Abstract
MicroRNA(miRNA)s have been identified as an emerging class for therapeutic interventions mainly due to their extracellularly stable presence in humans and animals and their potential for horizontal transmission and action. However, treating Type 2 diabetes mellitus using this technology has yet been in a nascent state. MiRNAs play a significant role in the pathogenesis of Type 2 diabetes mellitus establishing the potential for utilizing miRNA-based therapeutic interventions to treat the disease. Recently, the administration of miRNA mimics or antimiRs in-vivo has resulted in positive modulation of glucose and lipid metabolism. Further, several cell culture-based interventions have suggested beta cell regeneration potential in miRNAs. Nevertheless, few such miRNA-based therapeutic approaches have reached the clinical phase. Therefore, future research contributions would identify the possibility of miRNA therapeutics for tackling T2DM. This article briefly reported recent developments on miRNA-based therapeutics for treating Type 2 Diabetes mellitus, associated implications, gaps, and recommendations for future studies.
Keywords: miRNA, therapeutic, miRNA-mimics, antimiRs, metabolism, beta-cell differentiation
Introduction
The discovery of the microRNA (miRNA)s, a class of non-coding RNAs with an average of 22 nucleotides, has dramatically impacted modern disease-related molecular biological experiments. The miRNAs regulate the cellular pathways by activating or suppressing a gene related to that pathway.1 As suggested, the miRNAs could activate a gene by targeting its promoter region. Alternatively, miRNA could mitigate the gene expression by controlling the translational expression degrading the corresponding messenger RNA.2 According to Bartell,3 the miRNAs act as a guide for the RNA-induced silencing complex (RISC), which splices the mRNA partially or completely. On many occasions, in animals, the 7 nucleotide sequence from 2 to 7 in miRNAs starting from the 5′ end, which is denominated as the seed sequence, binds with the 3′ end UTR of the target mRNA to guide the RISCs.1,4 This gene suppression or activation process induced by various miRNAs is related to many diseases as contributing factor to the etiology and pathogenesis of those diseases.
Accordingly, differential expression of specific miRNAs and their role in the respective disease pathology has been widely experimented with in the past 2 decades.5-7 As suggested, the respective miRNAs act within the cells/tissues or are excreted from specific donor cells to extracellular biofluids such as serum, plasma, saliva, or milk.8 These extracellular miRNAs are typically arranged in RNA-binding proteins (Argonaute 2 and nucleophosmin 1), exosomes, exosome-like vesicles, and micro-vesicles. Those extracellular miRNAs contribute to vesicle or micro-particle mediated cell-to-cell communication and modulate the cellular functions of another set of cells. A miRNA released from a donor cell could transfer extracellularly, enter another cell type, and exacerbate its regulatory action9, which can happen in paracrine or endocrine.9 Along these lines, the disease-specific extracellular miRNA expressions and their abeyant utility as disease biomarkers and therapeutic targets were widely studied.
Because of the regulatory function and the ability to horizontally transfer from one cell type to another specific cell type, modern experiments have been centered on the potential of miRNAs to be used as “therapeutic drugs” to treat diseases by reversing or modulating the dysregulated miRNAs.10,11 Some therapeutic approaches have reached even the clinical phase successfully.12,13 There were 2 main approaches to restoring the functions of disease-related cellular pathways. Either the related downregulated miRNA/s can be restored by synthetic miRNA-mimics or inhibit the upregulation and function of a miRNA by using “chemically modified antisense oligonucleotides, that is, antimiRs or antagomiRs.”14 (Briefed below).
Since the cellular pathways dysregulated in T2DM were evident to govern by many different miRNAs, these approaches were potentially valuable for treating type 2 diabetes (T2DM). Therefore, researchers have drawn attention to using miRNAs as therapeutics in treating T2DM, and comprehensive in-vitro and in-vivo studies have shown promising results.15,16 MiRNAs play a significant role, both intracellular and extracellular, in the pathophysiology of T2DM. During the past few years, the focus on the miRNA signature in the pathophysiology of diabetes mellitus, especially in T2DM, has been immensely researched.17-20 Recent studies have demonstrated that the miRNAs play a crucial role in altering the cellular pathways, that is, β-cell survival, function, and peripheral insulin signaling, which are involved in the onset and progression of diabetes mellitus, mainly T2DM.17,21,22
Moreover, distinct profiles of miRNAs were observed in the extracellular fluids of T2DM patients, such as plasma and serum, which could be used as biomarkers for detection, diagnosis, and prediction.23,24 Arbitrarily, the discovery of the regulatory role of miRNAs in T2DM has revolutionized the modern clinical research platform, creating a solid background for exploring miRNA utilization in clinical perspectives as therapeutics. In the past few decades, the exponential increment of T2DM was recorded in low- and middle-income countries (LMICs). Despite this, most of the deaths related to T2DM occurred in LMICs more than in high-income countries (HICs).25,26 Hence, it would be interesting to discuss miRNAs’ potential availability and applicability as therapeutics.
In this review, we briefly summarized the potential utility of miRNAs as therapeutics for T2DM, highlighting the current in vivo and clinical approaches. Further, the implications and issues for future research were discussed. Several attempts succeeded in reestablishing the normal expression patterns of miRNAs in-vitro and in-vivo, resulting in a positive impact in terms of glycemic control, obesity control, and regenerating β-cells. (Tables 1 and 2).
Table 1.
In-vivo animal experiments on miRNA therapeutics related to the modulation of glucose and lipid metabolism.
| Target miRNA | Duration of the treatment | Frequency | Host organ/cell type | Delivery method | Dose (mg/kg) | Technology | Outcome compared to control | Reference |
|---|---|---|---|---|---|---|---|---|
| miR-21 | 18 wk | Once a week | Db/db C57BL/6 mice | Intraperitoneal injection | 10 | Locked nucleic acid modified anti-miR-21 | Obesity ↓ Adipocyte size of WAT ↓ Pericardial fat↓ | van Rooij and Olson11 |
| No change in HbA1c level. | ||||||||
| Let-7 | 8 wk | Once a week | High-fat diet-induced C57BL/6 mice | Subcutaneous injection | 20 | DNA/LNA antimiR-7 with saline. | Lean mass↑ Muscle weight ↑ | Kim et al98 |
| Slow increment in the fat mass following the HFD. Impaired liver fat deposition | ||||||||
| Insulin sensitivity in peripheral tissues↑ | ||||||||
| miR-181b | 6 wk | Twice a week | High-fat diet-induced C57BL/6 mice | Intravenous injection | 0.6 | miR-181b mimic | Glucose tolerance ↑Adipose tissue Insulin sensitivity ↑ | Keller et al97 |
| miR-208a | 6 wk | Three consecutive days and then once a week | C57BL/6 mice | Subcutaneous injection | 10 | Locked nucleic acid modified antimiR-208 | Lean mass↑, Slow increment in the fat mass following the HFD. Smaller adipose tissues, Serum triglyceride and cholesterol ↓, Normal glucose response, Fasting insulin level, Leptin level↓ | Zhu et al109 |
| miR-103/107 | 12 wk | Two consecutive days | C5BL/6J mice (Wild type and HFD.) | Tail vein | 15 | 2′-O-methyl modified antimiRs-103 & 107 encapsulated in liver targeting lipid nanoparticles | Increased insulin signaling in liver and adipose tissues. | Trajkovski et al91 |
Table 2.
In-vitro studies on using miRNAs to differentiate stem cells to functional pancreatic cells or insulin producing pancreatic like cells.
| Target miRNA | Specie or source | Cell type | Vector or transferring method | Reference |
|---|---|---|---|---|
| miR-21 | Chicken embryo | Nestin-positive pancreatic progenitor cells | Anti-miR-21 (vector not specified) | Tups et al36 |
| miR-07 | Human embryo | Human embryonic stem cells | miR-7 mimic (vector not specified) | Schinner et al37 |
| miR-375 | Human bone marrow | Human mesenchymal stem cells | miR-375 mimic and anti-miR-9 in lentiviral packaging plasmids | Janssen et al13 |
| miR-9 |
MiRNA Mimics, Antisense Oligonucleotides, and Their Stability
miRNA therapeutics could be used to either restore the downregulated miRNAs (using miRNA mimics) or ablate the upregulation and related action of a specific miRNA/s (using antimiRs or miRNA inhibitors). Once a dysregulated miRNA was identified and experimentally validated, the up-regulation can be manipulated by introducing miRNA inhibitors. Moreover, if the specific miRNA is downregulated in a specific cell or tissue type owing to a particular disease condition, that can be modulated by the successful transfection of miRNA mimics.14
miRNA mimics were designed to imitate the functions of endogenous miRNAs. In miRNA mimic technology, the target miRNA was designed based on a pre-defined unique sequence of the target gene to ensure gene specificity.27 Often the miRNA mimics were developed as double-stranded and modified chemically to enhance their stability and cellular assimilation.14
The miRNA inhibitors suppress the activity of the miRNAs. miRNA inhibitors have the partial or complete reverse complementary sequence of the target mature miRNA, bind with it, and inhibit its ability to repress the target mRNA translation.14 According to van Rooij and Kauppinen,28 an antisense oligonucleotide that represses a specific miRNA was named an antagomir when it has the entire complementary sequence. AntimiR is the general term where the sequence is either complementary to the entire mature miRNA or just for the seed region.
Two critical aspects to be considered in miRNA therapeutics are the stability of the therapeutic miRNAs in the extracellular fluids once injected into the body and the delivery methods, that is, transfection methods. It is crucial to enhance the stability of the oligonucleotides because of the presence of RNase enzymes in the extracellular fluids inside the body. Mimics and antimiRs were often chemically modified by adding a phosphorothioate backbone to achieve stability.11 Several “antimiRs” were often modified by adding 2-OH residues to the ribose, such as 2′-O-methyl (2′-O-Me), 2′-O-methoxyethyl (2′-MOE), and locked nucleic acids (LNA), to improve stability and affinity.29 Another method was to conjugate the antimiRs to specific macromolecules such as cholesterol and tri-antennary N-acetyl galactosamine.28
On the other hand, several transfection methods are currently practised to administer these exogenous synthetic short oligonucleotides, that is, miRNA mimics and antimiRs. Those were injected directly or via a vector. The often-studied vectors were the liposome nanoparticles, atelocollagen, lentivirus, and adeno-associated viruses.30-32
MiRNAs in the Pathogenesis of T2DM
A clear understanding of the pathogenesis of T2DM and the regulatory role played by the miRNAs is required in figuring out the related therapeutic potential of miRNAs. Generally, T2DM is caused by impaired glucose homeostasis, either due to insulin resistance or inadequate secretion of insulin from the pancreas in response to glucose stimulation.33 According to the American Diabetes Association, T2DM is defined as a condition that occurs “due to progressive loss of adequate β-cell insulin secretion frequently on the background of insulin resistance.”34 The pancreatic β-cell functions and the insulin signaling pathway (IS) are the intracellular metabolic signaling pathways related to maintaining glucose homeostasis in the body.35,36 Thus, the alterations in the IS and β-cell survival & functions play a significant role in impaired glucose homeostasis5,6. The process of insulin resistance primarily appears in insulin-sensitive tissues, namely the liver, muscle, and adipose tissue, mainly as a consequence of the dysregulated IS.38 However, evidence implies that the insulin resistance of peripheral tissues enhances the demand for insulin which ultimately increases the functions of β-cells.39 It was theorized that the accretion in secreting insulin by the β-cells altered the glucose sensing, insulin synthesis, processing and secretion, proliferation, and survival of β-cells, that is, β-cell dysfunction.40 Dysfunction of these cells is known to prevail in T2DM. Depreciation of the β-cell mass and loss of regular function is underlying pathogenic factors related to pancreatic β-cell dysfunction.40
Recent studies have demonstrated that miRNAs have a crucial role in the pathogenesis of DM, mainly T2DM, by dysregulating the cellular signaling pathways. Some miRNAs highly expressed in pancreatic β-cells and peripheral insulin-sensitive cells can suppress the translation of proteins involved as signaling molecules of related pathways crucial for β-cell survival, β-cell function, and peripheral insulin sensitivity. Further, the observed specific miRNA profiles in the extracellular fluids of T2DM patients have permitted research on potential miRNA as biomarkers. Herewith, we have briefly summarized the recent findings related to miRNA biomarkers and the regulatory role played by different miRNAs on β-cell survival, function, and insulin sensitivity.
Role of miRNAs in β-cell Survival and Function
The ablation of β-cell survival and function was indirectly caused by impaired glucose homeostasis. Ample evidence suggests that the miRNA expression patterns in the pancreatic β-cells vary in T2DM patients compared to non-disease individuals. Those dysregulated miRNAs were related to β-cell survival and function.
The miR-375 was suggested to involve in β-cell proliferation and contributed to pancreatic islet development.41 The miR-486-5p and the miR-17-92 cluster regulate β-cell proliferation via targeting Phosphate and Tensin homolog (PTEN), which is a modulator of cell proliferation and apoptosis.42,43 Additionally, miR-127, miR-19a-3p, and miR-185 have been involved in β-cell proliferation by targeting Kinesin family member 3B, suppressor of cytokine signaling 3 (SOCS3) genes, respectively.44-46
The differentiation of Nestin-positive pancreatic progenitor cells into the insulin-producing cell was induced by introducing miR-21 mimics transfected through lentiviral vectors.16 Another study investigated the role of miR-375 and miR-9 in pancreatic differentiation of human bone marrow mesenchymal stem cells (hBM-MSCs) by using miR-375 mimics and anti-miR-9. They found that the upregulation of miR-375 and the downregulation of miR-9 synergistically induced the differentiation process.32 These in vivo experiments warrant more research on using miRNA to increase the generation ability of β-cells from precursor cells in the T2DM pancreas.
Apoptosis is the primary mode of pancreatic β-cell death where many related underlying processes have been documented to be mediated by miRNAs. miR-375 and miR-30a, and miR-34a have demonstrated the dysregulation of β-cell impairment.47 Based on a study conducted on β-cell lines of Mouse insulinoma 6 (MIN6) and β-TC-6 derived from mouse pancreas, it was found that the expression of p53 apoptosis effector related to Peripheral myelin protein-22 (PMP-22) (Perp) which is a Trp53 (a pro-apoptotic gene) effector, was induced by the downregulation of miR-299-5p.48
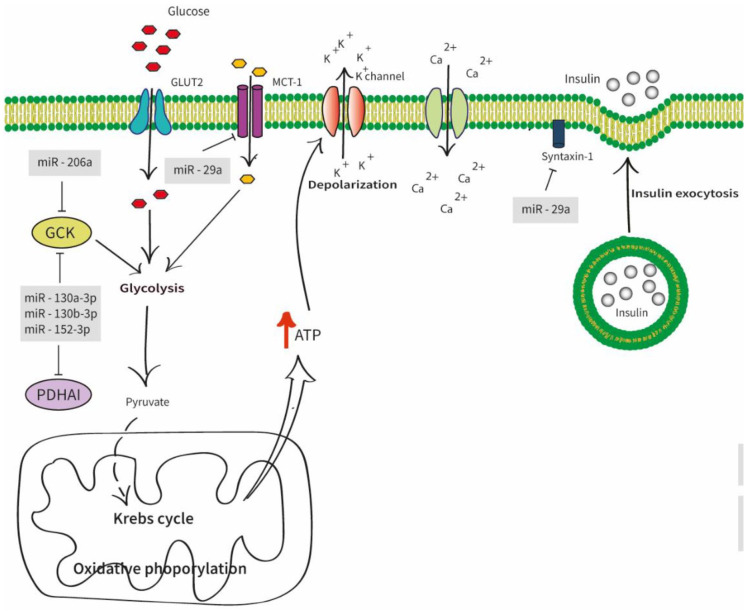
The primary function of pancreatic β-cells was insulin production and secretion (Figure 1).Many attempts argued that the miRNAs played a significant role in insulin secretion. It was found that the increased expression of miR-206 in pancreatic islets suppresses Gck and thereby controls the ATP production in β-cells, preventing the Ca2+ influx.49 The upregulation of 3 miRNAs; namely, miR-130a-3p, miR-130b-3p, and miR-152-3p, were identified to reduce the cytosolic ATP levels in INS-1 cell lines by partially targeting pyruvate dehydrogenase alpha 1 (PDHA1) and Gck.50 The t-SNARE protein syntaxin –1A(Stx-1A), one of the plasma membrane-localized SNARE proteins involved in insulin exocytosis, was also directly targeted by miR-29a.51 Further, miR-29a facilitates insulin secretion by suppressing the Monocarboxylate transporter-1 gene (Mct1), responsible for the transcription of monocarboxylate transporter-1 (MCT-1).52
Figure 1.
Diagram of different miRNAs involves in insulin secretion. The glucose uptake initiates the glucose-stimulated insulin secretion (GSIS) via the Glucose transporter 2 (GLUT2). The glucokinase (Gck) and other enzymes control glycolysis, Krebs cycle, and oxidative phosphorylation. This leads to high cytosolic adenosine triphosphate (ATP) concentration causing electrical excitation on the β-cell. The elevated ATP/adenosine diphosphate (ADP) ratio closes the K + ATP channels which ultimately aids the Ca2 + influx to the pancreatic β-cells which contributes to the insulin granule fusion from β-cells inducing insulin secretion. Abbreviations: GLUT2, glucose transporter 2; GCK, glucose kinase; PDHA1, pyruvate dehydrogenase alpha 1; ATP, adenosine triphosphate.
Role of miRNAs in the insulin signaling pathway
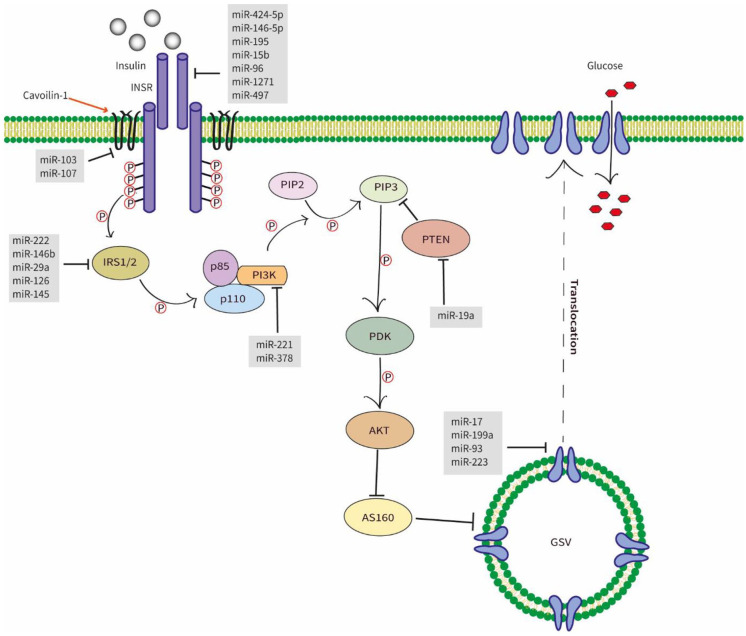
The insulin action on the peripheral tissues, such as the liver, muscles, and adipose, was exerted through the insulin signaling pathway. Insulin regulates the blood glucose level during hyperglycemic conditions. Most of the main signaling molecules of the IS pathway were under the regulation of miRNAs, namely, Insulin receptor (INSR), Insulin receptor substrate (IRS), phosphoinositide-3-kinases (PI3K), and glucose transporter 4 (GLUT4). Dysregulation of the IS pathway by the mentioned miRNAs has resulted in insulin resistance (Figure 2)
Figure 2.
Diagram of different miRNAs involves in the insulin signaling pathway. Once the insulin binds with the extracellular subunit of the INSR, intracellular subunits auto-phosphorylate, leading to the phosphorylation of IRS1/2. This leads to the activation of PI3K which, phosphorylating PIP2 to PIP3. The increased level of PIP3 activates the PDK-1 that activates the AKT which aids the translocation of GLUT4 to the cell membrane via the inactivation of AS160. The translocated GLUT4 can uptake glucose into the cell. Abbreviations: INSR, insulin signaling receptor; IRS-(1/2), insulin receptor substrate; PI3K, phosphoinositide 3-kinase; AKT2, serine/threonine kinase 2; PIP2, phosphaditylinositol bisphosphate; PIP3, phosphaditylinositol triphosphate; PTEN, phosphate and tensin homolog; PDK, phospoinositide dependent protein kinase-1; AKT, serine/threonine kinase; GLUT4, glucose transporter 4; GSV, GLUT4 storage vesicles.
Insulin receptor (INSR) is attached to the peripheral tissue cells, where the insulin binds and initiates the downstream signaling pathways responsible for glucose metabolism. It was found that miR-15b, miR-1271, miR-497, miR-424-5p, miR-146-5p, miR-96, and miR-195 suppressed the expression of INSR by targeting its 3′UTR sequence.53-58
The insulin receptor substrate is the immediate, intermediate substrate of the downstream signaling sequence of IS.59 The most predominant IRS proteins in the metabolic processes that were found to be regulated by miRNAs were IRS-1 and IRS-2, which are mainly found in skeletal muscles and liver, respectively.38 The in-vitro analysis demonstrated that miR-146b regulated glucose homeostasis by directly targeting IRS-1 in porcine primary pre-adipocyte cells.60 Yang et al suggested that miR-29a in diet-induced insulin-resistant L6 myocytes directly targeted the IRS-1, regulating glucose uptake.61 Similarly, another study conducted using primary hepatocytes isolated from the livers of male C57BL/6J mice showed overexpression of miR-222 attenuated IRS-1 through binding to its 3′UTR.62 In addition, miR-145 in hepatocellular carcinoma cell lines showed the same function.63
PI3K modulates the PIP2/PIP3 phosphorylation by binding to the SH-domain of IRS protein via the p85 subunit. This process is critical in PI3K/AKT/GLUT4 signaling pathway.64 The overexpression of miR-221 has downregulated the expression of PI3K in HepG2 cells,65 resulting in inhibited glucose uptake.
The GLUT4 is responsible for facilitated diffusion of glucose whenever insulin-induced translocation occurs.66 Thus, GLUT4 leads to accelerated glucose uptake into the cells. Notably, studies have documented several miRNAs which are responsible for the regulation of this transporter. miR-199a, miR-93, and miR-223 were documented as direct regulators.67-69 Besides that, miR-17, studied in rat skeletal muscle cell lines, had demonstrated direct inhibition of GLUT4 via negatively regulating the respective mRNA.70
Circulating miRNAs as biomarkers for T2DM
Despite the regulatory role of miRNAs in T2DM, recent research focused on the miRNA signature in extracellular fluids due to the disease conditions and the possibility of using those as biomarkers to diagnose and prognosticate. Several meta-analyses were conducted, suggesting different lists of miRNAs as potential biomarkers for T2DM patients. A comprehensive meta-analysis by Zhu and Leung on selected comparative miRNA profiling studies from 1993 to 2015 has suggested 40 miRNAs that were significantly dysregulated in T2DM patients. In this meta-analysis, they have suggested 8 potential circulating biomarkers, namely, miR-29a, miR-34a, miR-375, miR-103, miR-107, miR-132, miR-142-3p, and miR-144, and 2 potential tissue biomarkers; miR-199a-3p and miR-223.71 However, another meta-analysis conducted on case-control miRNA profiling studies from 1996 to 2016, explicitly considering the stress-related miRNA biomarkers related to T2DM, has suggested miR-148b, miR-223, miR-130a, miR-19a, miR-26b, and miR-27b as potential biomarkers for T2DM. Further, they have revealed 2 tissue-specific miRNA biomarkers in the cornea and heart, miR-146a and miR-21, respectively.72
However, the trend in finding new circulatory miRNA biomarkers has continued. Recently several new miRNA biomarkers emerged, expanding the research area.73-77 Effective prediction of T2DM or detecting the early onset of the disease was a promising research area77 because the efficacy of the presently utilized treatments would increase if implemented in the initial phase. The early medication would have the ability to prevent T2DM from leading the patients toward adverse complications such as renal failures, blindness, cardiovascular diseases, pulmonary respiratory diseases, and death. Standing on this phenomenon, the probability of utilizing miRNAs as biomarkers to predict the progression of T2DM has become a research interest. An association between the miR-483-5p with the early onset of T2DM was also recorded.78 The combination of different expressions of miR-486, miR-146b, and miR-15b in obese children have been suggested as potential biomarkers for predicting T2DM status in their adulthood.79 Another study suggested that the people with a lower level of miR-103, miR-28-3p, miR-29a miR-9, and a higher level of miR-30a-5p and miR-150 might have a significantly high potential to acquire T2DM in the future.23 Belongie et al80 suggested 6 miRNAs (miR-497, miR-25, let-7c,miR-205, miR-375, and miR-223) as early predictors of β-cell dysfunction.
Several studies have expanded their research on the possible associations between conventional biomarkers and the corresponding miRNA expressions. Mononen et al81 have found that the several whole blood miRNA levels have significantly associated with the glycemic status (miR-144-5p, miR-122-5p, miR-148a-3p, miR-589-5p, and let-7a-5p), glucose level (miR-144-5p and miR-122-5p), glycated albumin level (miR-148a-3p, miR-15b-3p, miR-93-3p, miR-146b-5p, miR-221-3p, miR-189-3p, miR-642a-5p, and miR-181-2-3p) insulin level and HOMA-IR levels (miR-144-5p,miR-122-5p,miR-184, and miR-339-3p).
MiRNA as Therapeutics for T2DM
The therapeutic potential of some of the above-mentioned regulatory miRNAs for T2DM was tried to treat animals with T2DM and obesity. Huge interest has been drawn to the ability to control the differentially expressed miRNAs that aggravate T2DM, that is, miRNA modulation.82 The main reason was the defects in the current drug solutions for T2DM. Some commonly used hypoglycemia-promoting drugs tend to exert side effects like increasing cardiovascular risk factors.83 Another common defect in the current therapeutic methods of T2DM was its inability to diminish the disease’s progression permanently. Although antidiabetic drugs can alter the insulin sensitivity in adipose tissues and muscle tissues or insulin secretion in liver cells, finding a perfect antidiabetic drug to address the disease has continued.84
The therapeutic effect of miRNA mimics and inhibitors have currently been experimented with at the preclinical stage on other diseases, that is, MRX34 for liver cancer85 and miravirsen for hepatitis C virus (HCV).86 Miravirsen was an antiviral intervention designed to diminish the action of miR-122, which was highly expressed in Hepatitis C virus (HCV)-infected livers of animals and humans. MiR-122 was implied to promote the action of the respective virus.87 Miravirsen is a locked nucleic acid-modified antisense oligonucleotide designed to complement the miR-122 expressed in the HCV-infected liver.88 The potency of utilizing these LNA-modified antimiRs as the antiviral intervention was studied in chimpanzees (in vivo) and has provided chronic suppression of HCV activity in their livers with fewer side effects.87 Later it was studied on HCV patients as a phase 2 clinical study, which produced a “dose-dependent reduction” of HCV RNA levels without side effects.13
MRX34 was considered the first miRNA mimic therapy to reach the clinical phase (Phase 1), which tried to replace miR-34 in patients with liver cancer. The amphoteric lipid nanoparticle was used as the vector.89 miR-34 family plays a significant role in tumor suppression (reviewed in Bader12) and is generally downregulated in cancer patients. Evidence implied that miR-34 targets a vast amount of oncogenes, and therapeutic ingestion of miR-34 as mimics exhibits lower tumor burden and less metastasis formation in vitro and in vivo (reviewed in Zhang et al89).
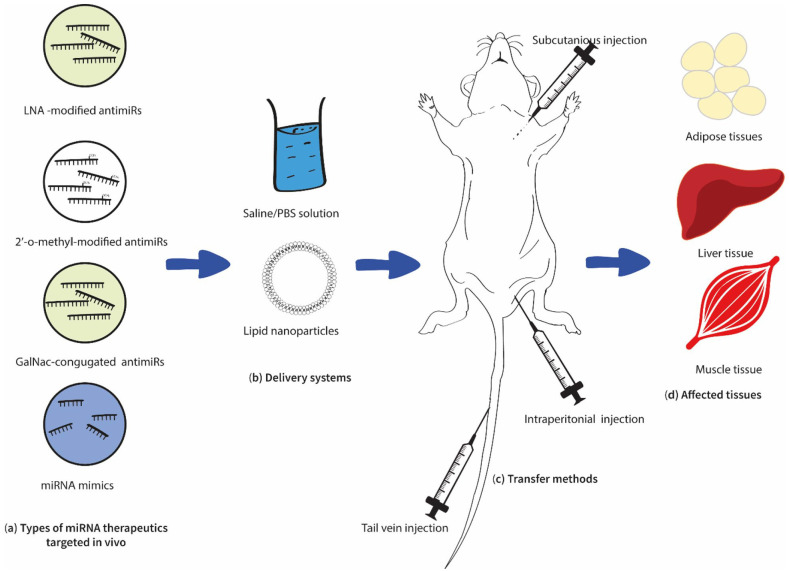
However, concerning T2DM, still, this technology has been restricted to in-vitro cell culture-based and in-vivo animal-based studies (Figure 3) with minimum clinical approaches. However, most of the cell culture studies conducted to evaluate the regulatory role of different miRNAs on T2DM have followed this technology by transfecting the required miRNA mimics and inhibitors to the isolated or seeded cell types such as murine islets MIN6 or INS-1 cells using adeno-associated viruses (AAV) or lipid encapsulated.90,91 In addition, several in vivo studies were also conducted on this phenomenon, which resulted in promising results.
Figure 3.
Summary of the current in vivo approaches in miRNA therapeutics for T2DM. Recent in vivo studies have evaluated the possibility of utilizing LNA-modified anitimiRs, 2′-o-methyl modified antimiRs, and miRNA mimics (a), delivered in saline/PBS (Phosphate buffer solution) or lipid nanoparticles (b), and subcutaneously, intraperitoneally or via tail vein (c). Treated subjects showed positive impacts on adipose, liver, and muscle tissues (d) (Table 1).
Studies on miRNA mimics and antimiRs suggested as therapeutics for T2DM
MiR-103/107 was considered the first miRNA family to be a clinical candidate for treating T2DM patients. The related product is RG-125 (AZD407) which is a GalNac-conjugated antimiR that targets the miR-103/107 family and will be tested on non-alcoholic steatohepatitis (NASH) patients with T2DM.92 Several in vitro and in vivo studies have proved the significant role played by this miRNA family, and in vivo administrations for non-human trials have shown positive results. For example, the tail vein administered 2′-O-methyl modified antimiRs for 12 weeks reduced the miR-103/107 expressions in diabetic-induced obese (DIO) and wild-type C57BL/6J mice91 (Table 1). Once treated, both mouse types showed improved glucose tolerance and insulin sensitivity. The association between the miR-103/107 family and glucose homeostasis has been immensely suggested in vitro and in vivo. Positive correlation between the HOMA index and the miR-103/107 expressions and previous studies that implied miR-103 & miR-107 were highly expressed in the livers of T2DM animals and humans93 provide the relationship between these miRNAs with T2DM.
In addition, several in-vivo studies have recorded positive results using antisense oligonucleotide and miRNA mimics transfection, on lipid and glucose metabolisms (Table 1). The miR-103/107 family and several other miRNAs were suggested in studies conducted on diet-induced mice treated with either antimiRs or miRNA mimics. Those treatments have restored or even increased their insulin sensitivity and glucose metabolism compared to non-treated controls.91,94,95 High-fat diet-induced mice treated with antimiRs of miR-21, miR-181b, miR-208a, and Let-7 have shown decreased adipogenesis, increased glucose tolerance, and lean mass increment.94-96
Long-term treatment of (intraperitoneally) the antisense oligonucleotides for miR-21 has positively regulated lipid metabolism in Db/db mice. It ablates the obesity and adipocyte size of the white adipose tissues yet results in no change in the HbA1c level.15 Recent studies have demonstrated that miR-21 acts on both glucose homeostasis and adipogenesis. Obese individuals have demonstrated high levels of miR-21 in their white adipose tissues compared to lean controls, while in vitro studies have implied its association with adipogenesis.97 In addition, several in vitro studies show that miR-21 targets several genes related to adipogenic differentiation.98 Moreover, some studies demonstrated the role of miR-21 on hepatic glucose metabolism99,100 and beta cell dysfunction,101,102 indicating its potential to become a therapeutic target of T2DM.
miR-181b was one of the anti-inflammatory regulators decreased in HFD mice models and T2DM patients.103,104 Moreover, higher levels of miR-181b in the adipose tissues improve insulin sensitivity and ameliorate inflammation advocating its potential as a therapeutic for T2DM. However, while experimenting with the miR-181b activity, Sun et al95 transfected miR-181b mimics to HFD mice models intravenously and improved glucose tolerance and insulin sensitivity. Further, they have observed less macrophage accumulation and macrophage-related markers in the white adipose tissues of miR-181b mimic treated mice exhibiting reduced inflammation.95
miR-208a is a cardiac-specific miRNA that affects cardiac function and was highly expressed in the hearts of individuals who suffered from myocardial infarction (MI).105 In vivo treatment of locked nucleic acid (LNA)-modified antimiR resulted in positive cardiac function and survival in rat models suffering from diastolic dysfunction.106 However, when HFD-induced mice models were treated with animiR-208a, they displayed high glucose response, less weight gain, less visceral and subcutaneous fats, and normal glucose response compared to control-antimiR treated HFD mice.107 As suggested, the antimiR-208a acts on the miR-208a expressed in those mice hearts inducing the MED13, a subunit in the mediator complex. It was the elevated MED13 that was responsible for the improved glucose homeostasis.107
The Let-7 family modulates glucose tolerance and pancreatic insulin secretion.108 Higher expression of Let-7 in the pancreas of mice has reduced insulin secretion and is suggested to regulate body weight, growth, and partly, the muscle IS.109 Let-7b-5p was overexpressed in T2DM patients and proposed to modulate pancreatic insulin secretion.110 However, the antimiR treatment for Let-7 has reduced the global Let-7 expression levels restoring the IS in mice muscles and livers.96 Further, the treatment tends to increase muscle mass while managing the fat mass as per control.96 Therefore, knocking down the Let-7 family using an antimiR as a treatment for T2DM might be another viable option, yet more studies must be conducted.
Another promising application currently studied was the usage of miRNA mimics and inhibitors to generate insulin-secreting cells from various sources such as nestin-positive pancreatic progenitor cells, human embryonic stem cells (hESC), and human bone marrow mesenchymal stem cells (hBM-MSCs) (Table 2). In the later stages of T2DM, patients suffer less beta-cell mass and function.33,111 Some alternative treatments for this issue were to either replace the pancreatic beta cells or increase the generation ability of beta cells from precursor cells.112,113 Suppose there is a possibility of administering miRNAs to promote the stem cells to differentiate into insulin-producing pancreatic beta cells; that would be an effective regenerative therapy. In-vitro studies (Table 2) have proved the potential of miRNAs in differentiating various stem cell types into functional pancreatic beta cells. However, only a few works of literature on in-vivo studies were found in this regard, catering to more research potential.
Recommendations and future directions for T2DM targeted miRNA therapeutics
However, certain obstacles must be tackled for T2DM-targeted miRNA therapeutics to reach from in-vitro to bedside. In the article, “Developing miRNA Therapeutics,” van Rooij et al14 discussed that the main challenge in this technology over in vitro approach was the production of off-targets and lack of effective delivery of miRNA mimics /inhibitors to live cells. Since the regulatory role of one specific miRNA is highly diversified, mere inhibition or encouragement using this technology to address one disease condition would cause another disease. For instance, the miR-144 was recorded to repress the growth of colorectal cancer cells and could be used as a suppressor to treat colorectal cancer.114 However, the same miRNA promotes T2DM by impairing the peripheral insulin signaling pathway.115 Thus, if miRNA-144 mimic were transfected to a specific colorectal patient, insulin resistance would occur in his/her body, leading to T2DM mainly in the longer run. However, this issue can be addressed by finding an effective delivery method to transfect the miRNA to the specific tissue that has been affected or tissue-specific administration. Another way is to target only disease-specific miRNAs whose expression pattern is ubiquitous in all related tissues, which is very challenging. Therefore, experiments must address this gap and cater to future technology delivering these synthetic oligonucleotides more effectively and tissue-specific. However, having multiple targets and the ability to affect numerous related signaling pathways in numerous tissues would also be beneficial when planning therapeutic interventions for complex diseases like T2DM.
Further, in many of these therapeutic approaches, the antimiRs that have been specifically designed were parenterally injected directly into the host using saline or PBS-like convey medium.94,96 However, treating humans would not be therapeutically feasible since the target tissues are peripheral, and the administration must be blood-based. So, viable vector-mediated therapeutic delivery possibilities must be evaluated in-vivo. Given the vast number of dysregulated miRNAs directly affecting insulin sensitivity, more miRNAs still need to be evaluated for their therapeutic potential.116 Identifying and effectively delivering miRNA candidates whose intonation directly affects only peripheral tissue insulin sensitivity in an in-vivo environment is still something to be worked out, which has a plausible future in improving miRNA therapeutic interventions to address T2DM.
Even though this therapeutic approach still lies on its bench level, future research contributions would identify the possibility of miRNA mimic/inhibitor transfection for tackling T2DM.
Footnotes
The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This research was funded by World Bank under the “Development-Oriented Research” scheme of the “Accelerating Higher Education and Expansion (AHEAD)” project, Ministry of Education, Sri Lanka, grant number AHEAD/DOR/STEM+HEMS No.78.
Author Contributions: Conceptualization, D.A.S., P.A.D.S and S.K.Y; investigation, P.A.D.S., and B.I.L.M.M.; resources, W.K.R.R.D.; data curation, P.A.D.S., and B.I.L.M.M; writing—original draft preparation, P.A.D.S.; writing—review and editing, D.A.S., S.K.Y., and W.Y.H.; visualiza-tion, P.A.D.S.; supervision, D.A.S., J.M.K.J.K.P., A.S.D., P.K., and U.S.; project administration, W.K.R.R.D.; funding acquisition, D.A.S., W.K.R.R.D, and J.M.K.J.K.P. All authors have read and agreed to the published version of the manuscript.
ORCID iD: PADS Palihaderu  https://orcid.org/0000-0003-0282-1868
https://orcid.org/0000-0003-0282-1868
References
- 1. Tang G, Tang X, Mendu V, et al. The art of microRNA: various strategies leading to gene silencing via an ancient pathway. Biochim Biophys Acta. 2008;1779:655-662. [DOI] [PubMed] [Google Scholar]
- 2. Eulalio A, Huntzinger E, Izaurralde E. Getting to the root of miRNA-Mediated gene silencing. Cell. 2008;132:9-14. [DOI] [PubMed] [Google Scholar]
- 3. Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281-297. [DOI] [PubMed] [Google Scholar]
- 4. Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15-20. [DOI] [PubMed] [Google Scholar]
- 5. Peng Y, Croce CM. The role of microRNAs in human cancer. Signal Transduct Target Ther. 2016;1:15004. doi: 10.1038/sigtrans.2015.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Zhang B, Pan X, Cobb GP, Anderson TA. MicroRNAs as oncogenes and tumor suppressors. Dev Biol. 2007;302:1-12. [DOI] [PubMed] [Google Scholar]
- 7. Saeedi Borujeni MJ, Esfandiary E, Taheripak G, Codoñer-Franch P, Alonso-Iglesias E, Mirzaei H. Molecular aspects of diabetes mellitus: resistin, microRNA, and exosome. J Cell Biochem. 2018;119(2):1257-1272. [DOI] [PubMed] [Google Scholar]
- 8. Sohel MH. Extracellular/circulating microRNAs: release mechanisms, functions and challenges. Achiev Life Sci. 2016;10:175-186. [Google Scholar]
- 9. Chen X, Liang H, Zhang J, Zen K, Zhang CY. Horizontal transfer of microRNAs: molecular mechanisms and clinical applications. Protein Cell. 2012;3:28-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Krützfeldt J. Strategies to use microRNAs as therapeutic targets. Best Pract Res Clin Endocrinol Metab. 2016;30:551-561. [DOI] [PubMed] [Google Scholar]
- 11. van Rooij E, Olson EN. MicroRNA therapeutics for cardiovascular disease: opportunities and obstacles. Nat Rev Drug Discov. 2012;11:860-872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Bader AG. MiR-34 – a microRNA replacement therapy is headed to the clinic. Front Genet. 2012;3:1-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Janssen HLA, Reesink HW, Lawitz EJ, et al. Treatment of HCV infection by targeting microRNA. New Engl J Med. 2013;368:1685-1694. [DOI] [PubMed] [Google Scholar]
- 14. van Rooij E, Purcell AL, Levin AA. Developing microRNA therapeutics. Circ Res. 2012;110:496-507. [DOI] [PubMed] [Google Scholar]
- 15. Seeger T, Fischer A, Muhly-Reinholz M, Zeiher AM, Dimmeler S. Long-term inhibition of miR-21 leads to reduction of obesity in db/db mice. Obesity. 2014;22:2352-2360. [DOI] [PubMed] [Google Scholar]
- 16. Bai C, Li X, Gao Y, et al. Role of microRNA-21 in the formation of insulin-producing cells from pancreatic progenitor cells. Biochim Biophys Acta. 2016;1859:280-293. [DOI] [PubMed] [Google Scholar]
- 17. Tang X, Tang G, Ozcan S. Role of microRNAs in diabetes. Biochim Biophys Acta. 2008;1779:697-701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. McClelland AD, Kantharidis P. MicroRNA in the development of diabetic complications. Clin Sci. 2014;126:95-110. [DOI] [PubMed] [Google Scholar]
- 19. Zhang Y, Sun X, Icli B, Feinberg MW. Emerging roles for microRNAs in diabetic microvascular disease: novel targets for therapy. Endocr Rev. 2017;38:145-168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Rosado JA, Diez-Bello R, Salido GM, Jardin I. Fine-tuning of microRNAs in type 2 diabetes mellitus. Curr Med Chem. 2019;26:4102-4118. [DOI] [PubMed] [Google Scholar]
- 21. Guay C, Roggli E, Nesca V, Jacovetti C, Regazzi R. Diabetes mellitus, a microRNA-related disease? Transl Res. 2011;157:253-264. [DOI] [PubMed] [Google Scholar]
- 22. Tiwari J, Gupta G, de Jesus Andreoli Pinto T, et al. Role of microRNAs (miRNAs) in the pathophysiology of diabetes mellitus. Panminerva Med. 2018;60:25-28. [DOI] [PubMed] [Google Scholar]
- 23. Sidorkiewicz I, Niemira M, Maliszewska K, et al. Circulating miRNAs as a predictive biomarker of the progression from prediabetes to diabetes: outcomes of a 5-year prospective observational study. J Clin Med. 2020;9:2184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Abbasi A, Sahlqvist AS, Lotta L, et al. A systematic review of biomarkers and risk of incident type 2 diabetes: an overview of epidemiological, prediction and aetiological research literature. PLoS One. 2016;11:e0163721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Noncommunicable diseases country profiles 2018. Geneva: World Health Organization; 2018. [Google Scholar]
- 26. Beran D, Colagiuri S, Gregg E, et al. Global Report on Diabetes. Vol. 978. World Health Organization; 2016. Accessed January 2, 2022. https://apps.who.int/iris/handle/10665/204871 [Google Scholar]
- 27. Wang Z. MicroRNA interference technologies. In: Wang Z, ed. MicroRNA Interference Technologies. Springer Berlin, Heidelberg; 2009;93-101. [Google Scholar]
- 28. van Rooij E, Kauppinen S. Development of micro RNA therapeutics is coming of age. EMBO Mol Med. 2014;6:851-864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Liu Z, Sall A, Yang D. MicroRNA: an emerging therapeutic target and intervention tool. Int J Mol Sci. 2008;9:978-999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Pramanik D, Campbell NR, Karikari C, et al. Restitution of tumor suppressor microRNAs using a systemic nanovector inhibits pancreatic cancer growth in mice. Mol Cancer Ther. 2011;10:1470-1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Takeshita F, Patrawala L, Osaki M, et al. Systemic delivery of synthetic microRNA-16 inhibits the growth of metastatic prostate tumors via downregulation of multiple cell-cycle genes. Mol Ther. 2010;18:181-187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Jafarian A, Taghikani M, Abroun S, et al. The generation of insulin producing cells from human mesenchymal stem cells by miR-375 and anti-miR-9. PLoS One. 2015;10:e0128650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Galicia-Garcia U, Benito-Vicente A, Jebari S, et al. Pathophysiology of type 2 diabetes mellitus. Int J Mol Sci. 2020;21:6275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Care D, Suppl SS. Classification and diagnosis of diabetes: standards of medical care in diabetes-2020. Diabetes Care. 2020;43:S14-S31. [DOI] [PubMed] [Google Scholar]
- 35. Röder PV, Wu B, Liu Y, Han W. Pancreatic regulation of glucose homeostasis. Exp Mol Med. 2016;48:e219-e219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Tups A, Benzler J, Sergi D, Ladyman SR, Williams LM. Central regulation of glucose homeostasis. Compr Physiol. 2017;7:741-764. [DOI] [PubMed] [Google Scholar]
- 37. Schinner S, Scherbaum WA, Bornstein SR, Barthel A. Molecular mechanisms of insulin resistance. Diabet Med. 2005;22:674-682. [DOI] [PubMed] [Google Scholar]
- 38. Wilcox G. Insulin and insulin resistance. Clin Biochem Rev. 2005;26:19-39. Accessed January 2, 2022. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC1204764/ [PMC free article] [PubMed] [Google Scholar]
- 39. Halban PA, Polonsky KS, Bowden DW, et al. β-Cell failure in type 2 diabetes: postulated mechanisms and prospects for prevention and treatment. J Clin Endocrinol Metab. 2014;99:1983-1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Weir GC, Laybutt DR, Kaneto H, Bonner-Weir S, Sharma A. Beta-cell adaptation and decompensation during the progression of diabetes. Diabetes. 2001;50:S154-S159. [DOI] [PubMed] [Google Scholar]
- 41. Joglekar MV, Joglekar VM, Hardikar AA. Expression of islet-specific microRNAs during human pancreatic development. Gene Expr Patterns. 2009;9:109-113. [DOI] [PubMed] [Google Scholar]
- 42. Tian H, Yang J, Xie Z, Liu J. MiR-486-5p regulates pancreatic β cell function in type 2 diabetes mellitus by targeting PTEN and FOXO1. Pharmazie. 2018;73:477-481. [Google Scholar]
- 43. Chen Y, Tian L, Wan S, et al. MicroRNA-17-92 cluster regulates pancreatic beta-cell proliferation and adaptation. Mol Cell Endocrinol. 2016;437:213-223. [DOI] [PubMed] [Google Scholar]
- 44. Shen Z, Jiang H, Hsu HT, et al. MicroRNA-127 inhibits cell proliferation via targeting kif3b in pancreatic β cells. Aging. 2019;11:1342-1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Li Y, Luo T, Wang L, Wu J, Guo S. MicroRNA-19a-3p enhances the proliferation and insulin secretion, while it inhibits the apoptosis of pancreatic β cells via the inhibition of SOCS3. Int J Mol Med. 2016;38:1515-1524. [DOI] [PubMed] [Google Scholar]
- 46. Bao L, Fu X, Si M, et al. MicroRNA-185 targets SOCS3 to inhibit beta-cell dysfunction in diabetes. PLoS One. 2015;10:e0116067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Li Y, Zhang T, Zhou Y, et al. A presenilin/notch1 pathway regulated by miR-375, miR-30a, and miR-34a mediates glucotoxicity induced-pancreatic beta cell apoptosis. Sci Rep. 2016;6:12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Huang Q, You Y, Sun Y, et al. Glucolipotoxicity-inhibited miR-299-5p regulates β-cell function and survival. Diabetes. 2018;67:2280-2292. [DOI] [PubMed] [Google Scholar]
- 49. Vinod M, Patankar JV, Sachdev V, et al. MiR-206 is expressed in pancreatic islets and regulates glucokinase activity. Am J Physiol Endocrinol Metab. 2016;311:E175-E185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Ofori JK, Salunkhe VA, Bagge A, et al. Elevated miR-130a/miR130b/miR-152 expression reduces intracellular ATP levels in the pancreatic beta cell. Sci Rep. 2017;7:1-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Bagge A, Dahmcke C, Dalgaard L. Syntaxin-1a is a direct target of miR-29a in insulin-producing β-cells. Horm Metab Res. 2013;45:463-466. [DOI] [PubMed] [Google Scholar]
- 52. Pullen TJ, da Silva Xavier G, Kelsey G, Rutter GA. MiR-29a and miR-29b contribute to pancreatic β-Cell-Specific silencing of monocarboxylate transporter 1 (Mct1). Mol Cell Biol. 2011;31:3182-3194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Yang WM, Jeong HJ, Park SY, Lee W. Saturated fatty acid-induced miR-195 impairs insulin signaling and glycogen metabolism in HepG2 cells. FEBS Lett. 2014;588:3939-3946. [DOI] [PubMed] [Google Scholar]
- 54. Yang WM, Jeong HJ, Park SW, Lee W. Obesity-induced miR-15b is linked causally to the development of insulin resistance through the repression of the insulin receptor in hepatocytes. Mol Nutr Food Res. 2015;59:2303-2314. [DOI] [PubMed] [Google Scholar]
- 55. Min KH, Yang WM, Lee W. Saturated fatty acids-induced miR-424–5p aggravates insulin resistance via targeting insulin receptor in hepatocytes. Biochem Biophys Res Commun. 2018;503:1587-1593. [DOI] [PubMed] [Google Scholar]
- 56. Yang WM, Min K-H, Lee W. Induction of miR-96 by dietary saturated fatty acids exacerbates hepatic insulin resistance through the suppression of INSR and IRS-1. PLoS One. 2016;11:e0169039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Wang X, Wang M, Li H, et al. Upregulation of miR-497 induces hepatic insulin resistance in E3 rats with HFD-MetS by targeting insulin receptor. Mol Cell Endocrinol. 2015;416:57-69. [DOI] [PubMed] [Google Scholar]
- 58. Yang WM, Min K-H, Lee W. MiR-1271 upregulated by saturated fatty acid palmitate provokes impaired insulin signaling by repressing INSR and IRS-1 expression in HepG2 cells. Biochem Biophys Res Commun. 2016;478:1786-1791. [DOI] [PubMed] [Google Scholar]
- 59. White MF. Regulating insulin signaling and β-cell function through IRS proteins. Can J Physiol Pharmacol. 2006;84:725-737. [DOI] [PubMed] [Google Scholar]
- 60. Zhu Y-L, Chen T, Xiong J-L, et al. MiR-146b inhibits glucose consumption by targeting IRS1 gene in porcine primary adipocytes. Int J Mol Sci. 2018;19:783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Yang WM, Jeong HJ, Park SY, Lee W. Induction of miR-29a by saturated fatty acids impairs insulin signaling and glucose uptake through translational repression of IRS-1 in myocytes. FEBS Lett. 2014;588:2170-2176. [DOI] [PubMed] [Google Scholar]
- 62. Ono K, Igata M, Kondo T, et al. Identification of microRNA that represses IRS-1 expression in liver. PLoS One. 2018;13:e0191553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Wang Y, Hu C, Cheng J, et al. MicroRNA-145 suppresses hepatocellular carcinoma by targeting IRS1 and its downstream Akt signaling. Biochem Biophys Res Commun. 2014;446:1255-1260. [DOI] [PubMed] [Google Scholar]
- 64. Hemmings BA, Restuccia DF. PI3K-PKB/Akt Pathway. Cold Spring Harb Perspect Biol. 2012;4:a011189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Huang F, Chen J, Wang J, Zhu P, Lin W. Palmitic acid induces MicroRNA-221 expression to decrease glucose uptake in HepG2 cells via the PI3K/AKT/GLUT4 pathway. Biomed Res Int. 2019;2019:1-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Brunetti A, Arcidiacono B, Foti DP, Semple RK. Editorial: transcriptional regulation of glucose metabolism: gaps and controversies. Front Endocrinol. 2019;10:1-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Yan ST, Li CL, Tian H, et al. MiR-199a is overexpressed in plasma of type 2 diabetes patients which contributes to type 2 diabetes by targeting GLUT4. Mol Cell Biochem. 2014;397:45-51. [DOI] [PubMed] [Google Scholar]
- 68. Chen YH, Heneidi S, Lee JM, et al. MiRNA-93 inhibits GLUT4 and is overexpressed in adipose tissue of polycystic ovary syndrome patients and women with insulin resistance. Diabetes. 2013;62:2278-2286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Chuang TY, Wu HL, Chen CC, et al. MicroRNA-223 expression is upregulated in insulin resistant human adipose tissue. J Diabetes Res. 2015;2015:1-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Xiao D, Zhou T, Fu Y, et al. MicroRNA-17 impairs glucose metabolism in insulin-resistant skeletal muscle via repressing glucose transporter 4 expression. Eur J Pharmacol. 2018;838:170-176. [DOI] [PubMed] [Google Scholar]
- 71. Zhu H, Leung SW. Identification of microRNA biomarkers in type 2 diabetes: a meta-analysis of controlled profiling studies. Diabetologia. 2015;58:900-911. [DOI] [PubMed] [Google Scholar]
- 72. Liang Y, Li J, Xiao H, He Y, Zhang L, Yan Y. Identification of stress-related microRNA biomarkers in type 2 diabetes mellitus: a systematic review and meta-analysis. J Diabetes. 2020;12:633-644. [DOI] [PubMed] [Google Scholar]
- 73. Ghai V, Kim T-K, Etheridge A, et al. Extracellular vesicle encapsulated microRNAs in patients with type 2 diabetes are affected by metformin treatment. J Clin Med. 2019;8:617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Yang ZM, Chen LH, Hong M, et al. Serum microRNA profiling and bioinformatics analysis of patients with type 2 diabetes mellitus in a Chinese population. Mol Med Rep. 2017;15:2143-2153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Avgeris M, Kokkinopoulou I, Maratou E, et al. Blood-based analysis of 84 microRNAs identifies molecules deregulated in individuals with type-2 diabetes, risk factors for the disease or metabolic syndrome. Diabetes Res Clin Pract. 2020;164:108187. [DOI] [PubMed] [Google Scholar]
- 76. de Candia P, Spinetti G, Specchia C, et al. A unique plasma microRNA profile defines type 2 diabetes progression. PLoS One. 2017;12:e0188980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Kraniotou C, Karadima V, Bellos G, Tsangaris GT. Predictive biomarkers for type 2 of diabetes mellitus: bridging the gap between systems research and personalized medicine. J Proteomics. 2018;188:59-62. [DOI] [PubMed] [Google Scholar]
- 78. Gallo W, Esguerra JLS, Eliasson L, Melander O. MiR-483-5p associates with obesity and insulin resistance and independently associates with new onset diabetes mellitus and cardiovascular disease. PLoS One. 2018;13:e0206974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Cui X, You L, Zhu L, et al. Change in circulating microRNA profile of obese children indicates future risk of adult diabetes. Metabolism. 2018;78:95-105. [DOI] [PubMed] [Google Scholar]
- 80. Belongie KJ, Ferrannini E, Johnson K, Andrade-Gordon P, Hansen MK, Petrie JR. Identification of novel biomarkers to monitor β-cell function and enable early detection of type 2 diabetes risk. PLoS One. 2017;12:e0182932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Mononen N, Lyytikäinen LP, Seppälä I, et al. Whole blood microRNA levels associate with glycemic status and correlate with target mRNAs in pathways important to type 2 diabetes. Sci Rep. 2019;9:8887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Thanikachalam PV, Ramamurthy S, Wong ZW, et al. Current attempts to implement microRNA-based diagnostics and therapy in cardiovascular and metabolic disease: a promising future. Drug Discov Today. 2018;23:460-480. [DOI] [PubMed] [Google Scholar]
- 83. Ferrannini E, DeFronzo RA. Impact of glucose-lowering drugs on cardiovascular disease in type 2 diabetes. Eur Heart J. 2015;36:2288-2296. [DOI] [PubMed] [Google Scholar]
- 84. Levina A, Lay PA. Metal-based anti-diabetic drugs: advances and challenges. Dalton Trans. 2011;40:11675-11686. [DOI] [PubMed] [Google Scholar]
- 85. Bouchie A. First microRNA mimic enters clinic. Nat Biotechnol. 2013;31:577. [DOI] [PubMed] [Google Scholar]
- 86. Gebert LF, Rebhan MA, Crivelli SE, Denzler R, Stoffel M, Hall J. Miravirsen (SPC3649) can inhibit the biogenesis of miR-122. Nucleic Acids Res. 2014;42:609-621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Lanford RE, Hildebrandt-Eriksen ES, Petri A, et al. Therapeutic silencing of microRNA-122 in primates with chronic hepatitis C virus infection. Science. 2010;327:198-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Ottosen S, Parsley TB, Yang L, et al. In vitro antiviral activity and preclinical and clinical resistance profile of miravirsen, a novel anti-hepatitis C virus therapeutic targeting the human factor miR-122. Antimicrob Agents Chemother. 2015;59:599-608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Zhang L, Liao Y, Tang L. MicroRNA-34 family: a potential tumor suppressor and therapeutic candidate in cancer. J Exp Clin Cancer Res. 2019;38:1-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Wang S, Wen X, Han XR, et al. MicroRNA-30d preserves pancreatic islet β-cell function through negative regulation of the JNK signaling pathway via SOCS3 in mice with streptozotocin-induced diabetes mellitus. J Cell Physiol. 2018;233:7343-7355. [DOI] [PubMed] [Google Scholar]
- 91. Trajkovski M, Hausser J, Soutschek J, et al. MicroRNAs 103 and 107 regulate insulin sensitivity. Nature. 2011;474:649-653. [DOI] [PubMed] [Google Scholar]
- 92. Regulus Therapeutics. RG-125 (AZD4076), a microRNA Therapeutic Targeting microRNA-103/107 for the Treatment of NASH in Patients with Type 2 Diabetes/Pre-Diabetes, Selected as Clinical Candidate by AstraZeneca. Inc., Regulus Therapeutics. 2015. Accessed December 30, 2021. http://www.regulusrx.com/
- 93. Luo M, Xu C, Luo Y, Wang G, Wu J, Wan Q. Circulating miR-103 family as potential biomarkers for type 2 diabetes through targeting CAV-1 and SFRP4. Acta Diabetol. 2020;57:309-322. [DOI] [PubMed] [Google Scholar]
- 94. Esau C, Davis S, Murray SF, et al. MiR-122 regulation of lipid metabolism revealed by in vivo antisense targeting. Cell Metab. 2006;3:87-98. [DOI] [PubMed] [Google Scholar]
- 95. Sun X, Lin J, Zhang Y, et al. MicroRNA-181b improves glucose homeostasis and insulin sensitivity by regulating endothelial function in white adipose tissue. Circ Res. 2016;118:810-821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Frost RJ, Olson EN. Control of glucose homeostasis and insulin sensitivity by the let-7 family of microRNAs. Proc Natl Acad Sci U S A. 2011;108:21075-21080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Keller P, Gburcik V, Petrovic N, et al. Gene-chip studies of adipogenesis-regulated microRNAs in mouse primary adipocytes and human obesity. BMC Endocr Disord. 2011;11:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Kim YJ, Hwang SJ, Bae YC, Jung JS. MiR-21 regulates adipogenic differentiation through the modulation of TGF-beta signaling in mesenchymal stem cells derived from human adipose tissue. Stem Cells. 2009;27:3093-3102. [DOI] [PubMed] [Google Scholar]
- 99. Luo A, Yan H, Liang J, et al. MicroRNA-21 regulates hepatic glucose metabolism by targeting FOXO1. Gene. 2017;627:194-201. [DOI] [PubMed] [Google Scholar]
- 100. Sekar D, Venugopal B, Sekar P, Ramalingam K. Role of microRNA 21 in diabetes and associated/related diseases. Gene. 2016;582:14-18. [DOI] [PubMed] [Google Scholar]
- 101. Ibrahim SM, Mirmira R, Anderson R, Sims E. MiR-21 contributes to Cytokine-Induced beta cell dysfunction via inhibition of mRNAs regulating beta cell identity. FASEB J. 2019;33:694.13-694.13. Accessed February 22, 2020. https://www.fasebj.org/doi/abs/10.1096/fasebj.2019.33.1_supplement.694.13 [Google Scholar]
- 102. Sims EK, Lakhter AJ, Anderson-Baucum E, Kono T, Tong X, Evans-Molina C. MicroRNA 21 targets BCL2 mRNA to increase apoptosis in rat and human beta cells. Diabetologia. 2017;60:1057-1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Dehghani M, Aghaei Zarch SM, Vahidi Mehrjardi MY, et al. Evaluation of miR-181b and miR-126-5p expression levels in T2DM patients compared to healthy individuals: relationship with NF-κB gene expression. Endocrinol Diabetes Nutr. 2020;67:454-460. [DOI] [PubMed] [Google Scholar]
- 104. Mohsen S, Zarch A, Yahya M, et al. MiR-181b expression levels as molecular biomarkers for type 2 diabetes. J Mazandaran Univ Med Sci. 2019;29:195-201. [Google Scholar]
- 105. Oliveira-Carvalho V, Carvalho VO, Bocchi EA. The emerging role of miR-208a in the heart. DNA Cell Biol. 2013;32:8-12. [DOI] [PubMed] [Google Scholar]
- 106. Montgomery RL, Hullinger TG, Semus HM, et al. Therapeutic inhibition of miR-208a improves cardiac function and survival during heart failure. Circulation. 2011;124:1537-1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Grueter CE, van Rooij E, Johnson BA, et al. A cardiac MicroRNA governs systemic energy homeostasis by regulation of MED13. Cell. 2012;149:671-683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Zhu H, Shah S, Shyh-Chang N, et al. Lin28a transgenic mice manifest size and puberty phenotypes identified in human genetic association studies. Nat Genet. 2010;42:626-630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Zhu H, Shyh-Chang N, Segrè A, et al. The lin28/Let-7 axis regulates glucose metabolism. Cell. 2011;147:81-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Aljaibeji H, Elemam NM, Mohammed AK, et al. Let7b-5p is upregulated in the serum of Emirati patients with type 2 diabetes and regulates insulin secretion in INS-1 cells. Exp Clin Endocrinol Diabetes. 2022;130:22-29. [DOI] [PubMed] [Google Scholar]
- 111. Zaccardi F, Webb DR, Yates T, Davies MJ. Pathophysiology of type 1 and type 2 diabetes mellitus: a 90-year perspective. Postgrad Med J. 2016;92:63-69. [DOI] [PubMed] [Google Scholar]
- 112. Scharfmann R. Alternative sources of beta cells for cell therapy of diabetes. Eur J Clin Invest. 2003;33:595-600. [DOI] [PubMed] [Google Scholar]
- 113. Shahjalal HM, Abdal Dayem A, Lim KM, Jeon TI, Cho SG. Generation of pancreatic β cells for treatment of diabetes: advances and challenges. Stem Cell Res Ther. 2018;9:355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Xiao R, Li C, Chai B. MiRNA-144 suppresses proliferation and migration of colorectal cancer cells through GSPT1. Biomed Pharmacother. 2015;74:138-144. [DOI] [PubMed] [Google Scholar]
- 115. Karolina DS, Armugam A, Tavintharan S, et al. MicroRNA 144 impairs insulin signaling by inhibiting the expression of insulin receptor substrate 1 in type 2 diabetes mellitus. PLoS One. 2011;6:e22839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Feng J, Xing W, Xie L. Regulatory roles of microRNAs in diabetes. Int J Mol Sci. 2016;17:1729. [DOI] [PMC free article] [PubMed] [Google Scholar]