Abstract
Background
Detection of seroconversion after SARS-CoV-2-infection or vaccination is relevant to discover subclinical cases and recognize patients with a possible immunity.
Objectives
Test performance, effects of age, time-point of seroconversion and immune status regarding neutralizing antibodies (NAbs) and T-cell-reactivity were investigated.
Study design
Two antibody assays (Viramed-Test for S/N-specific IgG, Roche-Test for N-specific IgA, -M, -G) were evaluated with classified samples. In total, 381 subjects aged 6-99 years, who had either recovered from the disease or had been vaccinated, were screened for SARS-CoV-2-specific antibodies. This screening was part of an open observational study with working adults. Additionally, children and adults were analyzed in a longitudinal COVID-19 study in schools. For immunity evaluation, virus neutralization tests and ELISpot tests were performed in a subgroup of subjects.
Results
Viramed revealed a slightly lower test performance than Roche, but test quality was equally well in samples from very young or very old donors. The time-point of seroconversion after the respective immunization detected by the two tests was not significantly different. N-specific antibodies, detected with Roche, highly correlated with NAbs in recovered subjects, whereas a positive Viramed-Test result was paralleled by a positive ELISpot result.
Conclusion
Viramed-Test was not as sensitive as Roche-Test, but highly specific and beneficial to distinguish between recovered and vaccinated status. For both tests correlations with humoral and cellular immunity were found. Of note, the expected early detection of IgA and IgM by the Roche-Test did not prove to be an advantage over IgG testing by Viramed.
Keywords: SARS-CoV-2-specific antibodies, Seroconversion, Test performance, Neutralizing antibodies, T-cell-reactivity
Abbreviations
- Ig
immune-globulin
- N
nucleocapsid protein
- n
number
- NAbs
neutralizing antibodies
- PCR
polymerase-chain-reaction
- RBD
receptor-binding-domain
- S
spike-glycoprotein
- SARS-CoV-2
Severe-Acute-Respiratory-Syndrome-Coronavirus-Type-2
- VNT
virus neutralization test
- T-cells
T-lymphocytes
- ELISpot-test
Enzyme-Linked-Immuno-Spot-test
- COI
cutoff index
1. Background
During the SARS-CoV-2 (Severe-Acute-Respiratory-Syndrome-CoronaVirus-Type-2)-pandemic, serological tests were used to detect SARS-CoV-2 asymptomatic infection, thus providing additional information about virus prevalence [1].
The SARS-CoV-2 structure proteins spike-glycoprotein (S) and nucleocapsid-protein (N) are immune dominant. S - mediating the virus cell-entry - consists of subunits S1 with the receptor-binding-domain (RBD) and S2, responsible for virus replication. Whereas S1- and N-antibodies are specific and sensitive for SARS-CoV-2, S2-antibodies are judged less appropriate to describe the immune response [2] due to its high sequence homology with the seasonal “common cold” CoVs and thus cross-reactivity. N is used to differentiate immune responses triggered by previous infection from those triggered by vaccination, as vector vaccines only contains the spike protein. S1 is assumed to represent the target for neutralizing antibodies (NAbs) elicited by currently used vaccines [3].
Generally, specific immune-globulins (Ig) correlate with disease activity, possibly compromising diagnostic accuracy [4]. IgA and -M represent an early defense line within a virus-related immune response before generation of specific IgG or T-lymphocytes (T-cells) [5], and rise three to six days after onset of COVID-19-typical symptoms [5], [6], [7] up to a maximum approximately six weeks after infection. RBD-specific IgM starts to decrease ∼60 days after symptoms to a low level of 22% [8]. In contrast, IgG-antibodies can be detected over several months. N-specific IgG was observed to decrease 120 days after infection onset to 15%, while S-specific IgG rests at a 80%-level in parallel to avidity maturation and presence of NAbs [8].
As a special challenge for diagnostic, antibody titers in children and adolescents (5–21 years) with SARS-CoV-2-infection were shown to be low compared to adults, even though 92.3% of these children produced NAbs [9]. A lower immune response was also reported for seniors aged 60+ years [10]. After vaccination, an S- and RBD-specific antibody reaction is expected, but protection level are not yet defined [11,12]. Different test principles are available for SARS-CoV-2 antibody testing with a possible impact on test performance [13]. Crucial parameters are the recombinant virus antigen, the targeted Ig class, and the sensitivity of the respective biotechnological principle used [14].
2. Objectives
Antibody screening was critical during SARS-CoV-2-pandemic to identify recovered or successfully vaccinated individuals in the general population including children and seniors. This study had two objectives. The first aim was to show, which serological assay was most suitable to detect SARS-CoV-2-specific antibodies after infection or vaccination in general: i) electro-chemiluminescence immunoassay from Roche Diagnostics (detecting N-specific IgM-, IgG-antibodies; well evaluated before, e.g. [15]), ii) microarray-based immunoassay from Viramed Biotech AG (detecting S1-, S2- and N-specific IgG-antibodies). Due to this, assay performance of these serological assays for detecting virus-specific antibodies were evaluated using serum samples from 381 subjects, aged 6–99 years, who had either recovered from the disease or had been vaccinated and therefore should show seroconversion. Since the very presence of virus-specific antibodies is not suitable to give insight concerning an assumed immunity to SARS-CoV-2 upon infection or vaccination, the study further investigated possible correlations between S- and N-specific antibodies and the occurrence of neutralizing antibodies (NAbs) and T-cell-reactivity. Thus, it should be investigated which of those assays, best represents reality - the second aim of this study.
3. Study design
3.1. Antibody detection tests and initial assay verification
The electro-chemiluminescence immunoassay Elecsys® Anti-SARS-CoV-2 (Roche Diagnostics, “Roche-Test”) detecting N-antigen-specific IgM/IgA-antibodies and the microarray-based enzyme immunoassay Anti-SARS-CoV-2 ViraChip® IgG (Viramed Biotech AG, “Viramed-Test”) detecting S1-, S2- and N-antigen-specific antibodies were used as described in supplemental material and elsewhere [1].
Roche- and Viramed-Test were initially evaluated regarding analytical sensitivity, specificity, and interassay precision with the following verification samples: i) 192 samples from German Red Cross blood donation center in Lower Saxony, Germany (collected before 2019), ii) 33 samples of SARS-CoV-2-antibody-positive persons after polymerase-chain-reaction (PCR)-confirmation (classified using Roche-Test) from Medical Care Center (MVZ)-Labor Dr. Limbach in Heidelberg, Germany, and iii) 30 samples of SARS-CoV-2-antibody-tested persons (classified using LIAISON SARS-CoV-2 S1/S2 IgG assay, DiaSorin; 12 positive, PCR-confirmed/18 negative samples) from MVZ-Labor Limbach, Lehrte, Germany. In three independent measurements 47 samples (18 positively classified samples, 29 negatively classified samples) were analyzed. Assay verification of both tests met acceptance criteria, since more than 93% of samples were correctly detected (Table S1, detailed Viramed-Test results for each antigen in Table S2).
3.2. ELISpot-test for T-cell-reactivity and test for virus-NAbs
Enzyme-Linked-Immuno-Spot (ELISpot)-test (Immunospot, Cleveland, US) was carried out in pre-coated 96-well plates (Mabtech) according to the manufacturer's instructions (supplement) with a response of >15 SFU/106 considered positive. Virus neutralization test (VNT) was also performed as described before [16,1,17].
3.3. Subject cohorts and ethics vote
Serum samples were collected in two large cohorts, subdivided into two subgroups each (recovered and vaccinated subjects). Fig. 1 and Table S3 give an overview of all subgroups analyzed. Antibody testing was offered to subjects at intervals of a few months. Clinical data were collected in a pseudonymized form after informed consent in parallel to serum collection [1,18]. While fresh serum was used for cohort 1, in cohort 2 test-laboratory was not located close to blood collection site. Therefore, after collection, frozen samples were brought to the laboratory, and had to be thawed overnight at 4 °C before testing.
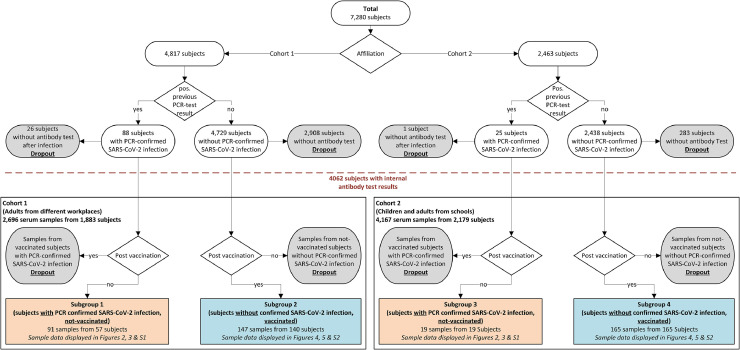
Fig. 1.
Schematic displaying subjects and samples classified into two cohorts and four subgroups based on their study affiliation and recovered or vaccination status. For this study (rectangular frames), either 110 samples from 76 not-vaccinated subjects with PCR-confirmed SARS-CoV-2 infection (subgroups 1 & 3, highlighted orange) or 312 samples from 305 vaccinated subjects without PCR-confirmed SARS-CoV-2 infection (subgroups 2 & 4, highlighted blue) were considered for subgroups. Details for exclusion from evaluation (dropout, gray bubbles): 27 subjects with a positive PCR-test did not participate in antibody screening; samples from subjects which had a PCR-confirmed SARS-CoV-2 infection and at least one shot of vaccine at date of sample collection; samples from not-vaccinated subjects without PCR-confirmed infection.
Serum samples of cohort 1 were collected during an open observational study in Lower Saxony, Germany, between August 2020 and June 2021 [1]. No study subject has given information on known immune dysfunctions and/or taking immunosuppressant drugs (Table S4). Subgroup 1 consisted of 57 subjects, recovered after a PCR-confirmed SARS-CoV-2 infection (predominantly the B.1.617.2 variant, determined by PCR melting curve analysis using the VirSNiP SARS-CoV-2 Spike Kits 484A 486 V (Delta) as well as S371L S373P (BA1 and BA2) from Tib MolBiol (Berlin, Germany)), and subgroup 2 included 140 subjects, that were analyzed after first shot of vaccination (Biontech/Pfizer: 120, Moderna: 3, AstraZeneca: 17) [19]. Since sample collection was scheduled in advance and vaccination was often performed spontaneously (especially during the early stages of vaccine availability), there was no set period between vaccination and serum collection. Within subgroup 1 and 2, 30 subjects were tested twice for seroconversion. VNT- and ELISpot-tests were performed in recovered (nVNT = 56; nELISpot = 45) and vaccinated subjects (nVNT = 36; nELISpot = 88) with seroconversion.
Cohort 2 included 1180 pupils aged 6–17 years, but also parents, and teachers, out of a longitudinal study in Hannover, Germany, between June 2020 and June 2021 [18]. Subgroup 3 consisted of 19 recovered subjects, subgroup 4 consisted of 165 subjects, among those 56 pupils aged 6–17 years, analyzed after first shot of vaccination with Biontech/Pfizer. Here, no subjects in subgroup 3 and 4 provided more than one serum sample.
Ethical approval was given for cohort 1 by ”Aerztekammer Niedersachsen” in August 2020 (No. Bo/30/2020; Bo/31/2020; Bo/32/2020; amendment for the vaccinated subjects in November 2021), for cohort 2 by Institutional Review Board of the Medical School Hannover (No. 9085_BO_S_2020).
3.4. Statistical methods
Descriptive statistics are indicated. Wilcoxon ranked test was used to compare negative and positive samples of different age groups and groups sorted by time point of diagnosis as well as for age- and time-point-sorted groups of negatively and positively tested subjects as indicated with p < 0.05 for significance.
4. Results
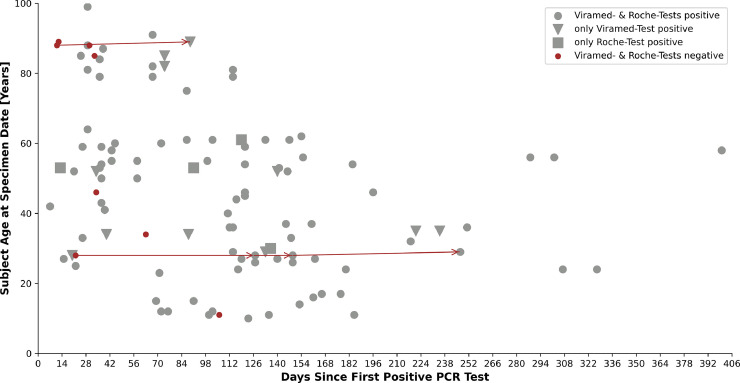
Analyzing 110 serum samples of 76 recovered subjects (subgroup 1: 57 subjects; subgroup 3: 19 subjects), the assays detected antibodies in all samples (Viramed-Test: 98 cases; Roche-Test: 91 cases). Both tests detected evidence of seroconversion between 7 and 400 days following initial positive PCR-test. N-specific seroconversion in recovered subjects was detected solely with N-specific Roche-Test until day 119 and solely with an S-specific Viramed-result until day 154. In contrast, a consistent positive test result was present until day 400 (Figs. 2 , 3 ). The time-point of test performance was highly significant for classification of result: negative results according to Roche-Tests (n = 19) were obtained at 76.8 ± 65.9 days after PCR-confirmed infection (Figure S1), positive Roche-results (n = 91) were measured significantly later (Fig. 2) (p < 0.001 according to Wilcoxon) at 111.6 ± 76.6 days after infection. According to Viramed-Test, negative results (n = 12) were found significantly earlier at 55.8 ± 45.5 days after a confirming PCR (Figure S1) than the 98 positive Viramed-results at 111.6 ± 76.5 days after infection (p < 0.001 according to Wilcoxon) (Fig. 2). Although Roche-Test also covers IgA and IgM, there was no significant difference in time-dependent test performance between the tests (not shown). Viramed-Test, which includes N- and S-specific antibodies, was even found to detect seroconversion as early as Roche (Fig. 2, tests indicated by different symbols).
Fig. 2.
All test results of both antibody tests in a time interval (in days) after PCR-confirmation of a SARS-CoV-2 infection. Red dots symbolize negative test results in both tests, gray dots positive results in both tests, triangles symbolize samples tested positive only with the Viramed-Test, and squares symbolize positive test results gained only with the Roche-Test. Arrow lines connect samples from two subjects that were initially tested negative with both test systems. However, both subjects displayed seroconversion during later tests. The six additional samples that tested negative with both test systems were obtained from subjects without additional samples provided.
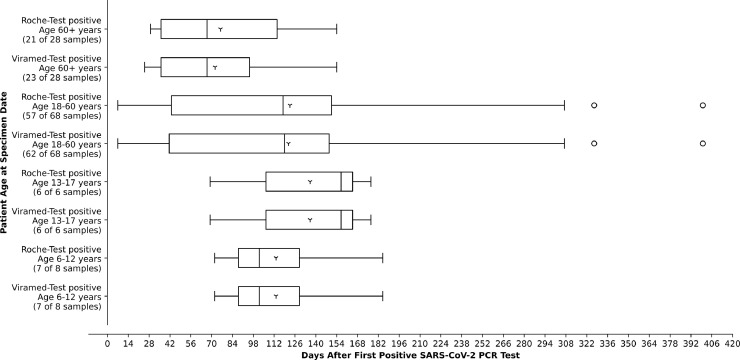
Fig. 3.
Age dependency of antibody detection in subjects with a PCR-confirmed SARS-CoV-2 infection. Age groups comprise 6–12 years, 13–17 years, 18 to 60 years, and over 60 years. They were sorted by the test used (Viramed or Roche). The number of positive samples out of all samples tested in each age group is indicated.
Both tests displayed a comparable result in very young (6–12 years) and juvenile subjects (12–17 years). In middle-aged subjects (18–60 years) and subjects aged 60+ years, Viramed detected seroconversion more often than Roche (Fig. 3). Viramed-Test identified seroconversion in age group 60+ years up to day 154 after immunization; seroconversion here occurred significantly earlier at 72.3 ± 40.0 days after PCR-confirmation than in all other tested subjects (123.81 ± 80.9 days; p < 0.003 according to Wilcoxon).
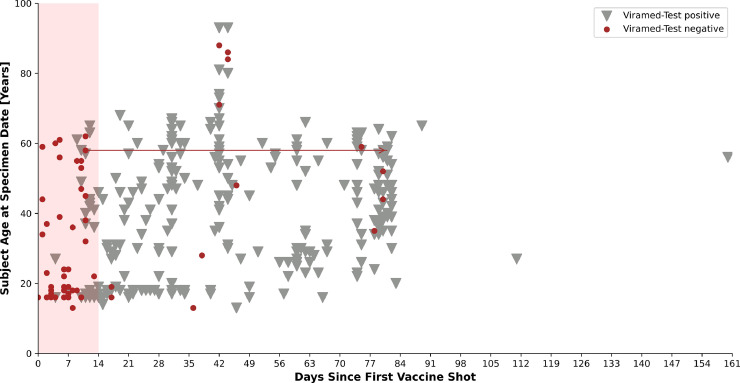
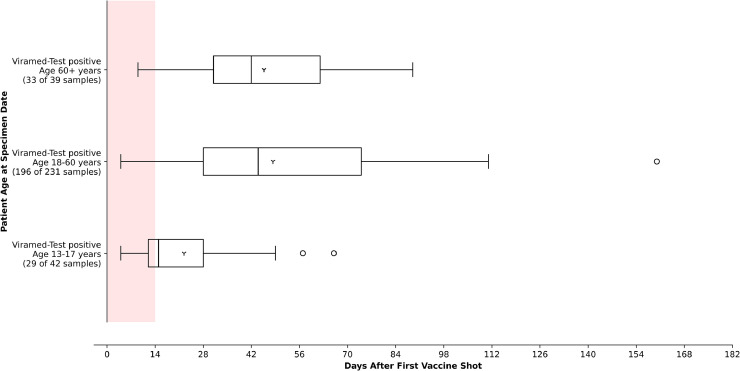
Viramed-Test identified a seroconversion in 252/305 vaccinated subjects of subgroups 2 and 4 (Fig. 4 ). In juvenile subjects up to 17 years, immune response occurred significantly earlier than expected (day 4–66 after vaccination, p < 0.01 according to Wilcoxon) than in all other groups (Fig. 5 ). In the juvenile group, 13/42 samples were tested negative, whereas this was the case in 35/231 samples of middle-aged subjects and only 6/39 samples in group 60+ years.
Fig. 4.
Time of test performance after the first shot of vaccination of the Viramed-Tests, with positive detection of S1/S2-specific antibodies until day 160. The colored area depicts the time interval in which, at least for Biontech-vaccinated, no humoral immunity to SARS-CoV-2 spike protein was expected (until day 14 after first vaccination) [19]. Distributed all over the age groups, there were samples with a negative test result belonging to 13 different subjects (age range 18–86 years, median 47 years) after this 14 day interval. The two samples connected by an arrow indicate results of a subject that was initially tested negative but showed seroconversion at a later test.
Fig. 5.
Age group-sorted positive Viramed-Test results (S1/S2-specific antibodies after first shot of vaccination). The colored area depicts the time interval in which, at least for Biontech-vaccinated, no seroconversion was expected (until day 14 after first vaccination) [19].
Most of the negative samples of vaccinated subjects aged 13–60 years were collected predominantly in a very early time interval, in which seroconversion after vaccination could not be expected [19]. In contrast, negative test results were observed for older subjects 60+ years, in a time interval up to 44 days after the first shot of vaccination (Figure S2). Negative Viramed-Test results were observed early in 8.8% of all recovered 60+ years individuals and later in recovered middle-aged subjects (17.9%; Figure S1). Negative results of N-specific Roche-Test were obtained in 16.2% of middle-aged and 25% of 60+ year subjects. A non-responder was observed out of eight subjects younger than 12 years with negative results in both tests 106 days after positive PCR. Above these observations, there was no significant influence of the factor age on the classification of results according to Wilcoxon (results not shown). We observed eight non-responders after infection and 13 non-responders after vaccination with antibody detection performed at least 14 days after the first vaccination shot (Table S5). In addition, a Cohens’ kappa correlation test indicating interrater-reliability between the different assays revealed a value of only 0.4 among all samples of all cohorts tested in this work.
Whether or not a positive result of either Viramed- or Roche-Test indicates a possible SARS-CoV-2-immunity by either infection or vaccination was tested upon determination of NAbs and T-cell-reactivity in seroconverted subjects of cohort 1; among those 73 subjects tested positive with Viramed (41 recovered, 32 vaccinated), and 38 recovered subjects tested positive with Roche (Table 1 ). Generally, T-cell-reactivity was present 15–302 days after immunization, and NAbs occurred 7–400 days (not shown). Depending on age, between 75% and 82% of all samples from these Viramed-defined 73 seroconverted subjects were positive for T-cell-reactivity, and above 90% were positive for NAbs (Table 1 and 2 ). S-specific antibodies were detected in 70/73 subjects, and 3/73 had solely N-specific antibodies according to Viramed-Test. Again, depending on age, Roche-Test recognized 78-90% of recovered subjects with signs of T-cell-reactivity and >92% of subjects with NAbs. Detection of a full pattern with N-, S1-, and S2-specific antibodies by the Viramed-Test more often correlated with T-cell-reactivity than with NAbs and was more frequent in subjects over 18 years (Table 2).
Table 1.
36 respective 32 either recovered or vaccinated subjects with comparable age structure were identified as seroconverted by antibody tests and were also tested by ELISpot for possible T-lymphocyte (T-cell) reactivity against SARS-CoV-2 and for Nabs (virus neutralization test, VNT). n.a.: not available (vaccination does not trigger N-antibody production).
| Status | Both assays | Viramed | Roche | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Number (subjects) | T-cell-reactivity | VNT | Number (subjects) | T-cell- reactivity | VNT | Number (subjects) | T-cell- reactivity | VNT | |
| Recovered | 36 | 80.56% | 97.22% | 41 | 78.05% | 90.24% | 38 | 73.17% | 94.74% |
| Age [years] (median; range) | 51; 22–99 | 52; 22–99 | 53; 22–99 | ||||||
| Vaccinated | 32 | 81.25% | 93.75% | 32 | 81.25% | 93.75% | n. a. | n. a. | n. a. |
| Age [years] (median; range) | 55; 26–93 | 55; 26–93 | n. a. | ||||||
Table 2.
Seropositive adult subjects in different age groups (median, range of years); percentage of additional T-lymphocyte (T-cell)-reactivity and virus neutralization test (VNT) in the number of subjects tested. n.a.: not available (vaccination does not trigger N-antibody production).
| Viramed | Roche | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Age group | Number of subjects (n; median age; range age) | Antibody signatures | T-cell- reactivity | VNT | Number of subjects (n; median age; range age) | Antibody signatures | T-cell- reactivity | VNT | |
| 18–60 years; recovered subjects | 29; 29; 22–59 |
18x N, S1, S2; 7x N, S1; 3x N; 1x S1, S2 |
79.31% | 89.66% | 27; 43; 22–59; |
18x N, S1,S2; 7x N, S1; 2x N; |
77.78% | 92.59% | |
| 18–60 years; subjects after vaccination | 21; 47; 26–59; |
18x S1,S2; 3x S1 |
80.95% | 95.24% | n.a. | n.a. | n.a. | n.a. | |
| 60+ years; recovered subjects | 12; 81.5; 61–99; |
8x N,S1,S2; 2x N,S1; 1x S1, S2; 1x S1 |
75,00% | 91.67% | 11; 79; 61–99 |
8x N, S1, S2; 2x N, S1; 1x N |
90% | 100% | |
| 60+ years; subjects after vaccination | 11; 73; 61–93; | 8x S1,S2; 3x S1 |
81.82% | 90.91% | n.a. | n.a. | n.a. | n.a. | |
34/110 samples (∼31%) obtained from recovered subjects showed discrepant test results concerning N-specific antibodies. In ten recovered subjects, this divergence persisted across all serum samples taken over time (Table S6), whereas in the other subjects, this divergence did not reappear in later analyses. Among the former, there were two subjects with no seroconversion according to Roche-Test (no N-specific antibodies). These also had no NAbs and one subject showed no signs of T-cell-reactivity. 7/8 subjects with a negative Viramed-Test but a positive Roche-Test for N-specific antibodies had positive VNTs, and 6/8 also had a positive T-cell-test.
5. Discussion
Altogether, this study showed comparatively higher sensitivity and specificity for both assays than reported by other groups [20], [21], [22], [23]. For Viramed-Test, this could be caused by the larger cohorts presented here compared to previously reported collectives. Interassay precision of Viramed-Test was comparably lower than of Roche-Test. The comparably more sensitive Roche-Test correlated better with positive NAbs results in a number of subjects tested, but Viramed-Test - also detecting S-specific antibodies - was highly associated with T-cell-reactivity [24].
Divergences concerning N-antibody detection were observed in 31% of all samples from recovered subjects. False-negative results of Viramed-Test may be due to the fact, that this test covers only IgG, not IgM or IgA. Another cause for this could be the different molecular technology principles used in the assays. The antigen of interest is initially free in solution in Roche-Test, whereas it is immobilized in Viramed-Test with a possible negative impact on its conformation and binding capacity of antibodies and thus test sensitivity. In all, we found more cases with only S-positivity than only N-positivity in Viramed-Test after infection; thus we assume this antibody signature as more sustainable [25].
The assays perform relatively equally in different age groups and time intervals after infection. Since detection windows were the same for both tests presented here, we can underline the observation of others that assays detecting also IgM and IgA have no added value for early diagnosis [26]. Possibly, due to its broader antigen-specificity, Viramed-Test detected seroconversion in more subjects than Roche-Test. According to Cohens´s kappa value, the interassay correlation was low at 0.4, which is explainable by the different immune globulin classes and antibody specificities covered by these two tests. In addition, this low correlation value confirms the observation of others that occurrence of N- and S-specific antibodies, as well as IgM and IgG, show high inter-subject differences after SARS-CoV-2-infection [8].
A limitation but also an opportunity is the random driven design of the presented study as part of an open observational study as well as of a longitudinal study. Subjects at different affiliations were invited for regular, voluntary PCR-tests [1,18]. Later on subjects participating the PCR-tests were invited for voluntary antibody-screenings at least at two different time points. As summarized in Fig. 1 3218/7280 subjects did not participate in the antibody-screening. Especially in the first month of SARS-CoV-2-pandemic the number of PCR-confirmed infections was low and thus the number of recovered subjects in this study was 76 (subgroup 1: 57 subjects; subgroup 3: 19 subjects). Furthermore, only 30 subjects in subgroup 1 and 2 and no subjects in subgroup 3 and 4 were tested more than once for seroconversion (Table S3), resulting in a total of 110 serum samples of 76 recovered subjects. But there was no standardized time period between sample collections throughout the study. Since sample collection was scheduled in advance and vaccination was often performed spontaneously (especially during the early stages of vaccine availability), there also was no set period between vaccination and serum collection. This increased variety of subjects and samples of this study. Due to the design of the longitudinal study at different schools (cohort 2) no VNT and ELISpot-tests were performed with samples of subgroups 3 and 4, thus this study is lacking assumptions to immunity to SARS-CoV-2 especially for children.
As expected, negative tests were obtained significantly earlier than positive tests, consistent with the period in which humoral immunity builds based on activation of virus-specific B-lymphocytes after SARS-CoV-2-infection. Of note, recovered subjects aged 60+ years had the fastest humoral immune response, but its sustainability is unclear, since these subjects received vaccine before further immune analyses could be performed. However, negative test results were observed until day 44 after infection in this age group, which may reflect immunosenescence (6). S-antibodies occurred comparatively earlier than N-specific antibodies in recovered subjects of all cohorts described in this work.
Viramed-Test performance detected seroconversion after vaccination within the expected periods [19]. Of note, juvenile vaccinated subjects showed seroconversion already within the first days after receiving first vaccine dose.
Antibody screening tests such as those presented here are often taken as possible predictors of general immunity. Therefore, we correlated antibody assays with VNT-results and ELISpot-testing in a subgroup analysis. ELISpot-testing does not reflect T-cell-subtypes and thus the precise quality of the immune response. Our observations of late NAbs coincide with a study by McCallum et al. [27], who described diverse antibodies derived from memory-B-cells interacting with the S-protein of SARS-CoV-2. S-specific antibodies also show a high timely correlation with T-cell-reactivity [24]. Although others have shown that N-specific antibodies drop down early within months [28], our results showed detectable N-specific antibodies up to 400 days after infection. N-specific antibodies (detected by Roche-Test) occurred in parallel to S1-specific antibodies (detected by Viramed-Test) and to NAbs, that are presumably represented by these S1-specific antibodies [8].
In conclusion, we recommend a complete analysis using antibody screening tests covering all antigen-specificities to be able to i) recognize seroconversion early after infection or vaccination, ii) distinguish solely vaccinated from recovered and vaccinated subjects, and iii) have a high probability of predicting sustainable immunity. The Viramed-Test evaluation presented in this work meets this recommendation.
Data statement
De-identified subject datasets will be available upon written request to the corresponding author following publication.
Funding
The MCA study (cohort 1) was financed by state funds from the Ministry of Economics of Lower Saxony. A.M., M.S., A.W., M.P. and P.-C. P. were additionally funded by the Ministry of Social Affairs, Health and Equality of Lower Saxony (cohort 2 study). The respective sponsor did not exert any influence or make any recommendation as to which groups of people should be tested. The offer of testing was requested by various institutions or groups of persons themselves.
The work at the Research Center for Emerging Infections and Zooneses was financially supported by the Ministry of Research and Culture of Lower Saxony (14-76103-184 CORONA-15/20) as well as COFONI and MWK for A.O. and M.G.H. G.S., and G.F.R. were further supported by the Alexander von Humboldt Foundation in the framework of the Alexander von Humboldt Professorship endowed by the German Federal Ministry of Education and Research. In addition, F.K.K. was funded by the DFG (German Research Foundation) - 398066876/GRK 2485/1 (VIPER); A.D.M.E.O. and G.F.R. were funded by RESIST cluster of excellence (EXC 2155, Project number 390874280), and the European Union's Horizon 2020 research and innovation program ISOLDA (Grant n. 848166).
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
A big thank you goes to Dr. Martin Kintrup and his team from Viramed Biotech AG for their advice and support during implementation of their test system in our observational study infrastructure. We thank several PhD students and coworkers at the Institutes of Technical Chemistry and Microelectronic Systems (Leibniz University Hannover, Germany) for intense work on both PCR and antibody testing, as well as technical and organizational support in this project. Furthermore, we thank the medical and dental students from the Medical School Hannover (MHH) and from the university in Goettingen for their support registering subjects and taking swabs. We thank Prof. Dr. Nils Hoppe and his team for their support of the study's ethical vote and procedures with regard to data protection (centre of Ethics and Law in the Life Sciences, Leibniz University, Hannover, Germany).
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.jcv.2022.105322.
Appendix. Supplementary materials
References
- 1.Jonczyk R., Stanislawski N., Seiler L.K., Blume H., Heiden S., Lucas H., et al. Combined prospective seroconversion and PCR data of selected cohorts indicate a high rate of subclinical SARS-CoV-2 infections—an open observational study in lower Saxony. Germany. Microbiol. Spectrum. 2022;10:e01512–e01521. doi: 10.1128/spectrum.01512-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stringhini S., Wisniak A., Piumatti G., Azman A.S., Lauer S.A., Baysson H., et al. 2020. Seroprevalence of Anti-SARS-CoV-2 IgG Antibodies in Geneva, Switzerland (SEROCoV-POP): a population-based study. The Lancet 396:313–319. doi:10.1016/S0140-6736(20)31304-0. [DOI] [PMC free article] [PubMed]
- 3.National Center for Immunization and Respiratory Diseases. 2022. Interim Guidance For Antigen Testing For SARS-CoV-2. https://stacks.cdc.gov/view/cdc/113645. Accessed May 4th.
- 4.Mazzini L., Martinuzzi D., Hyseni I., Benincasa L., Molesti E., Casa E., et al. Comparative analyses of SARS-CoV-2 binding (IgG, IgM, IgA) and neutralizing antibodies from human serum samples. J. Immunol. Methods. 2021;489 doi: 10.1016/j.jim.2020.112937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sette A., Crotty S. Adaptive immunity to SARS-CoV-2 and COVID-19. Cell. 2021;184:861–880. doi: 10.1016/j.cell.2021.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Guo L., Ren L., Yang S., Xiao M., de Chang, Yang F., et al. Profiling early humoral response to diagnose novel coronavirus disease (COVID-19) Clin. Infect. Dis. 2020;71:778–785. doi: 10.1093/cid/ciaa310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lou B., Li T.-.D., Zheng S.-.F., Su Y.-.Y., Li Z.-.Y., Liu W., et al. Serology characteristics of SARS-CoV-2 infection after exposure and post-symptom onset. Eur. Respir. J. 2020;56 doi: 10.1183/13993003.00763-2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Scheiblauer H., Nübling C.M., Wolf T., Khodamoradi Y., Bellinghausen C., Sonntagbauer M., et al. Antibody response to SARS-CoV-2 for more than one year - kinetics and persistence of detection are predominantly determined by avidity progression and test design. J. Clin. Virol. 2022;146 doi: 10.1016/j.jcv.2021.105052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Szépfalusi Z., Schmidthaler K., Sieber J., Kopanja S., Götzinger F., Schoof A., et al. Lessons from low seroprevalence of SARS-CoV-2 antibodies in schoolchildren: a cross-sectional study. Pediatr. Allergy Immunol. 2021;32:762–770. doi: 10.1111/pai.13459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Montecino-Rodriguez E., Berent-Maoz B., Dorshkind K. Causes, consequences, and reversal of immune system aging. J. Clin. Invest. 2013;123:958–965. doi: 10.1172/JCI64096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fehr A.R., Perlman S. Coronaviruses: an overview of their replication and pathogenesis. Methods Mol. Biol. 2015;1282:1–23. doi: 10.1007/978-1-4939-2438-7_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nam M., Seo J.D., Moon H.-.W., Kim H., Hur M., Yun Y.-.M. Evaluation of humoral immune response after SARS-CoV-2 vaccination using two binding antibody assays and a neutralizing antibody assay. Microbiol. Spectrum. 2021;9 doi: 10.1128/Spectrum.01202-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Loeffelholz M.J., Tang Y.-.W. Laboratory diagnosis of emerging human coronavirus infections - the state of the art. Emerg. Microbes Infect. 2020;9:747–756. doi: 10.1080/22221751.2020.1745095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pflüger L.S., Bannasch J.H., Brehm T.T., Pfefferle S., Hoffmann A., Nörz D., et al. Clinical evaluation of five different automated SARS-CoV-2 serology assays in a cohort of hospitalized COVID-19 patients. J. Clin. Virol. 2020;130 doi: 10.1016/j.jcv.2020.104549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jochum S., Kirste I., Hortsch S., Grunert V.P., Legault H., Eichenlaub U., et al. Clinical utility of Elecsys anti-SARS-CoV-2 S assay in COVID-19 vaccination: an exploratory analysis of the mRNA-1273 phase 1 trial. medRxiv. 2021 doi: 10.1101/2021.10.04.21264521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang X., Lam J.-.Y., Wong W.-.M., Yuen C.-.K., Cai J.-.P., Au S.W.-.N., et al. Accurate diagnosis of COVID-19 by a novel immunogenic secreted SARS-CoV-2 orf8 protein. MBio. 2020;11 doi: 10.1128/mBio.02431-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schulz C., Martina B., Mirolo M., Müller E., Klein R., Volk H., et al. 2021. SARS-CoV-2-specific antibodies in domestic cats during first COVID-19 wave, Europe. Emerg. Infect. Dis. 27:3115–3118. doi:10.3201/eid2712.211252. [DOI] [PMC free article] [PubMed]
- 18.Paulsen M., A Zychlinsky Scharff, de Cassan K, Sugianto R.I., Blume C., Blume H., et al. Children and adolescents’ behavioral patterns in response to escalating COVID-19 restriction reveal sex and age differences. J. Adolescent Health. 2022;70:378–386. doi: 10.1016/j.jadohealth.2021.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Amit S., Regev-Yochay G., Afek A., Kreiss Y., Leshem E. Early rate reductions of SARS-CoV-2 infection and COVID-19 in BNT162b2 vaccine recipients. The Lancet. 2021;397:875–877. doi: 10.1016/S0140-6736(21)00448-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chan C.W., Parker K., Tesic V., Baldwin A., Tang N.Y., van Wijk X.M.R., et al. Analytical and clinical evaluation of the automated Elecsys anti-SARS-CoV-2 antibody assay on the Roche cobas e602 analyzer. Am. J. Clin. Pathol. 2020;154:620–626. doi: 10.1093/ajcp/aqaa155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Favresse J., Eucher C., Elsen M., Tré-Hardy M., Dogné J.-.M., Douxfils J. Clinical performance of the Elecsys electrochemiluminescent immunoassay for the detection of SARS-CoV-2 total antibodies. Clin. Chem. 2020;66:1104–1106. doi: 10.1093/clinchem/hvaa131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jahrsdörfer B., Kroschel J., Ludwig C., Corman V.M., Schwarz T., Körper S., et al. Independent side-by-side validation and comparison of 4 serological platforms for SARS-CoV-2 antibody testing. J. Infect. Dis. 2021;223:796–801. doi: 10.1093/infdis/jiaa656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Krüttgen A., Cornelissen C.G., Dreher M., Hornef M., Imöhl M., Kleines M. Comparison of four new commercial serologic assays for determination of SARS-CoV-2 IgG. J. Clin. Virol. 2020;128 doi: 10.1016/j.jcv.2020.104394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Palacios-Pedrero M.Á., Jansen J.M., Blume C., Stanislawski N., Jonczyk R., Molle A., et al. Age-related signs of immunosenescence correlate with poor outcome of mRNA COVID-19 vaccination in older adults. Nat. Aging. 2022 doi: 10.1038/s43587-022-00292-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Atyeo C., Fischinger S., Zohar T., Slein M.D., Burke J., Loos C., et al. Distinct early serological signatures track with SARS-CoV-2 survival. Immunity. 2020;53:524–532. doi: 10.1016/j.immuni.2020.07.020. e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Coste A.T., Jaton K., Papadimitriou-Olivgeris M., Greub G., Croxatto A. Comparison of SARS-CoV-2 serological tests with different antigen targets. J. Clin. Virol. 2021;134 doi: 10.1016/j.jcv.2020.104690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McCallum M., de Marco A, Lempp F.A., Tortorici M.A., Pinto D., Walls A.C., et al. N-terminal domain antigenic mapping reveals a site of vulnerability for SARS-CoV-2. Cell. 2021;184:2332–2347. doi: 10.1016/j.cell.2021.03.028. e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lumley S.F., Wei J., O'Donnell D., Stoesser N.E., Matthews P.C., Howarth A., et al. The duration, dynamics, and determinants of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) antibody responses in individual healthcare workers. Clin. Infect. Dis. 2021;73:e699–e709. doi: 10.1093/cid/ciab004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.