Abstract
As commercial fed cattle consume large amounts of concentrate feedstuffs, hindgut health can be challenged. The objective of this study was to evaluate the effects of a commercially available Bacillus feed additive on cattle health outcomes and cecal microbiota of fed cattle at the time of harvest. Commercial cattle from a single feedlot were identified for characterization of cecal microbial communities using 16S ribosomal ribonucleic acid gene sequencing. All cattle were fed a common corn-based finishing diet. Control cattle (CON) were administered no treatment while treated cattle (TRT) were supplemented daily with 0.050 g of MicroSaf 4C 40 (2 billion colony forming units of Bacillus spp.; Phileo by Lesaffre, Milwaukee, WI). Immediately after harvest and evisceration, the cecal contents of cattle were sampled. After DNA extraction, amplification, and sequencing, reads from CON samples (N = 12) and TRT samples (N = 12) were assigned taxonomy using the SILVA 138 database. Total morbidity, first treatment of atypical interstitial pneumonia, and early shipments for harvest were decreased among TRT cattle compared to CON cattle (P ≤ 0.021). On average, cecal microbiota from TRT cattle had greater alpha diversity than microbiota from CON cattle as measured by Shannon diversity, Pielou’s evenness, and feature richness (P < 0.010). Additionally, TRT microbial communities were different (P = 0.001) and less variable (P < 0.001) than CON microbial communities when evaluated by unweighted UniFrac distances. By relative abundance across all samples, the most prevalent phyla were Firmicutes (55.40%, SD = 15.97) and Bacteroidetes (28.17%, SD = 17.74) followed by Proteobacteria (6.75%, SD = 10.98), Spirochaetes (4.54%, SD = 4.85), and Euryarchaeota (1.77%, SD = 3.00). Spirochaetes relative abundance in TRT communities was greater than that in CON communities and was differentially abundant between treatments by ANCOM testing (W = 11); Monoglobaceae was the only family-level taxon identified as differentially abundant (W = 59; greater mean relative abundance in TRT group by 2.12 percentage points). Half (N = 6) of the CON samples clustered away from all other samples based on principal coordinates and represented cecal dysbiosis among CON cattle. The results of this study indicated that administering a four-species blend of Bacillus positively supported the cecal microbial communities of finishing cattle. Further research is needed to explore potential mechanisms of action of Bacillus DFM products in feedlot cattle.
Keywords: Bacillus, cecum, feedlot, 16S ribosomal ribonucleic acid
Supplementing commercial feedlot cattle with four species of Bacillus indicated potential for improving hindgut microbial diversity and preventing dysbiosis. Differences in cecal microbial communities observed could be associated with decreased energy density of digesta, decreased pathogen challenges, and decreased methane production among cattle fed Bacillus.
Introduction
Bovine gastrointestinal microbes impact digestion, growth performance, and health of cattle (Myer, 2019). The relative abundance of microbial populations in the rumen has become well characterized (Hobson and Stewart, 1997; Petri et al., 2013; Henderson et al., 2015). However, less is known about the microbial communities of the hindgut. Divergence of microbial relative abundance has been demonstrated between the rumen and cecum (de Oliveira et al., 2013; Myer et al., 2015a; Bergmann, 2017). Additionally, grain feeding and acidosis have had greater impacts on the hindgut microbiota compared to the foregut microbiota (Khafipour et al., 2016). Dysbiosis of the hindgut has been identified as a factor contributing to overgrowth of organisms associated with negative health outcomes (Zeng et al., 2017; Simpson et al., 2018). For example, Clostridiaceae bacteria have been considered ubiquitous in the hindgut (Myer et al. 2015a, 2016; Freetly et al., 2020), yet Clostridium perfringens overgrowth has been associated with necrotic enteritis in growing and finishing cattle (Simpson et al., 2018; Yang et al., 2019). Direct-fed microbial (DFM) products have been investigated for modulating the rumen and fecal microbiota for potential health impacts (Krehbiel et al., 2003; McAllister et al., 2011; Elghandour et al., 2015). Proposed mechanisms of action of DFM products that could aid in preventing dysbiosis include competitive inhibition, immune stimulation, modulation of fermentation, and antimicrobial effects (McAllister et al., 2011). Few published studies have tested the effects of DFM products on cecal microbiota, especially in bovine models. The objective of this study was to evaluate the effects of a commercial Bacillus DFM product, MicroSaf 4C 40 (Phileo by Lesaffre, Milwaukee, WI), on the cecal microbiota of finishing cattle at the time of harvest.
Materials and Methods
Animal care and management protocols followed the recommendations of the Guide for the Care and Use of Agricultural Animals in Research and Teaching, 4th Edition. Since all cattle were harvested in a commercial processing facility and samples were collected postmortem, no IACUC approval was sought for this research.
Description of treatments, cattle management, and cattle selection
Commercial feedlot cattle from a single High Plains feedlot were used to evaluate the effect of Bacillus DFM supplementation on cattle health and postmortem cecal microbial communities. Cattle were managed in large pen conditions (170 ± 30.2 cattle per pen) and fed 153 d (SD = 12.2). After an adaptation period of 27 d, cattle were fed a high-concentrate basal diet (formulation and nutrient composition provided in Table 1). Cattle in the control group (CON) received no additional microbial feed additives. Cattle in the treated group (TRT) received diets formulated to provide each animal 0.050 g of a Bacillus supplement daily once acclimated to the finishing diet. The Bacillus supplement provided 2 billion colony forming units of a combination of Bacillus amyloliquefaciens, B. subtilis, B. pumilus, and B. lichenformis daily. Treatment was assigned based on pen location within the feedlot to minimize the effects of pen conditions. A total of 260 pens (42,495 cattle) were assigned to CON, and 51 pens (9,461 cattle) were assigned to TRT. Cattle assigned to TRT received Bacillus supplementation for no less than 90 d. Morbidity (total number of treatments, first treatments for atypical interstitial pneumonia, and first treatments for blot) and early shipments (cattle harvested before their home lot because of suspected metabolic disease) were recorded on an individual level and summarized as pen-level counts.
Table 1.
Ingredient formulation and chemical composition of the finishing diet
| Item | Value |
|---|---|
| Ingredient, % dry matter | |
| Steam flaked corn | 43.13 |
| High-moisture corn | 33.89 |
| Wet distillers grain | 9.61 |
| Corn silage | 6.01 |
| Supplement1 | 4.15 |
| Corn oil | 1.65 |
| Mixed hay | 1.56 |
| Diet composition2,3 | |
| Dry matter, % | 59.98 |
| Crude protein, % | 13.85 |
| Non-protein nitrogen, % | 1.01 |
| NEM, Mcal/kg | 2.22 |
| NEG, Mcal/kg | 1.53 |
Supplement provided dietary concentration of 0.01691 g/kg of monensin sodium (Rumensin, Elanco, Greenfield, IN).
All values except diet DM on a dry matter basis.
Tylosin phosphate (Tylan, Elanco) was fed through the micromachine to provide each animal with 0.075 g daily.
At the time of harvest, a subset of cattle was selected to characterize cecal microbial populations. For postmortem cecal microbial sampling, the study population was defined as cattle that weighed between 350 and 450 kg and arrived at the feedlot over a 9-d period beginning on October 12, 2019 and included 408 cattle. At feedlot arrival, CON cattle weighed 384 kg (SD = 25.8) and TRT cattle weighed 407 kg (SD = 36.6). At harvest, 30 cattle (15 each from CON and TRT lots) were randomly selected for cecal microbial sampling.
Microbial sampling
Cattle were harvested on a single day at a High Plains commercial beef processing plant. Immediately after evisceration, the ceca of 15 cattle from CON and the ceca of 15 cattle from TRT groups were sampled for microbial analysis. Each cecum was opened, and digesta was sampled with sterile PurFlock Ultra Regular Tip Swab (Puritan Medical Products, Guilford, ME). Following sampling, swabs were immediately transported on ice to the Center for Meat Safety and Quality at Colorado State University (Fort Collins, CO). Samples were stored at −80 °C.
DNA extraction, amplification, and sequencing
Extraction of DNA and library preparation were performed at the Metcalf Laboratory at Colorado State University consistent with the recommendations of Weinroth et al. (2022). Manufacturers’ protocols were used to extract DNA using the MO BIO MagAttract Powersoil DNA Extraction Kit (Qiagen, Carlsbad, CA) and a KingFisher Flex robot (Thermo Fisher Scientific, Waltham, MA). Cecal samples were loaded into a 96-well extraction plate by cutting the inoculated swab tip into the plate well with location randomly assigned to samples, uninoculated swabs (N = 5), negative controls (N = 7), and one positive control (ZymoBIOMICS Microbial Community Standard 6300; Zymo Research, Irvine, CA).
Amplicon library preparation was completed by polymerase chain reaction (PCR) with barcoded primers targeting the V4 region of the 16S rRNA gene. The barcode assay adapted for the Illumina MiSeq (Illumina; San Diego, CA) was used and included Illumina adaptor, barcode, spacer, and primer. Earth Microbiome Project (EMP) primers 515F and 806R were used for amplification (Caporaso et al., 2012; Apprill et al., 2015; Parada et al., 2016). Duplicate PCR runs were conducted using an Eppendorf Vapo.Protect MasterCycler Pro-S thermocycler (Eppendorf, Hauppauge, NY). Conditions for PCR followed EMP protocols and included initial denaturation at 94 °C for 3 min; 30 cycles of denaturation (94 °C, 45 s), annealing (50 °C, 60 s), and elongation (72 °C, 90 s); and a final 10-min extension at 72 °C (Gilbert et al. 2010, 2014; Thompson et al., 2017). Amplicons were subjected to agarose gel electrophoresis to visualize correct sizes of PCR products and the absence of signal from negative controls. Products were evaluated for effective amplification by agarose gel electrophoresis with expected band size of approximately 300 to 350 base pairs.
Concentration of amplicon products was determined by Quant-IT PicoGreen dsDNA Assay Kit (Thermo Fisher Scientific) read on a Fluoroskan (Thermo Fisher Scientific) plate reader. Amplicons were pooled to form the sequencing library with a target inclusion of 300 ng of DNA from each sample. No more than 50 μL from a single sample was added to maintain the integrity of the negative control. Pooled amplicons were cleaned using MinElute PCR Purification Kit (Qiagen) following manufacturer protocols. Cleaned libraries were evaluated for amplicon concentration by NanoDrop Lite spectrophotometer (Thermo Fisher Scientific).
The amplicon library was diluted to a loading concentration of 8 pM and combined with 15% PhiX control library. Paired-end sequencing (2 × 250 bp) was performed using the 500 cycle MiSeq Reagent Kit v2 (Illumina) on the Illumina MiSeq platform at the Next Generation Sequencing Core Laboratory at Colorado State University.
Sequence processing
Amplicon sequence data were bioinformatically processed in QIIME2 version 2020.8 (Bolyen et al., 2019) using the High-Performance Computing Center at Texas Tech University. Barcodes, forward, and reverse sequences were imported, demultiplexed, and filtered for quality using the q2-demux plugin (Hamday et al., 2008; Hamday and Knight, 2009CJML_BIB_J_0029CJML_BIB_J_0028). Sequences were denoised with DADA2 (Callahan et al., 2016) with forward reads trimmed from 15 to 220 base pairs and reverse reads trimmed from 12 to 155 base pairs. Taxonomy was assigned to amplicon sequence variants (ASV) with the q2-feature-classifier plugin and classify-sklearn naive Bayes classifier (Pedregosa et al., 2011; Bokulich et al., 2018). Reference sequences specific to the V4 region from Silva 138 database (Pruesse et al., 2007; Quast et al., 2013) were used to identify ASV at a 99% similarity threshold.
The sequencing depth of each negative control was evaluated to ensure cleanliness of extraction and library preparation; the number of reads generated by each control well before and after denoising was recorded. The sequencing depth of the positive control was similarly recorded. Additionally, the taxa relative abundance of the positive control was exported and visualized in R version 4.1.3 (R Core Team, 2022) using the geom_bar function of ggplot2 (Wickham, 2009) and compared to the known composition of the mock community for qualitative confirmation.
Reads classified as mitochondria and chloroplasts were removed from the data set. Additionally, all reads generated by controls and technical replicates were removed. All reads from six samples that yielded less than 500 denoised sequences were also removed. Features observed in only one sample were disregarded for further analysis. ASV were assigned phylogeny using SEPP methodology to construct an insertion tree with q2-fragment-insertion (Matsen et al. 2010, 2012; Janssen et al., 2018). Adequate sampling depth was justified by constructing a rarefaction curve with alpha diversity metrics. Sampling depth was standardized for diversity analysis by subsampling without replacement (Weiss et al., 2017) to 23,389 sequences per sample using q2-diversity.
Classification of dysbiosis
Dysbiosis was classified based on rarefied abundance data exported from QIIME2 as relative abundance. For each microbial family observed in greater than 5% relative abundance across all samples, the mean relative abundance ± 1 SD was calculated. By family, each sample was compared the respective range of the mean ± 1 SD. If an individual sample’s relative abundances were outside of this range for half or more of the family-level taxa, the microbial community was considered in a state of dysbiosis.
Statistical analysis
Health outcomes were analyzed by logistic regression of count data summarized by pen using R version 4.1.3 (R Core Team, 2022). Taxa differential abundance was evaluated using QIIME2 by ANCOM testing at both the phylum and family level from rarefied sequence counts (Mandal et al., 2015). Significance for differential abundance was evaluated as a W value indicating log-fold change against a model-determined threshold based on a bimodal distribution. Alpha diversity was measured by richness (the number of observed features), Pielou’s evenness (Pielou, 1966), and Shannon diversity index (Shannon, 1948). Beta diversity was measured as unweighted UniFrac distances (Lozupone and Knight, 2005). Principal Coordinate Analysis (PCoA) was used to spatially visualize samples (Vázquez-Baeza et al., 2017). Microbial relative abundance, alpha diversity measures, unweighted UniFrac distance matrix, and principal coordinates were exported from QIIME2 and imported into R using the qiime2R package (Bisanz, 2018). Differences in alpha diversity were evaluated between treatment groups with Kruskal-Wallis testing (Kruskal and Wallis, 1952). K-means clustering was used to group samples (Lloyd, 1957; MacQueen, 1967). Treatment groups and clusters were evaluated for beta diversity with PERMANOVA testing (Anderson, 2017) using the vegan package of R (Oksanen et al., 2022). The individual cecal microbial community was considered the experimental unit. Statistical significance was established at P < 0.050. All data were visualized in R version 4.1.3 (R Core Team, 2022) using the geom_boxplot and geom_bar function of ggplot2 (Wickham, 2009).
Results
Health performance
Total morbidity, first treatment of atypical interstitial pneumonia, and early shipments for harvest were decreased among TRT cattle compared to CON cattle (P ≤ 0.021; Table 2). No statistical differences were observed in morbidity during the last 60 d of the feeding period (P ≥ 0.208).
Table 2.
Health outcomes of cattle
| Treatment | ||||
|---|---|---|---|---|
| CON1 | TRT2 | SEM3 | P-value | |
| Total lots, N | 260 | 51 | ||
| Total cattle, N | 42,495 | 9,461 | ||
| Morbidity, whole feeding period (%) | ||||
| Total | 12.29 | 11.43 | 0.327 | 0.019 |
| AIP4, first treatment | 0.17 | 0.07 | 0.028 | 0.021 |
| Bloat, first treatment | 0.14 | 0.18 | 0.044 | 0.391 |
| Morbidity, last 60 DOF (%) | ||||
| Total | 2.21 | 2.35 | 0.156 | 0.402 |
| AIP | 0.11 | 0.06 | 0.026 | 0.208 |
| Bloat | 0.09 | 0.13 | 0.037 | 0.242 |
| Early shipments5 (%) | 0.08 | 0.01 | 0.013 | 0.007 |
Cattle not administered MicroSaf 4C 40.
Cattle administered MicroSaf 4C 40.
Standard error of the mean.
Atypical interstitial pneumonia.
Cattle shipped for harvest prior to shipment of lot because of suspected metabolic disease.
Sequencing results
A total of 1,744,899 sequence reads were generated for cecal samples, technical replicates, and controls. Following denoising, 1,218,231 reads remained. Denoised sequencing depths of positive controls and cecal samples were almost three magnitudes of order greater than negative controls (Table 3). Of the eight bacteria represented by the positive control, seven were identified at the genus level, and the remaining bacterium was identified at the family level. Once chloroplasts, mitochondria, features observed in only one sample, and controls were removed from the data set, samples from CON (N = 12) and TRT (N = 12) groups were retained representing 1,521 unique features with a mean sequencing depth of 39,229 (SD = 7,637). Only five sequencing reads classified as Bacillus were identified across all 24 samples retained in the data set.
Table 3.
Mean sequence counts of samples and controls
| N | Sequences | Denoised sequences | |
|---|---|---|---|
| Samples | 24 | 56,257 | 48,732 |
| Negative control, empty well | 7 | 151 | 56 |
| Negative control, swab1 | 5 | 550 | 54 |
| Positive control2 | 1 | 73,696 | 64,549 |
PurFlock Ultra Regular Tip Swabs (Puritan Medical Products, Guilford, ME).
ZymoBIOMICS Microbial Community Standard 6300 (Zymo Research, Irvine, CA).
Beta diversity
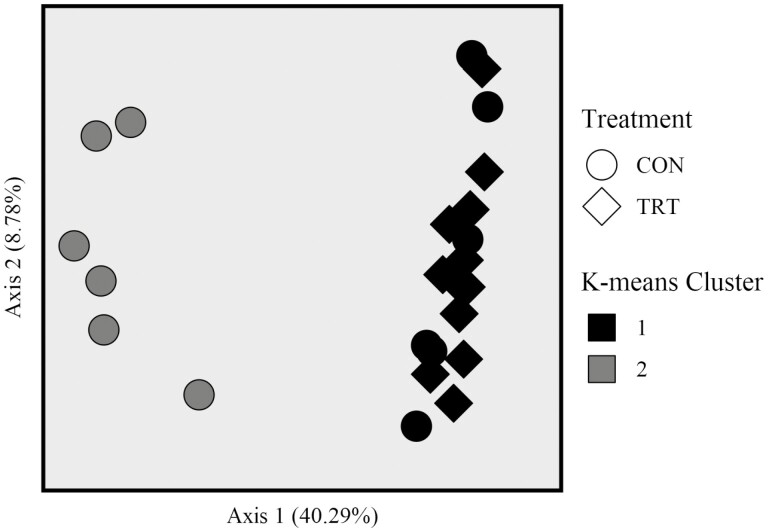
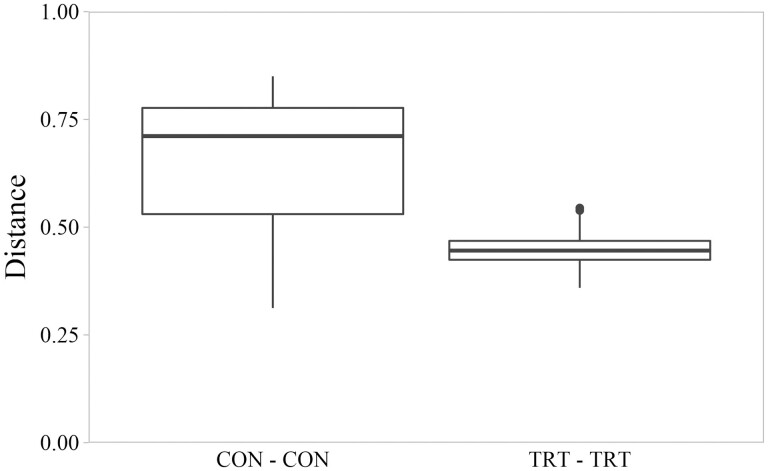
Based on PERMANOVA analysis of unweighted UniFrac distances, microbiota from cecal CON samples differed from microbiota of cecal TRT samples (Figure 1; P = 0.001, F = 4.235, permutations = 999). The phylogenetic distances among communities of CON cecal samples were greater than distances among communities of TRT cecal samples (Figure 2; P < 0.001, Kruskal-Wallis χ2 = 56.32). Greater distances between CON communities were caused by K-means cluster 2 which included six CON samples and segregated from K-means cluster 1 (which included 6 CON samples and all 12 TRT samples) by having a lesser value on the PCoA x-axis (Figure 1). All six samples in K-means cluster 2 were considered instances of dysbiosis; no samples in K-means cluster 1 were considered instances of dysbiosis (Supplementary Table S1).
Figure 1.
Principal coordinate analysis of cecal microbial communities from control cattle (CON, round marker) and cattle fed 0.050 g of MicroSaf 4C 40 daily (TRT, diamond marker). Microbiota of cecal CON samples differed from microbiota of cecal TRT samples based on PERMANOVA analysis of unweighted UniFrac distances (P = 0.001, F = 4.235, permutations = 999). Microbiota of cecal samples in K-means group 1 differed from microbiota of cecal samples in K-means group 2 based on PERMANOVA analysis of unweighted UniFrac distances (P = 0.001, F = 14.275, permutations = 999).
Figure 2.
Pairwise distances between cecal microbial communities based on unweighted UniFrac distance matrix as evaluated by Kruskal–Wallis testing. Box plots represent distances between control cattle (CON) and distances between cattle fed 0.050 g of MicroSaf 4C 40 daily (TRT; supplementation of 2-billion colony forming units of a combination of four species of Bacillus daily). Distances between CON communities were greater than distances between TRT communities (P < 0.001, χ2 = 56.32, df = 1).
Alpha diversity
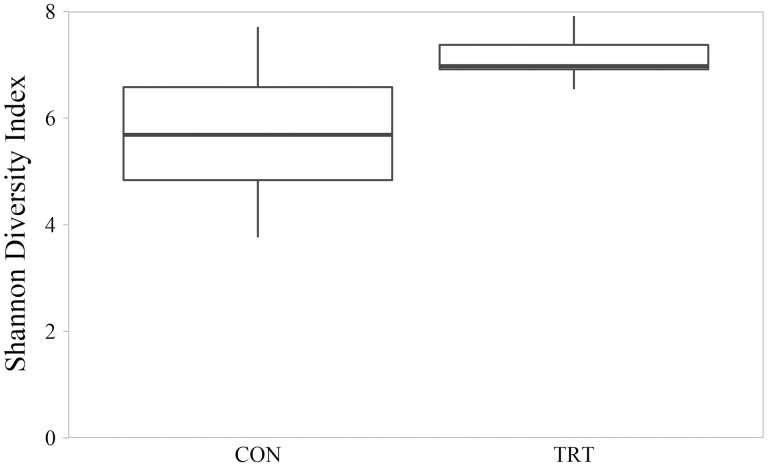
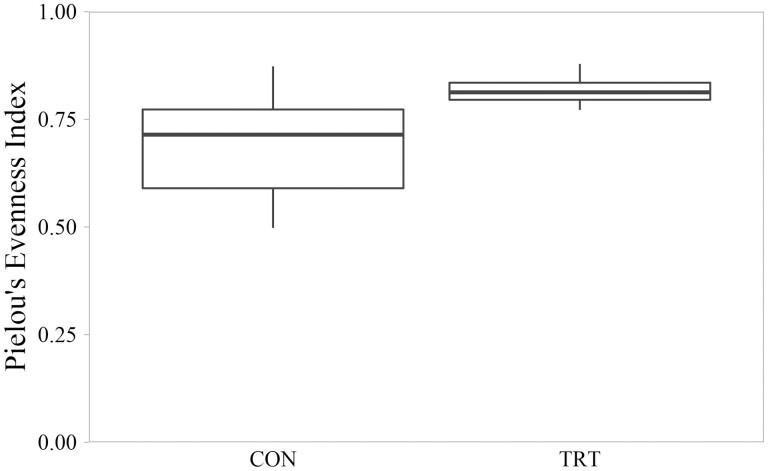
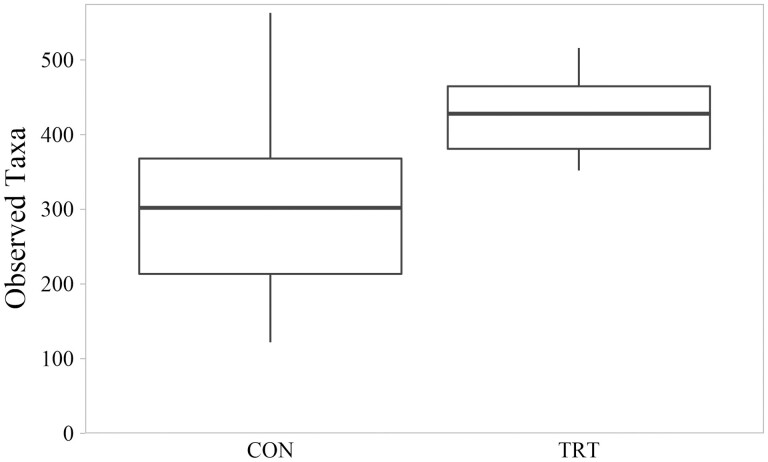
Microbial diversity measured by Shannon Index ranged from 3.76 to 7.92. Mean Shannon Index for CON communities was 5.67 (SD = 1.33); mean Shannon diversity for TRT communities was 7.12 (SD = 0.40). On average, TRT microbial communities were 26% more diverse that CON microbial communities (P = 0.005; Figure 3). Shannon diversity differences were associated with TRT communities having both greater evenness as measured by Pielou’s Index (P = 0.003; Figure 4) and greater richness as measured by number of observed features (P = 0.008; Figure 5). Within-group variation of Shannon diversity measured as SD was more than three-times greater for the CON group compared to TRT.
Figure 3.
Shannon diversity index of cecal microbial communities of control cattle (CON) and cattle fed 0.050 g of MicroSaf 4C 40 daily (TRT; supplementation of 2-billion colony forming units of a combination of four species of Bacillus daily). Shannon diversity index differed between CON and TRT groups (P = 0.005, H = 8.003, N = 24).
Figure 4.
Evenness of cecal microbial communities measured by Pielou’s Index of control cattle (CON) and cattle fed 0.050 g of MicroSaf 4C 40 daily (TRT; supplementation of 2-billion colony forming units of a combination of four species of Bacillus daily). Evenness differed between CON and TRT groups (P = 0.003, H = 8.670, N = 24).
Figure 5.
Richness of cecal microbial communities measured by the number of bacterial taxa observed of control cattle (CON) and cattle fed 0.050 g of MicroSaf 4C 40 daily (TRT; supplementation of 2-billion colony forming units of a combination of four species of Bacillus daily). Richness differed between CON and TRT groups (P = 0.008, H = 7.056, N = 24).
Variation between CON samples was caused by divergence of CON samples between K-means clusters. Mean Shannon Index for K-means cluster 1 communities was 6.93 (SD = 0.70); mean Shannon diversity for K-means cluster 2 communities was 4.78 (SD = 0.97). On average, K-means cluster 1 microbial communities were 45% more diverse that CON microbial communities (P = 0.001). Similarly, K-means cluster 1 had greater evenness and richness than K-means cluster 2 (P ≤ 0.001). Richness was decreased more than 52% in K-means cluster 2 compared to that of K-means cluster 1. Mean number of observed features for K-means cluster 1 communities was 421 (SD = 68.6); mean number of observed features for K-means cluster 2 communities was 202 (SD = 52.3).
Phylum-level taxonomy
Classification of the sequence reads using the SILVA database identified five phyla in greater than 1.5% relative abundance across all samples. The two most prevalent phyla were Firmicutes (55.40% relative abundance, SD = 15.97) and Bacteroidetes (28.17% relative abundance, SD = 17.74). The ratio of Firmicutes to Bacteroidetes was calculated for each sample; mean Firmicutes to Bacteroidetes ratio was 157.45 (range: 0.58 to 1,207.11) for CON microbial communities and 1.51 (range: 0.66 to 2.02) for TRT microbial communities. No difference in Firmicutes to Bacteroidetes ratio was detected based on nonparametric Kruskal-Wallis testing (P = 0.119, H = 2.43).
Other phyla observed included Proteobacteria (6.75% relative abundance, SD = 10.98), Spirochaetes (4.54% relative abundance, SD = 4.85), and Euryarchaeota (1.77% relative abundance, SD = 3.00). The phylum Spirochaetes composed 2.00% (SD = 4.09) of CON microbial communities and 7.34% (SD = 4.25) of TRT communities and was identified as differentially abundant between treatments by ANCOM testing (W = 11).
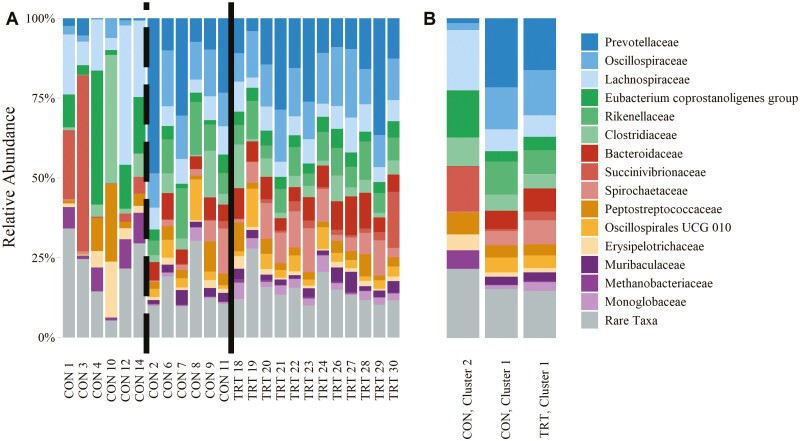
Family-level taxonomy
Fifteen family-level taxa were observed in greater than 1.5% relative abundance across all reads (Table 4). These families are visualized in Figure 6. Of all the families observed, Monoglobaceae was the only family identified as differentially abundant by ANCOM testing (W = 59). The phylum Firmicutes was represented by Oscillospiraceae (10.89% relative abundance, SD = 5.96), Lachnospiraceae (9.81% relative abundance, SD = 8.70), Eubacterium coprostanoligenes group (6.57% relative abundance, SD = 8.44), Clostridiaceae (5.63% relative abundance, SD = 8.26), Peptostreptococcaceae (4.44% relative abundance, SD = 5.01), Oscillospirales UCG-010 (3.11% relative abundance, SD = 3.41), Erysipelotrichaceae (2.27% relative abundance, SD = 3.45), and Monoglobaceae (1.66% relative abundance, SD = 1.64). The phylum Bacteroidetes was represented by the families Prevotellaceae (13.87% relative abundance, SD = 12.56), Rikenellaceae (6.43% relative abundance, SD = 4.92), Bacteroidaceae (5.08% relative abundance, SD = 3.65), and Muribaculaceae (1.94% relative abundance, SD = 1.65). The phylum Spirochaetes was represented by the Spirochaetaceae family (4.94% relative abundance, SD = 4.89) and the Treponema genus. The phylum Proteobacteria was represented by Succinivibrionaceae (5.03% relative abundance, SD = 11.95). The phylum Euryarchaeota was represented by Methanobacteriaceae (1.69% relative abundance, SD = 3.02). The genus Methanobrevibacter composed the majority of all Methanobacteriaceae reads. For 13 of the 15 family-level taxa observed, the SD of CON relative abundances were greater than those of TRT relative abundances.
Table 4.
Mean family-level taxa relative abundance of cecal microbial families observed in greater than 1.5% mean relative abundance across all reads
| Relative abundance, % ± SD | ||||
|---|---|---|---|---|
| Phylum | Family | Control1 (N = 12) |
Treated2 (N = 12) |
Treated:control |
| Bacteroidetes | Prevotellaceae | 11.55 ± 15.12 | 16.20 ± 9.45 | 1.40 |
| Rikenellaceae | 5.20 ± 6.35 | 7.67 ± 2.64 | 1.47 | |
| Bacteroidaceae | 2.88 ± 3.18 | 7.28 ± 2.67 | 2.53 | |
| Muribaculaceae | 1.26 ± 1.52 | 2.63 ± 1.54 | 2.09 | |
| Firmicutes | Oscillospiraceae | 7.66 ± 6.20 | 14.13 ± 3.59 | 1.85 |
| Lachnospiraceae | 12.89 ± 11.50 | 6.72 ± 2.28 | 0.52 | |
| Eubacterium coprostanoligenes group | 8.93 ± 11.66 | 4.20 ± 0.85 | 0.47 | |
| Clostridiaceae | 7.00 ± 11.22 | 4.27 ± 3.59 | 0.61 | |
| Peptostreptococcaceae | 5.37 ± 6.72 | 3.52 ± 2.32 | 0.66 | |
| Oscillospirales UCG-010 | 2.33 ± 3.74 | 3.89 ± 3.00 | 1.67 | |
| Erysipelotrichaceae | 3.16 ± 4.72 | 1.37 ± 0.93 | 0.43 | |
| Monoglobaceae | 0.60b ± 1.19 | 2.72a ± 1.33 | 4.53 | |
| Proteobacteria | Succinivibrionaceae | 7.40 ± 16.22 | 2.67 ± 4.82 | 0.36 |
| Spirochaetota | Spirochaetaceae | 2.33 ± 4.14 | 7.55 ± 4.25 | 3.24 |
| Euryarchaeota | Methanobacteriaceae | 2.99 ± 3.91 | 0.40 ± 0.30 | 0.13 |
Cecal microbiota of cattle not administered MicroSaf 4C 40.
Cecal microbiota of cattle administered MicroSaf 4C 40.
Family-level relative abundance differed by ANCOM testing.
Figure 6.
Relative abundance of family-level taxa composing the cecal microbial communities of control cattle (CON) and cattle fed 0.050 g of MicroSaf 4C 40 daily (TRT; supplementation of 2-billion colony forming units of a combination of four species of Bacillus daily). Rare taxa include all bacterial families observed in less than 1.5% average relative abundance across all samples. Microbial relative abundance by sample; solid line separates CON and TRT samples; the dashed line separates K-means clusters (A; as identified in Figure 1). Microbial relative abundance summarized by treatment and cluster (B).
Discussion
Diversity and dysbiosis
Microbial dysbiosis is related to pathologies of the gastrointestinal tract (Plaizier et al., 2014; Khafipour et al., 2016; Azad et al., 2019). Dysbiosis was observed in six CON samples that clustered away from all other samples (K-means cluster 2). Divergence of CON samples between K-means clusters drove greater variation in alpha diversity and beta diversity measures among CON samples (compared to TRT samples which all clustered together). The decreased variability between TRT microbial communities compared to that of CON communities was similar to the findings of Schofield et al. (2018) when B. amyloliquefaciens was fed to dairy calves and sheep. Together, these results suggested that feeding Bacillus could promote more consistent microbial communities between animals partly by decreasing the incidence of dysbiosis.
Overall, Shannon diversity values from the present study were intermediate to those identified within steer cecal microbiota by Freetly et al. (2020) and in steer colon microbiota by Myer et al. (2015a). Shannon values for the TRT group and K-means cluster 1 were similar to those previously reported from bovine fecal samples by Xu et al. (2014) and Durso et al. (2012). However, mean Shannon diversity value for K-means cluster 2 was approximately 30% less than previously reported values. This is further evidence that the communities in K-means cluster 2 were instances of dysbiosis. Decreased Shannon diversity values have been observed in the hindgut of cattle when dysbiosis coincided with a disease state (Fecteau et al., 2016). However, greater Shannon diversity of hindgut microbiota has been associated with improved feed efficiency (Welch et al., 2020). In models of other species, Bacillus spp. supplementation has increased intestinal alpha diversity and similarly resulted in improved daily weight gain and feed conversion (Sun et al., 2010; Hong et al., 2019; Li et al. 2019a, 2019b; Luo et al., 2020). Collectively, decreased alpha diversity in the CON communities represented in K-means cluster 2 was associated with dysbiosis; feeding Bacillus could mitigate instances of hindgut dysbiosis to improve digestive health and decrease morbidity and early shipments caused by metabolic disease.
Potential pathology
Clostridiaceae and related taxa were observed in numerically increased relative abundance in one sample of the current study. Control sample 10 (classified into K-means cluster 2) had 40.29% relative abundance of Clostridiaceae and 24.20% relative abundance of Peptostreptococcaceae. Clostridiaceae bacteria are known to be spore-forming bacteria that exist in greater relative abundance in the hindgut and contribute to digestion of carbohydrates and protein (Myer et al. 2015b, 2016; Freetly et al., 2020). However, the relative abundance observed in CON sample 10 could be approaching dominance of the microbial community. While 16S methodology does not measure viable cells, this finding could exemplify dysbiosis associated with a pathological state. Clostridium sensu stricto 1, the predominate genera of Clostridiaceae observed in this study, has been positively associated with Clostridium perfringens and necrotic enteritis (Yang et al., 2019). Peptostreptococcaceae is associated with the lumen of the hind gut (Mao et al., 2015). Romboutsia, the primary Peptostreptococcaceae genus observed in this study, is associated with Escherichia coli O157:H7 challenge (Mir et al., 2019), and the second most observed Peptostreptococcaceae genus in this study, Paeniclostridium, is closely related to pathogenic Clostridium (Rabi et al., 2017). With greater than 60% of the microbial community of CON sample 10 composed of Clostridiaceae and Peptostreptococcaceae, this cecal sample exemplifies a state of dysbiosis with pathological implications.
Prevention of overgrowth of Clostridiaceae among TRT communities may have been modulated by the four species of Bacillus that were fed. Bacillus spp. have previously exhibited antimicrobial properties. Bacillus subtilis supplemented to Holstein cows supported hindgut health and reduced loads of Clostridium (Song et al., 2014). Protective properties of Bacillus spp. from the pathogens Salmonella typhimurium and Escherichia coli have also been demonstrated (Broadway et al., 2020; Lin et al., 2020). Secretion of the antimicrobial peptide subtilosin and the protease subtilin have been identified as products of Bacillus subtilis bacteria that enable antibacterial and antifungal properties (Algburi et al., 2020; Lin et al., 2020; Luise et al., 2022). As such, findings indicated that Bacillus supplementation could have protected TRT microbial communities from Clostridiaceae overgrowth.
Indications of hindgut nutrient availability
Feed intake, rate of passage, and microbial communities are interrelated (Colucci et al., 1982; Okine and Mathison, 1991CJML_BIB_J_0017CJML_BIB_J_0070; Freetly et al., 2020). After energy extraction has occurred in the rumen and small intestine, differences in cecal energy abundance could reflect rate of passage and extent of nutrient digestion. Numerically, CON sample 1 and 3 had greater abundance of Succinivibrionaceae. This bacterial family has been prevalent in the rumen and less prevalent in the hindgut because Succinivibrionaceae thrive when starch is available as a substrate (Hespell, 1992). The CON communities enriched in Succinivibrionaceae indicated that greater concentration of starch was reaching the hindgut. Consistent with poorer efficiency of starch use when digested in the hindgut compared to the foregut, Myer et al. (2015b) observed an inverse relationship between cecal relative abundance of Succinivibrionaceae and feed efficiency. Increased cecal Succinivibrionaceae populations indicated greater quantities of starch were available for digestion in the hindgut. In the two instances in which Succinivibrionaceae was increased in the cecal microbiota (CON 1 and 3), the community was also considered to be in dysbiosis, potentially caused by greater starch flow to the hindgut.
Similarly, numerically greater Erysipelotrichaceae relative abundance among CON microbial communities could have indicated greater energy density of digesta reaching the cecum and greater potential for cecal lipid digestion. The Erysipelotrichaceae family has been associated with lipid metabolism, energy density, and inflammation in human and animal models (Kaakoush, 2015; Minaya et al., 2020). Additionally, increased relative abundance of the Erysipelotrichaceae genus Turicibacter has previously been associated with feeding of high concentrate diets (Liu et al., 2014). Numerically greater Erysipelotrichaceae relative abundance among CON microbial communities indicated greater flow of digestible nutrients to the hindgut.
Along with suspected greater energy density of the digesta, CON communities observed in dysbiosis (K-means cluster 2) had numerically greater relative abundance of Methanobacteriaceae. Specifically, the genus Methanobrevibacter, which is considered the primary methanogen of the hindgut, was numerically enriched in K-means cluster 2 samples (Kim and Whitman, 2014). Ramayo-Caldas et al. (2020) found a negative relationship between Succinivibrionaceae and methane, the product of Methanobacteriaceae fermentation. On the contrary, CON communities in the present study had greater mean relative abundance of both Succinivibrionaceae and Methanobacteriaceae bacteria compared to TRT microbiota. However, mean Succinivibrionaceae relative abundance is likely inflated among the CON group by samples 1 and 3 in which Succinivibrionaceae relative abundance was 21.6% and 55.2%. Consistent with the present study, Schofield et al. (2018) observed lesser Methanobrevibacter when ruminants were supplemented with B. amyloliquefaciens. Numerically decreased Methanobacteriaceae relative abundance among TRT samples indicated decreased hindgut methanogenesis and decreased nutrient utilization likely as a function of greater prececal extent of digestion.
Microbial abundances and fibrolytic families
Microbial relative abundances observed in this study were like previous results. Firmicutes and Bacteroidetes are commonly cited as the predominant phyla composing the gastrointestinal microbiota (Callaway et al., 2010; Durso et al., 2012; Liu et al., 2016). Proteobacteria, Spirochaetes, and Euryarchaeota have been reported as common phyla of the bovine digestive tract (Huebner et al., 2019; Andrade et al., 2020).
However, within the Firmicutes phylum and more specifically the Eubacteriales order, numerical trends in relative abundance of cellulolytic bacteria were observed. While CON communities had numerically greater Lachnospiraceae relative abundance, TRT communities had greater Monoglobaceae and numerically greater Oscillospiraceae and Oscillospirales UCG-010 relative abundance. Fibrolytic properties have been demonstrated for all these bacterial families. The Lachnospiraceae family has demonstrated butyrate production and cellulolytic activity (Cotta and Forster, 2006; Nyonyo et al., 2014; Bach et al., 2019). Monoglobaceae (which was solely represented by the genus Monoglobus, a pectinolytic bacterium) was also associated with capacity for fiber degradation (Kim et al., 2019). Oscillospiraceae, a basonym for Ruminococcaceae, has been identified as a cellulolytic family in ruminant gastrointestinal tracts (Deusch et al., 2017; Yoon et al., 2017). Oscillospiraceae has been identified in fecal microbiota of grazing beef cows when taxonomy was assigned using a SILVA database (Pruesse et al., 2007; Koester et al., 2020); others have identified Ruminococcaceae in the hindgut when taxonomy was assigned using Greengenes (DeSantis et al., 2006; Myer et al., 2015a; Freetly et al., 2020). Regardless, the relative abundance of Oscillospiraceae and Oscillospirales UCG-010 was likely associated with greater capacity for fiber degradation (Biddle et al., 2013).
Together, numerical differences in family relative abundance of taxa belonging to the Eubacteriales order between CON and TRT communities indicated redundancy in fibrolytic potential. The observed alteration of microbial taxa assumed to be digesting fiber in the cecum could have been modulated by the Bacillus treatment because B. amyloliquefaciens has demonstrated cellulase production (Lee et al., 2008; Sun et al., 2017). However, more targeted data on community function and enzyme presence is needed to determine if supplemental Bacillus has a meaningful impact on fibrolytic bacterial populations in the cecum of fed cattle.
Hindgut microbial communities and health
Improved richness and evenness of hindgut microbiota—such as that observed in the TRT microbial communities—could be associated with benefits to feedlot cattle health and growth performance (Gressley et al., 2011; Rodriguez-Jimenez et al., 2019; Sanz-Fernandez et al., 2020). As high-concentrate finishing diets are fed to feedlot cattle, risk of acidosis is increased (Owens et al., 1998). The US feeder and fed cattle supplies are influenced predominantly by Angus genetics because of premiums paid for black-hided cattle that yield well-marbled carcasses (Parish et al., 2012; Williams et al., 2012; McCabe et al., 2019). Angus genetics are associated with greater feed intake than that of other breeds (Retallick et al., 2017), and expected progeny differences for dry matter intake suggest that genetics within the Angus population will continue to increase feed intake (American Angus Association, 2022). Collectively, these factors have resulted in greater feed intake by feedlot cattle, which coincides with increased rate of passage and shifts a greater proportion of digestion to the hindgut (Church, 1988). As postruminal digestion increases, risk is increased for hindgut digestive challenges including acidosis, disruption of microbial communities, and overgrowth of pathogens.
The hindgut is suggested to be more vulnerable to acidic pH than the rumen. Absence of protozoal species in the hindgut has limited sequestration of rapidly fermentable carbohydrates (Hume, 1997) and no saliva secretions are observed (Erdman, 1988). Additionally, the linings of the rumen and hindgut are composed of different epithelial structures. The rumen epithelium is composed of four layers of cuboidal and squamous epithelium while the hindgut epithelium is composed of a monolayer of columnar epithelium covered by mucus (Church, 1988). In the event of acidotic stress, the barrier function of the gastrointestinal epithelium can be compromised and associated with leak of proinflammatory toxins such as lipopolysaccharide (Rodriguez-Jimenez et al., 2019). Since the intestinal content is highly immunogenic, Sanz-Fernandez et al. (2020) suggested that the local inflammation from the hindgut could greatly contribute to systemic inflammation. Widespread inflammation is a large energy cost (Sanz-Fernandez et al., 2020) and is implicated in disease complexes such as acute interstitial pneumonia (Loneragan and Gould, 2000). As such, prevention of hindgut acidosis and associated dysbiosis in feedlot cattle would positively affect cattle health and potentially growth performance.
Bacillus could have exhibited protection from gastrointestinal dysbiosis by modulating foregut fermentation and corresponding rate of passage. Interestingly, only five reads classified as Bacillus were observed in cecal communities in this study. This suggested that the primary mode of action of the Bacillus treatment was not as a dominant member of the cecal microbiota. Bacillus either enters vegetative growth at a more proximal location in the gastrointestinal tract (such as the rumen or small intestine), or its relative abundance was below the practical detectable limit of the sequencing depth of this analysis as demonstrated by Schofield et al.’s comparison of Bacillus recovery by qPCR and 16S sequencing (2018).
Implications
Results of this study indicated that cattle fed Bacillus exhibited less animal-to-animal variation of cecal microbiota compared to that of negative control cattle. No instances of cecal dysbiosis were observed in the microbial communities of cattle that were fed Bacillus, and greater alpha diversity was identified in TRT communities compared to CON communities. Under the management conditions of this study with a highly fermentable, high-moisture finishing diet, Bacillus protected against imbalances in the hindgut microbiota and improved cattle health. Future studies should evaluate rate of passage, extent of ruminal digestion, and culture confirmation of potentially pathogenic organisms. Additionally, well-replicated, large-pen studies should test differences in feed efficiency and growth performance.
Supplementary Material
Acknowledgments
We express appreciation to CattleTrail Inc., Metcalf Laboratory at Colorado State University, Colorado State University Next Generation Sequencing Core Laboratory, and Texas Tech High Performance Computing Center for assistance with this research.
Glossary
Abbreviations
- ASV
amplicon sequence variant
- DFM
direct-fed microbial
- EMP
Earth Microbiome Project
- PCoA
principal coordinate analysis
Contributor Information
Luke K Fuerniss, Department of Animal and Food Sciences, Texas Tech University, Lubbock, TX 79409, USA.
Kelly K Kreikemeier, Pioneer Feedyard, Foote Cattle Company, Oakley, KS 67748, USA.
Lynn D Reed, Phileo by Lesaffre, Milwaukee, WI 52404, USA.
Matt D Cravey, Phileo by Lesaffre, Milwaukee, WI 52404, USA.
Bradley J Johnson, Department of Animal and Food Sciences, Texas Tech University, Lubbock, TX 79409, USA.
Conflict of Interest Statement
Lynn D. Reed and Matt D. Cravey are employed by the manufacturer of the product used as a treatment in this study. All other authors declare no conflict of interest.
Literature Cited
- Algburi, A., Alazzawi S. A., Al-Ezzy A. I. A., Weeks R., Chistyakov V., and Chikindas M. L.. . 2020. Potential probiotics Bacillus subtilis KATMIRA1933 and Bacillus amyloliquefaciens B-1895 co-aggregate with clinical isolates of Proteus mirabilis and prevent biofilm formation. Probiotics Antimicrob. Proteins 12:1471–1483. doi: 10.1007/S12602-020-09631-0 [DOI] [PubMed] [Google Scholar]
- American Angus Association. 2022. Genetic Trend EPD/$Value by Birth Year. [accessed February 12, 2022]. https://www.angus.org/Nce/GeneticTrends.
- Anderson, M. J. 2017. Permutational multivariate analysis of variance (PERMANOVA). In: Wiley StatsRef: Statistics Reference Online. Chichester (UK): John Wiley & Sons; p. 1–15. doi: 10.1002/9781118445112.stat07841 [DOI] [Google Scholar]
- Andrade, B. G. N., Bressani F. A., Cuadrat R. R. C., Tizioto P. C., De Oliveira P. S. N., Mourão G. B., Coutinho L. L., Reecy J. M., Koltes J. E., Walsh P., . et al. 2020. The structure of microbial populations in Nelore GIT reveals inter-dependency of methanogens in feces and rumen. J. Anim. Sci. Biotechnol. 11:1–10. doi: 10.1186/s40104-019-0422-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apprill, A., McNally S., Parsons R., and Weber L.. . 2015. Minor revision to V4 region SSU rRNA 806R gene primer greatly increases detection of SAR11 bacterioplankton. Aquat. Microb. Ecol. 75:129–137. doi: 10.3354/ame01753 [DOI] [Google Scholar]
- Azad, E., Derakhshani H., Forster R. J., Gruninger R. J., Acharya S., McAllister T. A., and Khafipour E.. . 2019. Characterization of the rumen and fecal microbiome in bloated and non-bloated cattle grazing alfalfa pastures and subjected to bloat prevention strategies. Sci. Rep. 9:4272. doi: 10.1038/s41598-019-41017-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bach, A., Lã A., Gonzà O., Elcoso G., Fà bregas F., Chaucheyras-Durand F., Castex M., López-García A., González-Recio O., Elcoso G., . et al. 2019. Changes in the rumen and colon microbiota and effects of live yeast dietary supplementation during the transition from the dry period to lactation of dairy cows. J. Dairy Sci. 102:6180–6198. doi: 10.3168/jds.2018-16105 [DOI] [PubMed] [Google Scholar]
- Bergmann, G. T. 2017. Microbial community composition along the digestive tract in forage- and grain-fed bison. BMC Vet. Res. 13:1–9. doi: 10.1186/s12917-017-1161-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biddle, A., Stewart L., Blanchard J., and Leschine S... 2013. Untangling the genetic basis of fibrolytic specialization by Lachnospiraceae and Ruminococcaceae in diverse gut communities. Diversity 5(3):627–640. doi: 10.3390/d5030627 [DOI] [Google Scholar]
- Bisanz, J. E. 2018. qiime2R: importing QIIME2 artifacts and associated data into R sessions. https://github.com/jbisanz/qiime2R [Google Scholar]
- Bokulich, N. A., Kaehler B. D., Rideout J. R., Dillon M., Bolyen E., Knight R., Huttley G. A., and Caporaso J. G.. . 2018. Optimizing taxonomic classification of marker-gene amplicon sequences with QIIME 2’s q2-feature-classifier plugin. Microbiome. 6:90. doi: 10.1186/s40168-018-0470-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolyen, E., Rideout J. R., Dillon M. R., Bokulich N. A., Abnet C. C., Al-Ghalith G. A., Alexander H., Alm E. J., Arumugam M., Asnicar F., . et al. 2019. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 37:852–857. doi: 10.1038/s41587-019-0209-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broadway, P. R., Carroll J. A., Burdick Sanchez N. C., Callaway T. R., Lawhon S. D., Gart E. V., Bryan L. K., Nisbet D. J., Hughes H. D., Legako J. F., . et al. 2020. Bacillus subtilis PB6 supplementation in weaned Holstein steers during an experimental Salmonella challenge. Foodborne Pathog. Dis. 17(8):521–528. doi: 10.1089/FPD.2019.2757. [DOI] [PubMed] [Google Scholar]
- Callahan, B. J., McMurdie P. J., Rosen M. J., Han A. W., Johnson A. J. A., and Holmes S. P.. . 2016. DADA2: high-resolution sample inference from Illumina amplicon data. Nat. Methods 13:581. doi: 10.1038/nmeth.3869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callaway, T. R., Dowd S. E., Edrington T. S., Anderson R. C., Krueger N., Bauer N., Kononoff P. J., and Nisbet D. J.. . 2010. Evaluation of bacterial diversity in the rumen and feces of cattle fed different levels of dried distillers grains plus solubles using bacterial tag-encoded FLX amplicon pyrosequencing. J. Anim. Sci. 88:3977–3983. doi: 10.2527/jas.2010-2900 [DOI] [PubMed] [Google Scholar]
- Caporaso, J. G., Lauber C. L., Walters W. A., Berg-Lyons D., Huntley J., Fierer N., Owens S. M., Betley J., Fraser L., Bauer M., . et al. 2012. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J. 6:1621–1624. doi: 10.1038/ismej.2012.8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Church, D. C. 1988. The ruminant animal: digestive physiology and nutrition. Prospect Heights (IL): Waveland Press. [Google Scholar]
- Colucci, P. E., Chase L. E., and Van Soest P. J.. . 1982. Feed intake, apparent diet digestibility, and rate of particulate passage in dairy cattle. J. Dairy Sci. 65:1445–1456. doi: 10.3168/jds.S0022-0302(82)82367-9 [DOI] [Google Scholar]
- Cotta, M., and Forster R.. . 2006. The Family Lachnospiraceae, including the genera Butyrivibrio, Lachnospira and Roseburia. In: Dworkin M., Falkow S., Rosenberg E., Schleifer K. H., and Stackebrandt E., editors, The Prokaryotes. New York, NY: Springer; p. 1002–1021. doi: 10.1007/0-387-30744-3_35 [DOI] [Google Scholar]
- DeSantis, T. Z., Hugenholtz P., Larsen N., Rojas M., Brodie E. L., Keller K., Huber T., Dalevi D., Hu P., and Andersen G. L.. . 2006. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl. Environ. Microbiol. 72:5069–5072. doi: 10.1128/AEM.03006-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deusch, S., Camarinha-Silva A., Conrad J., Beifuss U., Rodehutscord M., and Seifert J.. . 2017. A structural and functional elucidation of the rumen microbiome influenced by various diets and microenvironments. Front. Microbiol. 8:1605. doi: 10.3389/fmicb.2017.01605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durso, L. M., Wells J. E., Harhay G. P., Rice W. C., Kuehn L., Bono J. L., Shackelford S., Wheeler T., and Smith T. P. L.. . 2012. Comparison of bacterial communities in faeces of beef cattle fed diets containing corn and wet distillers’ grain with solubles. Lett. Appl. Microbiol. 55:109–114. doi: 10.1111/j.1472-765X.2012.03265.x [DOI] [PubMed] [Google Scholar]
- Elghandour, M. M. Y., Salem A. Z. M., Castañeda J. S. M., Camacho L. M., Kholif A. E., and Chagoyán J. C. V.. . 2015. Direct-fed microbes: a tool for improving the utilization of low quality roughages in ruminants. J. Integr. Agric 14:526–533. doi: 10.1016/S2095-3119(14)60834-0 [DOI] [Google Scholar]
- Erdman, R. A. 1988. Dietary buffering requirements of the lactating dairy cow: a review. J. Dairy Sci. 71:3246–3266. doi: 10.3168/JDS.S0022-0302(88)79930-0 [DOI] [Google Scholar]
- Fecteau, M. E., Pitta D. W., Vecchiarelli B., Indugu N., Kumar S., Gallagher S. C., Fyock T. L., and Sweeney R. W.. . 2016. Dysbiosis of the fecal microbiota in cattle infected with Mycobacterium avium subsp. paratuberculosis. PLoS One 11(8):e0160353. doi: 10.1371/JOURNAL.PONE.0160353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freetly, H. C., Dickey A., Lindholm-Perry A. K., Thallman R. M., Keele J. W., Foote A. P., and Wells J. E.. . 2020. Digestive tract microbiota of beef cattle that differed in feed efficiency. J. Anim. Sci. 98:1–16. doi: 10.1093/jas/skaa008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert, J. A., Meyer F., Antonopoulos D., Balaji P., Titus Brown C., Brown C. T., Desai N., Eisen J. A., Evers D., Field D., . et al. 2010. Meeting report: the terabase metagenomics Workshop and the vision of an Earth Microbiome Project. Stand. Genomic Sci 3:243–248. doi: 10.4056/sigs.1433550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert, J. A., Jansson J. K., and Knight R.. . 2014. The Earth Microbiome project: successes and aspirations. BMC Biol. 12:69. doi: 10.1186/s12915-014-0069-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gressley, T. F., Hall M. B., and Armentano L. E.. . 2011. Ruminant nutrition symposium: productivity, digestion, and health responses to hindgut acidosis in ruminants. J. Anim. Sci. 89:1120–1130. doi: 10.2527/jas.2010-3460 [DOI] [PubMed] [Google Scholar]
- Hamday, M., and Knight R.. . 2009. Microbial community profiling for human microbiome projects: tools, techniques, and challenges. Genome Res. 19:1141–1152. doi: 10.1101/gr.085464.108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamday, M., Walker J. J., Harris J. K., Gold N. J., and Knight R.. . 2008. Error-correcting barcoded primers allow hundreds of samples to be pyrosequenced in multiplex. Nat. Methods 5:235–237. doi: 10.1038/nmeth.1184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson, G., Cox F., Ganesh S., Jonker A., Young W., Janssen P. H., Abecia L., Angarita E., Aravena P., Arenas G. N., . et al. 2015. Rumen microbial community composition varies with diet and host, but a core microbiome is found across a wide geographical range. Sci. Rep. 5:14567. doi: 10.1038/srep14567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hespell, R. B. 1992. The genera Succinivibrio and Succinimonas. In: Balows A., Trüper H. G., Dworkin M., Harder W., and Schleifer K. H., editors, The Prokaryotes. New York (NY): Springer; p. 3979–3982. doi: 10.1007/978-1-4757-2191-1_60 [DOI] [Google Scholar]
- Hobson, P. N., and Stewart C. S.. . 1997. The rumen microbial ecosystem. 2nd ed. New York (NY): Chapman & Hall. [Google Scholar]
- Hong, Y., Cheng Y., Li Y., Li X., Zhou Z., Shi D., Li Z., and Xiao Y.. . 2019. Preliminary study on the effect of Bacillus amyloliquefaciens TL on cecal bacterial community structure of broiler chickens. Biomed Res. Int. 2019:5431354. doi: 10.1155/2019/5431354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huebner, K. L., Martin J. N., Weissend C. J., Holzer K. L., Parker J. K., Lakin S. M., Doster E., Weinroth M. D., Abdo Z., Woerner D. R., . et al. 2019. Effects of a Saccharomyces cerevisiae fermentation product on liver abscesses, fecal microbiome, and resistome in feedlot cattle raised without antibiotics. Sci. Rep. 9:2559. doi: 10.1038/s41598-019-39181-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hume, I. D. 1997. Fermentation in the hindgut of mammals. In: Mackie R. I. and White B. A., editors. Gastrointestinal microbiology. New York (NY): Chapman & Hall; p. 84–115. doi: 10.1007/978-1-4615-4111-0_4 [DOI] [Google Scholar]
- Janssen, S., McDonald D., Gonzalez A., Navas-Molina J. A., Jiang L., Xu Z. Z., Winker K., Kado D. M., Orwoll E., Manary M., . et al. 2018. Phylogenetic placement of exact amplicon sequences improves associations with clinical information. N. Chia, editor. mSystems 3:e00021–e00018. doi: 10.1128/mSystems.00021-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaakoush, N. O. 2015. Insights into the role of Erysipelotrichaceae in the human host. Front. Cell. Infect. Microbiol. 5:84. doi: 10.3389/fcimb.2015.00084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khafipour, E., Li S., Tun H. M., Derakhshani H., Moossavi S., and Plaizier J. C.. . 2016. Effects of grain feeding on microbiota in the digestive tract of cattle. Anim. Front 6:13–19. doi: 10.2527/af.2016-0018 [DOI] [Google Scholar]
- Kim, W., and Whitman W. B.. . 2014. Methanogens. In: Batt C. A. and Tortorello M. L., editors, Encyclopedia of food microbiology. 2nd ed. Cambridge, MA: Academic Press; p. 602–606. doi: 10.1016/B978-0-12-384730-0.00204-4 [DOI] [Google Scholar]
- Kim, C. C., Healey G. R., Kelly W. J., Patchett M. L., Jordens Z., Tannock G. W., Sims I. M., Bell T. J., Hedderley D., Henrissat B., and Rosendale D. I.. . 2019. Genomic insights from Monoglobus pectinilyticus: a pectin-degrading specialist bacterium in the human colon. ISME J. 13(6):1437–1456. doi: 10.1038/s41396-019-0363-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krehbiel, C. R., Rust S. R., Zhang G., and Gilliland S. E.. . 2003. Bacterial direct-fed microbials in ruminant diets: performance response and mode of action. J. Anim. Sci. 81:E120–E132. doi: 10.2527/2003.8114_suppl_2E120x [DOI] [Google Scholar]
- Kruskal, W. H., and Wallis W. A.. . 1952. Use of ranks in one-criterion variance analysis. J. Am. Stat. Assoc. 47:583. doi: 10.2307/2280779 [DOI] [Google Scholar]
- Koester, L. R., Poole D. H., Serão N., and Schmitz-Esser S.. . 2020. Beef cattle that respond differently to fescue toxicosis have distinct gastrointestinal tract microbiota. PloS one 15(7):e0229192. doi: 10.1371/journal.pone.0229192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, Y. J., Kim B. K., Lee B. H., Jo K. I., Lee N. K., Chung C. H., Lee Y. C., and Lee J. W.. . 2008. Purification and characterization of cellulase produced by Bacillus amyoliquefaciens DL-3 utilizing rice hull. Bioresour. Technol. 99:378–386. doi: 10.1016/J.BIORTECH.2006.12.013 [DOI] [PubMed] [Google Scholar]
- Li, A., Jiang X., Wang Y., Zhang L., Zhang H., Mehmood K., Li Z., Waqas M., and Li J.. . 2019a. The impact of Bacillus subtilis 18 isolated from Tibetan yaks on growth performance and gut microbial community in mice. Microb. Pathog. 128:153–161. doi: 10.1016/j.micpath.2018.12.031 [DOI] [PubMed] [Google Scholar]
- Li, A., Wang Y., Pei L., Mehmood K., Li K., Qamar H., Iqbal M., Waqas M., Liu J., and Li J.. . 2019b. Influence of dietary supplementation with Bacillus velezensis on intestinal microbial diversity of mice. Microb. Pathog. 136:103671. doi: 10.1016/j.micpath.2019.103671 [DOI] [PubMed] [Google Scholar]
- Lin, L. Z., Zheng Q. W., Wei T., Zhang Z. Q., Zhao C. F., Zhong H., Xu Q. Y., Lin J. F., and Guo L. Q.. . 2020. Isolation and characterization of fengycins produced by Bacillus amyloliquefaciens JFL21 and its broad-spectrum antimicrobial potential against multidrug-resistant foodborne pathogens. Front. Microbiol. 11:579621. doi: 10.3389/FMICB.2020.579621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, J., Xu T., Zhu W., and Mao S.. . 2014. High-grain feeding alters caecal bacterial microbiota composition and fermentation and results in caecal mucosal injury in goats. Br. J. Nutr. 112:416–427. doi: 10.1017/S0007114514000993 [DOI] [PubMed] [Google Scholar]
- Liu, J., Zhang M., Zhang R., Zhu W., and Mao S.. . 2016. Comparative studies of the composition of bacterial microbiota associated with the ruminal content, ruminal epithelium and in the faeces of lactating dairy cows. Microb. Biotechnol. 9:257–268. doi: 10.1111/1751-7915.12345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd, S. P. 1957. Least squares quantization in PCM. Technical Report RR-5497, Bell Lab, September 1957. [Google Scholar]
- Loneragan, G. H., and Gould D. H.. . 2000. Acute interstitial pneumonia in feedlot cattle. Am. Assoc. Bov. Pract. Proc. Annu. Conf 33:129–132. doi: 10.21423/AABPPRO20005375 [DOI] [Google Scholar]
- Lozupone, C., and Knight R.. . 2005. UniFrac: a new phylogenetic method for comparing microbial communities. Appl. Environ. Microbiol. 71:8228–8235. doi: 10.1128/AEM.71.12.8228-8235.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luise, D., Bosi P., Raff L., Amatucci L., Virdis S., and Trevisi P.. . 2022. Bacillus spp. probiotic strains as a potential tool for limiting the use of antibiotics, and improving the growth and health of pigs and chickens. Front. Microbiol. 13:801827. doi: 10.3389/FMICB.2022.801827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo, L., Xu Q., Xu W., Li J., Wang C., Wang L., and Zhao Z.. . 2020. Effect of Bacillus megaterium-coated diets on the growth, digestive enzyme activity, and intestinal microbial diversity of Songpu mirror carp Cyprinus specularis Songpu. Biomed Res. Int. 2020:8863737. doi: 10.1155/2020/8863737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacQueen, J. B. 1967. Some methods for classification and analysis of multivariate observations. In Le Cam L. M. and Neyman, J. editors. Proceedings of the fifth Berkeley symposium on mathematical statistics and probability. Berkely (CA): University of California Press; p. 281–297. [Google Scholar]
- Mandal, S., Van Treuren W., White R. A., Eggesbø M., Knight R., and Peddada S. D.. . 2015. Analysis of composition of microbiomes: a novel method for studying microbial composition. Microb. Ecol. Health Dis. 26(1):27663. doi: 10.3402/mehd.v26.27663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao, S., Zhang M., Liu J., and Zhu W.. . 2015. Characterising the bacterial microbiota across the gastrointestinal tracts of dairy cattle: membership and potential function. Sci. Rep. 5:1–14. doi: 10.1038/srep16116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsen, F. A., Kodner R. B., and Armbrust E. V.. . 2010. pplacer: linear time maximum-likelihood and Bayesian phylogenetic placement of sequences onto a fixed reference tree. BMC Bioinf. 11:538. doi: 10.1186/1471-2105-11-538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsen, F. A., Hoffman N. G., Gallagher A., and Stamatakis A.. . 2012. A format for phylogenetic placements. PLoS One 7:1–4. doi: 10.1371/journal.pone.0031009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAllister, T. A., Beauchemin K. A., Alazzeh A. Y., Baah J., Teather R. M., and Stanford K.. . 2011. Review: the use of direct fed microbials to mitigate pathogens and enhance production in cattle. Can. J. Anim. Sci. 91:193–211. doi: 10.4141/cjas10047 [DOI] [Google Scholar]
- McCabe, E. D., King M. E., Fike K. E., Hill K. L., Rogers G. M., and Odde K. G.. . 2019. Breed composition affects the sale price of beef steer and heifer calves sold through video auctions from 2010 through 2016. Appl. Anim. Sci 35:221–226. doi: 10.15232/AAS.2018-01806 [DOI] [Google Scholar]
- Minaya, D. M., Turlej A., Joshi A., Nagy T., Weinstein N., DiLorenzo P., Hajnal A., and Czaja K.. . 2020. Consumption of a high energy density diet triggers microbiota dysbiosis, hepatic lipidosis, and microglia activation in the nucleus of the solitary tract in rats. Nutr. Diabetes 10:1–12. doi: 10.1038/s41387-020-0119-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mir, R. A., Schaut R. G., Allen H. K., Looft T., Loving C. L., Kudvaid I. T., and Sharmaid V. K.. . 2019. Cattle intestinal microbiota shifts following Escherichia coli O157:H7 vaccination and colonizationtravel. PLOS ONE 14(12):e0227403. doi: 10.1371/journal.pone.0226099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myer, P. R. 2019. Bovine genome-microbiome interactions: metagenomic frontier for the selection of efficient productivity in cattle systems. mSystems 4(3):e00103-19. doi: 10.1128/msystems.00103-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myer, P. R., Wells J. E., Smith T. P. L., Kuehn L. A., and Freetly H. C.. . 2015a. Microbial community profiles of the colon from steers differing in feed efficiency. Springerplus 4:454. doi: 10.1186/s40064-015-1201-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myer, P. R., Wells J. E., Smith T. P. L., Kuehn L. A., and Freetly H. C.. . 2015b. Cecum microbial communities from steers differing in feed efficiency. J. Anim. Sci. 93:5327–5340. doi: 10.2527/jas.2015-9415 [DOI] [PubMed] [Google Scholar]
- Myer, P. R., Wells J. E., Smith T. P. L., Kuehn L. A., and Freetly H. C.. . 2016. Microbial community profiles of the jejunum from steers differing in feed efficiency. J. Anim. Sci. 94:327–338. doi: 10.2527/jas.2015-9839 [DOI] [PubMed] [Google Scholar]
- Nyonyo, T., Shinkai T., and Mitsumori M.. . 2014. Improved culturability of cellulolytic rumen bacteria and phylogenetic diversity of culturable cellulolytic and xylanolytic bacteria newly isolated from the bovine rumen. FEMS Microbiol. Ecol. 88:528–537. doi: 10.1111/1574-6941.12318 [DOI] [PubMed] [Google Scholar]
- Okine, E. K., and Mathison G. W.. . 1991. Effects of feed intake on particle distribution, passage of digesta, and extent of digestion in the gastrointestinal tract of cattle. J. Anim. Sci. 69:3435–3445. doi: 10.2527/1991.6983435x [DOI] [PubMed] [Google Scholar]
- Oksanen, J., Simpson G. L., Blanchet F. G., Kindt R., Legendre P., Minchin P. R., O’Hara R. B., Solymos P., Stevens M. H. H., Szoecs E., Wagner H.et al.. 2022. vegan: Community Ecology Package. R package version 2.6-2. Available from: https://CRAN.R-project.org/package=vegan.
- de Oliveira, M. N. V., Jewell K. A., Freitas F. S., Benjamin L. A., Tótola M. R., Borges A. C., Moraes C. A., and Suen G.. . 2013. Characterizing the microbiota across the gastrointestinal tract of a Brazilian Nelore steer. Vet. Microbiol. 164:307–314. doi: 10.1016/j.vetmic.2013.02.013 [DOI] [PubMed] [Google Scholar]
- Owens, F. N., Secrist D. S., Hill W. J., and Gill D. R.. . 1998. Acidosis in cattle: a review. J. Anim. Sci. 76:275–286. doi: 10.2527/1998.761275x [DOI] [PubMed] [Google Scholar]
- Parada, A. E., Needham D. M., and Fuhrman J. A.. . 2016. Every base matters: assessing small subunit rRNA primers for marine microbiomes with mock communities, time series and global field samples. Environ. Microbiol. 18:1403–1414. doi: 10.1111/1462-2920.13023 [DOI] [PubMed] [Google Scholar]
- Parish, J. A., Bourg B. M., Marks M. L., Simmons N. B., and Smith T.. . 2012. Evaluation of different methods of cattle hip height data collection 1. Prof. Anim. Sci. 28:292–299. doi: 10.15232/S1080-7446(15)30358-2 [DOI] [Google Scholar]
- Pedregosa, F., Varoquaux G., Gramfort A., Michel V., Thirion B., Grisel O., Blondel M., Prettenhofer P., Weiss R., Dubourg V., . et al. 2011. Scikit-learn: machine learning in Python. J. Mach. Learn. Res. 12:2825–2830. doi: 10.5555/1953048.2078195 [DOI] [Google Scholar]
- Petri, R. M., Schwaiger T., Penner G. B., Beauchemin K. A., Forster R. J., McKinnon J. J., and McAllister T. A.. . 2013. Characterization of the core rumen microbiome in cattle during transition from forage to concentrate as well as during and after an acidotic challenge. X. Ren, editor. PLoS One 8:e83424. doi: 10.1371/journal.pone.0083424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pielou, E. C. 1966. The measurement of diversity in different types of biological collections. J. Theor. Biol. 13:131–144. doi: 10.1016/0022-5193(66)90013-0 [DOI] [Google Scholar]
- Plaizier, J. C., Li S., Gozho G., and Khafipour E.. . 2014. Minimizing the risk for rumen acidosis. In: Eastridge M., editor. 23rdRD Tri-State Dairy Nutrition Conference. Columbus, Ohio. p. 11–26.
- Pruesse, E., Quast C., Knittel K., Fuchs B. M., Ludwig W., Peplies J., and Glockner F. O.. . 2007. SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res. 35( 21):7188–7196. doi: 10.1093/nar/gkm864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quast, C., Pruesse E., Yilmaz P., Gerken J., Schweer T., Yarza P., Peplies J. J., Glöckner F. O., and Glockner F. O.. . 2013. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 41:D590–D596. doi: 10.1093/nar/gks1219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team. 2022. R: a language and environment for statistical computing. R Foundation for Statistical Computing. Available from: https://www.R-project.org/ [Google Scholar]
- Rabi, R., Turnbull L., Whitchurch C. B., Awad M., and Lyras D.. . 2017. Structural characterization of Clostridium sordellii spores of diverse human, animal, and environmental origin and comparison to Clostridium difficile spores. mSphere 2( 5):e00343-17. doi: 10.1128/msphere.00343-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramayo-Caldas, Y., Zingaretti L., Popova M., Estellé J., Bernard A., Pons N., Bellot P., Mach N., Rau A., Roume H., . et al. 2020. Identification of rumen microbial biomarkers linked to methane emission in Holstein dairy cows. J. Anim. Breed Genet. 137(1):49–59. doi: 10.1111/jbg.12427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Retallick, K. J., Bormann J. M., Weaber R. L., MacNeil M. D., Bradford H. L., Freetly H. C., Hales K. E., Moser D. W., Snelling W. M., Thallman R. M., . et al. 2017. Genetic variance and covariance and breed differences for feed intake and average daily gain to improve feed efficiency in growing cattle. J. Anim. Sci. 95:1444–1450. doi: 10.2527/jas.2016.1260 [DOI] [PubMed] [Google Scholar]
- Rodriguez-Jimenez, S., Horst E. A., Mayorga E. J., Kvidera S. K., Abeyta M. A., Goetz B. M., Carta S., and Baumgard L. H.. . 2019. The what, why, and physiologic cost of leaky gut syndrome. Am. Assoc. Bov. Pract. Proc. Annu. Conf 52:165–171. doi: 10.21423/AABPPRO20197129 [DOI] [Google Scholar]
- Sanz-Fernandez, M. V., Daniel J. B., Seymour D. J., Kvidera S. K., Bester Z., Doelman J., and Martín-Tereso J.. . 2020. Targeting the hindgut to improve health and performance in cattle. Anim 10:1–18. doi: 10.3390/ANI10101817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schofield, B. J., Lachner N., Le O. T., McNeill D. M., Dart P., Ouwerkerk D., Hugenholtz P., and Klieve A. V.. . 2018. Beneficial changes in rumen bacterial community profile in sheep and dairy calves as a result of feeding the probiotic Bacillus amyloliquefaciens H57. J. Appl. Microbiol. 124:855–866. doi: 10.1111/jam.13688 [DOI] [PubMed] [Google Scholar]
- Shannon, C. E. 1948. A mathematical theory of communication. Bell Syst. Tech. J. 27:623–656. doi: 10.1002/j.1538-7305.1948.tb00917.x [DOI] [Google Scholar]
- Simpson, K. M., Callan R. J., and Van Metre D. C.. . 2018. Clostridial abomasitis and anteritis in ruminants. Vet. Clin. North Am. Food Anim. Pract. 34:155–184. doi: 10.1016/j.cvfa.2017.10.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song, D. J., Kang H. Y., Wang J. Q., Peng H., and Bu D. P.. . 2014. Effect of feeding Bacillus subtilis natto on hindgut fermentation and microbiota of holstein dairy cows . Asian-Australasian J. Anim. Sci 27:495. doi: 10.5713/AJAS.2013.13522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, P., Wang J. Q., and Zhang H. T.. . 2010. Effects of Bacillus subtilis natto on performance and immune function of preweaning calves. J. Dairy Sci. 93:5851–5855. doi: 10.3168/jds.2010-3263 [DOI] [PubMed] [Google Scholar]
- Sun, L., Cao J., Liu Y., Wang J., Guo P., and Wang Z.. . 2017. Gene cloning and expression of cellulase of Bacillus amyloliquefaciens isolated from the cecum of goose. Anim. Biotechnol. 28:74–82. doi: 10.1080/10495398.2016.1205594 [DOI] [PubMed] [Google Scholar]
- Thompson, L. R., Sanders J. G., McDonald D., Amir A., Ladau J., Locey K. J., Prill R. J., Tripathi A., Gibbons S. M., Ackermann G., . et al. 2017. A communal catalogue reveals Earth’s multiscale microbial diversity. Nature 551:457–463. doi: 10.1038/nature24621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vázquez-Baeza, Y., Gonzalez A., Smarr L., McDonald D., Morton J. T., Navas-Molina J. A., and Knight R.. . 2017. Bringing the dynamic microbiome to life with animations. Cell Host Microbe 21:7–10. doi: 10.1016/j.chom.2016.12.009 [DOI] [PubMed] [Google Scholar]
- Weinroth, M. D., Belk A. D., Dean C., Noyes N., Dittoe D. K., Rothrock M. J., Ricke S. C., Myer P. R., Henniger M. T., Ramírez G. A., Oakley B. B., Summers K. L., Miles A. M., Ault-Seay T. B., Yu Z., Metcalf J. L., and Wells J. E.. . 2022. Considerations and best practices in animal science 16S ribosomal RNA gene sequencing microbiome studies. J. Anim. Sci. 100:1–18. doi: 10.1093/JAS/SKAB346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss, S., Xu Z. Z., Peddada S., Amir A., Bittinger K., Gonzalez A., Lozupone C., Zaneveld J. R., Vázquez-Baeza Y., Birmingham A., . et al. 2017. Normalization and microbial differential abundance strategies depend upon data characteristics. Microbiome 5:27. doi: 10.1186/s40168-017-0237-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welch, C. B., Lourenco J. M., Davis D. B., Krause T. R., Carmichael M. N., Rothrock M. J., Dean Pringle T., and Callaway T. R.. . 2020. The impact of feed efficiency selection on the ruminal, cecal, and fecal microbiomes of Angus steers from a commercial feedlot. J. Anim. Sci. 98:1–10. doi: 10.1093/jas/skaa230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickham, H. 2009. ggplot2: elegant graphics for data analysis. New York, NY: Springer. doi: 10.1007/978-0-387-98141-3 [DOI] [Google Scholar]
- Williams, G. S., Raper K. C., DeVuyst E. A., Peel D., and McKinney D.. . 2012. Determinants of price differentials in Oklahoma value-added feeder cattle auctions on JSTOR. J. Agric. Resour. Econ 37:114–127. 10.22004/ag.econ.122309 [DOI] [Google Scholar]
- Xu, Y., Dugat-Bony E., Zaheer R., Selinger L., Barbieri R., Munns K., McAllister T. A., and Selinger L. B.. . 2014. Escherichia coli O157:H7 super-shedder and non-shedder feedlot steers harbour distinct fecal bacterial communities. PLoS One 9:e98115. doi: 10.1371/journal.pone.0098115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, W. Y., Lee Y., Lu H., Chou C. H., and Wang C.. . 2019. Analysis of gut microbiota and the effect of lauric acid against necrotic enteritis in Clostridium perfringens and Eimeria side-by-side challenge model. PLoS One 14(5):e0205784. doi: 10.1371/journal.pone.0205784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon S. H., Ha S. M., Kwon S., Lim J., Kim Y., Seo H., Chun J.. . 2017. Introducing EzBioCloud: a taxonomically united database of 16S rRNA gene sequences and whole-genome assemblies. Int. J. Syst. Evol. Microbiol. 67(5):1613–1617. doi: 10.1099/ijsem.0.001755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng, M. Y., Inohara N., and Nuñez G.. . 2017. Mechanisms of inflammation-driven bacterial dysbiosis in the gut. Mucosal Immunol 10:18–26. doi: 10.1038/mi.2016.75 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.