SUMMARY
OBJECTIVE:
Chronic kidney disease (CKD) remains one of the major common health problems, and the number of people affected by the disease is progressively increasing in Turkey and worldwide. This study aimed to investigate molecular defects in Alport syndrome (AS) and other genes in patients with clinically suspected CKD using whole-exome sequencing (WES).
METHODS:
Patients with clinical suspicion of CKD were included in the study. Molecular genetic analyses were performed on genomic DNA by using WES.
RESULTS:
A total of 15 with 5 different pathogenic or likely pathogenic variants were identified in CKD patients, with a diagnostic rate of 30%. Eight variants of uncertain significance were also detected. In this study, 10 variants were described for the first time. As a result, we detected variants associated with CKD in our study population and found AS as the most common CKD after other related kidney diseases.
CONCLUSIONS:
Our results suggest that in heterogeneous diseases such as CKD, WES analysis enables accurate identification of underlying molecular defects promptly. Although CKD accounts for 10–14% of all renal dysfunction, molecular genetic diagnosis is necessary for optimal long-term treatment, prognosis, and effective genetic counseling.
KEYWORDS: Chronic kidney disease, Alport syndrome, Variant, Whole-exome sequencing
INTRODUCTION
Chronic kidney disease (CKD) is a major public health problem, with increasing incidence and prevalence 1 and affecting more than 10% of people worldwide 2 . CKD is associated with disease enclosing many disorders such as metabolic and end-stage renal failure and is a leading cause of death 3 , showing one of the highest increases in mortality for 10 years 4 . However, it is generally not recognized by people and specialists 5 . Moreover, clinical trials in nephrology are still insufficient for diagnosis, which has limited therapeutic options to alter disease progression and high costs for the health system 6 .
The description of hereditary (genetic) risk factors for CKD can enable early detection and increase our understanding of the pathogenesis of the disease. Genetic factors include both Mendelian (monogenic) and polygenic risk. Recent clinical sequencing studies have detected diagnostic variants in 10–25% of patients of various CKD populations, suggesting a high burden of Mendelian disorders among patients with nephropathy 7 .
Alport syndrome (AS) is the most common CKD after other related kidney diseases 8 . Recent and comprehensive studies have reported that most patients with CKD have variants in AS-related genes 9 . AS is a well-defined genetic disorder characterized by kidney failure, hematuria, hearing impairment, and vision abnormalities 10 and it is one of the hereditary causes of nephropathies and can genetically be transmitted from one generation to the next as a recessive dominant or X-linked 8 because of variants in the COL4A3, COL4A4, and COL4A5 genes, which encode, respectively, α3, α4, and α5 chains of collagen type IV that consist of glomerular basement membrane in the kidney 10 . With the development of technologies such as massively parallel sequencing, it is possible to treat complex diseases that are difficult to diagnose more cheaply and precisely 9 . Our study aimed to detect variant in AS genes in CKD and contribute new variants to the literature.
METHODS
A total of 50 unrelated patients with a clinical CKD were included in the study after obtaining informed consent from the patients and their parents for medical examination and genomic analysis. The procedures adhered to the Declaration of Helsinki, and the Ethics Committee approved the relevant studies of the Basaksehir Cam and Sakura City Hospital (E-96317027-000-8273 2021.03.02/KAEK). All the patients met the generally accepted clinical diagnostic criteria for abnormalities of kidney structure or function present more than 3 months.
Genomic DNA was extracted from peripheral blood, and whole-exome sequencing (WES) was performed by capturing the coding regions and splice sites of target genes (101 genes) via the Twist Human Comprehensive Exome kit panel. After library enrichment and quality control, the samples were sequenced on the BGI platform (Shenzhen, China) with 100-bp paired-end reads at an average sequencing depth of ×100. Annotation of detected variants was performed using www.genomize.com.tr (version 6.14.4), interval, Franklin, VarSome, ClinVar, OMIM, and Pubmed. Variants with a frequency higher than 0.1% were filtered out. dbNSFP (contains MetaLR, MetaSVM, MetaRNN, REVEL, SIFT, PolyPhen-2, LRT, Mutation Taster 4.2 versions) was used to predict variants’ pathogenicity (deleteriousness). Detected variants were classified as “pathogenic,” “likely pathogenic (LP),” or “variants of uncertain significance (VUS)” according to the international guidelines of the ACMG. These variants are listed in Tables 1 and 2.
Table 1. Characteristics of detected variations.
| P | G | Age | Gene | Variant | Variant type | Zygosity | ACMG classification | Patient frequency | Novelty |
|---|---|---|---|---|---|---|---|---|---|
| P1 | F | 37 | PKD2 | c.872T>C (p.Leu291Pro) | Missense mutation | Het | LP | N/A | Novel |
| P2 | M | 6 | LMX1B | c.237G>T (p.Glu79Asp) | Missense mutation | Het | P | N/A | Novel |
| P4 | F | 9 | MEFV | c.2080A>G (p.Met694Val) c.442G>C (p.Glu148Gln) |
Missense mutation Missense mutation |
Het Het |
P B |
ƒ = 0.000282 ƒ = 0.0711 |
rs61752717 rs3743930 |
| P5 | M | 2 | FN1 | c.4151T>C (p.Ile1384Thr) | Missense mutation | Het | V | ƒ = 0.0000159 | rs768047478 |
| P8 | M | 0 | PLCE1 | c.5410_5411del (p.Phe1804LeufsTer15) | Small deletion (frame shift) | Homo | P | N/A | Novel |
| P9 | M | 11 | COQ8B | c.1027C>T (p.Arg343Trp) | Missense mutation | Homo | LP | ƒ = 0.000004 | rs398122981 |
| P10 | F | 51 | PKD1 | c.7666C>T (p.Gln2556Ter) | Nonsense mutation | Het | P | N/A | rs1567184366 |
| P12 | M | 38 | PKD1 | c.8302G>A (p.Val2768Met) | Missense mutation | Het | P | ƒ = 0.0000081 | rs1456510041 |
| P13 | F | 6 | PKD1 | c.2308C>T (p.Arg770Trp) | Missense mutation | Het | V | ƒ = 0.0000147 | rs771288493 |
| P15 | M | 29 | PKD1 | c.6260_6263dup (p.Arg2089ProfsTer20) | Small insertion (frame shift) | Het | P | N/A | Novel |
| P17 | F | 58 | SLC34A1 | c.1325C>T (p.Pro442Leu) | Missense mutation | Het | V | ƒ = 0.0000159 | rs760010857 |
| P18 | F | 21 | PKD1 | c.7401del (p.Asn2468ThrfsTer152) | Small deletion (frame shift) | Het | P | N/A | Novel |
| P19 | F | 32 | PKD1 | c.4168C>T (p.Gln1390Ter) | Nonsense mutation | Het | P | N/A | Novel |
| P23 | M | 9 | CD2AP | c.902A>T (p.Lys301Met) | Missense mutation | Het | P | ƒ = 0.000191 | rs141778404 |
| P24 | M | 57 | PKD1 | c.3877G>A (p.Val1293Ile) | Missense mutation | Het | V | ƒ = 0.0000168 | rs774353552 |
P: patient; G: gender; F: female; M: male; N/A: not available. The genes, variant types, zygosities, pathogenicity, frequency, and database numbers of the detected variations were determined according to the ACMG guideline. Six variations were detected, which were novels.
Table 2. Clinical characteristics of the patients and detected Alport syndrome genes.
| P | G | Age | Clinical or pathological diagnosis before WES-based analysis | Spot urine protein/creatinine ratio (0–150 mg/g) | Serum albumin (35–52 g/L) | Serum creatinine (0.5–0.9 mg/dL) | Serum globulin (26–46 g/L) | Alport gene | Variant | Variant type | Zygosity | ACMG classification | Patient frequency | Novelty |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| P3 | F | 7 | SRNS | 190↑ | 25↓ | 0.51 | 17↓ | COL4A3 | c.3829G>A (p.Gly1277Ser) | Missense mutation | Het | P | ƒ = 0.000377 | rs190598500 |
| P7 | F | 17 | Pancytopenia, bleeding diathesis, NS | 178↑ | 34↓ | 0.43↓ | 23↓ | COL4A3 | c.4421T>C (p.Leu1474Pro) | Missense mutation | Het | P | ƒ = 0.0027 | rs200302125 |
| P11 | F | 9 | SRNS, FSGS | 4137↑ | 34↓ | 0.68↑ | 22↓ | COL4A3 | c.4153+1del | Splicing deletion | Homo | LP | N/A | Novel |
| P14 | F | 0 | NS | 130 | 26↓ | 1.48↑ | 15↓ | COL4A3 | c.4123C>T (p.Pro1375Ser) | Missense mutation | Het | LP | N/A | Novel |
| P22 | F | 10 | NS | 151↑ | 46 | 0.38↓ | 21↓ | COL4A3 | c.3182G>A (p.Gly1061Asp) | Missense mutation | Het | VUS | ƒ = 0.000197 | rs202078295 |
| P20 | M | 2 | FSGS, NS | 38157↑ | 23↓ | 0.19↓ | 20↓ | COL4A4 | c.665C>T (p.Pro222Leu) | Missense mutation | Het | VUS | ƒ = 0.000122 | rs773533313 |
| P6 | F | 5 | SRNS | 8812↑ | 38 | 0.18↓ | 21↓ | COL4A5 | c.4222A>G (p.Thr1408Ala) | Missense mutation | Het | VUS | N/A | Novel |
| P21 | F | 6 | NS | 7664↑ | 25↓ | 0.38↓ | 19↓ | COL4A5 | c.1061C>T (p.Thr354Ile) | Missense mutation | Het | VUS | N/A | Novel |
P: patient; G: gender; WES: Whole-Exome Sequencing; F: female; M: male; N/A: not available; SRNS: steroid-resistant nephrotic syndrome; NS: nephrotic syndrome; FSGS: focal segmental glomerulosclerosis. The genotype-phenotype correlation of the patients carrying the AS genes in which the variations were detected the most was calculated, and the damaging effects were examined according to the laboratory findings.
RESULTS
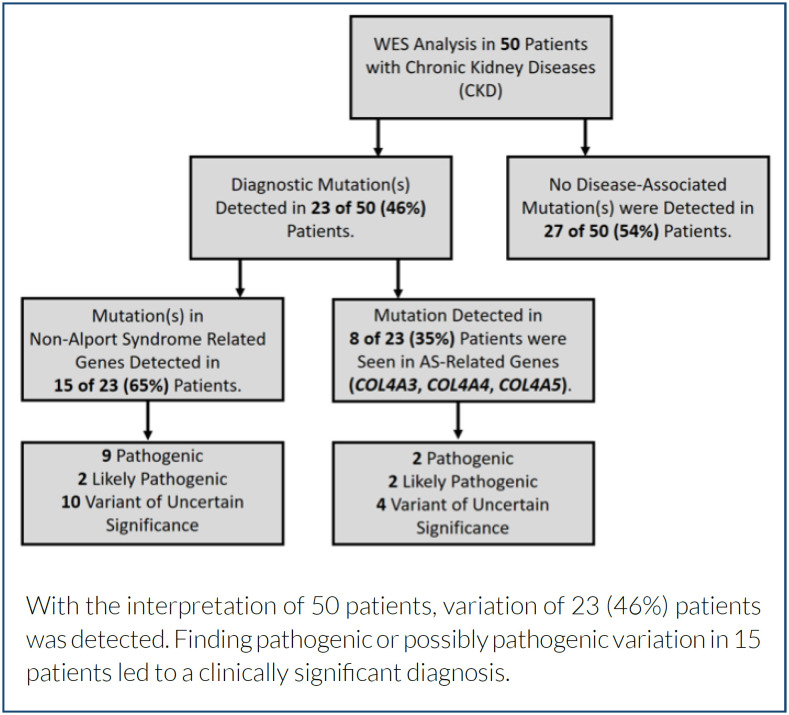
When 23 of 50 patients who underwent WES due to CKD were evaluated together, it was seen that 15 (65%) patients were older than 17 years and 8 (35%) were younger than 17 years (Tables 1 and 2). Of the 23 detected variants, 10 are novel types. Variants in the genes COL4A3 (c.4153+1del, c.4123C>T, c.4222A>G, c.1061C>T), PKD1 (c.6260_6263dup, c.7401del, c.4168C>T), PKD2 (c.872T>C), LMX1B (c.237G>T), and PLCE1 (c.5410_5411del) were described for the first time in this report (Tables 1 and 2). The genes with variants in 23 patients were COL4A3, COL4A4, COL4A5, COQ8B, FN1, LMX1B, MEFV, PKD1, PKD2, PLCE1, and SLC34A1. Variants were found in PKD1 in 7 of 23 patients, in COL4A3 in 5, in COL4A4 in 1, and in COL4A5 in 2 patients.
Within the scope of the study, the WES results of 50 patients diagnosed with CKD were evaluated. Pathogenic/LP variants were detected in 15 (30%) of 50 patients following ACMG criteria (Figure 1). Of these 15 patients, 4 (27%) had pathogenic/LP variants in genes other than AS genes (COL4A3, COL4A4, or COL4A5). Variants in genes associated with AS were detected in 8 (35%) of 23 patients (Table 2).
Figure 1. Flowchart of the Whole-Exome Sequencing analysis.
Variants in the AS-related genes COL4A3, COL4A4, and COL4A5 were found in 8 (35%) of 23 CKD patients who underwent WES (Table 2). Variants were detected in the COL4A3 gene in five of these eight patients, in the COL4A4 gene in one, and in the COL4A5 gene in two patients. One of the novel variants detected in the COL4A3 gene of two patients was homozygous and the other heterozygous, and both were not previously reported in the literature. These two novel variants were classified as LP according to the ACMG. Both heterozygous novel variants in the COL4A5 genes of two patients were not previously reported in the literature. These variants were classified as VUS, according to the ACMG. Heterozygous variant in the COL4A4 gene was detected in only one patient. The protein/creatinine ratio in the spot urine of patients with variants in the COL4A4 and COL4A5 genes was significantly higher than in patients with variants in the COL4A3 gene.
Variants were found in genes other than the AS-related genes in 15 (65%) of 23 patients who underwent WES due to CKD and were found to have the variants (Table 1).
DISCUSSION
Frequently, it is clinically challenging to distinguish CKD type. In general, CKD diagnosis can be made based on morphological, pathological, and genetic examination after complaints such as persistent alterations in kidney structure or function, filtration disorders, proteinuria, and hematuria 11 . The prevalence of all stages of CKD in studies varies between 7 and 12% in different regions of the world. Therefore, multiple patients with this monogenic form of CKD genetic testing emerge as an important tool in determining the cause of CKD, especially in children and young adults 12 . To date, 101 genes thought to be responsible for the CKD phenotype have been identified in the HPO database. Some gene groups responsible for the CKD phenotype stand out in the studies. Studies have shown that patients diagnosed with CKD have pathogenic variants, mostly in AS and polycystic disease (PD) genes 13 . The prominent gene groups in our study are genes related to AS and PD. Because of this, we focused on patients with detected variants in AS genes. In this study, the rate of patients diagnosed with AS gene was 35%. Furthermore, the frequency of disease-causing variants in CKD is likely to differ between different ethnic groups depending on age, gender, and the molecular analysis methods used for diagnosis 14 . Many CKD-associated genes have yet to be identified, and it is believed that unidentified CKD genes may play a role in populations with low detection rates 15 .
According to the U.S. Renal Data System (USRDS, https://www.usrds.org/), approximately 0.2% of adults and 3% of children with end-stage renal disease (ESRD) in the United States are diagnosed with AS. In this study, AS genes were found to be higher in patients with CKD and variant, with a rate of 35%. Also extensive studies, significant findings were observed at the rate of 24% as a diagnostic value 9 , but this rate was higher than the literature in our study (30% rate). In our study, dominant inheritance was more common than recessive and X-linked inheritance.
WES studies make it possible to identify new genes and detect significant variants from defined genes by rapidly and simultaneously examining genes related to monogenic CKD; the number of patients diagnosed with monogenic CKD can be expected to increase rapidly. In this study, we identified a possible pathogenic/pathogenic (i.e., high-risk) CKD-associated variant in 30% of our patients and provided a comprehensive description of these patients’ clinical and variational characteristics. A total of 15 variants in 23 patients were of the high-risk type, 5 (33.3%) patients had PKD1 variant, 4 (26.6%) had AS genes variant, and other genes (40.1%) had variant. For physicians, the most current challenge in the molecular genetic diagnosis of CKD is the evaluation of VUS, which perhaps represents a barrier to clinical interpretation. As a result, disaggregation analysis studies on family members will be particularly useful and, in some cases, even vital. In addition, extensive in silico analyses of the structural and functional impact on the protein product may be useful for some. We believe that our cases will contribute to developing the scientific literature on CKD. Clinically, there is no difference between patients with renal impairment (patients with VUS or no variant) and patients with confirmed CKD (i.e., patients with a probable pathogenic/pathogenic variant). However, among carriers of the AS gene variant, individuals with recessive inheritance and the COL4A5 variant had a higher protein/creatinine ratio (Table 2).
This study showed that for most patients, genetic diagnoses could be made clinically for patients. In this study, we manifest that our results highlight the potential of genetic findings to change the diagnostic value of consultation. For example, eight patients with the most detected variants of COL4A3, COL4A4, or COL4A5 did not have clinical diagnoses of classically associated with AS. Although these patients do not have eye and ear nose and throat findings, due to the detection of variants, patients with clinical nephropathy variant are diagnosed and treated according to the variant type 16 . In addition, it was determined how effective the variants detected in AS-related genes were in the clinic shown in Table 2. Consistent with recent studies, we identified autosomal and X-linked forms of AS among patients with a clinical diagnosis of kidney disease, supporting the variable phenotypic expression of variants in type IV collagen genes 9,17 .
To make a clinical diagnosis in nephrology, the need to “revise the disease ontology based on molecular classifiers” has been emphasized in some publications 18 . Our findings support the diagnostic utility of exome sequencing among the different clinical categories of kidney disease, the most detecting variant in AS-related genes. Therefore, looking at AS genes first in nephropathy patients in low-budget laboratories is recommended. It also highlights the potential of genetic testing to guide treatments for diseases accurately.
Large databases of controls, such as ExAC and gnomAD 19 , are of great value for interpreting detected variants and deciding clinical relevance based on their frequency in the population. New variant annotation algorithms that take into account genomic context to assess variant intolerance can facilitate variant classification in patients independent of previous clinical reports 20 and help standardize disease association interpretation. Therefore, many initiatives are currently examining the clinical relevance of genes and variants for diagnosing diseases (e.g., ClinGen (www.clinicalgenome.org) and will provide further evidence of the usefulness of genetic testing in various clinical settings 21 . When the novel variants detected in our study are analyzed according to population frequencies, and in silico analysis, it predicts that protein function may be impaired.
CONCLUSIONS
Large-scale genetic studies are needed to understand the genetic aspects of CKD in Turkey and other populations. Observational studies of CKD often reveal seemingly conflicting relationships between traditional risk factors and outcomes, making it difficult to predict outcomes in studies of CKD patients with normal kidney function. However, many completed CKD studies are limited by tentative results of uncertain clinical relevance or narrow eligibility criteria that limit external validity, and the implementation of proven treatments remains a challenge. Therefore, the nephrology community should capitalize on the recent interest in new approaches to experimental design, such as practical clinical trials 22 . Therefore, genetic approaches are important for treatment and diagnosis. Our study has shown that the most common CKD subtype is AS.
Footnotes
Funding: none.
REFERENCES
- 1.Eckardt KU, Coresh J, Devuyst O, Johnson RJ, Köttgen A, Levey AS, et al. Evolving importance of kidney disease: from subspecialty to global health burden. Lancet. 2013;382(9887):158–169. doi: 10.1016/S0140-6736(13)60439-0. [DOI] [PubMed] [Google Scholar]
- 2.Levin A, Tonelli M, Bonventre J, Coresh J, Donner JA, Fogo AB, et al. Global kidney health 2017 and beyond: a roadmap for closing gaps in care, research, and policy. Lancet. 2017;390(10105):1888–1917. doi: 10.1016/S0140-6736(17)30788-2. [DOI] [PubMed] [Google Scholar]
- 3.Naghavi M, Abajobir AA, Abbafati C, Abbas KM, Abd-Allah F, Abera SF, et al. Global, regional, and national age-sex specific mortality for 264 causes of death, 1980–2016: a systematic analysis for the Global Burden of Disease Study 2016. Lancet. 2017;390(10100):1151–1210. doi: 10.1016/S0140-6736(17)32152-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jha V, Garcia-Garcia G, Iseki K, Li Z, Naicker S, Plattner B, et al. Chronic kidney disease: global dimension and perspectives. Lancet. 2013;382(9888):260–272. doi: 10.1016/S0140-6736(13)60687-X. [DOI] [PubMed] [Google Scholar]
- 5.Chen TK, Knicely DH, Grams ME. Chronic kidney disease diagnosis and management: a review. JAMA. 2019;322(13):1294–1304. doi: 10.1001/jama.2019.14745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wuttke M, Li Y, Li M, Sieber KB, Feitosa MF, Gorski M, et al. A catalog of genetic loci associated with kidney function from analyses of a million individuals. Nat Genet. 2019;51(6):957–972. doi: 10.1038/s41588-019-0407-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carlassara L, Zanoni F, Gharavi AG. Familial aggregation of CKD: gene or environment? Am J Kidney Dis. 2021;77(6):861–862. doi: 10.1053/j.ajkd.2020.12.010. [DOI] [PubMed] [Google Scholar]
- 8.Torra R, Furlano M. New therapeutic options for Alport syndrome. Nephrol Dial Transplant. 2019;34(8):1272–1279. doi: 10.1093/ndt/gfz131. [DOI] [PubMed] [Google Scholar]
- 9.Lata S, Marasa M, Li Y, Fasel DA, Groopman E, Jobanputra V, et al. Whole-exome sequencing in adults with chronic kidney disease: a pilot study. Ann Intern Med. 2018;168(2):100–109. doi: 10.7326/M17-1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kruegel J, Rubel D, Gross O. Alport syndrome--insights from basic and clinical research. Nat Rev Nephrol. 2013;9(3):170–178. doi: 10.1038/nrneph.2012.259. [DOI] [PubMed] [Google Scholar]
- 11.Wang WJ, Cai GY, Chen XM. Cellular senescence, senescence-associated secretory phenotype, and chronic kidney disease. Oncotarget. 2017;8(38):64520–64533. doi: 10.18632/oncotarget.17327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Romagnani P, Remuzzi G, Glassock R, Levin A, Jager KJ, Tonelli M, et al. Chronic kidney disease. Nat Rev Dis Primers. 2017;3:17088–17088. doi: 10.1038/nrdp.2017.88. [DOI] [PubMed] [Google Scholar]
- 13.Vivante A, Hildebrandt F. Exploring the genetic basis of early-onset chronic kidney disease. Nat Rev Nephrol. 2016;12(3):133–146. doi: 10.1038/nrneph.2015.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Groopman EE, Marasa M, Cameron-Christie S, Petrovski S, Aggarwal VS, Milo-Rasouly H, et al. Diagnostic utility of exome sequencing for kidney disease. N Engl J Med. 2019;380(2):142–151. doi: 10.1056/NEJMoa1806891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hildebrandt F. Genetic kidney diseases. Lancet. 2010;375(9722):1287–1295. doi: 10.1016/S0140-6736(10)60236-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Canetta PAA, Radhakrishnan J. The evidence-based approach to adult-onset idiopathic nephrotic syndrome. Front Pediatr. 2015;3:78–78. doi: 10.3389/fped.2015.00078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Riedhammer KM, Braunisch MC, Gunthner R, Wagner M, Hemmer C, Strom TM, et al. Exome sequencing and identification of phenocopies in patients with clinically presumed hereditary nephropathies. Am J Kidney Dis. 2020;76(4):460–470. doi: 10.1053/j.ajkd.2019.12.008. [DOI] [PubMed] [Google Scholar]
- 18.Inrig JK, Califf RM, Tasneem A, Vegunta RK, Molina C, Stanifer JW, et al. The landscape of clinical trials in nephrology: a systematic review of Clinicaltrials.gov. Am J Kidney Dis. 2014;63(5):771–780. doi: 10.1053/j.ajkd.2013.10.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lek M, Karczewski KJ, Minikel EV, Samocha KE, Banks E, Fennell T, et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536(7616):285–291. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Petrovski S, Wang Q, Heinzen EL, Allen AS, Goldstein DB. Genic intolerance to functional variation and the interpretation of personal genomes. PLoS Genet. 2013;9(8):e1003709. doi: 10.1371/journal.pgen.1003709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vassy JL, Christensen KD, Schonman EF, Blout CL, Robinson JO, Krier JB, et al. The impact of whole-genome sequencing on the primary care and outcomes of healthy adult patients: a pilot randomized trial. Ann Intern Med. 2017;167(3):159–169. doi: 10.7326/M17-0188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Boer IH, Kovesdy CP, Navaneethan SD, Peralta CA, Tuot DS, Vazquez MA, et al. Pragmatic clinical trials in CKD: opportunities and challenges. J Am Soc Nephrol. 2016;27(10):2948–2954. doi: 10.1681/ASN.2015111264. [DOI] [PMC free article] [PubMed] [Google Scholar]