Abstract
Due to the exceptional laying performance of hens, the demand on lipid metabolism and oxidation in vivo is vigorous, resulting in excessive lipid accumulation in late-phase hens, which lowers the production performance. Bile acids regulate lipid metabolism and gut microbiota in humans and animals. However, the effect of porcine bile acids on lipid metabolism and cecal microbiota in laying hens in the late phase is still unclear. A total of 360 healthy 45-week-old laying hens were chosen for a 24-week feeding trial, where 0, 30, 60 and 90 mg/kg porcine bile acids were added to a basal diet, respectively. The results showed that dietary supplementation of 60 mg/kg bile acids increased egg production and feed conversion (P < 0.05). Also, 60 and 90 mg/kg porcine bile acids reduced abdominal fat percentage and body weight (P < 0.05). The levels of serum total cholesterol, triglyceride, and low-density lipoprotein cholesterol of hens decreased (P < 0.05) in bile acids supplement groups. As for cecal microbiota, bile acids supplementation did not affect the alpha diversity of cecal microbiota at the genus level. Moreover, dietary supplementation of 90 mg/kg bile acids resulted in an increase in the abundance of beneficial bacteria in the cecum, such as Lactobacillus, Bifidobacterium and Turicibacter. The changes in the cecal microbiota caused by bile acids supplementation correlated with serum lipid indexes. According to KEGG pathway analysis, dietary supplementation of 60 and 90 mg/kg bile acids promoted structural transformation of the cecal microbiota to down-regulate steroid biosynthesis, up-regulate fatty acid degradation and up-regulate unsaturated fatty acid biosynthesis. Meanwhile, bile acids bio-isomerization function of cecal microbiota was enhanced in 60 and 90 mg/kg bile acids treatment, and the short-chain fatty acid metabolism was also affected. In conclusion, the present study revealed dietary supplementation of porcine bile acids enriched probiotics in the gut and improved serum lipid metabolism of laying hens. These findings demonstrate that porcine bile acids can be a potential gut beneficial promoter for late-phase laying hens.
Keywords: Porcine bile acids, Lipid metabolism, Gut microbiota, Laying hens, 16S rRNA
1. Introduction
In the poultry industry, the laying performance and egg quality of laying hens in the late phase usually decline, with increased mortality on occasion (Liu et al., 2018). In the early and peak laying periods, fat synthesis of hepatic cells is in a delicate dynamic balance with the consumption of fat by cells by other organs (Kersten, 2001). However, steatosis occurs due to hepatic hypofunction during aging, with a symptomatic increase in serum triglyceride (TG) or low-density lipoprotein cholesterol (LDL-C) (Miller, 1987). These lipids are important biomarkers of cardiovascular disease (Ginsberg, 2002). Therefore, producers need to deal with this change by improving lipid metabolism of laying hens in order to deal with the performance loss of late-phase egg production.
Bile acids, specific products of cholesterol catabolism in the liver, are the principal components of human and animal bile (Marin et al., 2015). In the liver, bile acids have feedback role on cholesterol synthesis (Pandak et al., 1990). Bile acids regulate lipid metabolism via its receptors (Watanabe et al., 2004; Langhi et al., 2008; Chennamsetty et al., 2011). For instance, chenodesoxycholic acid (CDCA) raises peroxisome proliferator-activated receptor α by activating farnesoid X receptor (FXR, a bile acid nuclear receptor) to enhance the function of lipid metabolism (Pineda Torra et al., 2003). In the duodenum, bile acids play a crucial role in lipid absorption (Hofmann, 1978). Bile acids are also inextricably linked with intestinal microbiota simultaneously. In the posterior segment of gut, bile acids are isomerized under the action of intestinal bacterial enzymes into, for example, 7α-oxo cholic acid (Lucas et al., 2021). These bile acids isomers play an important role in maintaining intestinal immunity (Hang et al., 2019; Campbell et al., 2020; Song et al., 2020). Conversely, Islam et al. (2011) found that the abundance of intestinal microbiota decreased, Firmicutes increased and Bacteroidetes decreased in 5 mmol/kg cholic acid treated rats. Some populations of Bacteroidetes may be the “culprit” in the transfer of “lean” microbiota to “obese” microbiota (Ridaura et al., 2013). In response to the results of this study, some researchers put forward the “bile acid hypothesis” — It seems that the alteration in the gut microbiota caused by bile acids is related to high-fat diets (Yokota et al., 2012). Other evidence supported that the conjugate of cholic acid seemed to be able to alter the composition of the mouse gut microbiota towards a high-fat diet type (Zheng et al., 2017). Besides, changes in bile acids and gut microbiota are often accompanied by modulation of bile acid receptors. For instance, the use of metformin to treat obese mice can reduce the abundance of Bacteroides_fragilis and increase the content of glucoursodeoxycholic acid in the gut, thereby improving the outcome for metabolic diseases through intestinal FXR (Sun et al., 2018).
However, the study on the effects of exogenous bile acids on laying hens, especially on serum lipid metabolism and cecal microbiota, is rarely reported. For humans and birds, bile mainly includes CDCA and CA. However, the extraction of avian bile is difficult, due mainly to its low bile reserves. Porcine bile acids, composed of hyodeoxycholic acid and CDCA, are a potential substitute for bile acids due to its ease of extraction, stable properties and well-known composition. Hence, this experiment studied the effects of porcine bile acids on performance, serum lipid metabolism and cecal microbiota of laying hens, to provide a new means to alleviate the decline of hen performance in the late egg-laying phase.
2. Materials and methods
2.1. Animal ethics
The protocol was ratified by the Institutional Animal Care and Use Committee of China Agricultural University (grant No. AW16129102-2; Beijing, China). All animal experiments followed the ARRIVE guidelines (Kilkenny et al., 2010).
2.2. Experimental materials
The experimental porcine bile acids were produced by Shandong Longchang Co., Ltd (Dezhou, China). Bile acids used in this experiment were extracted from swine bile. According to the method described by John et al. (2014), the content of bile acids in the product was detected by liquid chromatography-tandem mass spectrometry. The product contained hyocholic acid 8.00%, hyodeoxycholic acid 70.67%, and chenodeoxycholic acid 19.61%.
2.3. Animals and management
A total of 360 healthy 45-week-old Hy-Line Grey laying hens with similar body weights were obtained from Deqingyuan Agricultural Technology Co., Ltd (Beijing, China). Birds were caged (3 birds per cage) and were allowed appropriate light (16 L:8 D). Feed and water are freely available during the experiment. The temperature and humidity in the house were kept at 26 °C and 65%, respectively. Birds were allowed to adapt to the cages for 7 days before the start of the experiment.
2.4. Experimental design and diet
Birds were distributed into 4 treatments with 6 replicates per treatment, and 15 hens per replicate. Corn-soybean meal diet was added with 0 (control), 30, 60, and 90 mg/kg porcine bile acids, respectively. The trial period was 24 weeks, which was the period from 45 to 69 weeks of age in laying hens. The diet was formulated according to the Nutrition Requirement of Chicken in China (NY/T 33-2004). The dietary composition and nutrient levels are shown in Table 1. Porcine bile acids were diluted and amplified step by step with corn flour, and then mixed with other raw materials.
Table 1.
Composition and nutrient level of basal diet (as-fed basis, %).
| Ingredients | Content | Nutrient levels1 | Content |
|---|---|---|---|
| Corn | 62.25 | ME, kcal/kg | 2,703.35 |
| Soybean meal | 26.00 | CP | 16.54 |
| Dicalcium phosphate | 1.70 | Digestible Met | 0.38 |
| Limestone | 8.20 | Digestible Lys | 0.78 |
| Sodium chloride | 0.30 | Ca | 3.60 |
| Choline chloride | 0.10 | Total P | 0.65 |
| DL-Met | 0.12 | Non-phytate P | 0.39 |
| Soybean oil | 1.00 | ||
| Vitamin premix2 | 0.03 | ||
| Mineral premix3 | 0.30 | ||
| Total | 100 |
ME = metabolic energy.
Nutrient levels are analyzed values.
The vitamin premix supplied the following per kilogram of diet: 8,500 IU of vitamin A (retinol acetate), 3,600 IU of vitamin D3, 21 IU of vitamin E (DL-α-tocopherol acetate), 4.2 mg of vitamin K3, 3.0 mg of vitamin B1, 10.2 mg of vitamin B2, 0.9 mg of folic acid, 15 mg of calcium pantothenate, 45 mg of niacin, 5.4 mg of vitamin B6, 24 μg of vitamin B12 and 150 μg of biotin.
The mineral premix provided the following per kilogram of diet: 6.8 mg of Cu (CuSO4·5H2O), 66 mg of Fe (FeSO4·7H2O), 83 mg of Zn (ZnSO4·7H2O), 80 mg of Mn (MnSO4·H2O), 1 mg of I (KI) and 0.3 mg of Se (Na2SeO3).
2.5. Sample collection
On the last day (the 168th day), one hen was selected per replicate for blood collection after a 12-h fast, and then was euthanized. Sub-wing vein blood was centrifuged in a centrifuge at 1,500 × g in 4 °C for 10 min to obtain serum. Abdominal adipose tissue and entire liver was taken out from hens and weighed. Cecal chyme was taken out, quick-frozen in liquid nitrogen, and stored at −80 °C.
2.6. Laying performance
At 15:00 every day during the experiment, number and weight of eggs were recorded. Feed intake was recorded every week. Then 15 hens in each treatment were randomly selected to measure their body weight every 4 weeks. Daily egg production, average egg weight, daily egg mass and daily feed intake (ADFI) were calculated according to replicate. Daily egg mass = daily egg production × egg weight. The feed conversion ratio (FCR) was calculated as feed to egg weight every week.
2.7. Serum biochemical parameters
An automatic biochemical analyzer (Hitachi 7600, Hitachi High-Technologies Corp., Tokyo, Japan) was used to determine serum TG, total cholesterol (TC), high-density lipoprotein cholesterol (HDL-C), and LDL-C. The ratio of LDL-C to HDL-C was calculated. The assay kits of TC (A111-1-1), TG (A110-1-1), LDL-C (A113-1-1), and HDL-C (A112-1-1) were purchased from Nanjing Jiancheng Bioengineering Institute (Nanjing, China).
2.8. Total community DNA extraction
Microbial DNA in the cecal samples was extracted using the QIAamp Fast DNA Stool Mini Kit (Qiagen, Germany) according to the instructions. Microbial community DNA was examined using NanoDrop 2000 spectrophotometer (Thermo Scientific, Wilmington, DE, USA) and 1% agarose gel electrophoresis to assess the quantity and quality.
2.9. 16S rRNA amplicon sequencing
The V3–V4 region of bacteria was amplified with 338 F (5′-ACTCCTACGGGAGGCAGCA-3′) and 806 R (5′-GGACTACHVGGGTWTCTAAT-3′) amplification primers. The PCR amplification program included: 4 μL 5 × FastPfu buffer, 2 μL 2.5 mM dNTPs, 0.8 μL 5 μM each primer, 0.4 μL FasPfu Polymerase and 10 ng template DNA. The obtained PCR products were purified using the AxyPrep DNA Gel Extraction Kit (Axygen Biosciences, Union City, CA, USA). Samples were sequenced on Illumina MiSeq PE300 platform (Illumina, San Diego, CA, USA) by Shanghai Majorbio Bio-pharm Technology Co., Ltd (Shanghai, China). The obtained raw paired-end reads were deposited in the NCBI Sequence Read Archive (Bioproject: PRJNA785366).
2.10. Statistical analysis
The performance and serum biochemical parameters data were analyzed using the SPSS Statistics 19.0 program (SPSS Inc., Chicago, IL, USA). ANOVA in GLM was used to analyze variance, and linear and quadratic P values were also calculated using orthogonal contrasts. The difference at P ≤ 0.05 was considered statistically significant. Tukey's multiple range test was used for post-hoc multiple comparisons. The replicate served as the experimental unit.
The obtained paired-end reads were spliced according to the overlap relationship, and the sequences were quality controlled and filtered by Fastp (0.19.6) and FLASH (1.2.11) software as fellows: average base quality score > 20; length > 50 bp; mismatch overlap region < 0.2. The obtained high-quality sequences were clustered into operational taxonomic units (OTUs) at 97% identity threshold by the Usearch software (7.0). Venn diagram and community composition bar were carried out by R (3.3.1). The Circos diagram at the genus level was made by Circos. Then, the alpha-diversity (Shannon, Simpson, ACE and Chao 1 index) was calculated by the Mothur software (1.30.2) and estimated by Student's t test. The sample Bray–Curtis distance was calculated by R (3.3.1) and principal coordinate analysis (PCoA) was performed. We calculated the beta-diversity distance matrix through Qiime (1.9.1). Then non-metric multidimensional scaling (NMDS) was achieved by vegan package in R (3.3.1), and Anosim method was used for significance test.
The Kruskal–Wallis H test was used for significance difference analysis, and false discovery rate procedure was used for multiple testing corrections. Linear discriminant analysis effect size (LEfSe) program provided by Segata et al. (2011) was used to carry out linear discriminant analysis (LDA) on samples according to the taxonomic composition, and present the communities that there were significant differences in sample division (LDA score >2.0).
Procrustes analysis based on PCoA at the genus level between metagenomics data and serum biochemical parameters was carried out using the vegan package in R (3.3.1), and goodness-of-fit statistics of the 2 ranking results were expressed by M2 as described by Gower (1975). The P-value of M2 was from 999 Monte-Carlo simulations. Spearman correlation coefficient of the genus and species level was brought out through the heatmap package in R (3.3.1), and P ≤ 0.05 is a significant correlation.
We used Tax4fun (0.3.1) package in R (3.3.1) provided by Aβhauer et al. (2015) for functional prediction. We transformed the 16S classification pedigree based on Silva database into the classification pedigree of prokaryotes in KEGG database, and then annotated the 16S RNA gene sequence. The abundance of 2 groups of different functional genes was compared using Student's ttest.
3. Results
3.1. Effects of dietary porcine bile acids on laying performance
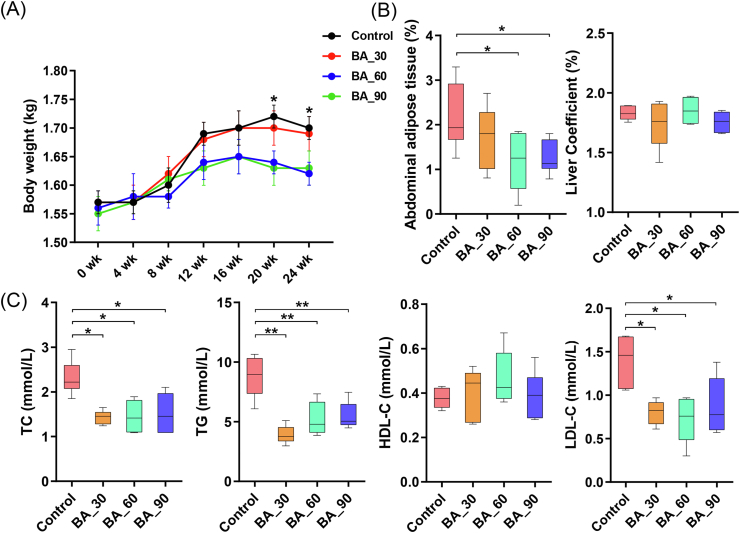
With increasing age, the body weight of hens also increased (Fig. 1A). However, dietary supplementation of 60 and 90 mg/kg bile acids slowed it down. Thus, the abdominal fat level in 60 and 90 mg/kg bile acids group decreased (P < 0.05; Fig. 1B), but the liver weight did not change (P > 0.05). Also, porcine bile acids did not affect ADFI (P > 0.05; Table 2). Importantly, dietary bile acids increased egg production quadratically (P < 0.05; Table 2) and dietary bile acids reduced average egg weight (P < 0.05). Supplementation of 60 mg/kg bile acids improved daily egg mass and FCR (P < 0.05), but not at 30 and 90 mg/kg bile acids (P > 0.05).
Fig. 1.
Effects of dietary porcine bile acids supplementation on body weight and serum biochemical indexes of laying hens. BA = bile acids; TC = total cholesterol; TG = triglyceride; HDL-C = high-density lipoprotein cholesterol; LDL-C = low-density lipoprotein cholesterol. (A) Body weight changes of laying hens during the experiment (n = 15). (B) Abdominal adipose tissue and liver weight as percentages of body weight. (C) Serum biochemical indexes of laying hens. Each box represents the mean value of 6 hens each treatment (n = 6). Whiskers represent maximum and minimum. ∗, P < 0.05; ∗∗, P < 0.01.
Table 2.
Effect of porcine bile acids on laying performance of laying hens1.
| Item | Control | Bile acids, mg/kg |
SEM |
P-value |
||||
|---|---|---|---|---|---|---|---|---|
| 30 | 60 | 90 | ANOVA | Linear | Quadratic | |||
| Daily egg production, % | 80.72c | 83.29b | 85.59a | 82.16bc | 0.51 | <0.01 | 0.20 | 0.02 |
| Average egg weight, g | 61.87a | 60.65b | 60.61b | 60.46b | 0.19 | 0.01 | <0.01 | 0.02 |
| Daily egg mass, g/d per bird | 49.45b | 50.51ab | 51.88a | 49.73b | 0.32 | 0.02 | 0.66 | 0.03 |
| ADFI, g/d per bird | 110.27 | 110.11 | 109.41 | 108.26 | 0.38 | 0.22 | 0.82 | 0.13 |
| FCR, g/g | 2.23a | 2.16ab | 2.15b | 2.17ab | 0.01 | 0.05 | 0.08 | 0.03 |
SEM = the standard error of the mean; ADFI = average daily feed intake; FCR = feed conversion ratio.
a, b, c Means within a row with no common superscripts differ significantly (P < 0.05).
Data represent the mean value of 6 replicates each treatment (n = 6).
3.2. Effects of dietary porcine bile acids on serum biochemical parameters
Dietary supplementation of bile acids decreased TC, TG and LDL-C concentrations in serum (P < 0.05, Fig. 1C). Compared with the control group, the content of HDL-C did not change with bile acids supplementation (P > 0.05), but the LDL-C/HDL-C ratio decreased (P < 0.05, Fig. S1).
3.3. Effects of dietary porcine bile acids on cecal microbiota composition
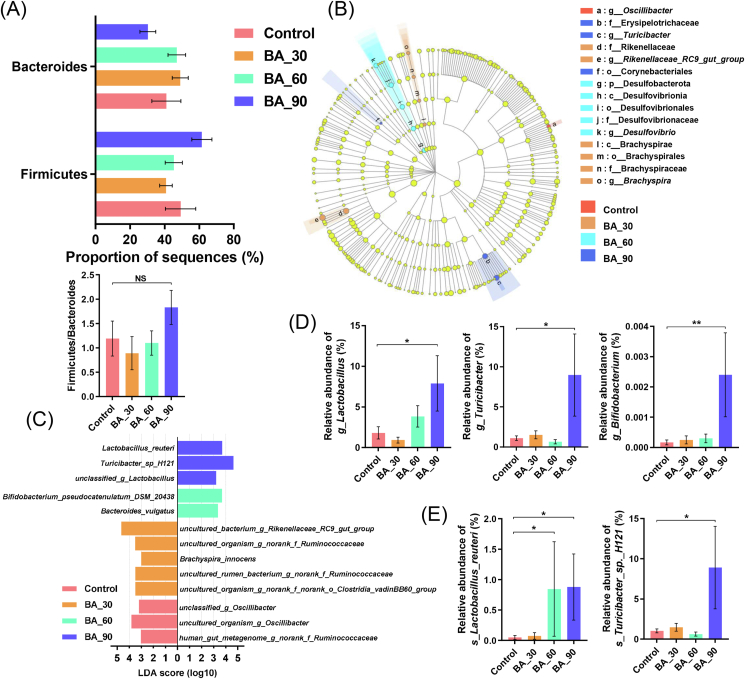
After optimization, there were 1,117,013 sequences with an average length of 417 bp. In general, 174 genera were the same in bile acids groups and the control (Fig. S2A). At the phylum level, the main populations were Firmicutes and Bacteroidetes followed by Actinobacteriota and Desulfobacterota (Fig. S2B). At the genus level, Bacteroides, Rikenellaceae_RC9_gut_group, unclassified_o_Bacteroidales, Romboutsia, Ruminococcus_torques_group, Lactobacillus were dominant (Fig. S2C). Noteworthily, there were more Lactobacillus and Turicibacter in the 90 mg/kg bile acids treatment group. At the species level, a substantial increase in Turicibacter_sp._H121 in 90 mg/kg bile acids treatment group (Fig. S2D).
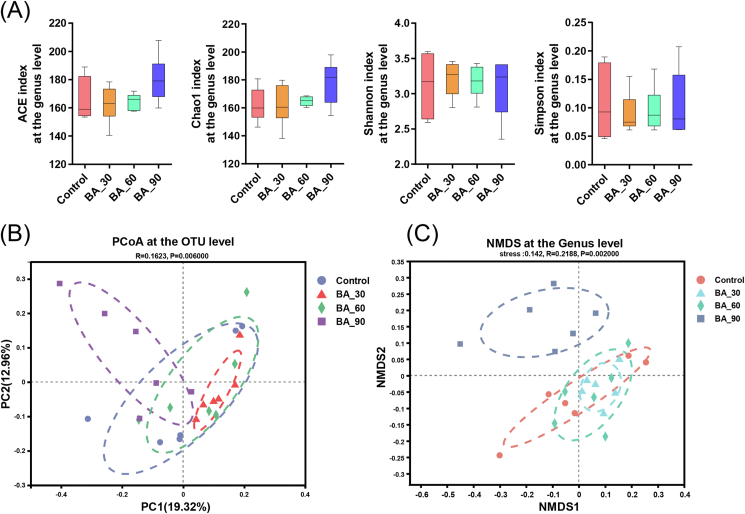
We selected 2 abundance indexes and 2 diversity indexes to compare the cecal microbiota of laying hens at the genus level. Compared with the control group, ACE and Chao 1 index of bile acids treatment groups had no noticeable change (Fig. 2A). There was also no significant change in the Shannon index and Simpson index of cecal microbiota. However, supplementation of 90 mg/kg bile acids seemed to clearly separated the cecal microbiota at the OTU level (Fig. 2B) as well as the genus level (Fig. 2C), while 30 and 60 mg/kg bile acids group did not. It demonstrated that although it did not affect alpha diversity, bile acids supplemented at 90 mg/kg markedly changed the distribution and composition of cecal microbiota in laying hens.
Fig. 2.
Effects of dietary porcine bile acids (BA) supplementation on alpha and beta diversity of cecal microbiota of laying hens. Samples from each treatment group included cecal contents samples from 6 birds (n = 6). (A) Alpha diversity at the genus level. Whiskers represent maximum and minimum. NS, P > 0.05. (B) Principal coordinate analysis (PCoA) based on Bray–Curtis distance at the OTU level (R = 0.1630, P = 0.034). (C) Non-metric multidimensional scaling (NMDS) at the genus level (stress = 0.14, R = 0.2387, P = 0.006). Points of different colors or shapes represent samples in different treatment groups.
In the Kruskal–Wallis H test of the phylum level, Firmicutes and Bacteroides as well as their ratio showed little change (P > 0.05; Fig. 3A). In LEfSe analysis at the phylum to genus level, a total of 15 bacterial phyla and genera showed significant changes (Fig. 3B). In the control, Oscillibacter significantly enriched. Changes in the populations of Rikenellaceae_RC9_gut_group and Brachyspira were most noticeable in the 30 mg/kg bile acids group. Compared with other groups, Desulfobacterota in the 60 mg/kg bile acids group were markedly enriched. In the 90 mg/kg bile acids group, Turicibacter became the most contributing genus. We also compared LDA at the level of genus and species and listed the populations with significant differences among groups (Fig. 3C). At the species level, Turicibacter_sp._H121, Lactobacillus_reuteri and unclassified_g_Lactobacillus in the 90 mg/kg bile acids group were most noticeable, whereas in the 60 mg/kg bile acids group, Bifidobacterium_pseudocatenulatum_DSM_20438 was highly enriched.
Fig. 3.
Effects of dietary porcine bile acids supplementation on differences in taxonomic abundance among groups. BA = bile acids. Samples from each treatment group included cecal contents samples from 6 birds (n = 6). (A) Firmicutes and Bacteroides abundance and their ratio in each treatment group. (B) Linear discriminant analysis effect size (LEfSe) analyses (LDA score ≥2.0) at the phylum level to the genus level. Different color nodes represent microbial groups that are significantly enriched in the corresponding treatment groups and have a significant impact on the differences among groups. (C) Comparison of LDA score of cecal microbiota from the genus level to the species level with significant changes among groups. Wilcoxon rank-sum test difference at P < 0.05 and LDA score (log10) > 3.0 was considered significant. (D-E) Relative abundance of cecal microbiota at the genus or species level with significant changes and specific functions. The data are expressed as mean ± SEM. NS, P > 0.05; ∗, P < 0.05; ∗∗, P < 0.01. Significance was calculated by Kruskal–Wallis H test.
Kruskal–Wallis H test was used to estimate the alterations of individual characteristics of microbial genus or species. Supplementation of 90 mg/kg bile acids increased the abundance of Lactobacillus and Turicibacter (P < 0.05) as is shown in Fig. 3D. Despite low abundance, the Bifidobacterium was markedly increased in the 90 mg/kg bile acids group (P < 0.01). Furthermore, the 60 mg/kg and 90 mg/kg bile acids groups increased Lactobacillus_reuteri (P < 0.05; Fig. 3E), whereas Turicibacter_sp._H121 increased in the 90 mg/kg bile acids group (P < 0.05).
3.4. Effects of dietary porcine bile acids on KEGG pathway of cecal microbiota
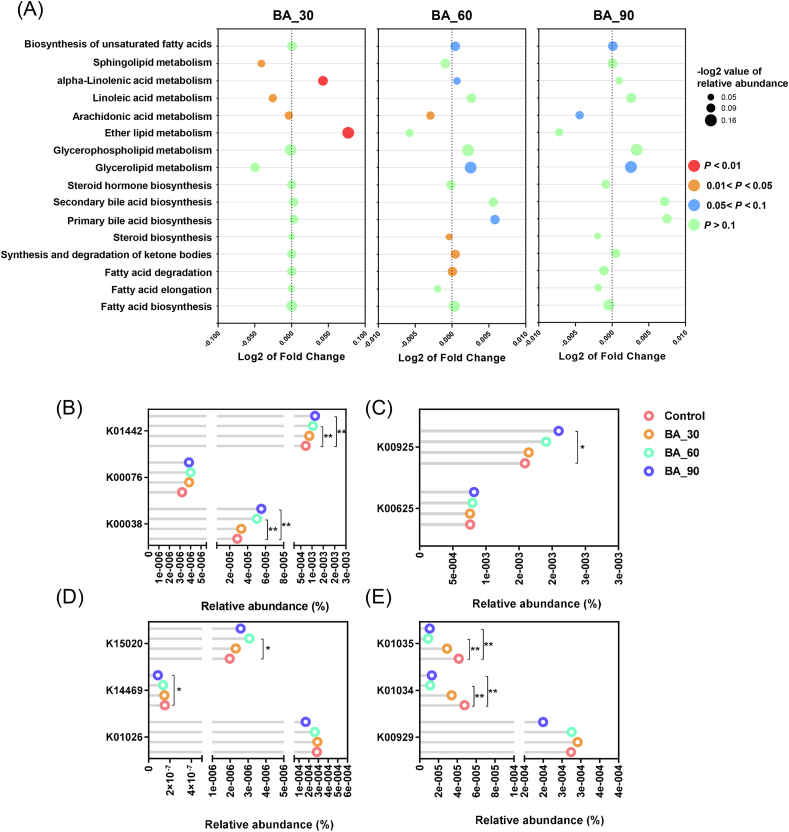
We used the Tax4fun prediction to predict the function of the KEGG pathway of cecal microbiota related to lipid metabolism (Fig. 4A). The result showed that function of sphingolipid, linoleic acid and arachidonic acid metabolism in the 30 mg/kg bile acids group decreased (P < 0.05), but alpha-linolenic acid and ether lipid metabolism increased (P < 0.05). In addition, the bacterial gene associated with fatty acid degradation increased in the 60 mg/kg bile acids group (P < 0.05) and the abundance of bacterial genes involved in the synthesis and degradation of ketone bodies also increased in this group (P < 0.05). The genes relevant to biosynthesis of unsaturated fatty acids, glycerolipid metabolism, alpha-linolenic acid metabolism and primary bile acid biosynthesis tended to increase (P < 0.1). In contrast, bacterial genes related to steroid biosynthesis and arachidonic acid metabolism showed a marked decrease (P < 0.05). In the 90 mg/kg bile acids group, the changes in bacterial genes were similar to those in the 60 mg/kg bile acids group, but the differences were smaller. Only the genes involved in biosynthesis of unsaturated fatty acids and glycerolipid metabolism tended to increase (P < 0.1), whereas that associated with arachidonic acid metabolism tended to decrease (P < 0.1).
Fig. 4.
Effects of dietary porcine bile acids supplementation on KEGG pathway and orthologous genes among groups, predicted by Tax4fun. BA = bile acids. (A) Changes in KEGG pathways associated with lipid metabolism. The color of bubbles represents the P-value of student's t test between bile acids treatment and the control. The size of bubbles represents the -log2 value of the functional proportion of the pathway. X-axis represents log2 value of fold change to the control group. (B) Changes of genes related to bile acid metabolism enzymes. K00038 is the KEGG number of the 3 alpha-hydroxysteroid dehydrogenase; K00076 is the KEGG number of the 7 alpha-hydroxysteroid dehydrogenase; K01442 is the KEGG number of the choloylglycine hydrolase. (C) Changes of genes related to acetic acid metabolism enzymes. K00625 is the KEGG number of phosphate acetyltransferase; K00925 is the KEGG number of acetate kinase. (D) Changes of genes related to propionic acid metabolism enzymes. K01026 is the KEGG number of the propionate CoA-transferase; K14469 is the KEGG number of the acrylyl-CoA reductase (NADPH); K15020 is the KEGG number of the acryloyl-CoA reductase. (E) Changes of genes related to butyric acid metabolism enzymes. K00929 is the KEGG number of the butyrate kinase; K01034 is the KEGG number of the acetate CoA-transferase alpha subunit; K01035 is the KEGG number of the acetate CoA-transferase beta subunit. ∗, 0.01 < P < 0.05; ∗∗, P < 0.01.
Given the fact that bile acids can be biotransformed and biomodificated under the action of gut microbiota, we also analyzed the relevant KEGG orthology (Fig. 4B–E). The abundance of 3 alpha-hydroxylsteroid dehydrogenase (HSDH, K00038) in the 60 and 90 mg/kg bile acids groups increased (P < 0.01) and the abundance of bile salt hydrolases (BSH, K01442) in the 60 and 90 mg/kg bile acids groups also significantly increased (P < 0.01), while 7 alpha-HSDH (K00076) remained unaffected.
We also evaluated bacterial genes for short-chain fatty acids (SCFA) associated with energy metabolism. In the acetic acid synthesis pathway, the acetate kinase level (K00925) in the 90 mg/kg bile acids group increased (P < 0.05), but the amount of phosphate acetyltransferase (K00625) only showed a numerical increase. For the propionic acid synthesis pathway, we selected 3 enzymes in this pathway for evaluation. Acryloyl-CoA reductase (K15020) reported by Hetzel et al. (2003) displayed an increase in the 60 mg/kg bile acids group (P < 0.05) whereas acrylyl-CoA reductase (with NADPH as acceptor; K14469) showed a decrease in the 90 mg/kg bile acids group (P < 0.05). However, the propionate CoA-transferase (K10026) described by Scott et al. (2006) was not affected by bile acids supplementation. Intriguingly, the abundance of bacterial genes related to the expression of acetate CoA-transferase alpha/beta subunit (K01034 and K01035) reported by Duncan et al. (2002) in butyric acid metabolism pathway decreased dramatically in the 60 and 90 mg/kg bile acids groups (P < 0.01), while butyrate kinase (K00929) reported by Louis et al. (2004) in another butyric acid metabolism pathway did not change.
3.5. Correlations between cecal microbiota and serum indicators
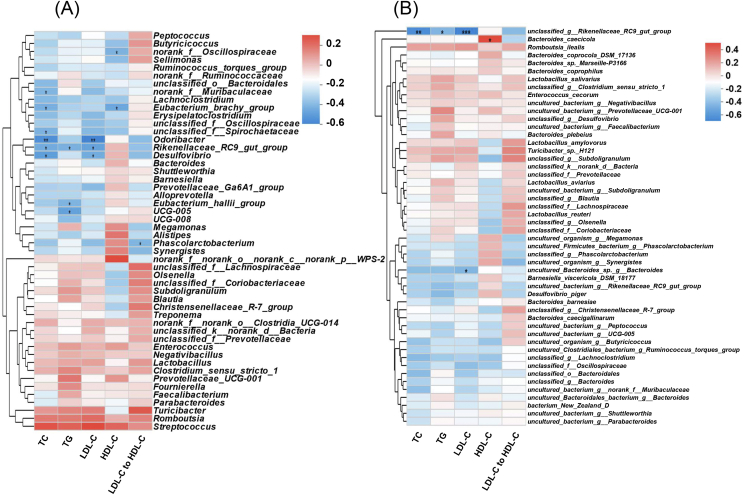
The result of Procrustes analysis indicated that there was a significant correlation between cecal microbiota and serum biochemical indexes of laying hens (M2 = 0.694, P = 0.011; Fig. S3). Spearman coefficient was used to evaluate the correlation between cecal microbiota and serum biochemical indexes. Correlations between cecal microbiota genus level and specific indicators were shown in Fig. 5A. Eubacterium_brachy_group, Rikenellaceae_RC9_gut_group, Desulfovibrio, Odoribacter, Eubacterium_hallii_group and UCG-005 were negatively correlated with serum lipid metabolism (TC, TG, or LDL-C).
Fig. 5.
Spearman's correlation analysis between serum biochemical indexes and cecal microbiota. TC = total cholesterol; TG = triglyceride; HDL-C = high-density lipoprotein cholesterol; LDL-C = low-density lipoprotein cholesterol (A) Correlation at the genus level. (B) Correlation at the species level. R-values are shown in different colors in the heatmap. ∗, 0.01 < P ≤ 0.05; ∗∗, 0.001 <P ≤ 0.01; ∗∗∗, P ≤ 0.001.
Correlations between cecal microbiota species level and serum biochemical indexes were shown in Fig. 5B. Overall, the correlation at the species level corresponds to the correlation at the genus level in our study. Interestingly, unclassified_g_Rikenellaceae_RC9_gut_group negatively correlated with serum TC, TG and LDL-C.
4. Discussion
Previous studies have focused on the interaction between bile acids and gut microbiota in different model animals (Islam et al., 2011; Xiong et al., 2018). Thus, to our best knowledge, the current study is the first to show the effect of porcine bile acids on the gut microbiota in birds. Bile acids are both hydrophobic and hydrophilic, a structural property that allows them to combine water and fat into a water-oil mixture (Fuchs, 2003). This change can increase the contact area of lipase with fat, leading to its increased hydrolysis. Also, lipase hydrolyzes fat only in combination with bile acids and co-lipase (Hofmann, 1978). The enhanced fat absorption capacity will improve the laying performance of laying hens (Barzegar et al., 2020). Porcine bile acids have been proved to play a role in fat absorption in broilers (Lai et al., 2018; Ge et al., 2019), which may be similarly responsible for the increase of laying hen performance. In a previous study investigating the safety of porcine bile acids in laying hens, we found that bile acids improved egg mass and they were safe to use at 10 times the recommended dose in laying hens (Yang et al., 2022). As the metabolites of cholesterol, bile acids play a feedback role of cholesterol synthesis (Salen et al., 1980; Pandak et al., 1990; Wang et al., 2003) and bile acids enhance low-density lipoprotein receptor gene expression on cell membrane, thereby improving the absorption and transformation of LDL-C (Nakahara et al., 2002; Langhi et al., 2008). As the current study demonstrated, bile acids are known to decrease serum TG (Bateson et al., 1978; Bilz et al., 2006; Watanabe et al., 2011; Jain et al., 2012), which may contribute to the reduction of adipose tissue weight.
Interaction between bile acids and gut microbiota is complex. In vitro, bile acids show a strong selective pressure to inhibit bacterial proliferation by destroying the membrane structure of bacteria (Floch et al., 1972; Kurdi et al., 2006). Islam et al. (2011) reported that the abundance of cecal microbiota in rats fed a CA supplemented diet decreased and the ratio of Firmicutes to Bacteroidetes increased significantly. Zhang et al. (2019) added obicholic acid, a bile acid derivative, to a high-fat diet of mice and found that the gut microbiota of mice changed, with the ratio of Firmicutes to Bacteroidetes increasing markedly. On the contrary, the diversity and the ratio of Firmicutes to Bacteroidetes of cecal microbiota in laying hens supplemented with bile acids supplemented diets did not change in the present study. It may be related to dietary energy or fat level (high-fat or normal diet) used in the present study. It has been reported that the Western diet (high-fat or high-sugar) mediated changes in the mouse gut microbiota (Turnbaugh et al., 2008; Schulz et al., 2014), which mimics the effect of bile acids in animal models fed high-fat diets. Yokota et al. (2012) proposed that bile acids may be a potential determinant of intestinal microbial alteration mediated by high-fat diets. It was reported that bile acids could change the composition of gut microbiota in mice fed a normal diet via FXR to transform it towards the type of gut microbiota found in animals or humans given high-fat diets (Zheng et al., 2017).
Bile acids are weak acids with detergent-like properties, which selectively inhibit or enrich the proliferation of certain bacteria (Foley et al., 2021). Lactobacilli harbor a variety of bile salt hydrolases (BSHs). BSHs help conjugated bile acids remove glycine or taurine. These BSHs are thought to reduce the toxicity of bile acids to some extent and promote Lactobacillus colonization (Moser and Savage, 2001). Furthermore, some species of Lactobacillus have been reported to be resistant to bile acids due to the phospholipid cardiolipin on their membranes (Kato et al., 2019). Our study found that the abundance of Lactobacillus increased significantly in the 90 mg/kg bile acids group, especially the Lactobacillus_reuteri species. Numerous studies have demonstrated the beneficial effect of Lactobacillus on serum lipid metabolism. Jones et al. (2012) reported that microencapsulated BSH-active Lactobacillus_reuteri lowered the serum TC, LDL-C and apoB-100, but did not affect the serum HDL-C in hypercholesterolaemic adults. Wu et al. (2017) used a meta-analysis to evaluate the efficacy of Lactobacillus on regulating serum lipid metabolism and showed that Lactobacillus_reuteri reduced serum TC and LDL-C significantly. The present study demonstrated that Bifidobacterium increased in the ceca of laying hens in the 90 mg/kg bile acids group. Bifidobacterium is regarded as a probiotic involved in lipid metabolism. Bernini et al. (2016) proved that Bifidobacterium_lactis reduced serum TC and LDL-C of patients with metabolic syndrome. Bifidobacteria can also protect intestinal health by producing acetic acid (Fukuda et al., 2011). In addition to the well-known probiotics Lactobacillus and Bifidobacterium, Turicibacter is also a genus of bacteria whose main secretion is lactic acid and is known to be beneficial to the host. According to the quantitative trait locus analysis, there is a certain interaction between Turicibacter and the content of endogenous bile acids, and its abundance is affected by the level of bile acids in the intestine (Kemis et al., 2019). Fung et al. (2019) reported that Turicibacter_sanguinis, a species of the genus Turicibacter, altered the expression of a variety of genes pathways in the host intestine, including lipid and steroid metabolism genes, thus reduced the whole-body TG levels and adipocyte size of the host. Furthermore, Gao et al. (2020) demonstrated that Turicibacter had a negative correlation with white adipocyte size, serum TC and LDL-C in mice fed a high-fat diet. Turicibacter also reduced blood glucose in obese rats, regulate immunity and inhibit inflammation (Jiao et al., 2018; Caslin et al., 2019; Zhou et al., 2019).
Our research also indicated that dietary supplementation of bile acids greatly affected the KEGG pathway of cecal microbiota on lipid metabolism. Enhancement of fatty acid degradation and weakening of steroid biosynthesis pathway directly affected the serum lipid metabolism function of the host. Unexpectedly, the pathway of unsaturated fatty acids biosynthesis was enhanced, which is beneficial to the gut microbiota as well as the host. Alpha-linolenic acid is an omega-3 essential fatty acid for humans, which can help reduce serum TC and LDL-C in both humans and animals (Aguilera et al., 2005; Burak et al., 2019). As to the function of bile acids biotransformation and biomodification, the present study proved that the bacterial gene involved in BSH (K01442) increased in bile acids groups, which corresponds to the increase of Lactobacilli mentioned above. BSH is the key enzyme for bile acids deconjugation, and the enhancement of this gene may be related to an increased amount of bile acids entering the gut. In addition, 3 alpha-HSDH (K00038) play a role in biotransformation of cholic acid to 3-oxoCA, CDCA to 3-oxoCDCA and DCA to 3-oxoDCA (Lucas et al., 2021). These oxo-bile acids preserve RORγ+ Treg cells homeostasis in mouse colon and ameliorate the symptoms of colitis, thus playing an indispensable role in gut immunity (Song et al., 2020).
Gut microbiota change may indicate a link between bile acids supplementation and SCFA production. Islam et al. (2011) showed that feeding cholic acid in rats decreased the contents of acetate and butyrate in the cecum, but not propionate. Chen et al. (2020) reported that Clostridium_butyricum reduced the abundance of bacteria biotransforming bile acids in the mouse cecum, thereby reducing bile acids excretion in the feces while increasing SCFA in the cecum. Combined with the results of our study, bile acids or bacteria that biotransform bile acids may compete or synergize with SCFA-producing bacteria, resulting in significant changes in SCFA-producing bacteria in the cecum following bile acids supplementation. This part needs to be further studied.
In the current study, Odoribacter was negatively correlated with serum TC and LDL-C. Odoribacter is a SCFA-producing member of the gut microbiota (Hiippala et al., 2020). The number of Odoribacter in the gut of mice fed a high-fat cholesterol diet decreased (Brandsma et al., 2019). Intriguingly, Rikenellaceae_RC9_gut_group, a bacteria group with mixed reviews, also showed a negative correlation with serum TC, TG, and LDL-C. Similar to the findings of our study, Rikenellaceae_RC9_gut_group abundance was negatively correlated with the serum TC and TG contents in the offspring of mice fed a high-fat diet supplemented with genistein (Zhou et al., 2018).
5. Conclusions
Supplementation of 60 mg/kg porcine bile acids increased the laying performance of late-phase hens, and at 60 and 90 mg/kg levels, bile acids decreased serum TC, TG, and LDL-C, leading to reduced abdominal adipose tissue. They also increased the abundance of beneficial microbiota in the cecum, such as Lactobacillus, Bifidobacterium and Turicibacter and promoted the structural transformation of the cecal microbiota which down-regulated steroid biosynthesis, up-regulated fatty acid degradation and unsaturated fatty acid biosynthesis. Meanwhile, bile acids bioisomerization function and butyric acid production function of cecal microbiota was enhanced. These findings demonstrate that porcine bile acids can be a potential gut microbiota promoter for late-phase laying hens.
Author contributions
Conceptualization, Bowen Yang and Qiugang Ma; methodology, Bowen Yang, Shimeng Huang, Guoxian Zhao, and Qiugang Ma; formal analysis and data curation, Bowen Yang and Shimeng Huang; writing - original draft preparation, Bowen Yang; writing - review and editing, Shimeng Huang, Guoxian Zhao, and Qiugang Ma; visualization and supervision Guoxian Zhao and Qiugang Ma; project administration and funding acquisition, Qiugang Ma. All authors have read and agreed to the published version of the manuscript.
Declaration of competing interest
We declare that we have no financial and personal relationships with other people or organizations that can inappropriately influence our work, and there is no professional or other personal interest of any nature or kind in any product, service and/or company that could be construed as influencing the content of this paper.
Acknowledgments
This study was supported by a special fund for China Agricultural Research System program (Grant No. CARS-40-K08), and the Modern Agricultural Industry Technology System Construction Project of Hebei Province (Grant No. HBCT2018150203).
Footnotes
Peer review under responsibility of Chinese Association of Animal Science and Veterinary Medicine.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.aninu.2022.08.003.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- Aguilera C.M., Ramirez-Tortosa C.L., Quiles J.L., Yago M.D., Martínez-Burgos M.A., Martínez-Victoria E., et al. Monounsaturated and omega-3 but not omega-6 polyunsaturated fatty acids improve hepatic fibrosis in hypercholesterolemic rabbits. Nutrition. 2005;21:363–371. doi: 10.1016/j.nut.2004.06.029. [DOI] [PubMed] [Google Scholar]
- Aßhauer K.P., Wemheuer B., Daniel R., Meinicke P. Tax4Fun: predicting functional profiles from metagenomic 16S rRNA data. Bioinformatics. 2015;31:2882–2884. doi: 10.1093/bioinformatics/btv287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barzegar S., Wu S.B., Choct M., Swick R.A. Implementation of net energy evaluating system in laying hens: validation by performance and egg quality. Poultry Sci. 2020;99:2624–2632. doi: 10.1016/j.psj.2020.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bateson M.C., Maclean D., Evans J.R., Bouchier I.A. Chenodeoxycholic acid therapy for hypertriglyceridaemia in men. Br J Clin Pharmacol. 1978;5:249–254. doi: 10.1111/j.1365-2125.1978.tb01632.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernini L.J., Simão A.N., Alfieri D.F., Lozovoy M.A., Mari N.L., de Souza C.H., et al. Beneficial effects of Bifidobacterium lactis on lipid profile and cytokines in patients with metabolic syndrome: a randomized trial. Effects of probiotics on metabolic syndrome. Nutrition. 2016;32:716–719. doi: 10.1016/j.nut.2015.11.001. [DOI] [PubMed] [Google Scholar]
- Bilz S., Samuel V., Morino K., Savage D., Choi C.S., Shulman G.I. Activation of the farnesoid X receptor improves lipid metabolism in combined hyperlipidemic hamsters. Am J Physiol Endocrinol Metab. 2006;290:E716–E722. doi: 10.1152/ajpendo.00355.2005. [DOI] [PubMed] [Google Scholar]
- Brandsma E., Kloosterhuis N.J., Koster M., Dekker D.C., Gijbels M.J.J., van der Velden S., et al. A proinflammatory gut microbiota increases systemic inflammation and accelerates atherosclerosis. Circ Res. 2019;124:94–100. doi: 10.1161/CIRCRESAHA.118.313234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burak C., Wolffram S., Zur B., Langguth P., Fimmers R., Alteheld B., et al. Effect of alpha-linolenic acid in combination with the flavonol quercetin on markers of cardiovascular disease risk in healthy, non-obese adults: a randomized, double-blinded placebo-controlled crossover trial. Nutrition. 2019;58:47–56. doi: 10.1016/j.nut.2018.06.012. [DOI] [PubMed] [Google Scholar]
- Campbell C., McKenney P.T., Konstantinovsky D., Isaeva O.I., Schizas M., Verter J., et al. Bacterial metabolism of bile acids promotes generation of peripheral regulatory T cells. Nature. 2020;581:475–479. doi: 10.1038/s41586-020-2193-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caslin B., Maguire C., Karmakar A., Mohler K., Wylie D., Melamed E. Alcohol shifts gut microbial networks and ameliorates a murine model of neuroinflammation in a sex-specific pattern. Proc Natl Acad Sci U S A. 2019;116:25808–25815. doi: 10.1073/pnas.1912359116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D., Jin D., Huang S., Wu J., Xu M., Liu T., et al. Clostridium butyricum, a butyrate-producing probiotic, inhibits intestinal tumor development through modulating Wnt signaling and gut microbiota. Cancer Lett. 2020;469:456–467. doi: 10.1016/j.canlet.2019.11.019. [DOI] [PubMed] [Google Scholar]
- Chennamsetty I., Claudel T., Kostner K.M., Baghdasaryan A., Kratky D., Levak-Frank S., et al. Farnesoid X receptor represses hepatic human APOA gene expression. J Clin Invest. 2011;121:3724–3734. doi: 10.1172/JCI45277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan S.H., Barcenilla A., Stewart C.S., Pryde S.E., Flint H.J. Acetate utilization and butyryl coenzyme A (CoA):acetate-CoA transferase in butyrate-producing bacteria from the human large intestine. Appl Environ Microbiol. 2002;68:5186–5190. doi: 10.1128/AEM.68.10.5186-5190.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Floch M.H., Binder H.J., Filburn B., Gershengoren W. The effect of bile acids on intestinal microflora. Am J Clin Nutr. 1972;25:1418–1426. doi: 10.1093/ajcn/25.12.1418. [DOI] [PubMed] [Google Scholar]
- Foley M., O'Flaherty S., Allen G., Rivera A.J., Stewart A.K., Barrangou R., et al. Lactobacillus bile salt hydrolase substrate specificity governs bacterial fitness and host colonization. Proc Natl Acad Sci U S A. 2021;118 doi: 10.1073/pnas.2017709118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs M. Bile acid regulation of hepatic physiology: III. Regulation of bile acid synthesis: past progress and future challenges. Am J Physiol Gastrointest Liver Physiol. 2003;284:G551–G557. doi: 10.1152/ajpgi.00468.2002. [DOI] [PubMed] [Google Scholar]
- Fukuda S., Toh H., Hase K., Oshima K., Nakanishi Y., Yoshimura K., et al. Bifidobacteria can protect from enteropathogenic infection through production of acetate. Nature. 2011;469:543–547. doi: 10.1038/nature09646. [DOI] [PubMed] [Google Scholar]
- Fung T.C., Vuong H.E., Luna C.D.G., Pronovost G.N., Aleksandrova A.A., Riley N.G., et al. Intestinal serotonin and fluoxetine exposure modulate bacterial colonization in the gut. Nat Microbiol. 2019;4:2064–2073. doi: 10.1038/s41564-019-0540-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao X., Chang S., Liu S., Peng L., Xie J., Dong W., et al. Correlations between α-linolenic acid-improved multitissue homeostasis and gut microbiota in mice fed a high-fat diet. mSystems. 2020;5 doi: 10.1128/mSystems.00391-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge X.K., Wang A.A., Ying Z.X., Zhang L.G., Su W.P., Cheng K., et al. Effects of diets with different energy and bile acids levels on growth performance and lipid metabolism in broilers. Poultry Sci. 2019;98:887–895. doi: 10.3382/ps/pey434. [DOI] [PubMed] [Google Scholar]
- Ginsberg H.N. New perspectives on atherogenesis: role of abnormal triglyceride-rich lipoprotein metabolism. Circulation. 2002;106:2137–2142. doi: 10.1161/01.cir.0000035280.64322.31. [DOI] [PubMed] [Google Scholar]
- Gower J.C. Generalized procrustes analysis. Psychometrika. 1975;40:33–51. [Google Scholar]
- Hang S., Paik D., Yao L., Kim E., Trinath J., Lu J., et al. Bile acid metabolites control T(H)17 and T(reg) cell differentiation. Nature. 2019;576:143–148. doi: 10.1038/s41586-019-1785-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hetzel M., Brock M., Selmer T., Pierik A.J., Golding B.T., Buckel W. Acryloyl-CoA reductase from Clostridium propionicum. An enzyme complex of propionyl-CoA dehydrogenase and electron-transferring flavoprotein. Eur J Biochem. 2003;270:902–910. doi: 10.1046/j.1432-1033.2003.03450.x. [DOI] [PubMed] [Google Scholar]
- Hiippala K., Barreto G., Burrello C., Diaz-Basabe A., Suutarinen M., Kainulainen V., et al. Novel Odoribacter splanchnicus strain and its outer membrane vesicles exert immunoregulatory effects in vitro. Front Microbiol. 2020;11 doi: 10.3389/fmicb.2020.575455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann A.F. Lipase, colipase, amphipathic dietary proteins, and bile acids: new interactions at an old interface. Gastroenterology. 1978;75:530–532. [PubMed] [Google Scholar]
- Islam K.B., Fukiya S., Hagio M., Fujii N., Ishizuka S., Ooka T., et al. Bile acid is a host factor that regulates the composition of the cecal microbiota in rats. Gastroenterology. 2011;141:1773–1781. doi: 10.1053/j.gastro.2011.07.046. [DOI] [PubMed] [Google Scholar]
- Jain A.K., Stoll B., Burrin D.G., Holst J.J., Moore D.D. Enteral bile acid treatment improves parenteral nutrition-related liver disease and intestinal mucosal atrophy in neonatal pigs. Am J Physiol Gastrointest Liver Physiol. 2012;302:G218–G224. doi: 10.1152/ajpgi.00280.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao N., Baker S.S., Nugent C.A., Tsompana M., Cai L., Wang Y., et al. Gut microbiome may contribute to insulin resistance and systemic inflammation in obese rodents: a meta-analysis. Physiol Genom. 2018;50:244–254. doi: 10.1152/physiolgenomics.00114.2017. [DOI] [PubMed] [Google Scholar]
- John C., Werner P., Worthmann A., Wegner K., Tödter K., Scheja L., et al. A liquid chromatography-tandem mass spectrometry-based method for the simultaneous determination of hydroxy sterols and bile acids. J Chromatogr A. 2014;1371:184–195. doi: 10.1016/j.chroma.2014.10.064. [DOI] [PubMed] [Google Scholar]
- Jones M.L., Martoni C.J., Parent M., Prakash S. Cholesterol-lowering efficacy of a microencapsulated bile salt hydrolase-active Lactobacillus reuteri NCIMB 30242 yoghurt formulation in hypercholesterolaemic adults. Br J Nutr. 2012;107:1505–1513. doi: 10.1017/S0007114511004703. [DOI] [PubMed] [Google Scholar]
- Kato S., Tobe H., Matsubara H., Sawada M., Sasaki Y., Fukiya S., et al. The membrane phospholipid cardiolipin plays a pivotal role in bile acid adaptation by Lactobacillus gasseri JCM1131(T) Biochim Biophys Acta Mol Cell Biol Lipids. 2019;1864:403–412. doi: 10.1016/j.bbalip.2018.06.004. [DOI] [PubMed] [Google Scholar]
- Kemis J.H., Linke V., Barrett K.L., Boehm F.J., Traeger L.L., Keller M.P., et al. Genetic determinants of gut microbiota composition and bile acid profiles in mice. PLoS Genet. 2019;15 doi: 10.1371/journal.pgen.1008073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kersten S. Mechanisms of nutritional and hormonal regulation of lipogenesis. EMBO Rep. 2001;2:282–286. doi: 10.1093/embo-reports/kve071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilkenny C., Browne W., Cuthill I.C., Emerson M., Altman D.G. Animal research: reporting in vivo experiments: the ARRIVE guidelines. Br J Pharmacol. 2010;160:1577–1579. doi: 10.1111/j.1476-5381.2010.00872.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurdi P., Kawanishi K., Mizutani K., Yokota A. Mechanism of growth inhibition by free bile acids in lactobacilli and bifidobacteria. J Bacteriol. 2006;188:1979–1986. doi: 10.1128/JB.188.5.1979-1986.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai W., Huang W., Dong B., Cao A., Zhang W., Li J., et al. Effects of dietary supplemental bile acids on performance, carcass characteristics, serum lipid metabolites and intestinal enzyme activities of broiler chickens. Poultry Sci. 2018;97:196–202. doi: 10.3382/ps/pex288. [DOI] [PubMed] [Google Scholar]
- Langhi C., Le May C., Kourimate S., Caron S., Staels B., Krempf M., et al. Activation of the farnesoid X receptor represses PCSK9 expression in human hepatocytes. FEBS Lett. 2008;582:949–955. doi: 10.1016/j.febslet.2008.02.038. [DOI] [PubMed] [Google Scholar]
- Liu Z., Sun C., Yan Y., Li G., Shi F., Wu G., et al. Genetic variations for egg quality of chickens at late laying period revealed by genome-wide association study. Sci Rep. 2018;8 doi: 10.1038/s41598-018-29162-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louis P., Duncan S.H., McCrae S.I., Millar J., Jackson M.S., Flint H.J. Restricted distribution of the butyrate kinase pathway among butyrate-producing bacteria from the human colon. J Bacteriol. 2004;186:2099–2106. doi: 10.1128/JB.186.7.2099-2106.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucas L.N., Barrett K., Kerby R.L., Zhang Q., Cattaneo L.E., Stevenson D., et al. Dominant bacterial phyla from the human gut show widespread ability to transform and conjugate bile acids. mSystems. 2021 doi: 10.1128/mSystems.00805-21. [DOI] [PubMed] [Google Scholar]
- Marin J.J., Macias R.I., Briz O., Banales J.M., Monte M.J. Bile acids in physiology, pathology and pharmacology. Curr Drug Metabol. 2015;17:4–29. doi: 10.2174/1389200216666151103115454. [DOI] [PubMed] [Google Scholar]
- Miller N.E. On the associations of body cholesterol pool size with age, HDL cholesterol and plasma total cholesterol concentration in humans. Atherosclerosis. 1987;67:163–172. doi: 10.1016/0021-9150(87)90276-0. [DOI] [PubMed] [Google Scholar]
- Moser S.A., Savage D.C. Bile salt hydrolase activity and resistance to toxicity of conjugated bile salts are unrelated properties in lactobacilli. Appl Environ Microbiol. 2001;67:3476–3480. doi: 10.1128/AEM.67.8.3476-3480.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakahara M., Fujii H., Maloney P.R., Shimizu M., Sato R. Bile acids enhance low density lipoprotein receptor gene expression via a MAPK cascade-mediated stabilization of mRNA. J Biol Chem. 2002;277:37229–37234. doi: 10.1074/jbc.M206749200. [DOI] [PubMed] [Google Scholar]
- Pandak W.M., Heuman D.M., Hylemon P.B., Vlahcevic Z.R. Regulation of bile acid synthesis. IV. Interrelationship between cholesterol and bile acid biosynthesis pathways. J Lipid Res. 1990;31:79–90. [PubMed] [Google Scholar]
- Pineda Torra I., Claudel T., Duval C., Kosykh V., Fruchart J.C., Staels B. Bile acids induce the expression of the human peroxisome proliferator-activated receptor alpha gene via activation of the farnesoid X receptor. Mol Endocrinol. 2003;17:259–272. doi: 10.1210/me.2002-0120. [DOI] [PubMed] [Google Scholar]
- Ridaura V.K., Faith J.J., Rey F.E., Cheng J., Duncan A.E., Kau A.L., et al. Gut microbiota from twins discordant for obesity modulate metabolism in mice. Science. 2013;341 doi: 10.1126/science.1241214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salen G., Colalillo A., Verga D., Bagan E., Tint G.S., Shefer S. Effect of high and low doses of ursodeoxycholic acid on gallstone dissolution in humans. Gastroenterology. 1980;78:1412–1418. [PubMed] [Google Scholar]
- Schulz M.D., Atay C., Heringer J., Romrig F.K., Schwitalla S., Aydin B., et al. High-fat-diet-mediated dysbiosis promotes intestinal carcinogenesis independently of obesity. Nature. 2014;514:508–512. doi: 10.1038/nature13398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott K.P., Martin J.C., Campbell G., Mayer C.D., Flint H.J. Whole-genome transcription profiling reveals genes up-regulated by growth on fucose in the human gut bacterium "Roseburia inulinivorans". J Bacteriol. 2006;188:4340–4349. doi: 10.1128/JB.00137-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segata N., Izard J., Waldron L., Gevers D., Miropolsky L., Garrett W.S., et al. Metagenomic biomarker discovery and explanation. Genome Biol. 2011;12:R60. doi: 10.1186/gb-2011-12-6-r60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song X., Sun X., Oh S.F., Wu M., Zhang Y., Zheng W., et al. Microbial bile acid metabolites modulate gut RORγ(+) regulatory T cell homeostasis. Nature. 2020;577:410–415. doi: 10.1038/s41586-019-1865-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L., Xie C., Wang G., Wu Y., Wu Q., Wang X., et al. Gut microbiota and intestinal FXR mediate the clinical benefits of metformin. Nat Med. 2018;24:1919–1929. doi: 10.1038/s41591-018-0222-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbaugh P.J., Bäckhed F., Fulton L., Gordon J.I. Diet-induced obesity is linked to marked but reversible alterations in the mouse distal gut microbiome. Cell Host Microbe. 2008;3:213–223. doi: 10.1016/j.chom.2008.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D.Q., Tazuma S., Cohen D.E., Carey M.C. Feeding natural hydrophilic bile acids inhibits intestinal cholesterol absorption: studies in the gallstone-susceptible mouse. Am J Physiol Gastrointest Liver Physiol. 2003;285:G494–G502. doi: 10.1152/ajpgi.00156.2003. [DOI] [PubMed] [Google Scholar]
- Watanabe M., Horai Y., Houten S.M., Morimoto K., Sugizaki T., Arita E., et al. Lowering bile acid pool size with a synthetic farnesoid X receptor (FXR) agonist induces obesity and diabetes through reduced energy expenditure. J Biol Chem. 2011;286:26913–26920. doi: 10.1074/jbc.M111.248203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe M., Houten S.M., Wang L., Moschetta A., Mangelsdorf D.J., Heyman R.A., et al. Bile acids lower triglyceride levels via a pathway involving FXR, SHP, and SREBP-1c. J Clin Invest. 2004;113:1408–1418. doi: 10.1172/JCI21025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y., Zhang Q., Ren Y., Ruan Z. Effect of probiotic Lactobacillus on lipid profile: a systematic review and meta-analysis of randomized, controlled trials. PLoS One. 2017;12 doi: 10.1371/journal.pone.0178868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong F., Wu S.G., Zhang J., Jakovlić I., Li W.X., Zou H., et al. Dietary bile salt types influence the composition of biliary bile acids and gut microbiota in grass carp. Front Microbiol. 2018;9:2209. doi: 10.3389/fmicb.2018.02209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang B., Huang S., Li S., Feng Z., Zhao G., Ma Q. Safety evaluation of porcine bile acids in laying hens: effects on laying performance, egg quality, blood parameters, organ indexes, and intestinal development. Front Vet Sci. 2022;9 doi: 10.3389/fvets.2022.895831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokota A., Fukiya S., Islam K.B., Ooka T., Ogura Y., Hayashi T., et al. Is bile acid a determinant of the gut microbiota on a high-fat diet? Gut Microbes. 2012;3:455–459. doi: 10.4161/gmic.21216. [DOI] [PubMed] [Google Scholar]
- Zhang D.Y., Zhu L., Liu H.N., Tseng Y.J., Weng S.Q., Liu T.T., et al. The protective effect and mechanism of the FXR agonist obeticholic acid via targeting gut microbiota in non-alcoholic fatty liver disease. Drug Des Devel Ther. 2019;13:2249–2270. doi: 10.2147/DDDT.S207277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng X., Huang F., Zhao A., Lei S., Zhang Y., Xie G., et al. Bile acid is a significant host factor shaping the gut microbiome of diet-induced obese mice. BMC Biol. 2017;15:120. doi: 10.1186/s12915-017-0462-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou L., Xiao X., Zhang Q., Zheng J., Li M., Yu M., et al. Improved glucose and lipid metabolism in the early life of female offspring by maternal dietary genistein is associated with alterations in the gut microbiota. Front Endocrinol (Lausanne) 2018;9:516. doi: 10.3389/fendo.2018.00516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou W., Xu H., Zhan L., Lu X., Zhang L. Dynamic development of fecal microbiome during the progression of diabetes mellitus in zucker diabetic fatty rats. Front Microbiol. 2019;10:232. doi: 10.3389/fmicb.2019.00232. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.