Abstract
Trypanosomatids belong to a remarkable group of unicellular, parasitic organisms of the order Kinetoplastida, an early diverging branch of the phylogenetic tree of eukaryotes, exhibiting intriguing biological characteristics affecting gene expression (intronless polycistronic transcription, trans-splicing, and RNA editing), metabolism, surface molecules, and organelles (compartmentalization of glycolysis, variation of the surface molecules, and unique mitochondrial DNA), cell biology and life cycle (phagocytic vacuoles evasion and intricate patterns of cell morphogenesis). With numerous genomic-scale data of several trypanosomatids becoming available since 2005 (genomes, transcriptomes, and proteomes), the scientific community can further investigate the mechanisms underlying these unusual features and address other unexplored phenomena possibly revealing biological aspects of the early evolution of eukaryotes. One fundamental aspect comprises the processes and mechanisms involved in the acquisition and loss of genes throughout the evolutionary history of these primitive microorganisms. Here, we present a comprehensive in silico analysis of pseudogenes in three major representatives of this group: Leishmania major, Trypanosoma brucei, and Trypanosoma cruzi. Pseudogenes, DNA segments originating from altered genes that lost their original function, are genomic relics that can offer an essential record of the evolutionary history of functional genes, as well as clues about the dynamics and evolution of hosting genomes. Scanning these genomes with functional proteins as proxies to reveal intergenic regions with protein-coding features, relying on a customized threshold to distinguish statistically and biologically significant sequence similarities, and reassembling remnant sequences from their debris, we found thousands of pseudogenes and hundreds of open reading frames, with particular characteristics in each trypanosomatid: mutation profile, number, content, density, codon bias, average size, single- or multi-copy gene origin, number and type of mutations, putative primitive function, and transcriptional activity. These features suggest a common process of pseudogene formation, different patterns of pseudogene evolution and extant biological functions, and/or distinct genome organization undertaken by those parasites during evolution, as well as different evolutionary and/or selective pressures acting on distinct lineages.
Keywords: genomics, trypanosomatids, homology search, sequence alignment
Significance.
Pseudogenes are molecular corpses of once-living genes damaged by random mutations in their DNA sequences, disrupting their coding potential, therefore losing their original biological role. These genomic relics offer an essential record of the evolutionary history of current genes and clues about the dynamics and evolution of the hosting genomes. However, some of them had been “brought back from the dead,” exhibiting biological functions primarily in the regulation of gene expression. Adopting functional protein sequences as proxies to reveal intergenic regions with protein-coding features, relying on a customized threshold to distinguish statistically and biologically significant sequence similarities, we scanned the genome sequences of three human parasites, representatives of an important branch of the phylogenetic tree of eukaryotes, causing disease and death in millions annually: Leishmania major, Trypanosoma brucei, and Trypanosoma cruzi. In our investigations, we found different pseudogenic patterns in this group of organisms, suggesting common processes of pseudogene formation, distinct origin, evolution, and transcriptional activity, contributing to clarifying the processes and mechanisms involved in the acquisition and loss of genes and biological functions throughout the evolutionary history of these living beings, shedding more light on the biology of early eukaryotes.
Introduction
The progress of large-scale sequencing technology has increased the speed and efficiency with which entire genomes are sequenced, stimulating more and more initiatives to obtain complete genomic sequences of countless species, especially model organisms and pathogens of importance in public health (McCombie et al. 2019). Ironically, although sequencing has become more accessible and cheaper, the process of extracting knowledge from genomic data has become more challenging. Due to the massive number of reads produced per run, the small size of these sequences, and/or the relatively high sequencing error rates, obtaining a high-quality assembly, a crucial first step to successfully annotate a genome, is tricky. However, recent technological advances have contributed to improving the quality of assemblies. Besides, the annotation process itself is intrinsically complicated, combining a range of computational and experimental methods in its execution (Griesemer et al. 2018).
As manual annotation is exceptionally laborious, expensive, and time-consuming, automated systems have been developed to perform this task efficiently. However, fully automatic annotation pipelines, though essential, can introduce and propagate inconsistent and incorrect annotations. Although genome annotation tools and resources are developing rapidly, as the scientific community is becoming increasingly dependent on this type of information for all aspects of biological research, some relevant genomic elements, such as pseudogenes, are primarily dismissed from genomic analyses and functional screenings (Poliseno 2014; Xiao et al. 2016; Couso and Patraquim 2017; Plaza et al. 2017). Even when these elements are considered, the criteria applied in separate investigations to recognize and characterize them are not uniform, creating inconsistencies when compared (Xiao et al. 2016).
Pseudogenes are described as segments of DNA that originate from genes that have been altered, losing their original function due to an interruption in one of the steps that commonly lead a DNA-coding sequence to a fully functional molecular product: transcription, pre-mRNA processing, translation, and folding. These genomic relics offer an essential record of the evolutionary history of functional genes, as well as clues about the dynamics and evolution of the hosting genomes, and are characterized by their similarity to known genes, displaying accumulated substitution(s) and/or insertion(s)/deletion(s) disrupting their open reading frames (ORFs; Harrison and Gerstein 2002; Liu et al. 2004; Chen et al. 2020). On the other hand, in distinct unicellular and multicellular organisms, it has been shown that several pseudogenes have independent (from other genes) transcriptional activity, indicating that these sequences are not necessarily lacking function (Zheng and Gerstein 2007; Muro et al. 2011; Pink et al. 2011; Pink and Carter 2013; Kovalenko and Patrushev 2018). Experimental evidence suggests the involvement of pseudogenes in the regulation of gene expression, forming non-coding RNAs, or regulating the messenger RNA stability of functional counterparts. Also, the conserved transcription profile shared between species separated by millions of years of evolution, tissue-specific transcription patterns, and differential expression during cell differentiation or caused by diseases suggest that transcripts derived from pseudogenes may play significant biological roles (Pink and Carter 2013; Singh et al. 2020).
In this work, we analyze the genomes of three important pathogens belonging to the genera Leishmania and Trypanosoma (family Trypanosomatidae, order Kinetoplastida), which together cause disease and death in millions of human beings and many infections in other mammals annually: Leishmania major (Lm), one of several species responsible for cases of leishmaniasis in numerous locations on the planet (Ivens et al. 2005); Trypanosoma brucei (Tb), the etiological agent of African trypanosomiasis (sleeping sickness in humans and Nagana in animals; Berriman et al. 2005); and Trypanosoma cruzi (Tc), causing American trypanosomiasis (Chagas disease; El-Sayed et al. 2005). Besides their importance in public health, these three trypanosomatids (TriTryps) belong to a remarkable group of living beings, including vertebrates, invertebrates, and even plant parasites (Acosta et al. 2014). They exhibit several peculiar and intriguing biological characteristics, such as: (1) complex mechanisms to control gene expression, involving the organization of most genes in large groups that are simultaneously transcribed as polycistronic units, undergoing a process of maturation that includes the phenomenon of trans-splicing, in addition to other complicated and energetically expensive mechanisms related to the mitochondrial RNA editing processes, as well as the almost complete absence of introns in their genomes; (2) unusual aspects of their metabolism, their surface molecules, as well as their organelles, such as the compartmentalization of glycolysis, the evasion of the host’s immune system through the variation of its surface molecules and mitochondrial DNA with an entirely unique architecture; and (3) complex cell biology and life cycle, including the ability to escape cell damage by migrating outside phagocytic vacuoles, and exhibiting intricate patterns of cell morphogenesis during their life cycles (Simpson et al. 2006).
Genomic sequences of several representatives of the family Trypanosomatidae have been published since 2005, with TriTrypDB <https://tritrypdb.org/tritrypdb/> becoming the most relevant resource for trypanosomatid genomic research since its first version, launched in 2009, contributing to a better understanding not only of the biology of these pathogens but also of relevant aspects of the evolution of their genomes (Maslov et al. 2019). This integrative database represents a collaborative effort between the VEuPathDB (Amos et al. 2022) team at the Universities of Pennsylvania and Georgia, the GeneDB (Logan-Klumpler et al. 2012) group at the Wellcome Trust Sanger Institute, and researchers at the Seattle Biomedical Research Institute, providing the scientific community access to complex querying tools and genomic-scale data (genomes, transcriptomes, and proteomes) of numerous trypanosomatids (Aslett et al. 2010).
Hence, implementing a genome-wide homology-based approach, we scanned these parasites' genomes, using functional protein sequences as proxies to reveal intergenic regions (termed as such due to the absence of predicted coding sequences (CDSs)) with protein-coding features, applying a similarity threshold determined by a shuffle-based statistical significance evaluation. Thousands of non-overlapping intergenic regions displaying statistically significant similarity to functional proteins—both manually annotated (Swiss-Prot database entries; Bairoch 2000) and/or computationally predicted (trypanosomatids' annotated protein sequences from TriTrypDB; Aslett et al. 2010)—were found in each of these TriTryps' genomes. Most of these sequences are pseudogenes, DNA segments resembling functional protein(s), with substitution(s) and/or insertion(s)/deletion(s) disrupting the ORF. However, several intergenic regions similar or entirely identical to functional protein(s), with preserved initiation and termination codons, were also found in these genomes, reasonably corresponding to non-annotated CDSs.
Results
The Repertoire of Novel Pseudogenes and Protein-coding Sequences in TriTryps
We found in TriTryps' genomes 8,520 intergenic sequences with 300 nucleotides or more in length, with no observable DNA strand preference, displaying statistically significant sequence similarity to functional proteins (as defined in Materials and Methods section Genome-wide homology-based searching): Lm (1,121), Tb (2,114), and Tc (5,285). Most of these regions (8,009/94.0%) encode pseudogenes—Lm (1,071/95.5%), Tb (1,969/93.1%), and Tc (4,969/94.0%)—operationally defined in this work as DNA segments globally or locally related to functional protein(s), showing substitution(s) and/or insertion(s)/deletion(s) disrupting the ORF, and/or presenting loss of initiation/termination codon(s) and/or displaying internal termination codons. Several intergenic sequences, similar—admitting non-disruptive substitution(s) and/or insertion(s)/deletion(s)—or entirely identical to functional protein(s), with preserved initiation and termination codons, were also found in these genomes, corresponding to non-annotated CDSs (511/6.0%): Lm (50/4.5%), Tb (145/6.9%), and Tc (316/6.0%; table 1 and supplementary material S1, Supplementary Material online). These cryptic, non-annotated CDSs are termed intergenic CDSs throughout the rest of this work.
Table 1.
The Repertoire of Pseudogenes (y) and Intergenic CDSs (iCDS) Annotated in TriTryps’ Genomes According to the Source of the Most Similar Protein Sequence in our Reference Data set: Swiss-Prot or TriTrypDB
| Organism | Source of the most similar protein sequence | |||||
|---|---|---|---|---|---|---|
| Swiss-Prot | TriTrypDB | Total | ||||
| iCDS | Y | iCDS | Y | iCDS | y | |
| Lm | 0 | 19 | 50 | 1,052 | 50 | 1,071 |
| Tb | 0 | 6 | 145 | 1,963 | 145 | 1,969 |
| Tc | 1 | 17 | 315 | 4,952 | 316 | 4,969 |
Pseudogenes and intergenic CDSs are randomly dispersed across chromosomes and contigs, with numbers roughly corresponding to chromosomes or contigs length (data not shown) and are unevenly distributed among TriTryps. However, no correlation with genome size or gene content was observed (see Discussion). A large (though variable) number of these intergenic elements have proteins encoded in these parasite genomes and/or in other representatives of the same species comprising our reference data set as their most similar functional sequence: Lm (498/46.5%), Tb (1,059/53.8%), and Tc (3,064/61.7%) for pseudogenes; Lm (34/68%), Tb (75/51.7%), and Tc (177/55.6%) for intergenic CDSs (supplementary material S1, Supplementary Material online). While these organisms have comparable coding densities (the number of nucleotides in CDS regions divided by the genome length in base pairs)—Lm (49.95%), Tb (45.96%), and Tc (47.54%)—pseudogene densities (the number of nucleotides in pseudogene regions divided by the genome length in base pairs)—Lm (2.2%), Tb (3.4%), and Tc (14.5%)—are remarkably distinct, as well as the proportion of pseudogenes relative to CDSs (pseudogene content)—Lm (11.0%), Tb (6.8%), and Tc (22.4%). Intergenic CDSs' density displayed similar characteristics: Lm (0.07%), Tb (0.20%), and Tc (0.40%).
TriTryp’s Pseudogenes and Intergenic Protein-coding Sequences Features
TriTryps displayed different size distribution of pseudogenes and intergenic CDSs, with most sequences (∼90%) ranging from 300 to 900 nucleotides in Lm, 300–1,800 in Tb, and 300–2,100 in Tc (data not shown). The average number of mutations per sequence and average pseudogene sequence length also varied substantially among the TriTryps: Lm (66.79/659.7), Tb (115.74/859.9), and Tc (64.75/1,553.3). On the other hand, in Tb and Tc, pseudogenes display comparable mutation profiles, that is, the relative frequency of each type of mutation (table 2 and supplementary material S1, Supplementary Material online), Lm presents a remarkably different pattern, especially if we compare the frequency of pseudogenes exclusively bearing one or two predominant types of mutation: loss of the stop codon (detected in 90–98% of all TriTryp's pseudogenes)—Lm (51.63%), Tb (30.78%), and Tc (37.7%)—and loss of the start and stop codons altogether (with loss of start codon affecting 30–37% of all pseudogenes in TriTryps)—Lm (28.20%), Tb (16.46%), and Tc (11.78%). Transcriptionally active pseudogenes present quite similar characteristics (supplementary material S1, Supplementary Material online).
Table 2.
TriTryp's Mutation Profiles. Absolute Numbers and Relative Frequencies of Mutations (top) or Pseudogenes Bearing Individual or Multiple Types of Mutations (Bottom)
| Mutation | Lm | Tb | Tc |
|---|---|---|---|
| Number and frequency (%) of each type of mutation in pseudogenes | |||
| Frameshift | 201 (10.21) | 1,009 (20.31) | 2,436 (49.02) |
| Deletiona | 80 (7.47) | 373 (18.94) | 904 (18.19) |
| Insertion | 69 (6.44) | 431 (21.89) | 899 (18.09) |
| Substitution | 189 (17.65) | 912 (46.32) | 2,182 (43.91) |
| Internal_stop | 98 (9.15) | 673 (34.18) | 1,136 (22.86) |
| Lost_start | 395 (36.88) | 719 (36.52) | 1,532 (30.83) |
| Lost_stop | 1,054 (98.41) | 1,782 (90.50) | 4,588 (92.33) |
| Number and frequency (%) of pseudogenes bearing exclusively one (loss of the start or stop codon) or two (loss of start and stop codons) predominant types of mutation among pseudogenes | |||
| Lost_stop | 553 (51.63) | 606 (30.78) | 1,873 (37.70) |
| Lost_start + lost_stop | 302 (28.20) | 324 (16.46) | 585 (11.78) |
| Lost_start | 15 (1.40) | 30 (1.52) | 75 (1.51) |
| Others | 201 (18.77) | 1,009 (51.24) | 2,436 (49.01) |
Note.—aIn-frame deletions.
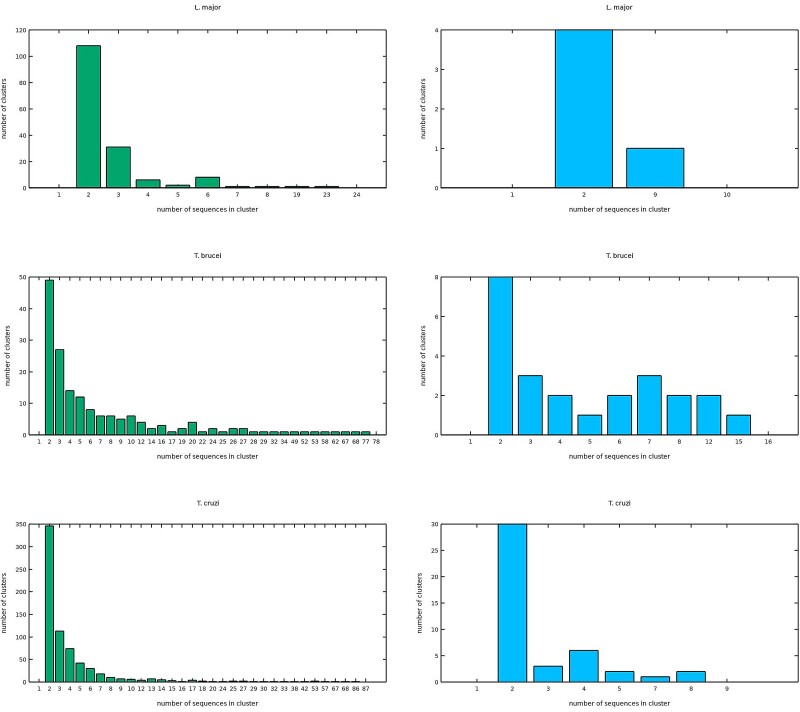
Clustering pseudogene's nucleotide sequences applying an 80% global identity threshold yielded 3,270 groups, in which 2,233 (68.2%) of them are singletons: Lm (555/729 = 76.1%), Tb (128/303 = 42.2%), and Tc (1,554/2,253 = 69%; supplementary material S2, Supplementary Material online). Multi-sequence families containing two or more pseudogenes from the same species constitute the remaining groups (1,037, representing 31.8%): Lm (174/729 = 23.9%), Tb (175/303 = 57.8%), and Tc (699/2,253 = 31.0%; fig. 1). Only 15 groups (0.46% of the clusters, comprising 1.4% of all pseudogenes) contain pseudogenes shared between two TriTryps (Lm/Tc: 14; Tb/Tc: 1), none of them consisting of sequences from the three genomes. Similarly, intergenic CDSs were grouped in 315 clusters at 80% global sequence identity, mostly singletons (242, corresponding to 76.8%)—Lm (33/38 = 86.8%), Tb (19/43 = 44.2%), and Tc (190/234 = 81.2%)—with scarce multi-sequence families (73, comprising 23.2%)—Lm (5/38 = 13.2%), Tb (24/43 = 55.8%), and Tc (44/234 = 18.8%; fig. 1) and no shared intergenic CDSs (supplementary material S2, Supplementary Material online).
Fig. 1.
Bar graphs displaying the number of sequences (y axis) in multi-sequence clusters comprising ≥ two sequences (x axis) of pseudogenes (bars in left panels) and intergenic protein-coding sequences (bars in right panels) exclusively encoded in Lm's, Tb's, and Tc's genomes.
The codon usage of intergenic CDSs and pseudogenes (supplementary material S3, Supplementary Material online) display a very slight deviation from the organisms' genomic codon usage in Lm as revealed by the Pearson's correlation coefficient: r = 0.921 (intergenic CDSs) and r = 0.910 (pseudogenes). However, in Tb, we detected a high codon usage deviation for both intergenic CDSs (r = 0.545) and pseudogenes (r = 0.434), Tc presents a considerably weaker deviation for intergenic CDSs (r = 0.841) and a pronounced deviation for pseudogenes (r = 0.614).
Protein's Functional Classes Affected by Pseudogenization in TriTryps
We used the classification of functional proteins in our data set as a proxy to infer the potential primitive function of pseudogenes and the possible biological function of intergenic protein-coding sequences, transferring the sequence annotation of the most similar functional protein (with the highest score density) to these genomics elements (table 1 and supplementary material S4, Supplementary Material online). However, only a limited fraction of intergenic protein-coding sequences (38/7.4%) and pseudogenes (506/6.3%) could have their original function inferred based on inherited classifications of proteins encoded in the parasite's genome or related species—Lm (0), Tb (3/2.1%), and Tc (34/10.7%) for intergenic CDSs; Lm (36/3.4%), Tb (31/1.6%), and Tc (413/8.3%) for pseudogenes—and of protein entries in Swiss-Prot as well—Lm (0), Tb (0), and Tc (1/0.3%) for intergenic CDSs; Lm (11/1.0%), Tb (6/0.3%), and Tc (9/0.2%) for pseudogenes.
TriTrypDB's and Swiss-Prot's annotations of protein sequences comprising our reference data set revealed pseudogenic and intergenic CDS counterparts of numerous known multigenic families in trypanosomatids (Avelar et al. 2020) and several other protein classes. However, most pseudogenes and intergenic CDSs identified remain unclassified as their most similar proteins in the reference group are annotated as hypothetical, unspecified, putative, pseudogene, or pseudogenic—Lm (1,074/95.8%), Tb (2,074/98.1%), and Tc (4,828/91.3%). As expected, the variant surface glycoproteins comprise a considerable proportion of pseudogenes in Tb (166/8.4%) (Berriman et al. 2005; supplementary material S1 and S4, Supplementary Material online). The 15 groups of clustered pseudogenes shared between Lm/Tc and Tb/Tc have their best hits annotated in TriTrypDB as hypothetical protein, unspecified product, or putative, except two: C3747_28g296-t42_1-p1 (40S_ribosomal_protein_S23) and BCY84_03604-t36_1-p1 (heat-shock_protein_hsp70).
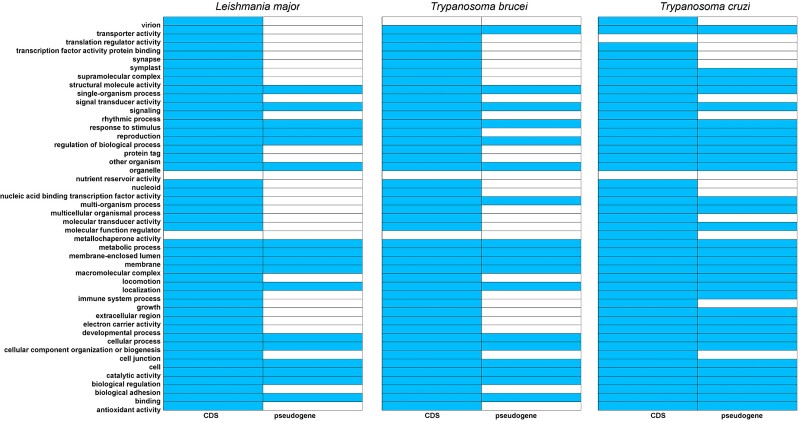
Nevertheless, this process showed that pseudogenization had affected similar proportions of the actual known functional space in Lm (17/44, 38.6%) and Tb (18/43, 41.8%) genomes, reaching approximately 40% of their functional classes, although in Tc (32/44, 72.7%), it corresponds to 73% of the recognized functional categories. Sixteen GO categories shared among the TriTryps account for >88% of the pseudogenized classes in Lm (16/17, 94.1%) and Tb (16/18, 88.9%), but much fewer classes in Tc (16/32, 50.0%; fig. 2).
Fig. 2.
Presence (colored) and absence (uncolored) matrix displaying the Gene Ontology functional classification of TriTryps' protein-coding genes (CDS) and pseudogenes.
Transcriptionally Active Pseudogenes and Intergenic Protein-coding Sequences in TriTryps
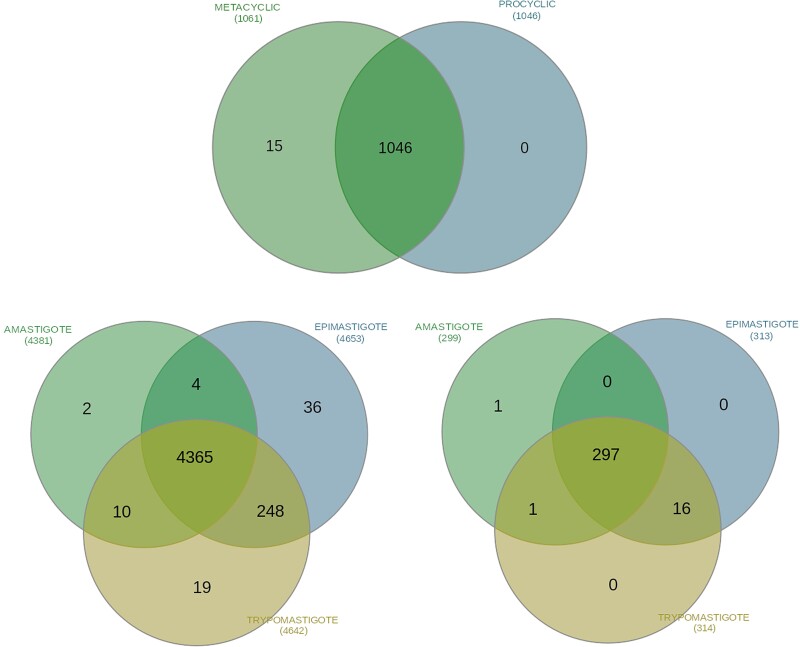
Examining transcriptomic data publicly available in SRA (table 5), we found numerous pseudogenes (7,325/91.5%) and intergenic CDSs (511/100%) unitarily encoded in mature mRNA sequences of each TriTryp (supplementary material S5, Supplementary Material online). Although the proportion of transcriptionally active pseudogenes is considerably different among them—Lm (1,061/91.1%), Tb (1,580/80.2%), and Tc (4,684/94.3%)—the majority of these elements are natively transcribed in these parasites. On the other hand, we observed distinct transcriptionally active pseudogenes and intergenic CDSs depending on the life-cycle stage in Lm and Tc (only bloodstream-form transcriptomes were available for Tb), with several pseudogenes transcriptionally active exclusively in the mammalian-infective form of Lm (15 pseudogenes in metacyclic promastigotes) and Tc (19 pseudogenes in trypomastigotes), as well as in the remaining two forms of Tc (36 pseudogenes in epimastigotes and 2 pseudogenes and one iCDS in amastigotes); a number of distinct pseudogenes and iCDS are exclusively shared between amastigotes and epimastigotes (four pseudogenes), amastigotes and trypomastigotes (10 pseudogenes and one iCDS), and epimastigotes and trypomastigotes (248 pseudogenes and 16 iCDS) among Tc's life-forms (fig. 3 and supplementary material S5, Supplementary Material online).
Table 5.
Summary Information about the 33 RNA-Seq Experiments Analyzed in This Work
| Organism | Project | Experiments | Layout | Strategy | Instrument | Life-cycle |
|---|---|---|---|---|---|---|
| Lm | PRJNA252769 | 20 | PAIRED | RNA-Seq | Illumina | Procyclic/Metacyclic promastigotes |
| Tb | PRJEB31609 | 7 | PAIRED | RNA-Seq | Illumina | Bloodstream form |
| Tc | SRA SUBMISSION | 6 | PAIRED | RNA-Seq | Illumina | Amastigotes/Epimastigotes/Trypomastigotes |
Note.—Source: SRA <https://www.ncbi.nlm.nih.gov/sra>.
Fig. 3.
Venn diagram displaying the distribution of transcriptionally active pseudogenes among life-cycle stages of Lm (top) and Tc (bottom left), and transcriptionally active intergenic CDSs among life-cycle stages of Tc (bottom right).
Discussion
Discrimination of Coding/non-coding DNA Segments and Pseudogene Annotation
Assigning putative functions to genes or transcripts is still a complex process that commonly yields incomplete and/or misannotations, which feed biological databases with artifacts that might be used as reference sequences for further annotation processes, therefore propagating errors in a vicious circle (Poptsova and Gogarten 2010). Besides, numerous putative protein-coding (and non-coding RNA) genes and transcripts remain uncharacterized, lacking functional assignments. The annotation of pseudogenes is even more challenging, as these elements can be mistakenly annotated as genes (and vice-versa), mainly when a fully automated annotation process is employed (Zheng and Gerstein 2006). However, pseudogene prediction is a necessary task in genome annotation as it is not feasible to directly recognize the functional state of each protein-coding region annotated in sequenced genomes, nor to disclosure remnant sequences using ab initio methods for gene prediction, as pseudogenes do not necessarily present the same features displayed by their functional counterparts, which are restrained by selective forces and comprise the data source for developing gene-finding algorithms (Xiao et al. 2016). As those molecular fossils offer an essential record of functional genes' evolutionary history and not rarely are brought back from the dead to play a part in post-transcriptional regulation processes, it is crucial to recognize these genomic elements (Singh et al. 2020).
In most cases, pseudogenes are identified using comparative approaches based on detected sequence similarities between functional proteins and intergenic regions of target genomes. However, as truly non-functional sequences are theoretically free from evolutionary constraints, accumulating deleterious mutations, sequence alignments producing low similarity scores are expected to occur, possibly reaching the limits of the existing methods for sequence comparison, in which they cannot distinguish between true and false positives, even displaying statistically significant similarities (Brenner et al. 1998; Rost 1999).
Hence, to discriminate pseudogenes and intergenic protein-coding regions from presumed non-coding DNA segments, we first determined the sequence similarity profile representing comparisons (1) between functional proteins, both manually annotated (Swiss-Prot) and computationally predicted (TriTrypDB), and (2) between these functional proteins and a data set of artificially created protein sequences with the average size and composition displayed by the group of functional sequences. Based on this profile, we defined a customized set of sequence similarity parameters to discriminate alignments without biological meaning in our searches, combining three similarity scores: (1) E-value ≤ 10e − 6, (2) alignment score above 120, and (3) protein sequence identity above 20% (see Materials and Methods section Shuffle-based statistical significance evaluation for a complete explanation).
Applying this genome-wide homology-based approach, we uncovered thousands of novel pseudogenes and intergenic protein-coding sequences in TriTryps' genomes, relying on functional proteins as proxies to reveal intergenic regions with coding features and on a customized threshold to distinguish statistically and biologically significant sequence similarities. The strategy can be equally applied to annotate pseudogenes in other species' genomes.
Unrecognized Pseudogenes and Protein-coding Sequences in TriTryps
When we compared our results to the annotation provided by TriTrypDB for pseudogenes, it became clear the relevance of automatizing and standardizing pseudogene annotations in trypanosomatids (and plausibly in numerous other organisms): Lm (194), Tb (5,349), and Tc (0) in version 44, compared with Lm (299), Tb (5,481), and Tc (1,878) in version 54. Merged pseudogenic regions in Lm (98) and Tc (1,748) match only a fraction of our predictions—Lm (1,071) and Tc (4,969)—whereas in Tb, all 5,259 pseudogenes with 300 nucleotides or more, obtained after this merging process, occur inside the 1,969 pseudogenic regions, we annotated in this organism's genome, showing that pseudogenes are mostly underestimated and misannotated in TriTryps (and likely in most genomes sequenced so far; supplementary material S1 and S6, Supplementary Material online).
Comparative Analysis of Pseudogenes and Intergenic Protein-coding Sequences in TriTryps
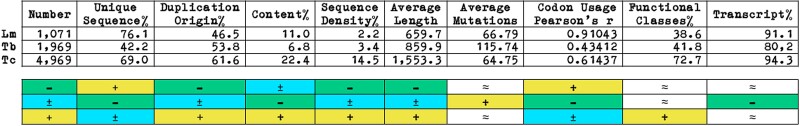
Pseudogenes (and intergenic CDSs) in TriTryps are randomly distributed in their genomes (supplementary material S1, Supplementary Material online), comprising shorter nucleotide sequences in Lm and average-size sequences in Tb and Tc (fig. 4), compared with the average length of encoded CDSs in their genomes—Lm (1,906), Tb (824), and Tc (1,471) (data not shown)—although in Tc the average pseudogenes' length is approximately twice as large than in Lm and Tb (fig. 4). The abundance of pseudogenes differs considerably among TriTryps, relative to genome size, CDS content, and genomic extent, with Lm showing a disproportionally low number, content, and density compared with the disproportionally high number, content and density found in Tc, and the pronounced number, and disproportionally low content and density displayed by Tb (fig. 4).
Fig. 4.
TriTryps' pseudogenes comparative charts summarizing (top) and representing (bottom) our results. Symbols in colored boxes indicate comparatively high (+), low (−), intermediate (±), or approximate (≈) values. See text for a full explanation.
Many of these intergenic sequences are unique, though markedly different proportions of pseudogenes' and intergenic CDSs' singleton clusters were observed among them (fig. 4, and supplementary material S2, Supplementary Material online). Multi-sequence groups containing two or more pseudogenes (or intergenic CDSs) of the same species are not rare, though notably increased in Tb (fig. 1, and supplementary material S2, Supplementary Material online). Although some of the most populated clusters contain pseudogenic counterparts of widely known multigenic protein families in trypanosomatids (data not shown), an in-depth analysis of multi-sequence pseudogene clusters in TriTryps is needed. Interestingly, a tiny fraction of pseudogenes (1.4%), with no recognizable primitive function in 96.3% of the instances, are shared between only two species among the TriTryps, Lm/Tc (14 clusters) and Tb/Tc (1 cluster; supplementary material S2, Supplementary Material online), showing that these intergenic sequences are predominately lineage specific, contrasting with nearly 94% of gene conservation previously observed among Lm, Tb, and Tc, in which most of the implicated genes are encoded in conserved syntenic regions (El-Sayed et al. 2005; Peacock et al. 2007).
Almost half of the pseudogenes in Lm, roughly half in Tb, and a higher proportion in Tc were unveiled by protein sequences encoded in their genomes or the genomes of close-related organisms (strains or isolates of the same species) comprising our reference data set, indicating predominant duplication origin in Tb and Tc, and de novo origin in Lm (fig. 4, and supplementary material S1, Supplementary Material online), although intergenic CDSs present different proportions (Results section TriTryp's pseudogenes and intergenic protein-coding sequences features).
Both pseudogenes' and intergenic CDSs' codon usage slightly deviate from the organisms' codon choice in Lm, whereas in Tb and Tc, these elements display a markedly distinct codon preference (fig. 4, and supplementary material S3, Supplementary Material online). The degree of erosion of pseudogenes, that is, the average number of mutations per sequence, places Tb and Lm/Tc on opposite sides, with Tb bearing pseudogenes with a disproportionally high number of mutations compared with Lm or Tc, which still carry pseudogenes with a pronounced number of mutations per sequence on average (fig. 4).
Comparing the predicted primitive functional profile of pseudogenes with the actual functional classes comprising the proteins encoded in each genome, we found a puzzling scenario, in which not all classes of proteins seem prone to pseudogenization in TriTryps' species (fig. 2, and supplementary material S4, Supplementary Material online). In Lm and Tb, pseudogenization has affected approximately 40% of their recognized functional space, whereas in Tc this process has reached a much higher proportion of 72.7%; 16 functional categories shared among the TriTryps account for most of the affected functional classes in Lm and Tb, except in Tc (fig. 2). Although these few protein classes represent a range of fundamental molecular functions (catalytic activity), biological processes (metabolic process, response to stimulus, and regulation of biological process), and cellular components (organelle, membrane, and macromolecular complex) in any organism, no reasonable explanation for that have been found yet. On the other hand, as numerous pseudogenes are transcriptionally active in TriTryps (fig. 4), varying according to the life-cycle stage at least in Lm and Tc (fig. 3, and supplementary material 5, Supplementary Material online), it would be interesting to investigate further the involvement of these molecules in post-transcriptional regulation processes, as already been (partially) done for Tb (Wen et al. 2011), as well as their putative participation in metacyclogenesis, a natural process of morphological and biochemical changes that occurs inside the invertebrate host of Lm (sandfly), Tb (tsetse fly), and Tc (triatomine insects), comprising a stepwise progression toward the differentiation of mammalian non-infective into mammalian-infective life-forms of these parasites (Amorim et al. 2017; Amiri-Dashatan et al. 2020; Toh et al. 2021).
Pseudogenes in trypanosomatids seem to originate from degradation of single-copy genes and native duplicated gene copies, contrasting with multicellular eukaryotes, in which retrotransposition and duplication of genomic segments are possible mechanisms responsible for pseudogene formation. Assuming pseudogenes are neutrally evolving, randomly accumulating mutations, Lm seems less prone to pseudogenization, bearing a limited amount, that is, disproportionally low number, content, and density, of recently acquired small-sized pseudogenes, as suggested by their slightly deviated codon usage (although displaying a considerable number of mutations per sequence), mainly arisen from the degradation of single-copy genes, as indicated by the disproportionally high frequency of singleton sequences and their restricted similarity with proteins encoded in its genome or the genomes of close-related organisms, affecting almost 40% of its functional landscape, with most of them transcriptionally active (fig. 4). On the other hand, in Tb, pseudogenes are larger and abundant, but with their relatively high number corresponding to a disproportionally low content, and density, and disproportionally high codon usage deviation and number of mutations per sequence, suggesting an ancestral origin, mainly arisen from duplication events but also from the degradation of single-copy genes, suggested by the moderate difference between the frequency of singleton sequences and similarity with proteins encoded in its genome or the genomes of close-related organisms, affecting >40% of its protein classes, with a remarkably lower fraction of them transcriptionally active (fig. 4). Tc presents a different scenario yet, with copious large-sized pseudogenes presented in disproportionally high number, content, density, and a pronounced codon usage deviation, with a considerable number of mutations per sequence, indicating they are relatively old, likely arising from duplication events or degradation of single-copy genes, considering the proportional frequency of singleton sequences and similarity with proteins encoded in its genome or the genomes of close-related organisms, affecting most of its functional categories, with the majority of them found transcriptionally active (fig. 4).
These characteristics, and the markedly distinct mutation profile exhibited by Lm (table 2 top, and supplementary material S1, Supplementary Material online), might indicate different patterns of pseudonization and extant biological functions, and/or distinct genome organization undertaken during the evolution of TriTryps, as well as different evolutionary and/or selective pressures acting on distinct lineages. However, the most frequent types of mutations found in TriTryps' pseudogenes suggest a shared process of pseudogene formation in which the sole loss of the stop codon most of the time triggers the disruption of the original ORF, followed by a predominant loss of the start codon by these sequences, and further erosion caused by several accumulating mutations of one or multiple types, with a negligible contribution of unique loss of the start codon (table 2 bottom, and supplementary material S1, Supplementary Material online). Extending the comparative analysis reported here, to include pseudogenes annotated in the remaining sequenced genomes of Kinetoplastida's species might clarify the processes and mechanisms involved in the acquisition and loss of genes and biological functions throughout the evolutionary history of this remarkable group of living beings and shed more light on the biology of early eukaryotes.
Conclusions
Pseudogene prediction is a necessary task in genome annotation processes, as it is not feasible to directly recognize the functional state of each protein-coding region annotated in sequenced genomes, nor to reveal remnant sequences using ab initio methods for gene prediction, as pseudogenes do not necessarily have similar statistical properties to protein-coding genes, on which prediction algorithms are based. As these molecular fossils offer an essential record of functional genes' evolutionary history, and several of them play relevant biological roles, it is crucial to recognize those genomic elements, inferring their origin and primitive function, and assessing their current functional state.
In mammals and other multicellular eukaryotes, retrotransposition and duplication of genomic segments seem to be the source of pseudogenes. Nevertheless, pseudogene formation in prokaryotes is distinct, originating from native duplicated gene copies, degradation of single-copy genes, and unsuccessful horizontal gene transfers (in which the transferred genes are not fixed as functional copies in the host's genome). In trypanosomatids, pseudogenes seem to arise either from decaying single-copy genes or duplicated counterparts by a sequential loss of their stop and start codons, affecting approximately 40–70% of their proteome's functional landscape. However, the possible contribution of undetected horizontal gene transfers needs to be investigated. Uncovering relics of functional protein-coding sequences in the remaining species of trypanosomatids may help us understand the main processes involved in gene acquisition and gene loss, shedding more light on these parasites' evolutionary history.
Altogether, it seems clear that scanning genomes adopting functional proteins as proxies to reveal intergenic regions with protein-coding features, relying on a customized threshold to distinguish statistically and biologically significant similarities, and reassembling scattered remnant sequences from their debris is a suitable approach to automatically annotate pseudogenes in trypanosomatids, especially when compared with the semi-automatic annotation of pseudogenes originally provided for Tb (Müller et al. 2018). Further testing of this approach in other species has the potential to establish this as a practical large-scale approach to annotate pseudogenes.
Applying this strategy to analyze the genome sequences of three major species of this group—Lm, Tb, and Tc—revealed distinct pseudogenic profiles of mutation, number, content, density, codon bias, average size, single- or multi-copy gene origin, number and type of mutations, putative primitive function, and transcriptional activity, suggesting different processes of pseudogene formation, evolution, and possibly yet unknown biological roles played by pseudogenes in these parasites.
Materials and Methods
TriTryps' Sequences
Genomic and protein sequences in FASTA format, and annotation files in GFF3 format of Lm Friedlin (Ivens et al. 2005), Tb Lister427 (Müller et al. 2018), and Tc Dm28c (Berná et al. 2018), were obtained from TriTrypDB database version 44 and 54 (only GFF3 files; Aslett et al. 2010; table 3). Trypanosoma brucei Lister427 and Tc Dm28c genomes were obtained with single-molecule real-time sequencing, developed by Pacific BioSciences (PacBio), yielding high-quality long-reads assemblies, which has only recently accomplished for Lm Friedlin (Camacho et al. 2021). However, as already shown by Camacho and coworkers (2021), we observed very few detectable differences between (genome size, number of Ns, number of gaps, number of chromosomes, among others) this newer assembly and the one already publicly available in TriTrypDB, a high-quality assembly obtained with Sanger technology, yielding basically the same assembly results as those obtained with the “reference” genome (data not shown).
Table 3.
Lm, Tb, and Tc Genomes Data Information
| Organism | Version | Assembly | Chr | Contigs | Size (Mb) |
CDS (v44) | CDS (v54) | y (v44) | y (v54) |
|---|---|---|---|---|---|---|---|---|---|
| Lm Friedlin | 2016 | GCA_000002725.2 | 36 | 0 | 32.8 | 8,606 | 8,563 | 194 | 299 |
| Tb Lister427 | 2018 | GCA_900497135.1 | 44 | 21 | 50.08 | 11,388 | 21,008 | 5,349 | 5,481 |
| Tc Dm28c | 2018 | GCA_003177105.1 | 0 | 1,028 | 53.27 | 17,197 | 17,197 | 0 | 1,878 |
Note.— Chr, chromosomes; Y, pseudogenes. Sources: <https://tritrypdb.org/tritrypdb/app/downloads/release-44/> and <https://tritrypdb.org/tritrypdb/app/downloads/release-54/>.
Data Set of Functional Proteins
We selected all entries in the Swiss-Prot database version 2019_05 (REF; 560,292 entries) and annotated protein sequences of the following trypanosomatids (332,216 CDSs) to comprise our data set of functional proteins (table 4).
Table 4.
Trypanosomatids Species and the Corresponding Number of Protein Sequences Comprising the Reference Data set of Functional Proteins in our Investigation
| Organism | Proteins |
|---|---|
| Leishmania aethiopica L147 | 8,722 |
| Leishmania amazonensis MHOMBR71973M2269 | 8,031 |
| Leishmania braziliensis MHOMBR75M2903 | 9,269 |
| Leishmania braziliensis MHOMBR75M2904 | 8,354 |
| Leishmania donovani BPK282A1 | 8,023 |
| Leishmania donovani CLSL | 8,632 |
| Leishmania donovani LV9 | 8,245 |
| Leishmania infantum JPCM5 | 8,591 |
| Leishmania major Friedlin | 8,519 |
| Leishmania major LV39c5 | 8,971 |
| Leishmania major SD751 | 8,818 |
| Leishmania mexicana MHOMGT2001U1103 | 8,246 |
| Leishmania panamensis MHOMCOL81L13 | 8,665 |
| Leishmania panamensis MHOMPA94PSC1 | 7,933 |
| Leishmania tropica L590 | 8,824 |
| Leishmania turanica LEM423 | 8,608 |
| Trypanosoma brucei GambienseDAL972 | 7,988 |
| Trypanosoma brucei Lister427 | 8,844 |
| Trypanosoma brucei Lister427_2018 | 16,869 |
| Trypanosoma brucei TREU927 | 11,203 |
| Trypanosoma congolense IL3000 | 11,585 |
| Trypanosoma cruzi Dm28c2014 | 11,348 |
| Trypanosoma cruzi Dm28c2017 | 20,350 |
| Trypanosoma cruzi Dm28c2018 | 17,197 |
| Trypanosoma cruzi SylvioX101 | 20,619 |
| Trypanosoma cruzi SylvioX101_2012 | 10,876 |
| Trypanosoma cruzi TCC | 27,155 |
| Trypanosoma cruzi marinkelleiB7 | 10,228 |
| Trypanosoma evansi STIB805 | 10,109 |
| Trypanosoma vivax Y486 | 11,394 |
Note.—Source: <https://tritrypdb.org/tritrypdb/app/downloads/release-44/>.
Functional Classification of TriTryps’ Protein-coding Sequences
We compared the set of proteins encoded in TriTryps' genomes with (1) proteins deposited in the Swiss-Prot database, using BLAST version 2.7.1 (Altschul et al. 1997), and with (2) protein family profiles in Pfam database version 31.0 (Finn et al. 2016), using HMMER version 3.1b1 (Finn et al. 2011), applying default sequence alignment parameters and customized BLAST output format (outfmt “6 qseqid sseqid evalue”). The result of these comparisons was used as input to the Argot2.5 server version 2.5 (Lavezzo et al. 2016) for functional inference, selecting the following parameters: Score (≥ 0): 200; Semantic similarity metrics: simGIC. Proteins were classified in three fundamental categories—molecular function, cellular component, and biological process—based on sequence and semantic similarity (homology inference and gene ontology assignment, respectively) with a controlled vocabulary, following the functional transfer criteria defined by the Gene Ontology (GO) Consortium (Lavezzo et al. 2016). We used GNU Octave version 5.2.0 to create the presence/absence matrix displaying the GO classification result.
Shuffle-based Statistical Significance Evaluation
To build a null-model representing protein sequences without any detectable biological meaning, we created a set of 10,000 artificial proteins with compositional characteristics and lengths similar to functional sequences in our data set (Swiss-Prot and TriTrypDB). Starting with a single computer-generated amino acid sequence with the average size and composition displayed by the functional sequences comprising our data set, we unbiasedly permuted the artificial sequence with the Fisher–Yates' algorithm implemented in the List::Util set of subroutines (shuffle function) of the Perl language version 5.10.1, saving the newly generated sequence in FASTA format, repeating this process 10,000 times. Next, we performed a pairwise sequence comparison between each functional sequence against each artificial sequence using the BLAST version 2.7.1 (Altschul et al. 1997) with the following parameters: blastp -word_size 3 -gapopen 11 -gapextend 1 -matrix BLOSUM62 -threshold 13 -comp_based_stats 2 -seg yes -soft_masking true -lcase_masking -evalue 10 -max_target_seqs 1000000. Finally, we parsed these results, searching for the most statistically significant sequence similarity and the highest BLAST (BLOSUM62) score obtained comparing functional proteins with randomly generated sequences, establishing a customized sequence similarity threshold to discriminate false similarities when scanning the genomes for intergenic protein-coding sequences: (1) E-value (10,000) ≤ 10e − 6, as the most significant E-value obtained comparing functional with artificially generated sequences was 1.7e − 6; (2) alignment score above 120, calculated with BLOSUM 62 substitution matrix, because the maximum similarity score obtained with the group of artificial sequences was 119; (3) protein sequence identity above 20%, as roughly 10% of evolutionarily related proteins can still be detected with local alignment algorithms below 25% identity (Rost 1999), and 20% is the expected average frequency of an amino acid in any protein sequence assuming all 61 codons have equal chance to occur, meaning that this percentage of amino acid identity is expected to happen by chance alone when comparing protein sequences.
Genome-wide Homology-based Searching
We used BEDTools version 2.29.2 (Quinlan and Hall 2010) to extract intergenic regions of each genome (including segments already described as pseudogene), based on the coordinates of annotated genomic elements (obtained from their respective genomic GFF file), then compared the set of functional protein sequences with these intergenic regions with BLAST, respectively: bedtools complement with default parameters; tblastn -word_size 3 -gapopen 11 -gapextend 1 -matrix BLOSUM62 -threshold 13 -comp_based_stats 2 -seg yes -soft_masking true -lcase_masking -evalue 10 -max_target_seqs 1000000. For each alignment obtained in the previous step, we re-aligned the pair of sequences with SSEARCH version 36.3.8 g <https://fasta.bioch.virginia.edu/> to calculate the best rigorous (dynamic programming) alignment between them, retaining only statistically significant pairwise alignments for subsequent analyzes with the following thresholds: protein identity >20%, score >120, and E-value ≤ 1e − 6. To estimate the statistical significance of each pairwise alignment in this step, we set SSEARCH to shuffle the functional sequence of the pair randomly, then align this artificially created sequence with the similar intergenic sequence of the pair, and repeat this process 500 times, calculating the E-value for the original pair of sequences based on an effective (apparent) database size of 10,000 sequences and the distribution of alignment scores obtained by these random sequences. The complete set of parameters used in this step was ssearch36 -b 1 -d 1 -k 500 -f -11 -g 1 -m 10 -q -s BL62 -z 11 -Z 10000 (-b: number of best scores to show, -d: number of best alignments to show, -k: specify the number of shuffles for statistical parameter estimation, -f: penalty for opening a gap, -g: penalty for additional residues in a gap, -m: alignment display options, -q: quiet option, -s: specify substitution matrix, -z: specify statistical calculation, -Z: set the apparent database size used for expectation value calculations). Subsequently, we merged adjacent and overlapping hits in single intergenic regions with BEDTools, obtaining the coordinates of all non-overlapping intergenic segments displaying statistically significant similarity to functional proteins in our data set: bedtools merge -c 2,4,4,4,5 – count, mean, min, max, collapse.
Parsing the SSEARCH alignment outputs previously obtained for each of these merged regions, we calculated the score density produced by each functional protein hitting one or more DNA segments inside a merged region as the summation of the scores obtained by their high scoring pairs (HSPs), divided by the summation of their sequences' length, obtaining normalized scores. We then selected the functional protein sequence producing the highest score density as the unique parent functional protein, simultaneously reconstructing and annotating these intergenic regions. Next, having the HSPs of each reconstructed/annotated intergenic region aligned with their parent functional protein, we scanned the pairwise alignment using the functional protein as a reference, searching for pseudogene-like features in the intergenic sequence, that is, disablements rendered by non-synonymous substitutions, in-frame insertions/deletions, frameshifts, loss of initiation and/or termination codons, and/or internal termination codons, calculating the total number of mutations as the summation of all disablements. Finally, as in silico approaches to annotate genomic sequences traditionally consider a lower limit of 300 nucleotides (100 codons) for an ORF to be claimed as a putative gene (Frith et al. 2006), we decided to apply the same cutoff in our data set of putative novel intergenic CDSs and pseudogenes. All these steps were performed with in-house scripts, using Perl language version 5.10.1.
Codon Usage Comparison
Codon usage tables were computed for CDSs', pseudogenes', and intergenic CDSs' nucleotide sequences encoded in TriTryps' genomes with EMBOSS package version 6.5.7 (Rice et al. 2000), employing the program cusp with default parameters. Pearson's correlation coefficient test was performed with GNU Octave version 5.2.0, comparing the frequencies of 64 codons between pseudogenes and CDSs, and intergenic CDSs and CDSs for each TriTryps' genomes.
Sequence Clustering
We clustered all pseudogene and intergenic CDS sequences separately with CD-HIT version 4.8.1 (Huang et al. 2010), applying a similarity threshold of 80% global identity (number of identical nucleotides in alignment divided by the entire length of the shorter sequence) to find potentially duplicated sequences or multi-sequence families. The clustering algorithm in cd-hit-est is a greedy incremental clustering algorithm described by developers: “…sequences are first sorted in order of decreasing length. The longest sequence becomes the representative of the first cluster. Then, each remaining sequence is compared with the representatives of existing clusters. If the similarity with any representative is above a given threshold, it is grouped into that cluster. Otherwise, a new cluster is defined with that sequence as the representative. For each sequence comparison, short word filtering is applied to the sequences to confirm whether the similarity is below the clustering threshold. If this cannot be confirmed, an actual sequence alignment is performed…” (Huang et al. 2010). Histograms representing the distribution of multi-sequence clusters were produced with GNU Octave version 5.2.0.
Transcriptional Activity Investigation
We obtained RNA-Seq data for Lm, Tb, and Tc from the Sequence Read Archive (SRA) <https://www.ncbi.nlm.nih.gov/sra>, selecting experiments performed with mature mRNAs and informed life-cycle stage of the parasites' sequenced transcriptome. Table 5 describes the 79 experiments we found with these characteristics.
The samples' quality control was performed with FastQC version 0.11.9 <https://www.bioinformatics.babraham.ac.uk/projects/fastqc/> with default parameters. Reads with Phred score >30 were trimmed to remove adapters with Trimommatic version 0.39 (Bolger et al. 2014), applying the following parameters: LEADING: 3, TRAILING: 3, and SLIDINGWINDOW: 4:30. Next, we mapped these reads against the corresponding reference genome sequence with Bowtie2 v2.3.5.1, extracted the uniquely aligning reads (those lacking the tag XS:i, an optional field indicating an alignment score for the best-scoring alignment found other than the alignment reported, in Bowtie2's SAM format output files), and then we used the program featureCounts (Liao et al. 2014) of the Subread package version 2.0.3 <http://subread.sourceforge.net/> to select/count reads mapping the annotated pseudogenes and intergenic CDSs with 100% coverage, applying default parameters in both analyses. The Venn diagram displaying the distribution of transcriptionally active pseudogenes and intergenic CDSs was obtained with InteractiVenn (Heberle et al. 2015).
Supplementary Material
Acknowledgments
This work was supported by the Programa de Apoio à Pesquisa Estratégica em Saúde [PAPES-VI/FIOCRUZ 407654/2012-7]; and the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior [CAPES-UDELAR 046/2013].
Contributor Information
Mayla Abrahim, Laboratório de Tecnologia Imunológica, Instituto de Tecnologia em Imunobiológicos, Vice-Diretoria de Desenvolvimento Tecnológico, Bio-Manguinhos, Fundação Oswaldo Cruz (FIOCRUZ), Rio de Janeiro, RJ, Brazil.
Edson Machado, Laboratório de Biologia Molecular Aplicada a Micobactérias, Instituto Oswaldo Cruz, Fiocruz, Brazil.
Fernando Alvarez-Valín, Unidad de Genómica Evolutiva, Sección Biomatemática, Universidad de la República del Uruguay, Montevideo, Uruguay.
Antonio Basílio de Miranda, Laboratório de Genética Molecular de Microrganismos, Instituto Oswaldo Cruz, Fiocruz, Brazil.
Marcos Catanho, Laboratório de Genética Molecular de Microrganismos, Instituto Oswaldo Cruz, Fiocruz, Brazil.
Supplementary Material
Supplementary material is available at Genome Biology and Evolution online.
Author Contributions
M.C., A.B.M., F.A.V.: study conception, experimental design
M.C., M.A.: investigation, computation
M.C., M.A., F.A.V.: formal analysis
M.C., M.A.: manuscript writing
A.B.M., F.A.V., E.M.: manuscript critical review
Data Availability
The data underlying this article are available in the article and its online supplementary material.
Literature Cited
- Acosta IDCL, Da Costa AP, Gennari SM, Marcili A. 2014. Survey of Trypanosoma and Leishmania in wild and domestic animals in an Atlantic rainforest fragment and surroundings in the state of Espírito Santo, Brazil. J Med Entomol. 51:686–693. [DOI] [PubMed] [Google Scholar]
- Altschul SF, et al. . 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amiri-Dashatan N, Rezaei-Tavirani M, Zali H, Koushki M, Ahmadi N. 2020. Quantitative proteomic analysis reveals differentially expressed proteins in Leishmania major metacyclogenesis. Microb Pathog. 149:104557. [DOI] [PubMed] [Google Scholar]
- Amorim JC, et al. . 2017. Quantitative proteome and phosphoproteome analyses highlight the adherent population during Trypanosoma cruzi metacyclogenesis. Sci Rep. 7:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amos B, et al. . 2022. VEuPathDB: the eukaryotic pathogen, vector and host bioinformatics resource center. Nucleic Acids Res. 50:D898–D911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aslett M, et al. . 2010. TriTrypDB: a functional genomic resource for the Trypanosomatidae. Nucleic Acids Res. 38:457–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avelar GST, et al. . 2020. Diversity and genome mapping assessment of disordered and functional domains in trypanosomatids. J Proteomics. 227:103919. [DOI] [PubMed] [Google Scholar]
- Bairoch A. 2000. The SWISS-PROT protein sequence database and its supplement TrEMBL in 2000. Nucleic Acids Res. 28:45–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berná L, et al. . 2018. Expanding an expanded genome: long-read sequencing of Trypanosoma cruzi. Microb Genom. 4(5):e000177. 10.1099/mgen.0.000177. Epub 2018 Apr 30. PMID: 29708484; PMCID: PMC5994713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berriman M, Ghedin E, Hertz-fowler C. 2005. The genome of the African trypanosome, Trypanosoma brucei. Science 309:416–422. [DOI] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, Usadel B. 2014. Genome analysis Trimmomatic: a flexible trimmer for Illumina sequence data 30:2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner SE, Chothia C, Hubbard TJP. 1998. Assessing sequence comparison methods with reliable structurally. Proc Natl Acad Sci U S A. 95:6073–6078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camacho E, et al. . 2021. Gene annotation and transcriptome delineation on a de novo genome assembly for the reference Leishmania major Friedlin strain. Genes (Basel) 12:1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, et al. . 2020. Re-recognition of pseudogenes: from molecular to clinical applications. Theranostics 10:1479–1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couso JP, Patraquim P. 2017. Classification and function of small open reading frames. Nat Rev Mol Cell Biol. 18:575–589. [DOI] [PubMed] [Google Scholar]
- El-Sayed NM, et al. . 2005. Comparative genomics of trypanosomatid parasitic protozoa. Science 309:404–409. [DOI] [PubMed] [Google Scholar]
- Finn RD, et al. . 2016. The Pfam protein families database: towards a more sustainable future. Nucleic Acids Res. 44:D279–D285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn RD, Clements J, Eddy SR. 2011. HMMER Web server: interactive sequence similarity searching. Nucleic Acids Res. 39:29–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frith MC, et al. . 2006. Discrimination of non-protein-coding transcripts from protein-coding mRNA. RNA Biol. 3:40–48. [DOI] [PubMed] [Google Scholar]
- Griesemer M, Kimbrel JA, Zhou CE, Navid A, D'haeseleer P. 2018. Combining multiple functional annotation tools increases coverage of metabolic annotation. BMC Genomics 19(1):948. 10.1186/s12864-018-5221-9. PMID: 30567498; PMCID: PMC6299973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison PM, Gerstein M. 2002. Studying genomes through the aeons: protein families, pseudogenes and proteome evolution. J Mol Biol. 318:1155–1174. [DOI] [PubMed] [Google Scholar]
- Heberle H, Meirelles GV, da Silva FR, Telles GP, Minghim R. 2015. Interactivenn: a web-based tool for the analysis of sets through Venn diagrams. BMC Bioinformatics 16:169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y, Niu B, Gao Y, Fu L, Li W. 2010. CD-HIT Suite: a web server for clustering and comparing biological sequences. Bioinformatics 26:680–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivens AC, et al. . 2005. The genome of the kinetoplastid parasite, Leishmania major. Science 309:436–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovalenko TF, Patrushev LI. 2018. Pseudogenes as functionally significant elements of the genome. Biochemistry 83:1332–1349. [DOI] [PubMed] [Google Scholar]
- Lavezzo E, Falda M, Fontana P, Bianco L, Toppo S. 2016. Enhancing protein function prediction with taxonomic constraints – the Argot2.5 web server. Methods 93:15–23. [DOI] [PubMed] [Google Scholar]
- Liao Y, Smyth GK, Shi W. 2014. Featurecounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30:923–930. [DOI] [PubMed] [Google Scholar]
- Liu Y, Harrison PM, Kunin V, Gerstein M. 2004. Comprehensive analysis of pseudogenes in prokaryotes: widespread gene decay and failure of putative horizontally transferred genes. Genome Biol. 5:R64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logan-Klumpler FJ, et al. . 2012. GeneDB – an annotation database for pathogens. Nucleic Acids Res. 40:D98–D108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maslov DA, et al. . 2019. Recent advances in trypanosomatid research: genome organization, expression, metabolism, taxonomy and evolution. Parasitology 146:1–27. [DOI] [PubMed] [Google Scholar]
- McCombie WR, McPherson JD, Mardis ER. 2019. Next-generation sequencing technologies. Cold Spring Harb Perspect Med. 9(11):a036798. 10.1101/cshperspect.a036798. PMID: 30478097; PMCID: PMC6824406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller LSM, et al. . 2018. Genome organization and DNA accessibility control antigenic variation in trypanosomes. Nature 563:121–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muro EM, Mah N, Andrade-Navarro MA. 2011. Functional evidence of post-transcriptional regulation by pseudogenes. Biochimie 93:1916–1921. [DOI] [PubMed] [Google Scholar]
- Peacock CS, et al. . 2007. Comparative genomic analysis of three Leishmania species that cause diverse human disease. Nat Genet. 39:839–847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pink RC, et al. . 2011. Pseudogenes: pseudo-functional or key regulators in health and disease ? RNA 17:792–798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pink RC, Carter DRF. 2013. Pseudogenes as regulators of biological function. Essays Biochem. 54:103–112. [DOI] [PubMed] [Google Scholar]
- Plaza S, Menschaert G, Payre F. 2017. In search of lost small peptides. Annu Rev Cell Dev Biol. 33:391–416. [DOI] [PubMed] [Google Scholar]
- Poliseno L. 2014. Pseudogenes functions and protocols. 1. New York, NY: Humana. 10.1007/978-1-0716-1503-4. [DOI] [Google Scholar]
- Poptsova MS, Gogarten JP. 2010. Using comparative genome analysis to identify problems in annotated microbial genomes. Microbiology 156:1909–1917. [DOI] [PubMed] [Google Scholar]
- Quinlan AR, Hall IM. 2010. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26:841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice P, Longden I, Bleasby A. 2000. EMBOSS: The European Molecular Biology Open Software Suite. Trends Genet. 16:276–277. [DOI] [PubMed] [Google Scholar]
- Rost B. 1999. Twilight zone of protein sequence alignments. Protein Eng. 12:85–94. [DOI] [PubMed] [Google Scholar]
- Simpson AGB, Stevens JR, Lukeš J. 2006. The evolution and diversity of kinetoplastid flagellates. Trends Parasitol. 22:168–174. [DOI] [PubMed] [Google Scholar]
- Singh RK, Singh D, Yadava A, Srivastava AK. 2020. Molecular fossils “pseudogenes” as functional signature in biological system. Genes Genomics. [DOI] [PubMed] [Google Scholar]
- Toh JY, et al. . 2021. Identification of positive and negative regulators in the stepwise developmental progression towards infectivity in Trypanosoma brucei. Sci Rep. 11:1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen Y-Z, et al. . 2011. Pseudogene-derived small interference RNAs regulate gene expression in African Trypanosoma brucei. Proc Natl Acad Sci U S A. 108:8345–8350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao J, et al. . 2016. Pseudogenes and their genome-wide prediction in plants. Int J Mol Sci. 17(12):1991. 10.3390/ijms17121991. PMID: 27916797; PMCID: PMC5187791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng D, Gerstein MB. 2006. A computational approach for identifying pseudogenes in the ENCODE regions. Genome Biol. 7(Suppl 1):S13.1-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng D, Gerstein MB. 2007. The ambiguous boundary between genes and pseudogenes: the dead rise up, or do they? Trends Genet. 23:219–224. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data underlying this article are available in the article and its online supplementary material.