In this study, Naranjo et al. describe optimized conditions to continuously expand murine alveolar type 2 (AT2) cells, a prominent cell of origin for LUAD, in organoid culture. They used this system to develop flexible and versatile immunocompetent organoid-based models of KRAS, BRAF, and ALK mutant LUAD, and demonstrate that this organoid platform is a powerful, versatile new model system to study LUAD.
Keywords: AT2 cells, lung cancer, organoids, stem cells
Abstract
Lung cancer is the leading cause of cancer-related death worldwide. Lung adenocarcinoma (LUAD), the most common histological subtype, accounts for 40% of all cases. While existing genetically engineered mouse models (GEMMs) recapitulate the histological progression and transcriptional evolution of human LUAD, they are time-consuming and technically demanding. In contrast, cell line transplant models are fast and flexible, but these models fail to capture the full spectrum of disease progression. Organoid technologies provide a means to create next-generation cancer models that integrate the most advantageous features of autochthonous and transplant-based systems. However, robust and faithful LUAD organoid platforms are currently lacking. Here, we describe optimized conditions to continuously expand murine alveolar type 2 (AT2) cells, a prominent cell of origin for LUAD, in organoid culture. These organoids display canonical features of AT2 cells, including marker gene expression, the presence of lamellar bodies, and an ability to differentiate into the AT1 lineage. We used this system to develop flexible and versatile immunocompetent organoid-based models of KRAS, BRAF, and ALK mutant LUAD. Notably, organoid-based tumors display extensive burden and complete penetrance and are histopathologically indistinguishable from their autochthonous counterparts. Altogether, this organoid platform is a powerful, versatile new model system to study LUAD.
Organoids are cultured miniorgans derived from healthy stem cells that proliferate continuously while retaining their physiological identity (Sato et al. 2009; Huch et al. 2013; Karthaus et al. 2014; Schutgens et al. 2019). Organoid technology has enabled the creation of models that both are practical and faithfully recapitulate clinical disease for several cancer types, including colorectal and pancreatic cancer (Boj et al. 2015; Roper et al. 2017). Unfortunately, technical limitations have prevented significant advancements in modeling LUAD with murine AT2-based organoids.
Previous murine AT2 organoid systems have displayed limited expansion capacity and/or lineage infidelity (Barkauskas et al. 2013; Lee et al. 2013, 2014; Shiraishi et al. 2019; Weiner et al. 2019). Moreover, many systems require coculture with feeder cells and/or an air–liquid interface (ALI; as opposed to being submerged in culture medium), making them technically challenging to use. Two groups have recently developed improved AT2 organoid platforms. The first is a feeder-free, ALI-based system that supports long-term passaging but yields organoids containing mixtures of AT1 and AT2 cells (Choi et al. 2021). The other is a feeder-free, submerged culture system that continuously expands pure AT2 organoids (Katsura et al. 2020).
The only described orthotopic organoid transplant model of LUAD uses an ALI and feeder-based system with limited expansion capacity (Lee et al. 2014; Dost et al. 2020). This model provides an important first step toward new, organoid-based LUAD models. In this context, introduction of oncogenic mutations rapidly induced transcriptional and morphological changes consistent with the profile of late stage LUAD (Winslow et al. 2011). Therefore, this system would seem to preclude the study of earlier stages of this disease. Furthermore, tumor penetrance and disease burden were not explored in this study, and it remains unclear whether these transformed organoids can form tumors in immunocompetent mice.
Here, we describe an optimized murine AT2 organoid platform that uses chemically defined media, circumvents feeder cells and ALI systems, and can be used to faithfully model LUAD in vivo. We demonstrate that these organoids can be expanded continuously and maintain the canonical anatomical and transcriptional characteristics of AT2 cells. Importantly, we show that activating mutations in Kras, Braf, or Alk in combination with Trp53 (TP53 in humans) loss—three common driving events in human LUAD—permit organoid proliferation in the absence of growth factors. These transformed organoids form tumors that display extensive burden and multistage progression with complete penetrance in immunocompetent hosts.
Results
Development of an optimized feeder-free, submerged culture system for long-term expansion of AT2 organoids
To build organoid-based models of LUAD, we undertook a systematic approach to define a cocktail of growth factors that could expand adult murine AT2 cells embedded in Matrigel and submerged in culture medium. We hypothesized that combining activators of alveolar regeneration and proliferation (Fgfr2 [Dorry et al. 2019], c-Met [Yamamoto et al. 2007], and Wnt [Frank et al. 2016; Nabhan et al. 2018]) with inhibitors of pathways that antagonize these processes (Bmp [Chung et al. 2018], Tgf-β [Riemondy et al. 2019], and p38 MAPK [Ventura et al. 2007]) would accomplish this goal.
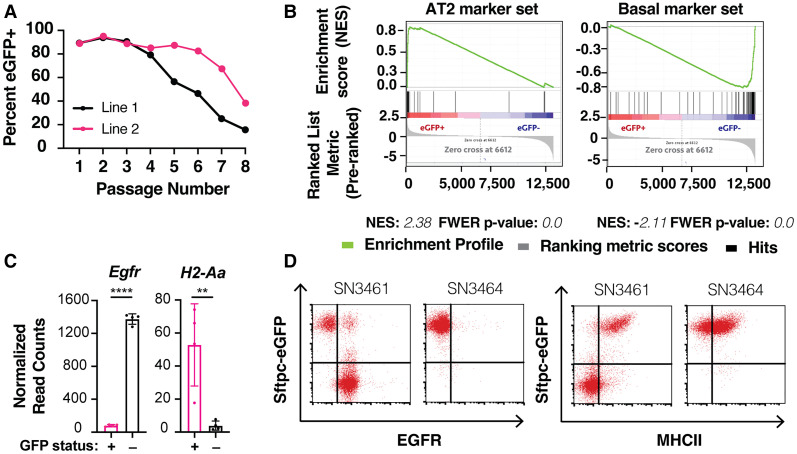
To test this hypothesis, we embedded freshly dissociated bulk lung cell suspensions from transgenic Sftpc-eGFP mice (Vanderbilt et al. 2015) in Matrigel droplets and overlaid F7NHCSA (FGF7, NOGGIN, HGF, CHIR99021, SB202190, and A-83-01) medium. The Sftpc-eGFP transgene fluorescently labels cells expressing the canonical AT2 marker gene, Sftpc. At each passage, we performed flow cytometry on bulk dissociated organoids from each line to determine the percentage of eGFP+ cells. Initially, we observed robust expansion of eGFP+ cells, indicating maintenance of the AT2 state. However, eGFP− cells progressively took over the cultures (Fig. 1A). To define the two populations transcriptionally, we performed bulk RNA sequencing (RNA-seq) on sorted eGFP+ and eGFP− cells. Among the most differentially expressed genes between the two groups were established markers of AT2 and pulmonary basal cells (Supplemental Fig. S1A). Consistently, gene set enrichment analysis (GSEA) (Mootha et al. 2003; Subramanian et al. 2005) of signatures from the major epithelial lung cell types confirmed that genes from AT2 cell (Treutlein et al. 2014) and basal cell (Dvorak et al. 2011) transcriptional signatures were the most highly enriched in the eGFP+ and eGFP− groups, respectively (Fig. 1B; Supplemental Fig. S1B). These results demonstrate that eGFP+ and eGFP− cells in our culture system likely represent AT2 and basal cells, respectively.
Figure 1.
Preliminary method for culturing alveolar organoids results in basal outgrowth. (A) Flow cytometry-based quantification of Sftpc-eGFP-expressing cells over time in organoid culture under preliminary culture conditions. A total of n = 2 independent lines was established from Sftpc-eGFP mice. (B) GSEA enrichment plots of AT2 and basal cell signatures comparing Sftpc-eGFP-positive and Sftpc-eGFP-negative organoids. N = 4 independent lines, sorted based on eGFP. Enrichment scores and FWER P-values are shown below their corresponding plots. (C) Egfr and H2-Aa (a component of MHCII) expression by RNA sequencing in Sftpc-eGFP-positive (pink) and Sftpc-eGFP-negative (black) organoids. Data are expressed as mean values ± the standard deviation. Statistical analysis was performed using a two-tailed Student's t-test. (****) P-value < 0.0001, (**) P-value = 0.0079. (D) Surface expression of EGFR (left) and MHCII (right) in Sftpc-eGFP-expressing organoids in representative phenotypically unstable (SN3461) or stable (SN3464) lines by flow cytometry.
By analyzing transcriptional data from these sorted organoids, we identified MHCII as a candidate surface marker for the eGFP+ AT2 population and Egfr as a candidate surface marker for the eGFP− basal population (Fig. 1C), consistent with previous studies (Shaykhiev et al. 2013; Hasegawa et al. 2017; Travaglini et al. 2020).Using flow cytometry, we validated that EGFR was enriched in the eGFP− cells while MHCII was enriched in the eGFP+ cells (Fig. 1D). When used in tandem, these two surface proteins are valuable markers for rapid and quantitative assessment of AT2 cell state fidelity in our system. Importantly, they can be applied to organoid cultures without an Sftpc reporter. Additionally, the AT2 and basal organoids have distinct morphologies that could be used to assess stability (Supplemental Fig. S1C).
Next, we tested a modified protocol to achieve expansion of AT2 organoids that retain their identity over time. We hypothesized that the presence of multiple cell types in the initial culture contributed to outgrowth of basal organoids. Thus, instead of bulk dissociated lung cells, we seeded freshly sorted AT2 cells as a starting population using a previously established sorting method (Hasegawa et al. 2017). Furthermore, we used F7NHCS medium (Fig. 2A), which lacks A-83-01, as preliminary data suggested that this factor exacerbated the growth of eGFP− organoids (Supplemental Fig. S1D).
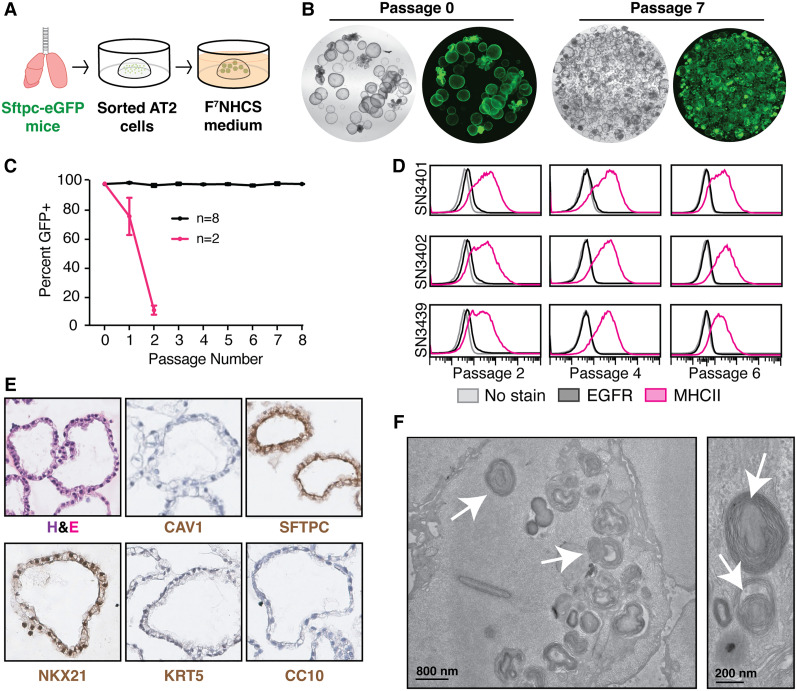
Figure 2.
Optimized culturing method enables long-term expansion of AT2 cells. (A) Method schematic of culture of alveolar organoids from normal lungs. (B) Representative images of Sftpc-eGFP organoids at an early (0) and late (7) passage; 2× magnification. (C) Flow cytometry-based quantification of Sftpc-eGFP-expressing cells over time in organoid culture. A total of n = 10 independent lines was established from Sftpc-eGFP mice and tested over three different experiments. All data are expressed as mean values ± the standard deviation. (D) Surface expression of EGFR (black) and MHCII (pink) in three independent, stable organoid lines at various passages by flow cytometry. (E) Hematoxylin and eosin (H&E) and immunohistochemical staining of organoids for canonical lung cell markers including SFTPC (AT2 cells), NKX2.1 (lung lineage), CAV1 (AT1 cells), KRT5 (basal cells), and CC10 (club cells). N = 2 lines assayed at passage 6 or later. Representative images are shown. (F) Electron microscopy images of organoids showing lamellar body structures. Arrows highlight some of the lamellar bodies. N = 3 lines assayed at passage 8 or later. Representative images are shown.
This optimized protocol supported long-term expansion (at least eight passages, with ∼1 wk between passages) of AT2 organoids in eight out of 10 lines tested (Fig. 2B,C). Importantly, lines that maintained alveolar identity throughout the first two passages remained stable long term (Fig. 2C). We also derived organoids from three mice lacking the Sftpc-eGFP reporter and demonstrated that they remained positive for MHCII and negative for EGFR for at least six passages (Fig. 2D). Notably, the endpoints for these experiments were preselected, and we have seen sustained lineage identity as late as 16 passages (Supplemental Fig. S2A,B). Collectively, these results demonstrate that our optimized protocol robustly expands AT2 cells that retain their identity over time.
AT2 organoids retain key phenotypes of their in vivo counterparts
Consistent with maintenance of the AT2 state, these organoids expressed SFTPC, as indicated by immunohistochemical staining, but lacked expression of markers of other major pulmonary epithelial cell types (Fig. 2E). Likewise, the cells comprising these organoids were densely packed with lamellar bodies, an organelle that stores and secretes pulmonary surfactant in primary AT2 cells (Fig. 2F).
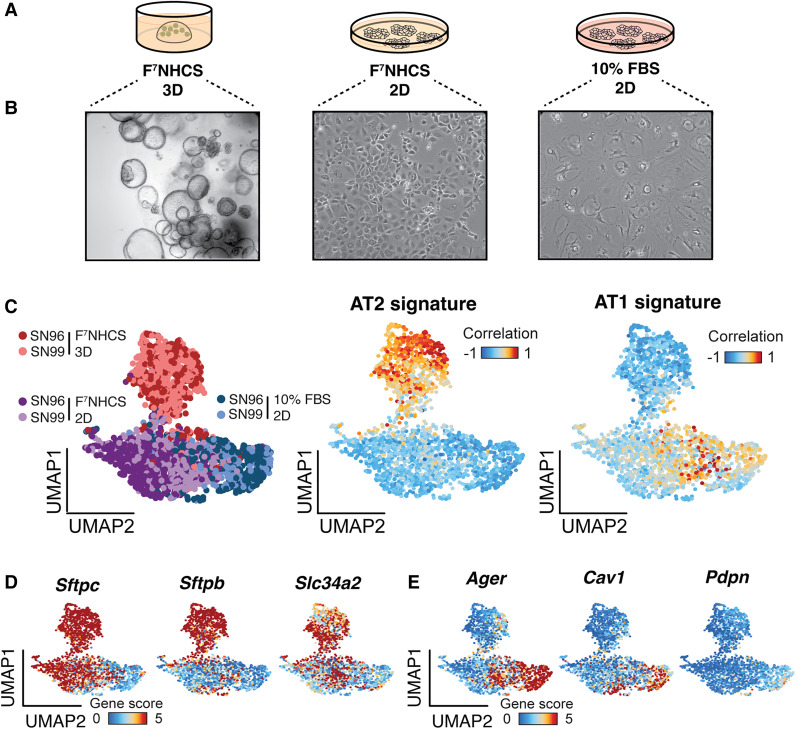
We next tested the ability of these organoids to differentiate into an alveolar type 1 (AT1) state, a characteristic feature of AT2 cells. We compared the morphology of organoids cultured in normal (F7NHCS 3D) culture conditions against two experimental monolayer conditions using either complete media (F7NHCS 2D) or differentiation media (10% FBS 2D) (Fig. 3A). The latter two conditions were based on previously established protocols for AT1 differentiation (Dobbs Williams and Brandt 1985; Salahudeen et al. 2020). Cells grown under the F7NHCS 2D condition maintained a morphology consistent with the F7NHCS 3D organoids (Fig. 3B, middle). In stark contrast, cells grown in the 10% FBS 2D condition adopted an elongated and flat morphology that is characteristic of AT1 cells (Fig. 3B, right).
Figure 3.
Optimized alveolar organoids retain the ability to differentiate into AT1 cells. (A) Schematic of culture conditions for in vitro differentiation experiment. (B) Bright-field images of organoid cells after 1 wk in the specified culture conditions (10×, 20×, and 20× magnification, from left to right). (C, left) UMAP visualization of sci-ATAC-seq profiling of organoids (n = 2) grown in the indicated culture conditions. (Red) Organoids grown in F7NHCS 3D , (purple) organoids grown in F7NHCS 2D, (blue) organoids grown in 10% FBS 2D. Correlation of chromatin profiles in single organoid cells to established chromatin accessibility signatures for primary AT2 (middle) and AT1 (right) cells. Cells are colored by their Pearson's differential correlation coefficients. (D) UMAP highlighting single-cell gene scores for three canonical AT2 markers in organoids cultured in the three conditions indicated in Figure 2A. (E) UMAP highlighting single-cell gene scores for three canonical AT1 markers in organoids cultured in the three conditions indicated in A.
To comprehensively test whether our experimental conditions induced differentiation toward the AT1 lineage at the molecular level, we performed single-cell combinatorial indexing assay for transposase-accessible chromatin sequencing (sci-ATAC-seq) (Cusanovich et al. 2015), which measures genome-wide chromatin accessibility in single cells across a population. We visualized these data using uniform manifold approximation and projection (UMAP) (https://arxiv.org/abs/1802.03426v3).
Our analyses showed that the majority of cells in the F7NHCS 3D condition displayed unique chromatin accessibility profiles relative to those in the other two conditions, which overlapped significantly (Fig. 3C, left). Cells in the F7NHCS 3D condition had high chromatin accessibility at genes comprising an epigenomic signature derived from primary AT2 cells (Fig. 3C, middle; LaFave et al. 2020) and at canonical AT2 marker genes (Fig. 3D). Cells in the F7NHCS 2D and 10% FBS 2D groups exhibited moderate and high accessibility, respectively, at genes comprising a primary AT1 epigenomic signature (Fig. 3C, right; LaFave et al. 2020) and at AT1 marker genes (Fig. 3E). Last, we confirmed higher SFTPC expression with F7NHCS 3D compared with 10% FBS 2D as well as equivalent PDPN expression across the two conditions, as predicted by the sci-ATAC-seq analysis (Supplemental Fig. S3D).
These results led us to hypothesize that the 2D culture environment promotes partial differentiation into the AT1 state, while the F7NHCS medium promotes retention of AT2 features. This hypothesis is supported by the heterogenous accessibility at the individual AT2 maker loci (Fig. 3D) for cells in the F7NHCS 2D condition. In contrast, the 10% FBS-supplemented medium induced more complete differentiation into the AT1 state (Fig. 3C,E). Importantly, none of these conditions promoted chromatin accessibility profiles that correlated significantly with signatures of other major lung epithelial cell types (Supplemental Fig. S3A).
We next performed bulk RNA-seq to examine whether the transcriptional states of these organoids were consistent with their epigenomic profiles. As expected, GSEA confirmed that the cells in the F7NHCS 3D condition correlated most strongly with an AT2 state, while the cells in the 10% FBS 2D condition correlated most strongly with an AT1 state (Supplemental Fig. S3B,C).
Improved AT2 organoid platform enables flexible LUAD modeling
We next used our optimized organoid system to model clinically relevant LUAD subtypes. KRAS and BRAF are mutated in ∼30% and ∼10% of LUAD patients, respectively, whereas ALK fusions are observed in ∼1% of clinical cases (The Cancer Genome Atlas Research Network 2014). TP53 is mutated in ∼50% of patient lung adenocarcinomas (The Cancer Genome Atlas Research Network 2014). Importantly, KRASG12D, BRAFV600E, and EML4-ALK alone initiate lung tumors in mice (Jackson et al. 2001; Dankort et al. 2007; Maddalo et al. 2014). While Trp53 loss alone is not a robust driver for LUAD initiation, it does accelerate disease in Kras-, Braf-, and Alk-driven LUAD GEMMs (Jackson et al. 2005; Dankort et al. 2007; Blasco et al. 2014). We therefore modeled KRASG12D, BRAFV600E, or EML4-ALK alterations in combination with TP53 loss.
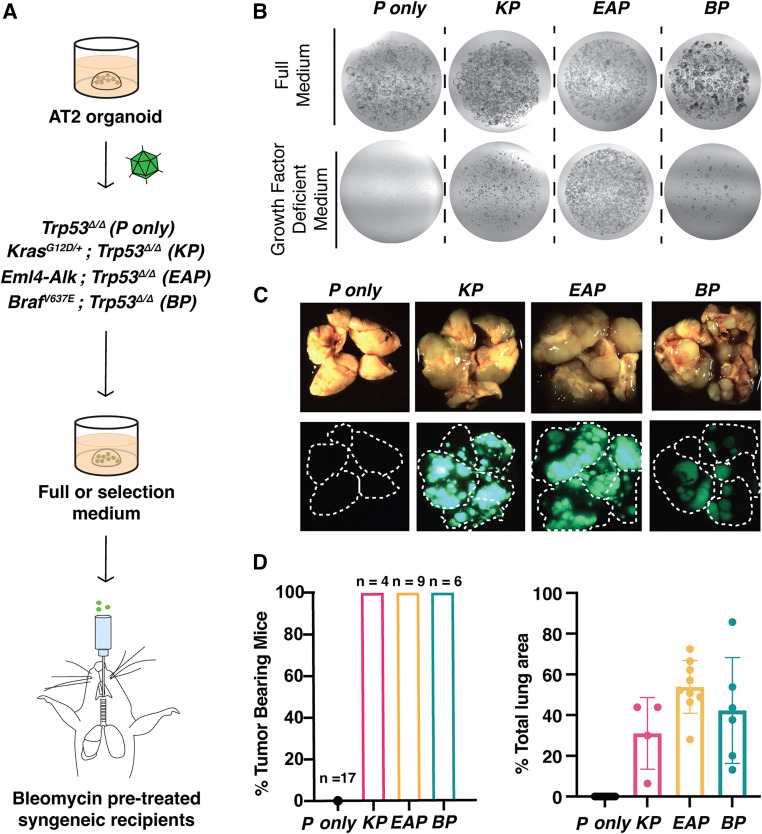
To test whether these prevalent genetic insults could transform wild-type AT2 organoids, we created Kras mutant, Trp53-deficient (KP) organoids by infecting KrasLSL-G12D/+;Trp53fl/fl;Rosa26LSL-Cas9-eGFP/LSL-Cas9-eGFP organoids (N = 1 independent line) with an adenovirus expressing Cre recombinase (Ad5-Cre). Likewise, we generated Eml4-Alk mutant, Trp53-deficient (EAP) organoids by transducing Trp53fl/fl;Rosa26LSL-Cas9-eGFP/LSL-Cas9-eGFP organoids (N = 2 independent lines) with an adenovirus expressing Cre and sgRNAs that were previously shown to induce a clinically relevant oncogenic fusion between Eml4 and Alk (Ad5-U6-sgEml4-U6-sgAlk-CBh-Cre) (Maddalo et al. 2014). Finally, we produced Braf mutant, Trp53-deficient (BP) organoids by infecting BrafLSL-V637E/+; Rosa26LSL-Cas9-eGFP/LSL-Cas9-eGFP organoids (N = 2 independent lines) with a lentivirus expressing Cre and sgRNAs targeting Trp53 (LV-U6-sgTrp53-EFS-Cre). As a control, we transduced Trp53fl/fl;R26LSL-Cas9-eGFP/LSL-Cas9-eGFP organoids with Ad5-Cre to create Trp53-deficient-only organoids (P only; N = 2 independent lines) (Fig. 4A). In each case, the recombined R26LSL-Cas9-eGFP allele provided a fluorescent tag that allowed us to easily visualize mutant organoids. PCR analysis demonstrated efficient recombination of the KrasLSL-G12D/+ and BrafLSL-V637E/+ alleles as well as induction of Eml4-Alk inversion (Supplemental Fig. S4A–C).
Figure 4.
Development of genetically diverse in vivo organoid transplant models of LUAD. (A) Strategy to introduce the indicated sets of oncogenic mutations and test their effect on AT2 organoids in vitro and in vivo (see the text for specific details). (B) Representative images of normal (P-only) and transformed (KP, EAP, and BP) organoids in full or selection medium, which lacks growth factors. (C) Representative gross bright-field (top) and fluorescent (bottom) images of lungs from recipient mice ∼4 mo (KP, BP, and P-only) and 2 mo (EAP) after transplantation. (D) Quantification of penetrance for tumor formation (left) and tumor burden (right). (Black) P-only, (pink) KP, (yellow) EAP, (teal) BP. P-only controls from all three cohorts (KP, BP, and EAP) have been combined.
We subsequently cultured these lines in complete (F7NHCS) and growth factor-depleted media (selection [CS] media) to assay for oncogenic transformation. P-only, KP, EAP, and BP organoids expanded in complete media without any obvious morphological differences (Fig. 4B; Supplemental Fig. S4D). In stark contrast, only the organoids carrying activated oncogenes were able to grow in media lacking growth factors and Noggin (growth factor-deficient media) (Fig. 4B).
To test the tumorigenic potential of these lines in vivo, we orthotopically transplanted them into syngeneic, immunocompetent mice (C57BL/6 R26LSL-Cas9-eGFP/LSL-Cas9-eGFP mice in all cases). Prior to transplantation, we intratracheally dosed recipients with low-dose bleomycin to improve engraftment efficiency (Stevens Arnal-Estapé and Nguyen 2018). Notably, KP, EAP, and BP organoids gave rise to macroscopic tumors in 100% of recipient mice and yielded extensive tumor burden (Fig. 4C,D).
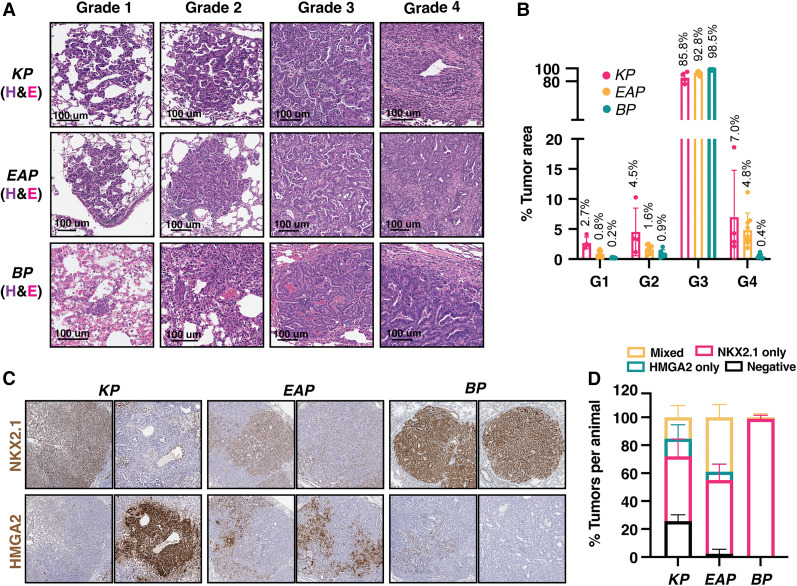
Next, we probed the ability of this system to recapitulate LUAD histopathology. KP, BP, and EAP organoid tumors displayed no obvious histopathological differences from their autochthonous counterparts. These tumors recapitulated the full spectrum of tumor progression, including hyperplasias (grade 1), adenomas (grade 2), adenocarcinomas (grade 3), and invasive adenocarcinomas (grade 4) (Fig. 5A,B). The majority of tumors across all genotypes were grade 3, consistent with the late time point at which the recipients were analyzed (KP ∼16 wk, BP ∼14 wk, and EAP ∼8 wk) (Fig. 5B). Importantly, late stage tumors in the autochthonous KP model display a similar grade distribution, further supporting the validity of this organoid platform (DuPage Dooley and Jacks 2009).
Figure 5.
Tumors from transplanted organoids recapitulate autochthonous tumor progression. (A) Hematoxylin and eosin (H&E) staining of representative tumors across all grades from transplanted KP (top row), EAP (middle row), and BP (bottom row) organoids. (B) Quantification of tumor burden by grade. (Pink) KP, (yellow) EAP, (teal) BP. Average value of tumor area per grade over total tumor area (per mouse) is listed above each bar. (C) Immunohistochemical staining of organoid-derived tumors for canonical markers of early (Nkx2.1) and late (Hmga2) stage lung adenocarcinoma. (D) Quantification of the number of tumor lesions that stained positive for NKX2.1, HMGA2, both, or neither via immunohistochemical staining.
Nkx2.1 and Hmga2 are robust markers of benign and malignant lung tumor progression stages, respectively, in the autochthonous KP model (Winslow et al. 2011). To assess tumor progression at a molecular level, we measured NKX2.1 and HMGA2 protein expression in these tumors. Most KP and EAP tumors (∼75% and ∼98%, respectively) expressed NKX2.1, HMGA2, or a mixture of both (Fig. 5C,D). Individual tumors that were classified as “mixed” predominately contained areas that were positive for one marker but not the other. On the other hand, BP tumors were virtually all exclusively positive for NKX2.1 (Fig. 5D). These data agree with the observation that NKX2.1 is required for tumorigenesis in autochthonous BP tumors (Zewdu et al. 2021). Furthermore, in contrast to Kras-driven tumors, Braf-driven tumors are not accelerated by loss of NKX2.1 after tumor formation (Snyder et al. 2013; Zewdu et al. 2021). These results further demonstrate that our organoid models recapitulate fundamental histopathological and molecular features of diverse genetic subtypes of LUAD throughout multiple stages of disease progression.
Antigen-expressing organoids can be transplanted to track tumor-immune interactions
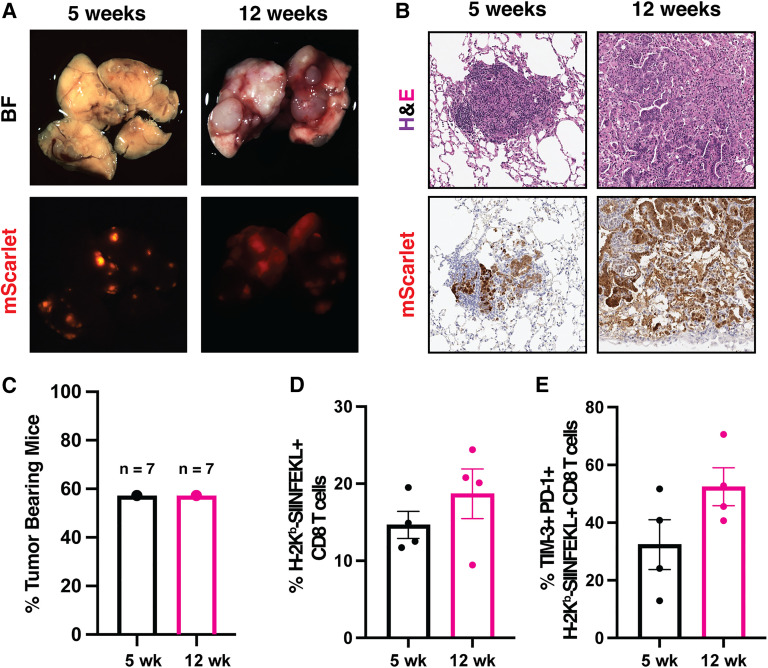
Finally, to take advantage of our ability to transplant into immunocompetent recipients, we sought to develop an immunogenic orthotopic organoid transplant model of LUAD. We transduced transformed KP organoids with a lentivirus containing the fluorescent reporter mScarlet fused to SIINFEKL, a MHC class I-restricted peptide antigen derived from chicken ovalbumin (OVA257–264), to generate KP-mScarlet-SIINFEKL (KP-SIIN) organoids. mScarlet can then be used as a proxy for sustained antigen expression in organoids and tumors (Freed-Pastor et al. 2021; Jaeger et al. 2022). KP-SIIN organoids were transplanted into bleomycin-pretreated, immunocompetent syngeneic recipients. More than 50% of recipients developed macroscopic tumors with robust mScarlet expression, even at late time points (Fig. 6A–C). While this penetrance was significantly lower than our nonimmunogenic models, it was in line with orthotopic transplant organoid models of PDAC-expressing mScarlet and SIINFEKL in which penetrance was reduced as a result of immune rejection (Freed-Pastor et al. 2021). Furthermore, we identified lung-infiltrating, antigen-specific CD8 T cells at all time points using peptide-MHC class I tetramers (Fig. 6D). As observed in previous autochthonous studies of immunogenic LUAD, there was a noticeable, though not statistically significant, up-regulation of the coinhibitory receptors PD-1 and TIM-3 in antigen-specific CD8 T cells over time (Burger et al. 2021; Schenkel et al. 2021). Together, these results affirm the use of this organoid transplant model for studies of tumor-immune interactions and, potentially, T cell dysfunction in LUAD.
Figure 6.
Tracking tumor-immune interactions in tumors from transplanted antigen-expressing organoids. (A) Bright-field (top) and fluorescent (bottom) images of representative KPSIIN tumors across tumor progression. (B) Hematoxylin and eosin (H&E) (top) and immunohistochemical staining for mScarlet (bottom) in KPSIIN organoid-derived tumors. (C) Quantification of KPSIIN tumor penetrance at each time point. (D) Quantification of the percent of antigen-specific CD8 T cells in the lung CD8 T cell infiltrate. N = 4 per time point. (E) Flow cytometry quantification of TIM3- and PD1-expressing CD8 T cells in the lung, antigen-specific CD8 T cell infiltrate. N = 4 per time point.
Discussion
Organoids provide a powerful platform to model cancer. Unfortunately, lack of stable AT2 organoid culture systems has precluded the application of this technology to develop next-generation models of LUAD. Here, we designed and validated a chemically defined growth factor cocktail that supports sustained growth and expansion of murine organoids displaying cellular and molecular canonical features of AT2 cells. Furthermore, we demonstrate that AT2 organoids can be genetically manipulated using a combination of Cre/loxP and CRISPR/Cas9 technologies followed by selection and orthotopic transplantation to rapidly develop clinically relevant organoid models of LUAD. As proof of concept, we used this system to rapidly develop Kras-, Braf-, and Alk-driven LUAD models. We show that mutant organoids can be transplanted to produce tumors that are highly faithful to LUAD pathology and indistinguishable from their autochthonous, “gold-standard” counterparts. Finally, we developed an orthotopic organoid LUAD model expressing the model antigen SIINFEKL and demonstrate a robust antigen-specific CD8 T cell response and evidence of immune dysfunction over time.
Our organoid culture system significantly improves on previous platforms. Rapid expansion capability and cell state stability are indispensable features that enabled us to model complex genotypes and expand our organoids for transplantation. The system developed by Katsura et al. (2020) is similarly fast (10- to 14-d expansion period) and maintains AT2 marker expression for at least six passages. Our media formulation differs from that of Katsura et al. (2020) in two major ways: While we used HFG to stimulate the c-MET pathway, they used IL-1β and EGF to activate inflammatory signaling and the EGFR cascade, respectively. Therefore, our platform provides an equally powerful yet distinct way to support alveolar organoid growth compared with the current state-of-the-art technology.
The observation that 20% of organoid lines are not phenotypically stable is not surprising given similar observations in other systems. For example, feeder cell-dependent transdifferentiation into basal cells has been observed in human AT2 organoids (Kathiriya et al. 2022). Given that the starting population in our platform consists of sorted AT2 cells, we believe that a subset of these cells could be transdifferentiating into pulmonary basal cells. Future studies could shed light on what causes this phenotypic shift, which might be relevant in diseases such as idiopathic pulmonary fibrosis, which in humans is thought to be caused by AT2-to-basal cell transdifferentiation in response to aberrant mesenchyme (Richeldi et al. 2017; Adams et al. 2020; Kathiriya et al. 2022). Nevertheless, the majority of our lines (∼80%) maintain their alveolar identity for the entirety of their propagation. Last, for practical usage of our system and to allow organoid derivation from mice with any desired genotype, we identified surface markers (MHCII and EGFR) to rapidly assess phenotypic stability over time.
Notably, our organoid-based cancer models provide significant advances in LUAD modeling. First, our system permits rapid interrogation of diverse genetic lesions. In this study, we illustrated rapid engineering of a chromosomal inversion that leads to an oncogenic fusion between Eml4 and Alk using CRISPR/Cas9. While our proof of concept experiments used Cre recombinase or the Cas9 nuclease, we anticipate that other genome-editing technologies, such as base editing (Komor et al. 2016) or prime editing (Anzalone et al. 2019), or transcriptional modulation technologies, such as CRISPR-based gene activation (Konermann et al. 2015) or repression (Gilbert et al. 2013), can be seamlessly deployed to engineer these organoids at any desired scale. This level of flexibility should accelerate efforts centered on functional characterization of genetic alterations that continue to be identified in patient tumors through large-scale genomic (The Cancer Genome Atlas Research Network 2014) and transcriptomic (Laughney et al. 2020) profiling studies. Likewise, combining genetic screens with genetically defined normal and transformed organoid pairs can help uncover tumor-specific and even genotype-specific vulnerabilities.
Moreover, we demonstrate the first organoid-based LUAD models with immunocompetent hosts. In contrast to previous work (Dost et al. 2020), our models demonstrate multistage progression, extensive burden, complete penetrance, and the ability to stimulate tumor-specific immune responses by the inclusion of a model antigen. These combined features will enable studies of immune dysfunction, emerging immunotherapies in lung cancer, and potentially, the function of MHCII in LUAD, a topic that remains poorly understudied.
Beyond modeling cancer, our data strongly indicate that our system expands AT2 organoids recapitulating various critical aspects of normal AT2 biology. Notably, we describe culture conditions that induce full and partial differentiation into the AT1 state, which can serve as a powerful model to study the molecular processes governing differentiation and regeneration. With the feasibility of genetic engineering and comprehensive molecular profiling at the single-cell level in organoids, these processes can be dissected in a high-throughput manner with chemical or genetic screens that use sci-ATAC-seq or single-cell RNA-seq as a readout; e.g., Perturb-ATAC (Rubin et al. 2019). Similar approaches could be applied to any other aspect of AT2 biology using our system.
Given the significant advances of our culture system and its flexibility to rapidly and faithfully model lung cancers of diverse genotypes, we envision transformative applications in normal alveolar stem cell biology and tumor evolution.
Materials and methods
Tissue processing for organoid culture
Mice 8–17 wk old were sacrificed and their lungs were transferred into 500 µL of dispase and minced. Digestion buffer (3–5 mL) containing advanced DMEM/F-12, penicillin/streptomycin, amphotericin B, 1 mg/mL collagenase (Sigma C9407-500MG), 40 U/mL DNase I (Roche 10104159001), 5 µM HEPES, and 0.36 mM CaCl2 was added for a 20- to 60-min incubation at 37°C in a rotating oven. The resulting suspension was incubated in 1 mL of ACK lysis buffer (Thermo A1049201) for 3–5 min at room temperature to lyse red blood cells. Samples were then washed twice with FACS buffer (1× PBS with 1 mM EDTA, 0.1% BSA) and filtered through 40-µm mesh to remove chunks. Samples were resuspended in 150 µL of FACS buffer, and CD45 cells were depleted using the EasySep mouse CD45-positive selection kit (Stem Cell Technologies 18945) and keeping the flowthrough. The suspension was washed one additional time and resuspended in 2–4 mL of FACS buffer for sorting (see below).
Organoid culture
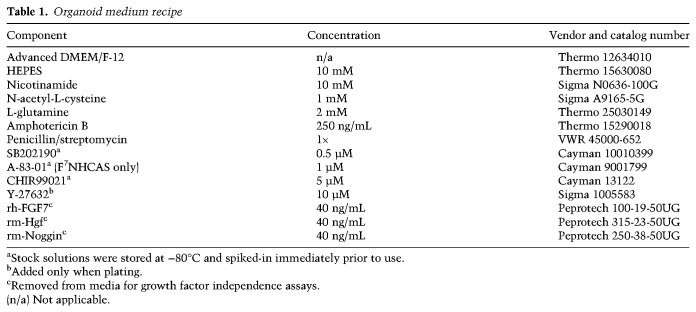
AT2 cells were sorted from dissociated lung cell suspensions as previously described (Hasegawa et al. 2017). Briefly, cells were resuspended in FACS buffer containing 1× PBS, 0.1% BSA, and 2 mM EDTA; stained with antimouse CD31-APC (1:500; Biolegend 102507), CD45-APC (1:500; BD Biosciences 559864), EpCAM-PE (1:500; Biolegend 118206), and MHCII-APC-eFluor-780 (1:500; Thermo 47-5321-82) for 30 min on ice; and then resuspended in FACS buffer containing 1 µg/mL DAPI (Thermo D1306). The suspensions were then sorted for DAPI−, CD31−, CD45−, EpCAM+, and MHCII+ cells. Approximately 20,000 sorted AT2 cells were mixed with growth factor-reduced Matrigel (Corning) at a ratio of 1:9 and seeded onto multiwell plates as 20-µL drops. The drops were incubated for 15 min to 3 h at 37°C to allow them to solidify and then overlaid with F7NHCAS or F7NHCS medium supplemented with Y-27632 (see below). The cultures were maintained in a humidified 37°C/5% CO2 incubator at ambient O2 pressure. Media was replenished every 3–4 d using F7NHCAS or F7NHCS medium without Y-27632, and organoids were passaged 6–12 d after plating. For passaging, Matrigel drops were dissolved in TrypLE Express (Sigma 12604-013) and incubated for 7–15 min at 37°C. The organoid suspensions were then dissociated into single cells by vigorous pipetting, washed twice, resuspended in 1× PBS, and plated as described above. We typically plated 10,000 cells per 20-µL drop. For components used in the organoid medium, their concentrations, and sources, see Table 1.
Table 1.
Organoid medium recipe
Flow cytometric analysis
For longitudinal tracking of SFTPC expression, the percentage of cells expressing SFTPC-eGFP was determined via flow cytometry at each passage. For staining cell type-specific surface markers on murine organoids, cells were incubated with either antimouse MHCII APC-eFluor 780 (1:1000; Thermo 47-5321-82) or biotinylated-EGF Alexa fluor 647 (200 ng/mL; Thermo E35351) for 20 min on ice. Flow cytometric analysis was performed on a Guava EasyCyte flow cytometer or a BD FACS Canto. Data analysis was performed using FlowJo software.
For differentiation experiments, bulk lung and organoid samples were stained with anti-PDPN (1:200; Thermo 12-5381-82) for 20 min on ice, followed by staining with Zombie NIR L/D APC-Cy7 (1:500; Biolegend 423105) for 15 min at room temperature. Cells were then fixed in 1% paraformaldehyde for 15 min on ice. The remaining steps were performed in 1× permeabilization buffer (Invitrogen 00833356). Cells were stained with anti-Prosurfactant Protein C (1:500; Abcam ab90716) for 30 min on ice and finally with goat antirabbit IgG (H+L) cross-adsorbed secondary antibody and Alexa fluor 647 (1:500; Thermo A-21244) for 30 min on ice. Flow cytometric analysis was performed on a BD LSRFortessa cell analyzer. Data analysis was performed using FlowJo software.
Differentiation experiments
Organoids at passage 9 or later were used for all experiments. For the F7NHCS 3D condition, organoids were cultured as described above (104 cells in 20-µL Matrigel drops). For the F7NHCS 2D and 10% FBS 2D conditions, 5 × 105 to 7.5 × 105 cells were plated per well. Ten percent FBS medium consisted of 10% FBS in advanced DMEM/F-12. Media was replenished every 3–5 d. A week after plating, cells were dissociated and processed for sci-ATAC-seq, RNA-seq, or flow cytometry.
CD8 T cell profiling
Mice were injected retro-orbitally with anti-CD45 (1:200; BD Biosciences 562420) 3 min prior to euthanization to label circulating immune cells. Following euthanasia, tumor-bearing lungs were split approximately equally, and half of the lung mass was processed for histology while the other was processed for T cell profiling. Tissue dissociation and staining were carried out as previously described (Schenkel et al. 2021). Briefly, ∼0.5 × 106 to 1 × 106 cells were stained for 1 h at 4°C in 96-well U-bottom plates (BD Biosciences) with directly conjugated antibodies (below) diluted in 1× DPBS without calcium and magnesium (VWR) supplemented with 2% heat-inactivated FBS. SIINFEKL-Kb tetramer was prepared using streptavidin-PE (Invitrogen S21388) or Streptavidin-APC (Invitrogen S32362) and SIINFEKL-Kb monomer from the National Institutes of Health Tetramer Core and used at 1:400. After staining, cells were fixed with a fixation/permeabilization kit (eBioscience). Intracellular cytokine and transcription factor staining was performed overnight at 4 °C in permeabilization buffer (eBioscience). Analysis of tissue-infiltrating antigen-specific CD8+ T cells (i.v.negCD8a+CD44hiSIINFEKL−PE+ SIINFEKL−APC+) was performed on a LSR Fortessa (BD) with 355-, 405-, 488-, 561-, and 640-nm lasers. Data analysis was performed using FlowJo software. Antibodies used were anti-CD8a (1:400; BD Biosciences 563786), anti-CD4 (1:400; BD Biosciences 564933), anti-CD44 (1:400; Biolegend 103047), anti-CD44 (1:400; BD Biosciences 563736), anti-PD-1 (1:400; Biolegend 135241), anti-Tim-3 (1:200; Biolegend 119716), and anti-Tim-3 (1:200; Biolegend 119721).
Organoid viral transduction
Viral transduction of organoids was carried out using the previously described “mix and seed” method (Wang et al. 2014). Briefly, organoids were processed for passaging, resuspended in concentrated virus, mixed with Matrigel, and plated. We do not recommend using spinfection-based protocols described in the literature because these organoid cells tend to attach to most surfaces, even those that have not been treated for tissue culture. Adenoviral transductions were performed at an MOI of 100–300. Lentiviral transductions were performed using nontitered viruses. The Ad5-sgEA-Cbh-Cre virus was purchased from ViraQuest, Inc. The Ad5-CMV-Cre virus was purchased from the Viral Vector Core at University of Iowa Carver College of Medicine.
Animal studies
Mice were housed at the animal facility at the Koch Institute for Integrative Cancer Research at Massachusetts Imstitute of Technology. All animal studies described here were approved by the Massachusetts Institute of Technology Institutional Animal Care and Use Committee. KrasLSL-G12D/+ (Jackson et al. 2001), Trp53fl/fl (Marino et al. 2000), Rosa26LSL-Cas9-eGFP (Platt et al. 2014), BrafLSL-V637E/+ (Dankort et al. 2007), and Sftpc-eGFP (Vanderbilt et al. 2015) mice have been previously described and were maintained on a pure C57BL/6 background.
Organoid transplants
P, KP, BP, and EAP transplants
Recipient mice were intratracheally inoculated with 40–50 µL of 0.1 mg/kg bleomycin (Cayman 13877). Three days later, 104 whole organoids were harvested with dispase (Corning 354235), suspended in advanced DMEM/F-12 (Thermo Fisher 12634010), and transplanted orthotopically into the bleomycin-pretreated, syngeneic recipients via intratracheal delivery. An approximately even distribution of male and female transplant recipients at 8–20 wk of age was used for all experiments, except for BP organoids, as they were derived from male donors and needed to be transplanted into male recipients.
KP-mScarlet-SIINFEKL transplants
Recipient mice were intratracheally inoculated with 40–50 µL of 0.1 mg/kg bleomycin (Cayman 13877). Three days later, 7.5 × 104 single cells were harvested with TrypLE Express (Thermo Fisher 12604013) and transplanted via intratracheal delivery into preconditioned syngeneic recipients (C57BL/6).
Tumor analysis
Mice receiving KP, BP, and EAP organoids were sacrificed 16, 14, and 8 wk after transplantation, respectively. Lungs were immediately scrutinized for green fluorescent nodules using a dissecting microscope. A positive score was assigned to mice bearing at least one fluorescent lesion. Histopathological analysis was conducted using a deep neural network developed by Aiforia Technologies and the Jacks laboratory, with consultation from the veterinarian pathologist, Dr. Roderick Bronson (LaFave et al. 2020; Romero et al. 2020).
Histology
Organoids were gently dissociated with a wide-bore pipette tip, washed in 1× PBS, and pelleted at 100g for 5 min. Organoid pellets were resuspended gently in 4% PFA for 2 h at 4°C, transferred to 70% EtOH, and embedded in paraffin. Lung tissues were fixed in zinc formalin (Polysciences 21516) overnight at room temperature and maintained in 70% EtOH before being processed for paraffin embedding. Lungs were inflated with zinc formalin immediately after sacrifice and imaging, incubated overnight at room temperature, and maintained in 70% EtOH before being processed for paraffin embedding.
Immunohistochemistry (IHC)
Sectioned organoids or tissues were stained with hematoxylin and eosin (H&E) or IHC-stained with the following antibodies: for organoids, anti-TTF1 (1:500; Abcam ab76013), rabbit anti-SFTPC (1:5000; Millipore ABC99), rabbit anti-Caveolin-1 (1:5000; Thermo C3237), rabbit anti-CCSP (1:5000; Millipore 07-623), and rabbit anti-Keratin 5 (1:5000; Biolegend 905501); for tissues, rabbit anti-TTF1 (1:500; Abcam ab76013) and rabbit anti-HMGA2 (1:400; Cell Signaling Technology 8179). For IHC staining of lung and organoid sections, antigen retrieval was performed in IHC retrieval solution (pH 6.0; Thermo TA250) for 20 min at 97°C. Slides were processed using a Thermo Scientific Autostainer 360 with the following run conditions: endogenous peroxidases (Thermo TA125H2O2q) blocking for 10 min, protein block (Biocare RBM96961L) for 30 min, primary antibody for 60 min, and labeled polymer (Biocare RMR622L) for 30 min with a 5-min DAB (Thermo TA125HDX) exposure.
Vector cloning
The Lenti-U6-Trp53-EFS-Cre vector was generated by digesting a Lenti-U6-filler-EFS-Cre (Tammela et al. 2017) with BsmBI (NEB, discontinued), and sgTrp53 (Maresch et al. 2016) was cloned into the cut vector as previously described (Ran et al. 2013).
Cell lines and cell culture
HEK293T cells were maintained in DMEM (Corning 10-013-CV) supplemented with 10% fetal bovine serum, 2 mM L-glutamine (Gibco 25030), and penicillin–streptomycin (VWR 45000-652).
Lentivirus production
Lentiviral vectors were produced by cotransfecting HEK293T cells with lentiviral and packaging plasmids (psPAX2 and pMD2.G). Viral supernatant was harvested 48 and 72 h after transfection, concentrated by centrifugation at 25,000 rpm for 2 h at 4°C, resuspended in OptiMEM (Gibco 31985-062), and frozen at −80°C.
Bulk transcriptome analysis
Total RNA was isolated from sorted cells using the TRIzol Plus RNA purification kit (Invitrogen 12183555). Briefly, cells were sorted directly into TRIzol. Following lysis and phase separation, total RNA was purified from the aqueous phase using the PureLink RNA mini kit (included in the TRIzol Plus RNA purification kit) according to the manufacturer's specifications. RNA-seq libraries were prepared from 300 ng of total RNA using the Kapa Hyperprep kit (Roche) with 14 cycles of PCR. RNA-seq libraries were quality-controlled using fragment analysis (Agilent) and qPCR and pooled for sequencing. The libraries were sequenced with single-end 75-bp reads on an Illumina NextSeq instrument. Sequence reads were trimmed to eliminate 3′ adapter traces of Illumina TruSeq Read 1 adapter using CutAdapt (v1.16) (Martin 2011). For comparison of eGFP+ and eGFP− organoids, trimmed reads were aligned to the mouse genome (mm9 build, UCSC annotation; https://www.genome.ucsc.edu) with STAR (v2.5.3a) (Dobin et al. 2013). Reads per feature were quantified using the featureCounts utility in the Subread package (v1.6.2) (Liao Smyth and Shi 2014). For the differentiation experiment, reads were mapped to the USCC mouse transcriptome (mm9 build, UCSC annotation; https://www.genome.ucsc.edu) using bowtie v1.2.3 (Langmead et al. 2009), and gene counts were quantified using RSEM v1.3.1 (Li and Dewey 2011). Differential analyses were performed using DESeq2 (v1.28.1) (Love Huber and Anders 2014) on raw feature/estimated counts with default normalization. Enrichment analyses were performed using GSEA (v2.0.13) in the “preranked” mode using DESeq2 reported log2 fold change values as the ranking metric. The c2 (curated, v7.2) collection from MSigDB (https://www.gsea-msigdb.org/gsea/msigdb) was used with the customized addition of four gene sets: AT2 markers from Treutlein et al. (2014), AT1 markers from Treutlein et al. (2014), club markers from Treutlein et al. (2014), and pulmonary basal cell markers from Dvorak et al. (2011). Differential analysis were performed in the R language for statistical computing (https://www.R-project.org).
Processing and analysis for sci-ATAC-seq
Cells were prepared for sci-ATAC-seq as previously described (LaFave et al. 2020; Del Priore et al. 2021).
Sci-ATAC-seq count generation and QC
Using the generated peak list, the number of reads for each peak window was determined for each barcode tag. This generated a matrix that associated ATAC reads in peaks for each single cell. Only cells with FRIP ≥ 0.3 and 1000 unique nuclear peaks were retained for downstream analysis. After this quality check, 2444 single cells were retained for further analysis.
Single-cell visualization
The matrix of k-mer accessibility deviation Z-scores was first column-scaled and centered (using the scale function in R v.3.5.3; https://www.R-project.org), and then run through a principal component analysis (PCA) dimensionality reduction. The uniform manifold approximation and projection (UMAP) algorithm (https://arxiv.org/abs/1802.03426v3) was then applied to project single cells in two dimensions using the k-mer PC scores for the first 20 PCs (implemented using the uwot package [v0.1.4] in R with the following nondefault clustering parameters: n_neighbors = 20, min_dist = 0.4, and metric = “cosine”).
Gene scoring
Single-cell gene scores were determined as previously described (LaFave et al. 2020). Scores were then normalized to the mean gene score per cell for further use in downstream analysis. For visualization of the gene scores in single cells, the gene scores were smoothed based on their nearest-neighbors (k = 30) in PC space. To assess correlation of AT2, AT1, basal, and club signatures to each individual single cell, AT1 and AT2 signatures were used from normal single-cell ATAC sequencing data previously described (LaFave et al. 2020) or from published signatures (Dvorak et al. 2011; Treutlein et al. 2014). Genes listed were intersected with the gene score matrix. Signature genes were mean-normalized, and Z-scores were visualized on the UMAP.
Primer and sgRNA sequences
Primer and sgRNA sequences used in this study were as follows: Eml4 fwd (GGTCAGCTACGGCTGAAGAC) (this study), Alk rev (CTTCACCATGGGAAACCAGT) (this study), Eml4 rev (GGCAGTTTGGGCTACACAGT) (this study), sgAlk (GTCCTGGCATGTCTATCTGTA) (Maddalo et al. 2014), sgEml4 (GTTTGTCGGGGTCTACTAGGA) (Maddalo et al. 2014), Kras fwd (GTCTTTCCCCAGCACAGTGC) (Jackson et al. 2001), Kras rev (CTCTTGCCTACGCCACCAGCTC) (Jackson et al. 2001), LSL fwd (AGCTAGCCACCATGGCTTGAGTAAGTCTGCA) (Jackson et al. 2001), Braf fwd (TGAGTATTTTTGTGGCAACTGC) (Dankort et al. 2007), Braf rev (CTCTGCTGGGAAAGCGGC) (Dankort et al. 2007), and sgTrp53 (GACACTCGGAGGGCTTCACT) (Maresch et al. 2016).
Data availability
The data for this study are available under GEO accession number GSE213975.
Supplementary Material
Acknowledgments
We thank Carla Concepcion, Sheng Rong Ng, Tuomas Tammela, Francisco J. Sánchez-Rivera, Grissel Jaramillo, Alex Jaeger, Caterina Colon, Demi Sandel, William Rideout III, Kim Mercer, Megan Burger, William Freed-Pastor, and the rest of the extended Jacks laboratory family for helpful discussions and technical assistance; George Eng, Jonathan Braverman, and Omer Yilmaz for helpful advice regarding organoid culture; the Massachusetts Institute of Technology BioMicro Center for performing high-throughput sequencing; and the Koch Institute's Robert A. Swanson (1969) Biotechnology Center for technical support, specifically the Hope Babette Tang (1983) Histology Facility, the Flow Cytometry Core Facility, the Animal Imaging and Preclinical Testing Core, and the Peterson (1957) Nanotechnology Materials Core Facility. This work was supported by the Howard Hughes Medical Institute, the Koch Institute Support Grant (P30-CA14051) from the National Cancer Institute, and the Koch Institute Frontier Research Program through gifts from Upstage Lung Cancer. S.N. was supported by the Howard Hughes Medical Institute Gilliam Fellowship Program and the David H. Koch Graduate Fellowship Fund. S.N., C.M.C., and R.R. were supported in part by the National Institutes of Health Predoctoral Training Grant (T32GM007287). D.Y. was supported by a Damon Runyon Cancer Research Foundation Postdoctoral Fellowship (DRG-2238-18).
Author contributions: S.N. and T.J. designed the study and wrote the manuscript with comments from all coauthors. C.M.C. contributed to the design of experiments and the writing of the manuscript. S.N. and C.M.C. conducted the majority of the experiments. L.M.L. and I.D.P. conducted sci-ATAC-seq experiments and analyses. P.M.K.W. performed histopathological tumor analyses and contributed significantly to the writing of the manuscript. R.R. conducted immunohistochemistry on organoid sections. A.G. and A.B. performed all bulk RNA sequencing bioinformatic analyses. L.Z.L. performed experiments and contributed significantly in the beginning stages of this project. S.-L.S. and C.M.C conducted antigenic model experiments. J.M.S. contributed to the differentiation experiment conceptualization and flow cytometric data analysis. D.Y. designed and generated viral vectors for tumor modeling and provided significant support in the beginning stages of this project.
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.349659.122.
Competing interest statement
T.J. is a member of the Board of Directors of Amgen and Thermo Fisher Scientific. He is also a cofounder of Dragonfly Therapeutics and T2 Biosystems. T.J. serves on the Scientific Advisory Board of Dragonfly Therapeutics, SQZ Biotech, and Skyhawk Therapeutics. He is the President of Break Through Cancer. None of these affiliations represent a conflict of interest with respect to the design or execution of this study or interpretation of data presented in this manuscript. T.J.’s laboratory currently also receives funding from the Johnson & Johnson Lung Cancer Initiative and the Lustgarten Foundation for Pancreatic Cancer Research, but this funding did not support the research described here.
References
- Adams TS, Schupp JC, Poli S, Ayaub EA, Neumark N, Ahangari F, Chu SG., Raby BA, DeIuliis G, Januszyk M, et al. 2020. Single-cell RNA-seq reveals ectopic and aberrant lung-resident cell populations in idiopathic pulmonary fibrosis. Sci Adv 6: eaba1983. 10.1126/sciadv.aba1983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anzalone AV, Randolph PB, Davis JR, Sousa AA, Koblan LW, Levy JM, Chen PJ, Wilson C, Newby GA, Raguram A, et al. 2019. Search-and-replace genome editing without double-strand breaks or donor DNA. Nature 576: 149–157. 10.1038/s41586-019-1711-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barkauskas CE, Cronce MJ, Rackley CR, Bowie EJ, Keene DR, Stripp BR, Randell SH, Noble PW, Hogan BLM. 2013. Type 2 alveolar cells are stem cells in adult lung. J Clin Invest 123: 3025–3036. 10.1172/JCI68782.ing [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blasco RB, Karaca E, Ambrogio C, Cheong T-C, Karayol E, Minero VG, Voena C, Chiarle R. 2014. Simple and rapid in vivo generation of chromosomal rearrangements using CRISPR/Cas9 technology. Cell Rep 9: 1219–1227. 10.1016/j.celrep.2014.10.051 [DOI] [PubMed] [Google Scholar]
- Boj SF, Hwang CI, Baker LA, Chio IIC, Engle DD, Corbo V, Jager M, Ponz-Sarvise M, Tiriac H, Spector M, et al. 2015. Organoid models of human and mouse ductal pancreatic cancer. Cell 160: 324–338. 10.1016/j.cell.2014.12.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burger ML, Cruz AM, Crossland GE, Gaglia G, Ritch CC, Blatt SE, Bhutkar A, Canner D, Kienka T, Tavana SZ, et al. 2021. Antigen dominance hierarchies shape TCF1+ progenitor CD8 T cell phenotypes in tumors. Cell 184: 4996–5014.e26. 10.1016/J.CELL.2021.08.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Cancer Genome Atlas Research Network. 2014. Comprehensive molecular profiling of lung adenocarcinoma. Nature 511: 543–550. 10.1038/nature13385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi J, Jang YJ, Dabrowska C, Iich E, Evans KV, Hall H, Janes SM, Simons BD, Koo B-K, Kim J, et al. 2021. Release of Notch activity coordinated by IL-1β signalling confers differentiation plasticity of airway progenitors via Fosl2 during alveolar regeneration. Nat Cell Biol 23: 953–966. 10.1038/s41556-021-00742-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung M-I, Bujnis M, Barkauskas CE, Kobayashi Y, Hogan BLM. 2018. Niche-mediated BMP/SMAD signaling regulates lung alveolar stem cell proliferation and differentiation. Development 145: dev163014. 10.1242/dev.163014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cusanovich DA, Daza R, Adey A, Pliner HA, Christiansen L, Gunderson KL, Steemers FJ, Trapnell C, Shendure J. 2015. Multiplex single-cell profiling of chromatin accessibility by combinatorial cellular indexing. Science 348: 910–914. 10.1126/SCIENCE.AAB1601/SUPPL_FILE/PAP.PDF [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dankort D, Filenova E, Collado M, Serrano M, Jones K, McMahon M. 2007. A new mouse model to explore the initiation, progression, and therapy tumors. Genes Dev 21: 379–384. 10.1101/gad.1516407.GENES [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Priore I, Ma S, Strecker J, Jacks T, LaFave LM, Buenrostro JD. 2021. Protocol for single-cell ATAC sequencing using combinatorial indexing in mouse lung adenocarcinoma. STAR Protoc 2: 100583. 10.1016/J.XPRO.2021.100583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobbs LG, Williams MC, Brandt AE. 1985. Changes in biochemical characteristics and pattern of lectin binding of alveolar type II cells with time in culture. Biochim Biophys Acta 846: 155–166. 10.1016/0167-4889(85)90121-1 [DOI] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. 2013. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29: 15–21. 10.1093/bioinformatics/bts635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorry SJ, Ansbro BO, Ornitz DM, Mutlu GM, Guzy RD. 2019. FGFR2 is required for AEC2 homeostasis and survival after bleomycin-induced lung injury. Am J Respir Cell Mol Biol 62: 608–621. 10.1165/rcmb.2019-0079oc [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dost AFM, Moye AL, Vedaie M, Tran LM, Fung E, Heinze D, Villacorta-Martin C, Huang J, Hekman R, Kwan JH, et al. 2020. Organoids model transcriptional hallmarks of oncogenic KRAS activation in lung epithelial progenitor cells. Cell Stem Cell 27: 663–678.e8. 10.1016/j.stem.2020.07.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DuPage M, Dooley AL, Jacks T. 2009. Conditional mouse lung cancer models using adenoviral or lentiviral delivery of Cre recombinase. Nat Protoc 4: 1064–1072. 10.1038/nprot.2009.95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dvorak A, Tilley AE, Shaykhiev R, Wang R, Crystal RG. 2011. Do airway epithelium air-liquid cultures represent the in vivo airway epithelium transcriptome? Am J Respir Cell Mol Biol 44: 465–473. 10.1165/rcmb.2009-0453OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank DB, Peng T, Zepp JA, Snitow M, Vincent TL, Penkala IJ, Cui Z, Herriges MJ, Morley MP, Zhou S, et al. 2016. Emergence of a wave of Wnt signaling that regulates lung alveologenesis by controlling epithelial self-renewal and differentiation. Cell Rep 17: 2312–2325. 10.1016/j.celrep.2016.11.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freed-Pastor WA, Lambert LJ, Ely ZA, Pattada NB, Bhutkar A, Eng G, Mercer KL, Garcia AP, Lin L, Rideout WM, et al. 2021. The CD155/TIGIT axis promotes and maintains immune evasion in neoantigen-expressing pancreatic cancer. Cancer Cell 39: 1342–1360.e14. 10.1016/j.ccell.2021.07.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert LA, Larson MH, Morsut L, Liu Z, Brar GA, Torres SE, Stern-Ginossar N, Brandman O, Whitehead EH, Doudna JA, et al. 2013. XCRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes. Cell 154: 442–451. 10.1016/J.CELL.2013.06.044/ATTACHMENT/A70D42C5-00A7-4421-A953-A0BBD6AE49E2/MMC1.PDF [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasegawa K, Sato A, Tanimura K, Uemasu K, Hamakawa Y, Fuseya Y, Sato S, Muro S, Hirai T. 2017. Fraction of MHCII and EpCAM expression characterizes distal lung epithelial cells for alveolar type 2 cell isolation. Respir Res 18: 1–13. 10.1186/s12931-017-0635-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huch M, Bonfanti P, Boj SF, Sato T, Loomans CJM, Van De Wetering M, Sojoodi M, Li VSW, Schuijers J, Gracanin A, et al. 2013. Unlimited in vitro expansion of adult bi-potent pancreas progenitors through the Lgr5/R-spondin axis. EMBO J 32: 2708–2721. 10.1038/emboj.2013.204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson EL, Willis N, Mercer K, Bronson RT, Crowley D, Montoya R, Jacks T, Tuveson DA. 2001. Analysis of lung tumor initiation and progression using conditional expression of oncogenic K-ras. Genes Dev 15: 3243–3248. 10.1101/gad.943001.GENES [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson EL, Olive KP, Tuveson DA, Bronson R, Crowley D, Brown M, Jacks T. 2005. The differential effects of mutant p53 alleles on advanced murine lung cancer. Cancer Res 65: 10280–10288. 10.1158/0008-5472.CAN-05-2193 [DOI] [PubMed] [Google Scholar]
- Jaeger AM, Stopfer LE, Ahn R, Sanders EA, Sandel DA, Freed-Pastor WA, Rideout WM, Naranjo S, Fessenden T, Nguyen KB, et al. 2022. Deciphering the immunopeptidome in vivo reveals new tumour antigens. Nature 607: 149–155. 10.1038/s41586-022-04839-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karthaus WR, Iaquinta PJ, Drost J, Gracanin A, Van Boxtel R, Wongvipat J, Dowling CM, Gao D, Begthel H, Sachs N, et al. 2014. Identification of multipotent luminal progenitor cells in human prostate organoid cultures. Cell 159: 163–175. 10.1016/j.cell.2014.08.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kathiriya JJ, Wang C, Zhou M, Brumwell A, Cassandras M, Le Saux CJ, Cohen M, Alysandratos K-D, Wang B, Wolters P, et al. 2022. Human alveolar type 2 epithelium transdifferentiates into metaplastic KRT5+ basal cells. Nat Cell Biol 24: 10–23. 10.1038/s41556-021-00809-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsura H, Sontake V, Tata A, Kobayashi Y, Edwards CE, Heaton BE, Konkimalla A, Asakura T, Mikami Y, Fritch EJ, et al. 2020. Human lung stem cell-based alveolospheres provide insights into SARS-CoV-2-mediated interferon responses and pneumocyte dysfunction. Cell Stem Cell 27: 890–904.e8. 10.1016/j.stem.2020.10.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komor AC, Kim YB, Packer MS, Zuris JA, Liu DR. 2016. Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature 533: 420–424. 10.1038/nature17946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konermann S, Brigham MD, Trevino AE, Joung J, Abudayyeh OO, Barcena C, Hsu PD, Habib N, Gootenberg JS, Nishimasu H, et al. 2015. Genome-scale transcriptional activation by an engineered CRISPR-Cas9 complex. Nature 517: 583–588. 10.1038/NATURE14136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaFave LM, Kartha VK, Ma S, Meli K, Del Priore I, Lareau C, Naranjo S, Westcott PMK, Duarte FM, Sankar V, et al. 2020. Epigenomic state transitions characterize tumor progression in mouse lung adenocarcinoma. Cancer Cell 38: 212–228.e13. 10.1016/j.ccell.2020.06.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL. 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10: R25. 10.1186/gb-2009-10-3-r25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laughney AM, Hu J, Campbell NR, Bakhoum SF, Setty M, Lavallée VP, Xie Y, Masilionis I, Carr AJ, Kottapalli S, et al. 2020. Regenerative lineages and immune-mediated pruning in lung cancer metastasis. Nat Med 26: 259–269. 10.1038/s41591-019-0750-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JH, Kim J, Gludish D, Roach RR, Saunders AH, Barrios J, Woo AJ, Chen H, Conner DA, Fujiwara Y, et al. 2013. Surfactant protein-C chromatin-bound green fluorescence protein reporter mice reveal heterogeneity of surfactant protein C-expressing lung cells. Am J Respir Cell Mol Biol 48: 288–298. 10.1165/rcmb.2011-0403OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JH, Bhang DH, Beede A, Huang TL, Stripp BR, Bloch KD, Wagers AJ, Tseng Y-H, Ryeom S, Kim CF. 2014. Lung stem cell differentiation in mice directed by endothelial cells via a BMP4–NFATc1–thrombospondin-1 axis. Cell 156: 440–455. 10.1016/j.cell.2013.12.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Dewey CN. 2011. RSEM: accurate transcript quantification from RNA-seq data with or without a reference genome. BMC Bioinformatics 12: 1–16. 10.1186/1471-2105-12-323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y, Smyth GK, Shi W. 2014. Featurecounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30: 923–930. 10.1093/bioinformatics/btt656 [DOI] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15: 550. 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddalo D, Manchado E, Concepcion CP, Bonetti C, Vidigal JA, Han Y-C, Ogrodowski P, Crippa A, Rekhtman N, de Stanchina E, et al. 2014. In vivo engineering of oncogenic chromosomal rearrangements with the CRISPR/Cas9 system. Nature 516: 423–427. 10.1038/nature13902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maresch R, Mueller S, Veltkamp C, Öllinger R, Friedrich M, Heid I, Steiger K, Weber J, Engleitner T, Barenboim M, et al. 2016. Multiplexed pancreatic genome engineering and cancer induction by transfection-based CRISPR/Cas9 delivery in mice. Nat Commun 7: 10770. 10.1038/ncomms10770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marino S, Vooijs M, Van Der Gulden H, Jonkers J, Berns A. 2000. Induction of medulloblastomas in p53-null mutant mice by somatic inactivation of Rb in the external granular layer cells of the cerebellum. Genes Dev 14: 994–1004. 10.1101/gad.14.8.994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin M. 2011. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.Journal 17: 10. 10.14806/ej.17.1.200 [DOI] [Google Scholar]
- Mootha VK, Lindgren CM, Eriksson KF, Subramanian A, Sihag S, Lehar J, Puigserver P, Carlsson E, Ridderstråle M, Laurila E, et al. 2003. PGC-1α-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet 34: 267–273. 10.1038/ng1180 [DOI] [PubMed] [Google Scholar]
- Nabhan AN, Brownfield DG, Harbury PB, Krasnow MA, Desai TJ. 2018. Single-cell Wnt signaling niches maintain stemness of alveolar type 2 cells. Science 359: 1118–1123. 10.1126/science.aam6603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platt RJ, Chen S, Zhou Y, Yim MJ, Swiech L, Kempton HR, Dahlman JE, Parnas O, Eisenhaure TM, Jovanovic M, et al. 2014. CRISPR-Cas9 knockin mice for genome editing and cancer modeling. Cell 159: 440–455. 10.1016/j.cell.2014.09.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA, Zhang F. 2013. Genome engineering using the CRISPR–Cas9 system. Nat Protoc 8: 2281–2308. 10.1038/nprot.2013.143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richeldi L, Collard HR, Jones MG. 2017. Idiopathic pulmonary fibrosis. Lancet 389: 1941–1952. 10.1016/S0140-6736(17)30866-8 [DOI] [PubMed] [Google Scholar]
- Riemondy KA, Jansing NL, Jiang P, Redente EF, Gillen AE, Fu R, Miller AJ, Spence JR, Gerber AN, Hesselberth JR, et al. 2019. Single-cell RNA sequencing identifies TGF-β as a key regenerative cue following LPS-induced lung injury. JCI Insight 4: e123637. 10.1172/jci.insight.123637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero R, Sánchez-Rivera FJ, Westcott PMK, Mercer KL, Bhutkar A, Muir A, González Robles TJ, Lamboy Rodríguez S, Liao LZ, Ng SR, et al. 2020. Keap1 mutation renders lung adenocarcinomas dependent on Slc33a1. Nat Cancer 1: 589–602. 10.1038/s43018-020-0071-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roper J, Tammela T, Cetinbas NM, Akkad A, Roghanian A, Rickelt S, Almeqdadi M, Wu K, Oberli MA, Sánchez-Rivera FJ, et al. 2017. In vivo genome editing and organoid transplantation models of colorectal cancer and metastasis. Nat Biotechnol 35: 569–576. 10.1038/nbt.3836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubin AJ, Parker KR, Satpathy AT, Qi Y, Wu B, Ong AJ, Mumbach MR, Ji AL, Kim DS, Cho SW, et al. 2019. Coupled single-cell CRISPR screening and epigenomic profiling reveals causal gene regulatory networks. Cell 176: 361–376.e17. 10.1016/j.cell.2018.11.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salahudeen AA, Choi SS, Rustagi A, Zhu J, van Unen V, de la O SM, Flynn RA, Margalef-Català M, Santos AJM, Ju J, et al. 2020. Progenitor identification and SARS-CoV-2 infection in human distal lung organoids. Nature 588: 670–675. 10.1038/s41586-020-3014-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato T, Vries RG, Snippert HJ, Van De Wetering M, Barker N, Stange DE, van Es JH, Abo A, Kujala P, Peters PJ, et al. 2009. Single Lgr5 stem cells build crypt–villus structures in vitro without a mesenchymal niche. Nature 459: 262–265. 10.1038/nature07935 [DOI] [PubMed] [Google Scholar]
- Schenkel JM, Herbst RH, Canner D, Li A, Hillman M, Shanahan SL, Gibbons G, Smith OC, Kim JY, Westcott P, et al. 2021. Conventional type I dendritic cells maintain a reservoir of proliferative tumor-antigen specific TCF-1+ CD8+ T cells in tumor-draining lymph nodes. Immunity 54: 2338–2353.e6. 10.1016/j.immuni.2021.08.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schutgens F, Rookmaaker MB, Margaritis T, Rios A, Ammerlaan C, Jansen J, Gijzen L, Vormann M, Vonk A, Viveen M, et al. 2019. Tubuloids derived from human adult kidney and urine for personalized disease modeling. Nat Biotechnol 37: 303–313. 10.1038/s41587-019-0048-8 [DOI] [PubMed] [Google Scholar]
- Shaykhiev R, Zuo WL, Chao IW, Fukui T, Witover B, Brekman A, Crystal RG. 2013. EGF shifts human airway basal cell fate toward a smoking-associated airway epithelial phenotype. Proc Natl Acad Sci 110: 12102–12107. 10.1073/pnas.1303058110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiraishi K, Shichino S, Ueha S, Nakajima T, Hashimoto S, Yamazaki S, Matsushima K. 2019. Mesenchymal–epithelial interactome analysis reveals essential factors required for fibroblast-free alveolosphere formation. IScience 11: 318–333. 10.1016/j.isci.2018.12.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snyder EL, Watanabe H, Magendantz M, Hoersch S, Chen TA, Wang DG, Crowley D, Whittaker CA, Meyerson M, Kimura S, et al. 2013. Nkx2-1 represses a latent gastric differentiation program in lung adenocarcinoma. Mol Cell 50: 185–199. 10.1016/J.MOLCEL.2013.02.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens LE, Arnal-Estapé A, Nguyen DX. 2018. Pre-conditioning the airways of mice with bleomycin increases the efficiency of orthotopic lung cancer cell engraftment. J Vis Exp 2018: e56650. 10.3791/56650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, et al. 2005. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci 102: 15545–15550. 10.1073/pnas.0506580102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tammela T, Sanchez-Rivera FJ, Cetinbas NM, Wu K, Joshi NS, Helenius K, Park Y, Azimi R, Kerper NR, Wesselhoeft RA, et al. 2017. A Wnt-producing niche drives proliferative potential and progression in lung adenocarcinoma. Nature 545: 355–359. 10.1038/nature22334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Travaglini KJ, Nabhan AN, Penland L, Sinha R, Gillich A, Sit RV, Chang S, Conley SD, Mori Y, Seita J, et al. 2020. A molecular cell atlas of the human lung from single-cell RNA sequencing. Nature 587: 619–625. 10.1038/S41586-020-2922-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treutlein B, Brownfield DG, Wu AR, Neff NF, Mantalas GL, Espinoza FH, Desai TJ, Krasnow MA, Quake SR. 2014. Reconstructing lineage hierarchies of the distal lung epithelium using single-cell RNA-seq. Nature 509: 371–375. 10.1038/nature13173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanderbilt JN, Gonzalez RF, Allen L, Gillespie AM, Leaffer D, Dean WB, Chapin C, Dobbs LG. 2015. High-efficiency type II cell-enhanced green fluorescent protein expression facilitates cellular identification, tracking, and isolation. Am J Respir Cell Mol Biol 53: 14–21. 10.1165/rcmb.2014-0348MA [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ventura JJ, Tenbaum S, Perdiguero E, Huth M, Guerra C, Barbacid M, Pasparakis M, Nebreda AR. 2007. P38α MAP kinase is essential in lung stem and progenitor cell proliferation and differentiation. Nat Genet 39: 750–758. 10.1038/ng2037 [DOI] [PubMed] [Google Scholar]
- Wang N, Zhang H, Zhang B-Q, Liu W, Zhang Z, Qiao M, Zhang H, Deng F, Wu N, Chen X, et al. 2014. Adenovirus-mediated efficient gene transfer into cultured three-dimensional organoids. PLoS One 9: e93608. 10.1371/journal.pone.0093608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiner AI, Jackson SR, Zhao G, Quansah KK, Farshchian JN, Neupauer KM, Littauer EQ, Paris AJ, Liberti DC, Scott Worthen G, et al. 2019. Mesenchyme-free expansion and transplantation of adult alveolar progenitor cells: steps toward cell-based regenerative therapies. NPJ Regen Med 4: 1–10. 10.1038/s41536-019-0080-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winslow MM, Dayton TL, Verhaak RGW, Kim-Kiselak C, Snyder EL, Feldser DM, Hubbard DD, DuPage MJ, Whittaker CA, Hoersch S, et al. 2011. Suppression of lung adenocarcinoma progression by Nkx2-1. Nature 473: 101–104. 10.1038/nature09881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto H, Jun Yun E, Gerber HP, Ferrara N, Whitsett JA, Vu TH. 2007. Epithelial-vascular cross talk mediated by VEGF-A and HGF signaling directs primary septae formation during distal lung morphogenesis. Dev Biol 308: 44–53. 10.1016/j.ydbio.2007.04.042 [DOI] [PubMed] [Google Scholar]
- Zewdu R, Mehrabad EM, Ingram K, Fang P, Gillis KL, Camolotto SA, Orstad G, Jones A, Mendoza MC, Spike BT, et al. 2021. An NKX2-1/ERK/WNT feedback loop modulates gastric identity and response to targeted therapy in lung adenocarcinoma. Elife 10: e66788. 10.7554/ELIFE.66788 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data for this study are available under GEO accession number GSE213975.