Abstract
SARS-CoV-2 spread in humans results in continuous emergence of new variants, highlighting the need for vaccines with broad-spectrum antigenic coverage. Using inter-lineage chimera and mutation-patch strategies, we engineered a recombinant monomeric spike variant (STFK1628x) that contains key regions and residues across multiple SAR-CoV-2 variants. STFK1628x demonstrated high immunogenicity and mutually complementary antigenicity to its prototypic form (STFK). In hamsters, a bivalent vaccine composed of STFK and STFK1628x elicited high titers of broad-spectrum neutralizing antibodies to 19 circulating SARS-CoV-2 variants, including Omicron sublineages BA.1, BA.1.1, BA.2, BA.2.12.1, BA.2.75, and BA.4/5. Furthermore, this vaccine conferred robust protection against intranasal challenges by either SARS-CoV-2 ancestral strain or immune-evasive Beta and Omicron BA.1. Strikingly, vaccination with the bivalent vaccine in hamsters effectively blocked within-cage virus transmission of ancestral SARS-CoV-2, Beta variant, and Omicron BA.1 to unvaccinated sentinels. Thus, our study provided insight and antigen candidates for the development of next-generation COVID-19 vaccines.
Keywords: SARS-CoV-2, COVID-19 vaccine, Omicron variants, variant of concern, broad-spectrum antibodies, virus transmission blocking
Graphical abstract
Wu et al. provided an approach to expand the cross-variants antigenic coverage of a recombinant SARS-CoV-2 spike protein. The bivalent vaccine derived from this approach elicited broad antibody responses against all 19 variants in an animal model, which conferred protection from variant challenge and reduced viral transmission.
Introduction
The ongoing coronavirus disease 2019 (COVID-19) pandemic caused by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) poses an unbearable public health burden. The SARS-CoV-2 spike mainly contains three immunogenic domains that act as targets of neutralizing antibody (nAb), i.e., the N-terminal domain (NTD), the receptor-binding domain (RBD), and the subunit 2 (S2), thereby serving as the essential antigen of COVID-19 vaccines. Although several COVID-19 vaccines are available, the constant emergence of SARS-CoV-2 variants is challenging the vaccine effectiveness (VE). Viral genome mutations may alter the biological phenotypes of SARS-CoV-2 in many aspects, such as viral infectivity, pathogenicity, and antigenicity. Critically, the amino-acid substitutions at the antigenic sites of the spike protein may enable viruses to escape from naturally acquired or vaccine-induced immunity (Harvey et al., 2021b). Among the variants currently identified as variants of concern (VOCs) or variants of interest (VOIs), many are able to cause immune escape. The Beta (B.1.351) variant, originally identified in South Africa, was found to cause a 6.5- to 8.6-fold decrease in nAb titers raised by existing mRNA vaccines (Wang et al., 2021b). Besides, the Gamma (P.1), Delta (B.1.617.2), and Mu (B.1.621) variants also caused a 3.8-, 4.8-, 2.9-, and 9.1-fold nAb decrease, respectively, according to previous reports (Uriu et al., 2021; Wang et al., 2021a). The Omicron (B.1.1.529) variant with distinct sublineages, including BA.1, BA.1.1, BA.2, BA.2.12.1, BA.4, and BA.5, has caused the resurgence of outbreaks worldwide due to the dramatically increased infectivity and immune escape compared with former variants (Cao et al., 2022; Cele et al., 2022; Cui et al., 2022; Dejnirattisai et al., 2022b; Garcia-Beltran et al., 2022a; Iketani et al., 2022; Liu et al., 2022; Wang et al., 2022; Yu et al., 2022).
Most of the currently licensed COVID-19 vaccines were designed based on the SARS-CoV-2 prototype spike; their VE appeared to be compromised in countering variants with immune evasion. The phase 3 clinical trial of AZD1222 indicated that this vaccine was ineffective against mild-to-moderate COVID-19 disease caused by the Beta variant (Madhi et al., 2021). In addition, a recent study showed that the efficacy of two doses of BNT162b2 against symptomatic illnesses caused by the Omicron variant was only about 30%, whereas AZD1222 did not show a significant protective effect against Omicron (Andrews et al., 2022). Nevertheless, a vaccine candidate composed of Delta-Omicron chimeric RBD-dimer was demonstrated recently to elicit broad sera neutralization of SARS-CoV-2 variants (Xu et al., 2022). An ideal goal is to develop an antigen providing a broad-spectrum coverage for all SARS-CoV-2 variants that resist nAbs raised by the prototypic spike. However, it remains hugely challenging to achieve this goal.
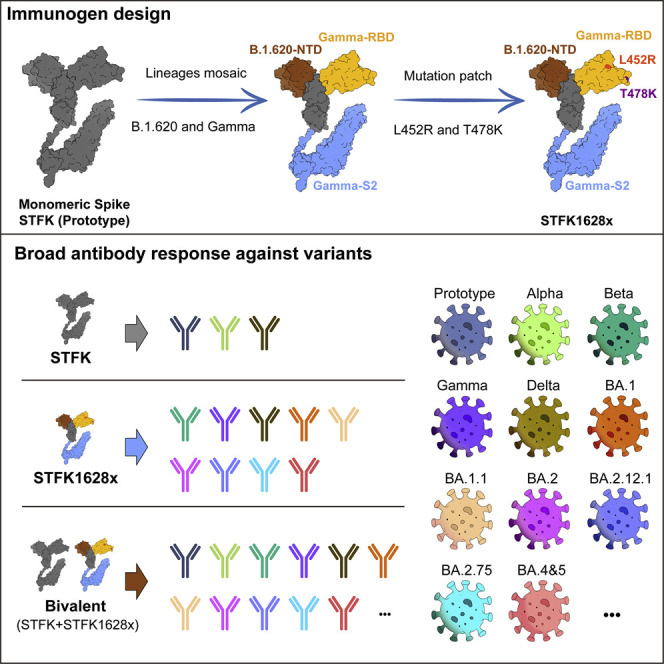
In this study, based on inter-lineage chimera and mutation-patch strategies, we generated a serial of monomeric spike ectodomain proteins harboring multi-site mutations from different VOCs/VOIs. Our evaluations demonstrated that a chimeric spike protein of STFK1628x, containing NTD from B.1.620 lineage, RBD-S2 from the Gamma variant, and additional RBD mutation patches from the Delta variant, and had mutually complementary antigenicity to the ancestral spike-derived monomeric protein (STFK). The bivalent vaccine composed of STFK and STFK1628x elicited high titers of broad-spectrum neutralizing antibodies to protect against in vivo challenges with the ancestral SARS-CoV-2, Beta, and Omicron variants in hamsters. More importantly, our study also provided evidence that this bivalent vaccine could block the within-cage virus transmission from vaccinated hamsters to unvaccinated sentinels. Overall, our findings shed light on the understandings of antigenic and immunogenic characteristics of SARS-CoV-2 spike variants, as well as provided the antigen candidates for developing next-generation COVID-19 vaccines.
Results
Monomeric spike ectodomain STFK protein is highly immunogenic in rodents and nonhuman primates
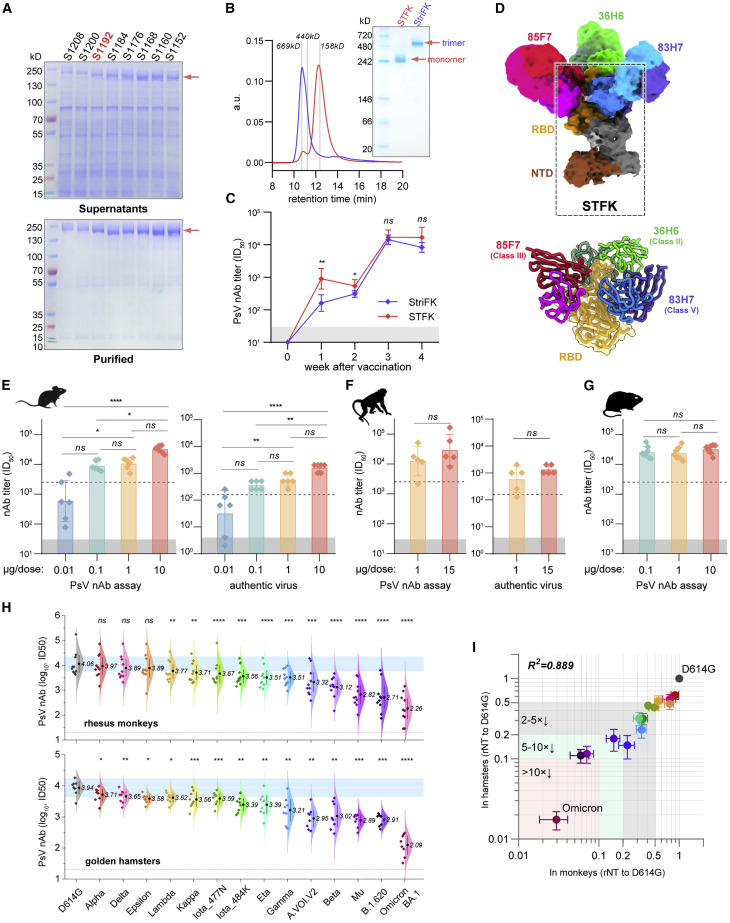
Several studies have demonstrated the immunogenicity of recombinant SARS-CoV-2 spike ectodomain protein in trimeric forms (Brouwer et al., 2021; Liang et al., 2021; Tian et al., 2021; Wu et al., 2021b). However, introducing the exogenous trimerization domain may elicit an unexpected immune response (Liang et al., 2021). Moreover, the recombinant trimer protein may dissociate during the cell cultures and downstream purification process, which decreases the yield of homogeneous trimeric proteins for vaccine production. Therefore, we tried to design and produce monomeric spike proteins based on Wuhan-Hu-1 strain (GenBank: MN908947.3) for the COVID-19 vaccine to address these issues. Although numerous high-resolution structures of SARS-CoV-2 spike trimers have been reported, the detailed structure of the S2 C terminus, particularly for those after amino acid (aa) 1,146, remains elusive. We tested eight constructs of furin site mutated spike ectodomain with progressively truncated C terminus in Chinese hamster ovary (CHO) cells. Interestingly, the C-terminal truncation to various positions between aa 1,152 and aa 1,192 lead to higher expression levels and purification yields than the construct encompassing the entire ectodomain (S1208) (Figure 1 A). In addition, the C-terminal truncated spike proteins presented comparable or even better ACE2 binding activity with the trimeric spike protein (StriFK) (Wu et al., 2021b; Figure S1A). To minimize the potential epitope loss associated with C-terminal truncation, we finally chose the construct of S1192 encompassing aa 1–1,192 (hereafter designated STFK) as an immunogen candidate for further study. As expected, the STFK was presented in monomeric form, as evidenced by the size exclusion chromatography (SEC)-HPLC and native-PAGE analyses (Figure 1B). In contrast to the trimeric StriFK, the STFK elicited significantly higher nAb titers against the pseudotyped virus (PsV) in mice at weeks 1 (p = 0.002) and 2 (p = 0.028) after the 1st dose, suggesting the advantage of the STFK for induction of rapid humoral response. After the 2nd dose immunization, STFK- and StriFK-based vaccines induced comparable nAb titers (Figure 1C).
Figure 1.
The monomeric STFK is highly immunogenic in rodents and nonhuman primates
(A) Reduced SDS-PAGE analyses for supernatants (top panel) and purified proteins (bottom panel) produced from constructs encoding progressive truncations from the C terminus of the furin site with mutated spike ectodomain in CHO cells. S1208, aa 1–1,208; S1200, aa 1–1,200; S1192, aa 1–1,192; S1184, aa 1–1,184; S1176, aa 1–1,176; S1168, aa 1–1,168; S1160, aa 1–1,160; S1152, and aa 1–1,152.
(B) Analyses of the monomeric STFK (aa 1–1,192) and trimeric StirFK by SEC-HPLC (left panel) and native-PAGE (right panel).
(C) Comparison of the PsV nAb titers elicited by the STFK and StriFK in mice. BALB/c mice were immunized twice with 1 μg antigen at weeks 0 and 2.
(D) 3.81 Å cryo-EM density map and corresponding atomic model of the STFK in complex of nAbs 36H6, 83H7, and 85F7. The black dotted box highlights the monomeric STFK protein.
(E and F) Serum nAb titers against pseudotyped (left panel) and authentic SARS-CoV-2 viruses (right panel) of (E) BALB/c mice (n = 6) or (F) Rhesus monkeys (n = 5) received 2 shots of STFK vaccinations at different antigen doses.
(G) Serum nAb titers against VSV-based PsV of hamsters vaccinated at 0.1 (n = 8), 1 (n = 7), or 10 μg (n = 8) of STFK per dose. The immunization schedule was week 0/3 for (E) and (G) and week 0/4 for (F). Sera were analyzed at weeks 4 (E), 6 (F), and 5 (G). The dotted lines show the PsV nAb titers of WHO International Standard for anti-SARS-CoV-2 immunoglobulin using the same assays (NIBSC 20/136).
(H) The nAb titers of sera from STFK-vaccinated rhesus monkeys (pooled of 1- and 15-μg groups, top panel) and hamsters (10-μg group, bottom panel) against lentiviral-pseudotyped SARS-CoV-2 spike variants compared with that against the ancestral D614G strain. The numbers showed the nAb GMT (log10) values.
(I) Comparison of the cross-neutralizing activities of vaccinated hamsters (x axis) and rhesus monkeys (y axis) against various lentiviral-pseudotyped SARS-CoV-2 variants. The relative nAb titer (rNT) was calculated as its ID50 ratio against a variant to the D614G control for each sample. Data in (D)–(H) were plotted as the geometric mean with SD. Dark shadows in (D)–(G) indicate the limit of detection (LOD). The dotted line in (H) indicates the LOD. Blue shadows in (H) represent the range of 50%–200% (within 2-fold changes) of the nAb GMT against D614G (as white line indicated). Uncorrected Kruskal-Wallis test (D, E, and G), Mann-Whitney U test (F), or Dunnett's multiple comparison test (H) were used for intergroup statistical comparisons. Asterisks indicate statistical significance (∗∗∗∗p < 0.0001; ∗∗∗p < 0.001; ∗∗p < 0.01; ∗p < 0.05; ns, not significant). Silhouettes indicating the species in (E)–(G) were from PhyloPic.org and available under the Public Domain Dedication 1.0 license.
See also Figure S1.
To determine the structural basis for the excellent immunogenicity of the STFK, we resolved the cryoelectron microscopy (cryo-EM) structure of the STFK in complex with three previously reported nAbs 36H6, 83H7, and 85F7 (Zhang et al., 2021, 2022). Following the previous classifications of the nAb target epitopes (classes I–V) (Barnes et al., 2020; Starr et al., 2021), the 36H6, 83H7, and 85F7 were categorized into classes II, V, and III, according to their binding modes, respectively (Figure S1B). The 3.81 Å resolution structure of the immune-complex confirmed the monomeric form of the STFK, which could interact with three antigen-binding fragments (Fabs) of nAbs simultaneously (Figures 1D and S1C; Table S1). The STFK is structurally similar to the monomeric form dissociated from a spike trimer (Wrobel et al., 2020). Due to the conformational flexibility in the monomeric form, the S2 subunit was not visualized in the reconstruction. However, in contrast to the trimeric spike, the STFK presents a more exposed RBD and NTD, thereby making the nAb epitopes more accessible which may contribute to its advantage for eliciting rapid nAb response.
Next, we evaluated the dose-dependent immunogenicity of the STFK-based vaccine with FH002C adjuvant in the BALB/c mice, rhesus monkeys, and golden hamsters. In our previous study, FH002C, a risedronate-modified adjuvant, showed potent immunostimulatory effects for hormonal and cellular immune responses (Wu et al., 2021b). In BALB/c mice, STFK vaccinations generated a dose-dependent response of anti-spike IgG, anti-RBD IgG (Figure S1D), and neutralizing antibodies at dose of 0.01–10 μg (Figure 1E). Two injections of STFK at a dose level as low as 0.1 μg induced a potent nAb response with a geometric mean titer (GMT) of 3.9 log10 against the PsV and 362 against the authentic virus [200 Median Tissue Culture Infectious Dose (TCID50)], which were 3.5- and 2.3-fold higher than that of an anti-SARS-CoV-2 standard (1,000 IU/mL) from National Institute for Biological Standards and Control (NIBSC, code: 20/136) in the corresponding assays, respectively (Figure 1E). In rhesus monkeys, STFK vaccinations at 1 or 15 μg dose level also elicited strong humoral immune responses (Figures 1F and S1E), high nAb titers were observed in immunized animals against both PsV (GMT = 4.1 and 4.5 log10 for 1 and 15-μg groups, respectively) and authentic virus (GMT = 588 and 1,351 for 1 and 15-μg groups, respectively). In addition, hamsters that received STFK vaccines of 0.1–10 μg per dose showed comparable and >4.0 log10 of nAb titers at week 2 after the boost dose (Figures 1G and S1F). Overall, the nAb titers (referred to as PsV nAb GMTs) elicited by 1 μg of FH002C-adjuvanted STFK vaccine in mice, monkeys, and hamsters were about 4.2- to 9.3-fold higher than that of the NIBSC 20/136 standard. Apart from humoral immunity, vaccinated mice also presented potent spike-specific T cell responses (p < 0.001) (Figure S1G). These data demonstrated a promising immunogenicity of the monomeric STFK recombinant protein in rodents and nonhuman primates.
Engineered STFK variant protein provides broad antigenic coverage which compensates for prototypic spike
We investigated the impacts of 14 VOCs/VOIs on nAbs raised by the prototypic STFK in animals. Notably, all sera from ten monkeys and eight hamsters at week 2 after 2-dose vaccinations showed detectable nAbs against all PsVs bearing VOC/VOI spike variants, including Omicron BA.1 (Figure 1H). However, by contrast to that against D614G lineage, STFK-elicited nAb titers in monkeys were markedly decreased (>5-fold) for Beta (6.5×), Mu (B.1.621) (14×), B.1.620 (17×), Omicron (34×); were mildly to moderately reduced (2- to 5-fold) for A.VOI.V2 (4.4×), Gamma (2.9×), Eta (B.1.525) (3.2×), Iota_484K (B.1.526) (2.8×), Iota_477N (B.1.526) (2.4×), and Kappa (B.1.617.1) (2.0×) (Figures 1H and 1I). For Alpha, Delta, Epsilon (B.1.429), and Lambda (C.37) variants, the nAb titers only slightly changed (<2-fold). Moreover, sera from immunized hamsters presented highly similar (R2 = 0.889, p < 0.001) cross-neutralizing profiles to that from monkeys (Figure 1I). These results are consistent with findings in humans that the E484K-harboring variants and Omicron may markedly evade nAbs raised by the prototypic spike.
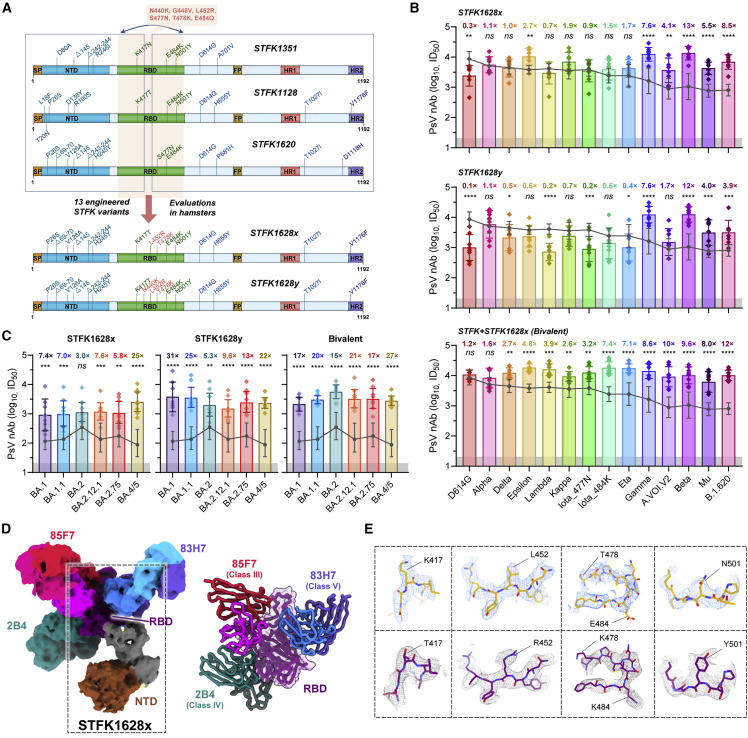
Following the approach as graphically depicted in Figure 2 A, we aimed to develop a modified STFK antigen providing complementary antigenic coverage to the prototypic protein to address the concerns for the evasive variants (Figure 2A). As the Mu and Omicron variants had not emerged when our experiment started, we tested mutated STFK antigens based on the spikes of Beta (STFK1351), Gamma (STFK1128), and B.1.620 (STFK1620) variants (Figure S2A). Compared with those immunized with STFK, hamsters vaccinated with STFK1351, STFK1128, and STFK1620 showed 1.0–3.0×, 1.3–6.2×, and 1.7–5.5× increased nAb titers (GMTs) in neutralizing four immune-escape variants (Gamma, A.VOI.V2, Beta, and B.1.620) (Figures S2B and S2C). The STFK1128 exhibited better immunogenicity than the other two, as it raised ∼4.0 log10 of nAb GMT in neutralizing its parental virus (Gamma) (Figures S2B and S2C). In contrast, Beta variant-derived STFK1351 was poorly immunogenic.
Figure 2.
nAb responses elicited by engineered STFK variants in hamsters
(A) Schematic of the progressive approach to enlarge the cross-variants antigenic covering of a recombinant spike protein via inter-lineage chimera and mutation-patch strategies.
(B and C) The nAb titers of sera from hamsters (n = 8) receiving vaccination of STFK1628x, STFK1628y, or a bivalent version of STFK+STFK1628x to neutralize lentiviral-pseudotyped SARS-CoV-2 former variants (B) and Omicron sublineages (C). Dark gray lines indicate the nAb GMTs induced by the prototypic STFK vaccine against the corresponding variants. Gray shadows indicate the LOD (ID50 = 20) of the assay. Data were plotted as the geometric mean with SD. The value of fold increase in neutralization for each variants relative to prototypic STFK vaccine is shown at the top of each bar. Uncorrected Fisher's LSD tests were used for statistical comparisons (STFK versus modified STFK vaccine). Asterisks indicate statistical significance (∗∗∗∗p < 0.0001; ∗∗∗p < 0.001; ∗∗p < 0.01; ∗p < 0.05; ns, not significant).
(D) 3.88 Å cryo-EM density map and corresponding atomic model of the STFK1628x in complex of nAbs 83H7, 85F7, and 2B4. The black dotted box highlights the monomeric STFK1628x protein.
(E) Comparisons of representative density maps of residues involved in RBD mutations on the STKF (top panel) and STFK1628x proteins (bottom panel).
See also Figures S2 and S3.
We then introduced circulating RBD mutations absent in the Gamma variant but present in other VOC/VOI viruses into the STFK1128 backbone for the following engineering. We used the mutations of L452R (noted in Delta, Kappa, and Epsilon), S477N (presented in Iota_477N and B.1.620), T478K (Delta-derived), and E484Q (Kappa-derived, to replace the E484K in STFK1128) to generate six "patched" antigens (Figure S2D). Hamster immunization tests revealed that the STFK1128e (L452R/S477N/E484Q), STFK1128f (L452R/T478K/E484K), and STFK1128g (L452R/T478K/E484Q) displayed improvements on the cross-neutralization spectrum in comparison to the STFK1128 (Figures S2E and S2F). As the Delta became the dominant variant worldwide since June 2021, we selected the STFK1128f exhibiting higher titers of nAbs to neutralize both Beta and Delta viruses as a candidate for further optimization.
Besides RBD mutations, NTD deletions presented in several VOCs/VOIs may also contribute to their immune-escape potentials. To cover the mutated NTD epitopes, we designed two inter-lineage chimeric constructs of STFK1328x and STFK1628x (Figure S3A); the former included the NTD of STFK1351 (Beta), whereas the latter had the NTD of STFK1620 (B.1.620). Both constructs shared the RBD-S2 domain of STFK1128f, except a K417N in STFK1328x. Remarkably, the STFK1628x could elicit a broad and potent nAb response in hamsters (Figure S3B), which showed 4.1–12.8× increased nAb titers (GMT) compared with prototypic STFK in neutralizing the A.VOI.V2 (4.1×), Gamma (7.6×), Beta (12.8×), and B.1.620 (8.5×). In contrast to its parental STFK1128f, the STFK1628x also exhibited higher nAb titers against most VOCs/VOIs (Figures 2B and S3C). These data supported the STFK1628x as a promising antigen candidate for the updated COVID-19 vaccine. As our previous study have suggested, the aa 439–448 was another hotspot region in addition to aa 484 (Zhang et al., 2022), we further made two modified STFK1628x versions, designated STFK1628y and STFK1628z, including N440K and G446V, respectively (Figures 2B and S3A). The STFK1628y and STFK1628z displayed distinct antigenic profiles in hamsters (Figures S3B and S3C). Following these data, we formulated a bivalent vaccine using the STFK1628x and the prototypic STFK at a mass ratio of 1:1. For most of the VOCs/VOIs, hamsters immunized with the bivalent vaccine showed significantly (p < 0.05) increased nAb titers than that elicited by the prototypic antigen (Figure 2B). Strikingly, the bivalent vaccine also elicited significantly higher (p < 0.05) nAb against Omicron sublineages, including BA.1 (GMT: 2,130), BA.1.1 (GMT: 3,023), BA.2 (GMT: 5,671), BA.2.12.1 (GMT: 3,318), BA.2.75 (GMT: 3,353), and BA.4/5 (GMT: 2,788) (Figure 2C). In addition, authentic virus neutralization assay demonstrated the broad-spectrum coverage of the bivalent vaccine against both ancestral SARS-CoV-2 and Omicron BA.1 (Figure S3D). Similar results were also confirmed on spike-binding antibodies (Figure S3E). Taken together, STFK plus STFK1628x provided a full-spectrum neutralization coverage to all VOCs/VOIs.
We also obtained a 3.88 Å cryo-EM structure of the STFK1628x in complexed with three nAbs 83H7, 85F7, and 2B4 (Figures 2D and S3F; Table S1). As the T478K abolishes the activity of 36H6 nAb, we replaced the 36H6 with a class IV mAb of 2B4 with cross-SARS-CoV-1/2 neutralization potency (Figure S3G; Zhang et al., 2021). As expected, the STKF1628x presented a similar structure to STFK but showed distinguished densities on the mutation sites, such as aa 417, 452, 478, 484, and 501, corresponding to its alternative antigenic profile (Figure 2E).
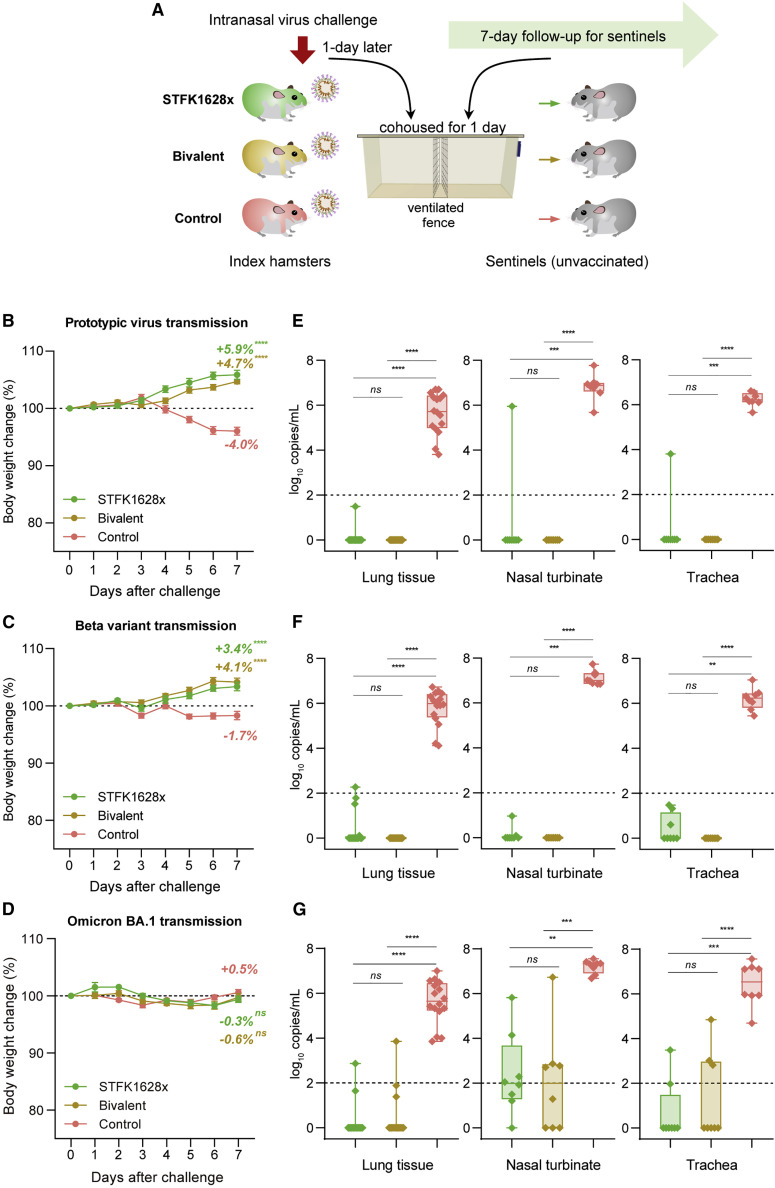
The bivalent vaccine protects hamsters against intranasal SARS-CoV-2 variants challenges
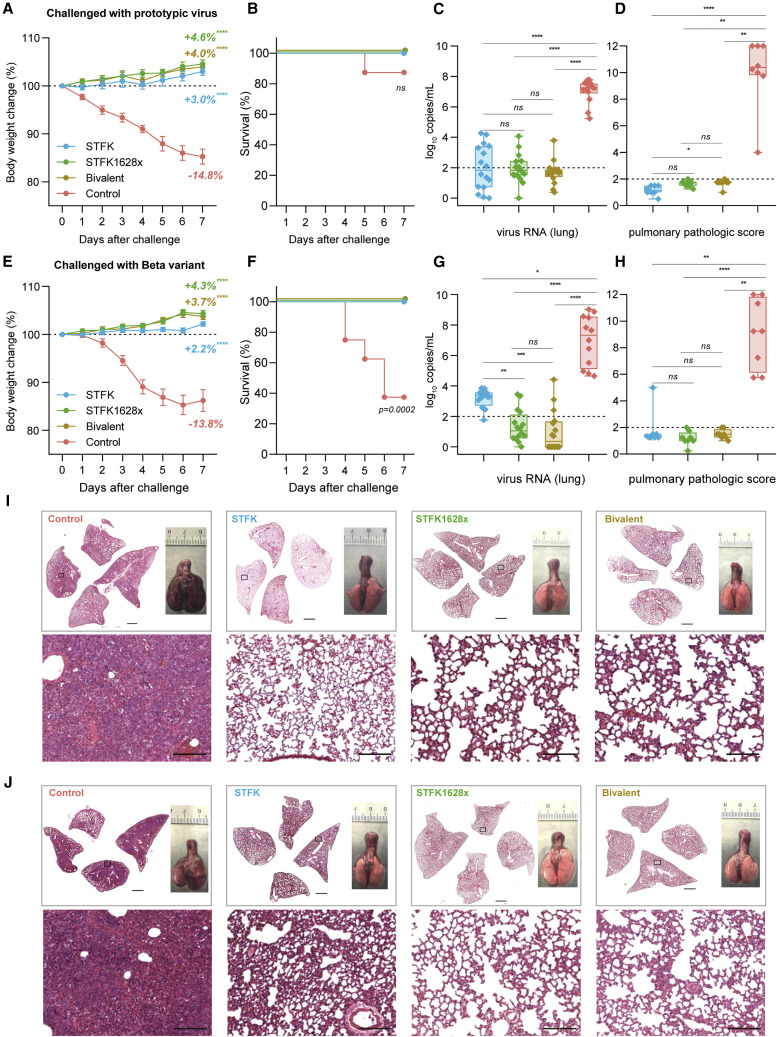
To assess the ability of the STFK-based vaccine to mediate protection against SARS-CoV-2 variants, we intranasally challenged hamsters that received STFK, STFK1628x, or the bivalent vaccine (Figure 3 ). For either challenge with the ancestral strain (Figures 3A–3D and 3I) or Beta variant (Figures 3E–3H and 3J), vaccinated hamsters showed an average of 2.2%–4.6% weight increase to their baseline levels by the end of a 7-day follow-up (Figures 3A and 3E). By contrast, unvaccinated animals showed a maximum weight loss of 14.8% and 13.8% at 7 days post-infection (dpi) in the ancestral strain and Beta variant challenges, respectively. Moreover, 1 of 8 and 5 of 8 unvaccinated animals died from ancestral SARS-CoV-2 and beta variant infections, respectively, but none in the vaccinated groups (Figures 3B and 3F). At 7 dpi, the median viral RNA levels of control hamsters challenged by the prototypic virus were 7.26 (range 5.24–7.78) log10 in the lung, 6.88 (range 6.12–7.32) log10 in the nasal turbinate, and 6.29 (range 5.04–6.67) log10 copies/mL in the trachea (Figures 3C and S4A). By contrast, hamsters that received vaccinations of STFK, STFK1628x, or the bivalent version showed significant (p < 0.01 for each comparison) viral RNA reductions by >5.0 log10, 2.0–3.0 log10, 3.0–4.0 log10 copies/mL in tissues of the lung, nasal turbinate, and trachea, respectively (Figures 3C and S4A). For protection against the prototypic virus challenge, the three vaccine candidates had comparable efficacy (p > 0.05) in decreasing viral loads in the respiratory tract tissues (Figures 3C and S4A). In the Beta variant challenges, control hamsters also showed high levels of viral RNA similar to those infected with the prototypic virus in their respiratory tract tissues. For vaccinated animals, the medians of pulmonary viral loads were 1.05, 0.35, and 3.36 log10 copies/mL in the STFK1628x, bivalent and STFK groups, corresponding to reductions of 6.27 (p < 0.0001), 6.96 (p < 0.0001), and 3.96 (p = 0.01) log10 to controls, respectively (Figure 3G). In tissues of nasal turbinate and trachea, no statistically significant difference in viral RNA suppression was observed among the 3 vaccination groups (Figure S4B), but hamsters immunized with the bivalent vaccine presented relatively lower viral loads than the others. Immumohistochemical (IHC) staining of the lung sections showed the presence of SARS-CoV-2 N proteins in all control animals but not in any vaccinated animals (Figures S4C and S4D).
Figure 3.
The STFK, STFK1628x, and bivalent vaccines offer protection against ancestral SARS-CoV-2 and Beta variant intranasally challenged in hamsters
(A–J) A total of 64 hamsters were used for two independent tests. Each group included eight hamsters (4 males and 4 females) that received 2-dose vaccinations (10 μg/dose, 3-week apart) before virus challenges. Hamsters were intranasally challenged with 1 × 104 PFU of ancestral SARS-CoV-2 (A–D) or Beta variant (E–H). After a 7-day weight monitoring follow-up, animals were euthanized for tissue analyses. Weight changes (A and E), survival curves (B and F), lung viral RNA levels (in two independent lung tissues, LOD = 2 log10 copies/mL) (C and G), and pulmonary pathological scores (D and H) of hamsters challenged by ancestral SARS-CoV-2 (A–D) or Beta variant (E–H) were shown. (I and J) Representative H&E-stained lung sections from ancestral SARS-CoV-2 (I) or Beta variant (J) challenged hamsters. Views of the whole lung lobes (four independent sections) and the gross observations of lung tissues were presented in the top panel (scale bars, 2 mm), areas in the black box were enlarged in the bottom panel (scale bars, 200 μm). Data in (A) and (E) were plotted as means± SEM. Data in (C), (D), (G), and (H) were shown as box and whisker plots; the median, first quartile, third quartile, minimum, and maximum values were plotted. Dunnett's multiple comparison test (A and E), two-sided log-rank test (B and F), or uncorrected Kruskal-Wallis test (C, D, G, and H) were used for intergroup statistical comparisons. Asterisks indicate statistical significance (∗∗∗∗p < 0.0001; ∗∗∗p < 0.001; ∗∗p < 0.01; ∗p < 0.05; ns, not significant).
See also Figure S4.
In addition to mediating virological suppressions, the three vaccines also protected hamsters from lung disease caused by SARS-CoV-2 infections. Pathological examinations revealed that most unvaccinated hamsters presented severe pulmonary diseases at 7 dpi regardless of the challenged virus type (Figures 3D and 3H). In contrast, gross lung observations and pulmonary pathology scorings demonstrated that all vaccinated animals were free from moderate-to-severe pneumonia, with the only exception noted in one from the STFK group challenged with Beta variant (Figures 3I and 3J).
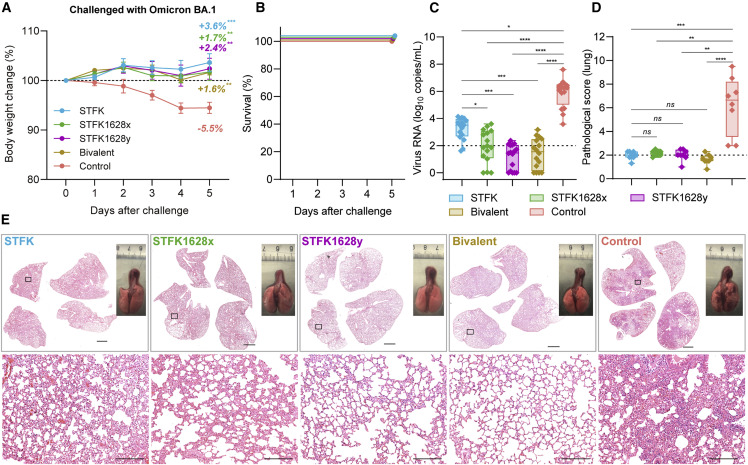
We further assessed the protective effects of STFK, STFK1628x, STFK1628y, and the bivalent vaccines against Omicron BA.1 variant in hamsters (Figure 4 ). Unvaccinated hamsters showed a relatively modest weight loss with an average 5.5% weight loss on 5 dpi and all survived after Omicron BA.1 challenge (Figures 4A and 4B), whereas the vaccinated hamsters showed an average of 1.6%–3.6% weight increase to their baseline levels by the end of a 5-day follow-up (Figure 4A). By the end of the experiment, control hamsters had detectable viral RNA with a median of 6.25 (range 3.58–7.60) log10 copies/mL in lung (Figure 4C). In comparison, animals vaccinated with STFK (3.51 log10 copies/mL, p = 0.0142), STFK1628x (1.93 log10 copies/mL, p < 0.0001), STFK1628y (1.46 log10 copies/mL, p < 0.0001), and the Bivalent vaccine (1.55 log10 copies/mL, p < 0.0001) exhibited significantly lower viral RNA load in lung after Omicron BA.1 challenge (Figure 4C). Consistent with the nAb results, STFK1628x (38-fold reduction, p = 0.0182), STFK1628y (111-fold reduction, p = 0.0002), and the bivalent (90-fold reduction, p = 0.001) showed significantly lower viral RNA loads than STFK (Figure 4C). IHC staining demonstrated that the presence of SARS-CoV-2 N proteins in lung tissues of unvaccinated hamsters that were challenged with Omicron variant. By contrast, viral N protein was rarely detected in lung tissues from vaccinated animals, particular for those who received the bivalent vaccine (Figure S4E). Histopathological examination showed mild-to-moderate pulmonary consolidation, alveolar destruction, and diffuse inflammation in most controls (Figures 4D and 4E). In contrast, the pathological severity scores of vaccinated hamsters were significant low than that of controls.
Figure 4.
The monovalent and the STFK/STFK1628x-combined bivalent vaccines offer protection against SARS-CoV-2 Omicron variant intranasally challenged in hamsters
(A–E) A total of 40 hamsters were used for challenge. Each group included eight hamsters (4 males and 4 females) that received 2-dose vaccinations (10 μg/dose, 3-week apart) before virus challenges. Hamsters were intranasally challenged with 1 × 105 PFU of SARS-CoV-2 Omicron BA.1. After a 5-day weight monitoring follow-up, animals were euthanized for tissue analyses. Weight changes (A), survival curves (B), lung viral RNA levels (in two independent lung tissues, LOD = 2 log10 copies/mL) (C), and pulmonary pathological scores (D) of challenged hamsters were shown.
(E) Representative gross observations and H&E-stained lung sections from SARS-CoV-2 Omicron BA.1 challenged hamsters. Views of the whole lung lobes (four independent sections) and the gross observations of lung tissues were presented in the top panel (scale bars, 2 mm), areas in the black box were enlarged in the bottom panel (scale bars, 200 μm). Data in (A) were plotted as means ± SEM. Data in (C) and (D) were shown as box and whisker plots. Dunnett's multiple comparison test (A), or uncorrected Kruskal-Wallis test (C and D) were used for intergroup statistical comparisons. Asterisks indicate statistical significance (∗∗∗∗p < 0.0001; ∗∗∗p < 0.001; ∗∗p < 0.01; ∗p < 0.05; ns, not significant).
See also Figure S4.
These data indicated that the STFK, STFK1628x, and the bivalent vaccines effectively protected hamsters against the SARS-CoV-2 challenge. Moreover, the STFK1628x and the bivalent vaccine showed a better protective efficacy against the Beta and Omicron variant challenge than STFK in hamsters.
The bivalent vaccine blocks SARS-CoV-2 transmissions in hamsters
In addition to protecting vaccine recipients from SARS-CoV-2 infection and pathological changes, an ideal COVID-19 vaccine should also reduce viral shedding and transmission by vaccinated individuals exposed to the virus. For assessments, index hamsters immunized with either STFK1628x or the bivalent vaccine were challenged intranasally with ancestral SARS-CoV-2, Beta variant, or Omicron BA.1. One day later, naive hamsters as sentinels were cohoused with index animals for 24 h (Figure 5 A). After a subsequent 7-day follow-up, sentinels of the unvaccinated-index hamsters showed an average weight loss of 4.0% and 1.7% in the ancestral virus and beta variant challenges, respectively. In contrast, sentinels of vaccinated indexes receiving either the STFK1628x or the bivalent version exhibited gradually increased weights (Figures 5B and 5C). However, due to the mild pathogenicity of Omicron BA.1, the sentinels of vaccinated and unvaccinated hamsters did not experience significant weight loss (Figure 5D). By the end of the experiment, all sentinels of the unvaccinated indexes had detectable viral RNA with approximate levels of 5.0–7.0 log10 copies/mL in the respiratory tract tissues (Figures 5E–5G). By contrast, 1/8 (12.5%), 4/8 (50%), and 7/8 (87.5%) sentinels cohoused with STFK1628x-vaccinated hamsters, when challenged with the ancestral SARS-CoV-2, Beta variant, and Omicron BA.1, respectively, showed detectable viral RNA in tissue from the lung, nasal turbinate, or trachea. Remarkably, no sentinel hamster of the indexes immunized with the bivalent vaccine showed detectable viral RNA in any tissues from the lung, nasal turbinate, and trachea after ancestral SARS-CoV-2 or Beta variant challenge (Figures 5E and 5F), 3/8 (37.5%) of sentinel hamsters were completely protected from the Omicron BA.1 transmission (Figure 5G). The significant increase in spike-specific IgA antibodies in bronchoalveolar lavage (BAL) fluid observed in mice after vaccinations, suggested that respiratory mucosal immunity against SARS-CoV-2 induced by the vaccine may contribute to virus transmission blocking (Figure S5). These data supported the protection from SARS-CoV-2 transmission conferred by the bivalent vaccine in hamsters.
Figure 5.
The monovalent STFK1628x and the STFK/STFK1628x-combined bivalent vaccines prevent SARS-CoV-2 transmission among hamsters
(A) Schematic diagram of the experimental design. Vaccinated- (STFK1628x or bivalent vaccine, 10 μg/dose, 2 doses and 3-week apart) and unvaccinated-index hamsters (n = 4) were intranasally challenged with 1 × 104 PFU of ancestral SARS-CoV-2 or Beta variant, or 1 × 105 PFU of Omicron BA.1. One day later, each 2 index hamsters were cohoused with 4 naive sentinels for one day (separated by a double-layer ventilated fence in the same cage). Sentinel hamsters (n = 8) were followed for 7-day and then euthanized for tissue analyses.
(B–G) Weight changes (B–D) and viral RNA levels in the respiratory tract tissues (E–G) of sentinels after cohoused with index hamsters challenged by ancestral SARS-CoV-2 (B and E), beta variant (C and F), or Omicron BA.1 (D and G) were shown. Viral RNA levels in tissues of the lung (in two independent tissues), nasal turbinate, and trachea were measured by qRT-PCR (LOD = 2 log10 copies/mL). Data in (B)–(D) were plotted as means ± SEM. Data in (E)–(G) were shown as box and whisker plots; median, first quartile, third quartile, minimum value, and maximum value were plotted. Dunnett's multiple comparison test (B–D), or uncorrected Kruskal-Wallis test (E–G) were used for intergroup statistical comparisons, and asterisks indicate statistical significance (∗∗∗∗p < 0.0001; ∗∗∗p < 0.001; ∗∗p < 0.01; ∗p < 0.05; ns, not significant).
See also Figure S5.
Discussion
With the widespread SARS-CoV-2 infections in large populations of humans and susceptible animals, the emergence of antigenic drift variants seems inevitable. The Omicron variant with >30 spike mutations brought great challenges to established immunity by vaccination or resolved infections. Recent studies revealed that the Omicron had led to widespread escape from nAbs acquired from infections by the ancestral SARS-CoV-2 or other VOCs, including Alpha, Beta, Gamma, and Delta (Dejnirattisai et al., 2022a; van der Straten et al., 2022), exhibiting a distinctly altered antigenicity. This variant is also highly resistant against nAbs elicited by a booster dose of the authorized COVID-19 vaccines (Hoffmann et al., 2022). Although some studies have found that the 3rd dose of vaccination may increase the nAb titer against Omicron, it is still lower than that against other variants (e.g., Delta) by several folds (Ai et al., 2021; Garcia-Beltran et al., 2022b). Next-generation COVID-19 vaccines with pan-variants protection are urgently required to deal with the growing threat of Omicron-like immune-escape variants.
Adding another spike antigen of an immune-escape variant is the most common strategy for developing an updated COVID-19 vaccine. Before the emergence of Omicron, the Beta and Gamma were two noticeable VOCs with increased immune evasion concerns; therefore, most previous studies used spikes from one of these two variants as the added antigen. The mRNA-1273.351, a Beta variant-based mRNA vaccine developed by Moderna, could elicit a potent nAb response against the Beta variant in mice but was inferior to the prototype-based mRNA-1273 in neutralizing the Gamma and Epsilon variants (Wu et al., 2021a). Neutralizing antibodies elicited by vaccines formulated with either Beta- or Gamma-derived spikes in animals appeared to be less effective against the Delta variant (Ying et al., 2022). Consistent findings were also noted in the presented study (Figure S2). Moreover, in our data, neither spike antigens from variants of Beta (STFK1351), Gamma (STFK1128), nor B.1.620 (STFK1620) induced markedly improved nAb against all VOCs in hamsters (Figure S2). Therefore, it is unlikely to generate broad-spectrum antigenic coverage by using a pre-existing naturally occurring variant.
Here, we provided a progressive approach to enlarge the cross-variants antigenic coverage of a recombinant spike protein. This approach followed inter-lineage chimera and mutation-patch strategies. As NTD, RBD, and S2 contain nAb-targeting epitopes, inter-lineage chimeric spike antigens might confer cross-variants antibody responses (Martinez et al., 2021). The engineered STFK1628x, providing mutually complementary antigenic coverage to the prototypic spike, was a chimeric construct of the B.1.620-derived NTD and the Gamma-derived RBD-S2. The B.1.620 NTD harbored amino-acid deletions of Δ69–70, Δ145, and Δ242–244, correspond to changed epitopes on the N2, N3, and N5 loops (Harvey et al., 2021a). Notably, Alpha, Eta, and Omicron variants shared the Δ69–70 and Δ145, whereas the Beta variant contained Δ242-244. The RBD-S2 of the two engineered antigens were derivates of the Gamma spike with additional antigenic mutation patches of L452R and T478K. Our study, along with several others, suggested that the Gamma-derived spike was highly immunogenic to elicit a potent nAb response against E484K-harboring variants, such as Beta and Gamma (Sharma et al., 2021). As two Delta-derived characteristic mutations, the introduction of L452R and T478K showed added value to the improvement of the neutralization response against the Delta variant (Figure S2). In contrast to STFK, the monovalent STFK1628x- and the STFK/STFK1628x-combined bivalent vaccines conferred improved protection against viral infection and transmission of SARS-CoV-2 Beta and Omicron variant in hamsters (Figures 3, 4, and 5). Given the potent cross-variants neutralization response elicited by the STFK/STFK1628x-combined bivalent vaccine in hamsters, broad-spectrum protection of this vaccine is to be expected.
Remarkably, the STFK1628x and its derivative STFK1628y elicited significantly higher nAb titers against the Omicron sublineages than the prototypic STFK (Figure 2C), which was possibly attributed to the shared mutations of Δ69–70, K417N/T, T478K, E484K/A, and N501Y. In comparison to STFK1628x, the STFK1628y has an additional N440K mutation but presented a higher nAb titer to neutralize Omicron (Figure 2C), suggesting the N440K plays an essential role in changing the antigenicity of this variant. Notably, as both antigens were generated before Omicron emergence, these results were encouraging and suggested the possibilities of prospective antigen design and vaccine preparations against an unknown future variant. Timely and comprehensive assessments for the antigenic influence of newly emerged spike mutations are essential for guiding vaccine antigen design.
Although trimerized spike antigens were commonly used in current COVID-19 vaccines, our study found that the monomeric spike protein is also highly immunogenic in rodents and nonhuman primates. The CHO-derived C-terminal truncated STFK proteins maintained comparable ACE2 binding activity with the trimeric StriFK, conferred potent immune responses and complete protection against SARS-CoV-2 in hamsters. The monomeric form with more exposed protein surfaces may enable the cryptic epitopes to be more accessible and thereby facilitate the rapid and diverse antibody response. Removing the additional motif required for protein trimerization eliminates unexpected immune responses targeting the trimeric domain. In addition, the high yield of STFK (800–1,000 mg/L from stable cell lines) and STFK1628x (>1,000 mg/L from stable cell lines) in the CHO-cell expression system is beneficial to reduce manufacturing costs and deal with the global shortage of COVID-19 vaccines.
In summary, our study provided a strategy to design antigens for next-generation COVID-19 vaccines aiming to confer broad-spectrum protection.
Limitations of the study
All vaccine candidates in this study were tested in naive animals, whereas most of the people currently have pre-existing immunity to SARS-CoV-2 from natural infection or/and vaccination. Due to the original antigenic sin (Aydillo et al., 2021; Pillai, 2022; Röltgen et al., 2022; Wheatley et al., 2021), pre-existing immunity may affect the boosting effect for the vaccines designed in the present study, such as antibody breadth. Therefore, it would be valuable in the future to study the antibody responses and protective effects induced by these vaccines in previously infected or vaccinated individuals.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Mouse RBD mAb 36H6 | Zhang et al., 2022 | N/A |
| Mouse RBD mAb 83H7 | Zhang et al., 2022 | N/A |
| Mouse RBD mAb 2B4 | Zhang et al., 2022 | N/A |
| Mouse RBD mAb 85F7 | Zhang et al., 2022 | N/A |
| Goat Anti-Mouse IgA alpha chain (HRP) | Abcam | Cat# ab97235; RRID: AB_10681186 |
| Goat Anti-Syrian Hamster IgG H&L (HRP) | Abcam | Cat# ab6892; RRID: AB_955427 |
| Goat Anti-Monkey IgG H&L (HRP) | Abcam | Cat# ab112767; RRID:AB_10866625 |
| HRP-conjugated anti-human IgG (H+L) | Thermo Fisher Scientific | Cat# A18805; RRID:AB_2535582 |
| Biological Samples | ||
| International Standard for anti-SARS-CoV-2 immunoglobulin (NIBSC code: 20/136) | NIBSC | Code: 20/136 |
| Recombinant proteins and peptides | ||
| StriFK protein | Wu et al., 2021b | N/A |
| S1152 (His Tag) | This paper | N/A |
| S1160 (His Tag) | This paper | N/A |
| S1168 (His Tag) | This paper | N/A |
| S1176 (His Tag) | This paper | N/A |
| S1184 (His Tag) | This paper | N/A |
| S1192 (His Tag) | This paper | N/A |
| S1200 (His Tag) | This paper | N/A |
| S1208 (His Tag) | This paper | N/A |
| STFK protein | This paper | N/A |
| STFK1351 protein | This paper | N/A |
| STFK1128 protein | This paper | N/A |
| STFK1620 protein | This paper | N/A |
| STFK1128b protein | This paper | N/A |
| STFK1128c protein | This paper | N/A |
| STFK1128d protein | This paper | N/A |
| STFK1128e protein | This paper | N/A |
| STFK1128f protein | This paper | N/A |
| STFK1128g protein | This paper | N/A |
| STFK1628x protein | This paper | N/A |
| STFK1628y protein | This paper | N/A |
| STFK1628z protein | This paper | N/A |
| STFK1328x protein | This paper | N/A |
| Recombinant human ACE2 (human Fc tag) | This paper | N/A |
| SARS-CoV-2 B.1.351 Spike S1+S2 trimer Protein (ECD, His tag) | Sino Biological inc | Cat# 40589-V08H13 |
| SARS-CoV-2 P.1 Spike S1+S2 Protein (ECD, His Tag) | Sino Biological inc | Cat# 40589-V08B10 |
| SARS-CoV-2 B.1.1.529 (Omicron) S1+S2 trimer Protein ( ECD, His Tag) | Sino Biological inc | Cat# 40589-V08H26 |
| SARS-CoV-2 (BA.2) Spike S1+S2 trimer Protein (ECD, His Tag) | Sino Biological inc | Cat# 40589-V08H28 |
| SARS-CoV-2 (BA.2.12.1) Spike S1+S2 trimer Protein (ECD, His Tag) | Sino Biological inc | Cat# 40589-V08H34 |
| SARS-CoV-2 (BA.4) Spike S1+S2 trimer Protein (ECD, His Tag) | Sino Biological inc | Cat# 40589-V08H32 |
| SARS-CoV-2 Spike Glycoprotein peptides | GenScript | Cat# RP30020 |
| Virus strains | ||
| Authentic SARS-CoV-2 virus (hCoV-19/China/AP8/2020) | Guan’s lab | GISAID access number: EPI_ISL_1655937 |
| Authentic SARS-CoV-2 B.1.351 virus (hCoV-19/China/AP100/2021) | Guan’s lab | GISAID access number: EPI_ISL_2779639 |
| SARS-CoV-2 wildtype pseudovirus (VSV) | Xiong et al., 2020 | N/A |
| SARS-CoV-2 D614G pseudovirus (LV) | Zhang et al., 2022 | N/A |
| SARS-CoV-2 B.1.1.7 variant pseudovirus (LV) | Zhang et al., 2022 | N/A |
| SARS-CoV-2 P.1 variant pseudovirus (LV) | Zhang et al., 2022 | N/A |
| SARS-CoV-2 B.1.351 variant pseudovirus (LV) | Zhang et al., 2022 | N/A |
| SARS-CoV-2 B.1429 variant pseudovirus (LV) | Zhang et al., 2022 | N/A |
| SARS-CoV-2 B.1.525 variant pseudovirus (LV) | Zhang et al., 2022 | N/A |
| SARS-CoV-2 B.1.526 variant (S477N) pseudovirus (LV) | Zhang et al., 2022 | N/A |
| SARS-CoV-2 B.1.526 variant (E484K) pseudovirus (LV) | Zhang et al., 2022 | N/A |
| SARS-CoV-2 B.1.617.1 variant pseudovirus (LV) | Zhang et al., 2022 | N/A |
| SARS-CoV-2 B.1.617.2 variant pseudovirus (LV) | Zhang et al., 2022 | N/A |
| SARS-CoV-2 A.VOI.V2 variant pseudovirus (LV) | Zhang et al., 2022 | N/A |
| SARS-CoV-2 C.37 variant pseudovirus (LV) | Zhang et al., 2022 | N/A |
| SARS-CoV-2 B.1.621 variant pseudovirus (LV) | Zhang et al., 2022 | N/A |
| SARS-CoV-2 B.1.620 variant pseudovirus (LV) | Zhang et al., 2022 | N/A |
| SARS-CoV-2 B.1.1.529 BA.1 variant pseudovirus (LV) | This paper | N/A |
| SARS-CoV-2 B.1.1.529 BA.1.1 variant pseudovirus (LV) | This paper | N/A |
| SARS-CoV-2 B.1.1.529 BA.2 variant pseudovirus (LV) | This paper | N/A |
| SARS-CoV-2 B.1.1.529 BA.2.12.1 variant pseudovirus (LV) | This paper | N/A |
| SARS-CoV-2 B.1.1.529 BA.2.75 variant pseudovirus (LV) | This paper | N/A |
| SARS-CoV-2 B.1.1.529 BA.4 variant pseudovirus (LV) | This paper | N/A |
| Critical Commercial Assays or Kits | ||
| Ni Sepharose 6 Fast Flow | Cytiva | Cat# 17-5318-03 |
| MabSelect SuRe resin | Cytiva | Cat# 17-5474-02 |
| ExpiFectamine™ CHO Transfection Kit | Thermo Scientific | Cat# A29129 |
| 4-12% SurePAGE | Genscript | Cat# M00653 |
| Native Sample Buffer | BIO-RAD | Cat# 1610738 |
| 7.5% Mini-Protean TGX Precast Protein Gels | BIO-RAD | Cat# 4561024 |
| SARS-CoV-2 RT-PCR Kit | Wantai | Cat# WS-1248 |
| QIAamp Viral RNA Mini Kit | Qiagen | Cat# 52906 |
| haematoxylin | Maxim Biotechnology | Cat# CTS-1096 |
| eosin | Maxim Biotechnology | Cat# CTS-4094 |
| Mouse IFN-γ ELISpot Kit | Dakewe Biotech | Cat# 2210005 |
| Deposited data | ||
| Cryo-EM structure of STKF:36H6:83H7:85F7 | This paper | Protein Data Bank: 7WP6 |
| Cryo-EM structure of STFK1628x:83H7:85F7:2B4 | This paper | Protein Data Bank: 7WP8 |
| EM density map of STKF:36H6:83H7:85F7 | This paper | Protein Data Bank: EMD-32676 |
| EM density map of STFK1628x:83H7:85F7:2B4 | This paper | Protein Data Bank: EMD-32678 |
| Experimental Models: Cell Lines | ||
| ExpiCHO-S cells | Thermo Scientific | Cat# A29127 RRID: CVCL_5J31 |
| H1299-huACE2 (human ACE2) | Zhang et al., 2022 | N/A |
| HBK21-hACE2 | Xiong et al., 2020 | N/A |
| Experimental models: Animals | ||
| BALB/c mice | SLAC Laboratory | N/A |
| C57BL/6 mice | SLAC Laboratory | N/A |
| LVG Syrian hamsters | Charles River | Cat# 501 |
| Rhesus monkey | Key Laboratory of Technical Evaluation of Fertility Regulation for Non-human Primate | N/A |
| Recombinant DNA | ||
| Plasmid: EIRBsMie-S1152 | This paper | N/A |
| Plasmid: EIRBsMie-S1160 | This paper | N/A |
| Plasmid: EIRBsMie-S1168 | This paper | N/A |
| Plasmid: EIRBsMie-S1176 | This paper | N/A |
| Plasmid: EIRBsMie-S1184 | This paper | N/A |
| Plasmid: EIRBsMie-S1192 | This paper | N/A |
| Plasmid: EIRBsMie-S1200 | This paper | N/A |
| Plasmid: EIRBsMie-S1208 | This paper | N/A |
| Plasmid: pGS01b-STFK1351 | This paper | N/A |
| Plasmid: pGS01b-STFK1128 | This paper | N/A |
| Plasmid: pGS01b-STFK1620 | This paper | N/A |
| Plasmid: pGS01b-STFK1128b | This paper | N/A |
| Plasmid: pGS01b-STFK1128c | This paper | N/A |
| Plasmid: pGS01b-STFK1128d | This paper | N/A |
| Plasmid: pGS01b-STFK1128e | This paper | N/A |
| Plasmid: pGS01b-STFK1128f | This paper | N/A |
| Plasmid: pGS01b-STFK1128g | This paper | N/A |
| Plasmid: pGS01b-STFK1628x | This paper | N/A |
| Plasmid: pGS01b-STFK1628y | This paper | N/A |
| Plasmid: pGS01b-STFK1628z | This paper | N/A |
| Plasmid: pGS01b-STFK1328x | This paper | N/A |
| Plasmid: EIRBsMie-hACE2 | This paper | N/A |
| Software and Algorithms | ||
| Columbus Analysis system (version 2.5.0) | PerkinElmer | https://www.perkinelmer.com/ |
| GraphPad Prism (version 8.0.1) | Graphpad | https://www.graphpad.com/ |
| Origin 2021 | OriginLab | https://www.originlab.com/2021 |
| Discovery Studio software | Accelrys | https://www.3dsbiovia.com |
| MotionCor2 | Zheng et al., 2017 | http://msg.ucsf.edu/em/software/motioncor2.html |
| Gctf | Zhang, 2016 | ==https://en.wikibooks.org/w/index.php?title=Software_Tools_For_Molecular_Microscopy&stable=0#Gctf |
| Cryosparc V3 | Punjani et al., 2017 | https://cryosparc.com |
| ResMap | Kucukelbir et al., 2014 | http://resmap.sourceforge.net |
| Chimera | Pettersen et al., 2004 | http://www.cgl.ucsf.edu/chimera |
| Coot | Emsley and Cowtan, 2004 | http://www2.mrc-lmb.cam.ac.uk/personal/pemsley/coot |
| PHENIX | Adams et al., 2010 | http://phenix-online.org |
| Molprobity | Chen et al., 2010 | http://molprobity.biochem.duke.edu |
| ChimeraX | Pettersen et al., 2021 | https://www.cgl.ucsf.edu/chimerax/ |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Ningshao Xia (nsxia@xmu.edu.cn).
Materials availability
All requests for resources and reagents should be directed to the lead contact author. All reagents, which includes antibodies, proteins, plasmids, and virus, will be made available on request after completion of a Materials Transfer Agreement for non-commercial usage.
Experimental model and subject details
Animals
Six to eight-week-old female BALB/c and C57BL/6 mice were purchased from Shanghai SLAC Laboratory Animal Co., Ltd. Lakeview Golden (LVG) Syrian hamsters were purchased from Charles River Laboratories (Beijing). The animals were fed in Specific-pathogen-free circumstances. The mouse and hamster studies were carried out in strict accordance with the recommendations of the Guide for the Care and Use of Laboratory Animals under the approval of the Institutional Animal Care and Use Committee of Xiamen University. Ten rhesus monkeys (Macaca mulatta) aged 4.5-7.5 years were divided into two groups, each containing 2 males and 3 females, to evaluate the immunogenicity of the vaccine. The rhesus monkey experiment was conducted at the Key Laboratory of Technical Evaluation of Fertility Regulation for Nonhuman Primate Inc in Fujian province, and was authorized by the Institutional Animal Care and Use Committee of Fujian Maternity and Child Health Hospital (approval number: 2021-01).
Cell lines and virus
The H1299 cells expressing human ACE2 (H1299-huACE2) were culture in Dulbecco's modified Eagle medium (DMEM) supplemented with 10% fetal bovine serum (FBS) and penicillin-streptomycin. Vero cells were culture in DMEM supplemented with 10% FBS and penicillin-streptomycin. ExpiCHO-S (Thermo Scientific) cells were maintained in ExpiCHO Expression Medium (Thermo Scientific). The authentic SARS-CoV-2 virus, including prototype (hCoV-19/Wuhan/AP8/2020, GISAID access number: EPI_ISL_1655937), Beta variant (BetaCoV/Jiangsu/JS02/2020, EPI_ISL_411952), Beta variant (hCoV-19/China/AP100/2021, GISAID access number: EPI_ISL_2779639), Omicron BA.1 variant (share an identical sequence with EPI_ISL_8182026), were used in this study.
Method details
Constructs, protein expressions, and purifications
Trimeric S-ectodomain proteins of SARS-CoV-2 (StriFK) were expressed and purified as previously described (Wu et al., 2021b). Expression cassettes encoding monomeric S-ectodomain proteins (containing mutated furin-site, RRAR to GSAS) involved in the study were generated by site-directed mutagenesis via PCR cloning based on the parental codon-optimized StriFK (Wu et al., 2021b). The expression cassettes of S1152 to S1208 with C-terminal 8×His-tag were cloned into the EIRBsMie vector (Zhang et al., 2022). The tag-free STFK and STFK variants were constructed into a pGS01b vector, modified from the pCGS3 vector containing glutamine synthetase (GS) selection marker (Sigma Aldrich). All STFK constructs had an N1192M modification to reduce potential protein aggregation, possibly attributed the K933-N1192 interaction (Xia et al., 2020). As previously described, transient protein expressions were performed using the ExpiCHO expression system (Thermo Fisher Scientific). Stable cell lines expressing the STFK, STFK1628x, and STKF1628y were generated via transfections of the PGS01b-vectored constructs into CHOZN GS-/- cells (Sigma Aldrich) following GS selections and single-cell clonings. Polyhistidine-tagged proteins (S1152 to S1208) purified from culture supernatants were collected on day 7 after transfection using Ni Sepharose 6FF (Cytiva). The tag-free STFK proteins were purified by using Q-FF Sepharose ion-exchange chromatography (Cytiva). Recombinant human ACE2 (human Fc tag, rACE2) protein also was produced in ExpiCHO-S cells and purified by protein-A affinity chromatography column (Cytiva) as previously described (Wu et al., 2021b).
SDS-polyacrylamide gel electrophoresis (SDS-PAGE) and Native-PAGE
SDS-PAGE analyses were performed using 4-12% SurePAGE (Genscript), the precast mini polyacrylamide gels. For native-PAGE, the protein samples were mixed with the Native Sample Buffer (BIO-RAD) in equal volume and then subjected to electrophoresis using the 7.5% Mini-Protean TGX Precast Protein Gels (Bio-RAD) in a non-denaturing buffer. Gel images were captured using FUSION FX7 Spectra multispectral imaging system (Vilber).
Size exclusion chromatography (SEC-HPLC)
The SEC-HPLC analysis shown in Figure 1B was performed using a TSK-GEL G3000PWXL column on an HPLC system (Waters Alliance) and conducted as described previously (Wu et al., 2021b).
ACE2 binding assays
A capture antibody 45C3 (developed in our laboratory) recognizes spike S2 domain was coated in 96-well microplates at 200 ng per well. Plates were incubated overnight at 4 °C and then blocked with ELISA-blocking buffer (Wantai BioPharm). The STFK proteins were twofold serially diluted from 10 μg mL−1 to 9.8 ng mL−1 in duplicate and then added to wells (100 μL). After incubation for 1 hour at 25°C followed by washing with PBST buffer, rACE2 protein solution (100 μL per well, 1 μg mL−1) was added to the wells. Subsequently, the microplates were incubated for 1 hour at 25°C. After washing five times, HRP-conjugated anti-human IgG (Thermo Fisher Scientific) solutions were added and incubated for 1 hour at 25°C. Following washing five times, tetramethylbenzidine chromogen (TMB) solution (Wantai BioPharm) was added into microplates 100 μL per well. After a further 10 minutes of incubation at 25°C, 2 M H2SO4 was added to stop the chromogen reaction, and the optical density (OD450-630) value was measured. The half-maximal effective concentration (EC50) was calculated by the 4-parameter logistic (4PL) regression using GraphPad Prism 8 software.
Vaccine preparations
Recombinant spike protein subunit vaccines used in this study were composed of spike proteins and a nitrogen bisphosphonate-modified zinc-aluminum hybrid adjuvant (FH002C), which was described detailly in our previous study (Wu et al., 2021b). Briefly, the proteins were mixed with an equal volume of 2× concentration FH002C adjuvant to achieve the final desired concentration of antigen in the final formulation. All vaccine formulations were mixed well and stored at 2-8 °C until use.
Mouse immunizations
Six to eight-week-old BALB/c mice (n=6 per group) were immunized with STFK vaccines at 0.01, 0.1, 1, or 10 μg per dose in 150 μL through intramuscular injection (IM) following a two-dose schedule 2 (Figure 1C) or 3 (Figure 1E) weeks apart. For T cell response assessment, six to eight-week-old C57BL/6 mice were immunized with STFK vaccine single dose at 10 μg per dose in 150 μL through intramuscular injection. Immunized mice were sacrificed on day 7 after immunization to collect splenocytes for further assay. For BAL IgA alaysis, six to eight-week-old BALB/c mice (n=6 per group) were unvaccinated or immunized with 2 dose of indicated vaccine (10 μg per dose in 150 μL through IM) at week 0 and 3. BAL fluid were collected two weeks after the booster as previously described (Chen et al., 2022).
Rhesus monkey immunizations
Ten rhesus monkeys were allocated randomly into two groups (three females and two males per group). Groups of monkeys were injected with 1 μg or 15 ug of STFK vaccine per dose (150 μL) via the intramuscular route for 2 doses. All monkeys were vaccinated at weeks 0 and 4. Two weeks after boosting, serum samples were collected for antibody analyses, including measurement of anti-spike IgG, anti-RBD IgG, pseudovirus neutralizing antibody, and authentic neutralizing antibody titers.
Hamster immunizations
Six to eight-week-old hamsters were used to evaluate the immunogenicity and protective effect of the vaccine candidates. Each group contained four males and four females. Groups of hamsters were immunized intramuscularly twice with the vaccine candidates at 10 μg per dose in 200 μl, three weeks apart. All serum samples were collected at week-2 after the 2nd dose via retro-orbital bleeding to measure the antibody titers.
Pseudovirus (PsV) neutralization assays
The nAb titers against the ancestral spike-pseudotyped virus presented in Figures 1C and 1E–1G were determined by using a vesicular stomatitis virus (VSV) system as previously described (Xiong et al., 2020). The International Standard for anti-SARS-CoV-2 immunoglobulin (NIBSC code: 20/136) was obtained from National Institute for Biological Standards and Control, UK (Knezevic et al., 2022).
The lentiviral-based pseudovirus (LV) neutralization assay was used to determine vaccine-elicited nAbs against circulating SARS-CoV-2 variants (Figures 1H, 2, and S5–S7). The lentiviral pseudoviruses bearing spikes from SARS-CoV-2 variants were generated as described previously (Chang et al., 2021; Zhang et al., 2022), including D614G (site-directed mutagenesis), Alpha (B.1.1.7, GISAID accession number: EPI_ISL_601443), Beta ( B.1.351, EPI_ISL_700428), Gamma (P.1, EPI_ISL_792680), Delta (B.1.617.2, EPI_ISL_1662451), Omicron BA.1 (B.1.1.529, EPI_ISL_6704867), Omicron BA.1.1 (B.1.1.529, EPI_ISL_9640036), Omicron BA.2 (B.1.1.529, EPI_ISL_8253179), BA.2.12.1 (B.1.1.529, EPI_ISL_11704386), BA.2.75 (B.1.1.529, EPI_ISL_13623453), BA.4 (B.1.1.529, EPI_ISL_11550739), Iota_484K (B.1.526_484K, EPI_ISL_1009654), Iota_477N (B.1.526_477N, EPI_ISL_995145), Epsilon (B.1.429, EPI_ISL_873881), Eta (B.1.525, EPI_ISL_762449), Kappa (B.1.617.1, EPI_ISL_1595904), A.VOI.V2 (EPI_ISL_1347941), Lambda (C.37, EPI_ISL_2921532), Mu (B.1.621, EPI_ISL_3933281) and B.1.620 (EPI_ISL_1620228). The PsV nAb measurements were performed as previously described using Opera Phenix or Operetta CLS High-Content Analysis System (PerkinElmer) (Zhang et al., 2022).
Authentic SARS-CoV-2 neutralization assay
The nAb titers of sera from immunized animals against authentic SARS-CoV-2 were detected using a cytopathic effect (CPE)-based microneutralization assay as previously described (Li et al., 2021). The ancestral virus (BetaCoV/Jiangsu/JS02/2020, EPI_ISL_411952) and Omicron BA.1 (share an identical sequence with EPI_ISL_8182026) were used. Briefly, serum samples were twofold serially diluted from 1:4 to 1:8192 in duplicate with DMEM medium. All prepared samples were mixed with the virus of 200 TCID50 and incubated for 2 hours at 37 °C. The mixtures (150 μL per well) were added to a monolayer of Vero cells in a 96-well plate and incubated at 37 °C supplying with 5% CO2. Three-day later, the cytopathic effect was assessed with microscopic examinations. The neutralizing titer of serum was expressed as the reciprocal of the maximal sample dilution that protects at least 50% of cells from CPE.
Enzyme-linked immunospot (ELISpot) assay
According to the manufacturer's instructions, the assays were performed with mouse IFN-γ ELISpot plates kits (Dakewe Biotech, 2210005). In brief, single-cell suspensions were obtained from mouse spleen (106 cells per well) through grinding in 70 μm cell strainers and were seeded in anti-mouse IFN-γ antibody precoated ELISpot plates. Then, cells were incubated with pooled peptides of SARS-CoV-2 spike (15-mer peptides with 11aa overlap covering the entire spike protein; GenScript) and cultured at 37°C with 5% CO2 for 20 hours. Spots were counted and analyzed by using CTL-ImmunoSpot S5 (Cellular Technology Limited). The numbers of IFN-γ-secreting cells were calculated by subtracting phosphate-buffered saline (PBS)-stimulated wells from spike peptide pool-stimulated wells.
Anti-RBD IgG, anti-spike IgG, and anti-spike IgA measurements
For serum IgG antibody against SARS-CoV-2 Wuhan-1 strain, microplates pre-coated with recombinant antigens of RBD or spike ectodomain were provided by Beijing Wantai Biological Pharmacy. For serum IgG antibody against SARS-CoV-2 variants and BAL IgA antibody, microplates were coated with 100 ng/well of indicated recombinant spike (Sino Biological inc). The measurements were performed following previously described procedures (Wu et al., 2021b), with the only difference that the cutoff (CO) value was set as 0.1 (OD450-630). The IgG and IgA titer of each sample was determined as the cutoff index (OD450-630/CO) at the dilution limit multiplied by the maximum dilution folds. Representative data from technical replicates were performed at least twice for plotting.
Cryo-EM sample preparation and data collection
Fabs of 36H6, 83H7, 2B4, and 85F7 were prepared by papain digestion of the mAbs and further purified with MabSelect SuRe (Cytiva). Aliquots (3 μl) of 3.5 mg/mL mixtures of purified STFK or STFK1628x proteins in complex with excess Fab fragments of nAbs were incubated in 0.01% (v/v) Digitonin (Sigma) and then loaded onto glow-discharged (60 s at 20 mA) holey carbon Quantifoil grids (R1.2/1.3, 200 mesh, Quantifoil Micro Tools) using a Vitrobot Mark IV (ThermoFisher Scientific) at 100% humidity and 4°C. Data were acquired using the SerialEM software on an FEI Tecnai F30 transmission electron microscope (ThermoFisher Scientific) operated at 300 kV and equipped with a Gatan K3 direct detector. Images were recorded in the 36-frame movie mode at a nominal 39,000× magnification at super-resolution mode with a pixel size of 0.339 Å. The total electron dose was set to 60 e− Å−2 , and the exposure time was 4.5 s.
Image processing and 3D reconstruction
Drift and beam-induced motion correction were performed with MotionCor2 (Zheng et al., 2017) to produce a micrograph from each movie. Contrast transfer function (CTF) fitting and phase-shift estimation were conducted with Gctf (Zhang, 2016). Micrographs with astigmatism, obvious drift, or contamination were discarded before reconstruction. The following reconstruction procedures were performed by using Cryosparc V3 (Punjani et al., 2017). In brief, particles were automatically picked by using the "Blob picker" or "Template picker". Several rounds of reference-free 2D classifications were performed, and the selected good particles were then subjected to ab-initio reconstruction, heterogeneous refinement and final non-uniform refinement. The resolution of all density maps was determined by the gold-standard Fourier shell correlation curve, with a cutoff of 0.143. Local map resolution was estimated with ResMap (Kucukelbir et al., 2014).
Atomic model building, refinement, and 3D visualization
The initial models of nAbs were generated from homology modeling by Accelrys Discovery Studio software (available from: https://www.3dsbiovia.com). The structure from the prototypic trimeric spike (PDB no. 6VSB) (Wrapp et al., 2020) was used as the initial modes of our proteins. We initially fitted the templates into the corresponding final cryo-EM maps using Chimera (Pettersen et al., 2004), and further corrected and adjusted them manually by real-space refinement in Coot (Emsley and Cowtan, 2004). The resulting models were then refined with phenix.real_space_refine in PHENIX (Adams et al., 2010). These operations were executed iteratively until the problematic regions, Ramachandran outliers, and poor rotamers were either eliminated or moved to favored regions. The final atomic models were validated with Molprobity (Chen et al., 2010; Robert and Gouet, 2014). All figures were generated with Chimera or ChimeraX (Pettersen et al., 2021).
SARS-CoV-2 virus challenges in hamster
Two weeks after boosting, hamsters were challenged with the ancestral SARS-CoV-2 of hCoV-19/China/AP8/2020 (EPI_ISL_1655937) or Beta variant of hCoV-19/China/AP100/2021 (EPI_ISL_2779639) or Omicron BA.1 variant (share an identical sequence with EPI_ISL_8182026). For the intranasal challenge, hamsters were challenged with 1×104 PFU (ancestral strain and Beta variant) or 1×105 PFU (Omicron BA.1) of SARS-CoV-2 virus (diluted in 100 μL of PBS, 50 μL in each nare) through the intranasal route under anesthesia. For virus transmission-blocking study, vaccinated- or unvaccinated-hamsters were intranasally inoculated with 1×104 PFU (ancestral strain and Beta variant) or 1×105 PFU (Omicron BA.1) as indexes. One day post-infection, index hamsters were cohoused with naïve sentinels for one day (Figure 5A). The daily diet was limited to 7 g per 100 g of body weight to prevent animals from overeating. All hamsters were monitored for body weight until being humanely euthanized on day 7 or 5 after challenge or exposure. The respiratory tissues of hamsters were collected for viral RNA quantification or histopathological assessments. All challenge experiments were conducted in the Animal Biosafety Level 3 (ABSL-3) facility.
SARS-CoV-2 RNA quantification
Viral RNA levels in tissues from hamsters were determined by SARS-CoV-2 RT-PCR Kit (Wantai BioPharm). For each animal, two pieces (separately) of lung tissue (0.1∼0.2 g each), one piece of the trachea tissue (0.1∼0.2 g), and a half of nasal turbinate (0.8∼1.2 g) were respectively homogenized with TissueLyser II (Qiagen) in 1 ml PBS. Viral RNA in tissue lysates was extracted using the QIAamp Viral RNA Mini Kit (Qiagen) and subjected to qRT-PCR assays by using primers and probes in the kit targeting ORF1ab of genomic RNA. Representative data from technical replicates were obtained from at least two independent experiments for plotting.
Histopathology
The lung tissues from challenged hamsters were fixed with neutral buffered formalin for 48 hours and processed routinely into paraffin blocks. Then tissues were sectioned to 3 μm by microtome (Leica). Next, the fixed lung sections were stained with hematoxylin and eosin to analyze lung histopathology or were stained with a mouse anti–SARS-CoV-2 NP-specific antibody to evaluate the expression and distribution of viral protein in lung tissues. Whole-slide images of the lung sections were captured with the EVOS M7000 Images System (Thermo Fisher Scientific). Microscopic evaluation of pathological lung lesions was performed blindly by pathologists following a semiquantitative scoring system with the inclusion of three indicators (Yuan et al., 2021): (i) alveolar septum thickening and consolidation; (ii) hemorrhage, exudation, pulmonary edema, and mucous; and (iii) recruitment and infiltration of inflammatory immune cells. For each hamster, three or four lobes of the lung were assessed independently, and the average score was calculated to indicate the overall pathological severity.
Quantification and statistical analysis
Number of animals (n) and statistical details of experiments can be found in the figure legends. The Mann-Whitney U test was used for the comparison between two independent samples. The uncorrected Kruskal-Wallis test, Dunnett's Multiple Comparison test, or uncorrected Fisher's LSD test was applied to analyze differences among more than two groups. A two-sided log-rank test was applied to compare the difference in survival. Statistical differences were considered to be significant for two-tailed P values of < 0.05. All statistical analyses were conducted in GraphPad Prism 8 software.
Acknowledgments
This study was supported by National Natural Science Foundation of China grants 81991491 (to N.X.), 81871316 (to Q.Y.), 31730029 (to N.X.), U1905205 (to Q.Y.), 81902057 (to Y.Z.), 32170943 (to T.Z.); the Science and Technology Major Project of Xiamen (3502Z2021YJ013 to Y.Z.); funding for Guangdong-Hongkong-Macau Joint Laboratory grant 2019B121205009 (to Y.G.) and EKIH Pathogen Research Institute grant HZQB-KCZYZ-2021014 (to Y.G.); Fujian Natural Science Foundation for Distinguished Young Scholars grant 2020J06007 (to T.Z.). We thank Dr. Huirong Pan, Dr. Daning Wang from Xiamen Innovax Biotech; and Dr. Baofu Ni, Dr. Jinghua Zhao, Dr. Min You, Dr. Chunfeng Huang from Antibody Institute of Zhejiang Yangshengtang Biotech Co., Ltd. for helping in the expression and purification of recombinant proteins.
Author contributions
H. Zhu, Q.Y., Y.G., and N.X. conceptualized and designed the study. S.W., K.W., Y.W., Y.Z., M.W., and J. Xiao designed the clones and produced and characterized the proteins. Y.W., L.Y., J.M., M.Z., C.L., and W.G. performed the animal experiments. H.C., Z.L., H. Zhang, X.W., and Y.W. performed the rhesus monkey experiments. Z.W., Y.H., D.C., M.L., Huilin Guo, J.G., and H.X. performed the ELISA and neutralization assays. J. Xu and Huiyu Guo performed enzyme-linked immunospot (ELISpot) assays. J.M. and P.C. performed the SARS-CoV-2 RNA quantification. M.N. and Y.W. formulated the vaccines. Y.S. and W.H. prepared mAbs. Q. Zheng carried out the cryo-EM studies. Q.Y., Y.W., S.W., Q. Zheng, and Z.H. wrote the paper. S.L., T.C., T.W., Y.C., Q. Zhao, J.Z., T.Z., H. Zhu, Y.G., and N.X. revised the manuscript. All authors read and approved the final version of the manuscript.
Declaration of interests
Q.Y., Y.W., S.W., Y.Z., M.W., K.W., Z.W., J. Xiao, T.Z., J.Z., and N.X. are coinventors on a patent in the application for the spike constructs and their applications described in this study (202210078434.8).
Published: October 18, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.chom.2022.10.011.
Supplemental information
Data and code availability
Structure coordinates are deposited in the Protein Data Bank (PDB) under accession codes: STKF:36H6:83H7:85F7 (PDB: 7WP6), STFK1628x:83H7:85F7:2B4 (PDB: 7WP8). The corresponding EM density maps have been deposited in the Electron Microscopy Data Bank (EMBD) under accession number: STKF:36H6:83H7:85F7 (EMD-32676), STFK1628x:83H7:85F7:2B4 (EMD-32678).
This paper does not report any original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- Adams P.D., Afonine P.V., Bunkóczi G., Chen V.B., Davis I.W., Echols N., Headd J.J., Hung L.W., Kapral G.J., Grosse-Kunstleve R.W., et al. Phenix: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ai J., Zhang H., Zhang Y., Lin K., Zhang Y., Wu J., Wan Y., Huang Y., Song J., Fu Z., et al. Omicron variant showed lower neutralizing sensitivity than other SARS-CoV-2 variants to immune sera elicited by vaccines after boost. Emerg. Microbes Infect. 2021:1–24. doi: 10.1080/22221751.2021.2022440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews N., Stowe J., Kirsebom F., Toffa S., Rickeard T., Gallagher E., Gower C., Kall M., Groves N., O'Connell A.M., et al. Covid-19 vaccine effectiveness against the omicron (B.1.1.529) variant. N. Engl. J. Med. 2022;386:1532–1546. doi: 10.1056/NEJMoa2119451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aydillo T., Rombauts A., Stadlbauer D., Aslam S., Abelenda-Alonso G., Escalera A., Amanat F., Jiang K., Krammer F., Carratala J., Garcia-Sastre A. Immunological imprinting of the antibody response in COVID-19 patients. Nat. Commun. 2021;12:3781. doi: 10.1038/s41467-021-23977-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes C.O., Jette C.A., Abernathy M.E., Dam K.A., Esswein S.R., Gristick H.B., Malyutin A.G., Sharaf N.G., Huey-Tubman K.E., Lee Y.E., et al. SARS-CoV-2 neutralizing antibody structures inform therapeutic strategies. Nature. 2020;588:682–687. doi: 10.1038/s41586-020-2852-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brouwer P.J.M., Brinkkemper M., Maisonnasse P., Dereuddre-Bosquet N., Grobben M., Claireaux M., de Gast M., Marlin R., Chesnais V., Diry S., et al. Two-component spike nanoparticle vaccine protects macaques from SARS-CoV-2 infection. Cell. 2021;184:1188–1200.e19. doi: 10.1016/j.cell.2021.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y., Yisimayi A., Jian F., Song W., Xiao T., Wang L., Du S., Wang J., Li Q., Chen X., et al. BA.2.12.1, BA.4 and BA.5 escape antibodies elicited by Omicron infection. Nature. 2022;608:593–602. doi: 10.1038/s41586-022-04980-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cele S., Jackson L., Khoury D.S., Khan K., Moyo-Gwete T., Tegally H., San J.E., Cromer D., Scheepers C., Amoako D.G., et al. Omicron extensively but incompletely escapes Pfizer BNT162b2 neutralization. Nature. 2022;602:654–656. doi: 10.1038/s41586-021-04387-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang L., Hou W., Zhao L., Zhang Y., Wang Y., Wu L., Xu T., Wang L., Wang J., Ma J., et al. The prevalence of antibodies to SARS-CoV-2 among blood donors in China. Nat. Commun. 2021;12:1383. doi: 10.1038/s41467-021-21503-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J., Wang P., Yuan L., Zhang L., Zhang L., Zhao H., Chen C., Wang X., Han J., Chen Y., et al. A live attenuated virus-based intranasal COVID-19 vaccine provides rapid, prolonged, and broad protection against SARS-CoV-2. Sci. Bull. 2022;67:1372–1387. doi: 10.1016/j.scib.2022.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen V.B., Arendall W.B., 3rd, Headd J.J., Keedy D.A., Immormino R.M., Kapral G.J., Murray L.W., Richardson J.S., Richardson D.C. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui Z., Liu P., Wang N., Wang L., Fan K., Zhu Q., Wang K., Chen R., Feng R., Jia Z., et al. Structural and functional characterizations of infectivity and immune evasion of SARS-CoV-2 Omicron. Cell. 2022;185:860–871.e13. doi: 10.1016/j.cell.2022.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dejnirattisai W., Huo J., Zhou D., Zahradník J., Supasa P., Liu C., Duyvesteyn H.M.E., Ginn H.M., Mentzer A.J., Tuekprakhon A., et al. SARS-CoV-2 Omicron-B.1.1.529 leads to widespread escape from neutralizing antibody responses. Cell. 2022;185:467–484.e15. doi: 10.1016/j.cell.2021.12.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P., Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Garcia-Beltran W.F., St Denis K.J., Hoelzemer A., Lam E.C., Nitido A.D., Sheehan M.L., Berrios C., Ofoman O., Chang C.C., Hauser B.M., et al. mRNA-based COVID-19 vaccine boosters induce neutralizing immunity against SARS-CoV-2 Omicron variant. Cell. 2022;185:457–466.e4. doi: 10.1016/j.cell.2021.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harvey W.T., Carabelli A.M., Jackson B., Gupta R.K., Thomson E.C., Harrison E.M., Ludden C., Reeve R., Rambaut A., COVID-19 Genomics UK (COG-UK) Consortium SARS-CoV-2 variants, spike mutations and immune escape. Nat. Rev. Microbiol. 2021;19:409–424. doi: 10.1038/s41579-021-00573-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann M., Krüger N., Schulz S., Cossmann A., Rocha C., Kempf A., Nehlmeier I., Graichen L., Moldenhauer A.-S., Winkler M.S., et al. The Omicron variant is highly resistant against antibody-mediated neutralization – implications for control of the COVID-19 pandemic. Cell. 2022;185:447–456.e11. doi: 10.1016/j.cell.2021.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iketani S., Liu L., Guo Y., Liu L., Chan J.F., Huang Y., Wang M., Luo Y., Yu J., Chu H., et al. Antibody evasion properties of SARS-CoV-2 Omicron sublineages. Nature. 2022;604:553–556. doi: 10.1038/s41586-022-04594-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knezevic I., Mattiuzzo G., Page M., Minor P., Griffiths E., Nuebling M., Moorthy V. WHO International Standard for evaluation of the antibody response to COVID-19 vaccines: call for urgent action by the scientific community. Lancet Microbe. 2022;3:e235–e240. doi: 10.1016/S2666-5247(21)00266-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kucukelbir A., Sigworth F.J., Tagare H.D. Quantifying the local resolution of cryo-EM density maps. Nat. Methods. 2014;11:63–65. doi: 10.1038/nmeth.2727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Hui A., Zhang X., Yang Y., Tang R., Ye H., Ji R., Lin M., Zhu Z., Türeci Ö., et al. Safety and immunogenicity of the SARS-CoV-2 BNT162b1 mRNA vaccine in younger and older Chinese adults: a randomized, placebo-controlled, double-blind phase 1 study. Nat. Med. 2021;27:1062–1070. doi: 10.1038/s41591-021-01330-9. [DOI] [PubMed] [Google Scholar]
- Liang J.G., Su D., Song T.Z., Zeng Y., Huang W., Wu J., Xu R., Luo P., Yang X., Zhang X., et al. S-trimer, a COVID-19 subunit vaccine candidate, induces protective immunity in nonhuman primates. Nat. Commun. 2021;12:1346. doi: 10.1038/s41467-021-21634-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L., Iketani S., Guo Y., Chan J.F., Wang M., Liu L., Luo Y., Chu H., Huang Y., Nair M.S., et al. Striking antibody evasion manifested by the Omicron variant of SARS-CoV-2. Nature. 2022;602:676–681. doi: 10.1038/s41586-021-04388-0. [DOI] [PubMed] [Google Scholar]
- Madhi S.A., Baillie V., Cutland C.L., Voysey M., Koen A.L., Fairlie L., Padayachee S.D., Dheda K., Barnabas S.L., Bhorat Q.E., et al. Efficacy of the ChAdOx1 nCoV-19 Covid-19 vaccine against the B.1.351 Variant. N. Engl. J. Med. 2021;384:1885–1898. doi: 10.1056/NEJMoa2102214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez D.R., Schäfer A., Leist S.R., De la Cruz G., West A., Atochina-Vasserman E.N., Lindesmith L.C., Pardi N., Parks R., Barr M., et al. Chimeric spike mRNA vaccines protect against Sarbecovirus challenge in mice. Science. 2021;373:991–998. doi: 10.1126/science.abi4506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen E.F., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M., Meng E.C., Ferrin T.E. UCSF Chimera--a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- Pettersen E.F., Goddard T.D., Huang C.C., Meng E.C., Couch G.S., Croll T.I., Morris J.H., Ferrin T.E. UCSF ChimeraX: structure visualization for researchers, educators, and developers. Protein Sci. 2021;30:70–82. doi: 10.1002/pro.3943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillai S. SARS-CoV-2 vaccination washes away original antigenic sin. Trends Immunol. 2022;43:271–273. doi: 10.1016/j.it.2022.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Punjani A., Rubinstein J.L., Fleet D.J., Brubaker M.A. cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods. 2017;14:290–296. doi: 10.1038/nmeth.4169. [DOI] [PubMed] [Google Scholar]
- Robert X., Gouet P. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 2014;42:W320–W324. doi: 10.1093/nar/gku316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Röltgen K., Nielsen S.C.A., Silva O., Younes S.F., Zaslavsky M., Costales C., Yang F., Wirz O.F., Solis D., Hoh R.A., et al. Immune imprinting, breadth of variant recognition, and germinal center response in human SARS-CoV-2 infection and vaccination. Cell. 2022;185:1025–1040.e14. doi: 10.1016/j.cell.2022.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma S., Vercruysse T., Sanchez-Felipe L., Kerstens W., Rasulova M., Abdelnabi R., Foo C., Lemmens V., Looveren D., Maes P. 2021. Updated vaccine protects from infection with SARS-CoV-2 variants, prevents transmission and is immunogenic against Omicron in hamsters. Preprint at bioRxiv. [DOI] [Google Scholar]
- Starr T.N., Czudnochowski N., Liu Z., Zatta F., Park Y.J., Addetia A., Pinto D., Beltramello M., Hernandez P., Greaney A.J., et al. SARS-CoV-2 RBD antibodies that maximize breadth and resistance to escape. Nature. 2021;597:97–102. doi: 10.1038/s41586-021-03807-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian J.H., Patel N., Haupt R., Zhou H., Weston S., Hammond H., Logue J., Portnoff A.D., Norton J., Guebre-Xabier M., et al. SARS-CoV-2 spike glycoprotein vaccine candidate NVX-CoV2373 immunogenicity in baboons and protection in mice. Nat. Commun. 2021;12:372. doi: 10.1038/s41467-020-20653-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uriu K., Kimura I., Shirakawa K., Takaori-Kondo A., Nakada T.A., Kaneda A., Nakagawa S., Sato K., Genotype to Phenotype Japan (G2P-Japan) Consortium Neutralization of the SARS-CoV-2 mu variant by convalescent and vaccine serum. N. Engl. J. Med. 2021;385:2397–2399. doi: 10.1056/NEJMc2114706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Straten K., Guerra D., van Gils M.J., Bontjer I., Caniels T.G., van Willigen H.D.G., Wynberg E., Poniman M., Burger J.A., Bouhuijs J.H., et al. 2022. Mapping the antigenic diversification of SARS-CoV-2. Preprint at medRxiv. [DOI] [Google Scholar]
- Wang P., Casner R.G., Nair M.S., Wang M., Yu J., Cerutti G., Liu L., Kwong P.D., Huang Y., Shapiro L., Ho D.D. Increased resistance of SARS-CoV-2 variant P.1 to antibody neutralization. Cell Host Microbe. 2021;29:747–751.e4. doi: 10.1016/j.chom.2021.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P., Nair M.S., Liu L., Iketani S., Luo Y., Guo Y., Wang M., Yu J., Zhang B., Kwong P.D., et al. Antibody resistance of SARS-CoV-2 variants B.1.351 and B.1.1.7. Nature. 2021;593:130–135. doi: 10.1038/s41586-021-03398-2. [DOI] [PubMed] [Google Scholar]
- Wang Q., Guo Y., Iketani S., Nair M.S., Li Z., Mohri H., Wang M., Yu J., Bowen A.D., Chang J.Y., et al. Antibody evasion by SARS-CoV-2 Omicron subvariants BA.2.12.1, BA.4 and BA.5. Nature. 2022;608:603–608. doi: 10.1038/s41586-022-05053-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheatley A.K., Fox A., Tan H.X., Juno J.A., Davenport M.P., Subbarao K., Kent S.J. Immune imprinting and SARS-CoV-2 vaccine design. Trends Immunol. 2021;42:956–959. doi: 10.1016/j.it.2021.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrapp D., Wang N., Corbett K.S., Goldsmith J.A., Hsieh C.L., Abiona O., Graham B.S., McLellan J.S. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367:1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrobel A.G., Benton D.J., Hussain S., Harvey R., Martin S.R., Roustan C., Rosenthal P.B., Skehel J.J., Gamblin S.J. Antibody-mediated disruption of the SARS-CoV-2 spike glycoprotein. Nat. Commun. 2020;11:5337. doi: 10.1038/s41467-020-19146-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu K., Choi A., Koch M., Elbashir S., Ma L., Lee D., Woods A., Henry C., Palandjian C., Hill A., et al. Variant SARS-CoV-2 mRNA vaccines confer broad neutralization as primary or booster series in mice. Vaccine. 2021;39:7394–7400. doi: 10.1016/j.vaccine.2021.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y., Huang X., Yuan L., Wang S., Zhang Y., Xiong H., Chen R., Ma J., Qi R., Nie M., et al. A recombinant spike protein subunit vaccine confers protective immunity against SARS-CoV-2 infection and transmission in hamsters. Sci. Transl. Med. 2021;13:eabg1143. doi: 10.1126/scitranslmed.abg1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia S., Liu M., Wang C., Xu W., Lan Q., Feng S., Qi F., Bao L., Du L., Liu S., et al. Inhibition of SARS-CoV-2 (previously 2019-nCoV) infection by a highly potent pan-coronavirus fusion inhibitor targeting its spike protein that harbors a high capacity to mediate membrane fusion. Cell Res. 2020;30:343–355. doi: 10.1038/s41422-020-0305-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong H.L., Wu Y.T., Cao J.L., Yang R., Liu Y.X., Ma J., Qiao X.Y., Yao X.Y., Zhang B.H., Zhang Y.L., et al. Robust neutralization assay based on SARS-CoV-2 S-protein-bearing vesicular stomatitis virus (VSV) Pseudovirus and ACE2-overexpressing BHK21 cells. Emerg. Microbes Infect. 2020;9:2105–2113. doi: 10.1080/22221751.2020.1815589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu K., Gao P., Liu S., Lu S., Lei W., Zheng T., Liu X., Xie Y., Zhao Z., Guo S., et al. Protective prototype-Beta and Delta-Omicron chimeric RBD-dimer vaccines against SARS-CoV-2. Cell. 2022;185:2265–2278.e14. doi: 10.1016/j.cell.2022.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ying B., Whitener B., VanBlargan L.A., Hassan A.O., Shrihari S., Liang C.Y., Karl C.E., Mackin S., Chen R.E., Kafai N.M., et al. Protective activity of mRNA vaccines against ancestral and variant SARS-CoV-2 strains. Sci. Transl. Med. 2022;14:eabm3302. doi: 10.1126/scitranslmed.abm3302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J., Collier A.Y., Rowe M., Mardas F., Ventura J.D., Wan H., Miller J., Powers O., Chung B., Siamatu M., et al. Neutralization of the SARS-CoV-2 omicron BA.1 and BA.2 Variants. N. Engl. J. Med. 2022;386:1579–1580. doi: 10.1056/NEJMc2201849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan L., Zhu H., Zhou M., Ma J., Chen R., Chen Y., Chen L., Wu K., Cai M., Hong J., et al. Gender associates with both susceptibility to infection and pathogenesis of SARS-CoV-2 in Syrian hamster. Signal Transduct. Target. Ther. 2021;6:136. doi: 10.1038/s41392-021-00552-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K. Gctf: real-time CTF determination and correction. J. Struct. Biol. 2016;193:1–12. doi: 10.1016/j.jsb.2015.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Wang S., Wu Y., Hou W., Yuan L., Shen C., Wang J., Ye J., Zheng Q., Ma J., et al. Virus-free and live-cell visualizing SARS-CoV-2 cell entry for studies of neutralizing antibodies and compound inhibitors. Small Methods. 2021;5:2001031. doi: 10.1002/smtd.202001031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Wei M., Wu Y., Wang J., Hong Y., Huang Y., Yuan L., Ma J., Wang K., Wang S., et al. Cross-species tropism and antigenic landscapes of circulating SARS-CoV-2 variants. Cell Rep. 2022;38:110558. doi: 10.1016/j.celrep.2022.110558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng S.Q., Palovcak E., Armache J.P., Verba K.A., Cheng Y., Agard D.A. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods. 2017;14:331–332. doi: 10.1038/nmeth.4193. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Structure coordinates are deposited in the Protein Data Bank (PDB) under accession codes: STKF:36H6:83H7:85F7 (PDB: 7WP6), STFK1628x:83H7:85F7:2B4 (PDB: 7WP8). The corresponding EM density maps have been deposited in the Electron Microscopy Data Bank (EMBD) under accession number: STKF:36H6:83H7:85F7 (EMD-32676), STFK1628x:83H7:85F7:2B4 (EMD-32678).
This paper does not report any original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.