Abstract
Uncoupling protein 3 (UCP3) is known to regulate energy dissipation, proton leakage, fatty acid oxidation, and oxidative stress. To identify the putative protein regulators of UCP3, we performed yeast two-hybrid screens. Here we report that UCP3 interacted with HS-1 associated protein X-1 (Hax-1), an anti-apoptotic protein that was localized in the mitochondria, and is involved in cellular responses to Ca2+. The hydrophilic sequences within loop 2, and the matrix-localized hydrophilic domain of mouse UCP3, were necessary for binding to Hax-1 at the C-terminal domain, adjacent to the mitochondrial inner membrane. Interestingly, interaction of these proteins occurred in a calcium-dependent manner. Moreover, the NMR spectrum of the C-terminal domain of Hax-1 was dramatically changed by removal of Ca2+, suggesting that the C-terminal domain of Hax-1 underwent a Ca2+-induced conformational change. In the Ca2+-free state, the C-terminal Hax-1 tended to unfold, suggesting that Ca2+ binding may induce protein folding of the Hax-1 C-terminus. These results suggested that the UCP3-Hax-1 complex may regulate mitochondrial functional changes caused by mitochondrial Ca2+.
Keywords: Mitochondria, Uncoupling protein 3, Calcium ion, Folding
1. Introduction
Mitochondria play an important role in calcium cycling, ATP production, generation of reactive oxygen species (ROS), and apoptosis within most mammalian cells. It has been proposed that elevated Ca2+ levels in the mitochondrial matrix within the physiological range lead to increased ATP formation and reduced ROS production caused by activation of manganese superoxide dismutase (MnSOD) [1,2]. The mitochondrial ATP production stimulated by Ca2+ was attributed to activation of several matrix dehydrogenases, including pyruvate dehydrogenase, isocitrate dehydrogenase, and oxoglutarate dehydrogenase [3]. However, the influx of high levels of Ca2+ into the mitochondrial matrix in pathological conditions promoted the permeability transition pore (PTP) opening, leading to ROS production and decreased ATP production [4].
Uncoupling proteins (UCPs) comprise a subfamily of mitochondrial inner membrane anion carrier proteins that dissipate the mitochondrial proton gradient [5]. In mice, UCP1 which played a physiologically important role in regulation of heat production, was almost exclusively expressed in brown adipose tissues, whereas UCP3 that was first identified as a UCP1 homolog in 1997 was predominantly expressed in skeletal muscle and heart [6]. Previous studies suggested that UCP3 regulated mitochondrial ROS production and fatty acid oxidation as well as proton leakage [7]. Although UCP3 was also reported to contribute to mitochondrial Ca2+ uptake [8], a recent study suggested that the Ca2+ uptake rate in the isolated mitochondria from UCP3-deficient mice was the same as that from wild type mice [9]. It had however been reported that UCP3 had a cardioprotective effect through the inhibition of mitochondrial PTP opening that was induced by mitochondrial Ca2+ [10]. Therefore, these results suggested the possibility that UCP3 could influence mitochondrial functions associated with Ca2+, but not directly regulate Ca2+ uptake.
Close interactions between the endoplasmic reticulum (ER)/sarcoplasmic reticulum (SR) and mitochondria, termed the “mitochondria-associated membranes” (MAM), play an important role in rapid and sustained Ca2+ uptake by mitochondria, thereby regulating intracellular calcium homeostasis [11]. However in the heart, the sarcoplasmic reticulum Ca2+ ATPase (SERCA) and its regulatory protein phospholamban (PLN) are involved in facilitating calcium uptake into the ER/SR [12]. Recent studies have shown that HS-1-associated protein X-1 (Hax-1), known as an anti-apoptotic protein, interacted with PLN, suggesting that Hax-1 maintained calcium homeostasis and cell survival by regulation of ER/SR Ca2+ concentrations [12,13]. Heterozygous mice, with 36% Hax-1 expression levels in the heart, showed significant increases in the rate of calcium transport to the SR, as compared to wild type mice [13]. However, some studies in CoS-7 and striated muscle cells suggested that Hax-1 proteins localized at the mitochondria as well as at the ER/SR [14,15].
In the present study, we report that UCP3 binds with Hax-1 in the mitochondria. The interaction regions between UCP3 and Hax-1 were identified by mutation studies, and consisted of the matrix-localized hydrophilic intermembrane loop (IML) 2 domain of UCP3 and the C-terminal domain of Hax-1. Additionally, our data suggested that Hax-1 induced structural folding in the presence of Ca2+, resulting in the binding to UCP3.
2. Materials and Methods
2.1. Yeast two-hybrid screening
Yeast two-hybrid screening was performed as described previously [16]. The hydrophilic sequence in the matrix-located IML2 domain of UCP3 (nucleotides encoding amino acids 134–176) was used as bait. From this screen, we identified a cDNA encoding Hax-1 that was characterized by sequencing using the BLAST algorithm.
2.2. Plasmid DNA constructs
The loop2 domain truncation mutants of UCP3 (UCP3 Δloop2A134–146, Δloop2B147–162, and Δloop2C163–176), the loop3 domain truncation mutants of UCP3 (UCP3 Δloop3A235–243, Δloop3B247–251, and Δloop3C252–266), and the truncated forms of GST-Hax-1 were constructed using the modified polymerase chain reaction (PCR) technique of Stratagene Cloning Systems (Stratagene, La Jolla, CA, USA) as described previously [16].
2.3. Cell culture
HEK293 cells were cultured in DMEM containing 10% fetal bovine serum (FBS) and a penicillin–streptomycin solution (Nacalai Tesque, Kyoto, Japan) at 37 °C with 5% CO2. HEK293 cells were transiently transfected using FuGENE6 HD transfection reagent (Roche, Tucson, AZ, USA) according to the manufacturer’s instructions. Cells were allowed to incubate for 24 h prior to experiments.
2.4. Isolation of mitochondria and submitochondrial localization assay
Mitochondria from HEK293 cells were prepared according to previously established protocols [16]. Water-soluble digitonin (Sigma–Aldrich, St. Louis, MO, USA) was used for permeabilization. Following 5 μg/mL proteinase K treatment, 100 μM PMSF was added to stop the enzyme activity. Anti-mitofusin 2 (Abcam, Cambridge, MA, USA), anti-Smac (Cell Signaling Technology, Danvers, MA, USA), and anti-HSP60 (Enzo Life Sciences, Plymouth Meeting, PA, USA) primary antibodies were used as markers of the outer membrane, the mitochondrial intermembrane space, and matrix, respectively.
2.5. GST pull-down assay
Purified GST-tagged proteins were prepared as previously described [17]. Five μg of each purified GST fusion protein or GST protein alone was added to 100 μg of cell extracts from HEK293 cells and incubated in GST lysis buffer with or without Ca2+ (20 mM Tris–HCl, pH 7.4, 0.5 mM DTT, 150 mM NaCl, 0.5% Triton X-100, 2.5 mM CaCl2 or 5 mM EDTA, plus protease inhibitors) for 1.5 h at 4 °C. After addition of 20 μL of glutathione Sepharose 4B (GE Healthcare, Uppsala, Sweden), the binding was allowed to continue for an additional 16 h at 4 °C. The bound beads were washed four times each with lysis buffer and analyzed by SDS-PAGE.
2.6. Immunoblotting and immunoprecipitation
Immunoblotting and immunoprecipitation analyses were performed as described previously [16]. The following antibodies were used: anti-V5 (Invitrogen, Grand Island, NY, USA), anti-myc (Up-state Biotechnology, Lake Placid, New York, USA), anti-UCP3 (Abcam), anti-mitofusin2 (Abcam), anti-Hax-1 (BD Biosciences, Franklin Lakes, NJ, USA), anti-GAPDH (Santa Cruz Biotechnology, Santa Cruz, CA, USA), anti-β-actin (Calbiochem, San Diego, CA, USA), anti-Smac/Diablo, and anti-HSP60 antibodies.
2.7. Immunofluorescence staining
Cells were cultured on coverslips and loaded with 50 nM MitoTracker® Red CMXROS (Invitrogen) for 30 min at 37 °C in the dark, then fixed in 2% paraformaldehyde for 10 min at 25 °C, and permeabilized with 0.2% Triton X-100 for 10 min at 4 °C. The cells were incubated with anti-V5 or anti-myc antibodies, followed by secondary anti-mouse Alexa 568, anti-mouse Alexa 488, or anti-rabbit Alexa 488 (Invitrogen). Cells were further incubated with Hoechst-33342 stain (Dojindo, Kumamoto, Japan) for 10 min at room temperature in the dark after being washed. All fluorescent images were acquired with the BIOTRVO BZ-9000 (Keyence, Osaka, Japan).
2.8. 2D 1H–15N HSQC nuclear magnetic resonance (NMR) spectroscopy
Recombinant C-terminal amino acids 211–280 of Hax-1 (Hax-1211–280) protein, uniformly labeled with 15N, was obtained by growing bacterial cells in M9 minimal media containing 15NH4Cl as the sole nitrogen source. GST-tagged HAX-1211–280 was obtained using the above-mentioned procedure (“GST Pull-down Assay”). In GST-tagged HAX-1211–280, the GST tag was removed by digestion with PreScission protease (GE Healthcare), on the column in the storage buffer (20 mM Tris–HCl, pH 7.5,150 mM NaCl, 1 mM DTT, 1 mM CaCl2, 1% CHAPS) for 2 days at 10 °C. The protein was further purified by affinity chromatography for removal of PreScission protease, anion exchange chromatography with HiTrap™ Q-HP (GE Healthcare), and cation exchange chromatography with HiTrap™ SP-HP (GE Healthcare). Finally, the protein solution was applied to a size exclusion Superdex 75 10/300 GL column (GE Healthcare) equilibrated with the storage buffer. Purified proteins were concentrated at 4 °C in a VIVASPIN® 15R 2000 cut-off membrane concentrator (Sartorius Stedim Biotech, Goettingen, Germany) and used for NMR experiments.
All NMR samples were dissolved in either Ca2+-containing buffer (20 mM Tris–HCl, pH 7.5,150 mM NaCl, 1 mM DTT, 1 mM CaCl2,1% CHAPS, 5% D2O) or Ca2+-free buffer (20 mM Tris–HCl, pH 7.5,150 mM NaCl, 1 mM DTT, 1% CHAPS, 5% D2O). To investigate the Ca2+-binding properties of HAX-1211–280, we recorded the 1H–15N HSQC spectrum of 0.25 mM 15N-labeled HAX1211–280 in the Ca2+-free state. Ca2+ was removed from 15N-labeled samples by incubation in 10 mM EDTA for 1 h at 4 °C, and then the EDTA-treated samples were dialyzed with a Ca2+-free buffer for 20 h to remove EDTA.
All NMR experiments were performed at 298 K. Data collection was done on a Bruker AVANCE II 800 MHz NMR spectrometer (Bruker, Billerica, MA, USA) equipped with a cryoprobe, which was operated at 800 MHz for 1H resonance frequency. NMR data analyses were carried out using SPARKY 3.114.
3. Results
3.1. UCP3 associated with Hax-1 in mitochondria
To identify novel UCP3 regulatory mechanisms, yeast two-hybrid screens were performed using a heart cDNA library with bait of a DNA coding sequences corresponding to the IML2 of the mouse UCP3 protein (subcloned into the pGBKT7 yeast expression vector; Clontech, Mountain View, CA, USA). We identified Hax-1 as a candidate binding partner with the IML2 hydrophilic domain consisting of UCP3 (data not shown). To confirm the yeast two-hybrid results, we performed co-immunoprecipitation (IP) assays in HEK293 cells. Cell lysates from empty vector transfectants were used as negative controls for antibody specificity (mock, Fig. 1A, left lane). Unlike in mock transfectants, in the cells coexpressing UCP3–V5 and Hax-1-myc, anti-myc antibody (Hax-1-myc) co-immunoprecipitated UCP3–V5 (immunoblots, IB: anti-V5) (Fig. 1A, upper panel). The control samples, using IgG as the antibody, did not exhibit such co-immunoprecipitation. Expressions of the transfected proteins and equal protein loading were confirmed by immunoblotting for V5, myc, and β-actin in whole cell lysates from the indicated transfectants (Fig. 1A, lower three panels). We also performed immunostaining in HEK293 cells co-transfected with UCP3 and Hax-1. As expected, both UCP3–V5 and Hax-1-myc each colocalized (Merge) in a punctate, perinuclear fashion with the mitochondrial dye MitoTracker® Red (Mito) (Fig. 1B, upper and middle panels). Moreover, coimmunostaining for UCP3 (V5) and Hax-1 (myc) showed that the expressed proteins interacted in an overlapping mitochondrial pattern (Fig. 1B, lower panels).
Fig. 1.
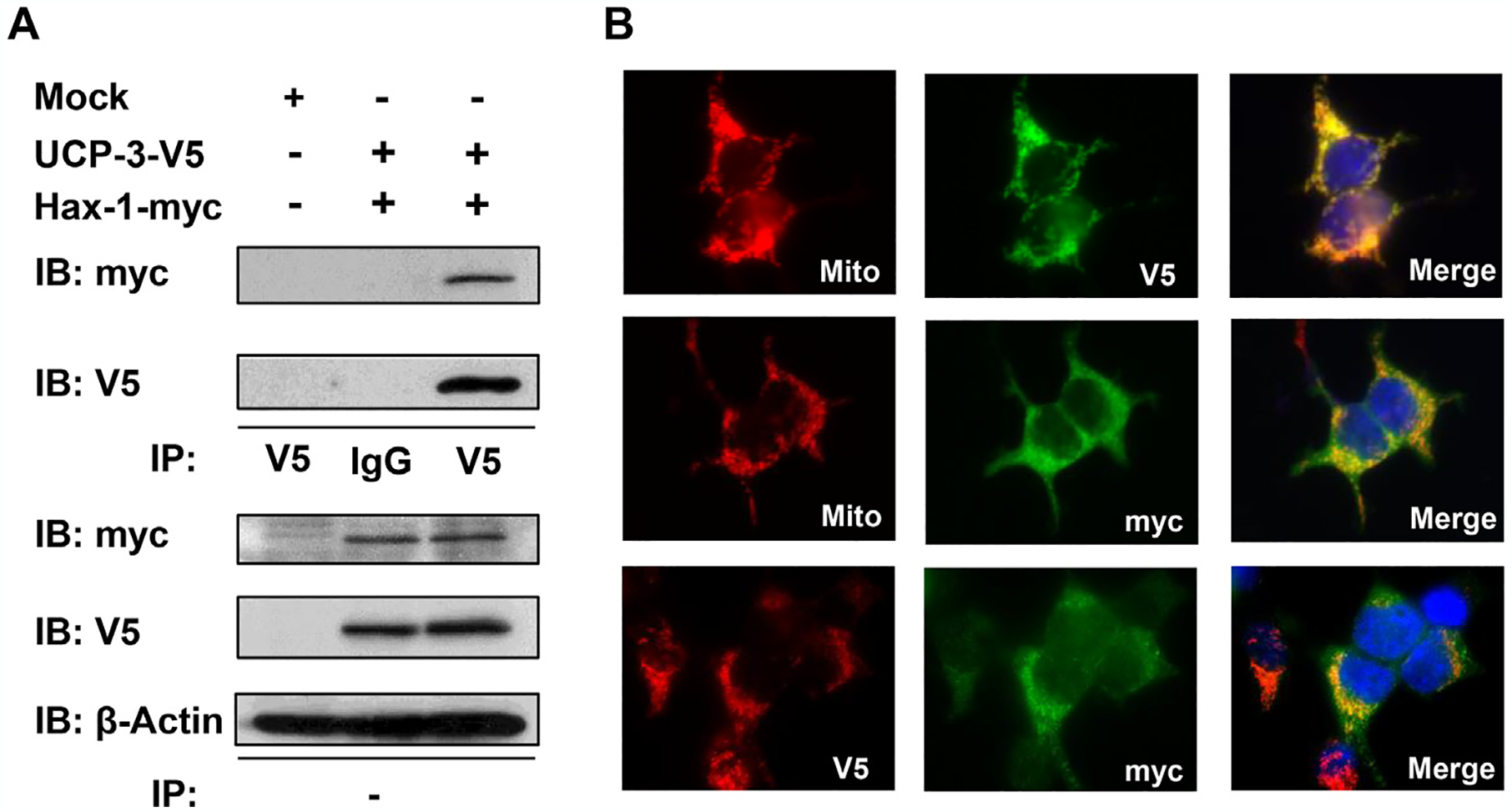
Interaction of UCP3 and Hax-1 in mitochondria. (A) HEK293 cell extracts transfected with empty vector (Mock), myc-tagged Hax-1 (Hax-1-myc), and V5-tagged UCP3 (UCP3–V5) were immunoprecipitated (IP) with anti-IgG or anti-V5 antibodies and analyzed by immunoblotting to detect V5 (UCP3) and myc (Hax-1). β-actin was used as an internal standard. (B) HEK293 cells transfected with UCP3–V5 (top) or Hax-1-myc (middle) were immunostained with V5 (red) or myc (green) antibodies and co-stained with the mitochondrial indicator MitoTracker® Red (red), as indicated. HEK293 cells cotransfected with UCP3–V5 and Hax-1-myc (bottom) were co-immunostained with V5 (red) and myc (green) antibodies. The overlay shows that UCP3 colocalized with Hax-1.
3.2. Hax-1 binding to UCP3 localized at the mitochondrial inner membrane
Hax-1 has been known to localize in the ER and cytoplasm as well as in mitochondria [15]; therefore we determined the submitochondrial localization of Hax-1 and its interaction with UCP3. First, we examined Hax-1 localization in the mitochondrial and cytosolic fractions from HEK293 cells. Hax-1 was localized in the mitochondrial fraction containing the mitochondrial mitofusin 2 protein, but was not in the cytosolic fraction containing the cytosolic marker GAPDH (Fig. 2A, IB: Hax-1, MFN2). To further define the mitochondrial localization of Hax-1, a submitochondrial localization assay was done using varying concentrations of digitonin (DIG) in combination with protease K (Pro K) treatment. Immune reactivity of Hax-1 and UCP3 (V5) were more resistant to DIG when compared with the outer membrane (OM) and the intermembrane space (IMS) resident Smac (Fig. 2B, lanes 1–5), indicating that the UCP3 and Hax-1 complex was located in the surrounding IMS. In contrast, the matrix-localized HSP60 required increased DIG concentrations for proteolysis (Fig. 2B, lanes 1–7).
Fig. 2.
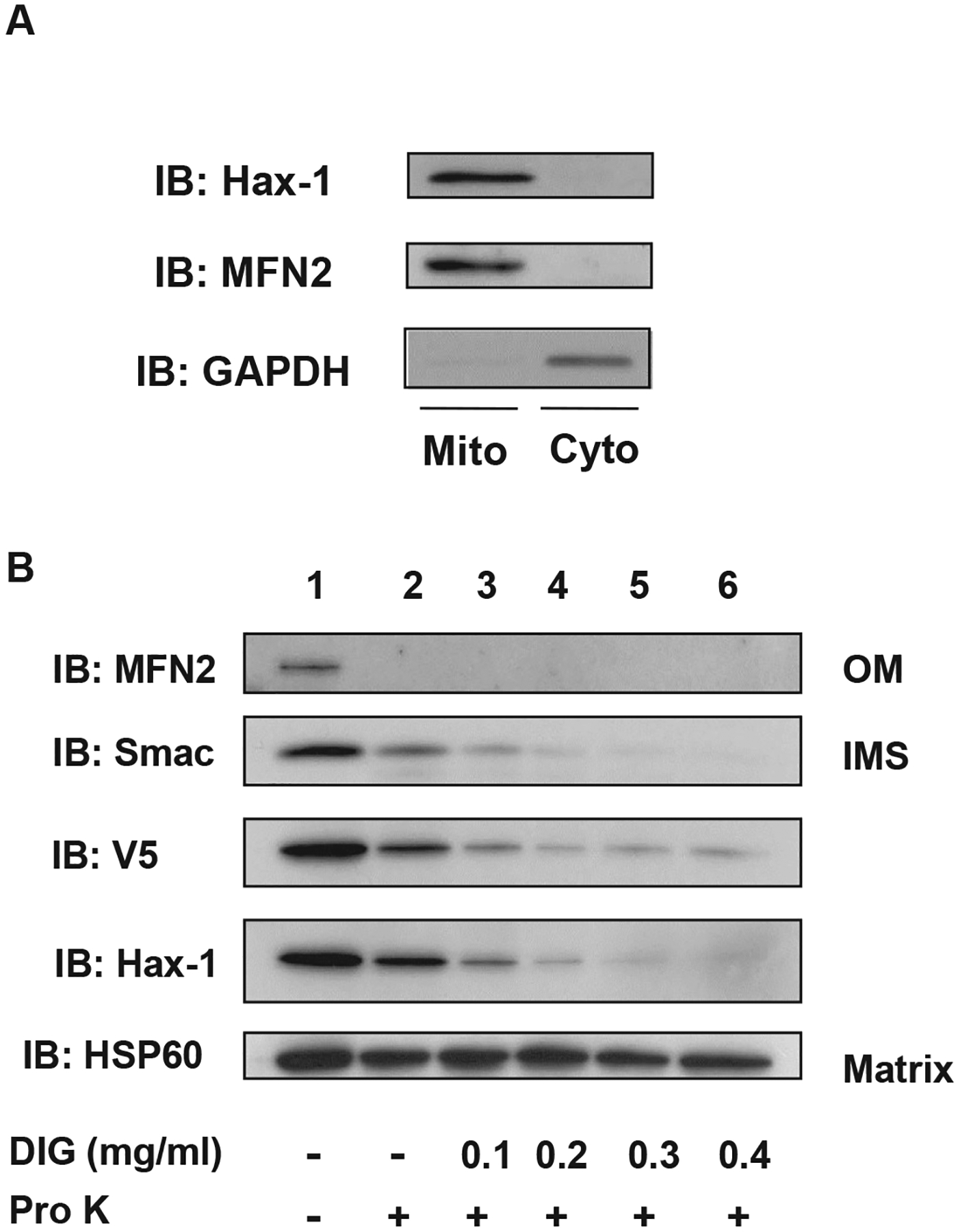
Mitochondrial localization of UCP3 and Hax-1. (A) Mitochondria and cytosolic fractions were isolated from HEK293 cells. Mitochondrial and cytosolic extracts (20 μg/lane) were subjected to SDS-PAGE followed by immunoblotting with anti-myc, anti-mitofusin 2 (MFN2), and anti-GAPDH antibodies. (B) Mitochondria isolated from HEK293 cells transfected with UCP3–V5 were subjected to mitochondrial sublocalization assays, as described in the Materials and Methods. Mitochondrial pellets and supernatants were resuspended in SDS-PAGE buffer, and then loaded onto SDS-PAGE gels. Immunoblots show the presence of the outer membrane (OM) resident MFN2, the intermembrane space (IMS) resident Smac, and the matrix resident HSP60 in mitochondria that were untreated or treated with proteinase K (Pro K) (lanes 2–6) and increasing concentrations of digitonin (DIG) (lanes 3–6, 0.1 mg/mL to 0.4 mg/mL DIG).
3.3. The UCP3 IML2 domain associated with the C-Terminus of Hax-1
As described above, the matrix-localized IML2 hydrophilic domain of UCP3 was predicted to mediate complex formation with Hax-1. To determine the role of the UCP3 IML2 domain in Hax-1 binding, we constructed UCP3 loop2 domain truncation mutants of UCP3 (UCP3 Δloop2A134–146, Δloop2B147–162, and Δloop2C163–176), and UCP3 loop3 domain truncation mutants of UCP3 (UCP3 Δloop3A235–243, Δloop3B247–251, and Δloop3C252–266) (Fig. 3A). Unlike full-length UCP3 and the UCP3 truncation mutants UCP3Δloop2B147–162 and UCP3 loop3, the UCP3 Δloop2A134–146 and Δloop2C163–176 truncation mutants that exhibited appropriate mitochondrial localization lost the ability to bind Hax-1 (Fig. 3B and C). These results confirmed the yeast two-hybrid data and indicated that the IML2 hydrophilic domain of UCP3 was required for Hax-1 binding.
Fig. 3.
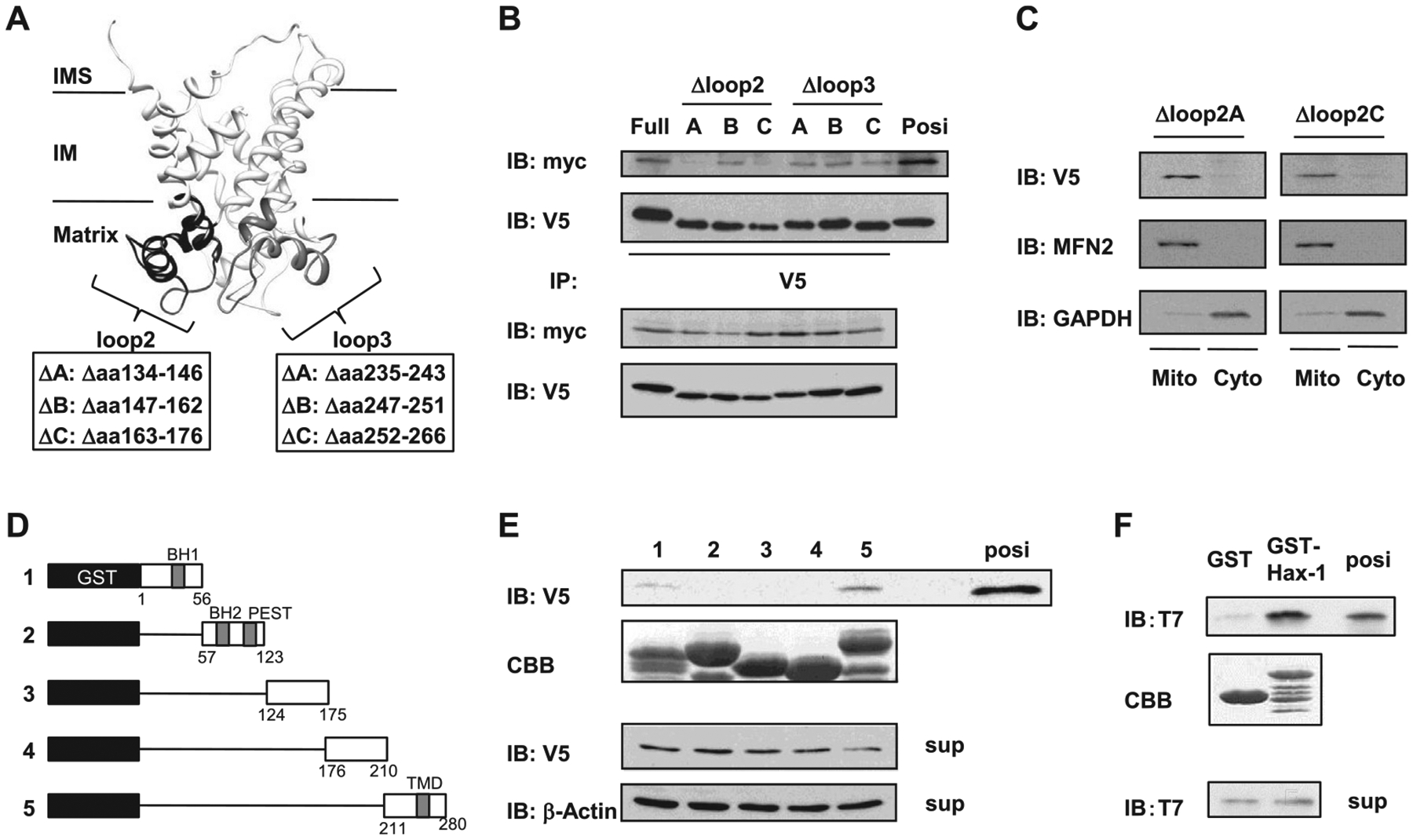
Mutational mapping of the UCP3-Hax-1 binding domain. (A) Speculative structure of mouse UCP3 was rendered with UCSF chimera (17). The UCP3 loop2 domain truncation mutants of UCP3 (UCP3 Δloop2A134–146, Δloop2B147–162, and Δloop2C163–176), and the UCP3 loop3 domain truncation mutants of UCP3 (UCP3 Δloop3A235–243, Δloop3B247–251, and Δloop3C252–266) were constructed as negative controls to determine the binding domain to Hax-1. (B) The loop 2 hydrophilic domain of UCP3 required for binding to Hax-1 was defined using mutagenesis and immunoprecipitation. Cell extracts (100 μg) were immunoprecipitated with anti-V5 antibodies and analyzed by immunoblotting with anti-myc antibodies. (C) Mitochondria and cytosolic extracts (20 μg/lane) were isolated from HEK293 cells transfected with UCP3 mutations and were subjected to SDS-PAGE followed by immunoblotting with anti-V5, anti-mitofusin 2 (MFN2), and anti-GAPDH antibodies. (D) Diagram for expression constructs of (1) GST-Hax-1 residues 1–56, (2) GST-Hax-1 residues 57–123, (3) GST-Hax-1 residues 124–175), (4) GST-Hax-1 residues 176–210), and (5) GST-Hax-1 residues 211–280. (E) The residues of Hax-1 required for direct binding to UCP3 were determined with GST pull-down assays. GST-fusion proteins were purified and stained by Coomassie Brilliant Blue (CBB) as indicated. In HEK293 cell extracts expressing UCP3–V5, the ability of the truncated Hax-1 fusion proteins to bind to UCP3 was analyzed by immunoblotting against V5. (F) Direct protein interactions were determined by incubating purified recombinant T7-tagged UCP3 IML2 domain with GST-Hax-1 or GST. The samples from the GST pull-down assays were subjected to SDS-PAGE and immunoblotting against T7. Purified protein of the UCP3 IML2 domain was used as a positive control.
Because UCP3 was shown to bind specifically to Hax-1, we constructed and bacterially expressed and purified a variety of serial mutants of Hax-1 in frame with N-terminal GST, using glutathione beads (constructs are shown in Fig. 3D). We tested the ability of wild type Hax-1 and its mutant derivatives to bind UCP3–V5 from transfected whole cell extracts (WCE) using GST pull-down assays. As revealed by UCP3 immunoblotting, the Hax-1 C-terminal construct (GST-Hax-1 amino acids 211–280) bound to UCP3 (Fig. 3E, lane 5). However, the Hax-1 mutants coding the amino acids 1–210 could not bind to UCP3 (Fig. 3E, lanes 1–4). A positive control (WCE) for UCP3 immunoreactivity, a Coomassie Brilliant Blue-stained gel (CBB) that confirmed construct expression, and an internal control are also shown (Fig. 3E). These results indicated that the C-terminal amino acids 211–280 of Hax-1 (Hax-1211–280) were both necessary and sufficient for binding to the IML2 hydrophilic domain of UCP3.
To test whether these were direct interactions, we performed in vitro binding reactions using purified recombinant GST, GST-Hax-1211–280, and T7-tagged UCP3 IML2 proteins. As shown in Fig. 3E, recombinant GST-Hax-1211–280 bound efficiently to UCP3 IML2-T7 (IB: T7).
3.4. UCP3 interacted with Hax-1 in a calcium-dependent manner
We examined whether Ca2+ was required for the binding of UCP3 and Hax-1. Unlike GST alone, GST-Hax-1211–280 was bound to UCP3 in the presence of Ca2+ (Fig. 4A). As expected, GST-Hax-1211–280 failed to bind to UCP3 in the presence of EDTA. Moreover, treatment with a calcium ionophore strongly increased the UCP3-Hax-1 interaction in UCP3 and in Hax-1 cotransfected HEK293 cells (Fig. 4B). These results suggested that Ca2+ was required for UCP3 to interact with Hax-1.
Fig. 4.
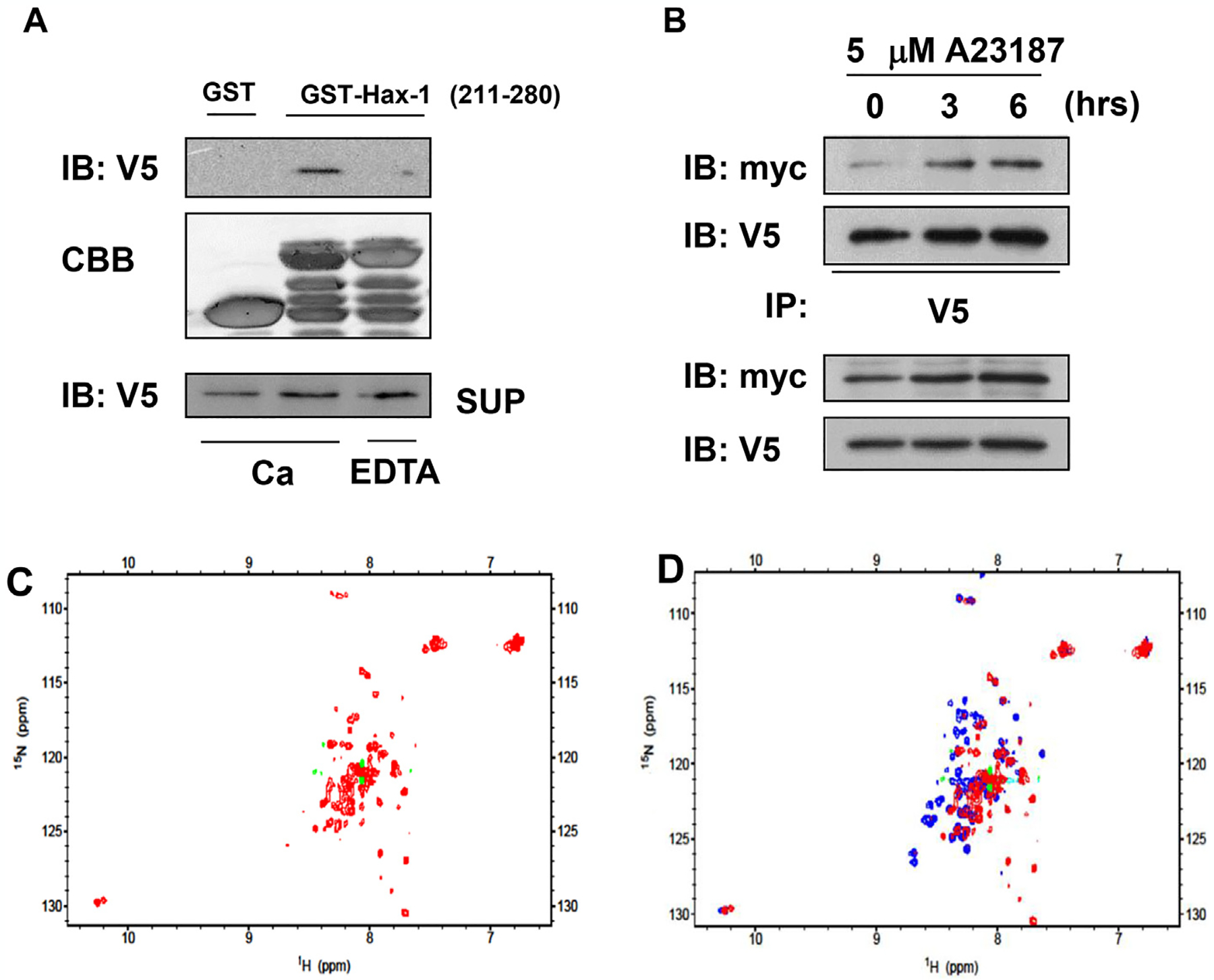
Calcium-dependent interaction of UCP3 and Hax-1. (A) The calcium-dependent protein interactions were determined by incubating HEK293 cell extracts expressing UCP3–V5 with GST alone or the C-terminal Hax-1 construct (GST-Hax-1 residues 211–280) in the presence of 2.5 mM CaCl2 or 5 mM EDTA. The samples from the GST pull-down assays were subjected to SDS-PAGE and immunoblotting against V5. (B) HEK293 cells expressing Hax-1-myc and UCP3–V5 were treated with calcium ionophore (5 μM A23187) for 0, 3, or 6 h. Cell extracts (100 μg) were immunoprecipitated with anti-V5 antibodies and analyzed by immunoblotting to detect V5 (UCP3) and myc (Hax-1). The NMR spectra of Hax-1211–280 in the presence and absence of Ca2+. (C) 1H–15N HSQC spectrum of 15N labeled- Hax-1211–280 without Ca2+. (D) Overlay of the spectra from Fig. 4D (1 mM Ca2+, blue) and Fig. 4C (without Ca2+, red).
To further understand the role of Ca2+ in the formation of the HAX-1-UCP3 complex, we measured the 1H–15N HSQC spectrum of Hax-1211–280 in the Ca2+-free state (Fig. 4C). The superposition of the two spectra (Fig. 4D) showed that many signals dramatically shifted when Ca2+ was removed. Significant conformational changes of Hax-1211–280 were caused by the removal of Ca2+. Interestingly, in the spectrum for the Ca2+-free state, we observed broad peaks with little dispersion in the center of the spectrum, which likely reflected the partial unfolding of the protein. These results suggested that Ca2+ may induce the structural stabilization and folding of HAX-1211–280. Our observations were consistent with the reversal of the Ca2+-induced destabilization of Hax-1211–280 during the purification (data not shown).
4. Discussion
The results of this study demonstrated that the interaction of UCP3 and Hax-1 was regulated by calcium ion concentration. In addition, we have provided direct biological and biochemical evidence that UCP3 interacted with Hax-1 in the mitochondria via the matrix-localized hydrophilic domain of UCP3 and the C-terminus of Hax-1. We showed that the Hax-1211–280 bound to the UCP3 in the presence of Ca2+ (Fig. 4A). Moreover, the 1H–15N HSQC spectrum of Hax-1211–280 was dramatically changed by removal of Ca2+ (Fig. 4D), suggesting that Hax-1211–280 underwent a Ca2+-induced conformational change. In the Ca2+-free states, Hax-1211–280 tended to unfold.
We demonstrated that the Hax-1 C-terminal domain required Ca2+ to bind with UCP3. By using NMR analysis, we found that the structure of the Hax-1 C-terminus was dynamically altered by combination with Ca2+ (Fig. 4D). Generally, the EF-hand motif is found in calmodulin, troponin C, and S100 protein [18], and is characterized by a helix-loop-helix structure composed of approximately 30 amino acids [19]. This is the most common structural motif associated with Ca2+ binding. Perocchi et al. also reported that mitochondrial calcium uptake 1 (MICU1), a novel modulator of mitochondrial calcium uptake, had two canonical EF hands, and was involved in calcium sensing and mitochondrial calcium uptake at the mitochondrial inner membrane [20]. In spite of lacking an EF-hand motif, the structural alteration of Hax-1 also occurred in the presence of Ca2+. Therefore, we hypothesized that the binding of the Hax-1 C-terminus to Ca2+ was independent of an EF-hand domain. In non-EF hand proteins, it has been reported that both Asp and Glu residues are important amino acids for binding Ca2+ [21]. Importantly, both Asp and Glu residues are highly conserved in the Hax-1 C-terminus across biological species. Thus, these findings suggested that the EF-hand-independent structural alteration in the Hax-1 C-terminus mediated by Ca2+ played an important role in mitochondrial calcium sensing.
Kuriyan et al. reported that the spectrum of the isolated EF-hand of RasGRP1 dramatically changed upon the addition of Ca2+ [22]. In the absence of Ca2+, the NMR spectrum of the RasGRP1 EF-hand showed very broad resonances with poor chemical shift dispersion. Moreover, the circular dichroism (CD) spectra also suggested that a small increase in helicity was observed upon addition of calcium. They concluded that the RasGRP1 EF-hand was likely to contribute to activation through Ca2+-triggered conformational changes [22]. Consistent with this earlier report, in the present study we observed similar changes in the 1H–15N HSQC spectra. Notably, this study showed that Ca2+-induced conformational changes were involved in the formation of the UCP3-binding surface.
Hax-1 contains BH1 and BH2 domains that are characterized by an apparent homology with the anti-apoptotic protein, B cell lymphoma/leukemia-1 (bcl-2). Based on this profile, numerous studies have proposed that Hax-1 plays an important role in regulating apoptosis and cell survival [23–25]. Indeed, overexpression of Hax-1 improved the decrease in mitochondrial membrane potential stimulated by cisplatin, which caused apoptotic cell death [26]. It has been reported that overloaded mitochondrial calcium concentration led to the release of caspase cofactors, resulting in the initiation of apoptosis [27]. We found that Hax-1 was localized in the mitochondria and its conformation was altered by Ca2+. Therefore, these findings suggested that the interaction of UCP3 and Hax-1 in mitochondria decreased apoptosis. However, further studies are necessary to define this mechanism.
Acknowledgments
This work was supported in part by Grant No. 24700753, 26350148 (K.H.) from a Grant-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science, and Technology, Japan.
Abbreviations:
- UCP3
Uncoupling protein 3
- Hax-1
HS-1 associated protein X-1
- ROS
reactive oxygen species
- MnSOD
manganese superoxide dismutase
- PTP
permeability transition pore
- MAM
mitochondria-associated membranes
- PLN
phospholamban
- IML
intermembrane loop
Footnotes
Transparency document
Transparency document related to this article can be found online at http://dx.doi.org/10.1016/j.bbrc.2016.02.075.
References
- [1].Csordás G, Hajnóczky G, SR/ER-mitochondrial local communication: calcium and ROS, Biochim. Biophys. Acta 1787 (2009) 1352–1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Hopper RK, Carroll S, Aponte AM, et al. , Mitochondrial matrix phosphoproteome: effect of extra mitochondrial calcium, Biochemistry 45 (2006) 2524–2536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Patron M, Raffaello A, Granatiero V, et al. , The mitochondrial calcium uniporter (MCU): molecular identity and physiological roles, J. Biol. Chem 288 (2013) 10750–10758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Di Lisa F, Bernardi P, A CaPful of mechanisms regulating the mitochondrial permeability transition, J. Mol. Cell Cardiol 46 (2009) 775–780. [DOI] [PubMed] [Google Scholar]
- [5].Azzu V, Brand MD, The on-off switches of the mitochondrial uncoupling proteins, Trends Biochem. Sci 35 (2010) 298–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Boss O, Samec S, Paoloni-Giacobino A, et al. , Uncoupling protein-3: a new member of the mitochondrial carrier family with tissue-specific expression, FEBS Lett. 408 (1997) 39–42. [DOI] [PubMed] [Google Scholar]
- [7].Krauss S, Zhang CY, Lowell BB, The mitochondrial uncoupling-protein homologues, Nat. Rev. Mol. Cell. Biol 6 (2005) 248–261. [DOI] [PubMed] [Google Scholar]
- [8].Trenker M, Malli R, Fertschai I, et al. , Uncoupling proteins 2 and 3 are fundamental for mitochondrial Ca2+ uniport, Nat. Cell Biol 9 (2007) 445–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Brookes PS, Parker N, Buckingham JA, et al. , UCPs-unlikely calcium porters, Nat. Cell Biol 10 (2008) 1235–1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Chen Y, Liu J, Zheng Y, et al. , Uncoupling protein 3 mediates H2O2 preconditioning-afforded cardioprotection through the inhibition of MPTP opening, Cardiovasc. Res 105 (2015) 192–202. [DOI] [PubMed] [Google Scholar]
- [11].Hayashi T, Rizzuto R, Hajnoczky G, et al. , MAM: more than just a housekeeper, Trends Cell Biol. 19 (2009) 81–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Vafiadaki E, Sanoudou D, Arvanitis DA, et al. , Phospholamban interacts with HAX-1, a mitochondrial protein with anti-apoptotic function, J. Mol. Biol 367 (2007) 65–79. [DOI] [PubMed] [Google Scholar]
- [13].Zhao W, Waggoner JR, Zhang ZG, et al. , The anti-apoptotic protein HAX-1 is a regulator of cardiac function, Proc. Natl. Acad. Sci. U. S. A 106 (2009) 20776–20781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Suzuki Y, Demoliere C, Kitamura D, et al. , HAX-1, a novel intracellular protein, localized on mitochondria, directly associates with HS1, a substrate of Src family tyrosine kinases, J. Immunol 158 (1997) 2736–2744. [PubMed] [Google Scholar]
- [15].Yap SV, Vafiadaki E, Strong J, et al. , HAX-1: a multifaceted antiapoptotic protein localizing in the mitochondria and the sarcoplasmic reticulum of striated muscle cells, J. Mol. Cell Cardiol 48 (2010) 1266–1279. [DOI] [PubMed] [Google Scholar]
- [16].Hirasaka K, Lago CU, Kenaston MA, et al. , Identification of a redox-modulatory interaction between uncoupling protein 3 and thioredoxin 2 in the mitochondrial intermembrane space, Antioxid. Redox Signal 15 (2011) 2645–2661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Psarra AM, Hermann S, Panayotou G, et al. , Interaction of mitochondrial thioredoxin with glucocorticoid receptor and NF-kappaB modulates glucocorticoid receptor and NF-kappaB signalling in HEK-293 cells, Biochem. J 422 (2009) 521–531. [DOI] [PubMed] [Google Scholar]
- [18].Moncrief ND, Kretsinger RH, Goodman M, Evolution of EF-hand calcium-modulated proteins. I. Relationships based on amino acid sequences, J. Mol. Evol 30 (1990) 522–562. [DOI] [PubMed] [Google Scholar]
- [19].Kretsinger RH, Nockolds CE, Carp muscle calcium-binding protein. II. Structure determination and general description, J. Biol. Chem 248 (1973) 3313–3326. [PubMed] [Google Scholar]
- [20].Perocchi F, Gohil VM, Girgis HS, et al. , MICU1 encodes a mitochondrial EF hand protein required for Ca (2+) uptake, Nature 467 (2010) 291–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Kirberger M, Wang X, Deng H, Statistical analysis of structural characteristics of protein Ca2+-binding sites, J. Biol. Inorg. Chem 13 (2008) 1169–1181. [DOI] [PubMed] [Google Scholar]
- [22].Iwig JS, Vercoulen Y, Das R, et al. , Structural analysis of autoinhibition in the Ras-specific exchange factor RasGRP1, eLife 2 (2013) e00813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Bouchier-Hayes L, Oberst A, McStay GP, et al. , Characterization of cytoplasmic caspase-2 activation by induced proximity, Mol. Cell 35 (2009) 830–840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Chao JR, Parganas E, Boyd K, et al. , Hax1-mediated processing of HtrA2 by Parl allows survival of lymphocytes and neurons, Nature 452 (2008) 98–102. [DOI] [PubMed] [Google Scholar]
- [25].Han Y, Chen YS, Liu Z, et al. , Overexpression of HAX-1 protects cardiac myocytes from apoptosis through caspase-9 inhibition, Circ. Res 99 (2006) 415–423. [DOI] [PubMed] [Google Scholar]
- [26].Cilenti L, Soundarapandian MM, Kyriazis GA, et al. , Regulation of HAX-1 anti-apoptotic protein by Omi/HtrA2 protease during cell death, J. Biol. Chem 279 (2004) 50295–50301. [DOI] [PubMed] [Google Scholar]
- [27].Pinton P, Giorgi C, Siviero R, et al. , Calcium and apoptosis: ER-mitochondria Ca2+ transfer in the control of apoptosis, Oncogene 27 (2008) 6407–6418. [DOI] [PMC free article] [PubMed] [Google Scholar]


