Introduction
Dolutegravir (DTG), an integrase strand transfer inhibitor (INSTI)-based HIV-1 therapy, is widely recommended in first-line and second-line regimens.1,2 Integrase strand transfer inhibitor resistance mutations associated with DTG-containing regimens have been well described, most often occurring after DTG monotherapy or in INSTI-experienced patients.3Although rare, emergence of these mutations has also been described in patients on DTG-containing triple-drug regimens4 and INSTI-naïve patients.5,6 The R263K mutation is commonly associated with the emergence of DTG resistance but reduces viral fitness and DNA integration.7,8 Here we describe a case of very slow viral decline (~42 months) in a treatment-experienced, INSTI-naïve patient on a DTG-based triple therapy regimen.
Case
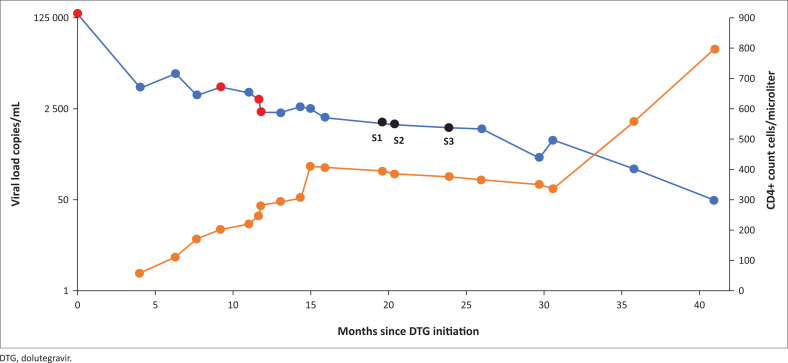
A 43-year-old man was diagnosed with HIV-1 in 2012 and commenced on an antiretroviral treatment (ART) regimen consisting of tenofovir disoproxil fumarate (TDF), emtricitabine (FTC) and efavirenz (EFV). Initial poor adherence to treatment and virologic failure was reported for 2012–2015 (viral load range 14 269 copies/mL – 173 455 copies/mL), and due to concerns of having possibly acquired resistance to this regimen, treatment was empirically switched according to national guidelines to lamivudine (3TC), zidovudine (AZT) and lopinavir/ritonavir (LPV/r). He remained on this regimen with his family practitioner from 2015 until early 2018, when he was referred back to our hospital for persistently high HIV viral loads. He admitted to poor adherence secondary to LPV/r-associated diarrhoea, and was subsequently switched to 3TC, AZT and DTG. Patient clinical data and treatment history are shown in Table 1. After initial viral load decline to 6500 copies/mL the viral load returned to 11 700 copies/mL and non-adherence was suspected. Although admitting poor adherence during 2012–2018 the patient claimed perfect adherence since starting his DTG regimen. Adherence was confirmed initially by means of directly observed therapy, and then by unannounced random (same-day unscheduled clinic visit requests) 3TC and DTG drug concentration testing on two and four occasions (3TC 1.24 μg/mL – 4.45 μg/mL, DTG 0.583 μg/mL – 1.354 μg/mL in plasma). Drug resistance testing on multiple occasions (Figure 1) identified M184V which, results in resistance to 3TC and increases susceptibility to AZT; K65R was also detected and also increases AZT susceptibility.9 Integrase sequencing indicated susceptibility to DTG from January 2018 until September 2019 (21 months), after which resistance to DTG (N155H and R263K) was detected (January 2020) in a single sample. Follow-up INSTI drug resistance testing could not confirm DTG resistance. Even though DTG resistance was detected at one time point, the viral load continued to decline (Figure 1) despite the patient remaining on a DTG-based regimen. Plasma samples from three time points and cells (buffy coat/peripheral blood mononuclear cells [PBMCs]) from two time points were collected (S1, S2 & S3) for further investigation (Table 1 and Figure 1). All samples used in this study were collected after DTG initiation, with the relevant regimen over sampling period S1 and S2 being 3TC/AZT/DRV/r and S3 being 3TC/AZT/DTG.
TABLE 1.
Patient clinical data and treatment regimens.
| Variable | Description |
|---|---|
| Age | 43 |
| Gender | Male |
| Diagnosis | 2012 |
| Samples (time after DTG initiation) and viral load (copies/mL) | S1 (~16 months): 1420 |
| S2 (~20 months): 1290 | |
| S3 (~24 months): 1128 | |
| Duration of detectable viraemia | ~42 months |
| Previous ART regimens and viral load range (copies/mL) | FTC/TDF/EFV (2012–2015): 29 271–173 455 |
| 3TC/AZT/LPV/r (2015–2018): 14 269–149 000 | |
| Study period ART regimen and viral load range (copies/mL) | 3TC/AZT/DTG (January 2018 – August 2019): 313–149 000 |
| 3TC/AZT/DRV/r (August 2019 – November 2019): 650 | |
| 3TC/AZT/DTG (November 2019 onward): 50–193 |
ART, antiretroviral treatment; TDF, tenofovir disoproxil fumarate; FTC, emtricitabine; EFV, efavirenz; AZT, zidovudine; DRV, darunavir; DTG, dolutegravir; LPV, lopinavir; r, ritonavir; PBMCs, peripheral blood mononuclear cells.
FIGURE 1.
Patient viral load (blue) and CD4 data (orange) graph displayed in months since dolutegravir initiation indicating samples available for further investigation (black dots) (S1, S2 & S3). Routine drug resistance testing performed on multiple occasions are indicated by red dots.
Molecular workup
PBMCs were isolated from freshly collected whole blood ethylenediaminetetraacetic acid specimens according to the HIV/AIDS Network Coordination Cross-Network Peripheral Blood Mononuclear Cell Processing Standard Operating Procedure (www.hanc.info/labs/labresources/procedures/Pages/pbmcSop.aspx). Cell-associated DNA and RNA were isolated from PBMCs and viral RNA was isolated from plasma as reported by Hong et al.10 Single genome amplification products covering the p6-protease-reverse transcriptase (p6PrRT) region were generated and Sanger sequencing was performed as described previously.11 Additionally, single genome amplicons covering the entire polymerase region were analysed for drug resistance mutations. Population and single genome nucleotide sequences were analysed for drug resistance mutations.
Ethical considerations
Ethical approval was obtained from the University of the Witwatersrand, where the patient was enrolled. Written informed consent was obtained from the patient.
Results
Patient p6PrRT nucleotide sequences from S1 (plasma RNA and buffy coat DNA), S2 (plasma RNA) and S3 (plasma RNA, cell-associated DNA and RNA) indicate drug resistance mutation M184V (Table 2).
TABLE 2.
Patient p6-protease-reverse transcriptase nucleotide sequences obtained from various sources for each sample with drug resistance mutations detected.
| Sample (time after DTG initiation) | Sequence source | Number of sequences obtained | Drug resistance mutations |
|
|---|---|---|---|---|
| NRTI | NNRTI | |||
| S1 (~16 months) | Plasma RNA | 8 | A62V, K65R, M184V | L100I, K103N |
| Buffy coat DNA | 24 | None | None | |
| S2 (~20 months) | Plasma RNA | 12 | A62V, K65R, M184V | L100I, K103N |
| S3 (~24 months) | Plasma RNA | 22 | A62V, K65R, M184V | L100I, K103N |
| PBMC DNA | 24 | None | None | |
| PBMC RNA | 21 | None | K103N | |
NNRTI, non-nucleoside reverse transcriptase inhibitor; NRTI, nucleoside reverse transcriptase inhibitor; PBMC, peripheral blood mononuclear cell; DTG, dolutegravir.
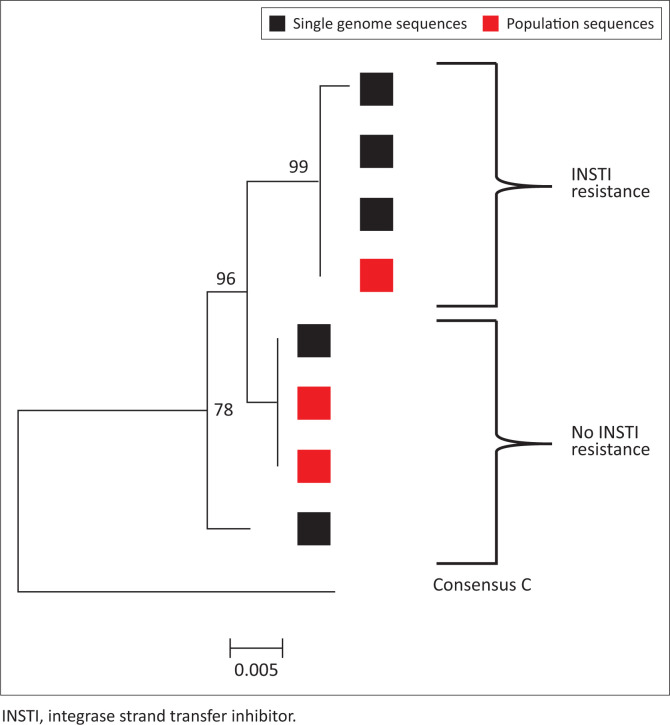
Mutations N155K and R263K were only detected once in S3 and were confirmed with single genome sequencing (Table 3). Drug resistance analyses of integrase population and single genome amplicons from plasma RNA indicate two distinct populations: INSTI resistant and INSTI non-resistant (Table 3). These two integrase populations are reflected in phylogenetic analysis (Figure 2).
TABLE 3.
Integrase population and single genome sequences indicating associated drug resistance mutations.
| Sequence origin | Time since DTG initiation | Genes | INSTI major | INSTI accessory |
|---|---|---|---|---|
| Routine integrase drug resistance population sequences | 12 months | IN | None | None |
| ~20 months | IN | None | None | |
| S3 (24 months) | IN | N155H, R263K | D232N | |
| Single genome sequences | S3 Sequence 1 (24 months) | IN | None | None |
| S3 Sequence 2 (24 months) | IN | None | None | |
| S3 Sequence 3 (24 months) | IN | N155H, R263K | D232N | |
| S3 Sequence 4 (24 months) | IN | N155H, R263K | D232N | |
| S3 Sequence 5 (24 months) | IN | N155H, R263K | D232N |
INSTI, integrase strand transfer inhibitor; DTG, dolutegravir.
FIGURE 2.
Neighbour joining p-distance phylogenetic tree of integrase nucleotide sequences generated from population and single genome amplicons.
At the time INSTI resistance was definitively confirmed, the patient’s HIV viral load had already declined to 50 copies/mL. After much discussion with the patient, the patient elected to continue on his current regimen (rather than switch to a non-DTG-based regimen) with regular HIV viral load monitoring.
Discussion
We report a patient with viral load suppression despite a mixed viral population that includes INSTI-resistant variants with N155H and R263K. The patient was treatment experienced but INSTI naïve when initiated on a DTG-based regimen. DTG-resistant mutations N155H and R263K were first detected 24 months after DTG initiation; however, despite this the HIV-1 viral load continued decline to 50 copies/mL after 42 months. It is unclear why the DTG-resistant variants did not outcompete DTG-susceptible variants, but the low viral fitness could have contributed. Mutations R263K, M184V and K65R are all associated with decreased viral fitness and here occurred on the same viral variant. The slow but continued viral load decline could be due to the remaining activity and hyper-susceptibility to AZT, in the presence of the M184V and K65R mutations.12,13,14,15 Moreover CD4+ cell recovery and associated improved cytotoxic T-cell responses could have contributed to immune reconstitution to HIV and resulting viral load decline.
Although previously thought to be rare, several cases of virological failure as a result of DTG resistance in INSTI-naïve patients have been described.4,16,17 The factors associated with DTG resistance are poorly understood. Whereas DTG resistance when used in first-line therapy is exceedingly rare, it is not unusual when DTG is administered to treatment-experienced patients,1,6 even when previously INSTI naïve. It is interesting that six of the nine cases of treatment-experienced, INSTI-naïve cases in the NADIA study1 with drug resistance received the same regimen as this patient, albeit that in our case this was given as a third-line regimen in contrast to NADIA where the patients received DTG in second-line treatment. Reported clinical risk factors associated with emergence of DTG resistance include poor treatment adherence, drug interactions and HIV factors such as a high baseline viral load.6 In this case we report good adherence but the high viral load (> 100 000 copies/mL) prior to DTG initiation could be a contributing risk factor, even though INSTI resistance was only detected after 24 months. The combination of N155H and R263K is associated with virologic failure on DTG-based regimens,4,18,19 in spite of the fitness cost of R263K.7 Even though N155H partially compensates for the R263K viral fitness cost, development of compensatory mutations to the N155H, R263K strain may be unlikely or slow despite drug pressure.20 No additional risk factors were identified in this case.
The presence of DTG resistance only partially explains the viraemia in this patient but offers no clarity on the susceptible sub-population. The detectable viraemia could be the result of intermittent adherence or compartmentalised replication or clonal viraemia.11,21,22 Random unannounced drug concentration testing on various occasions suggest that intermittent adherence is an unlikely cause of the viraemia. Both compartmentalised replication and clonal viraemia could explain the relatively high viral load for an extended period of time. We investigated clonal viraemia in this case as described by Halvas et al.,11 but we were not able to find a matching proviral clone to the monotypic plasma virus population (data not shown); further investigation could be performed to shed light on this.
This case report provides further evidence that DTG is not impervious to drug resistance, especially when used in treatment-experienced patients. It also suggests that patients with DTG resistance may not present with immediate viral rebound due to the fitness cost of mutations and could present with delayed viral load suppression or low-level viraemia. It is not known whether this patient’s HIV viral load will continue to remain low or whether additional compensatory mutations might allow the drug-resistant variant to regain fitness resulting in rebound. Continued vigilance is required when using DTG in treatment-experienced patients and sensitive drug resistance assays may be needed to detect drug resistance at low viral load levels.
Acknowledgements
The patient for participating in this study. The funding body that allowed us to investigate this case.
Competing interests
The authors declare that they have no financial or personal relationships that may have inappropriately influenced them in writing this article.
Authors’ contributions
Patient enrolment and sample collection was done by M.E. and J.N. J.C.B. and G.U.v.Z. designed the study. Data collection was performed by J.C.B. and K.S. J.C.B., K.S., J.N. and G.U.v.Z. analysed the data. J.C.B. wrote the article and all authors reviewed it. Supervision of the project was done by G.U.v.Z.
Funding information
The assay development was supported by the United States (US) National Institutes of Health and South African Medical Research Council through its US–South Africa Program for Collaborative Biomedical Research (National Cancer Institute grant no. U01CA200441). The funding body had no involvement in study design, data collection or manuscript writing.
Data availability
All figures and data are available within the article.
Disclaimer
The views and opinions expressed in this article are those of the authors and do not necessarily reflect the official policy or position of any affiliated agency of the authors.
Footnotes
How to cite this article: Botha JC, Steegen K, Edoo M, Nel J, Van Zyl GU. Low-level viraemia despite emergence of dolutegravir-resistant variants. S Afr J HIV Med. 2022;23(1), a1398. https://doi.org/10.4102/sajhivmed.v23i1.1398
References
- 1.Paton NI, Musaazi J, Kityo C, et al. Efficacy and safety of dolutegravir or darunavir in combination with lamivudine plus either zidovudine or tenofovir for second-line treatment of HIV infection (NADIA): Week 96 results from a prospective, multicentre, open-label, factorial, randomised, non-inferiority trial. Lancet HIV. 2022;9(6):e381–e393. 10.1016/S2352-3018(22)00092-3 [DOI] [PubMed] [Google Scholar]
- 2.World Health Organisation . Consolidated guidelines on the use of antiretroviral drugs for treating and preventing HIV infection [homepage on the Internet]. WHO; 2016. [cited 2020 Sep 28]. Available from: https://www.who.int/publications/i/item/9789241549684. [PubMed] [Google Scholar]
- 3.Rhee S-Y, Grant PM, Tzou PL, et al. A systematic review of the genetic mechanisms of dolutegravir resistance. J Antimicrob Chemother. 2019;74(11):3135–3149. 10.1093/jac/dkz256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cahn P, Pozniak AL, Mingrone H, et al. Dolutegravir versus raltegravir in antiretroviral-experienced, integrase-inhibitor-naive adults with HIV: Week 48 results from the randomised, double-blind, non-inferiority SAILING study. Lancet. 2013;382(9893):700–708. 10.1016/S0140-6736(13)61221-0 [DOI] [PubMed] [Google Scholar]
- 5.Pena MJ, Chueca N, D’Avolio A, Zarzalejos JM, Garcia F. Virological failure in HIV to triple therapy with dolutegravir-based firstline treatment: Rare but possible. Open Forum Infect Dis. 2019;6(1):ofy332. 10.1093/ofid/ofy332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cevik M, Orkin C, Sax PE. Emergent resistance to dolutegravir among INSTI-Naïve patients on first-line or second-line antiretroviral therapy: A review of published cases. Open Forum Infect Dis. 2020;7(6):ofaa202. 10.1093/ofid/ofaa202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mesplède T, Quashie PK, Osman N, et al. Viral fitness cost prevents HIV-1 from evading dolutegravir drug pressure. Retrovirology. 2013;10:22. 10.1186/1742-4690-10-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mesplède T, Leng J, Pham HT, et al. The R263K dolutegravir resistance-associated substitution progressively decreases HIV-1 integration. MBio. 2017;8(2):e00157–17. 10.1128/mBio.00157-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu TF, Shafer RW. Web resources for HIV type 1 genotypic-resistance test interpretation. Clin Infect Dis. 2006;42(11):1608–1618. 10.1086/503914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hong F, Aga E, Cillo AR, et al. Novel assays for measurement of total cell-associated HIV-1 DNA and RNA. J Clin Microbiol. 2016;54(4):902–911. 10.1128/JCM.02904-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Halvas EK, Joseph KW, Brandt LD, et al. HIV-1 viremia not suppressible by antiretroviral therapy can originate from large T cell clones producing infectious virus. J Clin Invest. 2020;130(11):5847–5857. 10.1172/JCI138099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Larder B, Kemp S, Harrigan P. Potential mechanism for sustained antiretroviral efficacy of AZT-3TC combination therapy. Science. 1995;269(5224):696–699. 10.1126/science.7542804 [DOI] [PubMed] [Google Scholar]
- 13.Melikian GL, Rhee S-Y, Taylor J, et al. Standardized comparison of the relative impacts of HIV-1 reverse transcriptase (RT) mutations on nucleoside RT inhibitor susceptibility. Antimicrob Agents Chemother. 2012;56(5):2305–2313. 10.1128/AAC.05487-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stephan C, Dauer B, Bickel M, et al. Intensification of a failing regimen with zidovudine may cause sustained virologic suppression in the presence of resensitising mutations including K65R. J Infect. 2010;61(4):346–350. 10.1016/j.jinf.2010.06.008 [DOI] [PubMed] [Google Scholar]
- 15.Wu NC, De La Cruz J, Al-Mawsawi LQ, et al. HIV-1 quasispecies delineation by tag linkage deep sequencing. PLoS One. 2014;9(5):e97505. 10.1371/journal.pone.0097505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lepik KJ, Harrigan PR, Yip B, et al. Emergent drug resistance with integrase strand transfer inhibitor-based regimens. AIDS. 2017;31(10):1425–1434. 10.1097/QAD.0000000000001494 [DOI] [PubMed] [Google Scholar]
- 17.Aboud M, Kaplan R, Lombaard J, et al. Dolutegravir versus ritonavir-boosted lopinavir both with dual nucleoside reverse transcriptase inhibitor therapy in adults with HIV-1 infection in whom first-line therapy has failed (DAWNING): An open-label, non-inferiority, phase 3b trial. Lancet Infect Dis. 2019;19(3):253–264. 10.1016/S1473-3099(19)30036-2 [DOI] [PubMed] [Google Scholar]
- 18.Blanco JL, Rojas J, Paredes R, et al. Dolutegravir-based maintenance monotherapy versus dual therapy with lamivudine: A planned 24 week analysis of the DOLAM randomized clinical trial. J Antimicrob Chemother. 2018;73(7):1965–1971. 10.1093/jac/dky093 [DOI] [PubMed] [Google Scholar]
- 19.Quashie PK, Mesplede T, Han Y-S, et al. Characterization of the R263K mutation in HIV-1 integrase that confers low-level resistance to the second-generation integrase strand transfer inhibitor dolutegravir. J Virol. 2012;86(5):2696–2705. 10.1128/JVI.06591-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Anstett K, Fusco R, Cutillas V, Mesplède T, Wainberg MA. Dolutegravir-selected HIV-1 containing the N155H and R263K resistance substitutions does not acquire additional compensatory mutations under drug pressure that lead to higher-level resistance and increased replicative capacity. J Virol. 2015;89(20):10482–10488. 10.1128/JVI.01725-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Marras D, Bruggeman LA, Gao F, et al. Replication and compartmentalization of HIV-1 in kidney epithelium of patients with HIV-associated nephropathy. Nat Med. 2002;8:522–526. 10.1038/nm0502-522 [DOI] [PubMed] [Google Scholar]
- 22.Li JZ, Gallien S, Ribaudo H, Heisey A, Bangsberg DR, Kuritzkes DR. Incomplete adherence to antiretroviral therapy is associated with higher levels of residual HIV-1 viremia. AIDS. 2014;28(2):181–186. 10.1097/QAD.0000000000000123 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All figures and data are available within the article.