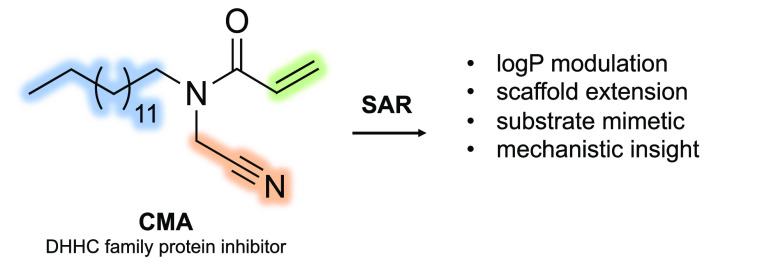
Abstract
Protein S-acylation is a dynamic and reversible lipid post-translational modification that can affect the activity, stability, localization, and interactions of target proteins. Lipid modification occurs on cysteine residues via a thioester bond and in humans is mediated by 23 Asp-His-His-Cys domain-containing protein acyltransferases (DHHC-PATs). The DHHC-PATs have well-known roles in physiology and disease, but much remains to be discovered about their biological function and therapeutic potential. We recently developed cyanomyracrylamide (CMA), an acrylamide-based DHHC inhibitor with key improvements over existing inhibitors. Here we conduct a structure–activity relationship (SAR) study of CMA and its acrylamide derivatives against zDHHC20, the most structurally characterized member of the human DHHC family, and validate the results against the homologous zDHHC2. This SAR maps out the limitations and potential of the acrylamide scaffold, underscoring the need for a bivalent inhibitor and identifying along the way three molecules with activity on par with CMA but with an improved logP.
Keywords: Protein S-acylation, DHHC family, structure−activity relationship, covalent inhibitors
S-acylation is a post-translational modification (PTM) that involves the covalent attachment of a fatty acid to a cysteine on a target protein.1 The increase in hydrophobic character conferred by the lipid, often 16C palmitate, can have diverse effects on target proteins, affecting for example the subcellular trafficking of the small GTPase Ras, the dimerization of the transcription factor RFX3, and the stability of the protein pump responsible for reuptake of the neurotransmitter dopamine, DAT.2−4 Central to the regulatory role of this PTM is its enzymatic reversibility. The addition of palmitate is catalyzed by the protein acyltransferase enzymes (PATs), while its removal is catalyzed by acyl protein thioesterases (APTs).5 Conserved from yeast to humans, the DHHC family of PAT proteins possess an aspartate–histidine–histidine–cysteine (DHHC) motif and have been associated with or directly linked to a variety of pathological conditions, including cancer, autoimmune disease, and neurodegenerative disease.6
While our understanding of DHHC-mediated proteome S-acylation and its clinical significance has increased in recent years, our ability to probe the individual and overlapping functional roles of the 23 mammalian family members remains limited. While classic genetic tools can be employed, functional redundancy between DHHCs and interfamily regulation often result in compensatory effects that obscure both molecular mechanisms and cellular phenotypes.7 Chemical inhibitors, which can rapidly interrupt protein activity, represent a promising approach for the study of this protein family, although no isoform-selective DHHC inhibitors have been developed to date. DHHC-catalyzed S-acylation involves the reaction of a catalytic cysteine with an acyl coenzyme A (CoA) lipid donor to form an acyl intermediate, followed by transfer of the lipid onto a target protein. 2-Bromopalmitate (2BP), the most widely used broad-spectrum DHHC inhibitor, disrupts this mechanism by intercalating the DHHC lipid binding pocket and alkylating the catalytic cysteine.8,9 However, 2BP inhibits not only the DHHCs but also the APTs, convoluting interpretation of acylation-mediated cellular events.10 We recently developed cyanomyracrylamide (CMA), a lipid-based covalent inhibitor featuring an acrylamide, a warhead known to react faster with cysteine thiols than serine hydroxyl groups and unlikely to form reactive acyl CoA intermediates, unlike the α-halo carboxylate of 2BP.11CMA demonstrated decreased cytotoxicity and improved potency relative to 2BP while also avoiding the off-target inhibition of the S-acylation eraser enzymes. While these improvements highlight the potential of the acrylamide in targeting DHHC family proteins, CMA retains the lipid-based structure of 2BP and thus is also reactive in cellulo with off-target lipidated and lipid-interacting proteins.11,12
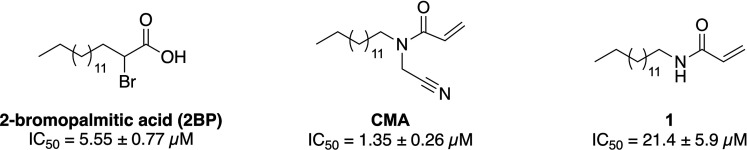
In designing the next generation of DHHC inhibitors, improving selectivity will likely mean reducing the lipophilicity and/or targeting additional binding motifs, such as the CoA binding pocket. In the present study, we report an expanded structure–activity relationship (SAR) study of CMA and its derivatives against zDHHC20 (Figure 1), the most structurally characterized family member,13−15 and the homologous zDHHC2, with an eye toward understanding the limits and potential of the acrylamide scaffold.
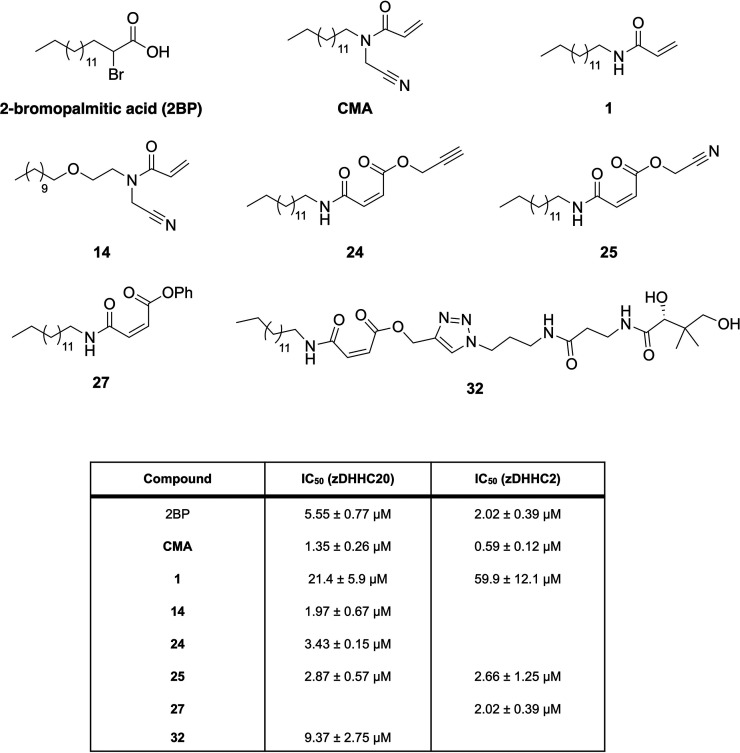
Figure 1.
Structures and IC50 values of the acrylamide-based zDHHC20 inhibitors that form the starting point of this work.
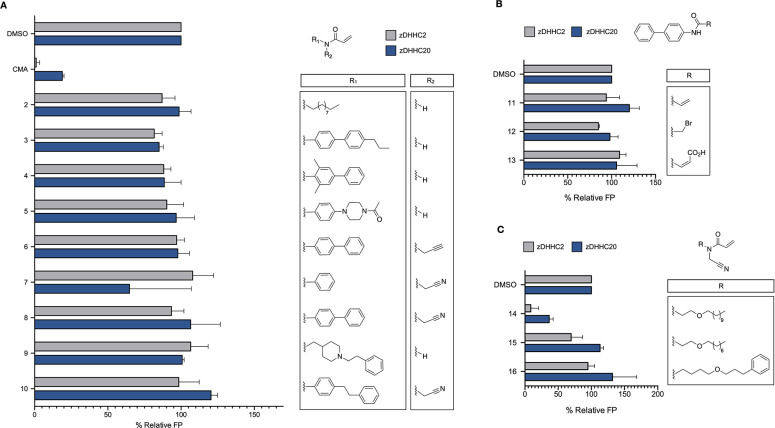
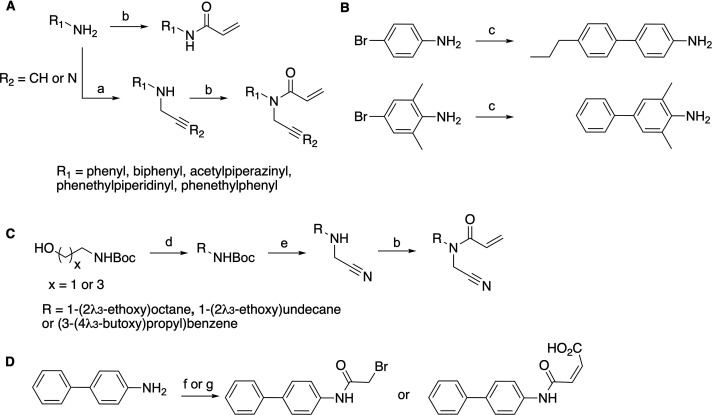
We began our exploration of the chemical space around CMA with the acyl chain. Given our previous observation that shortening the acyl chain (compound 2) abrogated the efficacy of acrylamide-based molecules against zDHHC20, we initially aimed to substitute it with pseudoisosteres (Figure 2A). We anticipated that exchange of the aliphatic chemical motif with another uncharged moiety possessing broadly similar properties would maintain its inhibitory capability. Therefore, we appended a series of biphenyl and methylated biphenyl groups to the acrylamide warheads of both 1 and CMA (Figure 2A). The synthesis of these derivatives was straightforward, requiring only one or two steps starting from the commercially available alkylated amines (Scheme 1A). As the substituted biphenyl compounds were not readily available, Suzuki coupling of the two phenyl moieties was performed to access the desired intermediates (Scheme 1B). With these compounds in hand, we used our previously reported adaptation of the acyl-cLIP assay to screen against zDHHC20. In this assay, DHHC-mediated acylation of a peptide provides a polarized fluorescence readout of enzyme activity.11,16 Screening of this panel of molecules (compounds 3–8) against zDHHC20 revealed that substitution of the N-alkyl chain abrogated the activity of all molecules (Figure 2A).
Figure 2.
SAR study of the aliphatic chain of CMA. (A) Fluorescence polarization (FP) screening of a panel of acrylamide-based molecules with N-alkyl chain substitutions against both zDHHC20 and zDHHC2, with activity normalized to DMSO. (B) FP screening of a panel of biphenyl molecules substituted with cysteine-reactive electrophiles against both zDHHC20 and zDHHC2, with activity normalized to DMSO. (C) FP screening of a panel of acrylamide-based molecules with ether substitutions. All of the compounds in (A–C) were screened at 10 μM. All data are presented as mean ± standard deviation (n = 3).
Scheme 1. Synthetic Route for the Aliphatic Analogues of 1 and CMA.
(A) Aliphatic chain replacement synthetic route. Conditions: (a) Propargyl bromide or bromoacetonitrile, K2CO3, DMF, 85 °C, 8–15 h. (b) Acryloyl chloride, NEt3, DCM, r.t., 2 h or acryloyl chloride, K2CO3, THF, r.t., 2 h 30 min. (B) Biphenyl amine intermediate synthetic route. Conditions: (c) Boronic acid, K2CO3, Pd(PPh)3, dioxane/water/MeOH (4:1:10), 80 °C, 3 h 30 min. (C) N-Ether-N-cyanomethyl acrylamide derivative synthetic route. Conditions: (d) RI, NaH, THF, 0 °C to r.t., 8–11 h. (e) 20% TFA in DCM, then bromoacetonitrile, K2CO3, ACN, 85 °C, 4–8 h. (b) Acryloyl chloride, K2CO3, THF, r.t., 2 h 30 min. (D) Warhead replacement of the biphenyl derivative. Conditions: (f) Bromoacetyl bromide, NEt3, DCM, 0 °C to r.t., 41 h. (g) maleic anhydride, toluene, 60 °C, 2 h 15 min.
Positing that the rigidity of these structures, in contrast to the lipid chain, impeded their reactivity, we next synthesized phenethylpiperidine (9) and bibenzyl (10) derivatives, both of which possess ethylene linkers for improved rotational flexibility. However, these compounds also showed marginal to no inhibitory activity (Figure 2A). The addition of more reactive electrophiles, including an α-brominated (12) and fumaric acid (13) amide, to the biphenyl group failed to rescue the activity of the biphenyl acrylamide (11) against zDHHC20 (Figure 2B and Scheme 1D). To further probe the sensitivity of the lipid tail, we next exchanged a methylene group of the aliphatic chain with an oxygen (14) (Figure 2C and Scheme 1C). The resulting compound showed both a potency on par with that of CMA (IC50 = 1.97 ± 0.67 μM) and an improved logP of 4.10 relative to CMA at 5.48 (Figure S1). However, this molecule was also resistant to acyl chain shortening, as seen with 15 (Figure 2C). Extension of the lipid chain with a benzyl moiety (16)—with the aim of improving the lipophilicity while maintaining lipid channel contacts—also failed to maintain potency (Figure 2C). Overall, the stringent requirement for the lipid chain shows that the steric and hydrophobic constraints of the zDHHC20 lipid-binding channel drive the potency of the acrylamide-based scaffold against DHHC family proteins (Figure 2).
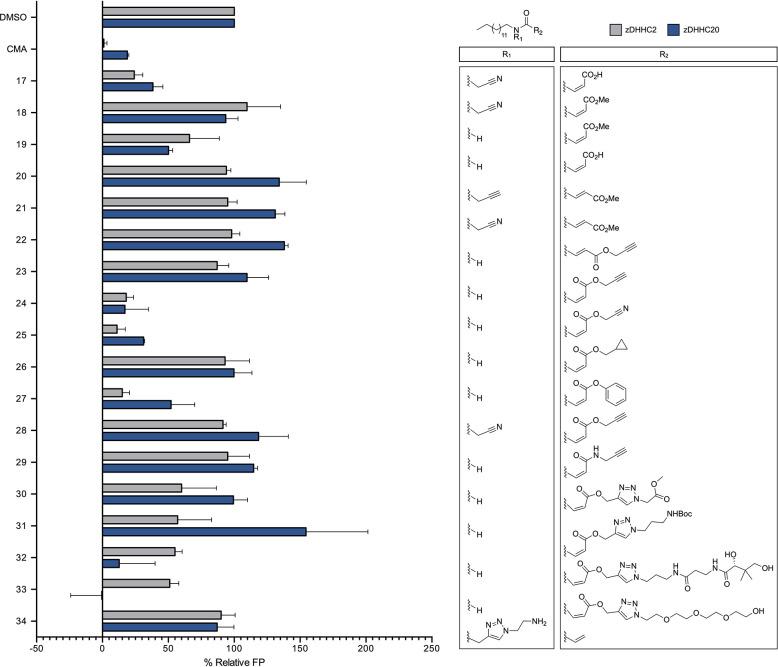
Given the recalcitrance of the lipid tail to modification, we next attempted to restyle the warhead (Figure 3). We hypothesized that extension of this warhead could increase protein contacts, thereby increasing the potency of this scaffold while also tuning its lipophilicity. To explore this potential, we began with the monomaleic acid amide derivative of CMA (17), a molecule with activity against zDHHC20 (IC50 = 3.45 ± 1.65 μM) and in possession of an acid suitable for extended modification.11 As the monomaleic acid warhead has increased steric hindrance, we first aimed to enhance its reactivity and potential potency via methyl esterification of the 17 carboxylate. We hypothesized that esterification would increase the electrophilicity of the olefinic carbon proximal to the amide and favor α cysteine thiolate addition, in contrast to thiolate β addition to CMA. Surprisingly though, this modification (18) extinguished the activity of 17, which suggested that this CMA derivative is a poor starting point for scaffold extension. However, despite our previous observation of the criticality of the cyanomethyl group paired with the acrylamide warhead, we found that 19, an acrylic acid methyl ester with a secondary amine, was more active than 18 (Figure 3).
Figure 3.
SAR study of the warhead. FP screening of a panel of acrylamide-based molecules with an extended warhead against both zDHHC20 and zDHHC2, with activity normalized to DMSO. Compounds were screened at 10 μM. Data are presented as mean ± standard deviation (n = 3).
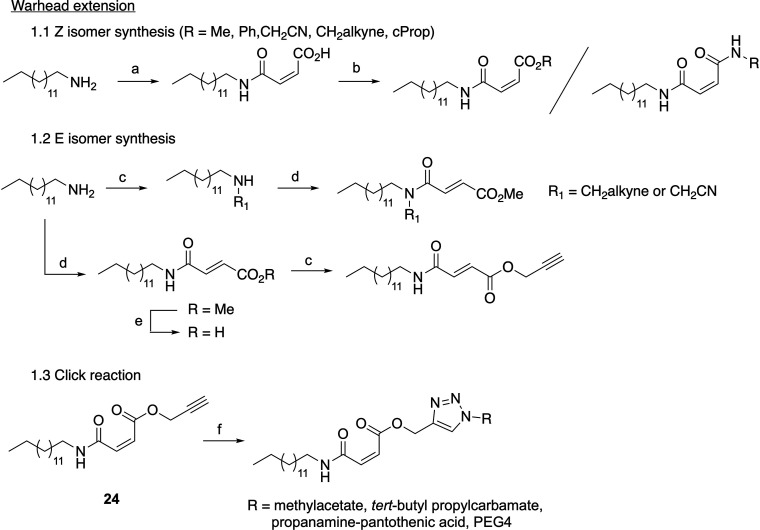
Therefore, from 20, a molecule accessible in one step in high yield and purity from tetradecylamine (Scheme 2), we synthesized a panel of extended or bulky ester derivatives with both cis- and trans-butenoic acid moieties. In synthesizing Z esters of 20, certain conditions yielded a majority of the E isomer (Table S1), emphasizing the importance of considering reaction conditions for stereochemical control.17 However, 23, the E isomer of 24, was readily accessed via peptide coupling of tetradecylamine with the monomethyl fumarate followed by deprotection of the acid and alkylation with propargyl bromide (Scheme 2); no isomerization was observed. In screening these esters against zDHHC20 (compounds 24–28), we observed a slight improvement over the potency of the methyl ester derivative for all compounds, but only 24 and 25 were significantly active (Figure 3). Further analysis demonstrated that these two molecules, which possess either an alkyne (24) or a methyl cyano ester (25), had IC50 values against zDHHC20 on par with CMA (3.43 ± 0.15 and 2.87 ± 0.57 μM, respectively) (Figure S1). Notably, 23 displayed no activity against zDHHC20, nor did any other E isomer derivatives (compounds 21 and 22). This result indicates that the inhibitor activity is strongly dependent on the reactivity of the warhead, which in turn suggests that modulating the alignment of the warhead with zDHHC20 could improve the potency and decrease the reactivity requirements. In addition, exchange of the 24 ester for an amide (29) in an attempt to improve potential in cellulo stability also resulted in a loss of activity against zDHHC20 (Figure 3).
Scheme 2. Synthetic Route for the Substituted Warhead Analogues of 1 and CMA.
Conditions: (a) Maleic anhydride, toluene, 60 °C, 1 h 30 min. (b) H2SO4, MeOH, 40 °C, 1 h 30 min or alkyl bromide, base, DMF, r.t., 5 h–4 days or phenol, DCC, pyridine, DCM, r.t., 2 h or propargylamine hydrochloride, Oxyma, DIC, DMF, r.t., 2 h. (c) Propargyl bromide or bromoacetonitrile, K2CO3, DMF, 85 °C, 8–15 h. (d) Monomethyl fumarate, Oxyma, DIC, DIPEA, DCM, r.t., 3 h. (e) LiOH (1.4 M), dioxane, r.t., 1 h. (f) R–N3, CuSO4·5H2O, sodium ascorbate, TBTA, THF/H2O, r.t., 17–25 h.
Given the potential of the 24 alkyne for rapid expansion via click chemistry, we decided to explore derivatives of this scaffold further. We aimed to mimic the native substrate of zDHHC20, acyl CoA, which interacts not only with the lipid-binding pocket but also with CoA-binding components, including an ADP-binding pocket. In fact, it has been shown that bivalent recognition is critical for DHHC–substrate binding.15 The CoA can be seen as three elements: a cysteamine, a pantothenic acid linker, and a 3′-phosphoadenosine diphosphate. As the acrylamide warhead of 24 can be seen as a proxy for the cysteamine, we next aimed to extend its scaffold via click-chemistry-mediated addition of pantothenic acid. We observed that the resulting compound 32—with pantothenic acid coupled to 24 via a triazole linker—was active against zDHHC20 (IC50 = 9.73 ± 2.75 μM), albeit to a lesser degree than 24 itself. This activity was retained when PEG-4 was substituted for the pantothenic acid (33) (Figure 3 and Scheme 2). In addition, both of these molecules had improved logP values (3.33 and 4.55, respectively). However, click-chemistry-derived analogues (30 and 31) with smaller linkers ending with either a hydrogen-bond donor or acceptor were inactive, as was an acrylamide lipid extended at the amine (34). Taken together, these results emphasize that accessing additional protein contacts, such as the charged ADP-binding pocket, will be critical in improving the potency of the acrylamide scaffold. They also highlight the potential of extending this scaffold at the warhead to reduce lipophilicity and thus potential lipid-binding off-targets.
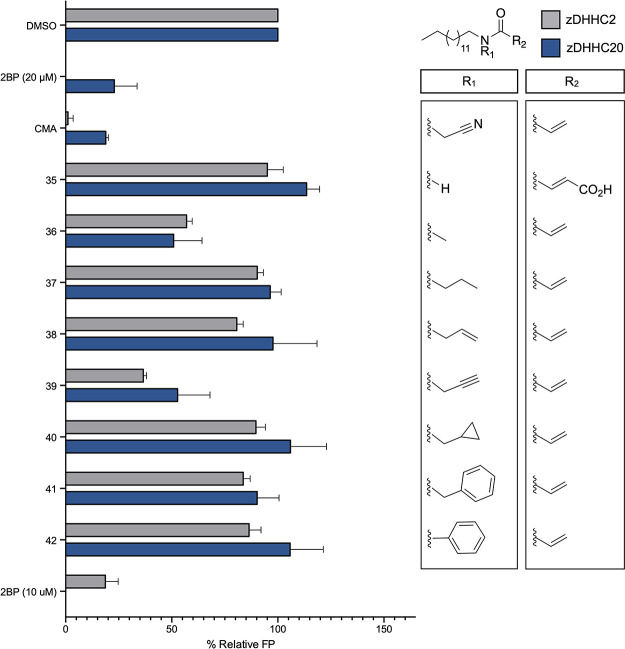
In order to validate these observations and test the generality of our SAR data, we next aimed to profile the activity of the library of inhibitors against zDHHC2, a DHHC family member that AlphaFold mapping suggests shares a high degree of similarity with zDHHC20 (Figure S2). Moreover, we have previously observed using activity based protein profiling (ABPP) that 39, the alkyne analogue of CMA, can label zDHHC2 and that CMA can engage with zDHHC2 in cellulo.11 To screen against zDHHC2, we adopted the FP-based acyl-cLIP assay previously deployed against purified zDHHC20.11,16 After validating the sensitivity of the assay to known inhibitors and its suitability for screening via Z-factor determination (Z′ = 0.67) (Figure S3), we screened our library of acrylamide-based compounds against zDHHC2. We found that the activity trends mostly paralleled those of zDHHC20, with CMA, 14, and 24 emerging as the most potent inhibitors of both zDHHC2 and zDHHC20 (Figures 1–4 and S4). Interestingly, 27, with the scaffold extended through a benzyl ester, demonstrated activity against zDHHC2 on par with CMA (IC50 = 2.02 ± 0.39 μM), although its activity against zDHHC20 was only 50% at 10 μM. In addition, while 32 and 33 were active against zDHHC20, they demonstrated negligible inhibition of zDHHC2 activity relative to CMA. This suggests that perhaps the extended acrylamide scaffold can be used to chemically target individual DHHC family members (Figures 3 and S4). Overall, these observations validated the chemical constraints on the acrylamide scaffold set by screening against zDHHC20.
Figure 4.
FP screening of a panel of acrylamide-based molecules against both zDHHC20 and zDHHC2, with activity normalized to DMSO. Compounds were screened at 10 μM. Data are presented as mean ± standard deviation (n = 3).
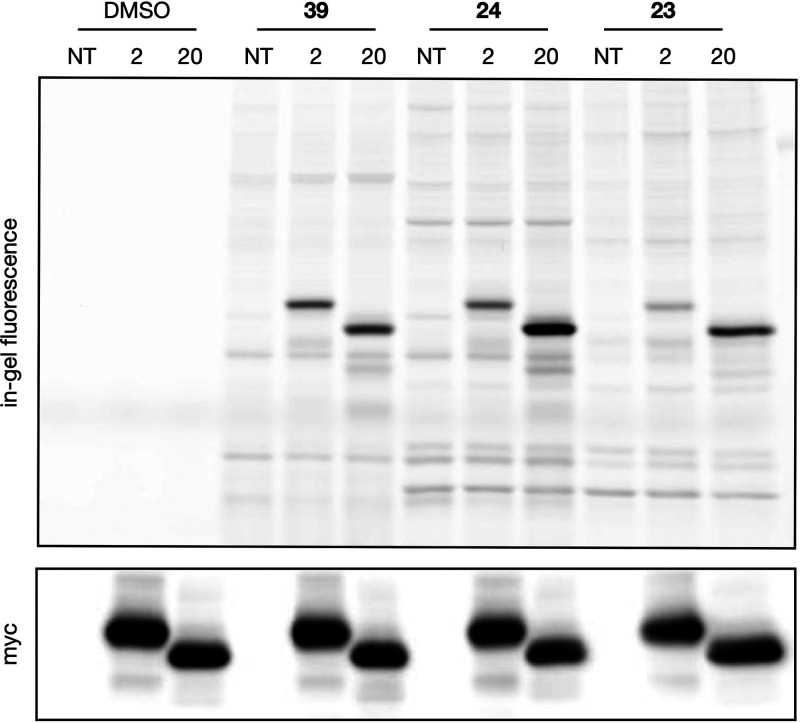
Notably, 24 possesses an alkyne, a group amenable to reporter conjugation via copper-catalyzed azide–alkyne “click” cycloaddition, which suggests that it can be used as an activity-based probe (ABP). Therefore, we next assessed whether 24 could be used to visualize DHHC family proteins and report protein–molecule engagement. HEK293T cells overexpressing zDHHC2 and cDHHC20 were treated with 24 and control molecules, followed by a lysate click reaction with TAMRA-azide. In-gel fluorescence analysis indicated that 24 successfully labeled both zDHHC2 and zDHHC20 (Figures 5 and S5). The labeling intensity suggests that 24 labels zDHHC2 similarly to 39, a previously validated probe for DHHC protein labeling.11 However, labeling of zDHHC20 by 24 was significantly more than that of 39, corresponding to their relative potencies in vitro (Figures 4–6 and S5). In addition, despite its lack of activity in vitro, 23 labeled both zDHHC2 and zDHHC20, albeit less than either compound 24 or 39. This suggests that additional regulatory mechanisms or E-to-Z isomerization are at play in cellulo, although isomerization under assay conditions was not observed in vitro (Figure S6). Overall, this experiment validates the application of 24 as an ABP.
Figure 5.
Compound engagement with DHHC family proteins. In-gel fluorescence analysis of myc-tagged zDHHC2 and zDHHC20 overexpressed in HEK293T cells labeled with 24, 39, or 23 (1 μM, 1 h). Cell lysates were subjected to click chemistry to conjugate the fluorescent TAMRA-azide to probe-modified proteins, and then the proteins were resolved via SDS-PAGE. Expression levels were visualized via anti-myc tag Western blot (n = 2).
Figure 6.
Structures and IC50 values of key molecules used in this work. Dose–response curves used to assess the IC50 values of CMA and compounds 14, 24, 25, and 32 against zDCCH20 can be found in Figure S1, while the dose–response curves used to assess the IC50 values of CMA and compounds 1, 25, and 27 against zDHHC2 can be found in Figure S4. All other IC50 values can be found in ref (11).
In conclusion, our exploration of the CMA chemical space around zDHHC20 reveals that the acyl chain is a critical component of its potency, with only an ether heteroatom substitution (compound 14) tolerated by the enzyme. We also determined that the warhead terminus is a potential node for improving not just the lipophilicity but also the potency and DHHC isomer selectivity of the acrylamide scaffold (compounds 33, 32, and 27). Finally, the narrow range of potency observed for all active molecules in this SAR makes clear the need for inhibitors that target both the lipid-binding pocket and other sites on the protein, such as the ADP-binding pocket.
Acknowledgments
This research was supported by the University of Chicago, the National Institute of General Medical Sciences of the National Institutes of Health (R35 GM119840 to B.C.D.), the National Institute of Diabetes and Digestive and Kidney Diseases of the National Institutes of Health (F30 DK125088 to S.-A.A.), and the Swiss National Science Foundation (P2BEP2_188250 to C.D.).
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsmedchemlett.2c00336.
Supporting figures, biological methods, synthetic methods, and molecular characterization (PDF)
Author Contributions
§ S.-A.A. and C.D. contributed equally to this work.
The authors declare no competing financial interest.
Supplementary Material
References
- Lanyon-Hogg T.; Faronato M.; Serwa R. A.; Tate E. W. Dynamic Protein Acylation: New Substrates, Mechanisms, and Drug Targets. Trends Biochem. Sci. 2017, 42 (7), 566–581. 10.1016/j.tibs.2017.04.004. [DOI] [PubMed] [Google Scholar]
- Busquets-Hernandez C.; Triola G. Palmitoylation as a Key Regulator of Ras Localization and Function. Front Mol. Biosci. 2021, 8, 659861. 10.3389/fmolb.2021.659861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen B.; Niu J.; Kreuzer J.; Zheng B.; Jarugumilli G. K.; Haas W.; Wu X. Auto-fatty acylation of transcription factor RFX3 regulates ciliogenesis. Proc. Natl. Acad. Sci. U. S. A. 2018, 115 (36), E8403–E8412. 10.1073/pnas.1800949115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rastedt D. E.; Vaughan R. A.; Foster J. D. Palmitoylation mechanisms in dopamine transporter regulation. J. Chem. Neuroanat. 2017, 83–84, 3–9. 10.1016/j.jchemneu.2017.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azizi S. A.; Kathayat R. S.; Dickinson B. C. Activity-Based Sensing of S-Depalmitoylases: Chemical Technologies and Biological Discovery. Acc. Chem. Res. 2019, 52 (11), 3029–3038. 10.1021/acs.accounts.9b00354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Main A.; Fuller W. Protein S-Palmitoylation: advances and challenges in studying a therapeutically important lipid modification. FEBS J. 2022, 289 (4), 861–882. 10.1111/febs.15781. [DOI] [PubMed] [Google Scholar]
- Gottlieb C. D.; Linder M. E. Structure and function of DHHC protein S-acyltransferases. Biochem. Soc. Trans. 2017, 45 (4), 923–8. 10.1042/BST20160304. [DOI] [PubMed] [Google Scholar]
- Davda D.; El Azzouny M. A.; Tom C. T.; Hernandez J. L.; Majmudar J. D.; Kennedy R. T.; Martin B. R. Profiling targets of the irreversible palmitoylation inhibitor 2-bromopalmitate. ACS Chem. Biol. 2013, 8 (9), 1912–7. 10.1021/cb400380s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tate E. W.; Kalesh K. A.; Lanyon-Hogg T.; Storck E. M.; Thinon E. Global profiling of protein lipidation using chemical proteomic technologies. Curr. Opin Chem. Biol. 2015, 24, 48–57. 10.1016/j.cbpa.2014.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedro M. P.; Vilcaes A. A.; Tomatis V. M.; Oliveira R. G.; Gomez G. A.; Daniotti J. L. 2-Bromopalmitate reduces protein deacylation by inhibition of acyl-protein thioesterase enzymatic activities. PLoS One 2013, 8 (10), e75232 10.1371/journal.pone.0075232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azizi S. A.; Lan T.; Delalande C.; Kathayat R. S.; Banales Mejia F.; Qin A.; Brookes N.; Sandoval P. J.; Dickinson B. C. Development of an Acrylamide-Based Inhibitor of Protein S-Acylation. ACS Chem. Biol. 2021, 16 (8), 1546–1556. 10.1021/acschembio.1c00405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan T.; Delalande C.; Dickinson B. C. Inhibitors of DHHC family proteins. Curr. Opin Chem. Biol. 2021, 65, 118–125. 10.1016/j.cbpa.2021.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rana M. S.; Kumar P.; Lee C. J.; Verardi R.; Rajashankar K. R.; Banerjee A. Fatty acyl recognition and transfer by an integral membrane S-acyltransferase. Science 2018, 359 (6372), eaao6326. 10.1126/science.aao6326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stix R.; Lee C. J.; Faraldo-Gomez J. D.; Banerjee A. Structure and Mechanism of DHHC Protein Acyltransferases. J. Mol. Biol. 2020, 432 (18), 4983–4998. 10.1016/j.jmb.2020.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C. J.; Stix R.; Rana M. S.; Shikwana F.; Murphy R. E.; Ghirlando R.; Faraldo-Gomez J. D.; Banerjee A. Bivalent recognition of fatty acyl-CoA by a human integral membrane palmitoyltransferase. Proc. Natl. Acad. Sci. U. S. A. 2022, 119 (7), e2022050119. 10.1073/pnas.2022050119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanyon-Hogg T.; Ritzefeld M.; Sefer L.; Bickel J. K.; Rudolf A. F.; Panyain N.; Bineva-Todd G.; Ocasio C. A.; O’Reilly N.; Siebold C.; Magee A. I.; Tate E. W. Acylation-coupled lipophilic induction of polarisation (Acyl-cLIP): a universal assay for lipid transferase and hydrolase enzymes. Chem. Sci. 2019, 10 (39), 8995–9000. 10.1039/C9SC01785B. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W.; Chen X.; Yi S. Kinetic Study on the Preparation of Fumaric Acid from Maleic Acid by Batch Noncatalytic Isomerization. ACS Omega 2019, 4 (5), 8274–8281. 10.1021/acsomega.9b00316. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.