Abstract
Biofilms are aggregates of bacteria, in most cases, which are resistant usually to broad-spectrum antibiotics in their typical concentrations or even in higher doses. A trend of increasing multi-drug resistance in biofilms, which are responsible for emerging life-threatening nosocomial infections, is becoming a serious problem. Biofilms, however, are at various sensitivity levels to environmental factors and are versatile in infectivity depending on virulence factors. This review presents the fundamental information about biofilms: formation, antibiotic resistance, impacts on public health and alternatives to conventional approaches. Novel developments in micro-biosystems that help reveal the new treatment tools by sensing and characterization of biofilms will also be discussed. Understanding the formation, structure, physiology and properties of biofilms better helps eliminate them by the usage of appropriate antibiotics or their control by novel therapy approaches, such as anti-biofilm molecules, effective gene editing, drug-delivery systems and probiotics.
Keywords: biofilms, bacterial resistance, prevention, prophylaxis, probiotics
1. Introduction
Biofilms are networks of aggregated living microorganisms that are formed either on biotic or abiotic moist surfaces [1],[2]. They are well-known predominating micro-ecosystems which are not only distributed widely in most natural environments but can be adapted to survive in extreme environmental conditions, such as UV radiation, high salinity, poor nutrients, high temperature, high pH and antibiotics [3],[4]. Because of their power of adaptation in extreme conditions, they are assumed to have developed self-protective mechanisms, creating an environment for their growth and survival, in addition to enhancing the communications among the microcolonies within the biofilms [3],[5]. Three-dimensional structure of biofilms is established by the layer which is formed by the aggregation and composition of the secretory insoluble substances produced by the associated microorganisms. The layer made of extracellular substances is collectively known as the extracellular polymeric substance (EPS), which has an indispensable role in the formation of biofilms [3],[6]. EPS also supports the transformation of biofilms from reversible to irreversible and establishes the biofilm on a suitable surface [7],[8]. Thus, the slimy EPS not only helps organisms adhere to each other but also protects biofilms from any adverse synthetic reactions [8],[9]. Biofilms are diverse, depending on organism types and environmental factors [9]. Most of the biofilms are aggregations of similar or different bacterial species, but they can also be composed of other organisms, such as fungi, algae, or protozoa [9]–[11]. They are usually tangled as a mass of prolonged and infectious organisms, along with their capacity to guard themselves against various antimicrobial drugs. Sugars, proteins and nucleic acids present in the EPS are used by some organisms for their metabolism, and their metabolic end products serve as the nutritional sources for other types of organisms living in the proximity bounded by the EPS [12],[13]. In addition, EPS supports cell-cell and cell-matrix communications, creating the synergistic interactions. The extracellular matrices of the biofilms give protection to the microbes from different factors in their surrounding environment [14],[15]. EPS, therefore, has a significant role in biofilm formation and its protection besides limiting the diffusion of antibiotics [16]. Interestingly, not all the organisms can be a part of the biofilm; only organisms which favor the association with other organisms are a part of a biofilm [2],[17]. The main function of the biofilms is the support for the propagation of diverse microorganisms and their survival [3],[14]. The biological and environmental factors in biofilms for microorganisms are substantially different from those in planktonic environment [18].
2. Biofilm formation
Biofilm formation is a dynamic complex process that constitutes a multifaceted nature [2],[19],[20], and it is triggered, modulated and dispersed by the microorganisms themselves which are associated with and participate in a biofilm community [3]. The planktonic phenotype of the microorganism, which appears during initial colonization on a new surface, is considered as the floating or the drifting type of microorganism that can be moved to new habitats at various depths. It can adhere to the surface, populate and become a sessile biofilm, depending on the environmental factors [21]. A sessile community is permanently or covalently bonded to a surface, forming a matrix phenotype or a buoyant. The acquired pellicle, however, is a unique first stage in biofilm formation on an aqueous surface which provides an ideal growth environment, supporting rapid biofilm growth within minutes, such as the pellicle formed closed to the enamel in the oral cavity [2],[22].
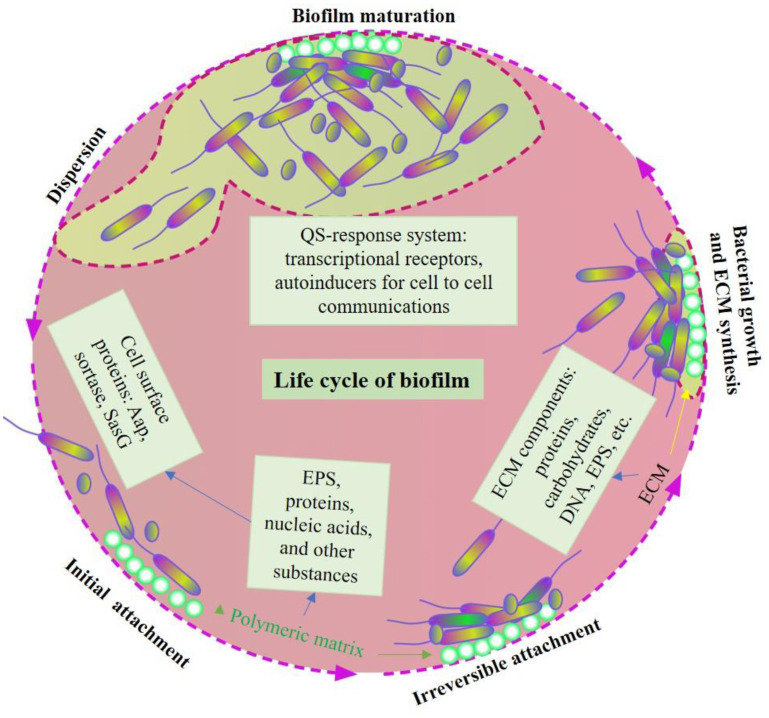
Regarding bacterial involvement, both Gram-positive and Gram-negative bacteria can form biofilms, such as Staphylococcus and Pseudomonas species. Formation of biofilm starts after some free moving microorganisms come into contact with a suitable surface and establish the communication roots to gather sources of nutrients and the production of a slimy material known as EPS [17],[23]. Overall, biofilm formation has five steps: 1) initial reversible attachment between the bacteria and the friendly surface; 2) irreversible attachment that leads to the formation of monolayer surface of extracellular matrix (ECM) comprising proteins, cellular debris, nucleic acids and polysaccharides; 3) maturation stage I, exhibiting biofilm growth and quorum sensing (QS); 4) maturation stage II, leading to formation of EPS to encase the biofilm and helping with cell-cell and cell-matrix interactions; and 5) dispersion stage, separating from parent biofilms and repeating the cycle in another place [3],[24]–[26]. (Figure 1).
Figure 1. Formation, maturation and dispersion of biofilm.
Bacterial attachment on the abiotic or biotic surface starts the biofilm formation. Initial reversible attachment is converted to irreversible attachment by binding of bacteria to the surface by the polymeric matrix, which is made of various exoproteins and substances released by the bacteria themselves. For example, exopolysaccharide substances (EPSs), proteins, nucleic acids and other substances help bacterial adherence on the surfaces. In addition, certain bacterial surface proteins, such as Aap, sortase and SasG, also play vital roles in irreversible attachment. After irreversible attachment, bacteria start division with the release of various extracellular matrix proteins (ECMs) for biofilm growth. Proteins, carbohydrates, DNA and EPS are some major components of ECMs that support bacterial growth during biofilm extension. Various QS response systems, with the help of transcription receptors and autoinducers, support cell-to-cell communications during biofilm maturation. Finally, because of nutritional deficiency and over-population, bacterial colonies start dispersion from the matured biofilm to move to a different place, ultimately leading to the expansion of biofilm.
The detailed explanation of biofilm formation is beyond the scope of this paper. Briefly, bacteria must be close enough to the surface (biotic or abiotic) which allows the attractive and repulsive force that ultimately accepts the interactions with the organisms through various physical mediators. Bacterial mediators, such as flagella, support the initial interactions with the surface, and type IV pili help twitch, adhere and form microcolonies [27],[28] (Figures 1 and 2). Certain human pathogens, such as S. epidermidis and S. aureus, have adhesive genes that recognize the body surface (matrix proteins) for attachments. The number of adhesive genes is variable. For example, S. aureus has over 20 genes and S. epidermidis has only 6 genes [29],[30]. The initial reversible stage changes to the irreversible stage by the strong physical and chemical forces that lead to the formation and maturation of biofilms [31],[32]. Once cells attach irreversibly to the surface, they begin cell division to form microcolonies and secrete EPS, which helps biofilm formation [17],[33] (Figures 1 and 2). Once the first layer of biofilm has been established, cells from the same or different species participate to extend the biofilm further. Biofilm grows outward into a mushroom-like or tower-like formation [34],[35]. Participation of a new bacterial species or strains in the biofilm adds additional substances, such as polysaccharide biopolymers, DNA and proteins in the biofilm, forming complex EPS, and supports biofilm survival in adverse conditions [7],[8],[36].
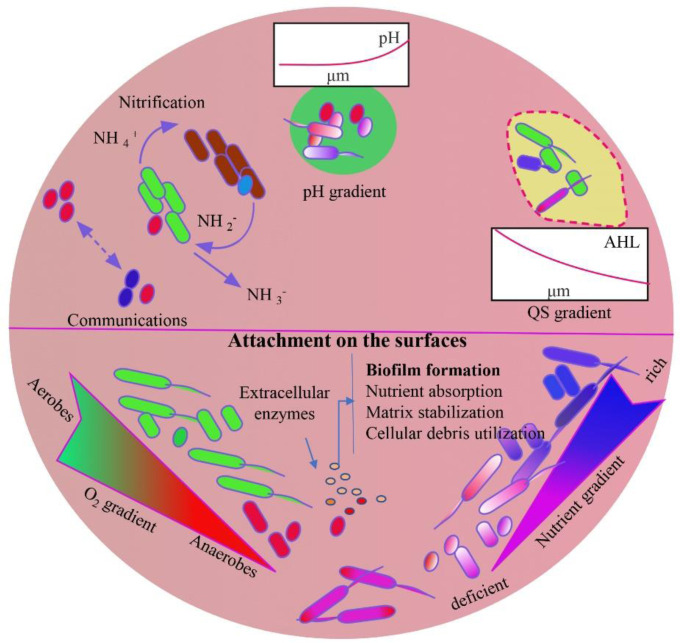
Figure 2. Biofilm microenvironment.
Microorganisms are coordinated with each other and can share their individual survival skills, adding several protective advantages [3],[36]. Development of a biofilm community can be completed within hours, but its size depends on the microorganism types, source of nutrients and biotic or abiotic contact surfaces [17],[37]. Nutritional deficiency leads to dispersal of biofilm organisms to another location, which is known as a seeding dispersal for the search for nutrients [38]–[41]. Biofilm formation is thus a defensive mechanism of the bacteria in a stressful environment [36].
The alternative lifestyle of microorganisms in biofilm not only prolongs lifespan in diverse environments but enables them to linger and persist for a longer period. Anaerobic bacteria can eventually join the thicker biofilm, which has over 100 layers, by entering its deeper layer [42],[43]. Mature biofilms turn eventually into a bacterial community. Active dispersal is a critical step not only to start biofilm at new sites but also to prevent nutritional deficiency and over-population in former established sites [41]. Many other non-organisms associated factors, such as pH, temperature, ionic strength and nutritional availability, play significant roles in affecting the biofilm formation rate [2],[14],[36].
As stated above, biofilms may have single or multiple microbial species. A single species biofilm assumes a temporary lifestyle where each organism participates in a group facilitating survivability in adverse conditions. Multiple species biofilm, such as dental biofilm, may have over 500 bacterial taxa, creating resistance with longer survivability compared to single species biofilms [44]–[46]. Within the biofilms, bacteria are encased by a self- produced EPS, forming microcolonies. Studies demonstrate that microcolonies not only include the same or different bacterial species but also have different organisms, such as fungi [47],[48]. EPS also contains water that helps regulate the flow of the substances and nutrients and maintains the hydrophilic and hydrophobic natures within the biofilms [49],[50]. The water channels separate microcolonies from each other, providing the heterogenous layers of biofilms [2],[51]. In addition, the polysaccharides present in EPS contribute to the ionic bonding with minerals, such as calcium and magnesium [8],[52]. However, the polysaccharides of Gram-positive bacteria display cationic properties [53]. Biofilms show polymorphic appearances with different growth patterns based on the availability of nutrients. For example, microcolonies grow exponentially when glucose is at a high level, facilitating a thick biofilm [2],[54]. Nitrifying bacteria in biofilms also supports biofilm adhesion on positively charged surfaces and increases the biofilm thickness within a short time [55],[56] (Figure 2). Microcolonies indicate a significant growth; however, the biofilms become thinner when there are not sufficient nutrients in their microenvironment [2],[57]. Importantly, EPS arranges in such a way that can shield microcolonies against negative environmental disturbances, such as UV radiation, and against abrupt changes in certain microenvironmental conditions, such as pH [52],[58] (Figure 2).
Biofilm formation starts after attachment of bacteria on the surfaces of biotic or abiotic substances, where they start nutrient absorption, cellular debris utilization and matrix stabilization for biofilm maturation. Gradients of oxygen, pH, QS and nutrition determine the biofilm microenvironment that influences its maturity stages, size and depth. Therefore, biofilm can be a suitable environment for the growth of various types of microorganisms, such as aerobic to anaerobic, alkaliphilic to acidophilic and normal to nutrient-deficient bacteria. Versatile types of bacterial populations in a biofilm contribute to complexity of the ECM, which has various types of enzyme-digested end products of substances, supporting the survival of bacteria in adverse environmental conditions.
3. Biofilm complexity
Biofilm is a complex and dynamic matrix network made of combinations of exopolysaccharides, proteins, lipids and environmental DNA (eDNA) synthesized by different associated microorganisms [2],[59]. The rigidity and structure of ECM can vary within different microbial species, such as Escherichia coli, Bacillus subtilis, Staphylococcus aureus and Vibrio cholerae [2],[14],[60]. Complexity of biofilms can be studied at molecular levels and demonstrated by spectroscopy [61],[62]. For example, E. coli, a biofilm creator, has been studied in detail and is capable of synthesizing a variety of autotransporter adhesins at the beginning of biofilm formation [63],[64]. Antigen 43, an autotransporter adhesin, is encoded by the gene agn43 and has two subunits: α and β. The α subunit is on the superficial surface of the cell and connects the β subunit, an integral outer membrane protein [65],[66]. The L-shaped proteins of α subunits and helical structures of β subunits are stabilized by hydrogen bonds, forming salt bridges. They connect with each other through physio-mechanical processes that are established by self-association and self-aggregation of microbes. A major component of E. coli biofilms is curli fibers [67],[68]. These fibers have two components, CsgB and CsgA, which are encoded by csgBAC [68]. The two major accessory proteins (CsgE and CsgF), and two components (CsgD and CsgG) are produced by csgDEFD. CsgD regulates cellulose and curli fibers, and CsgG plays a role in CsgA translocation [68]. The curli fibers are abundant in a β-sheet structure. The resulting protein network provides integrity of a biofilm, resisting denaturation. Cellulose, another major component of the biofilm matrix, is synthesized by cellulose synthase and is regulated by the bcsABZC [69]. Cellulose provides rigidity and protects from adverse environmental conditions [69]. Environmental DNA (eDNA), another biofilm component, has two layers: outer and inner, as seen in P. aeruginosa and S. aureus biofilms. The outer slime layer provides stability to the biofilm and helps transport nutrients through its pores [14],[70]. The inner layer increases antibiotic resistance by supporting the transfer of genetic information [71].
4. Biofilm communication
It is apparent that bacterial biofilms are pathogenic and can have detrimental effects on an individual's health in addition to resistance to certain drugs [72]. The nature of a biofilm depends on bacteria types, their strains and environmental conditions [14],[73]. Biofilms are composed of microbes, DNA and RNA, polysaccharides, proteins including enzymes and water [60],[74]. Cationic exopolysaccharides, which are suggested to be a polysaccharides rich in glucose, support the biofilm formation [75]. Pentasaccharides enhance the cross-linking between cells and between cells and the surfaces of both biotic and abiotic materials [75],[76]. Mycolic acid, on the other hand, is another vital molecule that protects the organisms in a biofilm by its immune resistance activity [77]. EPS is formed through the complex extra- and intra-cellular processes in microorganisms and plays an important role in biofilm shaping, cell communication (QS) and flow of nutrients [2],[8]. QS is extremely important in the microcolonies of the biofilm and accelerates bacterial gene expression, increasing the bacterial virulence and pathogenicity [2],[78]. It also helps with DNA transformation or plasmid transconjugation from one organism to another, facilitating and increasing the antibiotic resistance among different organisms in biofilm [79]. In addition, QS is widely known for regulating physiological activities of the bacteria through small diffusible signal molecules to increase the social interactions and biofilm pathogenicity [78]. In some bacteria, swarming motility appears to be responsible for the early development of a biofilm because of rapid surface motility [80]. Some components in EPS are exoenzymes that handle the modification, utilization and the formation of EPS layers by acting specifically on extracellular polysaccharides, proteins, lipids and eDNA [81],[82]. Not all bacteria or organisms form EPS, such as mycobacteria. They do not have the matrix structures seen in other biofilms but have other structures that are like EPS [79],[83].
5. Biofilm study through microsystems
Since biofilms are extremely sensitive to environmental factors, their physiologies and functions are variable, creating challenges for effective control or elimination. Microsystems have emerged to control the fluctuations of different environmental factors, hoping to minimize variance among repeated experiments. The optical, electrochemical and mechanical microsystems which are in use to characterize biofilms will be briefly discussed.
Optical microsystems have been standardized and optimized recently to study biofilms. The traditional optical microsystems, such as confocal microscopy, crystal violet staining and scanning electron microscopy (SEM), support end-point measurements of biofilms. Real time, non-invasive, non-destructive and label free optical systems have been developed to monitor the biofilm formation. For example, the modified confocal reflection microscopy (CRM) system allows the visualization of microbial growth in a biofilm over a period [84]. The dynamics of biofilms with high spatial resolution showing chemical bond signatures in biomolecules can be studied through synchrotron radiation-based Fourier transform infrared (SR-FITR) spectromicroscopy [85]. Mechanical properties of a biofilm, such as stiffness of the matrix, have been studied through the Brillouin micro-spectroscopy and Raman micro-spectroscopy [86]. In addition, biofilm physiology can be characterized by surface plasmon resonance (SPR) [87]. It is further advanced as electrochemical SPR, which can help study the formation of electroactive biofilms [88],[89].
The electrochemical biosensors are well-evaluated under three broad categories of operating principle: impedimetric (non-faradaic), potentiometric (faradaic), and amperometric (faradaic) transducers. Impedimetric transducers detect and quantify the bacteria in real time at a temperature by sensing biological samples via impedance [90]. Electrochemical impedance spectroscopy (EIS) extends the study about the adhesion properties of bacteria biofilms [91]. Measurements of concentrations of sodium and potassium ions, oxygen and pH levels within biofilms are possible through multi-electrode systems in a complex environment [92],[93]. Faradaic microsystems are useful in real-time sensing of biofilms and depend on the faradaic current generated because of the redox reaction [94]. Chemical distribution can be monitored through faradaic microsystems, which can characterize the biochemical and regulation processes in microcolonies [90],[95].
Mechanical microsystems, which usually use a thin-film piezoelectric material that can respond to electrical signals in real-time, are commonly used for biofilm sensing. Quartz crystal microbalances (QCMs), surface acoustic wave sensors and quartz tuning for oscillators are applicable to continuously monitoring the bacterial cell attachment, biofilm growth and complex cell-surface interactions [90],[96]–[98].
6. Biofilms in public health
Biofilms have both positive and negative impacts on public health issues [72]. They can be beneficial by protecting our bodies from certain harmful agents present in an environment through remediation of soil and groundwater. They are also helpful for plant protection, wastewater treatment and inhibition of corrosion [99]. However, biofilms are considered detrimental agents to our health. An example would be the formation of dental plaque, leading to serious oral and systemic health problems. Medical devices, such as pacemakers, catheters, prosthetic heart valves, contact lenses, breast implants and cerebrospinal fluid shunts, are the most common abiotic surfaces for biofilm formation. Biofilms can also grow on biotic surfaces, such as teeth, lungs and bone [2],[100]–[102]. Biofilms that grow in water systems supplying healthcare facilities are a serious problem, such as biofilms of Pseudomonas aeruginosa in metal water pipes [103],[104]. Such biofilms transferred to an individual are usually related to life-threatening infections, such as cystic fibrosis, periodontitis, infective endocarditis, otitis media, osteomyelitis and chronic wounds [105],[106]. Biofilms account for up to 80% of microbial infections according to the National Institutes of Health (NIH). Staphylococcal species are the leading cause of implantable device-associated infections [107],[108]. Biofilms play a significant role in various diseases, such as chronic respiratory infection, chronic lung disease, chronic obstructive pulmonary disease (COPD) and ventilator associated pneumonia [109],[110]. Secondary infections can sometimes cause severe bacteremia or septicemia after biofilm organisms enter into the blood through implanted devices [111],[112].
Scientists have realized the biofilm risks to public health. The health and safety issues can be divided into four categories: 1) resistance that leads to increased tolerance to most commonly used antibiotics [113], 2) persistent diseases favoring biofilm extension and modification with variable microcolonies and routinely shifted EPSs [8],[114], 3) failure of the host immune system to destroy biofilms [115] and 4) the cross contamination of clinical instruments that leads to a complex and novel biofilm depositing new and different EPS layers that protect organisms from adverse conditions [79],[116].
Biofilms are resistant to hosts' immune systems, and external stressors such as pH, osmolarity, nutrients and mechanical and shear forces [3],[36]. Resistance and survival, even in adverse conditions, help microbes colonize in various implantable materials, such as dental implants and catheters [100]. Most organisms in a biofilm are protected by the EPS layers and are resistant to various antimicrobial drugs [79],[117],[118]. The glycocalyx can trap antibacterial drugs at their maximum capacity, reducing the effectiveness of the drugs against the organisms present in a biofilm as well as plays an antimicrobial role [30],[118]. Some biofilms release enzyme-like substances that can transform certain antibacterial drugs into non-toxic materials that have no effect on the biofilm organisms [119],[120]. Certain bacteria in biofilms have metal degrading abilities that can convert active-state minerals like copper, zinc and silver to non-active states [121],[122].
7. Biofilm infections
Infections related to bacterial biofilms are categorized into two types: device and non-device related. Infections associated with different medical devices, such as catheters, contact lenses, pacemakers and prosthetics, are mild to severe and may include septicemia and keratitis [111],[123]. The most encountered cause of infection due to bacterial biofilm is often associated with indwelling catheters [22],[102]. Catheters used for less than 10 days have a high chance of biofilm formation on their external surfaces, while catheters used for over 30 days are more likely to develop biofilms in their lumens [2],[23]. Bacteria that grow in catheters depend on the fluid compositions administered through the catheter [22],[124]. The most common species are Gram-negative bacteria, such as P. aeruginosa, Enterobacter and Klebsiella species, which can easily grow in intravenous fluids [125]. The most commonly used catheter is the urinary catheter, which is inserted through the urethra into the bladder and is usually composed of latex or silicon, which favors bacterial attachment [126]. An open system catheter is more prone to bacterial contamination and, therefore, transfers of diseases, such as urinary tract infections (UTIs). E. coli, Enterococcus faecalis and Proteus mirabilis are among the most common pathogens in UTIs [127]. Contact lenses are also susceptible to bacterial biofilm growth because of the adhesive nature of both the lenses and the containers used to keep cleaning lens solution [128]. Bacteria, such as E. coli, S. aureus, Serratia and Proteus, are responsible for biofilm formation in contact lenses and cause severe infections, such as keratitis. In addition, P. aeruginosa may lead to blindness following severe keratitis [128],[129]. Other organisms, such as Streptococcus, Enterococcus and Candida, can form biofilms on mechanical heart valves and cause endocarditis [130],[131].
Non-device related diseases associated with biofilms occur when our own organs provide suitable biotic surfaces to organisms for attachment and support for the release of extracellular proteins which form EPS. For example, Fusobacterium nucleatum and P. aerobicus infect the gingiva and cause periodontitis when oral hygiene is poor [132],[133]. The biofilms established on the tooth surface interfere with the flow of calcium in the epithelial cells, ultimately forming plaque or tartar, which is a mineralized biofilm made of mainly calcium and phosphate ions [132],[134]. Osteomyelitis is another example of non-device related disease that can be caused by biofilms transported to the bone metaphysis through the bloodstream [135]. Immune reactions to the microorganism further deteriorate the bone tissue, leading to fracture [135]–[137]. In addition, the biofilms formed in open wounds of diabetic patients lead to chronic disease. Anaerobic bacteria colonize the interior while aerobic bacteria forms biofilms at the surfaces of deep wounds [138],[139].
Various studies have shown that biofilms are often responsible for chronic infections [72],[140],[141]. S. aureus and P. aeruginosa are most common biofilm organisms for chronic diseases in diabetic patients. Biofilms may delay treatment for years due to antibiotic resistance [139],[142]. Interestingly, the abnormal collection of water in some internal organs supports the reduction of tissue viscosity and accelerates biofilm formation [143]. For example, P. aeruginosa biofilms can cause serious lung infection in patients with cystic fibrosis (CF) that is often difficult to treat [144],[145]. Staphylococci, Streptococci, Gram-negative bacteria, and some fungi can form biofilms in intravenous catheters, and cause prosthetic valve endocarditis (PVE) and native valve endocarditis (NVE) in recipients. Both PVE and NVE can further extend to non-bacterial immune-related thrombotic endocarditis [72]. Coagulase-negative Staphylococcus (CoNS) can cause endocarditis, either by its circulation to the heart from local chronic wounds or by its direct transmission to the heart by contaminated cardiac valve prostheses [146]. Various Gram-positive and Gram-negative microorganisms are constant occupants in beds and often produce chronic polymicrobial infections, such as bed sores [147]. A few clinical circumstances exemplify the concern that biofilms resistant to pro-inflammatory reactions are responsible for severe diseases [148],[149]. The presence of microbial biofilms, which may go unnoticed during diagnosis and treatment phase, can lead to misdiagnosis and severe health consequences [140],[150].
8. Biofilm drug resistance
Diseases caused by microbial biofilms are serious public health issues because of the increasing antimicrobial resistance. It has been demonstrated that bacteria in biofilms are a thousand times more resistant to antibacterial drugs compared to those in the planktonic state [151]. The capsule-like outer EPS layer increases the resilience of organisms by restricting the entry of various antibacterial drugs, in addition to managing various regulatory systems. EPS can expel the harmful molecules out instead of allowing their entry into the biofilms according to the study by Mosaddad et al. [152]. It also acts as a barrier not allowing polar charged antibiotics [119]. The physiological components of a biofilm, such as nutrients and oxygen circulating through water channels in the EPS, also contribute to drug resistance [79],[120]. Over time, biofilms grow in size and shape, leading to the development of an interior anaerobic environment with minimal supply of nutrients and other molecules [42],[54]. Some antibiotics, such as β-lactams, fluoroquinolones and aminoglycosides, are ineffective to the inner anaerobic biofilm since they are inactive in the absence of oxygen and nutrients [153],[154]. Biofilms support gene transfer among microorganisms through their various connection channels, developing antibiotic resistance [118],[155].
Biofilms are structurally and dynamically complex, and they are resistant to common antibiotics [156]. In addition, biofilms have multiple tolerance mechanisms, such as impermeability, rapid growth, differential nutrient gradients and gene transfer [155]. Antibiotic penetration into biofilms primarily relies on the EPS structure that can confer impermeability to large molecules of aminoglycosides [52],[79]. In addition, EPS can quench antibiotic activities through certain mechanisms, such as diffusion-reaction inhibition and enzymatic degradation pathways [52],[157]. Even though EPS composition varies among the different biofilms, it usually contains lipopolysaccharide and alginate, both acting together as a barrier for antibacterial drug diffusion [155],[158]. Increased alginate in the EPS of the biofilm made of a mixed variant of P. aeruginosa helps overcome the immune system, ultimately extending the infection to the more chronic disease state [42],[159]. Mannuronic acid and guluronic acid residues present in the EPS of some biofilms act as potential virulence factors to prolong infections by suppressing immune responses and by protecting the biofilms from antibiotics, such as ciprofloxacin, gentamicin and ceftazidime.
Over time, bacteria can adapt to new adverse environments after horizontal gene transfer among microcolonies with no changes in biofilm [160]. P. aeruginosa is one of the bacteria that possesses a high level of antibiotic resistance. Because of its capacity for frequent spontaneous mutation, it can increase the production of β-lactamase, inactivating the β-lactam antibiotics [161]. Mutation also facilitates adjacent microcolonies in a biofilm taking up free DNA, making more antibiotic resistant biofilms [71]. eDNA increases biofilm resistance by two different mechanisms: 1) changing the outer membrane composition of the biofilm and 2) chelating cations, such as calcium and magnesium, as seen in P. aeruginosa, S. aureus, Enterococcus faecium, Klebsiella pneumoniae, Acinetobacter baumannii, Enterobacter species and Salmonella enterica [162],[163]. All pathogens are responsible for drug-resistant biofilm associated infections.
E. faecium causes biofilm mediated infections in patients implanted with medical devices. It releases eDNA for biofilm attachment and stability by autolysis of its subpopulation through the QS system, similar to S. aureus and S. epidermidis. This process is called fratricide and is mediated by autolysis or murein hydrolase [164],[165]. Low permeability of its cell wall to large molecule aminoglycosides and overexpression of low-affinity penicillin-binding proteins are responsible for antibacterial resistance in E. faecium to aminoglycosides and β-lactam antibiotics, respectively [166]. Rearrangement of the adherent biofilm cells in the monolayers because of genetic alternation in the presence of antibiotics-induced stress also leads to higher resistance [167]. Cell heterogeneity with different QS expression patterns with antibiotic persister cells in a biofilm of S. aureus leads to cell survival in the presence of antibiotics without development of resistance [168],[169]. Antibiotic persister S. aureus has low metabolic activity producing less ATP in the presence of low oxygen and nutrition, which eventually contributes to elevated antibiotic tolerance in biofilm [170]. Certain enzymes, such as proteases and nucleases produced by S. aureus, disperse in the biofilm, enhancing the survivability of the biofilm cells [171].
K. pneumoniae produces a thick layer of extracellular biofilm on both biotic and abiotic surfaces, increasing antibiotic tolerance [172]. Virulence factors, such as polysaccharides, lipopolysaccharides, fimbriae and outer membrane proteins participate in the biofilm formation [173]. K. pneumoniae is a greater biofilm producer than Enterobacter, which is moderate biofilm producer in vitro [174]. Similar to K. pneumoniae, A. baumannii in biofilm is more antibiotic resistant than Enterobacter and causes nosocomial infection after its entry through vascular catheters or Foley catheters. It adheres to the host cells with the help of fibronectin [175],[176]. The surface protein, AbOmpA, is present in the wall surface of A. baumannii. It is responsible for the pore formation in eukaryotic cells, supporting adhesion and biofilm formation along with other adhesion proteins and extracellular polymeric compounds [177]. The pathogens that cause nosocomial infections in hospitals usually form the biofilms which are resistant to multi-drug antibiotics. Among Gram-negative bacteria, E. coli (38%) is a predominant clinical isolate that forms biofilms, along with Acinetobacter species (20%), Klebsiella species (16%) and Pseudomonas species (12%) [178].
Metabolically active organisms release more enzymes and enhance the biofilm growth, influencing drug resistance, while slow-growing organisms have minimal or negligible concentrations of enzymes without affecting drug resistance [179]–[181]. The metabolically active organisms are, however, more susceptible to antibiotics compared to the less active organisms. For example, slow-growing organisms are less susceptible to antibiotics with more efficacy in drug resistance [182]. Most sessile organisms with negligible metabolic activity are categorized as persisters. Unlike mutants, they are at the state of no growth and represent a tiny part of the population with high resistance to antibiotics [156]. Some aerobic organisms, such as P. aeruginosa, become resistant to antibiotics (e.g., ciprofloxacin and tobramycin) when less oxygen is available in the microenvironment [183],[184].
Biofilm resistance and tolerance to antibiotics created by anti-penetration properties of the matrix, presence of polysaccharides, antibiotic-modifying enzymes, eDNA and bacteriophages have been well studied by Hall and Mah [185]. They also mentioned that physiological heterogeneity and hypoxic conditions increase the antibacterial resistance [185]. The heterogeneity creates a nutritional deficiency for certain bacterial populations, which favors more resistance [186]. Biofilm heterogeneity with both genotype and phenotype is one of the most important factors that provide resistance to the bacterial communities in biofilms against many antibacterial drugs [185],[187]. Antibiotic resistance in biofilms can be transferred among the biofilm organisms through external mechanisms such as QS and EPS. They can regulate the bacteria behavior through signaling cascades and gene expression [188],[189].
9. Biofilm control strategies
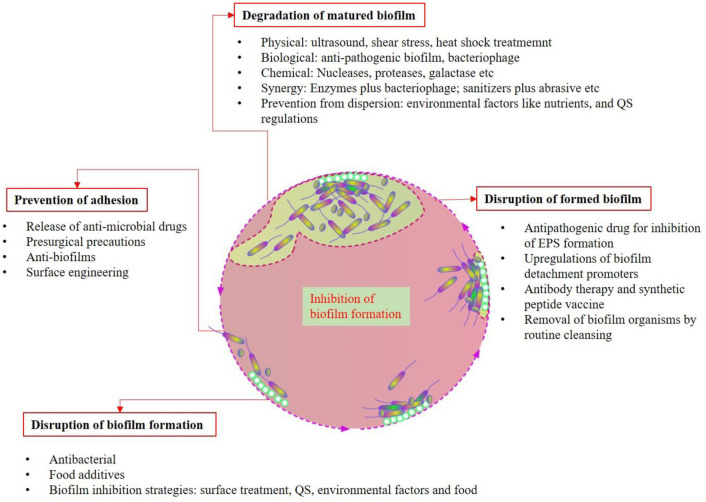
Finding an effective way to control biofilm related diseases is a challenge, but failure to do so will only increase mortality and morbidity rates [190]. Developing new strategies to control biofilms is important for healthcare facilities because of the misuse and overuse of antibiotics, favoring biofilms overcoming the current antibiotics and leading to recurrent infections [191]. In-depth knowledge and understanding of the mechanisms of biofilm matrix formation, its structural components and how communication occurs aid in developing better methods to impede colonization on medical or non-medical devices [69]. Constructing future novel alternative antibiotics or even vaccines targeting these areas can only minimize biofilm long-term infection [2]. Previous studies have demonstrated that combined traditional antibiotics have stronger antibiofilm potency [190]. Strategies depend on the stage of the biofilm, from its attachment to its dispersion, as shown in Figures 1 and 3. Prevention of bacterial adhesion can be accomplished through various strategies, such as coating antimicrobial drugs on implants, presurgical precautions and surface engineering that inhibits the microorganism attachment [192],[193]. Biofilm formation can be disrupted in the initial stage by various antibacterial molecules, food additives and surface treatment, in addition to alterations in other various environmental factors [119],[194] (Figure 3). Elimination of established biofilms is relatively challenging, but there are many approaches, such as EPS inhibitors, detachment promoters, vaccine therapy and mechanical removal of visible biofilms [195]–[198] (Figure 3). Degradation of mature biofilms through various physical, biological, chemical and synergistic methods has been applied in clinical practice [120],[199] (Figure 3).
Figure 3. Inhibition of biofilm formation.
Biofilm formation can be inhibited at any stage of biofilm formation, as described in Figure 3. Bacterial adhesion can be prevented by various approaches, such as release of antimicrobial drugs, presurgical precautions, anti-biofilms and surface engineering. Biofilm formation can be disrupted by using various antibacterial agents, food additives, surface treatments and alterations in environmental factors, etc. Disruption of formed biofilm is challenging compared to that of the immature biofilm. Different strategies have been applied for this purpose: for example, inhibition of EPS formation by application of antipathogenic drugs, upregulation of biofilm detachment promoters, antibody therapy and use of synthetic peptide vaccine and removal of biofilm organisms by routine cleansing procedures. There are various methods that can be applied for the degradation of matured biofilm: for example, physical (e.g., ultrasound, shear stress, heat shock treatment), biological (e.g., anti-pathogenic biofilm, bacteriophage), chemical (e.g., application of enzymes, such as nucleases, proteases and galactase) and synergistic approaches (e.g., enzymes plus bacteriophages and sanitizers plus abrasives) and prevention from dispersal (e.g., application of environmental factors to dysregulate nutrients and QS system).
10. Conventional antibiotics
The failure of classic antibiotic therapies has led to worsening situations globally, such as antibiotic resistance of biofilms in medical implants and catheters. Biofilms dynamically modify their EPS, which makes them impervious to various antibiotics (Figure 4). Orally administered β-lactams, quinolones, aminoglycosides and macrolide antibiotics are prime examples of treatment failures. β-lactams are hydrolyzed by bacterial β-lactamases, so they are ineffective and cannot penetrate biofilms [200]. Quinolones, aminoglycosides and macrolides are transported out of bacterial cells by efflux pumps [201]. In addition, quinolones are not more effective in the anaerobic environment of the biofilm [202]. Although difficult to treat with classic antibiotic therapy, biofilms have shown susceptibility to certain classes of drugs, when delivered correctly and in high doses. To treat these infections, many combination therapies, such as high dose topical treatments or antibiotic adjuvants, have been used [202]. Cephalosporins, aminoglycosides, monobactams, polymyxins, tetracyclines and glycylglycines have been proven to be effective against biofilms through topical administration in higher doses [203]. Biofilm infections in the lungs have been treated successfully through the inhalation of colistin, tobramycin, aztreonam, ciprofloxacin, levofloxacin and gentamicin. Some antibiotics, such as vancomycin, minocycline, linezolid, daptomycin, tigecycline, rifampicin and cephalosporins, applied directly to tubing or medical devices have been shown to prevent biofilm formation [202],[204].
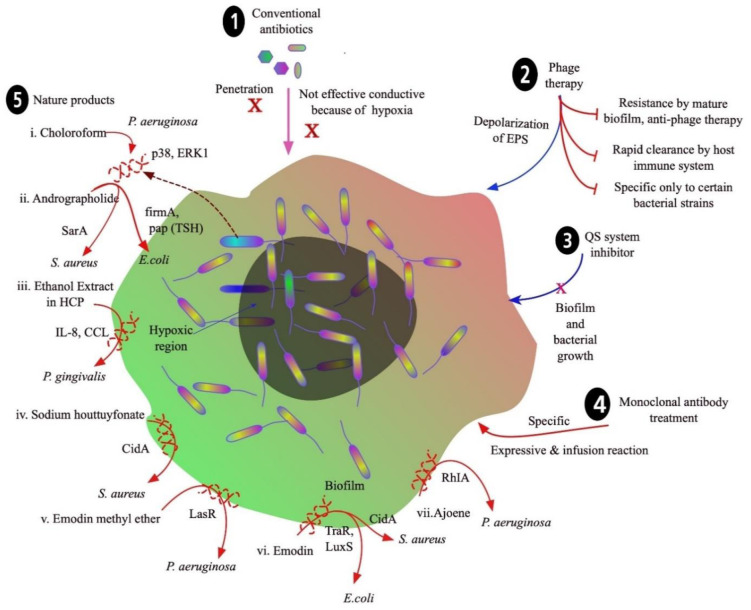
Figure 4. Conventional and new approaches to control biofilm formation.
11. Alternatives to conventional antibiotics
Many new methods of prevention and treatment are being studied and developed because of biofilm resistance to conventional antibiotics. There are various approaches to developing novel antibiofilm agents, such as small molecules capable of inhibiting some virulence factors, and enzymes targeting the matrix. DNase, proteinase K and trypsin are some enzymes that target matrix proteins and degrade eDNA, decreasing the rigidity and stability of the biofilm, as seen in S. epidermidis biofilm [119],[205]. Benzimidazole and N-acetylcysteine are chemicals that inhibit synthesis of EPS and support biofilm degradation [206],[207]. Microbial adherence can be inhibited by using certain chelators that bind ions, such as calcium and magnesium, preventing bacterial attachment to surfaces [208]. Bactericidal or bacteriostatic agents are used to coat the surfaces of medical devices. For example, metal implants are coated with broad-spectrum antibiotics, such as vancomycin, to inhibit biofilm formation. Studies showed effective results in inhibiting S. epidermidis biofilms, but some reports show that bacteria could develop resistance to vancomycin [209],[210]. Some bactericidal agents, such as silver, may inhibit biofilm formation by damaging DNA and microbial proteins [211]. Even though coating medical devices with silver nanoparticles showed good results, the use of high doses can also damage human cells because of their toxic effects [212]. The bio-inspired quercetin nanoparticles act as anti-adhesive agents against certain bacteria (e.g., Bacillus subtilis), preventing surface attachment [213]. Another coating antimicrobial agent is furanone, which shows good results in inhibiting biofilm formation by S. epidermidis [214]. Devices made of silicone rubber are usually coated by trimethoxysilyl propyldimethyloctadecyl ammonium chloride (QAS-30), which has bactericidal and bacteriostatic effects through its quaternary ammonium groups [215]. Another approach for inhibiting biofilm formation in medical devices and prosthetics is the repulsion of microbes from surfaces, which is accomplished by applying an anti-adhesion layer. The coating creates a rough texture or a charged surface, making it less compatible to proteins [216]. Some examples of these coating compounds are trimethylsilane (TMS), selenocyanatodiacetic acid (SCAA), and polymer brush [217]. Medical devices, such as implants, made of stainless steel and titanium coated with TMS showed significant inhibition of microbial biofilms, especially those produced by S. epidermidis. SCAA is an organo-selenium compound that releases superoxide radicals and inhibits biofilm formation in coated surfaces of hemodialysis catheters [218]. Another type of anti-adhesion is polymer brush coating made of polyethylene oxide (PEO), which creates a repulsive osmotic pressure to repel bacteria and proteins. This reduces microbial adhesion in vitro, but it shows no effect in vivo because of its weak attachable capacity to the moisturized medical devices since microbial flagella and pili help movement and adhesion [217]. Most opportunistic organisms that form biofilms can cause infection, but some actually protect against more infectious pathogens [219].
The novel antibiofilm agents are composed of both natural and synthetic molecules, which include halimane diterpenoids, imidazole and indole derivatives, plant extracts, peptides and polysaccharides [42]. Several degrading enzymes, drug delivery nanoparticles and cell-damaging photodynamic therapy (PDT) have also been recently investigated in treating biofilms [79]. Perhaps the most pervasive natural antibiofilm agents are halimane diterpenoids, which can be found in terrestrial plants, marine organisms and bacteria. They are versatile compounds and are used industrially for agricultural, pharmaceutical and beauty products, with their ability for antibacterial and antimycobacterial effects [220],[221]. 2-aminoimidazole, a natural imidazole derivative, has also been used to prevent and dissipate the biofilms formed by methicillin-resistant S. aureus (MRSA), A. baumannii, P. aeruginosa, V. cholerae, K. pneumonia, and Shigella boydii. Besides imidazole derivatives, indole derivatives have been shown to be effective against biofilms by affecting bacterial cell signaling and gene expression [42]. Interestingly, various natural plant extracts, including garlic, hordenine, limonoids and quercetin, are effective against biofilms by inhibiting the transcription of certain genes required for QS. Certain products, such as cranberry polyphenols, inhibit the production of virulence factors that are required for bacterial adhesion and bacterial structures needed for the formation of biofilms [222]. An antibiofilm peptide produced by the human body, called LL-37, stops bacteria from attaching and forming biofilms on mucosal surfaces. LL-37 has been shown to be effective against strains of P. aeruginosa, group A Streptococcus (GAS) and S. epidermidis. Larger polysaccharide molecules, such as galactan and galactose, can also have effects on biofilm formation. Galactan is produced by Kingella kingae and prevents other species of bacteria from forming biofilms [42],[223]. Ethylcholine, one of the novel cholines, inhibits biofilm formation without decreasing bacterial growth, as seen in P. aeruginosa [224]. Some other alternate products are outlined in the Figure 4.
Because of increasing biofilm resistance against currently available antibiotics, different new approaches have been studied, as shown in Figure 4. Conventional methods (1) usually contain antibiotics. Biofilms become more resistant because of the failure of most antibiotics to penetrate the biofilm EPS, and another reason could be a hypoxic environment that hinders effective distribution of antibiotics throughout the biofilm. Phage therapy (2) is another conventional method that disrupts the biofilm and its formation by depolarization of EPS; however, biofilms become resistant against phage therapy. Certain mature biofilms are resistant to phage therapy, and bacteria in biofilms can release anti-phage substances that neutralize the phage. In addition, the immune system clears the phage proteins rapidly from the body, so it is less effective on the biofilm. Another important limitation of phage therapy is that it is more specific to certain bacterial strains, so it is not applicable to other biofilm types. New, emerging strategies have been applied to disrupt biofilm. For example, QS system inhibitors (3), monoclonal antibody treatment (4) and use of nature products (5) are being studied extensively to control and eliminate biofilm. New nature products are under evaluation for their efficacy in eliminating the bacterial biofilms which are more resistant to commonly used broad-spectrum antibiotics. For example, chloroform, andrographolide, ethanol extract of Houttuynia cordata poultice (HCP), sodium houttuyfonate, emodin methyl ether, emodin and ajoene inhibit biofilm formation of different bacteria, such as P. aeruginosa and S. aureus, by suppressing various factors that are responsible for bacterial survival and virulence, as shown in Figure 4.
12. Micromolecules
Understanding microorganisms and their biofilm formation mechanisms might lead to the development of certain micromolecules. Such a strategy has been studied by dentists through the process of cavity formation by Streptococcus sobrinus and S. mutans [225]. This strategy has also been used in other fields to minimize the cases of biofilm infection. However, the rise of drug-resistant bacteria has led to the reassessment of the bacterial mechanisms and new developments by genetic alterations [196],[226].
13. Phage therapy
Phage therapy has been used as an alternate approach for controlling biofilm formation. It works by depolarizing the EPS, leading to the disruption of the biofilm. Mero et al. and Dickey et al. have described phage therapy as an alternative strategy to control biofilm [227],[228]. This method has certain limitations, including resistance by mature biofilms, rapid clearance by the host immune system, ineffectiveness against certain bacterial strains and fast development of phage-resistant subpopulations within a short time period [228]–[230] (Figure 4).
14. QS system inhibitors
QS is a microbial communication mechanism based on molecular signatures and is responsible for the regulation of biological functions, including the production of virulence factors and formation of biofilms [231] (Figure 4). QS system inhibitors are effective in inhibiting regulatory genes and virulence factors [232]. Commercially available anti-QS compounds could increase bacterial biofilm susceptibility to antibiotics both in vivo and in vitro, as demonstrated by Brackman et al. [233]. QS-inhibitors, such as N-(4-[4-fluoroanilno]butanoyl)-L-homoserine lactone (FABHL) and N-(4-[4-chlororoanilno]butanoyl)-L-homoserine lactone (CABHL), can effectively disable the QS system of Pseudomonas aeruginosa by down-regulating the expression levels of lasR and rhlR genes [234]. QS-inhibitors increase the efficacy of certain drugs by acting synergistically to control biofilm formation. This has been demonstrated by experiments with polypeptides [235], cephalosporins [236], aminoglycosides [237] and quinolones [238].
15. Monoclonal antibody treatment
Antibody-based antibiofilm therapy has been implemented and has shown promise in preclinical models that target several components present in biofilms. However, this method has been limited by target specificity and infusion reactions [239] (Figure 4). Pooled monoclonal antibody treatment significantly decreases biofilm formation and demonstrates promising therapeutic effects to prevent biofilm infection. For example, monoclonal antibodies 12C6, 12A1 and 3C1 were used against S. epidermidis, where they specifically bind to the bacterial accumulation-associated protein (AAP) to inhibit the biofilm formation [240]. A native human monoclonal antibody, TRL1068, binds to the DNABII proteins from both Gram-positive and Gram-negative bacteria and disturbs the biofilms of S. aureus and P. aeruginosa [241]. Together with antibiotics, TRL1068 would support the control of biofilm formation [241]. Monoclonal antibodies target specific antigens, such as adhesin proteins (ClFA, FnBPA, Can, SasG), to disrupt the formation of biofilms [242]–[245]. They also bind specifically with cell wall-modifying enzymes (Atl, Atl-Amd, Atl-Gmd, IsaA) and inactivate them to inhibit dynamic changes in the EPS [246]–[249]. Monoclonal antibodies against glycopolymers (WTA, CP and LTA) have been shown to promote opsonophagocytic killing (OPK) effects [250],[251]. Anti-matrix component antibodies such as PNAG and DNABII, immune evasion proteins such as Spa, toxins such as HIa and LukAB and proteins such as PhnD have also been studied [252].
16. Natural products
Biofilm formation by multi-drug resistant microorganisms is a major concern faced by clinicians and scientists in establishing potential antibacterial therapeutics. Several bioactive compounds exhibiting antibacterial activity have been extracted, purified and subjected to clinical testing to check their efficacy in inhibiting biofilms [253]. Natural products, such as chloroform extract, andrographolide, ethanol extract, sodium houttuyfonate, emodin methyl ether and emodin, inhibit the biofilms through various mechanisms that target bacterial cell signatures, as illustrated in figure 4. Chloroform extract of Andrographis paniculata has shown a significant reduction of the QS-controlled extracellular virulence factors in P. aeruginosa infections. It also has the potential to inhibit swarming motility and biofilm formation by decreasing p38 and ERK expression levels in the MAPK signal pathway [254] (Figure 4). Andrographolide disrupts the biofilm of S. aureus by inhibiting the transcriptional factor SarA and also works against E. coli by inhibiting fimA and pap (TSH) [255],[256]. In addition, natural product extracts in ethanol have been widely used for the control and elimination of anaerobic biofilms, such as in the suppression of P. gingivalis through the inhibition of IL-8 and CCL [257],[258]. Sodium houttuyfonate obtained from the oil of the plant Houttuynia cordata has been used for many years in clinical practices as an antimicrobial agent. It has been found to inhibit the biofilms of S. aureus and S. pneumoniae by suppressing the transcription levels of autolysis cidA [259]. Natural medicines, such as emodin and ajoene, have been shown to control the biofilms of various microorganisms (e.g., P. aeruginosa, E. coli and S. aureus) [260]–[263].
17. Probiotics
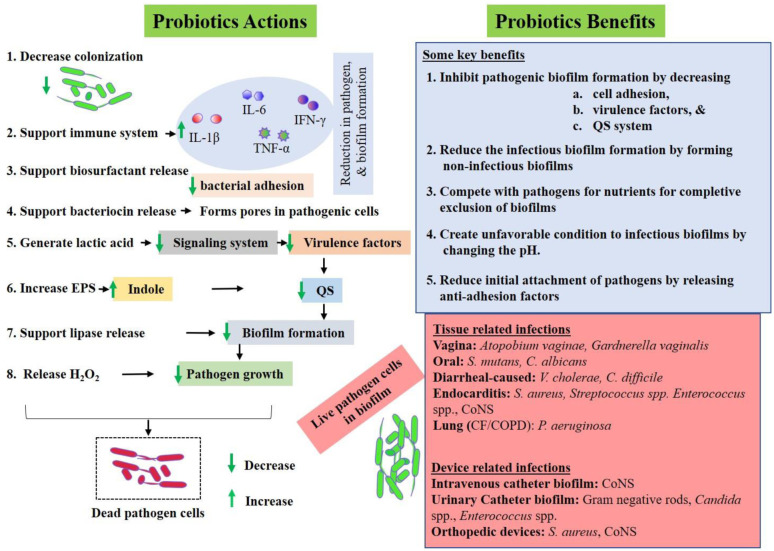
Probiotics are live microorganisms that confer health benefits to the host when they are administered in adequate amounts. Microorganisms, which show inhibitory activities against some pathogens, adherence to epithelial cells and resistance to certain concentrations of bile and acid, are usually considered as probiotics [264],[265]. In addition, they should be able to withstand antibiotics and bind to pathogens sufficiently to inactivate them [266]. Due to increased antibiotic resistance, probiotics that can inhibit biofilm formation have garnered attention. E. faecium WB2000, Bifidobacterium BB12 and Bifidobacterium adolescentis SPM1005 are some probiotics that have been considered for their role in inhibiting bacterial biofilm formation [267]–[269]. As shown in Figure 5, probiotics provide different molecules that help to suppress colonization [270],[271], reduce bacterial adhesion [271], boost the immune system [272], release bacteriocin to inhibit bacterial growth [273], maintain lactic acid in the environment that reduces bacterial virulence signaling systems [274], suppress EPS from pathogens [275] and increase the indole in the biofilm which down-regulates the QS system [276] and lipase inhibitor production [277]. Products of probiotics, such as bacteriocin, can lyse the pathogens by making pores into their cell walls [278]. In addition, hydrogen peroxide (H2O2) might be harmful to certain pathogenic microcolonies within the biofilm [279]. Probiotics, therefore, may play a vital role in inhibiting pathogenic biofilm from its initial stage of attachment and development to its dispersion. This occurs through various mechanisms, such as anti-adhesion activities, QS system suppressors and formation of non-infectious biofilms, that can compete with the nutrients and pH change, as illustrated in Figure 5 [280]–[282]. Probiotics can act as anti-pathogenic biofilms in both tissue-related and device-related infections [283]–[285].
Figure 5. Probiotic approaches to control biofilm formation.
Probiotics support pathogenic biofilm inhibition in various ways, as shown in the figure. Their colonization and release of biosurfactants and bacteriocin supports the inhibition of pathogenic bacteria adhesion and growth. Probiotics produce certain acids, such as lactic acid, that alter the pH in the biofilm microenvironment, suppressing various bacterial signaling systems which are responsible for the QS system, virulence and EPS secretion. In addition, probiotics signal host immune systems to release various inflammatory factors (e.g. IL-1B, IL-6, TNF, interferons) which help to inhibit bacterial growth and biofilm formation. Broadly, probiotics inhibit biofilm formation through various ways, such as reducing virulence of pathogenic bacteria, suppressing QS systems, producing anti-adhesion substances and altering biofilm optimal microenvironment by changing pH, nutrition and oxygen gradient, as shown in the figure. Probiotics would be effective for both tissue and device-related infections caused by various pathogenic biofilms. CoNS: Coagulase negative Staphylococci.
18. Gene editing techniques
Genetic alterations on biofilm pathogens might be possible through gene editing tools like short palindromic repeat-CRISPR- associated (CRISPR-Cas) systems, which could make them less virulent over time [286],[287]. CRISPR-Cas has been used in the engineering of bacterial genetics and the reversal of antibiotic resistance by targeting appropriate genes [288],[289]. The details of CRISPR-Cas gene editing techniques are beyond the scope of this review paper. This technique has been applied to minimize the resistance to various antibacterial drugs carried by plasmids of some infectious bacteria, such as E. coli, and also to target the specific genes in virulence and antibiotic resistance in bacterial populations [290]–[292]. Further, the CRISPR-Cas system has been used to neutralize the bacterial genes responsible for antibiotic resistance [291],[293]. Gene editing techniques are being developed to deliver polymeric nanoparticles effectively and target the virulent gene efficiently, with or without combination with other delivery systems, such as phage delivery or conjugative delivery [294]. Recently, anti-biofilm applications of gene editing techniques have been studied, such as by Zuberi et al. to inhibit E. coli biofilm formation [295], by Tang et al. to inhibit S. mutans biofilm formation [296], and by Garrido et al. to eliminate S. aureus biofilms in vivo [297].
19. Conclusion and future prospects
A less virulent pathogen becomes more virulent, acquiring more antibacterial resistance when it is part of a biofilm. Genetic transfer in a biofilm community modifies the bacterial population, enhancing secretion of various secretory substances and changing cell signatures, which increases the bacterial resistance. Treatment failure is due to acquired resistance through various secretory substances and cell signatures. The spread of multi-drug resistant microorganisms found in biofilms is worsening situations all around the globe and emerging as a new threat to public health. We have discussed how biofilms become more virulent, the roles of various factors responsible for biofilm formation, chronic infections related to biofilms, new strategies that are promising for treating the pathogenic biofilms and various other alternatives such as, natural medicines and probiotics, that act synergistically with antibacterial drugs to combat spreading biofilm infections. In addition, other alternative approaches, such as highly diffusible nano-antibodies or micromolecule formulations, novel anti-pathogenic biofilm agents and CRISPR gene editing technologies, might be future options to treat the multi-drug resistant biofilms. Furthermore, the development of microsystems that mimic the aerobic and anaerobic biofilm microenvironment could support microanalysis of environmental factors, such as pH, nutrients, temperatures and metabolites, that play roles in microbial sensing, virulence and resistance. Controlled guidance systems from microsystems would help to formulate new drugs and set a dose to eliminate the infectious biofilms effectively from both biotic and abiotic substrates.
Acknowledgments
This research received no specific grant from any funding agency in the public, commercial or not-for-profit sectors.
Footnotes
Contributions: G.R. conceived the idea of this review article. G.R. took the lead in writing the manuscript and prepared the illustrating figures. R.M, T.S and H.K. contributed to the writing. All other authors provided information related to the manuscript, provided critical feedback and contributed to the final version of the manuscript.
Ethics declarations: The authors declare no competing interests.
References
- 1.Silva VO, Soares LO, Silva Júnior A, et al. Biofilm formation on biotic and abiotic surfaces in the presence of antimicrobials by Escherichia coli isolates from cases of bovine mastitis. Appl Environ Microbiol. 2014;80:6136–6145. doi: 10.1128/aem.01953-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Donlan RM. Biofilms: microbial life on surfaces. Emerging Infect Dis. 2002;8:881–890. doi: 10.3201/eid0809.020063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yin W, Wang Y, Liu L, et al. Biofilms: The microbial “protective clothing” in extreme environments. Int J Mol Sci. 2019;20:3423. doi: 10.3390/ijms20143423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Koerdt A, Gödeke J, Berger J, et al. Crenarchaeal biofilm formation under extreme conditions. PLoS One. 2010;5:e14104. doi: 10.1371/journal.pone.0014104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen Y, Yan F, Chai Y, et al. Biocontrol of tomato wilt disease by Bacillus subtilis isolates from natural environments depends on conserved genes mediating biofilm formation. Environ Microbiol. 2013;15:848–864. doi: 10.1111/j.1462-2920.2012.02860.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Van Houdt R, Michiels CW. Biofilm formation and the food industry, a focus on the bacterial outer surface. J Appl Microbiol. 2010;109:1117–1131. doi: 10.1111/j.1365-2672.2010.04756.x. [DOI] [PubMed] [Google Scholar]
- 7.Di Martino P. Extracellular polymeric substances, a key element in understanding biofilm phenotype. AIMS Microbiol. 2018;4:274–288. doi: 10.3934/microbiol.2018.2.274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Decho AW, Gutierrez T. Microbial extracellular polymeric substances (EPSs) in ocean systems. Front Microbiol. 2017;8 doi: 10.3389/fmicb.2017.00922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Flemming HC. EPS-Then and Now. Microorganisms. 2016;4:41. doi: 10.3390/microorganisms4040041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kragh KN, Hutchison JB, Melaugh G, et al. Role of multicellular aggregates in biofilm formation. mBio. 2016;7:e00237–00216. doi: 10.1128/mBio.00237-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Taylor M, Ross K, Bentham R. Legionella, Protozoa and Biofilms: Interactions within complex microbial systems. Microbiol Ecol. 2009;58:538–547. doi: 10.1007/s00248-009-9514-z. [DOI] [PubMed] [Google Scholar]
- 12.de Alexandre Sebastião F, Pilarski F, Lemos MVF. Composition of extracellular polymeric substances (EPS) produced by flavobacterium columnare isolated from tropical fish in Brazil. Braz J Microbiol. 2013;44:861–864. doi: 10.1590/S1517-83822013005000058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McSwain BS, Irvine RL, Hausner M, et al. Composition and distribution of extracellular polymeric substances in aerobic flocs and granular sludge. Appl Environ Microbiol. 2005;71:1051. doi: 10.1128/AEM.71.2.1051-1057.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.López D, Vlamakis H, Kolter R. Biofilms. Cold Spring Harbor Perspect Biol. 2010;2:a000398. doi: 10.1101/cshperspect.a000398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nadell CD, Drescher K, Wingreen NS, et al. Extracellular matrix structure governs invasion resistance in bacterial biofilms. ISME J. 2015;9:1700–1709. doi: 10.1038/ismej.2014.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Davenport EK, Call DR, Beyenal H. Differential protection from tobramycin by extracellular polymeric substances from Acinetobacter baumannii and Staphylococcus aureus biofilms. Antimicrob Agents Chemother. 2014;58:4755–4761. doi: 10.1128/AAC.03071-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Donlan RM. Biofilm formation: A clinically relevant microbiological process. Clin Infect Dis. 2001;33:1387–1392. doi: 10.1086/322972. [DOI] [PubMed] [Google Scholar]
- 18.Lewis K. Persister cells. Annu Rev Microbiol. 2010;64:357–372. doi: 10.1146/annurev.micro.112408.134306. [DOI] [PubMed] [Google Scholar]
- 19.Rumbaugh KP, Sauer K. Biofilm dispersion. Nat Rev Microbiol. 2020;18:571–586. doi: 10.1038/s41579-020-0385-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rice SA, Tan CH, Mikkelsen PJ, et al. The biofilm life cycle and virulence of Pseudomonas aeruginosa are dependent on a filamentous prophage. ISME J. 2009;3:271–282. doi: 10.1038/ismej.2008.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Garrett TR, Bhakoo M, Zhang Z. Bacterial adhesion and biofilms on surfaces. Prog Nat Sci. 2008;18:1049–1056. doi: 10.1016/j.pnsc.2008.04.001. [DOI] [Google Scholar]
- 22.Donlan RM. Biofilms and device-associated infections. Emerg Infect Dis. 2001;7:277–281. doi: 10.3201/eid0702.010226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Donlan RM, Costerton JW. Biofilms: survival mechanisms of clinically relevant microorganisms. Clin Microbiol Rev. 2002;15:167–193. doi: 10.1128/cmr.15.2.167-193.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Armbruster CR, Parsek MR. New insight into the early stages of biofilm formation. Proc Natl Acad Sci. 2018;115:4317. doi: 10.1073/pnas.1804084115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Da Cunda P, Iribarnegaray V, Papa-Ezdra R, et al. Characterization of the different stages of biofilm formation and antibiotic susceptibility in a clinical acinetobacter baumannii Strain. Microb Drug Resist. 2019;26:569–575. doi: 10.1089/mdr.2019.0145. [DOI] [PubMed] [Google Scholar]
- 26.Rasamiravaka T, Labtani Q, Duez P, et al. The formation of biofilms by Pseudomonas aeruginosa: a review of the natural and synthetic compounds interfering with control mechanisms. Biomed Res Int. 2015;2015:759348. doi: 10.1155/2015/759348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Koczan JM, Lenneman BR, McGrath MJ, et al. Cell surface attachment structures contribute to biofilm formation and xylem colonization by Erwinia amylovora. Appl Environ Microbiol. 2011;77:7031–7039. doi: 10.1128/AEM.05138-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mandlik A, Swierczynski A, Das A, et al. Pili in Gram-positive bacteria: assembly, involvement in colonization and biofilm development. Trends Microbiol. 2008;16:33–40. doi: 10.1016/j.tim.2007.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Berne C, Ducret A, Hardy GG, et al. Adhesins involved in attachment to abiotic surfaces by Gram-Negative bacteria. Microbiol Spectrum. 2015;3:10. doi: 10.1128/microbiolspec.MB-0018-2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dunne WM., Jr Bacterial adhesion: seen any good biofilms lately? Clin Microbiol Rev. 2002;15:155–166. doi: 10.1128/cmr.15.2.155-166.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Renner LD, Weibel DB. Physicochemical regulation of biofilm formation. MRS Bull. 2011;36:347–355. doi: 10.1557/mrs.2011.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hinsa SM, Espinosa-Urgel M, Ramos JL, et al. Transition from reversible to irreversible attachment during biofilm formation by Pseudomonas fluorescens WCS365 requires an ABC transporter and a large secreted protein. Mol Microbiol. 2003;49:905–918. doi: 10.1046/j.1365-2958.2003.03615.x. [DOI] [PubMed] [Google Scholar]
- 33.Petrova OE, Sauer K. Sticky situations: key components that control bacterial surface attachment. J Bacteriol Res. 2012;194:2413–2425. doi: 10.1128/JB.00003-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Miller JK, Badawy HT, Clemons C, et al. Development of the Pseudomonas aeruginosa mushroom morphology and cavity formation by iron-starvation: a mathematical modeling study. J Theor Biol. 2012;308:68–78. doi: 10.1016/j.jtbi.2012.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Abebe GM. The role of bacterial biofilm in antibiotic resistance and food contamination. Int J Microbiol. 2020;2020:1705814. doi: 10.1155/2020/1705814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jefferson KK. What drives bacteria to produce a biofilm? FEMS Microbiol Lett. 2004;236:163–173. doi: 10.1111/j.1574-6968.2004.tb09643.x. [DOI] [PubMed] [Google Scholar]
- 37.Kurmoo Y, Hook AL, Harvey D, et al. Real time monitoring of biofilm formation on coated medical devices for the reduction and interception of bacterial infections. Biomater Sci. 2020;8:1464–1477. doi: 10.1039/c9bm00875f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Salgar-Chaparro SJ, Lepkova K, Pojtanabuntoeng T, et al. Nutrient level determines biofilm characteristics and subsequent impact on microbial corrosion and biocide effectiveness. Appl Environ Microbiol. 2020;86:e02885–02819. doi: 10.1128/AEM.02885-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wilson C, Lukowicz R, Merchant S, et al. Quantitative and qualitative assessment methods for biofilm growth: A mini-review. Res Rev J Eng Technol. 2017;6 Available from: http://www.rroij.com/open-access/quantitative-and-qualitative-assessment-methods-for-biofilm-growth-a-minireview-.pdf. [PMC free article] [PubMed] [Google Scholar]
- 40.Haney EF, Trimble MJ, Cheng JT, et al. Critical assessment of methods to quantify biofilm growth and evaluate antibiofilm activity of host defence peptides. Biomolecules. 2018;8:29. doi: 10.3390/biom8020029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kaplan JB. Biofilm dispersal: mechanisms, clinical implications, and potential therapeutic uses. J Dent Res. 2010;89:205–218. doi: 10.1177/0022034509359403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rabin N, Zheng Y, Opoku-Temeng C, et al. Biofilm formation mechanisms and targets for developing antibiofilm agents. Future Med Chem. 2015;7:493–512. doi: 10.4155/fmc.15.6. [DOI] [PubMed] [Google Scholar]
- 43.Sharpton TJ, Combrink L, Arnold HK, et al. Erratum to “Harnessing the gut microbiome in the fight against anthelminthic drug resistance”. Curr Opin Microbiol. 2021;53:26–34. doi: 10.1016/j.mib.2021.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Seth AK, Geringer MR, Hong SJ, et al. Comparative analysis of single-species and polybacterial wound biofilms using a quantitative, in vivo, rabbit ear model. PLoS One. 2012;7:e42897. doi: 10.1371/journal.pone.0042897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yang L, Liu Y, Wu H, et al. Current understanding of multi-species biofilms. Int J Oral Sci. 2011;3:74–81. doi: 10.4248/IJOS11027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lohse MB, Gulati M, Johnson AD, et al. Development and regulation of single- and multi-species Candida albicans biofilms. Nat Rev Microbiol. 2018;16:19–31. doi: 10.1038/nrmicro.2017.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lynch AS, Robertson GT. Bacterial and fungal biofilm infections. Annu Rev Med. 2008;59:415–428. doi: 10.1146/annurev.med.59.110106.132000. [DOI] [PubMed] [Google Scholar]
- 48.Seneviratne G, Zavahir JS, Bandara WMMS, et al. Fungal-bacterial biofilms: their development for novel biotechnological applications. World J Microbiol Biotechnol. 2007;24:739. doi: 10.1007/s11274-007-9539-8. [DOI] [Google Scholar]
- 49.Guo YS, Furrer JM, Kadilak AL, et al. Bacterial extracellular polymeric substances amplify water content variability at the pore scale. Front Environ Sci. 2018;6 doi: 10.3389/fenvs.2018.00093. [DOI] [Google Scholar]
- 50.Wingender J, Neu TR, Flemming HC, et al. Microbial extracellular polymeric substances: Characterization, structure and function. Berlin: Springer Berlin Heidelberg; 1999. What are bacterial extracellular polymeric substances? pp. 1–19. [DOI] [Google Scholar]
- 51.Wilking JN, Zaburdaev V, De Volder M, et al. Liquid transport facilitated by channels in Bacillus subtilis biofilms. Proc Natl Acad Sci U S A. 2013;110:848–852. doi: 10.1073/pnas.1216376110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Costa OYA, Raaijmakers JM, Kuramae EE. Microbial extracellular polymeric substances: ecological function and impact on soil aggregation. Front Microbiol. 2018;9:1636–1636. doi: 10.3389/fmicb.2018.01636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Navarre WW, Schneewind O. Surface proteins of gram-positive bacteria and mechanisms of their targeting to the cell wall envelope. Microbiol Mol Biol Rev. 1999;63:174. doi: 10.1128/MMBR.63.1.174-229.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Davey ME, O'Toole GA. Microbial biofilms: from ecology to molecular genetics. Microbiol Mol Biol Rev: MMBR. 2000;64:847–867. doi: 10.1128/mmbr.64.4.847-867.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hibiya K, Tsuneda S, Hirata A. Formation and characteristics of nitrifying biofilm on a membrane modified with positively-charged polymer chains. Colloids Surf, B. 2000;18:105–112. doi: 10.1016/S0927-7765(99)00141-1. [DOI] [Google Scholar]
- 56.Navada S, Knutsen MF, Bakke I, et al. Nitrifying biofilms deprived of organic carbon show higher functional resilience to increases in carbon supply. Sci Rep. 2020;10:7121. doi: 10.1038/s41598-020-64027-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hunt SM, Werner EM, Huang B, et al. Hypothesis for the role of nutrient starvation in biofilm detachment. Appl Environ Microbiol. 2004;70:7418–7425. doi: 10.1128/AEM.70.12.7418-7425.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Maddela NR, Zhou Z, Yu Z, et al. Functional determinants of extracellular polymeric substances in membrane biofouling: Experimental evidence from pure-cultured sludge bacteria. Appl Environ Microbiol. 2018;84:e00756–00718. doi: 10.1128/AEM.00756-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Stoodley P, Sauer K, Davies DG, et al. Biofilms as complex differentiated communities. Annu Rev Microbiol. 2002;56:187–209. doi: 10.1146/annurev.micro.56.012302.160705. [DOI] [PubMed] [Google Scholar]
- 60.Limoli DH, Jones CJ, Wozniak DJ. Bacterial extracellular polysaccharides in biofilm formation and function. Microbiol Spectrum. 2015;3:10. doi: 10.1128/microbiolspec.MB-0011-2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Magana M, Sereti C, Ioannidis A, et al. Options and limitations in clinical investigation of bacterial biofilms. Clin Microbiol Rev. 2018;31:e00084–16. doi: 10.1128/CMR.00084-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Denef VJ, Mueller RS, Banfield JF. AMD biofilms: using model communities to study microbial evolution and ecological complexity in nature. ISME J. 2010;4:599–610. doi: 10.1038/ismej.2009.158. [DOI] [PubMed] [Google Scholar]
- 63.Martinez-Gil M, Goh KGK, Rackaityte E, et al. YeeJ is an inverse autotransporter from Escherichia coli that binds to peptidoglycan and promotes biofilm formation. Sci Rep. 2017;7:11326. doi: 10.1038/s41598-017-10902-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Beloin C, Roux A, Ghigo JM. Escherichia coli biofilms. Curr Top Microbio Immunol. 2008;322:249–289. doi: 10.1007/978-3-540-75418-3_12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Klemm P, Hjerrild L, Gjermansen M, et al. Structure-function analysis of the self-recognizing Antigen 43 autotransporter protein from Escherichia coli. Mol Microbiol. 2004;51:283–296. doi: 10.1046/j.1365-2958.2003.03833.x. [DOI] [PubMed] [Google Scholar]
- 66.Müller D, Benz I, Tapadar D, et al. Arrangement of the translocator of the autotransporter adhesin involved in diffuse adherence on the bacterial surface. Infect Immun. 2005;73:3851–3859. doi: 10.1128/IAI.73.7.3851-3859.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Barnhart MM, Chapman MR. Curli biogenesis and function. Annu Rev Microbiol. 2006;60:131–147. doi: 10.1146/annurev.micro.60.080805.142106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Tursi SA, Tükel Ç. Curli-Containing enteric biofilms inside and out: Matrix composition, immune recognition, and disease implications. Microbiol Mol Biol Rev. 2018;82:e00028–18. doi: 10.1128/MMBR.00028-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hobley L, Harkins C, MacPhee CE, et al. Giving structure to the biofilm matrix: an overview of individual strategies and emerging common themes. FEMS Microbiol Rev. 2015;39:649–669. doi: 10.1093/femsre/fuv015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Chen XD, Zhang CK, Zhou Z, et al. Stabilizing effects of bacterial biofilms: EPS penetration and redistribution of bed stability down the sediment profile. J Geophys Res: Biogeosci. 2017;122:3113–3125. doi: 10.1002/2017JG004050. [DOI] [Google Scholar]
- 71.Ibáñez de Aldecoa AL, Zafra O, González-Pastor JE. Mechanisms and regulation of extracellular DNA release and its biological roles in microbial communities. Front Microbiol. 2017;8:1390–1390. doi: 10.3389/fmicb.2017.01390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Jamal M, Ahmad W, Andleeb S, et al. Bacterial biofilm and associated infections. J Chin Med Assoc. 2018;81:7–11. doi: 10.1016/j.jcma.2017.07.012. [DOI] [PubMed] [Google Scholar]
- 73.Toyofuku M, Inaba T, Kiyokawa T, et al. Environmental factors that shape biofilm formation. Biosci Biotechnol Biochem. 2016;80:7–12. doi: 10.1080/09168451.2015.1058701. [DOI] [PubMed] [Google Scholar]
- 74.Vu B, Chen M, Crawford RJ, et al. Bacterial extracellular polysaccharides involved in biofilm formation. Molecules (Basel,Switz) 2009;14:2535–2554. doi: 10.3390/molecules14072535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Skariyachan S, Sridhar VS, Packirisamy S, et al. Recent perspectives on the molecular basis of biofilm formation by Pseudomonas aeruginosa and approaches for treatment and biofilm dispersal. Folia Microbiol. 2018;63:413–432. doi: 10.1007/s12223-018-0585-4. [DOI] [PubMed] [Google Scholar]
- 76.Mann EE, Wozniak DJ. Pseudomonas biofilm matrix composition and niche biology. FEMS Microbiol Rev. 2012;36:893–916. doi: 10.1111/j.1574-6976.2011.00322.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zambrano MM, Kolter R. Mycobacterial biofilms: A greasy way to hold it together. Cell. 2005;123:762–764. doi: 10.1016/j.cell.2005.11.011. [DOI] [PubMed] [Google Scholar]
- 78.Li YH, Tian X. Quorum sensing and bacterial social interactions in biofilms. Sensors (Basel,Switz) 2012;12:2519–2538. doi: 10.3390/s120302519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Sharma D, Misba L, Khan AU. Antibiotics versus biofilm: an emerging battleground in microbial communities. Antimicrob Resist Infect Control. 2019;8:76. doi: 10.1186/s13756-019-0533-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Shrout JD, Chopp DL, Just CL, et al. The impact of quorum sensing and swarming motility on Pseudomonas aeruginosa biofilm formation is nutritionally conditional. Mol Microbiol. 2006;62:1264–1277. doi: 10.1111/j.1365-2958.2006.05421.x. [DOI] [PubMed] [Google Scholar]
- 81.Saygin H, Baysal A. Biofilm formation of clinically important bacteria on bio-based and conventional micro/submicron-sized plastics. Bull Environ Contam Toxicol. 2020;105:18–25. doi: 10.1007/s00128-020-02876-z. [DOI] [PubMed] [Google Scholar]
- 82.Fulaz S, Vitale S, Quinn L, et al. Nanoparticle-Biofilm interactions: The role of the EPS matrix. Trends Microbiol. 2019;27:915–926. doi: 10.1016/j.tim.2019.07.004. [DOI] [PubMed] [Google Scholar]
- 83.Chakraborty P, Kumar A. The extracellular matrix of mycobacterial biofilms: could we shorten the treatment of mycobacterial infections? Microb Cell (Graz,Austria) 2019;6:105–122. doi: 10.15698/mic2019.02.667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Yawata Y, Uchiyama H, Nomura N. Visualizing the effects of biofilm structures on the influx of fluorescent material using combined confocal reflection and fluorescent microscopy. Microbes Environ. 2010;25:49–52. doi: 10.1264/jsme2.me09169. [DOI] [PubMed] [Google Scholar]
- 85.Holman HY, Miles R, Hao Z, et al. Real-time chemical imaging of bacterial activity in biofilms using open-channel microfluidics and synchrotron FTIR spectromicroscopy. Anal Chem. 2009;81:8564–8570. doi: 10.1021/ac9015424. [DOI] [PubMed] [Google Scholar]
- 86.Mattana S, Alunni Cardinali M, Caponi S, et al. High-contrast brillouin and raman micro-spectroscopy for simultaneous mechanical and chemical investigation of microbial biofilms. Biophys Chem. 2017;229:123–129. doi: 10.1016/j.bpc.2017.06.008. [DOI] [PubMed] [Google Scholar]
- 87.Abadian PN, Tandogan N, Jamieson JJ, et al. Using surface plasmon resonance imaging to study bacterial biofilms. Biomicrofluidics. 2014;8:021804. doi: 10.1063/1.4867739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.McGoverin C, Vanholsbeeck F, Dawes JM, et al. Asia-pacific optical sensors conference: focus issue introduction. Opt Express. 2020;28:21745–21748. doi: 10.1364/OE.401277. [DOI] [PubMed] [Google Scholar]
- 89.Yuan Y, Guo T, Qiu X, et al. Electrochemical surface plasmon resonance fiber-optic sensor: In situ detection of electroactive biofilms. Anal Chem. 2016;88:7609–7616. doi: 10.1021/acs.analchem.6b01314. [DOI] [PubMed] [Google Scholar]
- 90.Subramanian S, Huiszoon RC, Chu S, et al. Microsystems for biofilm characterization and sensing-A review. Biofilm. 2019;2:100015. doi: 10.1016/j.bioflm.2019.100015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Stöckl M, Schlegel C, Sydow A, et al. Membrane separated flow cell for parallelized electrochemical impedance spectroscopy and confocal laser scanning microscopy to characterize electro-active microorganisms. Electrochim Acta. 2016;220:444–452. doi: 10.1016/j.electacta.2016.10.057. [DOI] [Google Scholar]
- 92.Estrada-Leypon O, Moya A, Guimera A, et al. Simultaneous monitoring of Staphylococcus aureus growth in a multi-parametric microfluidic platform using microscopy and impedance spectroscopy. Bioelectrochemistry. 2015;105:56–64. doi: 10.1016/j.bioelechem.2015.05.006. [DOI] [PubMed] [Google Scholar]
- 93.Huiszoon RC, Subramanian S, Ramiah Rajasekaran P, et al. Flexible platform for in situ impedimetric detection and bioelectric effect treatment of Escherichia coli biofilms. IEEE Trans Biomed Eng. 2019;66:1337–1345. doi: 10.1109/tbme.2018.2872896. [DOI] [PubMed] [Google Scholar]
- 94.Bayoudh S, Othmane A, Ponsonnet L, et al. Electrical detection and characterization of bacterial adhesion using electrochemical impedance spectroscopy-based flow chamber. Colloids Surf, A. 2008;318:291–300. doi: 10.1016/j.colsurfa.2008.01.005. [DOI] [Google Scholar]
- 95.Bellin DL, Sakhtah H, Zhang Y, et al. Electrochemical camera chip for simultaneous imaging of multiple metabolites in biofilms. Nat Commun. 2016;7:10535. doi: 10.1038/ncomms10535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Marcus IM, Herzberg M, Walker SL, et al. Pseudomonas aeruginosa attachment on QCM-D sensors: the role of cell and surface hydrophobicities. Langmuir. 2012;28:6396–6402. doi: 10.1021/la300333c. [DOI] [PubMed] [Google Scholar]
- 97.Olsson AL, van der Mei HC, Busscher HJ, et al. Influence of cell surface appendages on the bacterium-substratum interface measured real-time using QCM-D. Langmuir. 2009;25:1627–1632. doi: 10.1021/la803301q. [DOI] [PubMed] [Google Scholar]
- 98.Piasecki T, Guła G, Markwitz P, et al. Autonomous system for in situ assay of antibiotic activity on bacterial biofilms using viscosity and density sensing quartz tuning forks. Procedia Eng. 2016;168:745–748. doi: 10.1016/j.proeng.2016.11.267. [DOI] [Google Scholar]
- 99.Sfaelou S, Karapanagioti HK, Vakros J. Studying the formation of biofilms on supports with different polarity and their efficiency to treat wastewater. J Chem. 2015;2015:734384. doi: 10.1155/2015/734384. [DOI] [Google Scholar]
- 100.Khatoon Z, McTiernan CD, Suuronen EJ, et al. Bacterial biofilm formation on implantable devices and approaches to its treatment and prevention. Heliyon. 2018;4:e01067. doi: 10.1016/j.heliyon.2018.e01067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Revdiwala S, Rajdev BM, Mulla S. Characterization of bacterial etiologic agents of biofilm formation in medical devices in critical care setup. Crit Care Res Pract. 2012;2012:945805. doi: 10.1155/2012/945805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Trautner BW, Darouiche RO. Role of biofilm in catheter-associated urinary tract infection. Am J Infect Control. 2004;32:177–183. doi: 10.1016/j.ajic.2003.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Liu S, Gunawan C, Barraud N, et al. Understanding, monitoring, and controlling biofilm growth in drinking water distribution systems. Environ Sci Technol. 2016;50:8954–8976. doi: 10.1021/acs.est.6b00835. [DOI] [PubMed] [Google Scholar]
- 104.Maes S, Vackier T, Nguyen Huu S, et al. Occurrence and characterisation of biofilms in drinking water systems of broiler houses. BMC Microbiol. 2019;19:77. doi: 10.1186/s12866-019-1451-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Muhammad MH, Idris AL, Fan X, et al. Beyond risk: bacterial biofilms and their regulating approaches. Front Microbiol. 2020;11:928–928. doi: 10.3389/fmicb.2020.00928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Ashbolt NJ. Microbial contamination of drinking water and human health from community water systems. Curr Environ Health Rep. 2015;2:95–106. doi: 10.1007/s40572-014-0037-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Archer NK, Mazaitis MJ, Costerton JW, et al. Staphylococcus aureus biofilms: properties, regulation, and roles in human disease. Virulence. 2011;2:445–459. doi: 10.4161/viru.2.5.17724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Arciola CR, Campoccia D, Montanaro L. Implant infections: adhesion, biofilm formation and immune evasion. Nat Rev Microbiol. 2018;16:397–409. doi: 10.1038/s41579-018-0019-y. [DOI] [PubMed] [Google Scholar]
- 109.Boisvert AA, Cheng MP, Sheppard DC, et al. Microbial biofilms in pulmonary and critical care diseases. Ann Am Thorac Soc. 2016;13:1615–1623. doi: 10.1513/AnnalsATS.201603-194FR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Gnanadhas DP, Elango M, Datey A, et al. Chronic lung infection by Pseudomonas aeruginosa biofilm is cured by L-Methionine in combination with antibiotic therapy. Sci Rep. 2015;5:16043. doi: 10.1038/srep16043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Minasyan H. Sepsis: mechanisms of bacterial injury to the patient. Scand J Trauma Resusc Emerg Med. 2019;27:19. doi: 10.1186/s13049-019-0596-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.VanEpps JS, Younger JG. Implantable device-related infection. Shock (Augusta,Ga) 2016;46:597–608. doi: 10.1097/SHK.0000000000000692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Stewart PS. Mechanisms of antibiotic resistance in bacterial biofilms. Int J Med Microbiol. 2002;292:107–113. doi: 10.1078/1438-4221-00196. [DOI] [PubMed] [Google Scholar]
- 114.Vestby LK, Grønseth T, Simm R, et al. Bacterial biofilm and its role in the pathogenesis of disease. Antibiotics (Basel,Switz) 2020;9:59. doi: 10.3390/antibiotics9020059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.González JF, Hahn MM, Gunn JS. Chronic biofilm-based infections: skewing of the immune response. Pathog Dis. 2018;76:fty023. doi: 10.1093/femspd/fty023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Elias S, Banin E. Multi-species biofilms: living with friendly neighbors. FEMS Microbiol Rev. 2012;36:990–1004. doi: 10.1111/j.1574-6976.2012.00325.x. [DOI] [PubMed] [Google Scholar]
- 117.Hu X, Kang F, Yang B, et al. Extracellular polymeric substances acting as a permeable barrier hinder the lateral transfer of antibiotic resistance genes. Front Microbiol. 2019;10 doi: 10.3389/fmicb.2019.00736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Singh S, Singh SK, Chowdhury I, et al. Understanding the mechanism of bacterial biofilms resistance to antimicrobial agents. Open Microbiol J. 2017;11:53–62. doi: 10.2174/1874285801711010053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Roy R, Tiwari M, Donelli G, et al. Strategies for combating bacterial biofilms: A focus on anti-biofilm agents and their mechanisms of action. Virulence. 2018;9:522–554. doi: 10.1080/21505594.2017.1313372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Koo H, Allan RN, Howlin RP, et al. Targeting microbial biofilms: current and prospective therapeutic strategies. Nat Rev Microbiol. 2017;15:740–755. doi: 10.1038/nrmicro.2017.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Igiri BE, Okoduwa SIR, Idoko GO, et al. Toxicity and bioremediation of heavy metals contaminated ecosystem from tannery wastewater: A review. J Toxicol. 2018;2018:2568038. doi: 10.1155/2018/2568038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Singh R, Paul D, Jain RK. Biofilms: implications in bioremediation. Trends Microbiol. 2006;14:389–397. doi: 10.1016/j.tim.2006.07.001. [DOI] [PubMed] [Google Scholar]
- 123.Bispo PJM, Haas W, Gilmore MS. Biofilms in infections of the eye. Pathogens (Basel,Switz) 2015;4:111–136. doi: 10.3390/pathogens4010111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Nickel JC, Costerton JW. Bacterial biofilms and catheters: A key to understanding bacterial strategies in catheter-associated urinary tract infection. Can J Infect Dis. 1992;3:261–267. doi: 10.1155/1992/517456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Guynn JB, Jr., Poretz DM, Duma RJ. Growth of various bacteria in a variety of intravenous fluids. Am J Hosp Pharm. 1973;30:321–325. doi: 10.1093/ajhp/30.4.321. [DOI] [PubMed] [Google Scholar]
- 126.Ferrières L, Hancock V, Klemm P. Specific selection for virulent urinary tract infectious Escherichia coli strains during catheter-associated biofilm formation. FEMS Immunol Med Microbiol. 2007;51:212–219. doi: 10.1111/j.1574-695X.2007.00296.x. [DOI] [PubMed] [Google Scholar]
- 127.Delcaru C, Alexandru I, Podgoreanu P, et al. Microbial biofilms in urinary tract infections and prostatitis: Etiology, pathogenicity, and combating strategies. Pathogens (Basel,Switz) 2016;5:65. doi: 10.3390/pathogens5040065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Wiley L, Bridge DR, Wiley LA, et al. Bacterial biofilm diversity in contact lens-related disease: Emerging role of achromobacter, stenotrophomonas, and delftia. Invest Ophthalmol Visual Sci. 2012;53:3896–3905. doi: 10.1167/iovs.11-8762. [DOI] [PubMed] [Google Scholar]
- 129.Robertson DM, Parks QM, Young RL, et al. Disruption of contact lens-associated Pseudomonas aeruginosa biofilms formed in the presence of neutrophils. Invest Ophthalmol Visual Sci. 2011;52:2844–2850. doi: 10.1167/iovs.10-6469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Litzler PY, Benard L, Barbier-Frebourg N, et al. Biofilm formation on pyrolytic carbon heart valves: influence of surface free energy, roughness, and bacterial species. J Thorac Cardiovasc Surg. 2007;134:1025–1032. doi: 10.1016/j.jtcvs.2007.06.013. [DOI] [PubMed] [Google Scholar]
- 131.Piper C, Körfer R, Horstkotte D. Prosthetic valve endocarditis. Heart. 2001;85:590. doi: 10.1136/heart.85.5.590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Schaudinn C, Gorur A, Keller D, et al. Periodontitis: an archetypical biofilm disease. J Am Dent Assoc. 2009;140:978–986. doi: 10.14219/jada.archive.2009.0307. [DOI] [PubMed] [Google Scholar]
- 133.Naginyte M, Do T, Meade J, et al. Enrichment of periodontal pathogens from the biofilms of healthy adults. Sci Rep. 2019;9:5491. doi: 10.1038/s41598-019-41882-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Chandki R, Banthia P, Banthia R. Biofilms: A microbial home. J Indian Soc Periodontol. 2011;15:111–114. doi: 10.4103/0972-124X.84377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Brady RA, Leid JG, Calhoun JH, et al. Osteomyelitis and the role of biofilms in chronic infection. FEMS Immunol Med Microbiol. 2008;52:13–22. doi: 10.1111/j.1574-695X.2007.00357.x. [DOI] [PubMed] [Google Scholar]
- 136.Masters EA, Trombetta RP, de Mesy Bentley KL, et al. Evolving concepts in bone infection: redefining “biofilm”, “acute vs. chronic osteomyelitis”, “the immune proteome” and “local antibiotic therapy”. Bone Res. 2019;7:20. doi: 10.1038/s41413-019-0061-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Rochford ETJ, Sabaté Brescó M, Zeiter S, et al. Monitoring immune responses in a mouse model of fracture fixation with and without Staphylococcus aureus osteomyelitis. Bone. 2016;83:82–92. doi: 10.1016/j.bone.2015.10.014. [DOI] [PubMed] [Google Scholar]
- 138.Oates A, Bowling FL, Boulton AJM, et al. The visualization of biofilms in chronic diabetic foot wounds using routine diagnostic microscopy methods. J Diabetes Res. 2014;2014:153586. doi: 10.1155/2014/153586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Clinton A, Carter T. Chronic wound biofilms: Pathogenesis and potential therapies. Lab Med. 2015;46:277–284. doi: 10.1309/lmbnswkui4jpn7so. [DOI] [PubMed] [Google Scholar]
- 140.Bjarnsholt T. The role of bacterial biofilms in chronic infections. APMIS Suppl. 2013:1–51. doi: 10.1111/apm.12099. [DOI] [PubMed] [Google Scholar]
- 141.Burmølle M, Thomsen TR, Fazli M, et al. Biofilms in chronic infections-a matter of opportunity-monospecies biofilms in multispecies infections. FEMS Immunol Med Microbiol. 2010;59:324–336. doi: 10.1111/j.1574-695X.2010.00714.x. [DOI] [PubMed] [Google Scholar]
- 142.Percival SL, McCarty SM, Lipsky B. Biofilms and wounds: An overview of the evidence. Adv Wound Care. 2015;4:373–381. doi: 10.1089/wound.2014.0557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Abdel-Nour M, Duncan C, Low DE, et al. Biofilms: the stronghold of Legionella pneumophila. Int J Mol Sci. 2013;14:21660–21675. doi: 10.3390/ijms141121660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Høiby N, Ciofu O, Bjarnsholt T. Pseudomonas aeruginosa biofilms in cystic fibrosis. Future Microbiol. 2010;5:1663–1674. doi: 10.2217/fmb.10.125. [DOI] [PubMed] [Google Scholar]
- 145.Moreau-Marquis S, Stanton BA, O'Toole GA. Pseudomonas aeruginosa biofilm formation in the cystic fibrosis airway. Pulm Pharmacol Ther. 2008;21:595–599. doi: 10.1016/j.pupt.2007.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Lalani T, Kanafani ZA, Chu VH, et al. Prosthetic valve endocarditis due to coagulase-negative staphylococci: findings from the international collaboration on endocarditis merged database. Eur J Clin Microbiol Infect Dis. 2006;25:365–368. doi: 10.1007/s10096-006-0141-z. [DOI] [PubMed] [Google Scholar]
- 147.van Steenbergen TJM, van Winkelhoff AJ, de Graaff J. Pathogenic synergy: mixed infections in the oral cavity. Antonie van Leeuwenhoek. 1984;50:789–798. doi: 10.1007/BF02386241. [DOI] [PubMed] [Google Scholar]
- 148.Chen L, Deng H, Cui H, et al. Inflammatory responses and inflammation-associated diseases in organs. Oncotarget. 2017;9:7204–7218. doi: 10.18632/oncotarget.23208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Kany S, Vollrath JT, Relja B. Cytokines in inflammatory disease. Int J Mol Sci. 2019;20:6008. doi: 10.3390/ijms20236008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Hall-Stoodley L, Stoodley P, Kathju S, et al. Towards diagnostic guidelines for biofilm-associated infections. FEMS Immunol Med Microbiol. 2012;65:127–145. doi: 10.1111/j.1574-695X.2012.00968.x. [DOI] [PubMed] [Google Scholar]
- 151.Olson ME, Ceri H, Morck DW, et al. Biofilm bacteria: formation and comparative susceptibility to antibiotics. Can J Vet Res. 2002;66:86–92. [PMC free article] [PubMed] [Google Scholar]
- 152.Mosaddad SA, Tahmasebi E, Yazdanian A, et al. Oral microbial biofilms: an update. Eur J Clin Microbiol Infect Dis. 2019;38:2005–2019. doi: 10.1007/s10096-019-03641-9. [DOI] [PubMed] [Google Scholar]
- 153.Fair RJ, Tor Y. Antibiotics and bacterial resistance in the 21st century. Perspect Med Chem. 2014;6:25–64. doi: 10.4137/PMC.S14459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Hedberg M, Nord CE. Beta-lactam resistance in anaerobic bacteria: a review. J Chemother. 1996;8:3–16. doi: 10.1179/joc.1996.8.1.3. [DOI] [PubMed] [Google Scholar]
- 155.Macià MD, Rojo-Molinero E, Oliver A. Antimicrobial susceptibility testing in biofilm-growing bacteria. Clin Microbiol Infect. 2014;20:981–990. doi: 10.1111/1469-0691.12651. [DOI] [PubMed] [Google Scholar]
- 156.Lewis K. Riddle of biofilm resistance. Antimicrob Agents Chemother. 2001;45:999. doi: 10.1128/AAC.45.4.999-1007.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Kaplan JB, Mlynek KD, Hettiarachchi H, et al. Extracellular polymeric substance (EPS)-degrading enzymes reduce Staphylococcal surface attachment and biocide resistance on pig skin in vivo. PLoS One. 2018;13:e0205526. doi: 10.1371/journal.pone.0205526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Powell LC, Pritchard MF, Ferguson EL, et al. Targeted disruption of the extracellular polymeric network of Pseudomonas aeruginosa biofilms by alginate oligosaccharides. npj Biofilms Microbiomes. 2018;4:13. doi: 10.1038/s41522-018-0056-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Wozniak DJ, Wyckoff TJ, Starkey M, et al. Alginate is not a significant component of the extracellular polysaccharide matrix of PA14 and PAO1 Pseudomonas aeruginosa biofilms. Proc Natl Acad Sci U S A. 2003;100:7907–7912. doi: 10.1073/pnas.1231792100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Madsen JS, Burmølle M, Hansen LH, et al. The interconnection between biofilm formation and horizontal gene transfer. FEMS Immunol Med Microbiol. 2012;65:183–195. doi: 10.1111/j.1574-695X.2012.00960.x. [DOI] [PubMed] [Google Scholar]
- 161.Lister PD, Wolter DJ, Hanson ND. Antibacterial-resistant Pseudomonas aeruginosa: clinical impact and complex regulation of chromosomally encoded resistance mechanisms. Clin Microbiol Rev. 2009;22:582–610. doi: 10.1128/CMR.00040-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Wilton M, Charron-Mazenod L, Moore R, et al. Extracellular DNA acidifies biofilms and induces aminoglycoside resistance in Pseudomonas aeruginosa. Antimicrob Agents Chemother. 2015;60:544–553. doi: 10.1128/AAC.01650-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Mulcahy H, Charron-Mazenod L, Lewenza S. Extracellular DNA chelates cations and induces antibiotic resistance in Pseudomonas aeruginosa biofilms. PLoS Pathog. 2008;4:e1000213. doi: 10.1371/journal.ppat.1000213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Heilmann C, Hartleib J, Hussain MS, et al. The multifunctional Staphylococcus aureus autolysin aaa mediates adherence to immobilized fibrinogen and fibronectin. Infect Immunol. 2005;73:4793–4802. doi: 10.1128/iai.73.8.4793-4802.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Heilmann C, Hussain M, Peters G, et al. Evidence for autolysin-mediated primary attachment of Staphylococcus epidermidis to a polystyrene surface. Mol Microbiol. 1997;24:1013–1024. doi: 10.1046/j.1365-2958.1997.4101774.x. [DOI] [PubMed] [Google Scholar]
- 166.Ch'ng JH, Chong KKL, Lam LN, et al. Biofilm-associated infection by enterococci. Nat Rev Microbiol. 2019;17:82–94. doi: 10.1038/s41579-018-0107-z. [DOI] [PubMed] [Google Scholar]
- 167.Dale JL, Nilson JL, Barnes AMT, et al. Restructuring of Enterococcus faecalis biofilm architecture in response to antibiotic-induced stress. npj Biofilms Microbiomes. 2017;3:15. doi: 10.1038/s41522-017-0023-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Balaban NQ, Helaine S, Lewis K, et al. Definitions and guidelines for research on antibiotic persistence. Nat Rev Microbiol. 2019;17:441–448. doi: 10.1038/s41579-019-0196-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Schilcher K, Horswill AR. Staphylococcal biofilm development: Structure, regulation, and treatment strategies. Microbiol Mol Biol Rev. 2020;84:e00026–19. doi: 10.1128/MMBR.00026-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Waters EM, Rowe SE, O'Gara JP, et al. Convergence of Staphylococcus aureus persister and biofilm research: Can biofilms be defined as communities of adherent persister cells? PLoS Pathog. 2016;12:e1006012. doi: 10.1371/journal.ppat.1006012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Lister JL, Horswill AR. Staphylococcus aureus biofilms: recent developments in biofilm dispersal. Front Cell Infect Microbiol. 2014;4 doi: 10.3389/fcimb.2014.00178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Vuotto C, Longo F, Balice MP, et al. Antibiotic resistance related to biofilm formation in Klebsiella pneumoniae. Pathogens. 2014;3:743–758. doi: 10.3390/pathogens3030743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Nirwati H, Sinanjung K, Fahrunissa F, et al. Biofilm formation and antibiotic resistance of Klebsiella pneumoniae isolated from clinical samples in a tertiary care hospital, Klaten, Indonesia. BMC Proc. 2019;13:20. doi: 10.1186/s12919-019-0176-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Ramos-Vivas J, Chapartegui-González I, Fernández-Martínez M, et al. Biofilm formation by multidrug resistant Enterobacteriaceae strains isolated from solid organ transplant recipients. Sci Rep. 2019;9:8928. doi: 10.1038/s41598-019-45060-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Yang CH, Su PW, Moi SH, et al. Biofilm formation in acinetobacter baumannii: genotype-phenotype correlation. Molecules (Basel,Switz) 2019;24:1849. doi: 10.3390/molecules24101849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Smani Y, McConnell MJ, Pachón J, et al. Role of fibronectin in the adhesion of Acinetobacter baumannii to host cells. PLoS One. 2012;7:e33073. doi: 10.1371/journal.pone.0033073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Gaddy JA, Tomaras AP, Actis LA. The Acinetobacter baumannii 19606 OmpA protein plays a role in biofilm formation on abiotic surfaces and in the interaction of this pathogen with eukaryotic cells. Infect Immun. 2009;77:3150–3160. doi: 10.1128/iai.00096-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Dumaru R, Baral R, Shrestha LB. Study of biofilm formation and antibiotic resistance pattern of gram-negative Bacilli among the clinical isolates at BPKIHS, Dharan. BMC Res Notes. 2019;12:38. doi: 10.1186/s13104-019-4084-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Poulsen LK, Ballard G, Stahl DA. Use of rRNA fluorescence in situ hybridization for measuring the activity of single cells in young and established biofilms. Appl Environ Microbiol. 1993;59:1354–1360. doi: 10.1128/aem.59.5.1354-1360.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Sternberg C, Christensen BB, Johansen T, et al. Distribution of bacterial growth activity in flow-chamber biofilms. Appl Environ Microbiol. 1999;65:4108–4117. doi: 10.1128/aem.65.9.4108-4117.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Mitchison JM. Enzyme synthesis in synchronous cultures. Science. 1969;165:657–663. doi: 10.1126/science.165.3894.657. [DOI] [PubMed] [Google Scholar]
- 182.Gilbert P, Maira-Litran T, McBain AJ, et al. The physiology and collective recalcitrance of microbial biofilm communities. Adv Microb Physiol. 2002;46:202–256. doi: 10.1016/S0065-2911(02)46005-5. [DOI] [PubMed] [Google Scholar]
- 183.Walters MC, Roe F, Bugnicourt A, et al. Contributions of antibiotic penetration, oxygen limitation, and low metabolic activity to tolerance of Pseudomonas aeruginosa biofilms to ciprofloxacin and tobramycin. Antimicrob Agents Chemother. 2003;47:317–323. doi: 10.1128/aac.47.1.317-323.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Tresse O, Jouenne T, Junter GA. The role of oxygen limitation in the resistance of agar-entrapped, sessile-like Escherichia coli to aminoglycoside and beta-lactam antibiotics. J Antimicrob Chemother. 1995;36:521–526. doi: 10.1093/jac/36.3.521. [DOI] [PubMed] [Google Scholar]
- 185.Hall CW, Mah TF. Molecular mechanisms of biofilm-based antibiotic resistance and tolerance in pathogenic bacteria. FEMS Microbiol Rev. 2017;41:276–301. doi: 10.1093/femsre/fux010. [DOI] [PubMed] [Google Scholar]
- 186.Zheng Z, Stewart PS. Growth limitation of Staphylococcus epidermidis in biofilms contributes to rifampin tolerance. Biofilms. 2004;1:31–35. doi: 10.1017/S1479050503001042. [DOI] [Google Scholar]
- 187.Mah TFC, O'Toole GA. Mechanisms of biofilm resistance to antimicrobial agents. Trends Microbiol. 2001;9:34–39. doi: 10.1016/S0966-842X(00)01913-2. [DOI] [PubMed] [Google Scholar]
- 188.Zhao X, Yu Z, Ding T. Quorum-Sensing regulation of antimicrobial resistance in bacteria. Microorganisms. 2020;8:425. doi: 10.3390/microorganisms8030425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Shih PC, Huang CT. Effects of quorum-sensing deficiency on Pseudomonas aeruginosa biofilm formation and antibiotic resistance. J Antimicrob Chemother. 2002;49:309–314. doi: 10.1093/jac/49.2.309. [DOI] [PubMed] [Google Scholar]
- 190.Algburi A, Comito N, Kashtanov D, et al. Control of biofilm formation: Antibiotics and beyond. Appl Environ Microbiol. 2017;83:e02508–02516. doi: 10.1128/AEM.02508-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Jiao Y, Tay FR, Niu Ln, et al. Advancing antimicrobial strategies for managing oral biofilm infections. Int J Oral Sci. 2019;11:28. doi: 10.1038/s41368-019-0062-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Klemm P, Vejborg RM, Hancock V. Prevention of bacterial adhesion. Appl Microbiol Biotechnol. 2010;88:451–459. doi: 10.1007/s00253-010-2805-y. [DOI] [PubMed] [Google Scholar]
- 193.Veerachamy S, Yarlagadda T, Manivasagam G, et al. Bacterial adherence and biofilm formation on medical implants: A review. Proc Inst Mech Eng, Part H. 2014;228:1083–1099. doi: 10.1177/0954411914556137. [DOI] [PubMed] [Google Scholar]
- 194.Chow JY, Yang Y, Tay SB, et al. Disruption of biofilm formation by the human pathogen Acinetobacter baumannii using engineered quorum-quenching lactonases. Antimicrob Agents Chemother. 2014;58:1802–1805. doi: 10.1128/AAC.02410-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Brown HL, Reuter M, Hanman K, et al. Prevention of biofilm formation and removal of existing biofilms by extracellular DNases of campylobacter jejuni. PLoS One. 2015;10:e0121680. doi: 10.1371/journal.pone.0121680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Jiang Y, Geng M, Bai L. Targeting biofilms therapy: Current research strategies and development hurdles. Microorganisms. 2020;8 doi: 10.3390/microorganisms8081222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Gil C, Solano C, Burgui S, et al. Biofilm matrix exoproteins induce a protective immune response against Staphylococcus aureus biofilm infection. Infect Immun. 2014;82:1017–1029. doi: 10.1128/IAI.01419-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Jiang Q, Jin Z, Sun B. MgrA negatively regulates biofilm formation and detachment by repressing the expression of psm operons in Staphylococcus aureus. Appl Environ Microbiol. 2018;84 doi: 10.1128/aem.01008-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Fleming D, Rumbaugh KP. Approaches to dispersing medical biofilms. Microorganisms. 2017;5:15. doi: 10.3390/microorganisms5020015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Gallant CV, Daniels C, Leung JM, et al. Common beta-lactamases inhibit bacterial biofilm formation. Mol Microbiol. 2005;58:1012–1024. doi: 10.1111/j.1365-2958.2005.04892.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Reza A, Sutton JM, Rahman KM. Effectiveness of efflux pump inhibitors as biofilm disruptors and resistance breakers in Gram-Negative (ESKAPEE) bacteria. Antibiotics (Basel,Switz) 2019;8:229. doi: 10.3390/antibiotics8040229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Ciofu O, Rojo-Molinero E, Macià MD, et al. Antibiotic treatment of biofilm infections. APMIS. 2017;125:304–319. doi: 10.1111/apm.12673. [DOI] [PubMed] [Google Scholar]
- 203.Brooun A, Liu S, Lewis K. A dose-response study of antibiotic resistance in Pseudomonas aeruginosa biofilms. Antimicrob Agents Chemother. 2000;44:640–646. doi: 10.1128/aac.44.3.640-646.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Danese PN. Antibiofilm Approaches: Prevention of Catheter Colonization. Chem Biol. 2002;9:873–880. doi: 10.1016/S1074-5521(02)00192-8. [DOI] [PubMed] [Google Scholar]
- 205.Flemming HC, Ridgway H. Biofilm control: Conventional and alternative approaches. In: Flemming HC, Murthy PS, Venkatesan R, et al., editors. Marine and Industrial Biofouling. Heidelberg: Springer Berlin Heidelberg; 2009. pp. 103–117. [DOI] [Google Scholar]
- 206.Sambanthamoorthy K, Gokhale AA, Lao W, et al. Identification of a novel benzimidazole that inhibits bacterial biofilm formation in a broad-spectrum manner. Antimicrob Agents Chemother. 2011;55:4369. doi: 10.1128/AAC.00583-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Dinicola S, De Grazia S, Carlomagno G, et al. N-acetylcysteine as powerful molecule to destroy bacterial biofilms. A systematic review. Eur Rev Med Pharmacol Sci. 2014;18:2942–2948. Available from: https://europepmc.org/article/MED/25339490. [PubMed] [Google Scholar]
- 208.Abraham NM, Lamlertthon S, Fowler VG, et al. Chelating agents exert distinct effects on biofilm formation in Staphylococcus aureus depending on strain background: role for clumping factor B. J Med Microbiol. 2012;61:1062–1070. doi: 10.1099/jmm.0.040758-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Antoci V, Jr., Adams CS, Parvizi J, et al. The inhibition of Staphylococcus epidermidis biofilm formation by vancomycin-modified titanium alloy and implications for the treatment of periprosthetic infection. Biomaterials. 2008;29:4684–4690. doi: 10.1016/j.biomaterials.2008.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Ma Y, Chen M, Jones JE, et al. Inhibition of Staphylococcus epidermidis biofilm by trimethylsilane plasma coating. Antimicrob Agents Chemother. 2012;56:5923–5937. doi: 10.1128/aac.01739-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Sanyasi S, Majhi RK, Kumar S, et al. Polysaccharide-capped silver nanoparticles inhibit biofilm formation and eliminate multi-drug-resistant bacteria by disrupting bacterial cytoskeleton with reduced cytotoxicity towards mammalian cells. Sci Rep. 2016;6:24929. doi: 10.1038/srep24929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.AshaRani PV, Low Kah Mun G, Hande MP, et al. Cytotoxicity and genotoxicity of silver nanoparticles in human cells. ACS Nano. 2009;3:279–290. doi: 10.1021/nn800596w. [DOI] [PubMed] [Google Scholar]
- 213.Raie DS, Mhatre E, Thiele M, et al. Application of quercetin and its bio-inspired nanoparticles as anti-adhesive agents against Bacillus subtilis attachment to surface. Mater Sci Eng, C. 2017;70:753–762. doi: 10.1016/j.msec.2016.09.038. [DOI] [PubMed] [Google Scholar]
- 214.Hume EB, Baveja J, Muir B, et al. The control of Staphylococcus epidermidis biofilm formation and in vivo infection rates by covalently bound furanones. Biomaterials. 2004;25:5023–5030. doi: 10.1016/j.biomaterials.2004.01.048. [DOI] [PubMed] [Google Scholar]
- 215.Li H, Bao H, Bok KX, et al. High durability and low toxicity antimicrobial coatings fabricated by quaternary ammonium silane copolymers. Biomater Sci. 2016;4:299–309. doi: 10.1039/C5BM00353A. [DOI] [PubMed] [Google Scholar]
- 216.Trentin DS, Silva DB, Frasson AP, et al. Natural green coating inhibits adhesion of clinically important bacteria. Sci Rep. 2015;5:8287. doi: 10.1038/srep08287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Chen M, Yu Q, Sun H. Novel strategies for the prevention and treatment of biofilm related infections. Int J Mol Sci. 2013;14:18488–18501. doi: 10.3390/ijms140918488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Tran PL, Lowry N, Campbell T, et al. An organoselenium compound inhibits Staphylococcus aureus biofilms on hemodialysis catheters in vivo. Antimicrob Agents Chemother. 2012;56:972–978. doi: 10.1128/aac.05680-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Zhao X, Zhao F, Wang J, et al. Biofilm formation and control strategies of foodborne pathogens: food safety perspectives. RSC Adv. 2017;7:36670–36683. doi: 10.1039/C7RA02497E. [DOI] [Google Scholar]
- 220.de Sousa DG, Harvey LA, Dorsch S, et al. Interventions involving repetitive practice improve strength after stroke: a systematic review. J Physiother. 2018;64:210–221. doi: 10.1016/j.jphys.2018.08.004. [DOI] [PubMed] [Google Scholar]
- 221.Chappell TC, Nair NU. Engineered lactobacilli display anti-biofilm and growth suppressing activities against Pseudomonas aeruginosa. npj Biofilms Microbiomes. 2020;6:48. doi: 10.1038/s41522-020-00156-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Lu L, Hu W, Tian Z, et al. Developing natural products as potential anti-biofilm agents. Chin Med. 2019;14:11. doi: 10.1186/s13020-019-0232-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Bendaoud M, Vinogradov E, Balashova NV, et al. Broad-spectrum biofilm inhibition by Kingella kingae exopolysaccharide. J Bacteriol Res. 2011;193:3879–3886. doi: 10.1128/JB.00311-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Mi L, Licina GA, Jiang S. Nonantibiotic-Based Pseudomonas aeruginosa biofilm inhibition with osmoprotectant analogues. ACS Sustainable Chem Eng. 2014;2:2448–2453. doi: 10.1021/sc500468a. [DOI] [Google Scholar]
- 225.Kuang X, Chen V, Xu X. Novel approaches to the control of oral microbial biofilms. BioMed Res Int. 2018;2018:6498932. doi: 10.1155/2018/6498932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Chen H, Zhang B, Weir MD, et al. S. mutans gene-modification and antibacterial resin composite as dual strategy to suppress biofilm acid production and inhibit caries. J Dent. 2020;93:103278. doi: 10.1016/j.jdent.2020.103278. [DOI] [PubMed] [Google Scholar]
- 227.Pires DP, Melo LDR, Vilas Boas D, et al. Phage therapy as an alternative or complementary strategy to prevent and control biofilm-related infections. Curr Opin Microbiol. 2017;39:48–56. doi: 10.1016/j.mib.2017.09.004. [DOI] [PubMed] [Google Scholar]
- 228.Dickey J, Perrot V. Adjunct phage treatment enhances the effectiveness of low antibiotic concentration against Staphylococcus aureus biofilms in vitro. PLoS One. 2019;14:e0209390. doi: 10.1371/journal.pone.0209390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Pires DP, Vilas Boas D, Sillankorva S, et al. Phage therapy: a step forward in the treatment of Pseudomonas aeruginosa infections. J Virol. 2015;89:7449–7456. doi: 10.1128/jvi.00385-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Fu W, Forster T, Mayer O, et al. Bacteriophage cocktail for the prevention of biofilm formation by Pseudomonas aeruginosa on catheters in an in vitro model system. Antimicrob Agents Chemother. 2010;54:397–404. doi: 10.1128/aac.00669-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Paluch E, Rewak-Soroczyńska J, Jędrusik I, et al. Prevention of biofilm formation by quorum quenching. Appl Microbiol Biotechnol. 2020;104:1871–1881. doi: 10.1007/s00253-020-10349-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Singh VK, Mishra A, Jha B. Anti-quorum sensing and anti-biofilm activity of delftia tsuruhatensis extract by attenuating the quorum sensing-controlled virulence factor production in Pseudomonas aeruginosa. Front Cell Infect Microbiol. 2017;7 doi: 10.3389/fcimb.2017.00337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Brackman G, Defoirdt T, Miyamoto C, et al. Cinnamaldehyde and cinnamaldehyde derivatives reduce virulence in Vibrio spp. by decreasing the DNA-binding activity of the quorum sensing response regulator LuxR. BMC Microbiol. 2008;8:149. doi: 10.1186/1471-2180-8-149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Kalaiarasan E, Thirumalaswamy K, Harish BN, et al. Inhibition of quorum sensing-controlled biofilm formation in Pseudomonas aeruginosa by quorum-sensing inhibitors. Microb Pathog. 2017;111:99–107. doi: 10.1016/j.micpath.2017.08.017. [DOI] [PubMed] [Google Scholar]
- 235.Bulman ZP, Ly NS, Lenhard JR, et al. Influence of rhlR and lasR on polymyxin pharmacodynamics in Pseudomonas aeruginosa and implications for quorum sensing inhibition with azithromycin. Antimicrob Agents Chemother. 2017;61 doi: 10.1128/aac.00096-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Maura D, Rahme LG. Pharmacological inhibition of the Pseudomonas aeruginosa MvfR quorum-sensing system interferes with biofilm formation and potentiates antibiotic-mediated biofilm disruption. Antimicrob Agents Chemother. 2017;61 doi: 10.1128/aac.01362-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Stenvang M, Dueholm MS, Vad BS, et al. Epigallocatechin gallate remodels overexpressed functional amyloids in Pseudomonas aeruginosa and increases biofilm susceptibility to antibiotic treatment*. J Biol Chem. 2016;291:26540–26553. doi: 10.1074/jbc.M116.739953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Guo Q, Wei Y, Xia B, et al. Identification of a small molecule that simultaneously suppresses virulence and antibiotic resistance of Pseudomonas aeruginosa. Sci Rep. 2016;6:19141. doi: 10.1038/srep19141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Freire MO, Devaraj A, Young A, et al. A bacterial-biofilm-induced oral osteolytic infection can be successfully treated by immuno-targeting an extracellular nucleoid-associated protein. Mol Oral Microbiol. 2017;32:74–88. doi: 10.1111/omi.12155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Sun D, Accavitti MA, Bryers JD. Inhibition of biofilm formation by monoclonal antibodies against Staphylococcus epidermidis RP62A accumulation-associated protein. Clin Diagn Lab Immunol. 2005;12:93–100. doi: 10.1128/CDLI.12.1.93-100.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Estellés A, Woischnig AK, Liu K, et al. A high-affinity native human antibody disrupts biofilm from Staphylococcus aureus bacteria and potentiates antibiotic efficacy in a mouse implant infection model. Antimicrob Agents Chemother. 2016;60:2292–2301. doi: 10.1128/aac.02588-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Hall AE, Domanski PJ, Patel PR, et al. Characterization of a protective monoclonal antibody recognizing Staphylococcus aureus MSCRAMM protein clumping factor A. Infect Immun. 2003;71:6864–6870. doi: 10.1128/iai.71.12.6864-6870.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Visai L, Xu Y, Casolini F, et al. Monoclonal antibodies to CNA, a collagen-binding microbial surface component recognizing adhesive matrix molecules, detach Staphylococcus aureus from a collagen substrate. J Biol Chem. 2000;275:39837–39845. doi: 10.1074/jbc.M005297200. [DOI] [PubMed] [Google Scholar]
- 244.Rennermalm A, Li YH, Bohaufs L, et al. Antibodies against a truncated Staphylococcus aureus fibronectin-binding protein protect against dissemination of infection in the rat. Vaccine. 2001;19:3376–3383. doi: 10.1016/s0264-410x(01)00080-9. [DOI] [PubMed] [Google Scholar]
- 245.Belyi Y, Rybolovlev I, Polyakov N, et al. Staphylococcus aureus surface protein G is an immunodominant protein and a possible target in an anti-biofilm drug development. Open Microbiol J. 2018;12:94–106. doi: 10.2174/1874285801812010094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Haghighat S, Siadat SD, Sorkhabadi SM, et al. Cloning, expression and purification of autolysin from methicillin-resistant Staphylococcus aureus: potency and challenge study in Balb/c mice. Mol Immunol. 2017;82:10–18. doi: 10.1016/j.molimm.2016.12.013. [DOI] [PubMed] [Google Scholar]
- 247.Nair N, Vinod V, Suresh MK, et al. Amidase, a cell wall hydrolase, elicits protective immunity against Staphylococcus aureus and S. epidermidis. Int J Biol Macromol. 2015;77:314–321. doi: 10.1016/j.ijbiomac.2015.03.047. [DOI] [PubMed] [Google Scholar]
- 248.Varrone JJ, de Mesy Bentley KL, Bello-Irizarry SN, et al. Passive immunization with anti-glucosaminidase monoclonal antibodies protects mice from implant-associated osteomyelitis by mediating opsonophagocytosis of Staphylococcus aureus megaclusters. J Orthop Res. 2014;32:1389–1396. doi: 10.1002/jor.22672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.van den Berg S, Bonarius HP, van Kessel KP, et al. A human monoclonal antibody targeting the conserved Staphylococcal antigen IsaA protects mice against Staphylococcus aureus bacteremia. Int J Med Microbiol. 2015;305:55–64. doi: 10.1016/j.ijmm.2014.11.002. [DOI] [PubMed] [Google Scholar]
- 250.Liu B, Park S, Thompson CD, et al. Antibodies to Staphylococcus aureus capsular polysaccharides 5 and 8 perform similarly in vitro but are functionally distinct in vivo. Virulence. 2017;8:859–874. doi: 10.1080/21505594.2016.1270494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Weisman LE. Antibody for the prevention of neonatal noscocomial Staphylococcal infection: a review of the literature. Arch Pediatr. 2007;14 Suppl 1:S31–34. doi: 10.1016/s0929-693x(07)80008-x. [DOI] [PubMed] [Google Scholar]
- 252.Raafat D, Otto M, Reppschläger K, et al. Fighting Staphylococcus aureus biofilms with monoclonal antibodies. Trends Microbiol. 2019;27:303–322. doi: 10.1016/j.tim.2018.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Freire-Moran L, Aronsson B, Manz C, et al. Critical shortage of new antibiotics in development against multidrug-resistant bacteria-time to react is now. Drug Resist Updat. 2011;14:118–124. doi: 10.1016/j.drup.2011.02.003. [DOI] [PubMed] [Google Scholar]
- 254.Banerjee M, Moulick S, Bhattacharya KK, et al. Attenuation of Pseudomonas aeruginosa quorum sensing, virulence and biofilm formation by extracts of Andrographis paniculata. Microb Pathog. 2017;113:85–93. doi: 10.1016/j.micpath.2017.10.023. [DOI] [PubMed] [Google Scholar]
- 255.Zhang L, Bao M, Liu B, et al. Effect of andrographolide and its analogs on bacterial infection: A review. Pharmacology. 2020;105:123–134. doi: 10.1159/000503410. [DOI] [PubMed] [Google Scholar]
- 256.Rasool U, SP, Parveen A, et al. Efficacy of andrographis paniculata against extended spectrum β-lactamase (ESBL) producing E. coli. BMC Complementary Altern Med. 2018;18:244–244. doi: 10.1186/s12906-018-2312-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Carrol DH, Chassagne F, Dettweiler M, et al. Antibacterial activity of plant species used for oral health against Porphyromonas gingivalis. PLoS One. 2020;15:e0239316. doi: 10.1371/journal.pone.0239316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Gerits E, Verstraeten N, Michiels J. New approaches to combat Porphyromonas gingivalis biofilms. J Oral Microbiol. 2017;9:1300366. doi: 10.1080/20002297.2017.1300366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Liu G, Xiang H, Tang X, et al. Transcriptional and functional analysis shows sodium houttuyfonate-mediated inhibition of autolysis in Staphylococcus aureus. Molecules (Basel,Switz) 2011;16:8848–8865. doi: 10.3390/molecules16108848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Zhou J, Bi S, Chen H, et al. Anti-Biofilm and antivirulence activities of metabolites from plectosphaerella cucumerina against Pseudomonas aeruginosa. Front Microbiol. 2017;8:769. doi: 10.3389/fmicb.2017.00769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Rabin N, Zheng Y, Opoku-Temeng C, et al. Agents that inhibit bacterial biofilm formation. Future Med Chem. 2015;7:647–671. doi: 10.4155/fmc.15.7. [DOI] [PubMed] [Google Scholar]
- 262.Xiang H, Cao F, Ming D, et al. Aloe-emodin inhibits Staphylococcus aureus biofilms and extracellular protein production at the initial adhesion stage of biofilm development. Appl Microbiol Biotechnol. 2017;101:6671–6681. doi: 10.1007/s00253-017-8403-5. [DOI] [PubMed] [Google Scholar]
- 263.Jakobsen TH, van Gennip M, Phipps RK, et al. Ajoene, a sulfur-rich molecule from garlic, inhibits genes controlled by quorum sensing. Antimicrob Agents Chemother. 2012;56:2314–2325. doi: 10.1128/AAC.05919-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Balcázar JL, Vendrell D, de Blas I, et al. Characterization of probiotic properties of lactic acid bacteria isolated from intestinal microbiota of fish. Aquaculture. 2008;278:188–191. doi: 10.1016/j.aquaculture.2008.03.014. [DOI] [Google Scholar]
- 265.Didinen BI, Onuk EE, Metin S, et al. Identification and characterization of lactic acid bacteria isolated from rainbow trout (Oncorhynchus mykiss, Walbaum 1792), with inhibitory activity against Vagococcus salmoninarum and Lactococcus garvieae. Aquacult Nutr. 2018;24:400–407. doi: 10.1111/anu.12571. [DOI] [Google Scholar]
- 266.Ben Taheur F, Kouidhi B, Fdhila K, et al. Anti-bacterial and anti-biofilm activity of probiotic bacteria against oral pathogens. Microb Pathog. 2016;97:213–220. doi: 10.1016/j.micpath.2016.06.018. [DOI] [PubMed] [Google Scholar]
- 267.Lee DK, Park SY, An HM, et al. Antimicrobial activity of Bifidobacterium spp. isolated from healthy adult Koreans against cariogenic microflora. Arch Oral Biol. 2011;56:1047–1054. doi: 10.1016/j.archoralbio.2011.03.002. [DOI] [PubMed] [Google Scholar]
- 268.Schwendicke F, Horb K, Kneist S, et al. Effects of heat-inactivated Bifidobacterium BB12 on cariogenicity of Streptococcus mutans in vitro. Arch Oral Biol. 2014;59:1384–1390. doi: 10.1016/j.archoralbio.2014.08.012. [DOI] [PubMed] [Google Scholar]
- 269.Suzuki N, Yoneda M, Hatano Y, et al. Enterococcus faecium WB2000 inhibits biofilm formation by oral cariogenic Streptococci. Int J Dent. 2011;2011:834151. doi: 10.1155/2011/834151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.Sassone-Corsi M, Raffatellu M. No vacancy: how beneficial microbes cooperate with immunity to provide colonization resistance to pathogens. J immunol. 2015;194:4081–4087. doi: 10.4049/jimmunol.1403169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Tuomola EM, Ouwehand AC, Salminen SJ. The effect of probiotic bacteria on the adhesion of pathogens to human intestinal mucus. FEMS Immunol Med Microbiol. 1999;26:137–142. doi: 10.1111/j.1574-695X.1999.tb01381.x. [DOI] [PubMed] [Google Scholar]
- 272.Yan F, Polk DB. Probiotics and immune health. Curr Opin Gastroenterol. 2011;27:496–501. doi: 10.1097/MOG.0b013e32834baa4d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273.Hegarty JW, Guinane CM, Ross RP, et al. Bacteriocin production: a relatively unharnessed probiotic trait? F1000Res. 2016;5:2587. doi: 10.12688/f1000research.9615.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.Corcoran BM, Stanton C, Fitzgerald GF, et al. Survival of probiotic lactobacilli in acidic environments is enhanced in the presence of metabolizable sugars. Appl Environ Microbiol. 2005;71:3060–3067. doi: 10.1128/AEM.71.6.3060-3067.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.Vuotto C, Longo F, Donelli G. Probiotics to counteract biofilm-associated infections: promising and conflicting data. Int J Oral Sci. 2014;6:189–194. doi: 10.1038/ijos.2014.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276.Fang K, Jin X, Hong SH. Probiotic Escherichia coli inhibits biofilm formation of pathogenic E. coli via extracellular activity of DegP. Sci Rep. 2018;8:4939. doi: 10.1038/s41598-018-23180-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277.Jaffar N, Ishikawa Y, Mizuno K, et al. Mature biofilm degradation by potential probiotics: Aggregatibacter actinomycetemcomitans versus LactoBacillus spp. PLoS One. 2016;11:e0159466. doi: 10.1371/journal.pone.0159466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 278.Mathur H, Field D, Rea MC, et al. Fighting biofilms with lantibiotics and other groups of bacteriocins. npj Biofilms Microbiomes. 2018;4:9. doi: 10.1038/s41522-018-0053-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279.Hertzberger R, Arents J, Dekker Henk L, et al. H2O2 production in species of the LactoBacillus acidophilus group: a central role for a novel NADH-Dependent flavin reductase. Appl Environ Microbiol. 2014;80:2229–2239. doi: 10.1128/AEM.04272-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 280.Salas-Jara MJ, Ilabaca A, Vega M, et al. Biofilm forming LactoBacillus: New challenges for the development of probiotics. Microorganisms. 2016;4:35. doi: 10.3390/microorganisms4030035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 281.Jones SE, Versalovic J. Probiotic LactoBacillus reuteribiofilms produce antimicrobial and anti-inflammatory factors. BMC Microbiol. 2009;9:35. doi: 10.1186/1471-2180-9-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 282.Barzegari A, Kheyrolahzadeh K, Hosseiniyan Khatibi SM, et al. The battle of probiotics and their derivatives against biofilms. Infect Drug Resist. 2020;13:659–672. doi: 10.2147/IDR.S232982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 283.Miquel S, Lagrafeuille R, Souweine B, et al. Anti-biofilm activity as a health issue. Front Microbiol. 2016;7:592–592. doi: 10.3389/fmicb.2016.00592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 284.Tan L, Fu J, Feng F, et al. Engineered probiotics biofilm enhances osseointegration via immunoregulation and anti-infection. Sci Adv. 2020;6:eaba5723. doi: 10.1126/sciadv.aba5723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 285.Carvalho FM, Teixeira-Santos R, Mergulhão FJM, et al. The use of probiotics to fight biofilms in medical devices: A systematic review and meta-analysis. Microorganisms. 2020;9 doi: 10.3390/microorganisms9010027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 286.Gholizadeh P, Aghazadeh M, Asgharzadeh M, et al. Suppressing the CRISPR/Cas adaptive immune system in bacterial infections. Eur J Clin Microbiol Infect Dis. 2017;36:2043–2051. doi: 10.1007/s10096-017-3036-2. [DOI] [PubMed] [Google Scholar]
- 287.Yao R, Liu D, Jia X, et al. CRISPR-Cas9/Cas12a biotechnology and application in bacteria. Synth Syst Biotechnol. 2018;3:135–149. doi: 10.1016/j.synbio.2018.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 288.Jiang W, Bikard D, Cox D, et al. RNA-guided editing of bacterial genomes using CRISPR-Cas systems. Nat Biotechnol. 2013;31:233–239. doi: 10.1038/nbt.2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 289.Goren M, Yosef I, Qimron U. Sensitizing pathogens to antibiotics using the CRISPR-Cas system. Drug Resist Updat. 2017;30:1–6. doi: 10.1016/j.drup.2016.11.001. [DOI] [PubMed] [Google Scholar]
- 290.Touchon M, Charpentier S, Pognard D, et al. Antibiotic resistance plasmids spread among natural isolates of Escherichia coli in spite of CRISPR elements. Microbiology (Reading) 2012;158:2997–3004. doi: 10.1099/mic.0.060814-0. [DOI] [PubMed] [Google Scholar]
- 291.Hale CR, Majumdar S, Elmore J, et al. Essential features and rational design of CRISPR RNAs that function with the Cas RAMP module complex to cleave RNAs. Mol Cell. 2012;45:292–302. doi: 10.1016/j.molcel.2011.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 292.Bikard D, Euler CW, Jiang W, et al. Exploiting CRISPR-Cas nucleases to produce sequence-specific antimicrobials. Nat Biotechnol. 2014;32:1146–1150. doi: 10.1038/nbt.3043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 293.Vercoe RB, Chang JT, Dy RL, et al. Cytotoxic chromosomal targeting by CRISPR/Cas systems can reshape bacterial genomes and expel or remodel pathogenicity islands. PLoS Genet. 2013;9:e1003454. doi: 10.1371/journal.pgen.1003454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 294.Gholizadeh P, Köse Ş, Dao S, et al. How CRISPR-Cas system could be used to combat antimicrobial resistance. Infect Drug Resist. 2020;13:1111–1121. doi: 10.2147/IDR.S247271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 295.Zuberi A, Misba L, Khan AU. CRISPR interference (CRISPRi) inhibition of luxS gene expression in E. coli: An approach to inhibit biofilm. Front Cell Infect Microbiol. 2017;7 doi: 10.3389/fcimb.2017.00214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 296.Gong T, Tang B, Zhou X, et al. Genome editing in Streptococcus mutans through self-targeting CRISPR arrays. Mol Oral Microbiol. 2018;33:440–449. doi: 10.1111/omi.12247. [DOI] [PubMed] [Google Scholar]
- 297.Garrido V, Piñero-Lambea C, Rodriguez-Arce I, et al. Engineering a genome-reduced bacterium to eliminate Staphylococcus aureus biofilms in vivo. Mol Syst Biol. 2021;17:e10145. doi: 10.15252/msb.202010145. [DOI] [PMC free article] [PubMed] [Google Scholar]