SUMMARY
OBJECTIVE:
Obesity, which causes many serious diseases, is increasing exponentially in childhood across the world. Epigenetic changes, as well as genetics, play an important role in the process of adipogenesis. Therefore, we aimed to examine the expression levels of obesity-related MicroRNA-130b and MicroRNA-146b and the methylation status of hypoxia factor 3A and interleukin-6 genes associated with obesity in children.
METHODS:
This study was performed with 98 individuals (49 obese children and 49 controls) whose DNA was isolated from peripheral blood. Gene promoter methylations were analyzed by methylation-specific Polymerase chain reaction. In addition, expression levels of MicroRNAs were determined by quantitative real-time Polymerase chain reaction in 30 children (15 obese children and 15 controls).
RESULTS:
Methylation status of interleukin-6 gene was 93.9% in obese children (n=46/49) and 100% (n=49/49) in control group (p>0.05). There was no methylation for hypoxia factor 3A gene (p>0.05). As a result of the study, there was no statistically significant difference in terms of methylation status for hypoxia factor 3A and interleukin-6 genes in the obese group compared to the control group. However, we found that expression levels of MicroRNA-130b (p<0.01) and MicroRNA-146b (p<0.001) were higher in the obese group.
CONCLUSIONS:
Results support that MicroRNA-130b and MicroRNA-146b are potential biomarkers for the prevention and early diagnosis of obesity. This is the first study on childhood obesity in the Middle Black Sea region of Turkey. We believe that the results obtained by expanding the studies in our country and neighboring countries will be more decisive.
KEYWORDS: DNA methylation, Epigenomics, MicroRNA, Obesity
INTRODUCTION
Obesity is defined as abnormal or excessive fat accumulation that may impair health and cause morbidity and finally mortality by affecting many organs 1 . It is also an increasingly growing problem in childhood worldwide and it is a serious nutritional disorder that may lead to serious health problems in future if no measures are taken in early stage 2 . In 2016, approximately 40 million children under the age of 5, as well as 340 million children between the ages of 5 and 19 years, were affected by overweight or obesity 3 . According to COSI-TUR (Childhood Obesity Surveillance Initiative), the rate of overweight was 14.6% and that of obesity was 9.9% in primary school second-grade children in Turkey in 2016 4 . This rapid increase in obesity is not only due to genetic factors but also due to environmental factors that cause epigenetic effects 5 .
Hypoxia occurs due to the decrease in the amount of blood entering adipocytes with the growth of adipose tissue in obese subjects. Methylation of hypoxia factor 3A (HIF3A) gene is related to body mass index (BMI) and adiposity. It was also reported that obesity is associated with a low degree of chronic inflammation and increased interleukin-6 (IL-6) levels 5,6 . It is known that miRNAs play an important role in adipose tissue differentiation, proliferation, and lipid and glucose metabolism 7 . Identifying obesity-associated miRNAs that cause inadequate or overexpression of proteins in cells may be a therapeutic target for future treatment by using anti-miRNA oligonucleotides targeting these miRNAs. There are several other miRNAs associated with obesity; however, due to limited cost, we compared the expression levels of only two miRNAs in this study.
The tissue-specific epigenetic is subject to stunning changes during childhood development. A study has reported a correlation between methylation in adipose tissue and peripheral blood in adults, but it has not proved whether this holds true for children 7 . Therefore, we aimed to observe the methylation status of HIF3A and IL-6 genes and expression levels of miR-130b and miR-146b, which are thought to be associated with childhood obesity from peripheral blood.
As methylation is more tissue specific, it was necessary to use adipose tissue for most genes. In this study, blood sample was used in order to look for the methylation of obesity-related genes whose expression can be observed in the blood. Our study individuals involved adolescent in the Central Black Sea region and present previously unexplored data. It aims to support the data presented in previous studies in different populations. This is the first study to epigenetically examined HIF3A and IL-6 genes among Turkish obese children.
METHODS
Participants and peripheral blood collection
This study was carried out among children aged 6–12 years who were diagnosed with obesity (BMI>95th) before puberty period and who visited Ondokuz Mayıs University Faculty of Medicine Pediatric Endocrinology from the Middle Black Sea region of Turkey. A total of 98 children (49 obese children and 49 healthy controls) were selected to examine methylation status. The sample size was determined by statistical power analysis. To evaluate obese and control DNA methylations, 20 obese patients and 20 controls were selected, with 97% power to detect d=0.122 difference (with a maximum deviation of 0.11) at the 95% confidence interval. For miRNA expression analysis, a total RNA of 30 (15 obese subjects and 15 controls) samples were isolated from the whole peripheral blood samples of 98 volunteers. Again, 12 obese patients and 12 controls were calculated for 96% power to detect d=0.138 difference (with a maximum deviation of 0.15) at the 94% confidence interval to evaluate obese and control miRNA expression. This study was approved by Ethics Committee of the OMÜ KAEK (registration number 2016/301). In this study, chronic disease, chronic drug use, and endocrine, metabolic, or genetic short status were determined as exclusion criteria.
DNA isolation and bisulfite conversion and methylation-specific polymerase chain reaction
DNA was isolated from peripheral blood samples using Pure Link® Genomic DNA Mini Kit (Invitrogen, USA). EpiJET Bisulfite Conversion Kit (Thermo Fisher Scientific, Lithuania) was used for bisulfite modification of DNA. Specific primers for methylation-specific PCR analysis were designed with the “MethPrimer” database (Table 1). DreamTaq™ Hot Start DNA polymerase was used (Thermo Fisher Scientific). A 25-μl total volume of PCR reaction conditions was as follows:
Table 1. MSP sequences, hybridization temperatures, and expected amplicon sizes.
| Primer name | Base sequence 5′-3′ | Base number | Temp. (°C) | Amplicon size (bp) |
|---|---|---|---|---|
| IL-6 MSP-F | ATAGGTAAGATATTAGGTGAATCGA | 25 | 56 | 166 |
| IL-6 MSP-R | TTTCTAAAACTATTATAAAAATAAAACGTA | 30 | 53 | 166 |
| IL-6 USP-F | ATAGGTAAGATATTAGGTGAATTGA | 25 | 55 | 166 |
| IL-6 USP-R | TTTCTAAAACTATTATAAAAATAAAACATA | 30 | 52 | 166 |
| HIF-3A MSP-F | TAGGTTTGGCGTGGTATAGTTAATC | 25 | 60 | 259 |
| HIF-3A MSP-R | CCCGAAACGTTCTTAACTCG | 20 | 57 | 259 |
| HIF-3A USP-F | TTTAGGTTTGGTGTGGTATAGTTAATTG | 28 | 59 | 262 |
| HIF-3A USP-R | CCCCAAAACATTCTTAACTCAC | 22 | 57 | 262 |
HIF-3A: hypoxia factor 3A; MSP: methylation-specific primer; USP: unmethylated-specific primer; F: forward; R: reverse; IL-6: interleukin-6. Primers designed with the “MethPrimer” database http://www.urogene.org/cgi-bin/methprimer/methprimer.cgi.
For IL-6: initial denaturation was at 97°C for 5 min, followed by 40 cycles of 96°C for 45 s, 56°C for 45 s, and 72°C for 60 s. The final extension was at 72°C for 7 min.
For HIF3A: initial denaturation was at 98°C for 10 min, followed by 40 cycles of 97°C for 45 s, 60°C for 45 s, and 72°C for 70 s. The final extension was at 72°C for 7 min.
Agarose gel electrophoresis
A 2% of micropore (Prona, Nu micropor) agarose gel was prepared to display the MSP product. A 50-bp DNA marker (Thermo Fisher Scientific, Lithuania) was used as the marker and gel and viewed using UV transilluminator.
RNA extraction and quantitative real-time polymerase chain reaction
Total RNA was isolated from whole peripheral blood samples using Quick-RNA™ Whole Blood kit (Zymo Research, USA). Total RNA was reverse transcribed to cDNA using Ipsogen® RT Kit 33, V1 kit (QIAGEN, Germany). The reaction mixture was 15 μl. Incubation conditions were as follows: 16°C for 30 min, 42°C for 30 min, and 85°C for 5 min (Thermal Cycler GeneAmp PCR System 9700, Applied Biosystems, USA). For miR-146b and miR-130b qRT-PCR analysis, Taqman™ MicroRNA Assay (Applied Biosystems) and Premix Ex Taq™ master mix (Perfect Real Time, Takara, Japan) were used. RNA was normalized using the ABL housekeeping gene (İpsogen, BCR-ABL1 Mbcr IS-MMR) in each sample 8 . Using standards with a known number of molecules, one can establish a standard curve and determine the amount of target present in the test sample. To ensure accurate standard curves, we use four standard dilutions for ABL. The kit also includes an IS-MMR calibrator allowing conversion of results to the international scale. For ABL gene, raw CT values obtained from plasmid standard dilutions are plotted according to the log copy number. We compare the ABL gene with known copy number in the kit with our miRNAs whose copy number is unknown, and we obtain information about the copy number. We compare our ABL-normalized obese and control miRNAs with each other. As a result, with CT values, we obtained information about copy numbers increase or decrease of obese individuals compared to controls.
Rotor-gene Q 5 PLEX HRM (Germany, USA) device used for qRT-PCR. The miR-130b microRNA sequence is ACUCUUUCCCUGUUGCACUAC, while miR-146b microRNA sequence is UGAGAACUGAAUUCCAUAGGCUG (http://www.mirbase.org). The relative abundance of each miRNA transcript in each sample was determined using the comparative 2−ΔΔCt method and an endogenous housekeeping control gene. Cycle threshold (CT) values of participants (obese and control) and increase or decrease ratio of expression levels in miR-146b-5p/ABL and miR-130b/ABL were calculated according to the Livak’s formula 9 :
Statistical analysis
The relationship between parameters was evaluated using Fisher’s exact test using OpenEpi version 3.01 (last updated: May 6, 2013). SPSS software was used for data analyzing (version 22.0, SPSS Inc., Chicago, IL, USA). The statistical significance of miRNA expression was analyzed by the Student’s t-test, considering statistically significant at p<0.05.
RESULTS
Hypoxia factor 3A and interleukin-6 promoter methylation analysis results
The mean age of 49 obese groups (22 females, 27 males) whose methylation profile was analyzed was 10.0±1.9 years, and the mean age of 49 healthy controls (29 females, 20 males) was 9.2±2.0 years. HIF3A gene methylation was not detected in either group. IL-6 gene methylation was detected in 93.9% (46/49, p>0.05) of the obese group and 100% (49/49, p>0.05) of the control group. According to results of the tests, there was no statistical significance between the patient and control groups in terms of the promoter methylation profile of HIF-3A and IL-6 genes (p>0.05, Table 2).
Table 2. Distribution of hypoxia factor 3A and interleukin-6 genes promoter methylation profiles in the obese and control groups (obese n=49, control n=49).
| Gene | Group | Number (n) | Methylated (%) | Unmethylated (%) | p-value |
|---|---|---|---|---|---|
| HIF3A | Obese | 49 | 0 | 49 (100) | >0.05* |
| HIF3A | Control | 49 | 0 | 49(100) | |
| IL-6 | Obese | 49 | 46 (93.9) | 3 (6.1) | >0.05* |
| IL-6 | Control | 49 | 49 (100) | 0 |
HIF-3A: hypoxia factor 3A; IL-6: interleukin-6.
Statistically significant.
MicroRNA-146b and microRNA-130b quantitative real-time-polymerase chain reaction analysis results
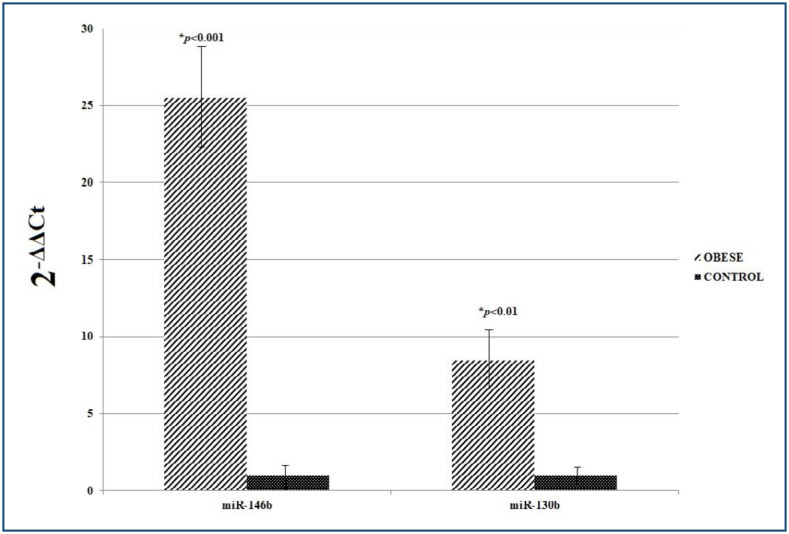
The expression of obese miR-146b according to the control group was increased in 14 of 15 children, with an average 25.5±17.3-fold (p<0.001). Similarly, the expression of miR-130b increased in 14 of 15 obese children, with an average 8.5±5.3-fold (p<0.01).
The qRT-PCR analysis results of the expression levels of miR-146b and miR-130b in obese and control groups were given in Table 3 and Figure 1.
Table 3. Expression levels of MicroRNA-130b and MicroRNA-146b in the obese and control groups.
| CT | ABL CT | ΔCT (Avg. miR CT−Avg. ABL CT) | ΔΔCT (Avg. ΔCT obese−Avg. ΔCT control) | Normalized miR Amount relative to control2−ΔΔCt | p-value | ||
|---|---|---|---|---|---|---|---|
| MicroRNA-130b | Obese | 26.5 | 20.6 | 5.8 | -3.1 | 8.5* | 0.008556 (*p<0.01) |
| Control | 31.4 | 22.4 | 8.9 | 0 | 1 | ||
| MicroRNA-146b | Obese | 20.9 | 19.1 | 1.7 | -4.6 | 25.5* | 0.000001 (*p<0.001) |
| Control | 25.5 | 19.0 | 6.4 | 0 | 1 |
CT: cycle threshold; ABL: abelson tyrosine-protein kinase.
Statistically significant.
Figure 1. Graphical representation of 2−ΔΔCt averages for the obese and control groups.
DISCUSSION
One of the most studied mechanisms as an epigenetic determinant of childhood obesity is DNA methylation 2 . In many studies, there was evidence showing that the HIF3A gene is associated with anthropometric parameters in childhood 2 . Previous studies have reported that methylation of the HIF3A gene is associated with BMI and adiposity 6,10 . In our study, we observed that there was no significant difference in methylation status of HIF3A genes in the obese group compared to the control group (p<0.01). The temporal sequence between epigenetic changes and the onset of childhood obesity is uncertain because epigenetics may be altered by a wide range of stimuli, including metabolic changes associated with obesity itself. Most of the studies included in the review use a cross-sectional design that makes it impossible to decipher the temporal order of events. Some DNA methylation studies have used longitudinal designs or statistical techniques such as Mendelian randomization to resolve this relationship 2 . In the ALSPAC (avon longitudinal study of parents and children) cohort, childhood BMI was associated with methylation at HIF3A gene in adolescence, but childhood methylation was not robustly associated with BMI in adolescence, and based on Mendelian randomization, HIF3A methylation did not play a causal role on BMI 2 . The children participating in our study were in pre-adolescent, so we may not have observed HIF3A gene methylation in our study.
Cytokines production causes an increase in white adipose tissue in obesity subjects 5 . In our study, there was no significant difference in methylation status of IL-6 gene in the obese group compared to the control group (p<0.01), similar to results obtained by Zhang et al. 11 . Na et al. 12 reported that IL-6 methylation can be used as a molecular biomarker for the assessment of obesity risk, finding which is in contrast to our results. The mechanism that can explain the increased IL-6 levels in obese can be attributed to the ability of fat tissue to produce and secrete IL-6 13 . In our study, DNA methylation may not reflect the actual methylation profile since DNA methylation was analyzed from peripheral blood, not directly from the affected adipose tissue. miRNAs, which are considered to be epigenetically important in regulating the expression of genes, play an active role in insulin secretion, cholesterol biosynthesis, fat metabolism, and adipogenesis.
In our study, miRNA profiles were examined in 30 children (15 obese and 15 control) whose proficiency was supported by power analysis due to project budget limitations. However, circulating mir-130b and mir-146b expression levels were found to be high in obese children, although the number of samples studied was limited. Mansego et al. 14 in 24 children (12 obese children and 12 controls) and Quyang et al. 15 in a total of 12 children (6 obese children and 6 controls) reported that the miRNA profiles were associated with early childhood obesity from peripheral blood.
Previous study showed that miR-130b expression in abdominal subcutaneous adipose tissue and plasma was low in obese. Lacomino and Siani 16 concluded that the level of miR-130b in the blood was found to be higher in obese children, as in our study (p<0.01). Wang et al. 17 determined miR-130b as a potential biomarker for overweight, hypertriglyceridemia, and metabolic syndrome. TGF-β signaling pathway is significant in the arrangement of energy homeostasis and studies suggest that miR-130b secreted from adipose tissues mediate the metabolic regulatory effect of TGF-β and TGF-β can stimulate miR-130b secretion from adipocytes 17 . This explains the 8.5±5.3-fold increase in miR-130b expression in the obese group in our study. Prtas-Puig et al. 18 proved that circulating miRNAs were unregulated in prepubescent obese children, which is similar to our results. Therefore, early detection of abnormal miRNA profile will be useful in the early diagnosis of obesity.
In addition, miR-146b expressed during adipogenic differentiation and its level increased in overweight, obese, and high fat diet mice 19 . The fact that the miR-146b expression level of obese individuals was 25.5±17.3-fold increase higher than the control group in our study supports this situation (p<0.001). Overexpression of Kruppel-like factor 7 (KLF7) inhibits expression of some adipogenic transcription factor genes and suppresses adipogenesis 19 . The inhibitory effects of miR-146b on KLF7 expression and experimentally demonstrated that KLF7 is the direct target of miR-146b 19 . Therefore, increased mir-146b expression in obesity supports adipogenesis indirectly. Sharma et al. 20 confirmed that the expression of miR-146a and miR-146b significantly correlated with BMI in adipose tissues. Studies indicate that miR-146b can make significant contributions to obesity formation in children 21 . Also, obese mice who were administered anti-miR-146b for 3 days to destroy miR-146b have shown losing weight 6 . These results indicate that miR-146b may be a potential target for the treatment of obesity.
CONCLUSION
The role of epigenetic pathways in the prevention and treatment of increasingly common obesity is one of the current research areas. More studies are needed in both larger and different populations to support the results.
ACKNOWLEDGMENT
This study was supported by Scientific Research Project Office of Ondokuz Mayıs University, Turkey (project no: PYO.TIP.1904.18.003).
Footnotes
Funding: Scientific Research Project Office of Ondokuz Mayıs University, Turkey (project no: PYO.TIP.1904.18.003).
REFERENCES
- 1.Gallardo-Escribano C, Buonaiuto V, Ruiz-Moreno MI, Vargas-Candela A, Vilches-Perez A, Benitez-Porres J, et al. Epigenetic approach in obesity: DNA methylation in a prepubertal population which underwent a lifestyle modification. Clin Epigenetics. 2020;12(1):144–144. doi: 10.1186/s13148-020-00935-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alfano R, Robinson O, Handakas E, Nawrot TS, Vineis P, Plusquin M. Perspectives and challenges of epigenetic determinants of childhood obesity: A systematic review. Obes Rev. 2022;23(Suppl 1):e13389. doi: 10.1111/obr.13389. [DOI] [PubMed] [Google Scholar]
- 3.Apperley LJ, Blackburn J, Erlandson-Parry K, Gait L, Laing P, Senniappan S. Childhood obesity: A review of current and future management options. Clin Endocrinol (Oxf) 2022 Mar;96(3):288–301. doi: 10.1111/cen.14625. [DOI] [PubMed] [Google Scholar]
- 4.Yılmazbaş P, Gökçay G. Childhood obesity and its prevention. J Child. 2018;18(3):103–112. doi: 10.5222/j.child.2018.59389. [DOI] [Google Scholar]
- 5.Cerdó T, García-Santos JA, Bermúdez MG, Campoy C. The role of probiotics and prebiotics in the prevention and treatment of obesity. Nutrients. 2019;11(3):635–635. doi: 10.3390/nu11030635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang S, Song J, Yang Y, Zhang Y, Wang H, Ma J. HIF3A DNA methylation is associated with childhood obesity and ALT. PLoS One. 2015;10(12):e0145944. doi: 10.1371/journal.pone.0145944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McGregor RA, Choi MS. microRNAs in the regulation of adipogenesis and obesity. Curr Mol Med. 2011;11(4):304–316. doi: 10.2174/156652411795677990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tuncer SB, Akdeniz D, Celik B, Kilic S, Sukruoglu O, Avsar M, et al. The expression levels of miRNA-15a and miRNA-16-1 in circulating tumor cells of patients with diffuse large B-cell lymphoma. Mol Biol Rep. 2019;46(1):975–980. doi: 10.1007/s11033-018-4554-4. [DOI] [PubMed] [Google Scholar]
- 9.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 10.Lee S, Kim HJ, Han S, Jeon JP, Park SI, Yu HY, et al. Positive correlation of cg16672562 methylation with obesity-related traits in childhood obesity, and its independence with underlying HIF3A (hypoxia-inducible factor 3a) genetic background. Oncotarget. 2017;8(40):67473–81. doi: 10.18632/oncotarget.18707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang JF, Fu WM, He ML, Wang H, Wang WM, Yu SC, et al. MiR-637 maintains the balance between adipocytes and osteoblasts by directly targeting Osterix. Mol Biol Cell. 2011;22(21):3955–3961. doi: 10.1091/mbc.E11-04-0356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Na YK, Hong HS, Lee WK, Kim YH, Kim DS. Increased methylation of interleukin 6 gene is associated with obesity in Korean women. Mol Cells. 2015;38(5):452–456. doi: 10.14348/molcells.2015.0005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wisse BE. The inflammatory syndrome: the role of adipose tissue cytokines in metabolic disorders linked to obesity. J Am Soc Nephrol. 2004;15(11):2792–2800. doi: 10.1097/01.ASN.0000141966.69934.21. [DOI] [PubMed] [Google Scholar]
- 14.Mansego ML, Garcia-Lacarte M, Milagro FI, Marti A, Martinez JA; GENOI members DNA methylation of miRNA coding sequences putatively associated with childhood obesity. Pediatr Obes. 2017;12(1):19–27. doi: 10.1111/ijpo.12101. [DOI] [PubMed] [Google Scholar]
- 15.Ouyang S, Tang R, Liu Z, Ma F, Li Y, Wu J. Characterization and predicted role of microRNA expression profiles associated with early childhood obesity. Mol Med Rep. 2017;16(4):3799–3806. doi: 10.3892/mmr.2017.7118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Iacomino G, Siani A. Role of microRNAs in obesity and obesity-related diseases. Genes Nutr. 2017;12:23–23. doi: 10.1186/s12263-017-0577-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang YC, Li Y, Wang XY, Zhang D, Zhang H, Wu Q, et al. Circulating miR-130b mediates metabolic crosstalk between fat and muscle in overweight/obesity. Diabetologia. 2013;56(10):2275–2285. doi: 10.1007/s00125-013-2996-8. [DOI] [PubMed] [Google Scholar]
- 18.Prats-Puig A, Ortega FJ, Mercader JM, Moreno-Navarrete JM, Moreno M, Bonet N, et al. Changes in circulating microRNAs are associated with childhood obesity. J Clin Endocrinol Metab. 2013;98(10):E1655–E1660. doi: 10.1210/jc.2013-1496. [DOI] [PubMed] [Google Scholar]
- 19.Cheng Z, Zheng L, Almeida FA. Epigenetic reprogramming in metabolic disorders: nutritional factors and beyond. J Nutr Biochem. 2018;54:1–10. doi: 10.1016/j.jnutbio.2017.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sharma NK, Varma V, Ma L, Hasstedt SJ, Das SK. Obesity associated modulation of miRNA and Co-regulated target transcripts in human adipose tissue of non-diabetic subjects. Microrna. 2015;4(3):194–204. doi: 10.2174/2211536604666151103121817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ahn J, Lee H, Jung CH, Jeon TI, Ha TY. MicroRNA-146b promotes adipogenesis by suppressing the SIRT1-FOXO1 cascade. EMBO Mol Med. 2013;5(10):1602–1612. doi: 10.1002/emmm.201302647. [DOI] [PMC free article] [PubMed] [Google Scholar]