Abstract
The reference method for immunoglobulin G (IgG) avidity determination includes reagent-consuming serum titration. Aiming at better IgG avidity diagnostics, we applied a logistic model for the reproduction of antibody titration curves. This method was tested with well-characterized serum panels for cytomegalovirus, Epstein-Barr virus, rubella virus, parvovirus B19, and Toxoplasma gondii. This approach for IgG avidity calculation is generally applicable and attains the diagnostic performance of the reference method while being less laborious and twice as cost-effective.
The diagnosis of acute viral and some other microbial infections often relies on the serological detection of immunoglobulin M (IgM) antibodies, but the available techniques have serious pitfalls that may lead to erroneous interpretations (3, 4, 9, 11). This problem is of particular importance for infections during the first trimester of pregnancy, which should be diagnosed as exactly as possible (8, 10, 18). The differential assay of high-avidity and low-avidity IgG antibodies can be used as an alternative or a complement to the IgM antibody assay and is gaining popularity as a diagnostic method for the assessment of the time of infection. In protein-denaturing avidity enzyme immunoassays (EIAs), the patient’s IgG (e.g., in serum) is allowed to bind to its antigen, followed by elution with or without a protein denaturant, such as urea. From the proportion of IgG remaining antigen bound, the time of primary infection can be deduced.
The most straightforward procedure for the calculation of avidity is a comparison of EIA absorbances in single (fixed) dilutions of serum. This procedure is quite sensitive and specific for the diagnosis of several different microbes (3, 6, 7, 13, 17, 18) but is affected to some extent by the concentration of specific IgG (7). The two-step avidity assay developed by Lecolier and Pucheu is based on the selection of working dilutions according to the level of specific IgG in each specimen (12). Another means of improved avidity calculation is based on the α method, in which the antibody titer is derived from a single dilution of serum by use of the formula log10 titer = α(ODβ), in which OD is optical density and α and β are constants specified by the assay manufacturer for each batch of kit reagents (5, 19). The avidity technique based on end-point titration of IgG (2, 8, 10, 11, 16, 17) is not influenced by IgG concentration but is relatively laborious and reagent consuming. Due to its excellent sensitivity and specificity, we consider this approach the reference method for several microbes (9). Aiming at low cost combined with high performance, we have applied a logistic procedure (14) for avidity calculation; here, we evaluate its diagnostic value.
MATERIALS AND METHODS
Serum panels.
The cytomegalovirus (CMV) panel contained sera from three groups. Group 1 comprised 91 sera from 48 patients with primary CMV infections verified by IgM-positive seroconversion of CMV IgG (Cytomegalovirus IgG EIA Kit; Labsystems, Helsinki, Finland). Group 2 contained 88 sera from 29 patients with exogenous reinfection or endogenous reactivation. The criteria were a ≥4-fold increase in the CMV IgG concentration in a serum pair, with a high avidity of CMV IgG (1) but no CMV IgM in the first sample. The 44 sera constituting group 3 were collected from asymptomatic members of the laboratory staff, all of which had tested CMV IgG positive at least 1 year earlier.
The Epstein-Barr virus (EBV) panel comprised 34 sera from 25 EBV IgM-positive (EBV IFA Test; Gull Laboratories, Salt Lake City, Utah) and EBV IgG-positive (Enzygnost Anti-EBV/IgG; Dade Behring, Marburg, Germany) patients. The cardinal symptoms were mononucleosis, tonsillitis, lymphadenitis, and fever. Controls were 19 sera from asymptomatic members of the laboratory staff shown to be EBV IgG positive at least 1 year earlier.
The rubella virus panel consisted of 121 samples from patients with primary infections and of 40 control samples shown to be seropositive at least 2 years earlier (7).
The parvovirus panel contained 128 sera from 68 patients with a B19 primary infection (17). Control samples were 56 sera from 34 B19 IgG-positive, B19 IgM-negative asymptomatic members of the laboratory staff with seropositivity documented for at least 2 years.
The 500 sera making up the Toxoplasma gondii serum panel had been studied at Ospedale S. Gerardo Di Monza, unlike the others described above. This panel was divided in two subgroups. The first subgroup consisted of 420 sera from 267 IgM-positive (Abbott) and IgG-positive (Labsystems) patients. The second subgroup comprised 80 control sera, 40 of which had, upon prior measurement, shown a high avidity and 40 of which had shown a low avidity.
In this study, IgG avidities for the respective pathogens were measured with the Cytomegalovirus IgG EIA Kit; the Enzygnost Anti-EBV/IgG kit with a mixture of viral capsid antigen (VCA), Epstein-Barr nuclear antigen (EBNA), and early antigen (EA) sequences as the antigen; and the Rubella IgG Avidity Kit (Labsystems). The avidity of B19 IgG was measured with the recombinant VP1 antigen EIA (17), and the avidity of toxoplasma IgG was measured with the Toxoplasma gondii IgG Avidity Kit (Labsystems). Titration curves were drawn (i) according to the reference method (8) with 4+4 dilutions per sample (i.e., one series of four dilutions washed with urea and another series of four dilutions washed without urea), (ii) from the same EIA data sets (4+4 dilutions per sample) with a curve-fitting software based on logistic functions (see below), (iii) with the same logistic model but with only 2+2 dilutions per sample, and (iv) from the same data sets with a log-log model. Avidity was calculated as the ratio of IgG titers: (titer with urea/titer without urea) × 100.
Logistic model.
A curve-fitting Win-95 computer program was developed for reproduction of the shapes of IgG titration curves. The fitting curve was, in its basic form, a so-called logistic function, f(x) = 1/[1 + exp(−x)], which was, however, parametrized into the form f(x) = (a + b)/{1 + exp[−(x − c)/d]}. Here, exp was the exponential function and x was the logarithm of the dilution ratio. The to-be-fitted variables a, b, c, and d were given certain limits beforehand that were based on avidity calculations performed earlier in our laboratory. For example, we know that f(x) = 0 for sufficiently large values of x. Also, from the absorbance value measured at the first dilution, an approximative limit for f(0) could be deduced. This value represents the theoretical maximum value of the titration curve with no dilution. Thus, only two parameters had to be fitted for each curve.
Log-log model.
For comparison with the logistic model, a piece of software which functions by linear interpolation of log(absorbance) versus log(dilution) values for native and denatured samples was developed. The end-point titer of both samples was calculated, and the ratio of end-point titers was taken to be the output avidity result. In fact, this process amounted to using the function log[f(x)] = a(x) + b for the curve fitting instead of the logistic function. As above, x was the logarithm of the dilution ratio.
RESULTS
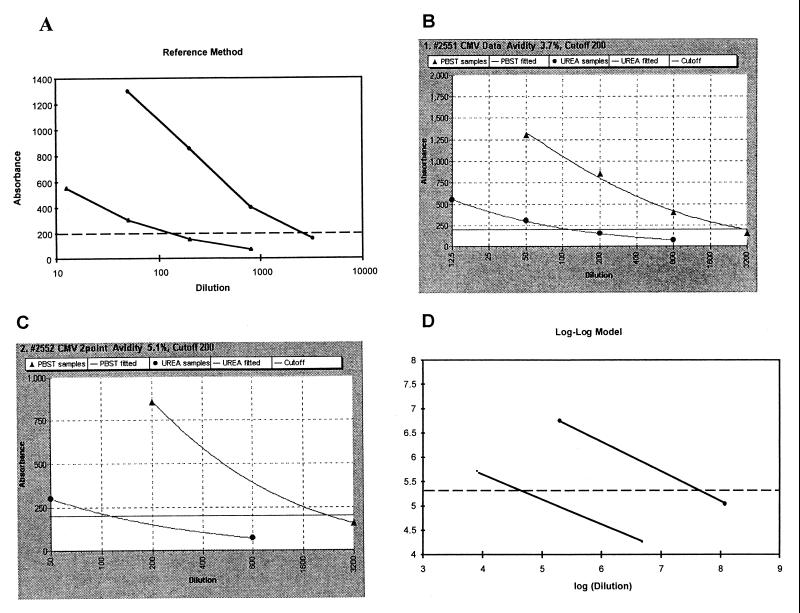
In the reference method, EIA absorbances were plotted against serum dilutions on a semilogarithmic scale, and the individual data points (4+4 dilutions per sample) were united by straight lines (Fig. 1A). Under the same conditions with 4+4 serum dilutions, the logistic model produced curvilinear, or “smooth,” IgG titration curves, which often bypassed individual data points (Fig. 1B). With 2+2 serum dilutions per sample, the same logistic model produced IgG titration curves that resembled those obtained with 4+4 serum dilutions (Fig. 1C) but, at the curve ends, met their data points precisely. The log-log model displayed linear IgG titration curves when both axes were linear (Fig. 1D).
FIG. 1.
End-point titration curves for CMV IgG avidity determinations. Views are as seen on the computer screen. In each curve pair, the upper one was obtained without urea and the lower one was obtained with urea. Calculations were done with the reference method and 4+4 data points (dilutions of serum) (A), with the logistic model and 4+4 data points (B) and 2+2 data points (C), and with the log-log model and 2+2 data points. This CMV IgG is of low avidity. PBST, phosphate-buffered saline containing 0.05% Tween 20.
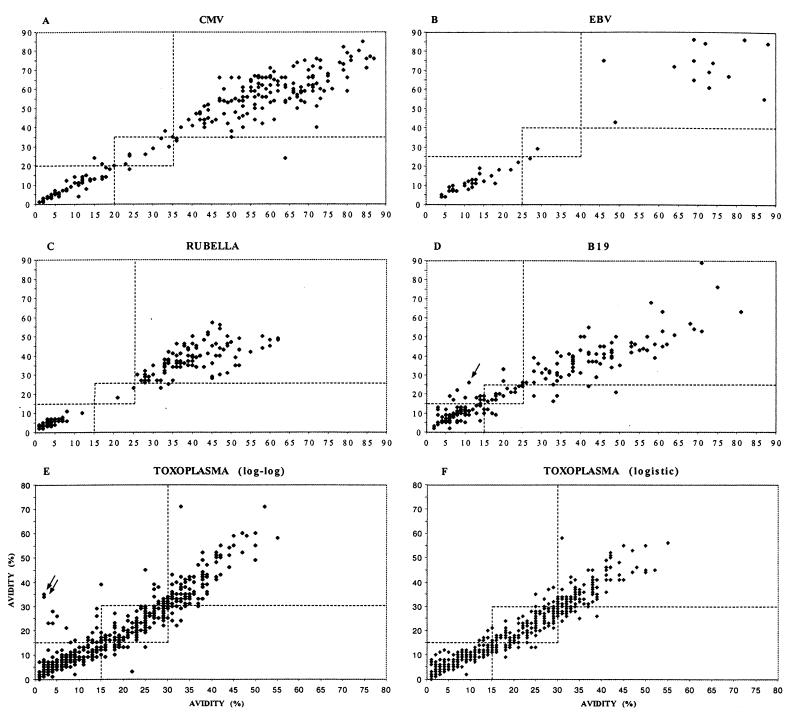
The logistic model operating with 2+2 dilutions per sample was tested with all the serum panels, and the results were compared with those obtained with the reference method (operating with 4+4 data points). Overall, the two methods showed excellent correlation; the correlation coefficients for all four viruses and the one protozoan were ≥0.94 (Fig. 2). Also illustrated are the domains (bordered by broken lines) in which the avidity values obtained could be allowed to move without a change in diagnosis. For the >1,000 samples studied, only once, in the parvovirus serum panel, was there disagreement between the two methods; the reference method produced a pathological value of low avidity (12%), whereas the 2+2 logistic method produced a nonpathological value of high avidity (26%). This single crossover was due to a deviant EIA data point caused by an apparent pipetting error; however, this error was well tolerated by the 4+4 logistic method, which produced a borderline-avidity result (17%).
FIG. 2.
Comparison of IgG avidity results calculated with the reference method (horizontal axis) and with the curve-fitting methods (vertical axis). (A to D and F) Results obtained with the logistic model. (E) Results obtained with the log-log model. (A) CMV (correlation coefficient [r], 0.96). (B) EBV (r, 0.96). (C) Rubella virus (r, 0.95). (D) Parvovirus B19 (r, 0.94). (E) T. gondii (r, 0.93). (F) T. gondii (r, 0.96). The broken lines mark the thresholds between low-borderline and borderline-high avidities. Crossovers from low to high avidity in panels D and E are marked by arrows.
The diagnostic value of the simple log-log model was determined with the large toxoplasma serum panel. As depicted in Fig. 2E, this model also corresponded fairly well to the reference method, yielding only two false high-avidity results. However, the logistic model (with 2+2 data points) was even more accurate (r, 0.96), producing no false avidity results and fewer crossovers to or from the borderline avidity zone (Fig. 2F).
DISCUSSION
We applied and evaluated curve-fitting methods for IgG avidity calculations. The diagnosis of four viruses (a nonenveloped single-stranded DNA virus, an enveloped single-stranded RNA virus, and two enveloped double-stranded DNA viruses) and one protozoan could be accomplished reliably with the logistic model and only 2+2 dilutions per sample. Success with T. gondii, an immunologically and structurally complex pathogen, is particularly noteworthy because simple indices obtained from single dilutions of serum are insufficient for its avidity determination (7, 8). Given previously published work (15), we were somewhat surprised to observe that the diagnostic performance of the log-log model lagged behind that of the more elaborate logistic model only slightly. The explanation may arise from the fact that in avidity determinations, two parallel (with and without a protein denaturant) titration curves are generated by the same model, the inherent errors of which are abolished when the two titration end-point values are divided.
In conclusion, the logistic approach for IgG avidity calculations is generally applicable, attains the diagnostic performance of the reference method, and is twice as cost-effective.
ACKNOWLEDGMENTS
We thank Lea Hedman for expert technical assistance.
This work was supported by the Helsinki University Central Hospital Research and Education Fund, the Finnish Technology Advancement Fund, the Center for International Mobility, and the Sohlberg Foundation.
REFERENCES
- 1.Aalto S M, Linnavuori K, Peltola H, Vuori E, Weissbrich B, Schubert J, Hedman L, Hedman K. Immunoreactivation of Epstein-Barr virus due to cytomegalovirus primary infection. J Med Virol. 1998;56:186–191. [PubMed] [Google Scholar]
- 2.Andersson A, Vetter V, Kreutzer L, Bauer G. Avidities of IgG directed against viral capsid antigen or early antigen: useful markers for significant Epstein-Barr virus serology. J Med Virol. 1994;43:238–244. doi: 10.1002/jmv.1890430308. [DOI] [PubMed] [Google Scholar]
- 3.Bodeus M, Feyder S, Goubau P. Avidity of IgG antibodies distinguishes primary from non-primary cytomegalovirus infection in pregnant women. Clin Diagn Virol. 1998;9:9–16. doi: 10.1016/s0928-0197(97)10016-2. [DOI] [PubMed] [Google Scholar]
- 4.de Ory F, Casas I, Domingo C J, Echevarria J M. Application of fluoroimmunoassay to the identification of low avidity specific IgG against pathogenic human viruses and Toxoplasma gondii. Clin Diagn Virol. 1995;3:323–332. doi: 10.1016/0928-0197(94)00045-v. [DOI] [PubMed] [Google Scholar]
- 5.Dopatka H D, Giesendorf B. Single point quantification of antibody by ELISA without need of a reference curve. J Clin Lab Anal. 1992;6:417–422. doi: 10.1002/jcla.1860060614. [DOI] [PubMed] [Google Scholar]
- 6.Grangeot-Keros L, Mayaux M J, Lebon P, Freymuth F, Eugene G, Stricker R, Dussaix E. Value of cytomegalovirus (CMV) avidity index for the diagnosis of primary CMV infection in pregnant women. J Infect Dis. 1997;175:944–946. doi: 10.1086/513996. [DOI] [PubMed] [Google Scholar]
- 7.Hedman K, Seppälä I. Recent rubella virus infection indicated by a low avidity of specific IgG. J Clin Immunol. 1988;8:214–221. doi: 10.1007/BF00917569. [DOI] [PubMed] [Google Scholar]
- 8.Hedman K, Lappalainen M, Seppälä I, Mäkelä O. Recent primary toxoplasma infection indicated by a low avidity of specific IgG. J Infect Dis. 1989;159:736–740. doi: 10.1093/infdis/159.4.736. [DOI] [PubMed] [Google Scholar]
- 9.Hedman K, Lappalainen M, Söderlund M, Hedman L. Avidity of IgG in serodiagnosis of infectious diseases. Rev Med Microbiol. 1993;4:123–129. [Google Scholar]
- 10.Jenum P, Stray-Pedersen B, Gundersen A-G. Improved diagnosis of primary Toxoplasma gondii infection in early pregnancy by determination of antitoxoplasma immunoglobulin G avidity. J Clin Microbiol. 1997;35:1972–1977. doi: 10.1128/jcm.35.8.1972-1977.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lappalainen M, Koskela P, Koskiniemi M, Ämmälä P, Hiilesmaa V, Teramo K, Raivio K O, Remington J, Hedman K. Toxoplasmosis acquired during pregnancy: improved serodiagnosis based on avidity of IgG. J Infect Dis. 1993;167:691–697. doi: 10.1093/infdis/167.3.691. [DOI] [PubMed] [Google Scholar]
- 12.Lecolier B, Pucheu B. Intérêt de l’étude de l’avidité des IgG pour le diagnostic de la toxoplasmose. Pathol Biol. 1993;41:155–158. [PubMed] [Google Scholar]
- 13.Lutz E, Ward K N, Szyldo R, Goldman J M. Cytomegalovirus antibody avidity in allogenic bone marrow recipients: evidence for primary or secondary humoral responses depending on donor immune status. J Med Virol. 1996;49:61–65. doi: 10.1002/(SICI)1096-9071(199605)49:1<61::AID-JMV10>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 14.Maciel R J. Standard curve fitting in immunodiagnostics: a primer. J Clin Immunol. 1985;8:98–106. [Google Scholar]
- 15.Plikaytis B D, Turner S H, Gheesling L L, Carlone G M. Comparisons of standard curve-fitting methods to quantitate Neisseria meningitidis group A polysaccharide antibody levels by enzyme-linked immunosorbent assay. J Clin Microbiol. 1991;29:1439–1445. doi: 10.1128/jcm.29.7.1439-1446.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sensini A, Pascoli S, Marchetti D, Castronari R, Marangi M, Sbaraglia G, Cimmino C, Favero A, Castelletto M, Mottola A. IgG avidity in the serodiagnosis of acute Toxoplasma gondii infection: a multicenter study. Clin Microbiol Infect. 1996;2:25–29. doi: 10.1111/j.1469-0691.1996.tb00196.x. [DOI] [PubMed] [Google Scholar]
- 17.Söderlund M, Brown C S, Cohen B J, Hedman K. Accurate serodiagnosis of B19 parvovirus infections by measurement of IgG avidity. J Infect Dis. 1995;171:710–713. doi: 10.1093/infdis/171.3.710. [DOI] [PubMed] [Google Scholar]
- 18.Thomas H I J, Morgan-Capner P. The use of antibody avidity measurements for the diagnosis of rubella. Rev Med Virol. 1991;1:41–50. [Google Scholar]
- 19.Weissbrich B. The use of semi-automated EBV IgG avidity determination for the diagnosis of infectious mononucleosis. J Med Virol. 1998;54:145–153. doi: 10.1002/(sici)1096-9071(199802)54:2<145::aid-jmv13>3.0.co;2-i. [DOI] [PubMed] [Google Scholar]