31.1, 31.0, 25.5, 19.7, 13.9; m/z [M + H] + = 525.6; HPLC (Shimadzu) tR = 2.33 min; SFC (Waters UPCC) tR = 2.33 min.
(2S,3S)-N-[(1S)-2-[[(1S)-1-(Cyclohexylmethyl)-2-(methylamino)-2-oxo-ethyl]amino]-2-oxo-1-(3-thienyl)ethyl]-3-phenyl-azetidine-2-carboxamide (90)
For detailed information about the chemical synthesis of 90, see the Supporting Information. The compound was obtained as a 65:35 mixture of epimers. 1H NMR (CD3OD, 400 MHz): δ 7.43–7.50 (m, 2H), 7.41 (s, 1H), 7.31–7.40 (m, 3H), 7.24–7.30 (m, 1H), 7.13–7.20 (m, 1H), 5.56–5.66 (m, 1H), 4.62 (d, J = 6.0 Hz, 1H), 4.41 (t, J = 8.0 Hz, 1H), 3.92–4.02 (m, 2H), 3.85–3.92 (m, 1H), 2.67 (s, 3H), 1.77 (d, J = 12.4 Hz, 1H), 1.70 (d, J = 10.8 Hz, 4H), 1.57–1.63 (m, 2H), 1.24–1.38 (m, 2H), 1.16–1.22 (m, 2H), 0.85–1.01 (m, 2H); m/z [M + H] + = 483.4; HPLC (Shimadzu) tR = 2.07 min (major epimer), 2.13 min (minor epimer); SFC (Waters UPCC) tR = 2.23 min (minor epimer), 2.52 min (major epimer).
(E,2R)-2-[[2-(Benzylamino)acetyl]amino]-N-[(1S)-2-(methylamino)-2-oxo-1-(p-tolylmethyl)ethyl]-5-phenyl-pent-4-enamide (91)
For detailed information about the chemical synthesis of 91, see the Supporting Information. 1H NMR (DMSO-d6, 400 MHz): δ 8.46–8.48 (d, J = 12.0 Hz, 1H), 8.34 (s, 1H), 7.96–7.98 (m, 1H), 7.27–7.33 (m, 7H), 7.20–7.21 (m, 3H), 7.12–7.19 (m, 2H), 7.04–7.10 (m, 2H), 6.14–6.18 (d, J = 16.0 Hz, 1H), 5.73–5.77 (m, 1H), 4.53–4.54 (m, 1H), 4.40–4.44 (m, 1H), 3.89 (s, 2H), 3.43 (s, 2H), 2.92–2.95 (m, 1H), 2.67–2.68 (m, 1H), 2.57–2.58 (m, 3H), 2.33–2.35 (m, 2H), 2.25–2.32 (m, 1H), 2.16 (s, 3H); m/z [M + H] + = 513; HPLC (Waters) tR = 0.893 min; SFC (Waters UPCC) tR = 4.06 min.
N-Methyl-2-[(2R)-3-(2-methyl-1H-indol-3-yl)-2-[[2-[2-(4-methylphenoxy)ethylamino]acetyl]amino]propanoyl]isoindoline-1-carboxamide (92)
For detailed information about the chemical synthesis of 92, see the Supporting Information. 1H NMR (CD3OD, 400 MHz) δ 7.57–7.62 (m, 1H), 7.35–7.46 (m, 1H), 7.34–7.34 (m, 1H), 7.25–7.30 (m, 1H), 7.07–7.22 (m, 2H), 7.01–7.05 (m, 3H), 6.99 (s, 1H), 6.86–6.93 (m, 1H), 6.74 (d, J = 8.4 Hz, 1H), 5.44 (d, J = 1.2 Hz, 1H), 5.08–5.15 (m, 1H), 4.61–4.68 (m, 1H), 4.07 (t, J = 5.2 Hz, 1H), 3.80–3.91 (m, 2H), 3.47–3.56 (m, 1H), 3.00–3.20 (m, 3H), 2.75–2.88 (m, 2H), 2.70 (s, 2H), 2.64 (s, 1H), 2.41 (s, 2H), 2.32 (s, 1H), 2.25 (s, 1H), 2.15 (s, 2H); m/z [M + H] + = 568.5; HPLC (Shimadzu) tR = 2.17 min; SFC (Waters UPCC) tR = 3.70 min.
Acknowledgments
This work was supported by Canadian Institutes of Health Research grants FDN-143277, PJT-178026, and PJT-180338 to F.S.; FDN-143301 to A.-C.G.; FRN-414829 to P.M.; and FDN-167277 to M.Tyers and was based upon research conducted at the Northeastern Collaborative Access Team beamlines, which are funded by the National Institute of General Medical Sciences from the National Institutes of Health (P30 GM124165). The Eiger 16M detector on 24-ID-E is funded by an NIH-ORIP HEI grant (S10OD021527). This research used resources of the Advanced Photon Source, a U.S. Department of Energy (DOE) Office of Science User Facility operated for the DOE Office of Science by Argonne National Laboratory under Contract No. DE-AC02-06CH11357. Proteomics work was performed at the Network Biology Collaborative Centre at the Lunenfeld-Tanenbaum Research Institute, a facility supported by Canada Foundation for Innovation funding, by the Government of Ontario and by Genome Canada and Ontario Genomics (OGI-139). NMR spectrometers were funded by the Canada Foundation for Innovation, and the NMR Core Facility was supported by the Princess Margaret Cancer Foundation. We thank Cassandra J. Wong, Brett Larsen, and Brendon Seale for their proteomics expertise; Dr. Geneviève Gasmi-Seabrook for technical expertise and access to the NMR Core Facility; Gerald Gish for initial production of degron peptides; and Almer van der Sloot for helpful discussions on GID4-small molecule co-structures. We dedicate this study to Richard Lerner, who was the founding father of DNA-encoding library development and a proponent of open academic access to these resources.
Glossary
Abbreviations
- ΔH
change in enthalpy
- ΔS
change in entropy
- ABI
abscisic acid insensitive
- ACN
acetonitrile
- aq
aqueous
- BSA
bovine serum albumin
- CETSA
cellular thermal shift assay
- Cmpd
compound
- CTLH
C-terminal to LisH
- DCM
dichloromethane
- DEL
DNA-encoded library
- DIPEA
N,N-diisopropylethylamine
- DMAP
4-dimethylaminopyridine
- DMEM
Dulbecco’s modified Eagle medium
- DMF
N,N-dimethylformamide
- DMSO
dimethylsulfoxide
- DSF
differential scanning fluorimetry
- EDC
N-(3-dimethylaminopropyl)-N′-ethylcarbodiimide
- EGFP
enhanced green fluorescent protein
- EtOAc
ethyl acetate
- EtOH
ethanol
- ESI
electrospray ionization
- FACS
fluorescence activated cell sorting
- FDR
false discovery rate
- Fmoc
fluorenylmethyloxycarbonyl
- FP
fluorescence polarization
- GFP
green fluorescent protein
- GID4
glucose-induced degradation protein 4 homolog
- HATU
[dimethylamino(triazolo[4,5-b]pyridin-3-yloxy)methylidene]-dimethylazanium;hexafluorophosphate
- HEPES
4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid
- HOBt
1-hydroxybenzotriazole
- HSQC
heteronuclear single quantum coherence
- ITC
isothermal titration calorimetry
- ITDRF-CETSA
isothermal dose–response fingerprinting cellular thermal shift assay
- Kd
dissociation constant
- MeOH
methanol
- MS
mass spectrometry
- N
stoichiometry
- NHEt2
diethylamine
- NMR
nuclear magnetic resonance
- PBS
phosphate-buffered saline
- PROTAC
proteolysis targeting chimera
- SAR
structure–activity relationship
- SDS-PAGE
sodium dodecyl sulfate polyacrylamide gel electrophoresis
- SEM
standard error of the mean
- SPR
surface plasmon resonance
- Tm
melting temperature
- TCEP
tris(2-carboxyethyl)phosphine
- TEA
triethylamine
- TFA
trifluoroacetic acid
- TIS
triisopropyl silane
- TMT
tandem mass tag
- TPD
targeted protein degradation
- TPP
thermal proteome profiling
- WT
wild type
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acs.jmedchem.2c00509.
Accession Codes
Authors will release the atomic coordinates and experimental data upon article publication for 7U3E (1), 7U3F (4), 7U3H (7), 7U3I (16), 7U3G (67), 7U3J (88), 7U3K (89), and 7U3L (91). Thermal proteome profiling raw data can be accessed in MassIVE using the accession MSV000089890 and password CTLHGID4.
Author Contributions
● P.M., G.P., and N.G.A.G contributed equally.
Author Contributions
The manuscript was written by C.K.C. and F.S., with input from all authors. C.K.C. purified proteins; prepared samples for NMR; and performed analog search, DSF, ITC, FP, crystallography experiment, and the DEL screen. P.M., G.P., and N.G.A.G. contributed equally. P.M. performed NMR data acquisition and analysis. G.P. designed and synthesized analogs. G.P. and S.M.O. performed docking analysis. N.G.A.G. performed CETSA and thermal proteome experiments. J.P. performed proximity induced degradation experiments. M.Tyers, D.J.S.-C., R.A.B., and A.-C.G. edited the manuscript. A.-C.G. and H.R. supervised N.G.A.G. R.A.B. supervised G.P. M.Taipale supervised J.P. D.F.C. prepared the fragment library. C.K.C. and P.M. performed SPR experiments not presented in this study. D.J.S.-C. prepared the chemical methods for the DEL molecules. V.P. provided guidance for ITC and structure determination. I.K. collected the X-ray data. X.T. generated the cell line expressing 3xFLAG-GID4. D.D., W.C., W.S., L.K., and R.S. performed deep sequencing of the DEL screen and synthesized molecules originating from the screen. F.S. advised throughout the project.
The authors declare the following competing financial interest(s): F.S. is a founder and consultant of Repare Therapeutics.
Supplementary Material
References
- ClinicalTrials.gov [Internet]. Bethesda, MD: National Library of Medicine (US). Accessed 2022. Feb 27.
- Mullard A. Targeted Protein Degraders Crowd into the Clinic. Nat. Rev. Drug Discov. 2021, 20, 247–250. 10.1038/d41573-021-00052-4. [DOI] [PubMed] [Google Scholar]
- Tan X.; Calderon-Villalobos L. I. A.; Sharon M.; Zheng C.; Robinson C. V.; Estelle M.; Zheng N. Mechanism of Auxin Perception by the TIR1 Ubiquitin Ligase. Nature 2007, 446, 640–645. 10.1038/nature05731. [DOI] [PubMed] [Google Scholar]
- Sakamoto K. M.; Kim K. B.; Kumagai A.; Mercurio F.; Crews C. M.; Deshaies R. J. Protacs: Chimeric Molecules That Target Proteins to the Skp1-Cullin-F Box Complex for Ubiquitination and Degradation. Proc. Natl. Acad. Sci. U. S. A. 2001, 98, 8554–8559. 10.1073/pnas.141230798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter G. E.; Buckley D. L.; Paulk J.; Roberts J. M.; Souza A.; Dhe-Paganon S.; Bradner J. E. DRUG DEVELOPMENT. Phthalimide Conjugation as a Strategy for in Vivo Target Protein Degradation. Science 2015, 348, 1376–1381. 10.1126/science.aab1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J.; Qian Y.; Altieri M.; Dong H.; Wang J.; Raina K.; Hines J.; Winkler J. D.; Crew A. P.; Coleman K.; Crews C. M. Hijacking the E3 Ubiquitin Ligase Cereblon to Efficiently Target BRD4. Chem. Biol. 2015, 22, 755–763. 10.1016/j.chembiol.2015.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zengerle M.; Chan K.-H.; Ciulli A. Selective Small Molecule Induced Degradation of the BET Bromodomain Protein BRD4. ACS Chem. Biol. 2015, 10, 1770–1777. 10.1021/acschembio.5b00216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishida T.; Ciulli A. E3 Ligase Ligands for PROTACs: How They Were Found and How to Discover New Ones. SLAS Discovery 2021, 26, 484–502. 10.1177/2472555220965528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schapira M.; Calabrese M. F.; Bullock A. N.; Crews C. M. Targeted Protein Degradation: Expanding the Toolbox. Nat. Rev. Drug Discov. 2019, 18, 949–963. 10.1038/s41573-019-0047-y. [DOI] [PubMed] [Google Scholar]
- Santt O.; Pfirrmann T.; Braun B.; Juretschke J.; Kimmig P.; Scheel H.; Hofmann K.; Thumm M.; Wolf D. H. The Yeast GID Complex, a Novel Ubiquitin Ligase (E3) Involved in the Regulation of Carbohydrate Metabolism. Mol. Biol. Cell 2008, 19, 3323–3333. 10.1091/mbc.e08-03-0328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherpa D.; Chrustowicz J.; Qiao S.; Langlois C. R.; Hehl L. A.; Gottemukkala K. V.; Hansen F. M.; Karayel O.; von Gronau S.; Prabu J. R.; Mann M.; Alpi A. F.; Schulman B. A. GID E3 Ligase Supramolecular Chelate Assembly Configures Multipronged Ubiquitin Targeting of an Oligomeric Metabolic Enzyme. Mol. Cell 2021, 81, 2445–2459.e13. 10.1016/j.molcel.2021.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regelmann J.; Schüle T.; Josupeit F. S.; Horak J.; Rose M.; Entian K.-D.; Thumm M.; Wolf D. H. Catabolite Degradation of Fructose-1,6-Bisphosphatase in the Yeast Saccharomyces Cerevisiae: A Genome-Wide Screen Identifies Eight Novel GID Genes and Indicates the Existence of Two Degradation Pathways. Mol. Biol. Cell 2003, 14, 1652–1663. 10.1091/mbc.e02-08-0456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menssen R.; Schweiggert J.; Schreiner J.; Kušević D.; Reuther J.; Braun B.; Wolf D. H. Exploring the Topology of the Gid Complex, the E3 Ubiquitin Ligase Involved in Catabolite-Induced Degradation of Gluconeogenic Enzymes. J. Biol. Chem. 2012, 287, 25602–25614. 10.1074/jbc.M112.363762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su A. I.; Wiltshire T.; Batalov S.; Lapp H.; Ching K. A.; Block D.; Zhang J.; Soden R.; Hayakawa M.; Kreiman G.; Cooke M. P.; Walker J. R.; Hogenesch J. B. A Gene Atlas of the Mouse and Human Protein-Encoding Transcriptomes. Proc. Natl. Acad. Sci. U. S. A. 2004, 101, 6062–6067. 10.1073/pnas.0400782101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binder J. X.; Pletscher-Frankild S.; Tsafou K.; Stolte C.; O’Donoghue S. I.; Schneider R.; Jensen L. J.. COMPARTMENTS: Unification and Visualization of Protein Subcellular Localization Evidence. Database 2014, 2014, 10.1093/DATABASE/BAU012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong C.; Zhang H.; Li L.; Tempel W.; Loppnau P.; Min J. Molecular Basis of GID4-Mediated Recognition of Degrons for the Pro/N-End Rule Pathway. Nat. Chem. Biol. 2018, 14, 466–473. 10.1038/s41589-018-0036-1. [DOI] [PubMed] [Google Scholar]
- Chrustowicz J.; Sherpa D.; Teyra J.; Loke M. S.; Popowicz G. M.; Basquin J.; Sattler M.; Prabu J. R.; Sidhu S. S.; Schulman B. A. Multifaceted N-Degron Recognition and Ubiquitylation by GID/CTLH E3 Ligases. J. Mol. Biol. 2022, 434, 167347 10.1016/j.jmb.2021.167347. [DOI] [PubMed] [Google Scholar]
- Chen S.-J.; Wu X.; Wadas B.; Oh J.-H.; Varshavsky A. An N-End Rule Pathway That Recognizes Proline and Destroys Gluconeogenic Enzymes. Science 2017, 355, eaal3655 10.1126/science.aal3655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong C.; Chen S.-J.; Melnykov A.; Weirich S.; Sun K.; Jeltsch A.; Varshavsky A.; Min J. Recognition of Nonproline N-Terminal Residues by the Pro/N-Degron Pathway. Proc. Natl. Acad. Sci. U. S. A. 2020, 117, 14158–14167. 10.1073/pnas.2007085117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hämmerle M.; Bauer J.; Rose M.; Szallies A.; Thumm M.; Düsterhus S.; Mecke D.; Entian K. D.; Wolf D. H. Proteins of Newly Isolated Mutants and the Amino-Terminal Proline Are Essential for Ubiquitin-Proteasome-Catalyzed Catabolite Degradation of Fructose-1,6-Bisphosphatase of Saccharomyces Cerevisiae. J Biol Chem 1998, 273, 25000–25005. 10.1074/jbc.273.39.25000. [DOI] [PubMed] [Google Scholar]
- Structural Genomics Consortium . PFI-7 Chemical probe for GID4, substrate-recognition subunit of the CTLH E3 ubiquitin-protein ligase complex.
- Poirson J.; Dhillon A.; Cho H.; Hiu M.; Lam Y.; Alerasool N.; Lacoste J.; Mizan L.; Taipale M.. Proteome-Scale Induced Proximity Screens Reveal Highly Potent Protein Degraders and Stabilizers .bioRxiv 2022, 10.1101/2022.08.15.503206. [DOI]
- Békés M.; Langley D. R.; Crews C. M. PROTAC Targeted Protein Degraders: The Past Is Prologue. Nat. Rev. Drug Discovery 2022, 21, 181–200. 10.1038/S41573-021-00371-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friesner R. A.; Banks J. L.; Murphy R. B.; Halgren T. A.; Klicic J. J.; Mainz D. T.; Repasky M. P.; Knoll E. H.; Shelley M.; Perry J. K.; Shaw D. E.; Francis P.; Shenkin P. S. Glide: A New Approach for Rapid, Accurate Docking and Scoring. 1. Method and Assessment of Docking Accuracy. J. Med. Chem. 2004, 47, 1739–1749. 10.1021/jm0306430. [DOI] [PubMed] [Google Scholar]
- Halgren T. A.; Murphy R. B.; Friesner R. A.; Beard H. S.; Frye L. L.; Pollard W. T.; Banks J. L. Glide: A New Approach for Rapid, Accurate Docking and Scoring. 2. Enrichment Factors in Database Screening. J. Med. Chem. 2004, 47, 1750–1759. 10.1021/jm030644s. [DOI] [PubMed] [Google Scholar]
- Friesner R. A.; Murphy R. B.; Repasky M. P.; Frye L. L.; Greenwood J. R.; Halgren T. A.; Sanschagrin P. C.; Mainz D. T. Extra Precision Glide: Docking and Scoring Incorporating a Model of Hydrophobic Enclosure for Protein-Ligand Complexes. J. Med. Chem. 2006, 49, 6177–6196. 10.1021/jm051256o. [DOI] [PubMed] [Google Scholar]
- Gironda-Martínez A.; Donckele E. J.; Samain F.; Neri D. DNA-Encoded Chemical Libraries: A Comprehensive Review with Succesful Stories and Future Challenges. ACS Pharmacol. Transl. Sci. 2021, 4, 1265–1279. 10.1021/acsptsci.1c00118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen J.; Brenner S.; Janda K. D. Synthetic Methods for the Implementation of Encoded Combinatorial Chemistry. J. Am. Chem. Soc. 1993, 115, 9812–9813. 10.1021/ja00074a063. [DOI] [Google Scholar]
- Brenner S.; Lerner R. A. Encoded Combinatorial Chemistry. Proc. Natl. Acad. Sci. U. S. A. 1992, 89, 5381–5383. 10.1073/pnas.89.12.5381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuai L.; O’Keeffe T.; Arico-Muendel C. Randomness in DNA Encoded Library Selection Data Can Be Modeled for More Reliable Enrichment Calculation. SLAS Discovery 2018, 23, 405–416. 10.1177/2472555218757718. [DOI] [PubMed] [Google Scholar]
- Ishii T.; Okai T.; Iwatani-Yoshihara M.; Mochizuki M.; Unno S.; Kuno M.; Yoshikawa M.; Shibata S.; Nakakariya M.; Yogo T.; Kawamoto T. CETSA Quantitatively Verifies in Vivo Target Engagement of Novel RIPK1 Inhibitors in Various Biospecimens. Sci. Rep. 2017, 7, 13000. 10.1038/s41598-017-12513-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jafari R.; Almqvist H.; Axelsson H.; Ignatushchenko M.; Lundbäck T.; Nordlund P.; Martinez Molina D. The Cellular Thermal Shift Assay for Evaluating Drug Target Interactions in Cells. Nat. Protoc. 2014, 9, 2100–2122. 10.1038/nprot.2014.138. [DOI] [PubMed] [Google Scholar]
- Savitski M. M.; Reinhard F. B. M.; Franken H.; Werner T.; Savitski M. F.; Eberhard D.; Molina D. M.; Jafari R.; Dovega R. B.; Klaeger S.; Kuster B.; Nordlund P.; Bantscheff M.; Drewes G. Tracking Cancer Drugs in Living Cells by Thermal Profiling of the Proteome. Science 2014, 346, 1255784. 10.1126/science.1255784. [DOI] [PubMed] [Google Scholar]
- Winter G. E.; Buckley D. L.; Paulk J.; Roberts J. M.; Souza A.; Dhe-Paganon S.; Bradner J. E. Selective Target Protein Degradation via Phthalimide Conjugation. Science 2015, 348, 1376. 10.1126/SCIENCE.AAB1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crew A. P.; Raina K.; Dong H.; Qian Y.; Wang J.; Vigil D.; Serebrenik Y. V.; Hamman B. D.; Morgan A.; Ferraro C.; Siu K.; Neklesa T. K.; Winkler J. D.; Coleman K. G.; Crews C. M. Identification and Characterization of von Hippel-Lindau-Recruiting Proteolysis Targeting Chimeras (PROTACs) of TANK-Binding Kinase 1. J. Med. Chem. 2018, 61, 583–598. 10.1021/acs.jmedchem.7b00635. [DOI] [PubMed] [Google Scholar]
- Han X.; Zhao L.; Xiang W.; Qin C.; Miao B.; Xu T.; Wang M.; Yang C. Y.; Chinnaswamy K.; Stuckey J.; Wang S. Discovery of Highly Potent and Efficient PROTAC Degraders of Androgen Receptor (AR) by Employing Weak Binding Affinity VHL E3 Ligase Ligands. J. Med. Chem. 2019, 62, 11218–11231. 10.1021/acs.jmedchem.9b01393. [DOI] [PubMed] [Google Scholar]
- Bancet A.; Raingeval C.; Lomberget T.; le Borgne M.; Guichou J. F.; Krimm I. Fragment Linking Strategies for Structure-Based Drug Design. J. Med. Chem. 2020, 63, 11420–11435. 10.1021/acs.jmedchem.9b01393. [DOI] [PubMed] [Google Scholar]
- Hung A. W.; Silvestre H. L.; Wen S.; Ciulli A.; Blundell T. L.; Abell C. Application of Fragment Growing and Fragment Linking to the Discovery of Inhibitors of Mycobacterium Tuberculosis Pantothenate Synthetase. Angew. Chem., Int. Ed. 2009, 48, 8452–8456. 10.1002/ANIE.200903821. [DOI] [PubMed] [Google Scholar]
- McCloskey K.; Sigel E. A.; Kearnes S.; Xue L.; Tian X.; Moccia D.; Gikunju D.; Bazzaz S.; Chan B.; Clark M. A.; Cuozzo J. W.; Guié M.-A.; Guilinger J. P.; Huguet C.; Hupp C. D.; Keefe A. D.; Mulhern C. J.; Zhang Y.; Riley P. Machine Learning on DNA-Encoded Libraries: A New Paradigm for Hit Finding. J. Med. Chem. 2020, 63, 8857–8866. 10.1021/acs.jmedchem.0c00452. [DOI] [PubMed] [Google Scholar]
- Delaglio F.; Grzesiek S.; Vuister G. W.; Zhu G.; Pfeifer J.; Bax A. NMRPipe: A Multidimensional Spectral Processing System Based on UNIX Pipes. J. Biomol. NMR 1995, 6, 277–293. 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- Vranken W. F.; Boucher W.; Stevens T. J.; Fogh R. H.; Pajon A.; Llinas M.; Ulrich E. L.; Markley J. L.; Ionides J.; Laue E. D. The CCPN Data Model for NMR Spectroscopy: Development of a Software Pipeline. Proteins 2005, 59, 687–696. 10.1002/prot.20449. [DOI] [PubMed] [Google Scholar]
- Kabsch W. XDS. Acta. Crystallogr., D: Biol. Crystallogr. 2010, 66, 125–132. 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCoy A. J.; Grosse-Kunstleve R. W.; Adams P. D.; Winn M. D.; Storoni L. C.; Read R. J. Phaser Crystallographic Software. J. Appl. Crystallogr. 2007, 40, 658–674. 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams P. D.; Afonine P. V.; Bunkóczi G.; Chen V. B.; Davis I. W.; Echols N.; Headd J. J.; Hung L.-W.; Kapral G. J.; Grosse-Kunstleve R. W.; McCoy A. J.; Moriarty N. W.; Oeffner R.; Read R. J.; Richardson D. C.; Richardson J. S.; Terwilliger T. C.; Zwart P. H. PHENIX: A Comprehensive Python-Based System for Macromolecular Structure Solution. Acta Crystallogr., D: Biol. Crystallogr. 2010, 66, 213–221. 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P.; Lohkamp B.; Scott W. G.; Cowtan K. Features and Development of Coot. Acta Crystallogr., D: Biol. Crystallogr. 2010, 66, 486–501. 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molina D. M.; Jafari R.; Ignatushchenko M.; Seki T.; Larsson E. A.; Dan C.; Sreekumar L.; Cao Y.; Nordlund P. Monitoring Drug Target Engagement in Cells and Tissues Using the Cellular Thermal Shift Assay. Science 2013, 341, 84–87. 10.1126/science.1233606. [DOI] [PubMed] [Google Scholar]
- Zinn N.; Werner T.; Doce C.; Mathieson T.; Boecker C.; Sweetman G.; Fufezan C.; Bantscheff M. Improved Proteomics-Based Drug Mechanism-of-Action Studies Using 16-Plex Isobaric Mass Tags. J. Proteome Res. 2021, 20, 1792–1801. 10.1021/acs.jproteome.0c00900. [DOI] [PubMed] [Google Scholar]
- Hughes C. S.; Moggridge S.; Müller T.; Sorensen P. H.; Morin G. B.; Krijgsveld J. Single-Pot, Solid-Phase-Enhanced Sample Preparation for Proteomics Experiments. Nat. Protoc. 2019, 14, 68–85. 10.1038/s41596-018-0082-x. [DOI] [PubMed] [Google Scholar]
- Erdjument-Bromage H.; Huang F.-K.; Neubert T. A. Sample Preparation for Relative Quantitation of Proteins Using Tandem Mass Tags (TMT) and Mass Spectrometry (MS). Methods Mol. Biol. 2018, 1741, 135–149. 10.1007/978-1-4939-7659-1_11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krieger J. R.; Wybenga-Groot L. E.; Tong J.; Bache N.; Tsao M. S.; Moran M. F. Evosep One Enables Robust Deep Proteome Coverage Using Tandem Mass Tags While Significantly Reducing Instrument Time. J. Proteome Res. 2019, 18, 2346–2353. 10.1021/ACS.JPROTEOME.9B00082. [DOI] [PubMed] [Google Scholar]
- Savitski M. M.; Zinn N.; Faelth-Savitski M.; Poeckel D.; Gade S.; Becher I.; Muelbaier M.; Wagner A. J.; Strohmer K.; Werner T.; Melchert S.; Petretich M.; Rutkowska A.; Vappiani J.; Franken H.; Steidel M.; Sweetman G. M.; Gilan O.; Lam E. Y. N.; Dawson M. A.; Prinjha R. K.; Grandi P.; Bergamini G.; Bantscheff M. Multiplexed Proteome Dynamics Profiling Reveals Mechanisms Controlling Protein Homeostasis. Cell 2018, 173, 260–274.e25. 10.1016/J.CELL.2018.02.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox J.; Mann M. MaxQuant Enables High Peptide Identification Rates, Individualized p.p.b.-Range Mass Accuracies and Proteome-Wide Protein Quantification. Nat. Biotechnol. 2008, 26, 1367–1372. 10.1038/NBT.1511. [DOI] [PubMed] [Google Scholar]
- Franken H.; Mathieson T.; Childs D.; Sweetman G. M. A.; Werner T.; Tögel I.; Doce C.; Gade S.; Bantscheff M.; Drewes G.; Reinhard F. B. M.; Huber W.; Savitski M. M. Thermal Proteome Profiling for Unbiased Identification of Direct and Indirect Drug Targets Using Multiplexed Quantitative Mass Spectrometry. Nat. Protoc. 2015, 10, 1567–1593. 10.1038/nprot.2015.101. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.