Abstract
Background
Many circular RNAs (circRNAs) have been identified as potential targets for cancer therapy. However, the role of circ_0002476 in non–small cell lung cancer (NSCLC) progression has not been explored.
Methods
The expression levels of circ_0002476, microRNA (miR)‐1182, and mitochondrial transcription factor A (TFAM) were detected by quantitative real‐time polymerase chain reaction. Cell functions were measured by cell counting kit 8 assay, EdU assay, colony formation assay, flow cytometry and transwell assay. Mitochondrial DNA (mtDNA) damage was assessed by measuring mtDNA copy number and transcript levels of ND1 and ATP6. Protein expression was examined by western blot. The interaction between miR‐1182 and circ_0002476 or TFAM was detected by dual‐luciferase reporter assay and RNA pull‐down assay. Animal experiments were performed to explore circ_0002476 role in vivo. Exosomes (Exs) were extracted and identified by transmission electron microscopy and nanoparticle tracking analysis.
Results
Circ_0002476 was overexpressed in NSCLC tissues and cells. Circ_0002476 knockdown suppressed NSCLC cell proliferation and invasion, while promoted apoptosis and mtDNA damage. Circ_0002476 could sponge miR‐1182, and miR‐1182 inhibitor reversed the influence induced by circ_0002476 knockdown. Moreover, TFAM was targeted by miR‐1182, and miR‐1182 hindered NSCLC cell progression by regulating TFAM. Additionally, circ_0002476 silencing could reduce NSCLC tumor growth by miR‐1182/TFAM. Further analyzed showed that Exs were involved in the transport of circ_0002476 between cells.
Conclusion
Taken together, our findings suggested that circ_0002476 might be a potential molecular target for NSCLC treatment.
Keywords: circ_0002476, miR‐1182, non–small cell lung cancer, TFAM
Circ_0002476 could promote proliferation, invasion, suppress apoptosis and mtDNA damage of NSCLC cells in vitro, as well as NSCLC tumor growth in vivo, through miR‐1182/TFAM axis, confirming that it might accelerate the malignant progression of NSCLC.

INTRODUCTION
Non–small cell lung cancer (NSCLC) is a common type of lung cancer, which is characterized by easy recurrence, micrometastasis, and poor prognosis. 1 , 2 Despite a lot of efforts, the treatment effect is still unsatisfactory for many NSCLC patients. 3 With the increasing understanding of molecular genetics, researchers have found that many genes play a vital role in NSCLC development, so various molecular targeted therapy drugs have been born. 4 At present, the application of molecular targeted drugs has significantly improved the prognosis of NSCLC patients. 5 , 6 Therefore, exploring the molecular mechanism affecting NSCLC process is expected to provide theoretical basis for seeking new molecular therapeutic targets.
Circular RNAs (circRNAs) have great potential to become molecular targets for cancer treatment. 7 , 8 In NSCLC, many circRNAs are confirmed to be involved in regulating NSCLC development and are considered to be potential therapeutic targets and prognostic markers for NSCLC, such as circ_101237 9 and circNDUFB2. 10 In this study, we used Gene Expression Omnibus database to find that circ_0002476 was upregulated in NSCLC tumor tissues. However, circ_0002476 role in NSCLC process has not been studied. It had been reported that circ_0002476 was overexpressed in bladder cancer and had a promotion effect on cancer cell growth and migration. 11 Therefore, we hypothesized that circ_0002476 might promote NSCLC progression and carried out a series of studies to confirm it.
CircRNA acts as a molecular sponge for microRNA (miRNA) to indirectly mediate downstream gene level. 12 , 13 Previous research suggested that miR‐1182 was underexpressed in NSCLC and it might play antitumor role in NSCLC. 14 , 15 Mitochondrial DNA (mtDNA) is an independent genome in mitochondria and its damage causes mitochondrial dysfunction, which in turn leads to human diseases, including cancer. 16 , 17 Mitochondrial transcription factor A (TFAM) is the first identified mtDNA transcription factor, which contains two high mobility protein domains. 18 TFAM had been shown to be highly expressed in NSCLC, and its knockdown reduced mtDNA copy numbers to influence cell function. 19
In our study, we found that circ_0002476 bound to miR‐1182, and miR‐1182 had binding sites with TFAM 3′UTR. Hence, we proposed the hypothesis that circ_0002476 regulated NSCLC processes through the miR‐1182/TFAM axis.
MATERIALS AND METHODS
Samples collection
Tumor tissues and adjacent normal tissues were obtained from NSCLC patients (n = 28, lung adenocarcinoma) that underwent surgical treatment at the Affiliated Xiangshan Hospital of Wenzhou Medial University. Tissues were stored at −80°C for further studies. All patients were followed up for 60 months to record overall survival. The clinicopathological features of NSCLC patients are listed in Table 1. This study was approved by the Ethics Committee of the Affiliated Xiangshan Hospital of Wenzhou Medial University.
TABLE 1.
Relationship between circ_0002476 expression and clinicopathological features of NSCLC patients
| Characteristics n = 28 | circ_0002476 Expression | p‐value a | ||
|---|---|---|---|---|
| Low (n = 14) | High (n = 14) | |||
| Gender | 0.4495 | |||
| Female | 15 | 9 | 6 | |
| Male | 13 | 5 | 8 | |
| Age (years) | >0.9999 | |||
| ≤60 | 15 | 8 | 7 | |
| >60 | 13 | 6 | 7 | |
| TNM grade | 0.0183* | |||
| I + II | 11 | 9 | 2 | |
| III + IV | 17 | 5 | 12 | |
| Lymph node metastasis | 0.0213* | |||
| Positive | 15 | 4 | 11 | |
| Negative | 13 | 10 | 3 | |
| Tumor size | 0.0461* | |||
| ≤3 cm | 10 | 8 | 2 | |
| >3 cm | 18 | 6 | 12 | |
Abbreviation: TNM, tumor‐node‐metastasis.
p < 0.05.
χ2 test.
Cell culture and transfection
Human NSCLC cells (H1299 and A549) and bronchial epithelial cells (16HBE) were bought from Procell and cultured in RPMI‐1640 medium (Gibco) containing 10% fetal bovine serum (FBS) (Gibco) and 1% penicillin–streptomycin (Invitrogen). NSCLC cells were transfected with circ_0002476 lentivirus short hairpin RNA (sh‐circ_0002476), miR‐1182 mimic or inhibitor (anti‐miR‐1182), plasmid cloning DNA (pcDNA) TFAM overexpression vector, and their controls (RiboBio) by Lipofectamine 3000 (Invitrogen).
Quantitative real‐time polymerase chain reaction
Total RNAs were extracted by TRIzol reagent (Invitrogen) and then used to synthesize complementary DNA (cDNA) by PrimeScript RT Reagent Kit (Takara). Quantitative real‐time polymerase chain reaction (qRT‐PCR) was performed with SYBR Green (Vazyme) and specific primers (Table 2). Data were analyzed by 2−ΔΔCt method. In addition, extracted RNA was treated with RNase R solution and then qRT‐PCR was used to confirm whether circ_0002476 could resist RNase R digestion.
TABLE 2.
Primer sequences used for qRT‐PCR
| Name | Primers for qRT‐PCR (5′–3′) | |
|---|---|---|
| hsa_circ_0002476 | Forward | GGGGAGATGATGCTCCCAAGGAAGAA |
| Reverse | GCTCCATAGAGCCCATACATCTCATC | |
| TFAM | Forward | TGATTCACCGCAGGAAAAGCTG |
| Reverse | CCTAACTGGTTTCCTGTGCCT | |
| miR‐1182 | Forward | GTATGAGGAGGGTCTTGGGAGGGAT |
| Reverse | CTCAACTGGTGTCGTGGAG | |
| β‐actin | Forward | CTCCATCCTGGCCTCGCTGT |
| Reverse | GCTGTCACCTTCACCGTTCC | |
| U6 | Forward | CTTCGGCAGCACATATACT |
| Reverse | AAAATATGGAACGCTTCACG |
Subcellular localization analysis
The cytoplasm and nuclear RNAs were isolated by PARIS Kit (Invitrogen) followed by qRT‐PCR to assess the distribution of circ_0002476. Briefly, NSCLC cells were treated with cell disruption buffer and then hatched with cell fractionation buffer to isolate nuclei and cytoplasm. After being centrifuged, the cytoplasmic fraction was separated from the nuclear pellet. Next, nuclear pellet was lysed by cell disruption buffer for RNA isolation. The cytoplasm RNA and nuclear RNA were used to measuring circ_0002476 expression using qRT‐PCR.
Cell proliferation detection
In cell counting kit 8 (CCK8) assay, NSCLC cells were cultured for 48 hours in 96‐well plates. CCK8 solution (Beyotime) was incubated with cells for 4 hours to measure cell viability at 450 nm using a microplate reader.
In EdU assay, NSCLC cells were stained by EdU solution, Apollo staining reaction, and DAPI solution in 96‐well plates according to the instructions of EdU Kit (RiboBio). The fluorescents were visualized under a fluorescent microscope to count EdU‐positive cell rate.
In colony formation assay, NSCLC cells were cultured for 14 days in 6‐well plates. After that, the colonies were counted under a microscope after fixed by paraformaldehyde and stained by crystal violet.
Cell apoptosis detection
After transfection for 48 hours, cells were collected and washed with phosphate buffered saline (PBS). Next, NSCLC cell suspensions were stained with annexin V‐FITC and propidium iodide (PI) in binding buffer (Vazyme). FACScan flow cytometry and CellQuest software were used to assess cell apoptosis rate.
Cell invasion detection
NSCLC cells suspended with RPMI‐1640 medium were seeded into the upper of tranwell chambers pre‐coated with Matrigel (BD Biosciences). The lower chamber was filled with completed medium. Twenty‐four hours later, cells on the bottom of chambers were fixed by paraformaldehyde, stained by crystal violet, and counted under a microscope.
Western blot analysis
Total protein was extracted using radioimmunoprecipitation assay (RIPA) buffer (Beyotime) and quantified by BCA Kit (Beyotime). The protein samples were subjected to sodium dodecyl sulfate‐polyacrylamide gel electrophoresis (SDS‐PAGE) gel and transferred onto polyvinylidene fluoride (PVDF) membranes. After blocked with non‐fat milk, membrane was treated with anti‐Bax (1:1000, ab32503), anti‐β‐actin (1:1000, ab8227), anti‐TFAM (1:1000, ab131607), anti‐cleaved‐caspase‐3 (1:500, ab2302), or anti‐Bcl‐2 (1:1000, ab32124) (Abcam). Following added with secondary antibody (1:50000, ab205718) at room temperature, membrane was treated with ECL reagent (Beyotime). Protein signal was detected and gray values were examined by ImageJ software.
mtDNA damage detection
EasyPure Genomic DNA Kit (TransGen Biotech) was used to isolate total DNA from NSCLC cells followed by performing qRT‐PCR to calculate mtDNA copy number. The transcript levels of mtDNA‐encoded ND1 and ATP6 were evaluated by qRT‐PCR.
Dual‐luciferase reporter assay
The wild‐type and mutant‐type of circ_0002476 or TFAM 3′UTR vectors were constructed by cloning these sequences into pmirGLO reporter vector. The WT/MUT‐circ_0002476 or WT/MUT‐TFAM 3′UTR vectors were transfected into NSCLC cells with miR‐1182 mimic/miR‐NC. Forty‐eight hours, Dual‐Luciferase Reporter Assay Kit (Vazyme) was used to measure relative luciferase activity (Firefly/Renilla).
RNA pull‐down assay
NSCLC cells were transfected with bio‐miR‐1182 probe and bio‐miR‐NC probe. Next, cell lysates were hatched with streptavidin magnetic beads (Invitrogen). After eluted magnetic beads‐RNA mixture, circ_0002476, and TFAM enrichments were analyzed using qRT‐PCR.
Mice xenograft models
A549 cells transfected with sh‐circ_0002476/sh‐NC were injected into BALB/c mice (right flank, n = 6 per group) (Vital River) subcutaneously. Tumor volume was detected every 3 days after 8 days. After 23 days, mice were sacrificed, and tumor tissues were harvested. In addition to performing qRT‐PCR and western blot (WB) analysis using tumor tissues, tumor tissues were used to prepare paraffin sections to carry out immunohistochemistry (IHC) staining using SP Kit (Invitrogen) and specific antibodies (anti‐TFAM, anti‐Ki67, anti‐Bax, and anti‐Bcl‐2). Animal experiments were approved by the Animal Ethics Committee of the Affiliated Xiangshan Hospital of Wenzhou Medial University.
Exosome isolation and identification
Exosomes (Exs) were extracted from the culture supernatants of cells using ExoQuick Exosome Precipitation Solution (SBI). Exs ultrastructure and particle size were detected by transmission electron microscopy (TEM) and nanoparticle tracking analysis (NTA). WB analysis was used to evaluate Exs marker (CD63 and CD9) protein using anti‐CD63 (1:1000, ab68418) and anti‐CD9 (1:500, ab223052).
Statistical analysis
Data were presented as mean ± standard deviation (SD). Student's t‐test or ANOVA was performed for comparisons between groups. GraphPad Prism 7.0 software was used for data analysis. p < 0.05 was considered as significant.
RESULTS
Circ_0002476 was upregulated in NSCLC tissues and cells
Through GSE158695 database (https://www.ncbi.nlm.nih.gov/geo/geo2r/?acc=GSE158695), we confirmed that circ_0002476 (ID number: hsa_circRNA_100900) had elevated expression in 3 paired NSCLC tumor tissues compared to normal tissues (Figure 1(a),(b)). In our research, we analyzed circ_0002476 expression in 28 paired NSCLC tumor tissues and adjacent normal tissues using qRT‐PCR and found that circ_0002476 was highly expressed in NSCLC tumor tissues (Figure 1(c)). In addition, high expression of circ_0002476 was associated with lower overall survival in patients with NSCLC (Figure 1(d)). We found that circ_0002476 expression was related to the tumor, node, and metastasis (TNM) grade, lymph node metastasis, and the tumor size of patients with NSCLC (Table 1). In NSCLC cells (H1299 and A549), circ_0002476 was markedly higher than that in 16HBE cells (Figure 1(e)). Moreover, circ_0002476 could resist the digestion of RNase R, which showed that circ_0002476 expression had not any changed and linear RNA β‐actin expression was significantly reduced in H1299 and A549 cells after RNase R treatment (Figure 1(f),(g)). Further analysis showed that circ_0002476 was mainly distributed in the cytoplasm of H1299 and A549 cells (Figure 1(h),(i)).
FIGURE 1.

Circ_0002476 Expression in NSCLC tissues and cells. (a) GSE158695 database screened the differentially expressed circRNA in three paired NSCLC tumor tissues and normal tissues. (b) Circ_0002476 expression in three paired NSCLC tumor tissues and normal tissues in GSE158695 database. (c) Circ_0002476 expression in NSCLC tumor tissues (n = 28) and adjacent normal tissues (n = 28) was analyzed by qRT‐PCR. (d) Kaplan–Meier analyzed the correlation between circ_0002476 expression and the overall survival of NSCLC patients. (e) Circ_0002476 expression in NSCLC cells (H1299 and A549) and 16HBE cells was detected by qRT‐PCR. (f),(g) after RNase R treatment, circ_0002476 and β‐Actin expression was analyzed by qRT‐PCR. (h),(i) Subcellular localization analysis was used to assess the distribution of circ_0002476 in cell cytoplasm and nuclear. **p < 0.01, ***p < 0.001, ****p < 0.0001
Knockdown of circ_0002476 suppressed NSCLC cell growth, invasion and promoted mtDNA damage
To reveal the role of circ_0002476 in NSCLC progression, circ_0002476 was silenced by sh‐circ_0002476 in H1299 and A549 cells (Figure 2(a)). Next, NSCLC cell proliferation, apoptosis, invasion, and mtDNA damage were evaluated. The results showed that circ_0002476 knockdown inhibited NSCLC cell viability, EdU‐positive cell rate, and the number of colonies, while enhanced cell apoptosis rate (Figure 2(b)–(e)). After circ_0002476 downregulation, the number of invasive NSCLC cells was remarkably reduced (Figure 2(f)). WB analysis was used to measure the protein expression of apoptosis marker Bax, cleaved‐caspase‐3, and anti‐apoptosis marker Bcl‐2. As shown in Figure 2(g),(h), circ_0002476 knockdown increased Bax and cleaved‐caspase‐3 protein expression, while it decreased Bcl‐2 protein expression. Furthermore, circ_0002476 silencing could strikingly restrain the mtDNA copy number and mtDNA transcript levels (ND1 and ATP6) in H1299 and A549 cells (Figure 2(i)–(k)). The above data confirmed that circ_0002476 might promote NSCLC growth, increase invasion, and alleviate mtDNA damage.
FIGURE 2.

Effects of sh‐circ_0002476 on NSCLC cell progression. H1299 and A549 cells were transfected with sh‐NC or sh‐circ_0002476. (a) Circ_0002476 expression was detected by qRT‐PCR. CCK8 assay (b), EdU assay (c), colony formation assay (d), flow cytometry (e), and transwell assay (f) measured cell proliferation, apoptosis, and invasion. (g),(h) WB analysis was used to examine protein expression. (i)–(k) The mtDNA copy number and mtDNA transcript levels (ND1 and ATP6) were measured to evaluate mtDNA damage. **p < 0.01, ***p < 0.001, ****p < 0.0001
Circ_0002476 could negatively regulate miR‐1182
The circinteractome software predicted that circ_0002476 had binding sites with miR‐1182 (Figure 3(a)). MiR‐1182 mimic was used to overexpress miR‐1182 in H1299 and A549 cells (Figure 3(b)). Next, dual‐luciferase reporter assay was performed and the results suggested that only the luciferase activity of WT‐circ_0002476 vector could be reduced by miR‐1182 mimic (Figure 3(c),(d)). Moreover, circ_0002476 enrichment had been confirmed to be increased in the bio‐miR‐1182 probe (Figure 3(e),(f)). The above results verified the interaction between circ_0002476 and miR‐1182. MiR‐1182 was underexpressed in NSCLC tumor tissues and its expression was negatively correlated with circ_0002476 expression (Figure 3(g),(h)). We detected the lowly expressed miR‐1182 in both NSCLC cells (H1299 and A549) compared to 16HBE cells (Figure 3(i)).
FIGURE 3.
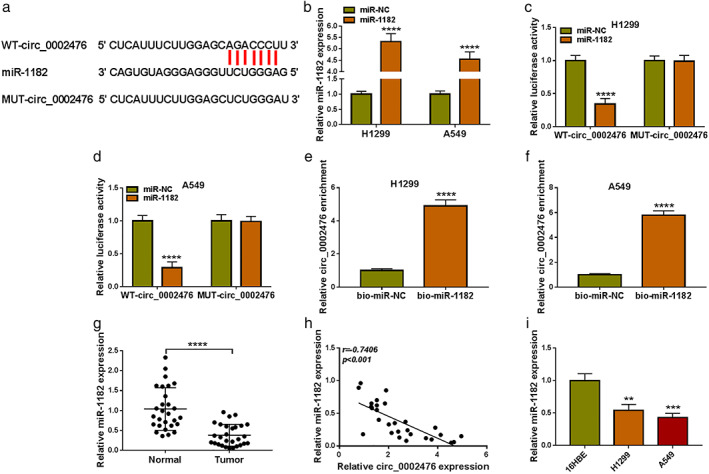
Circ_0002476 sponged miR‐1182. (a) The sequences of WT‐circ_0002476 and MUT‐circ_0002476 vectors were shown. (b) The transfection efficiency of miR‐1182 mimic was confirmed by qRT‐PCR. Dual‐luciferase reporter assay (c),(d) and RNA pull‐down assay (e),(f). (g) MiR‐1182 expression in NSCLC tumor tissues (n = 28) and adjacent normal tissues (n = 28) was analyzed by qRT‐PCR. (h) Pearson correlation analysis was used to evaluate the correlation between circ_0002476 and miR‐1182 expression. (i) MiR‐1182 expression in NSCLC cells (H1299 and A549) and 16HBE cells was detected by qRT‐PCR. **p < 0.01, ***p < 0.001, ****p < 0.0001
The influence of sh‐circ_0002476 on NSCLC cell progression could be abolished by anti‐miR‐1182
Afterward, sh‐circ_0002476 and anti‐miR‐1182 were co‐transfected into H1299 and A549 cells to further investigate whether circ_0002476 regulated NSCLC progression by sponging miR‐1182. The detection of miR‐1182 expression showed that circ_0002476 knockdown markedly enhanced miR‐1182 expression, whereas the addition of anti‐miR‐1182 reduced miR‐1182 expression (Figure 4(a)). Functional experiments revealed that anti‐miR‐1182 reversed the suppressive effects of circ_0002476 knockdown on NSCLC cell viability, EdU‐positive cell rate, the number of colonies, the number of invasive cells, and Bcl‐2 protein expression, as well as eliminated the promotion effects on cell apoptosis rate, Bax protein expression and cleaved‐caspase‐3 protein expression (Figure 4(b)–(i)). Moreover, the inhibitory effects of sh‐circ_0002476 on mtDNA copy number and mtDNA transcript levels (ND1 and ATP6) in H1299 and A549 cells also were overturned by miR‐1182 inhibitor (Figure 4(j)–(l)). Above all, we pointed out that circ_0002476 sponged miR‐1182 to promote NSCLC cell growth, invasion, and inhibit mtDNA damage.
FIGURE 4.
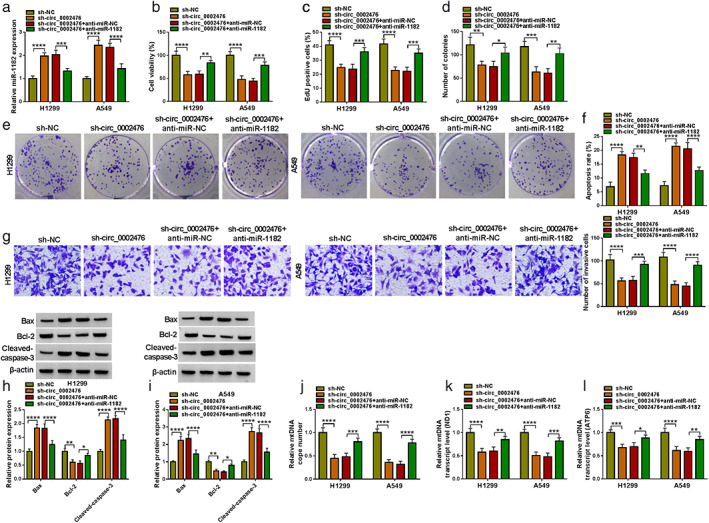
Effects of sh‐circ_0002476 and anti‐miR‐1182 on NSCLC cell progression. H1299 and A549 cells were transfected with sh‐circ_0002476 and anti‐miR‐1182. (a) MiR‐1182 expression was detected by qRT‐PCR. Cell proliferation, apoptosis and invasion were determined using CCK8 assay (b), EdU assay (c), colony formation assay (d),(e), flow cytometry (f), and transwell assay (g). (h),(i) Protein expression was examined using WB analysis. (j)–(l) The mtDNA damage was analyzed by measuring mtDNA copy number and mtDNA transcript levels (ND1 and ATP6). **p < 0.01, ***p < 0.001, ****p < 0.0001
MiR‐1182 directly interacted with TFAM
Targetscan software predicted that miR‐1182 could bind with the 3'UTR of TFAM (Figure 5(a)). The interaction between them was confirmed by dual‐luciferase reporter assay and RNA pull‐down assay, showing that miR‐1182 mimic significantly reduced the luciferase activity of WT‐TFAM 3'UTR vector and TFAM enrichment was markedly increased in the bio‐miR‐1182 probe (Figure 5(b)–(e)). The Cancer Genome Atlas (TCGA) database analyzed that TFAM was overexpressed in the tumor tissues of lung adenocarcinoma (LUAD, belongs to NSCLC) (Figure 5(f),(g)). Here, we observed highly expressed TFAM messenger RNA (mRNA) in NSCLC tumor tissues and found that there had negatively correlation between TFAM mRNA expression and miR‐1182 expression (Figure 5(h),(i)). At the protein levels, we confirmed that TFAM expression was upregulated in NSCLC tumor tissues and cells (Figure 5(j),(k)). In NSCLC co‐transfected with sh‐circ_0002476 and anti‐miR‐1182, the detection of TFAM protein expression showed that TFAM expression could be reduced by circ_0002476 knockdown, and miR‐1182 inhibiter could reverse this effect (Figure 5(l)). Therefore, we pointed out that circ_0002476 positively regulated TFAM by sponging miR‐1182.
FIGURE 5.
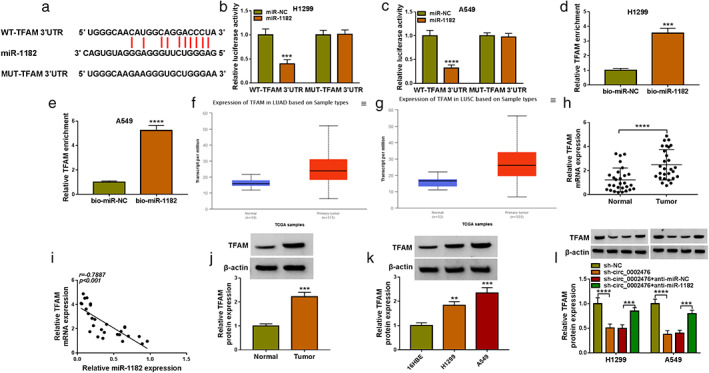
MiR‐1182 targeted TFAM. (a) The sequences of WT‐TFAM 3'UTR and MUT‐TFAM 3'UTR vectors were shown. Dual‐luciferase reporter assay (b),(c) and RNA pull‐down assay (d),(e). (f),(g) TCGA database analyzed TFAM expression in LUAD tumor tissues and normal tissues. (h) TFAM mRNA expression in NSCLC tumor tissues (n = 28) and adjacent normal tissues (n = 28) was analyzed by qRT‐PCR. (i) The correlation between miR‐1182 and TFAM expression was analyzed by Pearson correlation analysis. (j),(k) TFAM protein expression in NSCLC tissues and cells (H1299 and A549) was detected by WB analysis. (l) TFAM protein expression was tested by WB analysis in H1299 and A549 transfected with sh‐circ_0002476 and anti‐miR‐1182. **p < 0.01, ***p < 0.001, ****p < 0.0001
MiR‐1182 suppressed NSCLC cell progression by targeting TFAM
In the following study, H1299 and A549 cells were co‐transfected with miR‐1182 mimic and pcDNA TFAM overexpression vector to explore whether miR‐1182 regulated NSCLC cell progression by TFAM. Overexpressed miR‐1182 significantly inhibited TFAM protein expression, and this effect could be eliminated by pcDNA TFAM overexpression vector (Figure 6(a)). MiR‐1182 overexpression had an inhibitive effect on NSCLC cell viability, EdU‐positive cell rate, and the number of invasive cells, whereas these effects could be overturned by TFAM overexpression (Figure 6(b)–(e)). Moreover, miR‐1182 overexpression promoted NSCLC cell apoptosis, Bax protein expression, and cleaved‐caspase‐3 protein expression, while suppressed the number of invasive cells and Bcl‐2 protein expression. However, these effects also were reversed by overexpressing TFAM (Figure 6(f)–(i)). The mtDNA copy number and mtDNA transcript levels (ND1 and ATP6) in NSCLC cells could be decreased by miR‐1182 mimic, and TFAM overexpression could abolish this effect (Figure 6(j)–(l)). These results showed that miR‐1182 regulated NSCLC cell growth, invasion, and mtDNA damage by targeting TFAM.
FIGURE 6.

Effects of miR‐1182 and TFAM on NSCLC cell progression. H1299 and A549 cells were transfected with miR‐1182 mimic and pcDNA TFAM overexpression vector. (a) TFAM protein expression was detected by WB analysis. Cell proliferation, apoptosis, and invasion were assessed by CCK8 assay (b), EdU assay (c), colony formation assay (d),(e), flow cytometry (f), and transwell assay (g). (h),(i) WB analysis was performed to measure protein expression. (j)–(l) The mtDNA copy number and mtDNA transcript levels (ND1 and ATP6) were determined to analyze mtDNA damage. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001
Interference of circ_0002476 inhibited the tumor growth of NSCLC in vivo
Animal experiments were carried out to confirm the role of circ_0002476 in vivo. A549 cells transfected with sh‐circ_0002476 were injected into nude mice. After 23 days, we found that the tumor volume and weight were markedly reduced compared to the sh‐NC group (Figure 7(a),(b)). In the tumor tissues of sh‐circ_0002476 group, circ_0002476 expression was decreased, miR‐1182 expression was increased, whereas TFAM protein expression was reduced (Figure 7(c),(d)). Moreover, we detected that Bax protein expression was promoted and Bcl‐2 protein expression was inhibited in the tumor tissues of sh‐circ_0002476 group (Figure 7(e)). Additionally, IHC staining results showed that TFAM, Ki67 and Bcl‐2‐positive cells were significantly reduced, whereas Bax‐positive cells were enhanced in the tumor tissues of sh‐circ_0002476 group (Figure 7(f)). Above all, we confirmed that circ_0002476 could promote NSCLC tumor growth in vivo.
FIGURE 7.

Effects of sh‐circ_0002476 on the tumor growth of NSCLC in vivo. (a) Tumor volume was measured every 3 days after 8 days. (b) Tumor weight was detected after 23 days. (c) Circ_0002476 and miR‐1182 expression was assessed by qRT‐PCR. (d),(e) Protein expression was examined by WB analysis. (f) IHC staining assessed TFAM, Ki67, Bax, and Bcl‐2‐positive cells. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001
Exs could transport circ_0002476 in NSCLC cells
Exs were extracted from H1299 and A549 cells, and its ultrastructure was observed by TEM (Figure 8(a)). Through WB, we detected Exs markers CD63 and CD9, confirming that the extraction of Exs was successful (Figure 8(b)). Moreover, the particle size of Exs was analyzed by NTA (Figure 8(c)). The circ_0002476 expression was measured in the Exs of H1299 and A549 cells, and we confirmed that it was higher than that in 16HBE cells (Figure 8(d)). To confirm the transportation of Exs on circ_0002476, 16HBE cells were incubated with the Exs extracted from H1299 and A549 cells. By detecting circ_0002476 expression, we confirmed that circ_0002476 expression was significantly enhanced after Exs treatment (Figure 8(e),(f)). These data showed that Exs were involved in the transport of circ_0002476 between cells.
FIGURE 8.
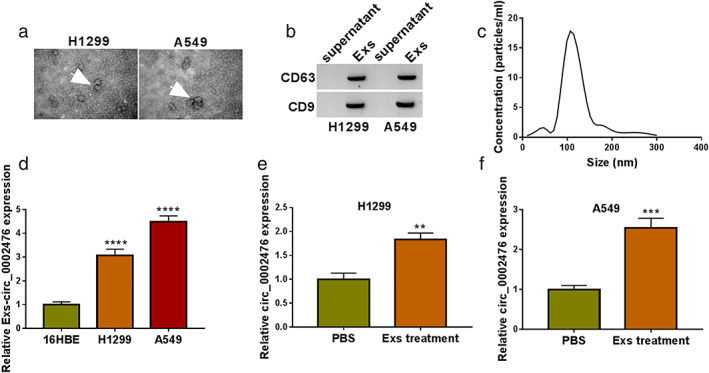
Exs transported circ_0002476 in NSCLC cells. (a) The ultrastructure of Exs was observed by TEM. (b) WB detected Exs markers CD63 and CD9. (c) The particle size of Exs was analyzed by NTA. (d) The circ_0002476 expression was measured in the Exs of 16HBE and NSCLC cells (H1299 and A549). (e),(f) Circ_0002476 expression was detected by qRT‐PCR in 16HBE cells treated with or without Exs from H1299 and A549 cells. **p < 0.01, ***p < 0.001, ****p < 0.0001
DISCUSSION
Many highly expressed circRNAs are believed to play a carcinogenic role in NSCLC progression. For example, circ_0000429 interference suppressed NSCLC proliferation and metastasis via regulating MAPK‐associated death domain‐containing protein (MADD) expression by sponging miR‐1197. 20 CircRNA_001010 had been confirmed to elevate NSCLC cell proliferation and metastasis through absorbing miR‐5112 to upregulate CDK4. 21 Moreover, circBIRC6 was confirmed to sponge miR‐145, thereby enhancing NSCLC cell growth and tumorigenesis. 22 Here, we explored the role of a new circRNA in the NSCLC process. Consistent with the results of high‐throughput sequencing, we confirmed the high expression of circ_0002476 in NSCLC tissues. In functional experiments, circ_0002476 silencing could restrain NSCLC cell proliferation, invasion, while increase apoptosis. It has been found that mtDNA copy number abnormality may increase mitochondrial oxidative stress and cell apoptosis, thereby aggravating NSCLC process. 23 In this, sh‐circ_0002476 reduced mtDNA copy number and mtDNA transcript level (ND1 and ATP6), confirming that circ_0002476 knockdown might exacerbate mtDNA damage. Additionally, we detected the increased circ_0002476 expression in 16HBE cells after incubated with Exs isolated from NSCLC cells, confirming that Exs mediated circ_0002476 transport. Not only that, downregulated circ_0002476 markedly reduced NSCLC tumor growth in vivo. The results indicated that circ_0002476 served as oncogene in NSCLC, similar to its role in bladder cancer. 11
To complete the mechanism of circRNA/miRNA/mRNA, 12 , 13 we performed bioinformatics analysis and pointed out that circ_0002476 might sponge miR‐1182. Previous data revealed that miR‐1182 could hinder ovarian cancer cell metastasis and proliferation, 24 and its overexpression attenuated gastric cancer proliferation and migration. 25 In NSCLC‐related study, miR‐1182 was confirmed to be downregulated in tumor tissues and had been suggested to suppress cancer cell proliferation, glycolysis, and metastasis. 14 , 15 , 26 Therefore, miR‐1182 played a tumor suppressor role in many cancers. Consistent with previous research, the lowly expressed miR‐1182 in NSCLC was confirmed in our study. MiR‐1182 could be absorbed by circ_0002476, and its knockdown reversed the function of sh‐circ_0002476 in NSCLC cells. We confirmed that miR‐1182 mimic not only repressed NSCLC cell proliferation and invasion, but also enhanced cell apoptosis and mtDNA damage. Above all, it was confirmed that circ_0002476 accelerated NSCLC development through miR‐1182.
TFAM has the ability of binding and unwinding mtDNA, which plays a vital role in regulating mtDNA replication, transcription, and damage repair. 27 In this, TFAM was confirmed to be targeted by miR‐1182. Many researches had suggested that TFAM was overexpressed in many tumors, such as glioma 28 and cervical cancer. 29 TFAM knockdown had been shown to change gastric cancer cell morphology and suppress cell proliferation by inactivating mtDNA transcription. 30 Moreover, TFAM silencing might reduce mtDNA copy number and mitochondrial function, therefore, inhibiting the invasion ability of colorectal cancer cells. 31 Similar to a previous study, 19 we confirmed that TFAM had elevated expression in NSCLC tissues. The rescue experiments showed that overexpressed TFAM overturned miR‐1182‐mediated cell function and mtDNA damage, verifying that miR‐1182 targeted TFAM to inhibit NSCLC development. Additionally, we pointed out that circ_0002476 positively regulated TFAM level, which completed the hypothesis of circ_0002476/miR‐1182/TFAM pathway.
There are some limitations to our study. Although we have confirmed that Exs are involved in the transport of circ_0002476, it is not clear whether exosomal circ_0002476 can be used as a diagnostic biomarker in patients with NSCLC. In the future, we need to collect blood from NSCLC patients to enrich our results. In addition, more clinical samples are needed to further validate our conclusions in the future.
In conclusion, our study pointed to a new regulatory axis that mediated NSCLC progression. Our research showed that circ_0002476 regulated NSCLC cell growth, invasion, and mtDNA damage through miR‐1182/TFAM pathway. The present study indicated that circ_0002476 might be a potential target for NSCLC therapy, a finding with important clinical significance.
FUNDING INFORMATION
This study was supported by Medical and health science and Technology project of Zhejiang Province (2022KY347) and Xiangshan County Science and Technology Project (2021XSX030016).
DISCLOSURE OF INTEREST
The authors declare that they have no conflicts of interest.
ACKNOWLEDGMENT
None.
Wang W, Sun H, Ma X, Zhu T, Zhang H. Circ_0002476 regulates cell growth, invasion, and mtDNA damage in non–small cell lung cancer by targeting miR‐1182/TFAM axis. Thorac Cancer. 2022;13(20):2867–2878. 10.1111/1759-7714.14631
Funding information Medical and health science and Technology project of Zhejiang Province, Grant/Award Number: 2022KY347; Xiangshan County Science and technology Project, Grant/Award Number: 2021XSX030016
REFERENCES
- 1. Sun J, Wu S, Jin Z, Ren S, Cho WC, Zhu C, et al. Lymph node micrometastasis in non‐small cell lung cancer. Biomed Pharmacother. 2022;149:112817. [DOI] [PubMed] [Google Scholar]
- 2. Baran K, Brzezianska‐Lasota E. Proteomic biomarkers of non‐small cell lung cancer patients. Adv Respir Med. 2021;89:419–26. [DOI] [PubMed] [Google Scholar]
- 3. Losanno T, Gridelli C. First‐line treatment of metastatic non‐small cell lung cancer in the elderly. Curr Oncol Rep. 2021;23:119. [DOI] [PubMed] [Google Scholar]
- 4. Sankar K, Gadgeel SM, Qin A. Molecular therapeutic targets in non‐small cell lung cancer. Expert Rev Anticancer Ther. 2020;20:647–61. [DOI] [PubMed] [Google Scholar]
- 5. Hill A, Gupta R, Zhao D, et al. Targeted therapies in non‐small‐cell lung cancer. Cancer Treat Res. 2019;178:3–43. [DOI] [PubMed] [Google Scholar]
- 6. Imyanitov EN, Iyevleva AG, Levchenko EV. Molecular testing and targeted therapy for non‐small cell lung cancer: current status and perspectives. Crit Rev Oncol Hematol. 2021;157:103194. [DOI] [PubMed] [Google Scholar]
- 7. Chen L, Shan G. CircRNA in cancer: fundamental mechanism and clinical potential. Cancer Lett. 2021;505:49–57. [DOI] [PubMed] [Google Scholar]
- 8. Lei M, Zheng G, Ning Q, Zheng J, Dong D. Translation and functional roles of circular RNAs in human cancer. Mol Cancer. 2020;19:30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Zhang ZY, Gao XH, Ma MY, Zhao CL, Zhang YL, Guo SS. CircRNA_101237 promotes NSCLC progression via the miRNA‐490‐3p/MAPK1 axis. Sci Rep. 2020;10:9024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Li B, Zhu L, Lu C, Wang C, Wang H, Jin H, et al. circNDUFB2 inhibits non‐small cell lung cancer progression via destabilizing IGF2BPs and activating anti‐tumor immunity. Nat Commun. 2021;12:295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zhang X, Liu X, Jing Z, Bi J, Li Z, Liu X, et al. The circINTS4/miR‐146b/CARMA3 axis promotes tumorigenesis in bladder cancer. Cancer Gene Ther. 2020;27:189–202. [DOI] [PubMed] [Google Scholar]
- 12. Panda AC. Circular RNAs act as miRNA sponges. Adv Exp Med Biol. 2018;1087:67–79. [DOI] [PubMed] [Google Scholar]
- 13. Lu Q, Liu T, Feng H, Yang R, Zhao X, Chen W, et al. Circular RNA circSLC8A1 acts as a sponge of miR‐130b/miR‐494 in suppressing bladder cancer progression via regulating PTEN. Mol Cancer. 2019;18:111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Zhao M, Ma W, Ma C. Circ_0067934 promotes non‐small cell lung cancer development by regulating miR‐1182/KLF8 axis and activating Wnt/beta‐catenin pathway. Biomed Pharmacother. 2020;129:110461. [DOI] [PubMed] [Google Scholar]
- 15. Yi S, Li Z, Wang X, du T, Chu X. Circ_0001806 promotes the proliferation, migration and invasion of NSCLC cells through miR‐1182/NOVA2 Axis. Cancer Manage Res. 2021;13:3067–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kopinski PK, Singh LN, Zhang S, Lott MT, Wallace DC. Mitochondrial DNA variation and cancer. Nat Rev Cancer. 2021;21:431–45. [DOI] [PubMed] [Google Scholar]
- 17. Filograna R, Mennuni M, Alsina D, Larsson NG. Mitochondrial DNA copy number in human disease: the more the better? FEBS Lett. 2021;595:976–1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Rubio‐Cosials A, Sidow JF, Jimenez‐Menendez N, et al. Human mitochondrial transcription factor A induces a U‐turn structure in the light strand promoter. Nat Struct Mol Biol. 2011;18:1281–9. [DOI] [PubMed] [Google Scholar]
- 19. Xie D, Wu X, Lan L, Shangguan F, Lin X, Chen F, et al. Downregulation of TFAM inhibits the tumorigenesis of non‐small cell lung cancer by activating ROS‐mediated JNK/p38MAPK signaling and reducing cellular bioenergetics. Oncotarget. 2016;7:11609–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wang JY, Zhang F, Hong L, Wei SJ. CircRNA_0000429 regulates development of NSCLC by acting as a sponge of miR‐1197 to control MADD. Cancer Manage Res. 2021;13:861–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wang Q, Kang PM. CircRNA_001010 adsorbs miR‐5112 in a sponge form to promote proliferation and metastasis of non‐small cell lung cancer (NSCLC). Eur Rev Med Pharmacol Sci. 2020;24:4271–80. [DOI] [PubMed] [Google Scholar]
- 22. Yang H, Zhao M, Zhao L, Li P, Duan Y, Li G. CircRNA BIRC6 promotes non‐small cell lung cancer cell progression by sponging microRNA‐145. Cell Oncol. 2020;43:477–88. [DOI] [PubMed] [Google Scholar]
- 23. Lin CS, Wang LS, Tsai CM, Wei YH. Low copy number and low oxidative damage of mitochondrial DNA are associated with tumor progression in lung cancer tissues after neoadjuvant chemotherapy. Interact Cardiovasc Thorac Surg. 2008;7:954–8. [DOI] [PubMed] [Google Scholar]
- 24. Hou XS, Han CQ, Zhang W. MiR‐1182 inhibited metastasis and proliferation of ovarian cancer by targeting hTERT. Eur Rev Med Pharmacol Sci. 2018;22:1622–8. [DOI] [PubMed] [Google Scholar]
- 25. Zhang D, Xiao YF, Zhang JW, Xie R, Hu CJ, Tang B, et al. miR‐1182 attenuates gastric cancer proliferation and metastasis by targeting the open reading frame of hTERT. Cancer Lett. 2015;360:151–9. [DOI] [PubMed] [Google Scholar]
- 26. Li C, Liu H, Niu Q, Gao J. Circ_0000376, a novel circRNA, promotes the progression of non‐small cell lung cancer through regulating the miR‐1182/NOVA2 network. Cancer Manage Res. 2020;12:7635–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Bruser C, Keller‐Findeisen J, Jakobs S. The TFAM‐to‐mtDNA ratio defines inner‐cellular nucleoid populations with distinct activity levels. Cell Rep. 2021;37:110000. [DOI] [PubMed] [Google Scholar]
- 28. Jiang J, Yang J, Wang Z, et al. TFAM is directly regulated by miR‐23b in glioma. Oncol Rep. 2013;30:2105–10. [DOI] [PubMed] [Google Scholar]
- 29. Wen Z, Lei Z, Jin‐An M, Xue‐Zhen L, Xing‐Nan Z, Xiu‐Wen D. The inhibitory role of miR‐214 in cervical cancer cells through directly targeting mitochondrial transcription factor A (TFAM). Eur J Gynaecol Oncol. 2014;35:676–82. [PubMed] [Google Scholar]
- 30. Lee WR, Na H, Lee SW, Lim WJ, Kim N, Lee JE, et al. Transcriptomic analysis of mitochondrial TFAM depletion changing cell morphology and proliferation. Sci Rep. 2017;7:17841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lin CS, Liu LT, Ou LH, Pan SC, Lin CI, Wei YH. Role of mitochondrial function in the invasiveness of human colon cancer cells. Oncol Rep. 2018;39:316–30. [DOI] [PubMed] [Google Scholar]


