Abstract
Noninfectious uveitis (NIU), an intraocular inflammation caused by immune-mediated reactions to eye antigens, is associated with systemic rheumatism and several autoimmune diseases. However, the mechanisms underlying the pathogenesis of uveitis are poorly understood. Therefore, we aimed to identify differentially expressed genes (DEGs) in individuals with NIU and to explore its etiologies using bioinformatics tools.
GSE66936 and GSE18781 datasets from the gene expression omnibus (GEO) database were merged and analyzed. Functional enrichment analysis was performed, and protein-protein interaction (PPI) networks were constructed.
A total of 89 DEGs were identified. Gene ontology (GO) enrichment analysis identified 21 enriched gene sets. Kyoto encyclopedia of genes and genomes (KEGG) pathway enrichment analysis identified four core enriched pathways: antigen processing and expression signaling, natural killer (NK) cell-mediated cytotoxicity signaling, glutathione metabolic signal transduction, and arachidonic acid metabolism pathways. PPI network analysis revealed an active component-target network with 40 nodes and 132 edges, as well as several hub genes, including CD27, LTF, NCR3, SLC4A1, CD69, KLRB1, KIR2DL3, KIR3DL1, and GZMK.
The eight potential hub genes may be associated with the risk of developing NIU. NK cell-mediated cytotoxicity signaling might be the key molecular mechanism in the occurrence and development of NIU. Our study provided new insights on NIU, its genetics, molecular pathogenesis and new therapeutic targets.
Keywords: differentially expressed genes, GEO database, noninfectious uveitis, Protein-protein interaction
1. Introduction
Uveitis is a vision-threatening eye disease[1] that occurs mostly in people over the age of 36 years.[2] Approximately 10% of blindness in Western countries is caused by uveitis, and 25% of legal blindness in developing countries is associated with uveitis.[3–5] Uveitis cases can be classified into infectious or noninfectious categories based on the etiology.[6] Non-infectious uveitis (NIU) is an intraocular inflammation caused by immune-mediated reactions to eye antigens. Approximately 41% to 55% of the cases are non-infectious uveitis and are associated with systemic rheumatism and several autoimmune diseases[6,7] such as ankylosing spondylitis (AS), Behcet disease (BD), Vogt–Koyanagi–Harada (VKH), and multiple sclerosis.[8,9] Different diseases result in the expression of different proteins.[10] Epidemiologically, uveitis exhibits geographical variations, with NIU being the leading cause of uveitis in industrialized countries affecting 121 in 100,000 people.[11,12] Higher incidences have been observed in some regional groups of the Chinese population,[13,14] but national epidemiological statistics are lacking. An epidemiological survey of uveitis in 8952 people over 40 years of age in Hengli Town, Dongguan City, Guangdong Province, China, suggested that the prevalence of uveitis was 904 per 100,000 individuals, and NIU accounting for 81.5% of the cases.[15] The clinical manifestations of NIU include red eyes, pain, and blurred vision. Vision loss often occurs if the lesions involve the choroid, retina, and macula. These manifestations are characterized by rapid onset, long course, easy recurrence, and many complications, and the blindness rate is higher than in infectious uveitis.[3]
The pathogenesis of uveitis is not yet fully understood, and disruption to the stability of the ocular immune microenvironment may be acritical mechanism associated with NIU.[16] Previous studies have revealed that interleukins (IL-17, IL-6, and IL-8) and cytokines (interferon-gamma [IFN-γ] and chemokines) play vital roles in the pathogenesis of intraocular inflammation.[17–19] Some immune-related pathways such as the interleukin (IL)-23/IL-17 pathway, reactive oxygen species signaling pathway, arachidonic acid signaling pathway, and natural killer (NK) cell-mediated cytotoxicity pathway play crucial roles in the pathogenesis of NIU.[20–24] Clear diagnosis and appropriate treatment are essential for preventing irreversible intraocular structural damage and blindness.
Although there are many clinical causes of NIU, the treatment is simple and mainly involves glucocorticoids and/or immune suppressants. In modern medicine, local and systemic corticosteroid therapy is generally used as first-line treatment. Immune modulators such as alkylating agents, antimetabolites, and T-cell inhibitor scan be used to treat patients with poor responses to corticosteroid therapy.[25] Therefore, individualized immunomodulatory treatment programs are required to make the treatment more accurate and effective and to reduce the toxicity and side effects of the drugs.
Recently, biological agents targeting specific molecules have been used to treat NIU. These have attracted attention as alternative therapies for refractory uveitis due to their safety and efficacy.[26] All molecules involved in the immune response, including cytokines, cell adhesion molecules, chemokines, and costimulatory molecules, may be potential targets for biotherapy.[27] Notably, a substantial proportion of the mechanism of protein expression associated with NIU pathogenesis is still lacking and one possible explanation is that unknown genes may partially contribute to the immune deficiency. Therefore, many related genes remain to be identified, these will help us better understand the pathogenesis of NIU and facilitate the discovery of new diagnostic markers or therapeutic targets.
Gene chip technology is a powerful tool for exploring regulatory patterns and molecular mechanisms of targets associated with NIU genesis and development. However, some limitations are associated with current studies based on chip technology. These limitations include the use of small sample sizes and experimental variability in different settings, even when studying the same disease. Integrated bioinformatic techniques are currently employed in biological research. These techniques may be used to identify protein expression and potential mechanisms underlying NIU and provide a strong basis for the precise diagnosis of NIU and the exploration of new drug targets.
In this study, transcriptomic data of patients with NIU and healthy individuals were downloaded from the gene expression omnibus (GEO) database, and multiple microchips were combined. The datasets were used to conduct a preliminary analysis of the potential mechanisms of action underlying the occurrence and development of NIU and provide a basis for experimental research on the pathogenesis of NIU and the identification of its novel therapeutic targets.
2. Materials and Methods
2.1. Data source
The GEO database (https://www.ncbi.nlm.nih.gov/) was accessed via the national center for biotechnology information website. “Uveitis” was taken as the keyword and used to search for related chip data. Original chip data files labeled GSE66936 and GSE18781 and chip gene annotation files labeled GPL570-55999 were retrieved. The GSE66936 dataset contains 21 samples, including 16from patients with uveitis (experimental group) and 5 from healthy individuals (control group). The GSE18781 dataset contains 55 samples, including 34 from patients with uveitis (experimental group) and 21 from healthy individuals (control group). The 2 sample groups were all patients with NIU. The GSE66936 chip data was generated at NEI Institute in Bethesda, USA, while the GSE18781 chip data was generated at Casey Eye Institute in Portland, USA.
2.2. Data collection and analysis of differentially expressed genes (DEGs)
Practical extraction and reporting language script was used to merge data with the same gene names in the GSE66936 and GSE18781 chips, and the Sva package in R software (https://www.r-project.org/, Version4.1.0) was used to correct for merged data in batches. The limma package was used to correct the filtered databased on |LogFC|≥1.5 and P < .05, and NIU-related differentially expressed genes were obtained. The heat map package was used to generate volcano plots and heat maps of all the DEGs.
2.3. Functional enrichment analysis of DEGs
Cluster Profiler, DOSE, and Enrichplot packages were used to conduct gene ontology (GO) and Kyoto encyclopedia of genes and genomes (KEGG) pathway enrichment analyses of the DEGs. P < .05 was used to select enriched gene sets and pathways.
2.4. Protein-protein interaction (PPI) network construction and analysis
A PPI network was constructed using the STRING database (http://www.string-db.org). An interaction score >0.4 was used as the threshold. The PPI network was imported into R for visualization, and the connectivity between nodes was calculated to predict the core genes.
2.5. Ethics and dissemination
Ethical approval is not required for this differentially expressed gene analysis as we did not use data related to individual patient. The final report of this paper will be published in a peer-reviewed scientific journal or at confer ences to provide evidence-based medical support on NIU, its genetics, molecular pathogenesis and new therapeutic targets for clinical workers, and dataset will be made freely available.
3. Results
3.1. Identification of DEGs
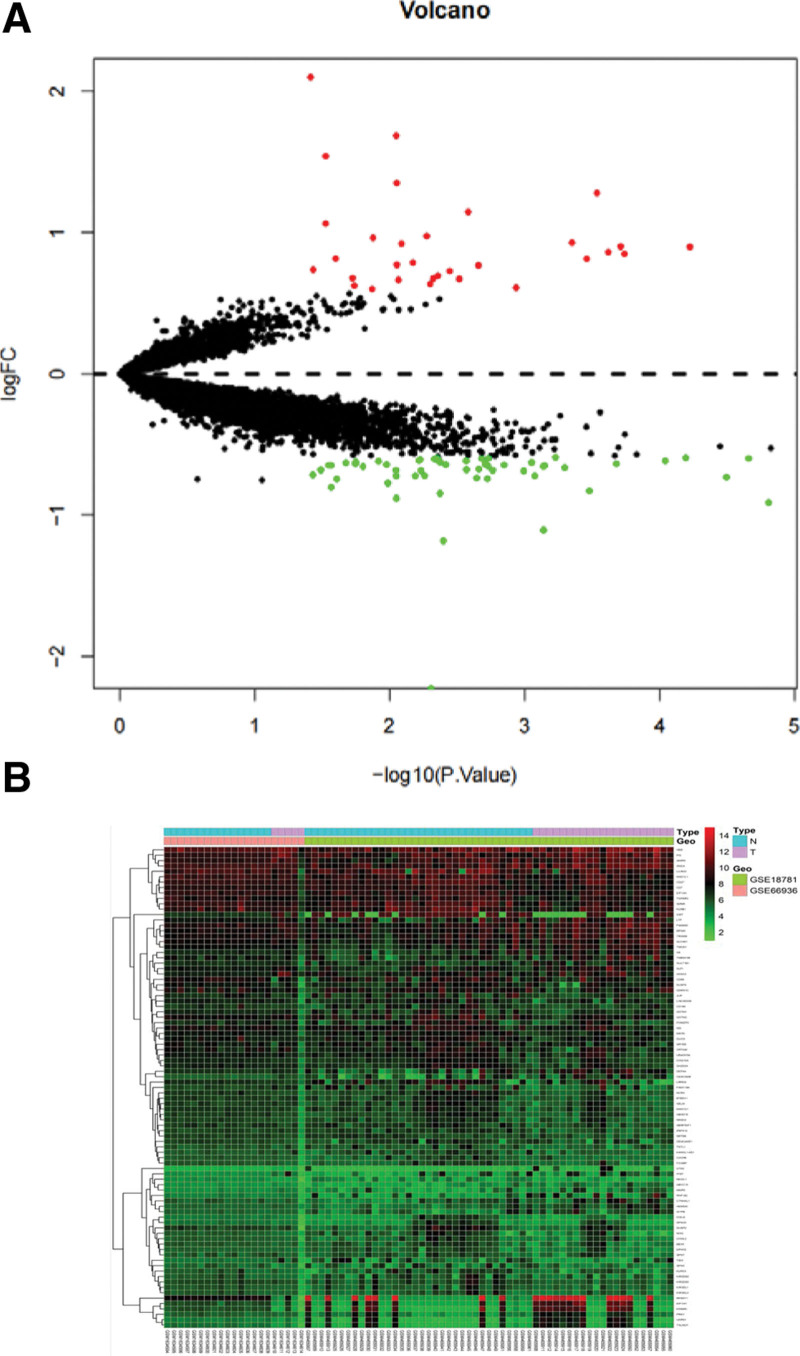
Analysis of GEO chip data identified genetic variations between healthy individuals and patients with uveitis (|LogFC|≥1.5 and P < .05) after batch correction for 89 different genes, including 31 upregulated genes (Table 1) and 58 downregulated genes (Table 2). R software was used to create a volcano plot (Fig. 1A) and heat map (Fig. 1B) of the DEGs.
Table 1.
NIU corresponds to up-regulated differential genes.
| ID | logFC | t | P value | B |
|---|---|---|---|---|
| HEMGN | 0.897971 | 4.245075 | 5.96E-05 | 0.95421 |
| FAM46C | 0.849353 | 3.929624 | .000182 | 0.142926 |
| SLPI | 0.900781 | 3.911032 | .000194 | 0.096235 |
| PI3 | 0.862023 | 3.849486 | .00024 | −0.05738 |
| DEFA4 | 1.279218 | 3.792935 | .000292 | −0.19723 |
| BPGM | 0.813999 | 3.741693 | .000347 | −0.32284 |
| XK | 0.927229 | 3.666386 | .000446 | −0.50551 |
| ABCC13 | 0.60886 | 3.372417 | .001161 | −1.19525 |
| ANXA3 | 0.767119 | 3.165726 | .002205 | −1.65642 |
| CEACAM8 | 1.145892 | 3.107911 | .002626 | −1.78169 |
| TRIM58 | 0.671613 | 3.057149 | .003057 | −1.89028 |
| MMP8 | 0.728421 | 3.002289 | .003596 | −2.00615 |
| SLC4A1 | 0.694066 | 2.933387 | .004397 | −2.14946 |
| GMPR | 0.674664 | 2.906066 | .004758 | −2.20558 |
| SULT1B1 | 0.636764 | 2.887372 | .005021 | −2.24376 |
| LTF | 0.974396 | 2.866734 | .005326 | −2.28568 |
| UTS2 | 0.787218 | 2.783662 | .006736 | −2.4521 |
| IFI27 | 0.921488 | 2.713577 | .008184 | −2.58952 |
| TMOD1 | 0.664582 | 2.694538 | .008623 | −2.62638 |
| GYPB | 0.771405 | 2.684869 | .008854 | −2.64502 |
| USP9Y | 1.349328 | 2.684208 | .00887 | −2.64629 |
| EIF1AY | 1.68577 | 2.680707 | .008956 | −2.65302 |
| RNF182 | 0.962246 | 2.53357 | .013289 | −2.92966 |
| SNCA | 0.600163 | 2.527753 | .013494 | −2.94034 |
| HBD | 0.623051 | 2.408884 | .018352 | −3.15411 |
| CTNNAL1 | 0.676558 | 2.397402 | .018896 | −3.17431 |
| PRKY | 0.815908 | 2.282359 | .025185 | −3.3722 |
| KDM5D | 1.539848 | 2.212397 | .029855 | −3.48851 |
| TXLNGY | 1.062751 | 2.211743 | .029902 | −3.48958 |
| TMEM158 | 0.737262 | 2.121928 | .037008 | −3.6343 |
| RPS4Y1 | 2.099179 | 2.103238 | .038659 | −3.66377 |
Table 2.
NIU corresponds to down-regulated differential genes.
| ID | logFC | t | P value | B |
|---|---|---|---|---|
| NCR3 | −0.90988 | −4.60725 | 1.56E-05 | 1.925831 |
| CD7 | −0.5967 | −4.51718 | 2.19E-05 | 1.680537 |
| ZNF514 | −0.73108 | −4.41521 | 3.20E-05 | 1.405688 |
| SETD6 | −0.59294 | −4.22519 | 6.40E-05 | 0.902048 |
| MATK | −0.61627 | −4.12682 | 9.11E-05 | 0.645975 |
| NKX3-1 | −0.63583 | −3.88983 | .000209 | 0.043158 |
| GZMK | −0.82873 | −3.75514 | .000331 | −0.28999 |
| COQ10A | −0.66393 | −3.62903 | .000505 | −0.59526 |
| CXCR6 | −0.58928 | −3.58162 | .000591 | −0.70829 |
| EIF1AX | −0.65107 | −3.52494 | .000711 | −0.84213 |
| NOG | −1.10609 | −3.51936 | .000724 | −0.85524 |
| CLIC3 | −0.65561 | −3.51793 | .000728 | −0.85859 |
| EPHX2 | −0.72139 | −3.4729 | .000842 | −0.96381 |
| ABHD15 | −0.62585 | −3.45785 | .000884 | −.99877 |
| FCGBP | −0.6273 | −3.45537 | .000891 | −1.00451 |
| SH2D2A | −0.68754 | −3.41413 | .001016 | −1.09972 |
| NR3C2 | −0.64623 | −3.30731 | .001424 | −1.34271 |
| GPX7 | −0.68275 | −3.24494 | .001729 | −1.48211 |
| KANSL1-AS1 | −0.59473 | −3.22329 | .001849 | −1.53006 |
| MAD1L1 | −0.65263 | −3.21857 | .001876 | −1.54047 |
| BEX5 | −0.74386 | −3.21596 | .001891 | −1.54623 |
| KIR2DS5 | −0.64108 | −3.20325 | .001966 | −1.57424 |
| TKTL1 | −0.6229 | −3.2024 | .001971 | −1.5761 |
| SERPINF1 | −0.59774 | −3.18403 | .002085 | −1.61643 |
| SELM | −0.67417 | −3.16808 | .002189 | −1.65128 |
| LINC00339 | −0.73869 | −3.15573 | .002273 | −1.6782 |
| KIR3DL1 | −0.67859 | −3.09797 | .002706 | −1.80305 |
| BTBD11 | −0.61627 | −3.09733 | .002711 | −1.80442 |
| CD27 | −0.61562 | −3.09611 | .002721 | −1.80705 |
| KIR3DL3 | −0.64353 | −2.97929 | .003847 | −2.05425 |
| LRRN3 | −1.18212 | −2.96509 | .00401 | −2.08383 |
| NR1D2 | −0.62313 | −2.94767 | .004218 | −2.11995 |
| KIR2DS2 | −0.84653 | −2.94571 | .004243 | −2.12401 |
| MAN1C1 | −0.61801 | −2.93756 | .004344 | −2.14084 |
| TGFBR3 | −0.59345 | −2.93034 | .004436 | −2.15573 |
| ID3 | −0.60108 | −2.91322 | .004661 | −2.19093 |
| XIST | −2.22804 | −2.89302 | .00494 | −2.23224 |
| COLQ | −0.72076 | −2.85404 | .005522 | −2.31135 |
| CHI3L2 | −0.68558 | −2.83249 | .005871 | −2.35475 |
| UBASH3A | −0.60699 | −2.82738 | .005956 | −2.36498 |
| CD160 | −0.61415 | −2.82059 | .006072 | −2.37859 |
| KLRC3 | −0.72178 | −2.79804 | .00647 | −2.42356 |
| DUSP2 | −0.88111 | −2.68178 | .00893 | −2.65097 |
| DUSP5 | −0.72236 | −2.68064 | .008957 | −2.65315 |
| JUP | −0.68365 | −2.67931 | .00899 | −2.65571 |
| POMZP3 | −0.77376 | −2.62854 | .010318 | −2.75253 |
| CEACAM21 | −0.64288 | −2.62054 | .010543 | −2.76767 |
| GSTM1 | −0.61873 | −2.56905 | .012099 | −2.86411 |
| CRTAM | −0.65701 | −2.46739 | .015794 | −3.04995 |
| GSTM2 | −0.62271 | −2.41806 | .017928 | −3.13791 |
| KLRB1 | −0.63632 | −2.41675 | .017988 | −3.14022 |
| GPX3 | −0.6292 | −2.35254 | .021159 | −3.25244 |
| CD69 | −0.74571 | −2.28999 | .024716 | −3.35933 |
| LILRA3 | −0.80325 | −2.25113 | .027183 | −3.4245 |
| CDKN1C | −0.64907 | −2.25081 | .027204 | −3.42503 |
| GPA33 | −0.64852 | −2.2335 | .028372 | −3.45375 |
| TSIX | −0.68071 | −2.1772 | .03248 | −3.54586 |
| FAM118A | −0.71538 | −2.11973 | .037199 | −3.63778 |
Figure 1.
Visualization of differentially expressed genes (DEGs). (A) Volcano map, the vertical coordinate represents the function of gene enrichment, the horizontal coordinate represents the ratio of the number of differential genes in gene enrichment to the total number of differential genes, and different colors represent the P value of the significance of enrichment. (B) Heat map, the vertical coordinate represents the function of gene enrichment, the horizontal coordinate represents the ratio of the number of differentially expressed genes to the total number of differentially expressed genes in the function of gene enrichment work set, and the different colors represent the P value of enrichment significance.
3.2. GO and KEGG enrichment analysis of DEGs
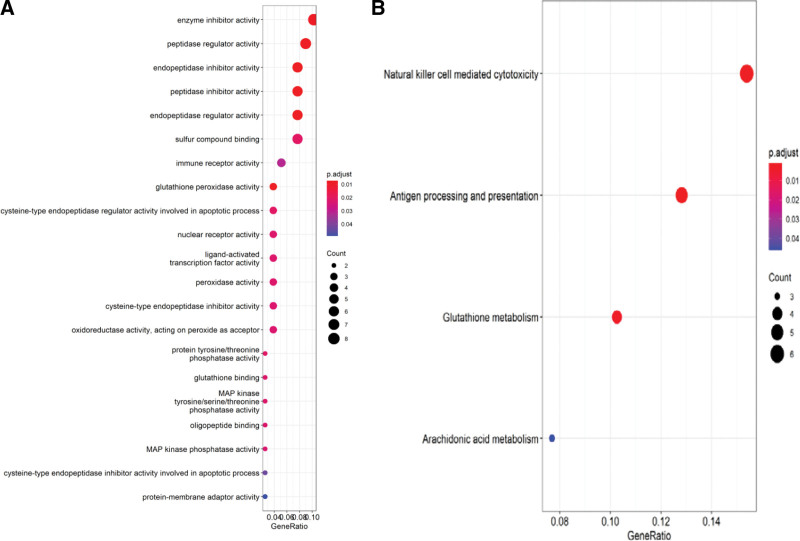
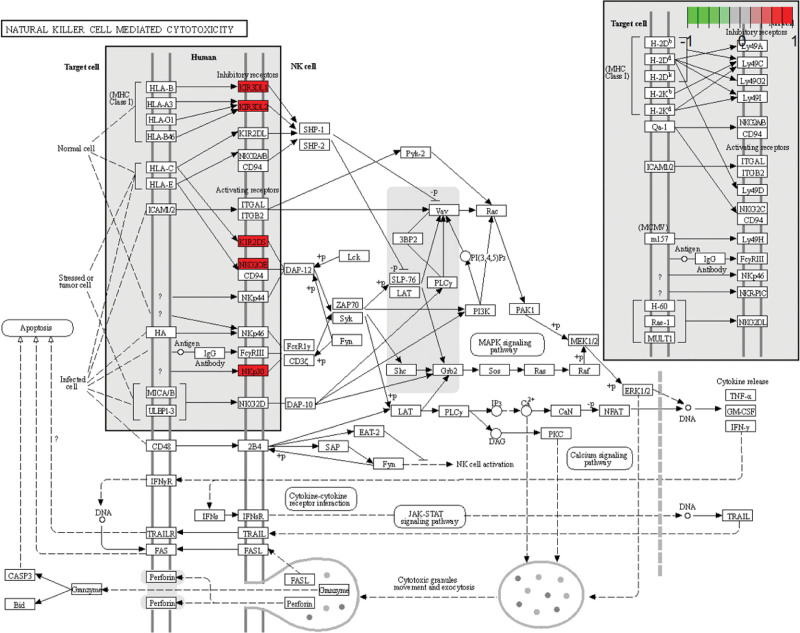
GO enrichment analysis helped identify 21 enriched core gene sets (P < .05) (Table 3 and Fig. 2A). KEGG pathway analysis of DEGs identified four enriched pathways (P < .05): antigen processing and expression, NK cell-mediated cytotoxicity, glutathione metabolism, and arachidonic acid pathways (Fig. 2B). In these pathways, the ones most closely related to the treatment of NIU were: NK cell-mediated cytotoxicity (Fig.3).
Table 3.
GO analysis results of DEGs (top 14 terms of BP category were listed). “Count” means how many DEGs are involved.
| ID | Description | p adjust | Count |
|---|---|---|---|
| GO:0004857 | Enzyme inhibitor activity | .008419 | 8 |
| GO:0061134 | Peptidase regulator activity | .007401 | 7 |
| GO:1901681 | Sulfur compound binding | .020612 | 6 |
| GO:0004866 | Endopeptidase inhibitor activity | .007401 | 6 |
| GO:0030414 | Peptidase inhibitor activity | .007401 | 6 |
| GO:0061135 | Endopeptidase regulator activity | .007401 | 6 |
| GO:0140375 | Immune receptor activity | .031426 | 4 |
| GO:0004869 | Cysteine-type endopeptidase inhibitor activity | .022549 | 3 |
| GO:0016684 | Oxidoreductase activity, acting on peroxide as acceptor | .022549 | 3 |
| GO:0043028 | Cysteine-type endopeptidase regulator activity involved in apoptotic process | .020612 | 3 |
| GO:0004602 | Glutathione peroxidase activity | .007401 | 3 |
| GO:0004879 | Nuclear receptor activity | .02193 | 3 |
| GO:0098531 | Ligand-activated transcription factor activity | .02193 | 3 |
| GO:0004601 | Peroxidase activity | .02193 | 3 |
Figure 2.
Results of functional enrichment analysis. (A) GO enrichment analysis. (B) KEGG enrichment analysis. KEGG = Kyoto encyclopedia of genes and genomes.
Figure 3.
Natural killer cell metabolism mediated cytotoxicity. Core targets are marked in red.
3.3. PPI network construction and analysis
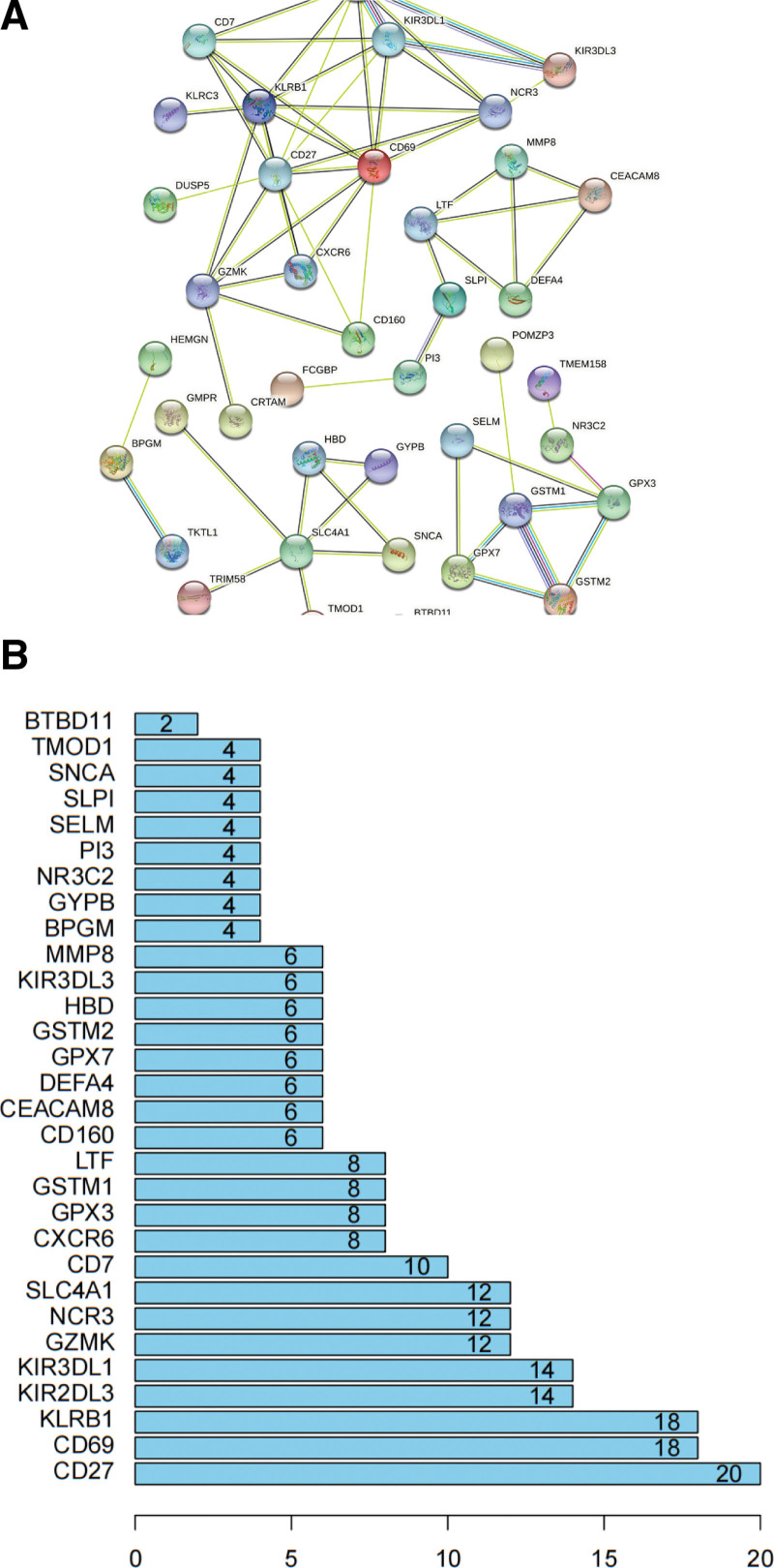
The network of interactions between proteins encoded by the DEGs was analyzed using the STRING database. The PPI network comprised of 40 nodes and 132 edges (Fig. 4A). Genes with connectivity ≥12 were selected as core genes. The most significant core gene was CD27, with connectivity of 20, followed by CD69, KLRB1, KIR2DL3, KIR3DL1, GZMK, NCR3, and SLC4A1 (Fig. 4B).
Figure 4.
Protein-protein interaction (PPI) network construction analysis.
4. Discussion
In this study, we analyzed NIU-related gene expression profiles in GSE66936 and GSE18781 microarray datasets containing 76 samples, including 50 samples from patients with uveitis (experimental group) and 26 samples from healthy individuals (control group). Data in the 2 chips were generated from peripheral blood samples collected from patients with NIU. Bioinformatics analysis-based screening for NIU-related genes helped identify 89 DEGs, including 31 upregulated and 58 downregulated genes. GO enrichment and KEGG pathway analyses revealed that the DEGs were enriched in biological processes such as peptidase regulatory activity, immune receptor activity, and cysteine endopeptidase inhibitor activity. Enriched pathways were mainly involved in antigen processing and presentation, NK cell-mediated cytotoxicity, glutathione metabolism, and arachidonic acid signaling. Among these, peptidase regulatory activity, immune receptor activity, and cysteine endopeptidase inhibitor activity are associated with immunity and inflammation.[28–30] The corresponding genes included CXCR6, KIR3DL1, CD160, CD27, SNCA, SLPI, NKX3-1, PI3, LTF, SERPINF1, and KIR2DS5. NK cell-mediated cytotoxicity leads to the expression of inhibitory signals on cell surfaces that are associated with uveitis. Immune balance can be achieved by regulating the secretion of cytokines and may contribute to the treatment of uveitis.[20] NK cells have activated and inhibitory receptors on their surface to regulate cell status,[31] CD94/NKG2A is one of the most important inhibitory molecules,[32] that CD94/NKG2A regulates the expression of in CD4 and CD8 + T cells, thereby regulating cytotoxic activity and cytokine production. Clinical studies have demonstrated that combined cyclosporine and prednisone therapy can down-regulate NK-like effector function of CD8 (bright) CD56 + T cells in patients with NIU.[33] Lipid mediators in the arachidonic acid signaling pathway are closely associated with intraocular inflammation, and drug intervention in the synthesis of lipid mediators may be of great significance in the treatment of uveitis.[21]
PPI network analysis of the DEGs performed using the STRING database suggested that CD27, CD69, KLRB1, KIR2DL3, KIR3DL1, GZMK, NCR3, and SLC4A1 were the core genes with the highest connectivity. CD27, a member of the tumor necrosis factor receptor superfamily, is a marker of NK cell maturation.[34] CD27expression was significantly enhanced in inflamed eye tissue of an experimental autoimmune uveitis model and was associated with uveitis pathogenesis.[20] CD69 expression in NK gene clusters is a marker of CD4 + T cell activation. Local antigen stimulation triggers CD4 + T cell activation, resulting in the regulation of the immune response through CD69 on the cell surface.[35] CD69 expression is upregulated during the inflammatory response in BD, AS, VKH, and other immune diseases.[36,37] Killer cell lectin-like receptor B1 (KLRB1; CD161) is expressed in NK cells, CD4 + and CD8 + T cell subsets, and NKT cells[38] and it is correlated with uveitis.[39] Bioinformatics analysis revealed that KLRB1 expression is also significantly upregulated in AS.[40] Therefore, we inferred that high KLRB1 levels during AS might increase the risk of NIU. Paule et al[41] reported that the CD4/CD8 ratio in the cerebrospinal fluid can be used to infer the etiology of intermediate and/or posterior uveitis, providing novel insights into the role of KLRB1 in NIU pathogenesis. Killer cell immunoglobulin-like receptor 3DL1 (KIR3DL1) and KIR3DL3, members of the killer cell immunoglobulin-like receptor (KIR) family, are NK and T cell subset modulators that protect target cells by inhibiting NK cell activation.[42,43] KIR3DL1 expression may be involved in the pathogenesis of BD, VKH, and AS.[44–46] KIR3DL3 is considered a susceptibility gene that is associated with BD. KIR3DL3 may affect the function of NK cells through genetic differentiation, thus participating in the pathogenesis of BD.[47] NCR3 encodes NKp30,[48] which can initiate an activation signal cascade in NK cells, leading to cytokine production or cytotoxicity.[49] NK cells are a subgroup of lymphocytes. Kucuksezer et al[23] confirmed that NK cells play a role in experimental autoimmune uveitis and may be a potential pathological factor. Considering that NCR3 is highly expressed in patients with NIU, inhibition of NCR3 activation may play a vital role in the treatment of NIU. SLC4A1 stimulates the functional activity of innate immune cells and is involved in immune responses, T cell activation, toll-like receptor-binding regulatory pathways, and granulocyte activation, all of which are closely associated with NIU.[50–53] Granzyme K (GZMK) is one of the pro-inflammatory members of the GZM family that can combine with IFN-γ to induce mouse fibroblasts to secrete inflammatory cytokines,[54] enhancing the inflammatory function of non-immune cells.[55] Its abnormal expression is associated with various auto immune diseases, including RA, AS, and scleroderma,[11] that can lead to the development of NIU. Additional studies on GZMK may provide new perspectives on the diagnosis, treatment, and pathogenesis of NIU.
In the current study, we discussed 8 potential crucial genes that are involved in the occurrence and development of NIU. These 8 crucial genes were involved in the activation and regulation of NK cells. NK cell-mediated cytotoxicity was the key pathway enriched in this study, suggesting that these genes may serve as potential biomarkers and therapeutic targets for NIU. However, the limitations of this study should be considered. First, it is difficult to consider some important factors such as region, race, and age. Considering that the etiology of NIU is complex, undetermined factors such as region and history should be evaluated in further studies. In addition, potential key genes need to be further validated by RT-qPCR in clinical samples. Finally, the mechanisms by which these genes work are not fully understood. More evidence is needed to investigate their biological foundation.
5. Conclusions
Understanding the cellular and biochemical events involved in NIU is essential for treating the disease. In this study, gene expression data were analyzed to identify core genes involved in NIU and to determine the underlying mechanism associated with the disease. Bioinformatics analyses revealed that the NK cell-mediated cytotoxicity pathway plays a vital role in the pathogenesis of NIU, and genes associated with this pathway included CD27, CD69, KLRB1, KIR3DL1, KIR3DL3, and NCR3. The results of this study provide novel insights for studying the underlying mechanisms and activation factors associated with NK cells in NIU. In addition, analysis of NIU core genes suggested that NIU is associated with several autoimmune diseases and that different diseases induce the release of diverse proteins. Thus, using bioinformatic techniques to identify the expression of genes and the potential mechanisms underlying NIU may provide a strong basis for its precise diagnosis and exploration of novel drug targets.
Acknowledgments
We would like to thank Editage (www.editage.cn) for English language editing.
Author contributions
Conceptualization: Jing Yao.
Data curation: Ning Zhang, Yan Wang, Qian Zhang, Jiadi Wang, Jing Yao.
Formal analysis: Ning Zhang.
Methodology: Jiadi Wang.
Software: Dandan Zhang, Qian Zhang.
Writing – original draft: Dandan Zhang, Ning Zhang, Jing Yao.
Writing – review & editing: Dandan Zhang, Ning Zhang.
Abbreviations:
- AS =
- ankylosing spondylitis
- BD =
- Behcet disease
- DEGs =
- differentially expressed genes
- GEO =
- gene expression omnibus
- GO =
- gene ontology
- KEGG =
- Kyoto encyclopedia of genes and genomes
- KLRB1 =
- killer cell lectin-like receptor B1
- NIU =
- noninfectious uveitis
- PPI =
- protein-protein interaction
- VKH =
- Vogt-Koyanagi-Harada
DDZ and NZ contributed equally to this work.
The datasets generated during and/or analyzed during the current study are not publicly available, but are available from the corresponding author on reasonable request.
The authors have no funding and conflicts of interest to disclose.
How to cite this article: Zhang D, Zhang N, Wang Y, Zhang Q, Wang J, Yao J. Analysis of differentially expressed genes in individuals with noninfectious uveitis based on data in the gene expression omnibus database. Medicine 2022;101:41(e31082).
Contributor Information
Dandan Zhang, Email: yayaguai19870611@163.com.
Ning Zhang, Email: yayaguai19870611@163.com.
Yan Wang, Email: wjd2005@126.com.
Qian Zhang, Email: yayaguai19870611@163.com.
Jiadi Wang, Email: wjd2005@126.com.
References
- [1].Krishna U, Ajanaku D, Denniston AK, et al. Uveitis: a sight- threatening disease which can impact all systems. Postgrad Med J. 2017;93:766–73. [DOI] [PubMed] [Google Scholar]
- [2].Çakar Özdal MP, Yazici A, Tüfek M, et al. Epidemiology of uveitis in areferral hospital in Turkey. Turk J Med Sci. 2014;44:337–42. [DOI] [PubMed] [Google Scholar]
- [3].Gritz DC, Wong IG. Incidence and prevalence of uveitis in NorthernCalifornia-The Northern California epidemiology of uveitis study. Ophthalmology. 2004;111:491–500; discussion 500. [DOI] [PubMed] [Google Scholar]
- [4].Suhler EB, Lloyd MJ, Choi D, et al. Incidence and prevalence of uveitis inveterans affairs medical centers of the Pacific Northwest. Am J Ophthalmol. 2008;146:890–6.e8. [DOI] [PubMed] [Google Scholar]
- [5].Muñoz-Fernández S, Martín-Mola E. Uveitis. Best Pract Res Clin Rheumatol. 2006;20:487–505. [DOI] [PubMed] [Google Scholar]
- [6].Leal I, Romão VC, Mano S, et al. A non-infectious uveitis multidisciplinary clinic in a tertiary referral center: clinical impact and added value. J Multidiscip Healthc. 2021;14:695–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Caspi RR. A look at autoimmunity and inflammation in the eye. J Clin Invest. 2010;120:3073–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Hsu YR, Huang JC, Tao Y, et al. Noninfectious uveitis in the Asia-Pacific region. Eye (Lond). 2019;33:66–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Gao F, Zhao C, Cheng G, et al. Clinical patterns of uveitis in a tertiary center in North China. Ocul Immunol Inflamm. 2017;25:S1–7. [DOI] [PubMed] [Google Scholar]
- [10].Bansal R, Gupta A. Protein biomarkers in uveitis. Front Immunol. 2020;11:610428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Thorne JE, Suhler E, Skup M, et al. Prevalence of noninfectious uveitis in the United States. JAMA Ophthalmol. 2016;134:1237–45. [DOI] [PubMed] [Google Scholar]
- [12].Anthony DA, Andrews DM, Watt SV, et al. Functional dissection of the granzyme family: cell death and inflammation. Immunol Rev. 2010;235:73–92. [DOI] [PubMed] [Google Scholar]
- [13].Han M, Chen Y, Nong L, et al. The effectiveness and safety of Chinese medicines for the treatment of uveitis: a protocol for systematic review and meta-analysis. Medicine (Baltim). 2020;99:20766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Yang P, Zhang Z, Zhou H, et al. Clinical patterns and characteristics of uveitis in a tertiary center for uveitis in China. Curr Eye Res. 2005;30:943–8. [DOI] [PubMed] [Google Scholar]
- [15].Zheng Y. Epidemiological Investigation of Uveitis in Hengli Town, Guangdong Province [D]. Chongqing Medical University. 2013. [Google Scholar]
- [16].Maleki A, Anesi SD, Look-Why S, et al. Pediatric uveitis: a comprehensive review. Surv Ophthalmol. 2021;21:0039–6257. [DOI] [PubMed] [Google Scholar]
- [17].Pohlmann D, Schlickeiser S, Metzner S, et al. Different composition of intraocular immune mediators in Posner-Schlossman-Syndrome and Fuchs’ Uveitis. PLoS One. 2018;13:e01993010199301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Chen YH, Lightman S, Calder VL. CD4+ T-cell plasticity in non-infectious retinal inflammatory disease. Int J Mol Sci. 2021;22:9584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Balamurugan S, Das D, Hasanreisoglu M, et al. Interleukins and cytokine biomarkers in uveitis. Indian J Ophthalmol. 2020;68:1750–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Lin W, Man X, Li P, et al. NK cells are negatively regulated by sCD83 in experimental autoimmune uveitis. Sci Rep. 2017;7:12895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Bazan NG, de Abreu MT, Bazan HE, et al. Arachidonic acid cascade and platelet-activating factor in the network of eye inflammatory mediators: therapeutic implications in uveitis. Int Ophthalmol. 1990;14:335–44. [DOI] [PubMed] [Google Scholar]
- [22].Zhong Z, Su G, Kijlstra A, et al. Activation of the interleukin-23/interleukin-17 signalling pathway in auto inflammatory and autoimmune uveitis. Prog Retin Eye Res. 2021;80:100866. [DOI] [PubMed] [Google Scholar]
- [23].Kucuksezer UC, Aktas-Cetin E, Bilgic-Gazioglu S, et al. Natural killer cells dominate a Th-1 polarized response in Behçet’s disease patients with uveitis. Clin Exp Rheumatol. 2015;33:24–9. [PubMed] [Google Scholar]
- [24].Hsu SM, Yang CH, Teng YT, et al. Suppression of the reactive oxygen response alleviates experimental autoimmune uveitis in mice. Int J Mol Sci. 2020;21:3261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Diwo E, Sève P, Trad S, et al. Therapeutic strategy for the treatment of non-infectious uveitis proposed by an expert panel. Rev Med Interne. 2018;39:687–98. [DOI] [PubMed] [Google Scholar]
- [26].Zhao C, Zhang M. Immunosuppressive treatment of non-infectious uveitis: history and current choices. Chin Med Sci J. 2017;32:48–61. [DOI] [PubMed] [Google Scholar]
- [27].Trivedi A, Katelaris C. The use of biologic agents in the management of uveitis. Intern Med J. 2019;49:1352–63. [DOI] [PubMed] [Google Scholar]
- [28].Dao D, Xie B, Nadeem U, et al. High-fat diet alters the retinal transcriptome in the absence of gut microbiota. Cells. 2021;10:2119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Galipeau HJ, Caminero A, Turpin W, et al. Novel fecal biomarkers that precede clinical diagnosis of ulcerative colitis. Gastroenterology. 2021;160:1532–45. [DOI] [PubMed] [Google Scholar]
- [30].Su W, Chapman NM, Wei J, et al. Protein prenylation drives discrete signaling programs for the differentiation and maintenance of effector treg cells. Cell Metab. 2020;32:996–1011.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Tassi I, Klesney-Tait J, Colonna M. Dissecting natural killer cell activation pathways through analysis of genetic mutations in human and mouse. Immunol Rev. 2006;214:92–105. [DOI] [PubMed] [Google Scholar]
- [32].Borrego F, Masilamani M, Marusina AI, et al. The CD94/NKG2 family of receptors: from molecules and cells to clinical relevance. Immunol Res. 2006;35:263–78. [DOI] [PubMed] [Google Scholar]
- [33].Ahn JK, Park YG, Park SW, et al. Combined low dose cyclosporine and prednisone down-regulate natural killer cell-like effector functions of CD8brightCD56+ T cells in patients with active Behçet uveitis. Ocul Immunol Inflamm. 2006;14:267–75. [DOI] [PubMed] [Google Scholar]
- [34].Held W, Jeevan-Raj B, Charmoy M. Transcriptional regulation of murine natural killer cell development, differentiation and maturation. Cell Mol Life Sci. 2018;75:3371–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Gorabi AM, Hajighasemi S, Kiaie N, et al. The pivotal role of CD69 in autoimmunity. J Autoimmun. 2020;111:102453. [DOI] [PubMed] [Google Scholar]
- [36].Bonacini M, Soriano A, Zerbini A, et al. Higher frequencies of lymphocytes expressing the natural killer group 2D receptor in patients with behçet disease. Front Immunol. 2018;9:2157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Bonacini M, Cimino L, De Simone L, et al. Vogt-Koyanagi-Harada patients show higher frequencies of circulating NKG2Dpos NK and NK T cells. Clin Exp Immunol. 2021;204:41–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Konduri V, Oyewole-Said D, Vazquez-Perez J, et al. CD8+CD161+ T-cells: cytotoxic memory cells with high therapeutic potential. Front Immunol. 2021;11:613204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Kang H, Wei Y, Liu M, et al. Flow cytometric analysis of T lymphocytes and cytokines in aqueous humor of patients with varicella zoster virus-mediated acute retinal necrosis. BMC Ophthalmol. 2021;21:193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Zheng Y, Cai B, Ren C, et al. Identification of immune related cells and crucial genes in the peripheral blood of ankylosing spondylitis by integrated bioinformatics analysis. PeerJ. 2021;9:e1212512125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Paule R, Denis L, Chapuis N, et al. Lymphocyte immune phenotyping and CD4/CD8 ratio in cerebrospinal fluid for the diagnosis of sarcoidosis-related uveitis. Ocul Immunol Inflamm. 2021;29:290–8. [DOI] [PubMed] [Google Scholar]
- [42].Boudreau JE, Mulrooney TJ, Le Luduec JB, et al. KIR3DL1 and HLA-B density and binding calibrate NK education and response to HIV. J Immunol. 2016;196:3398–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Moradi S, Stankovic S, O’Connor GM, et al. Structural plasticity of KIR2DL2 and KIR2DL3 enables altered docking geometries atop HLA-C. Nat Commun. 2021;12:2173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Castaño-Núñez A, Montes-Cano MA, García-Lozano JR, et al. Association of functional polymorphisms of KIR3DL1/DS1 with Behçet’s disease. Front Immunol. 2019;10:2755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Levinson RD, Yung M, Meguro A, et al. KIR and HLA genotypes implicated in reduced killer lymphocytes immunity are associated with Vogt-Koyanagi-Harada disease. PLoS One. 2016;11:e01603920160392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Vendelbosch S, Heslinga SC, John M, et al. Study on the protective effect of the KIR3DL1 gene in ankylosing spondylitis. Arthritis Rheumatol. 2015;67:2957–65. [DOI] [PubMed] [Google Scholar]
- [47].Kim SJ, Lee S, Park C, et al. Targeted resequencing of candidate genes reveals novel variants associated with severe Behçet’s uveitis. Exp Mol Med. 2013;45:49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Kruse PH, Matta J, Ugolini S, et al. Natural cytotoxicity receptors and their ligands. Immunol Cell Biol. 2014;92:221–9. [DOI] [PubMed] [Google Scholar]
- [49].Kinlein A, Janes ME, Kincer J, et al. Analysis of shark NCR3 family genes reveals primordial features of vertebrate NKp30. Immunogenetics. 2021;73:333–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Wang B, Tian Q, Guo D, et al. Activated γδ T cells promote dendritic cell maturation and exacerbate the development of experimental autoimmune uveitis (EAU) in mice. Immunol Invest. 2021;50:164–83. [DOI] [PubMed] [Google Scholar]
- [51].Hu DN, Zhang R, Yao S, et al. Cultured human uveal melanocytes express/secrete CXCL1 and CXCL2 constitutively and increased by lipopolysaccharide via activation of toll-like receptor 4. Curr Eye Res. 2021;46:1681–94. [DOI] [PubMed] [Google Scholar]
- [52].Weigand M, Hauck SM, Deeg CA, et al. Deviant proteome profile of equine granulocytes associates to latent activation status in organ specific autoimmune disease. J Proteomics. 2021;230:103989. [DOI] [PubMed] [Google Scholar]
- [53].Egwuagu CE, Alhakeem SA, Mbanefo EC. Uveitis: molecular pathogenesis and emerging therapies. Front Immunol. 2021;12:623725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Joeckel LT, Wallich R, Martin P, et al. Mouse granzyme K has pro-inflammatory potential. Cell Death Differ. 2011;18:1112–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Mogilenko DA, Shpynov O, Andhey PS, et al. Comprehensive profiling of an aging immune system reveals clonal GZMK+ CD8+ T cells as conserved hallmark of inflammaging. Immunity. 2021;54:99–115.e12. [DOI] [PubMed] [Google Scholar]