Abstract
Background
Both the tumor environment and the genomic landscape of lung cancer may shape patient responses to treatments, including immunotherapy, but their joint impacts on lung adenocarcinoma (LUAD) prognosis are underexplored.
Methods
RNA sequencing data and whole-exome sequencing results were downloaded from the TCGA database, and only LUAD-related data were included in this study. Based on gene expression data, the ESTIMATE algorithm was used to estimate stromal and immune scores, and CIBERSORT analysis was used for quantification of the relative abundances of immune cells. Somatic mutations were used for calculating tumor mutation burden (TMB). Specific mutations in genes involved in DNA damage repair (DDR) pathways were identified. The individual and joint associations of stromal and immune score, TMB, and DDR gene mutations with 5-year survival were analyzed by the Kaplan–Meier method and multivariate Cox model.
Results
LUAD patients with a high (>highest 25%) stromal or immune score had prolonged survival as compared to those with a low (<lowest 25%) score (log-rank P=0.05 and 0.035, respectively). Patients with both high stromal and immune scores had the most favorable survival. Although the survival differences between patients with high (>highest 25%) and low (<lowest 25%) TMB, or between patients with mutant- and wild-type DDR genes were not statistically significant, a survival benefit from high TMB or DDR gene mutations was observed in patients with high stromal or immune scores.
Conclusion
A comprehensive evaluation of transcriptomic signatures and genomic biomarkers may provide a novel avenue for improving prognosis stratification in LUAD.
1. Introduction
Lung cancer is the most commonly diagnosed cancer and the leading cause of cancer deaths worldwide [1]. In the past decades, significant advances have been made in the field of lung cancer, including screening for early detection and new agents for survival improvement. The treatment of lung cancer has evolved with the introduction of several lines of tyrosine kinase inhibitors in patients with EGFR, ALK, ROS1, and NTRK mutations [2]. More recently, immune checkpoint inhibitors (ICIs), particularly inhibitors of the programmed cell death 1 (PD-1) axis, have dramatically changed the landscape of lung cancer treatment. Clinical trials such as KEYNOTE [3], CheckMate [4], and POPLAR [5] have demonstrated survival benefits for patients with non-small cell lung cancer (NSCLC) who received either immunotherapy, monotherapy, or combined chemoimmunotherapy [2, 6]. Nonetheless, a large portion of NSCLC patients does not respond to ICIs. Even in the large phase III studies that evaluated ICIs combined with chemotherapy for NSCLC patients, overall response rates ranged from 47% to 63% at best [6–9]. Therefore, how to accurately identify a group of patients who will really benefit from immunotherapy poses a consistent clinical challenge. Reportedly, high expression of programmed cell death ligand 1 (PD-L1) in tumor tissues has been associated with a response to ICIs [10]. PD-L1 expression by immunohistochemistry (IHC) testing is an FDA-approved companion diagnostic test for pembrolizumab in NSCLC. However, the discriminatory potential of PD-L1 is doubted due to several limitations, such as intratumor heterogeneity, representativeness of biopsy samples, discordance when defining positive expression, and varied sensitivity of PD-L1 IHC assays [2, 10–13]. In reality, benefits are often seen in patients whose tumors do not express PD-L1, while many patients whose tumors do express PD-L1 expression do not derive benefits from PD-(L)1 blockade [4, 6]. Therefore, identification of high-performance biomarkers associated with response to ICIs and NSCLC prognostication is still an unmet need.
Malignant solid tumor tissues consist of not only tumor cells but also tumor-associated normal epithelial and stromal cells, immune cells, and vascular cells [14]. Stromal cells have important roles in tumor growth, disease progression, and drug resistance [14]. Generally, tumor tissue shapes the immune suppressive microenvironment to prevent T cells from infiltrating the tumor site, and enhancing T cell infiltration can improve cancer immunotherapy [15–17]. For example, an increase of CD8+tumor-infiltrating lymphocytes (TILs) has been reported to be associated with increased sensitivity to ICIs and favorable prognosis [15–17]. Yoshihara et al. recently developed a novel method, called ESTIMATE (estimation of stromal and immune cells in malignant tumor tissues using expression data), to infer the fraction of stromal and immune cells in tumor samples using gene expression signatures [14]. The potential of stromal and immune scores for predicting prognosis and response to immunotherapy has been suggested by recent studies conducted in patients with solid tumors, such as gastric [18], liver [19], renal [20], head, neck [21], and lung [22–27] cancers. In addition, more investigations have focused on the identification of predictive biomarkers for response to ICIs in lung adenocarcinoma (LUSD) patients, which may identify potential beneficiaries to guide clinic treatment. For example, LUSD patients with high levels of B2M protein were detected by more T and natural killer cells in their tumors and associated with an increased response to PD-1-based immunotherapy [28]. VTCN1 (B7-H4 gene), which negatively associates with granzyme B levels, contributes to immunosuppression for LUAD patients harboring EGFR-activating mutations [29]. Somatic copy number alterations (SCNAs) burden is negatively associated with ICIs progression-free survival [30]. In addition to gene expression signatures or SCNAs-based biomarkers, tumor mutation burden (TMB) has arisen as another potential indicator of response to ICIs, with the premise that an increase in TMB leads to an increased number of mutated proteins, or neoantigens, on the surface of tumor cells capable of eliciting an immune response [2, 6]. In addition, an association between mutation burden and sensitivity to ICIs is also evident in the hypermutated tumors of patients with deleterious alterations in DNA-repair genes such as MLH1, MSH2, MSH6, and PMS2, which are characterized by increased CD8+T-cell infiltrates, as well as malignancies with mutations in BRCA2, POLD1, and POLE [6]. Although several recent studies reported the prognostic value of stromal and immune scores in LUAD, the predominant histological subtype of NSCLC, by analyzing data from the Cancer Genome Atlas (TCGA) and/or the Gene Expression Omnibus (GEO) projects [22, 23, 25–27], none of these studies put both gene expression and mutation data in the same analytical framework.
In the current study, based on TCGA RNA sequencing data and whole-exome sequencing (WES) data, overall survival (OS) benefits were identified in LUAD patients with either a high stromal/immune score alone or in a combination of high TMB or mutant genes in DNA damage repair (DDR) pathways. Because the TCGA database lacks detailed information about treatments, including immunotherapy, the analyses in this study focused on assessing the prognosis stratification effects of these potential biomarkers.
2. Materials and Methods
2.1. Data Source
Data on gene expression and genomic variants, as well as clinicopathological information, were obtained from the TCGA database (https://tcga-data.nci.nih.gov/tcga/), which was jointly created by the National Cancer Institute and the National Human Genome Research Institute in 2006. It offers a comprehensive catalog of genomic, epigenomic, transcriptomic, and proteomic alterations that occur in 33 major cancer types [31]. Only LUAD-related data were extracted in this study. In brief, RNA sequencing data of tumor tissues were downloaded from 513 LUAD patients, and among them, 505 had clinical data. The TCGA WES data from 565 LUAD patients were used for mutation calling, and 479 of these patients also had gene expression and clinical data. After excluding 11 patients without information on time to death/last follow-up for survival analysis, the final dataset included 468 treatment-naïve LUAD patients.
2.2. Estimation of Stromal and Immune Scores
The ESTIMATE algorithm (R package “estimate”) was used to estimate the stromal and immune scores from the TCGA gene expression data [14]. The estimations were based on 141 stromal genes and other 141 different genes related to immune cell infiltration [14]. An analytical tool, named CIBERSORT (cibersort.stanford.edu), which characterizes the cell composition of complex tissues from their gene expression profiles [32], was used to quantify the relative abundances of 22 types of immune cells. LUAD patients were divided into high stromal/immune score (upper quartile) and low stromal/immune score groups (lower quartile).
2.3. Mutation Identification and TMB Quantification
The Mutect2 results from the TCGA WES data were used for identifying somatic mutations. The TMB of a tumor sample is calculated by the number of nonsynonymous somatic mutations (single nucleotide variants and small insertions/deletions) per megabase in coding regions [33, 34]. Moreover, stratified analyses according to mutant- and wild-type genes were conducted for those related to DDR pathways, including base excision repair (BER: POLE, MUTYH), checkpoint factors (ATM, ATR, CHEK1, CHEK2), Fanconi anemia (FA: BRCA2, BRIP1, FANCA, FANCC, FANCD2, PALB2, BLM), homologous recombination repair (HRR: BRCA1, MRE11A, RAD50, RAD51), and mismatch repair (MMR: MLH1, MSH2, MSH6, PMS2) [35].
2.4. Statistical Analysis
The distributions of stromal and immune scores were visualized using violin plots. A Wilcoxon test was used to compare the differences in medians between two comparison groups. OS was the clinical endpoint analyzed in this study. Survival curves were plotted using the Kaplan–Meier method, and survival differences were compared by a log-rank test [36]. The associations between prognostic factors and OS in LUAD patients were evaluated using the multivariate Cox model [37]. A detailed analytic procedure is shown in Figure 1. Statistical analyses were conducted using R software (version 3.6.1, packages “ggplot2,” “survival,” and “survminer”) [38, 39]. A P value of 0.05 (two-sided) was set as the cutoff point of statistical significance.
Figure 1.
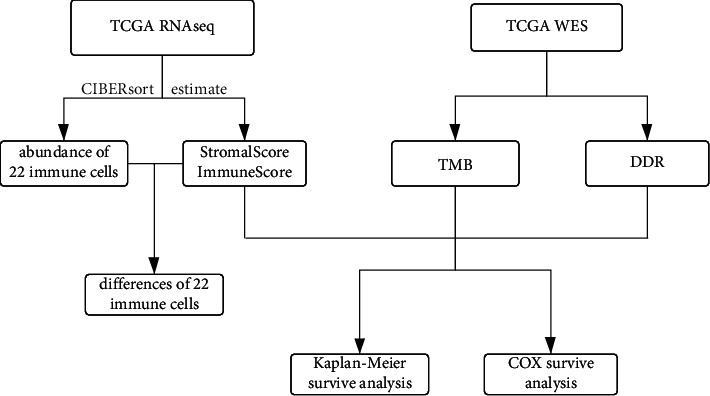
Flow chart of analytic procedure.
3. Results
3.1. Distribution of Stromal and Immune Scores
Among the 468 LUAD patients included in the analysis, 216 (46.2%) were males, 399 (85.3%) had a smoking history, and 114 of them were current smokers. There were 259, 108, 77, and 24 patients with stage I, II, III, and IV LUAD, respectively. The stromal and immune scores for each patient were estimated using the ESTIMATE algorithm. The score distributions according to different tumor stages are visualized in Figure 2. Patients with stage IV tumors had significantly lower stromal scores than those with either stage I or stage II tumors (P=0.007, 0.018, respectively). Similarly, patients with stage I tumors had significantly higher immune scores than those with advanced-stage tumors (P=0.008 when compared with stage III tumors and P=0.034 when compared with stage IV tumors). Furthermore, the relative abundances of 22 types of immune cells were quantified using CIBERsort software. Figure 3 depicts the abundances of each kind of immune cell between high or low stromal/immune score groups. Significant differences were observed in the abundances of T cells, B cells, and macrophages when comparing the high-score group with the low-score group. Specifically, there was more CD8+ T cell infiltration in the patients with a high immune score.
Figure 2.
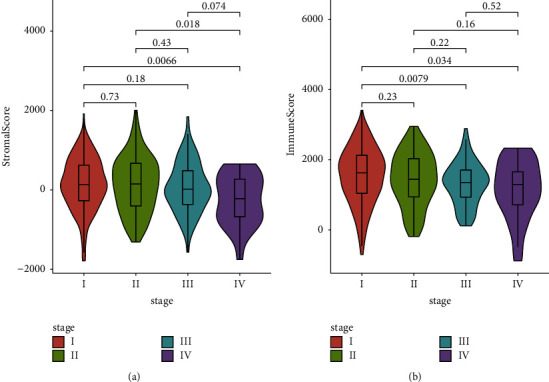
Violin plots to show the distributions of (a) stromal scores and (b) immune scores across tumor stages. The differences in medians between the two comparison groups were compared by the Wilcoxon test and P values were presented to indicate statistical significance. ∗P ≤ 0.05 and ∗∗P ≤ 0.01.
Figure 3.
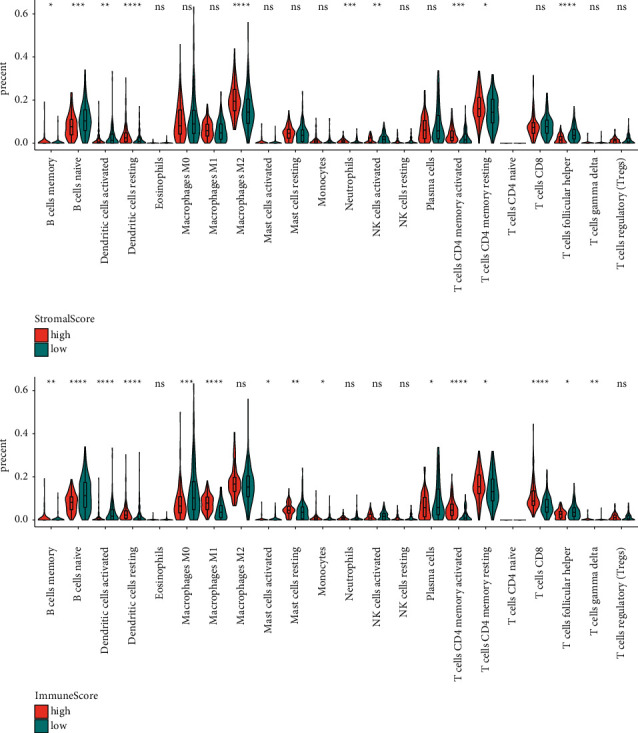
Relative abundances of immune cells in patients with high or low stromal/immune scores. ∗P ≤ 0.05, ∗∗P ≤ 0.01, ∗∗∗P ≤ 0.001, and ∗∗∗∗P ≤ 0.0001.
3.2. Survival Analysis Based on Stromal and Immune Scores
Patients were divided into three groups based on their stromal or immune score quartiles: high-score group with a score in the 4th quartile (>highest 25%), low-score group with a score in the 1st quartile (<lowest 25%), and medium-score group with a score in the 2nd or 3rd quartiles. The 5-year survival curves were plotted using the Kaplan–Meier method. As shown in Figure 4, LUAD patients with either a high stromal score or a high immune score had a prolonged survival as compared to those with a low score (log-rank P=0.05 and 0.035, respectively, in Figures 4(a) and 4(b)). To illuminate the joint effect of stromal and immune scores, the patients were further stratified into four groups: patients with both high stromal and immune scores (N = 72), patients with a high stromal score and a low immune score (N = 2), patients with a low stromal score and a high immune score (N = 0), and patients with both low stromal and immune scores (N = 78). Figure 4(c) shows that patients with both high stromal and immune scores had the most favorable survival.
Figure 4.
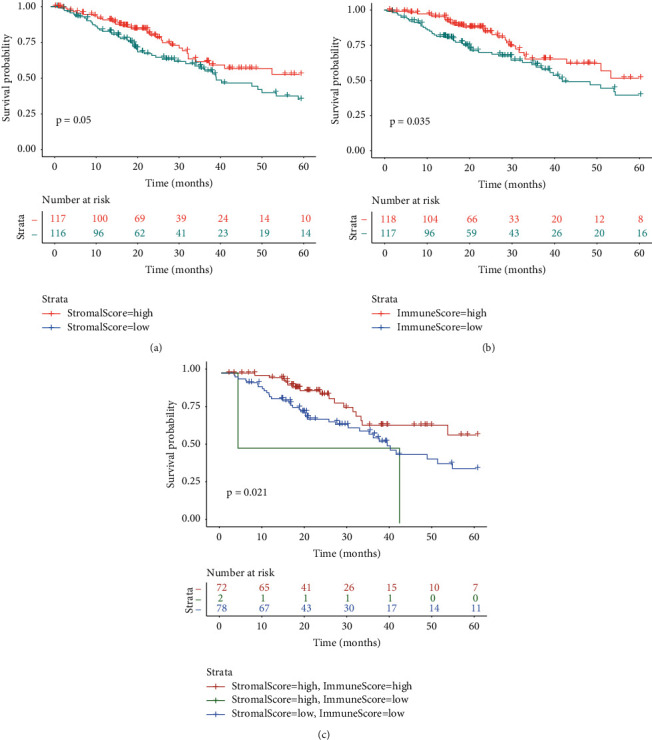
Kaplan–Meier survival curves to show differences in the 5-year survival (a) between patients with high and low stromal scores, (b) between patients with high and low immune scores, or (c) among different comparison groups which were defined according to both stromal and immune scores.
3.3. Survival Analysis in Combination with TMB
TMB was calculated on the basis of the Mutect 2 results from TCGA available WES data of LUAD. As shown in Figure 5(a), a slightly higher TMB was detected in patients with low stromal scores than in those with high stromal scores (P=0.011), but the TMB difference between patients with high and low immune scores was not statistically significant. LUAD patients were then stratified into high-TMB (>highest 25%), medium-TMB, and low-TMB (<lowest 25%) groups according to their TMB levels. The 5-year survival was similar between patients with high and low levels of TMB (log-rank P=0.40, Figure 5(b)). A comprehensive analysis was conducted by incorporating TMB levels into stromal or immune score-based analyses, similar to the patient stratification in the joint analyses of stromal and immune scores. Figures 5(c) and 5(d) indicate that OS benefits from high TMB were only observed in those with high stromal or immune scores.
Figure 5.
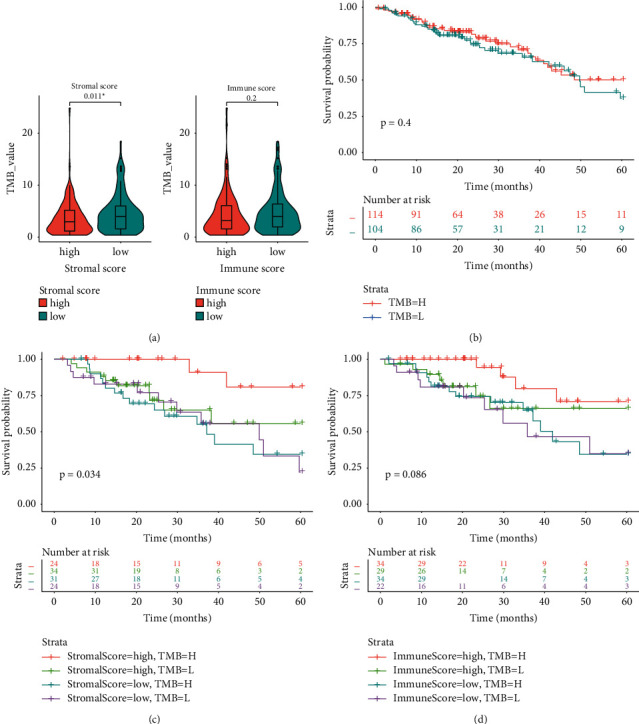
Integrative analyses of tumor mutation burden (TMB) and stromal/immune scores. (a) TMB levels in patients with high and low stromal/immune scores; ∗P ≤ 0.05. (b) The survival difference between patients with high and low levels of TMB. (c) The survival difference among four groups of patients classified according to the level of TMB and stromal score. (d) The survival difference among four groups of patients stratified according to the levels of TMB and immune score.
3.4. Survival Analysis in Combination with DDR Gene Mutations
Twenty-one genes involved in DDR pathway were analyzed. As shown in Figure 6, the top three mutant genes in these LUAD samples were ATM (7.6%), BRCA2 (5.7%), and POLE (4.6%). Patients with mutant DDR genes had significantly smaller stromal and immune scores than those with wild-type genes (P=0.024 and 0.015, respectively, Figure 7(a)). Although patients with mutant-type DDR genes had a relatively lower survival, the survival difference between those with mutant- and wild-type genes did not achieve statistical significance (log-rank P=0.22, Figure 7(b)). When conducting survival analyses by incorporating the mutation status of DDR genes into stromal and immune scores, the improved survival was dominated in patients with both high stromal/immune scores and mutant-type DDR genes (Figures 7(c) and 7(d)).
Figure 6.
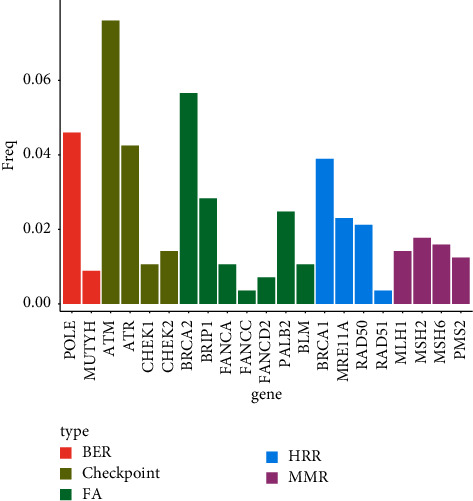
Frequencies of mutant genes involved in DDR pathways.
Figure 7.
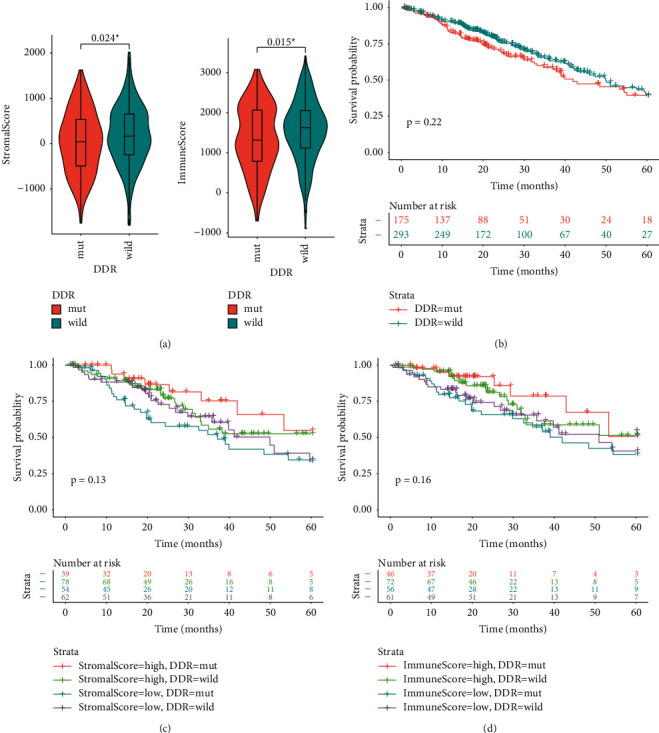
Integrative analyses of mutations in DNA damage repair (DDR) genes and stromal/immune scores. (a) Stromal and immune scores in patients with mutant- or wild-type DDR genes. (b) The survival difference between patients with mutant- and wild-type DDR genes; ∗P ≤ 0.05. (c) The survival difference among four groups of patients classified according to the status of DDR genes and stromal score. (d) The survival difference among four groups of patients stratified according to the status of DDR genes and immune score.
3.5. Multivariate Analysis in Combination of Stromal/Immune Scores, TMB, and DDR Gene Mutations
A multivariate Cox model including age, sex, smoking status, tumor stage, stromal score, immune score, TMB, and mutation status of the DDR gene was developed to evaluate the independent predictive value of these biomarkers. As shown in Figure 8, compared with late-stage patients, early-stage patients had a 58% (hazard ratio [HR] 0.42, P < 0.001) decreased risk of death. The death risk increased by 50% (HR 1.50, P=0.026) among patients with mutant-type DDR genes in comparison to those with wild-type genes. Although patients with a high immune score or a high TMB level had a lower risk of death, the associations were not independent of covariates.
Figure 8.
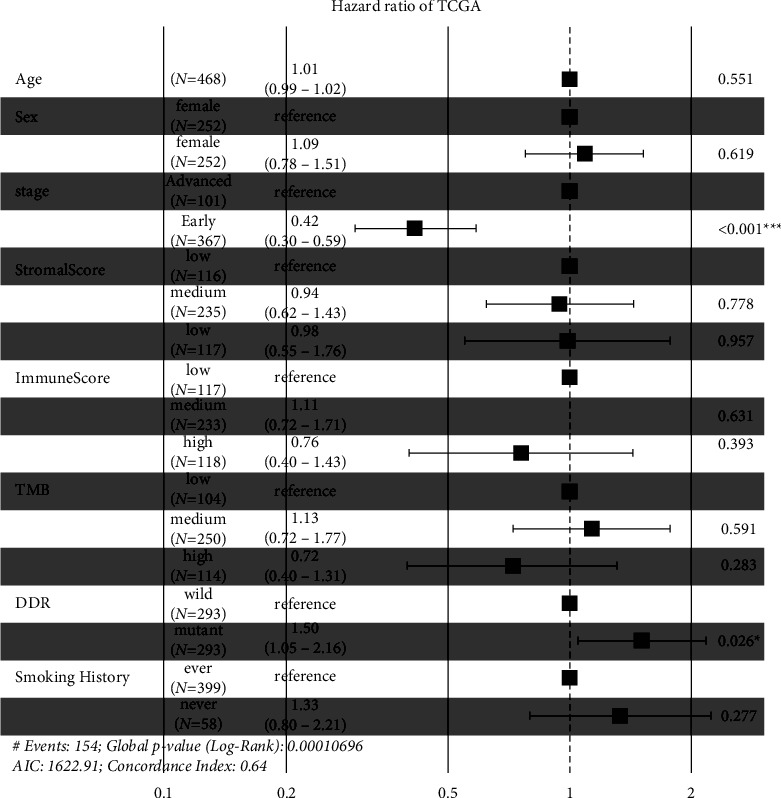
Multivariate Cox analyses in combination of stromal/immune scores, TMB, DDR gene mutations, and clinicopathological variables. TMB, tumor mutation burden; DDR, DNA damage repair; HR, hazard ratio; CI, confidence interval.
4. Discussion
Over the past several years, ICIs, which target inhibitory receptors on T cells and reinvigorate antitumor immune responses, have begun to transform clinical cancer care [10]. Because only a subset of patients derives clinical benefit from ICIs, it is critical to identify a specific biomarker or a group of biomarkers with high performance in discriminating potential responders from nonresponders. The determination of tumor PD-L1 expression by IHC has been extensively studied as a predictor of response to anti-PD1/PD-L1 agents, although it is sometimes inconclusive. Expression of PD-L1 on infiltrating immune cells in the tumor microenvironment has also been associated with clinical response to immunotherapy in cancers, including NSCLC [10, 15]. In particular, it has been recently mentioned that the combined use of the TTF1/PD-L1 score outperforms the gold standard PD-L1 biomarker for OS prediction in LUSD [40]. The characterization of the tumor microenvironment and its interaction with host genomics will help to optimize precision medicine and prognosis management. In this study, by analyzing data from the TCGA, the associations of gene-expression-based biomarkers and genomic-variant-based biomarkers with 5-year survival were evaluated individually and jointly. Significantly improved survival was observed in patients with high stromal and/or immune scores. Although using TMB level or DDR gene mutations alone could not differentiate patient risk for death, a combination of these biomarkers with stromal/immune scores has the potential to identify patients with OS benefits.
The tumor microenvironment consists of factors extrinsic to cancer cells, including various immune and stromal cells, vasculature, extracellular matrix, and cytokines that influence response to therapy [41]. The crosstalk between cancer cells and tumor stroma plays an important role in the progression of tumors and their metastasis [42]. The density of TILs in the tumor microenvironment confers a prognostic and predictive impact on some tumor types, including NSCLC, regardless of ICI therapy [10, 43, 44]. A metric known as the Immunescore, which involves quantification of CD8+ T cells at the center and periphery of a tumor, was reported to be a strong predictor of OS that can complement traditional TNM staging or microsatellite instability (MSI) status in colorectal cancer [45–47]. The T-cell-inflamed gene expression profile and immune gene expression signatures also represent emerging predictive biomarkers [48]. Stromal and immune scores are stromal tissue and ICIs related gene expression signature-based biomarkers recently developed by Yoshihara et al. [14] and have been associated with prognosis in lung cancer patients [22–27]. In this study, high stromal and immune scores, analyzed individually or jointly, were associated with improved 5-year survival, which was consistent with previous findings derived from the TCGA LUAD dataset [22, 25–27]. Moreover, CIBERSORT analysis identified more CD8+ T cells in the patients with a high immune score, highlighting the importance of the TIL phenotype in LUAD prognosis.
Cancer is a genetic disease, and neoplastic transformation results from the accumulation of somatic mutations in the DNA of affected cells [48]. Considering that high TMB correlates with a greater probability of displaying tumor neoantigens on human leukocyte antigen molecules on the surface of tumor cells, it is rational to hypothesize that the tumors with the highest TMB are more likely to respond to ICIs because greater mutation load would increase the likelihood of recognition by neoantigen-reactiveT cells [48]. Consistent with this hypothesis, several studies have demonstrated an association between high TMB and a response to ICIs in NSCLC [49–51]. Pembrolizumab, an anti-PD1 agent, was recently FDA-approved in TMB-high advanced solid cancers in response to results from the KEYNORE-158 trial [52]. Nevertheless, in the complex multiarm CheckMate227 trial in NSCLC, neither TMB nor PD-L1 expression could segregate therapy responsiveness [53]. In the present study, patients with high TMB had a slightly prolonged survival, but the difference in OS between patients with high and low levels of TMB was not statistically significant. This result was consistent with data from previous studies that demonstrated clinical benefit from high TMB with respect to objective response rate or progression-free survival, rather than OS [48]. The reason for not achieving OS improvement may be attributed to the fact that tumors with increased mutations and genomic instability can adapt more quickly to immune pressure, resulting in treatment resistance [6]. Furthermore, in this study, the survival advantage from high TMB was only observed in patients with high stromal or immune scores, suggesting an approach to improve prognosis stratification by using a biomarker panel in combination of gene expression and genomic variants.
In addition to the overall mutation burden, TMB-related specific mutations such as MGA [54], EPHA5, [55], CTNNA2 [56], and co-mutation of FAT3 and LRP1B [57] have been found as novel predictive biomarkers for ICIs response in nonsquamous NSCLC or LUSD. Hypermutation of driver genes may have a distinct impact on MSI in tumors [58]. The deficiency in DDR genes such as MLH1, MSH2, MSH6, PMS2, and POLE has high concordance to high MSI (MSI-H), and the concordance of MSI and MMR testing in prior studies was reported to be 92% [59, 60]. In the study of Rizvi, mutations of genes involved in the DDR pathways were enriched in patients who derived clinical benefits from anti-PD1 therapy [49]. A recent large study reported that, in MSI-H cases, the presence of DDR alterations correlated with a significantly higher TMB as compared with DDR-wild-type MSI-H cases [61]. Therefore, in this study, it was not surprising to observe an OS improvement in the patients with both high stromal/immune scores and mutant DDR genes, similar to the combined analysis with TMB. In addition, although DDR gene mutation was shown to be an independent prognostic factor in the multivariate analyses, due to the retrospective nature of this study, it is unclear whether the DDR alteration is causative of the higher TMB or a result of the high TMB phenotype, thus further investigations are needed for exploration.
This study comprehensively evaluated the prognostic values of transcriptomic signatures and genomic biomarkers and provided a novel avenue for improving prognosis stratification in LUAD. However, the current study has several limitations. First, the TCGA database lacks information on detailed treatments, so stratified analyses according to a given treatment such as immunotherapy are infeasible. Second, the biomarkers identified in this study could not be compared with other wildly used biomarkers such as tumor and immune cell PD-L1 expression due to the unavailability of these data. Third, in the combined analyses of stromal/immune scores, TMB, and DDR gene mutations, the statistical power may be insufficient due to a small number of patients in each subgroup. Fourth, the relative abundances of 22 types of immune cells were estimated using a bioinformatic tool, which may differ from the real situation in tumors. Finally, this was a retrospective analysis based on a publicly available database and no independent validation was conducted to further test the potential of these biomarkers. Large prospective studies with completed treatment-related data are warranted to confirm the findings in this study.
Acknowledgments
This study was supported by the Health Science and Technology Planning Project of Zhejiang Province (2021KY1149) and the Science and Technology Planning Project of Taizhou City (1801ky36).
Contributor Information
Dongping Wu, Email: wudp6666@163.com.
Shenpeng Ying, Email: 13957608158@139.com.
Data Availability
Publicly available datasets were analyzed in this study. All LUSD relevant RNA sequencing and whole-exome sequencing data can be downloaded from the TCGA database (https://tcga-data.nci.nih.gov/tcga/).
Ethical Approval
Because the analyses in this study were based on publicly available database, informed consent from was were waived.
Conflicts of Interest
The authors declare that the authors declare no conflicts of interest.
Authors' Contributions
WD conceived and designed the study project. WD, YS, HL, and MJ analyzed the data and drafted the manuscript. LJ, YW, XJ, ZW, and ZX performed data analysis and contributed in writing the manuscript. WD, YS, and HL revised the manuscript. All the authors have read and approved the final manuscript.
References
- 1.Bray F., Ferlay J., Soerjomataram I., Siegel R. L., Torre L. A., Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA: A Cancer Journal for Clinicians . 2018;68(6):394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Duma N., Santana-Davila R., Molina J. R. Non-small cell lung cancer: epidemiology, screening, diagnosis, and treatment. Mayo Clinic Proceedings . 2019;94(8):1623–1640. doi: 10.1016/j.mayocp.2019.01.013. [DOI] [PubMed] [Google Scholar]
- 3.Herbst R. S., Baas P., Kim D. W., et al. Pembrolizumab versus docetaxel for previously treated, PD-L1-positive, advanced non-small-cell lung cancer (KEYNOTE-010): a randomised controlled trial. The Lancet . 2016;387(10027):1540–1550. doi: 10.1016/s0140-6736(15)01281-7. [DOI] [PubMed] [Google Scholar]
- 4.Brahmer J., Reckamp K. L., Baas P., et al. Nivolumab versus docetaxel in advanced squamous-cell non-small-cell lung cancer. New England Journal of Medicine . 2015;373(2):123–135. doi: 10.1056/nejmoa1504627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fehrenbacher L., Spira A., Ballinger M., et al. Atezolizumab versus docetaxel for patients with previously treated non-small-cell lung cancer (POPLAR): a multicentre, open-label, phase 2 randomised controlled trial. The Lancet . 2016;387(10030):1837–1846. doi: 10.1016/s0140-6736(16)00587-0. [DOI] [PubMed] [Google Scholar]
- 6.Doroshow D. B., Sanmamed M. F., Hastings K., et al. Immunotherapy in non-small cell lung cancer: facts and hopes. Clinical Cancer Research . 2019;25(15):4592–4602. doi: 10.1158/1078-0432.ccr-18-1538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gandhi L., Rodríguez-Abreu D., Gadgeel S., et al. Pembrolizumab plus chemotherapy in metastatic non-small-cell lung cancer. New England Journal of Medicine . 2018;378(22):2078–2092. doi: 10.1056/nejmoa1801005. [DOI] [PubMed] [Google Scholar]
- 8.Socinski M. A., Jotte R. M., Cappuzzo F., et al. Atezolizumab for first-line treatment of metastatic nonsquamous NSCLC. New England Journal of Medicine . 2018;378(24):2288–2301. doi: 10.1056/nejmoa1716948. [DOI] [PubMed] [Google Scholar]
- 9.Paz-Ares L., Luft A., Vicente D., et al. Pembrolizumab plus chemotherapy for squamous non-small-cell lung cancer. New England Journal of Medicine . 2018;379(21):2040–2051. doi: 10.1056/nejmoa1810865. [DOI] [PubMed] [Google Scholar]
- 10.Havel J. J., Chowell D., Chan T. A. The evolving landscape of biomarkers for checkpoint inhibitor immunotherapy. Nature Reviews Cancer . 2019;19(3):133–150. doi: 10.1038/s41568-019-0116-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Büttner R., Gosney J. R., Skov B. G., et al. Programmed death-ligand 1 immunohistochemistry testing: a review of analytical assays and clinical implementation in non-small-cell lung cancer. Journal of Clinical Oncology . 2017;35(34):3867–3876. doi: 10.1200/jco.2017.74.7642. [DOI] [PubMed] [Google Scholar]
- 12.Hirsch F. R., McElhinny A., Stanforth D., et al. PD-L1 immunohistochemistry assays for lung cancer: results from phase 1 of the blueprint PD-L1 IHC assay comparison project. Journal of Thoracic Oncology . 2017;12(2):208–222. doi: 10.1016/j.jtho.2016.11.2228. [DOI] [PubMed] [Google Scholar]
- 13.Horvath L., Thienpont B., Zhao L., Wolf D., Pircher A. Overcoming immunotherapy resistance in non-small cell lung cancer (NSCLC)—novel approaches and future outlook. Molecular Cancer . 2020;19(1):p. 141. doi: 10.1186/s12943-020-01260-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yoshihara K., Shahmoradgoli M., Martínez E., et al. Inferring tumour purity and stromal and immune cell admixture from expression data. Nature Communications . 2013;4(1):p. 2612. doi: 10.1038/ncomms3612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Herbst R. S., Soria J. C., Kowanetz M., et al. Predictive correlates of response to the anti-PD-L1 antibody MPDL3280A in cancer patients. Nature . 2014;515(7528):563–567. doi: 10.1038/nature14011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ock C. Y., Keam B., Kim S., et al. Pan-cancer immunogenomic perspective on the tumor microenvironment based on PD-L1 and CD8 T-cell infiltration. Clinical Cancer Research . 2016;22(9):2261–2270. doi: 10.1158/1078-0432.ccr-15-2834. [DOI] [PubMed] [Google Scholar]
- 17.Fumet J. D., Richard C., Ledys F., et al. Prognostic and predictive role of CD8 and PD-L1 determination in lung tumor tissue of patients under anti-PD-1 therapy. British Journal of Cancer . 2018;119(8):950–960. doi: 10.1038/s41416-018-0220-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ren Q., Zhu P., Zhang H., et al. Identification and validation of stromal-tumormicroenvironment-based subtypes tightly associated with PD-1/PD-L1 immunotherapy and outcomes in patients with gastric cancer. Cancer Cell International . 2020;20(1):p. 92. doi: 10.1186/s12935-020-01173-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ge P. L., Li S. F., Wang W. W., et al. Prognostic values of immune scores and immune microenvironment-related genes for hepatocellular carcinoma. Aging (Albany NY) . 2020;12(6):5479–5499. doi: 10.18632/aging.102971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xu W. H., Xu Y., Wang J., et al. Prognostic value and immune infiltration of novel signatures in clear cell renal cell carcinoma microenvironment. Aging (Albany NY) . 2019;11(17):6999–7020. doi: 10.18632/aging.102233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang Z., Yuan H., Huang J., et al. Prognostic value of immune-related genes and immune cell infiltration analysis in the tumor microenvironment of head and neck squamous cell carcinoma. Head Neck . 2020;43 doi: 10.1002/hed.26474. [DOI] [PubMed] [Google Scholar]
- 22.Qi X., Qi C., Qin B., Kang X., Hu Y., Han W. Immune-stromal score signature: novel prognostic tool of the tumor microenvironment in lung adenocarcinoma. Frontiers in Oncology . 2020;10 doi: 10.3389/fonc.2020.541330.541330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Qu Y., Cheng B., Shao N., et al. Prognostic value of immune-related genes in the tumor microenvironment of lung adenocarcinoma and lung squamous cell carcinoma. Aging (Albany NY) . 2020;12(6):4757–4777. doi: 10.18632/aging.102871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li J., Li X., Zhang C., Zhang C., Wang H. A signature of tumor immune microenvironment genes associated with the prognosis of non-small cell lung cancer. Oncology Reports . 2020;43(3):795–806. doi: 10.3892/or.2020.7464. [DOI] [PubMed] [Google Scholar]
- 25.Tao Y., Li Y., Liang B. Comprehensive analysis of microenvironment-related genes in lung adenocarcinoma. Future Oncology . 2020;16(24):1825–1837. doi: 10.2217/fon-2019-0829. [DOI] [PubMed] [Google Scholar]
- 26.Xu Z. Y., Zhao M., Chen W., et al. Analysis of prognostic genes in the tumor microenvironment of lung adenocarcinoma. PeerJ . 2020;8 doi: 10.7717/peerj.9530.e9530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yuan J., Yuan B., Zeng L., et al. Identification and validation of tumor microenvironment-related genes of prognostic value in lung adenocarcinoma. Oncology Letters . 2020;20(2):1772–1780. doi: 10.3892/ol.2020.11735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhao Y., Cao Y., Chen Y., et al. B2M gene expression shapes the immune landscape of lung adenocarcinoma and determines the response to immunotherapy. Immunology . 2021;164(3):507–523. doi: 10.1111/imm.13384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lu Y., Wu F., Cao Q., et al. B7-H4 is increased in lung adenocarcinoma harboring EGFR-activating mutations and contributes to immunosuppression. Oncogene . 2022;41(5):704–717. doi: 10.1038/s41388-021-02124-6. [DOI] [PubMed] [Google Scholar]
- 30.Frigola J., Carbonell C., Iranzo P., et al. High levels of chromosomal aberrations negatively associate with benefit to checkpoint inhibition in NSCLC. Journal for ImmunoTherapy of Cancer . 2022;10(4) doi: 10.1136/jitc-2021-004197.e004197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang Z., Li H., Jiang S., et al. A survey and evaluation of Web-based tools/databases for variant analysis of TCGA data. Briefings in Bioinformatics . 2019;20(4):1524–1541. doi: 10.1093/bib/bby023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Newman A. M., Liu C. L., Green M. R., et al. Robust enumeration of cell subsets from tissue expression profiles. Nature Methods . 2015;12(5):453–457. doi: 10.1038/nmeth.3337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zehir A., Benayed R., Shah R. H., et al. Mutational landscape of metastatic cancer revealed from prospective clinical sequencing of 10, 000 patients. Nature Medicine . 2017;23(6):703–713. doi: 10.1038/nm.4333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xu Z., Dai J., Wang D., et al. Assessment of tumor mutation burden calculation from gene panel sequencing data. OncoTargets and Therapy . 2019;12:3401–3409. doi: 10.2147/ott.s196638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tian W., Shan B., Zhang Y., et al. Association between DNA damage repair gene somatic mutations and immune-related gene expression in ovarian cancer. Cancer Medicine . 2020;9(6):2190–2200. doi: 10.1002/cam4.2849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jager K. J., van Dijk P. C., Zoccali C., Dekker F. W. The analysis of survival data: the Kaplan-Meier method. Kidney International . 2008;74(5):560–565. doi: 10.1038/ki.2008.217. [DOI] [PubMed] [Google Scholar]
- 37.Christensen E. Multivariate survival analysis using cox’s regression model. Hepatology . 1987;7(6):1346–1358. doi: 10.1002/hep.1840070628. [DOI] [PubMed] [Google Scholar]
- 38.Wickham H. ggplot2: Elegant Graphics for Data Analysis . Berlin, Germany: Springer; 2016. [Google Scholar]
- 39.Kassambara A., Kosinski M., Biecek P. Package “Survminer,” Drawing Survival Curves Using “ggplot2”(R Package Version 03 1) 2017. [Google Scholar]
- 40.Galland L., Le Page A. L., Lecuelle J., et al. Prognostic value of thyroid transcription factor-1 expression in lung adenocarcinoma in patients treated with anti PD-1/PD-L1. OncoImmunology . 2021;10(1) doi: 10.1080/2162402x.2021.1957603.1957603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fares C. M., Van Allen E. M., Drake C. G., Allison J. P., Hu-Lieskovan S. Mechanisms of resistance to immune checkpoint blockade: why does checkpoint inhibitor immunotherapy not work for all patients? American Society of Clinical Oncology Educational Book . 2019;39:147–164. doi: 10.1200/edbk_240837. [DOI] [PubMed] [Google Scholar]
- 42.Bremnes R. M., Dønnem T., Al-Saad S., et al. The role of tumor stroma in cancer progression and prognosis: emphasis on carcinoma-associated fibroblasts and non-small cell lung cancer. Journal of Thoracic Oncology . 2011;6(1):209–217. doi: 10.1097/jto.0b013e3181f8a1bd. [DOI] [PubMed] [Google Scholar]
- 43.Brambilla E., Le Teuff G., Marguet S., et al. Prognostic effect of tumor lymphocytic infiltration in resectable non-small-cell lung cancer. Journal of Clinical Oncology . 2016;34(11):1223–1230. doi: 10.1200/jco.2015.63.0970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fridman W. H., Pagès F., Sautès-Fridman C., Galon J. The immune contexture in human tumours: impact on clinical outcome. Nature Reviews Cancer . 2012;12(4):298–306. doi: 10.1038/nrc3245. [DOI] [PubMed] [Google Scholar]
- 45.Galon J., Pagès F., Marincola F. M., et al. Cancer classification using the Immunoscore: a worldwild task force. Journal of Translational Medicine . 2012;10(1):p. 1. doi: 10.1186/1479-5876-10-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mlecnik B., Bindea G., Angell H. K., et al. Integrative analyses of colorectal cancer show immunoscore is a stronger predictor of patient survival than microsatellite instability. Immunity . 2016;44(3):698–711. doi: 10.1016/j.immuni.2016.02.025. [DOI] [PubMed] [Google Scholar]
- 47.Pagès F., Mlecnik B., Marliot F., et al. International validation of the consensus immunoscore for the classification of colon cancer: a prognostic and accuracy study. Lancet . 2018;391(10135):2128–2139. doi: 10.1016/S0140-6736(18)30789-X. [DOI] [PubMed] [Google Scholar]
- 48.Chan T. A., Yarchoan M., Jaffee E., et al. Development of tumor mutation burden as an immunotherapy biomarker: utility for the oncology clinic. Annals of Oncology . 2019;30(1):44–56. doi: 10.1093/annonc/mdy495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rizvi N. A., Hellmann M. D., Snyder A., et al. Cancer immunology. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science . 2015;348(6230):124–128. doi: 10.1126/science.aaa1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hellmann M. D., Nathanson T., Rizvi H., et al. Genomic features of response to combination immunotherapy in patients with advanced non-small-cell lung cancer. Cancer Cell . 2018;33(5) doi: 10.1016/j.ccell.2018.03.018.e844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hellmann M. D., Ciuleanu T. E., Pluzanski A., et al. Nivolumab plus ipilimumab in lung cancer with a high tumor mutational burden. New England Journal of Medicine . 2018;378(22):2093–2104. doi: 10.1056/nejmoa1801946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Marabelle A., Fakih M., Lopez J., et al. Association of tumour mutational burden with outcomes in patients with advanced solid tumours treated with pembrolizumab: prospective biomarker analysis of the multicohort, open-label, phase 2 KEYNOTE-158 study. The Lancet Oncology . 2020;21(10):1353–1365. doi: 10.1016/s1470-2045(20)30445-9. [DOI] [PubMed] [Google Scholar]
- 53.Hellmann M. D., Paz-Ares L., Bernabe Caro R., et al. Nivolumab plus ipilimumab in advanced non-small-cell lung cancer. New England Journal of Medicine . 2019;381(21):2020–2031. doi: 10.1056/nejmoa1910231. [DOI] [PubMed] [Google Scholar]
- 54.Sun L., Li M., Deng L., et al. MGA mutation as a novel biomarker for immune checkpoint therapies in non-squamousnon-small cell lung cancer. Frontiers in Pharmacology . 2021;12 doi: 10.3389/fphar.2021.625593.625593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Huang W., Lin A., Luo P., et al. EPHA5 mutation predicts the durable clinical benefit of immune checkpoint inhibitors in patients with lung adenocarcinoma. Cancer Gene Therapy . 2021;28(7-8):864–874. doi: 10.1038/s41417-020-0207-6. [DOI] [PubMed] [Google Scholar]
- 56.Wen Y., Lin A., Zhu W., et al. Catenin alpha-2 mutation changes the immune microenvironment in lung adenocarcinoma patients receiving immune checkpoint inhibitors. Frontiers in Pharmacology . 2021;12 doi: 10.3389/fphar.2021.645862.645862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhu M., Zhang L., Cui H., et al. Co-mutation of FAT3 and LRP1B in lung adenocarcinoma defines a unique subset correlated with the efficacy of immunotherapy. Frontiers in Immunology . 2021;12 doi: 10.3389/fimmu.2021.800951.800951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Campbell B. B., Light N., Fabrizio D., et al. Comprehensive analysis of hypermutation in human cancer. Cell . 2017;171(5) doi: 10.1016/j.cell.2017.09.048.1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shia J. Immunohistochemistry versus microsatellite instability testing for screening colorectal cancer patients at risk for hereditary nonpolyposis colorectal cancer syndrome part I. the utility of immunohistochemistry. Journal of Molecular Diagnostics . 2008;10:293–300. doi: 10.2353/jmoldx.2008.080031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Shia J., Stadler Z., Weiser M. R., et al. Immunohistochemical staining for DNA mismatch repair proteins in intestinal tract carcinoma: how reliable are biopsy samples? The American Journal of Surgical Pathology . 2011;35(3):447–454. doi: 10.1097/pas.0b013e31820a091d. [DOI] [PubMed] [Google Scholar]
- 61.Parikh A. R., He Y., Hong T. S., et al. Analysis of DNA damage response gene alterations and tumor mutational burden across 17, 486 tubular gastrointestinal carcinomas: implications for therapy. The Oncologist . 2019;24(10):1340–1347. doi: 10.1634/theoncologist.2019-0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Publicly available datasets were analyzed in this study. All LUSD relevant RNA sequencing and whole-exome sequencing data can be downloaded from the TCGA database (https://tcga-data.nci.nih.gov/tcga/).


