Abstract
Brucella abortus vaccine strain RB51 is a natural stable attenuated rough mutant derived from the virulent strain 2308. The genetic mutations that are responsible for the roughness and the attenuation of strain RB51 have not been identified until now. Also, except for an assay based on pulsed-field gel electrophoresis, no other simple method to differentiate strain RB51 from its parent strain 2308 is available. In the present study, we demonstrate that the wboA gene encoding a glycosyltransferase, an enzyme essential for the synthesis of O antigen, is disrupted by an IS711 element in B. abortus vaccine strain RB51. Exploiting this feature, we developed a PCR assay that distinguishes strain RB51 from all other Brucella species and strains tested.
Brucella abortus is one of six well-recognized species of the genus Brucella which infects cattle as well as a variety of other mammals including humans (1, 12). Infection with B. abortus leads to abortions and reduced fertility in cattle. Vaccination with live, attenuated B. abortus strains has been effective in preventing B. abortus infections and abortions in cattle. Until recently, strain 19 (S19), a naturally occurring smooth and attenuated strain of B. abortus, had been used as the vaccine for cattle brucellosis. Similar to virulent B. abortus strains, the lipopolysaccharide of S19 also contains O side chain, which is responsible for an immunodominant antibody response after vaccination or infection with field strains. S19 vaccination usually causes the appearance of a transient serologic titer of antibody to Brucella O antigen, and in some vaccinated cattle, these titers of antibody do persist (30). Hence, at least in a few cases, conventional serological techniques cannot be used to clearly distinguish field-infected from S19-vaccinated cattle. B. abortus vaccine strain RB51 is a stable, rough, and attenuated mutant that was derived from strain 2308, a smooth and virulent strain of B. abortus (25). B. abortus RB51 was approved in the United States in 1996 for use as a vaccine for cattle, replacing S19. Since the lipopolysaccharide of B. abortus RB51 is devoid of O side chain, antibodies induced by vaccination with this strain do not interfere with the conventional serology (27). The stability and vaccine efficacy of B. abortus RB51 have been well studied and documented (8, 9, 16, 18, 22). However, the genetic bases for the rough phenotype and attenuation in this strain are not known. Also, except for a pulsed-field gel electrophoresis-based assay (16), no other DNA-based method to distinguish B. abortus RB51 from its parent strain 2308 or similar field strains is available. Previously, we characterized the wboA gene of B. abortus that encodes glycosyltransferase, an enzyme essential in the biosynthesis of O antigen (19). We also demonstrated that disruption of the wboA gene in smooth strains B. abortus 2308, Brucella melitensis 16M, and Brucella suis biovar 4 resulted in conversion to a rough phenotype (19, 29 [in reference 29, the wboA gene was designated rfbU]). We have discovered that the wboA gene in B. abortus RB51 is disrupted by an IS711-like element. Based on this genetic feature, we have developed a PCR assay that can distinguish RB51 from other Brucella species and strains, including its parent, virulent strain 2308.
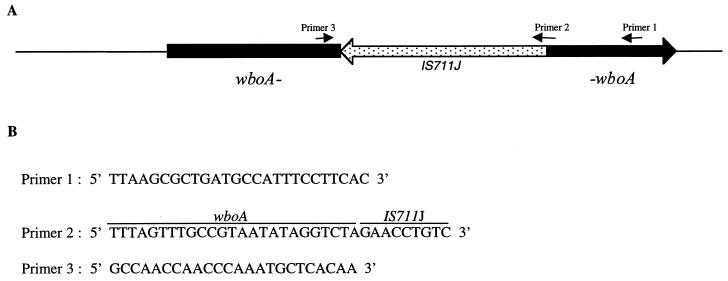
Interruption of the wboA gene by an IS711 element in B. abortus RB51.
The wboA gene along with the flanking nucleotide sequences was amplified by PCR from the genomic DNAs of B. abortus RB51 and 2308. B. abortus genomic DNAs were extracted and purified as described previously (14). The primers (forward primer, 5′ GGATGTCGACCAGCCCTCCACATCAATAGC 3′; reverse primer, 5′ TTGCGGATCCTTTACTCGTCCGTCTCTTAC 3′) used for the amplification were designed based on the previously described nucleotide sequence of the wboA gene from strain 2308 (19) (GenBank accession no. AF107768). PCR was performed with Ready-To-Go PCR beads (Pharmacia Biotech) and a thermal cycler (Hybaid). Each PCR tube contained 0.5 μM (each) primer and 5 ng of genomic DNA in a total volume of 25 μl. Amplification was performed for 40 cycles, each cycle comprising denaturation at 95°C for 1 min, annealing at 53°C for 30 s, and extension at 72°C for 1 min. The amplified products were separated by electrophoresis on a 0.8% agarose gel, stained with ethidium bromide, and viewed under UV light. The amplified product from RB51 genomic DNA was ∼3 kb in size, which was ∼900 bp larger than that from 2308 (data not shown). The PCR products were cloned in a pCR2.1 vector (Invitrogen, Inc.), and the nucleotide sequences of both strands were determined at the DNA Sequencing Facility of the Iowa State University (Ames). Computer analysis of the nucleotide sequence from RB51 revealed that the wboA gene was interrupted by an 842-bp fragment (Fig. 1). A BLAST search (2) (BLASTN program) indicated that the 842-bp fragment was almost identical to the previously described Brucella IS711 element (Fig. 2). IS711 is an insertion sequence of 842 bp initially found in Brucella ovis downstream to the gene encoding BCSP31 (13). This element was also discovered and sequenced by Ouahrani and colleagues (20), who designated the element IS6501. The element is present in five or more copies in Brucella spp. and appears to be quite stable in number and position in the chromosome (5, 6, 14). However, differences in the number of elements have been reported. B. abortus biovar 1 has at least six copies of IS711, but B. abortus 2308 and RB51 have tandem IS711 copies at one locus (6).
FIG. 1.
(A) Schematic diagram showing the interruption of the wboA gene by an IS711 element (IS711J) in B. abortus RB51 and the location of primers (small arrows) used in the PCR assay. (B) Nucleotide sequences of the primers used in the PCR assay.
FIG. 2.
Comparison of the sequences of IS711 (accession no. M94960) and the IS711-like element, IS711J, interrupting the wboA gene in B. abortus RB51. Ten base pairs of sequence flanking IS711J is also shown. Nucleotide residues varying between the two elements are indicated by lowercase, boldface, and underlining.
Sequence features of the IS711 element.
The IS711 element present in the wboA gene of B. abortus RB51 was designated IS711J. Comparison of IS711J with the IS711 element of B. ovis indicated 98.6% identity with specific nucleotide sequence differences (Fig. 2). The IS711J element is consistent with the IS711 elements with regard to insertion within the sequence 5′ CTAG 3′ and duplication of the sequence 5′ TA 3′ (13).
Minor sequence variation among the IS711 copies exists in B. ovis (13). The sequence variation occurs at specific loci within the element, with the ends of the elements being much more polymorphic than the coding regions (6a, 13). All the polymorphisms were at sites identified previously by sequencing the common copies of the element in brucellae (4, 6a). Only one of these sites, bp 747, differentiates the sequence of IS711J from that of other B. abortus IS711 copies. All the IS711 copies in B. abortus, including IS711J, are distinct from the other Brucella spp. IS711 elements because they have an A at positions 2 and 3 in one end of the element. All the rest of the elements have G or C at these positions. Transposition of the IS711 elements in brucellae does not appear to be limited to a specific copy or originate from a single locus, as unique copies of the element in B. ovis and IS711J of B. abortus vary in sequence.
B. abortus RB51-specific PCR assay.
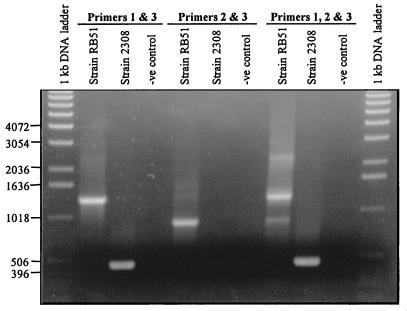
Exploiting the nature of wboA gene disruption by IS711J, we developed a PCR assay that can distinguish B. abortus RB51 from all other Brucella species and strains. Based on computer analysis (Primer Select program, LaserGene software; DNAStar Inc.), two primers, primers 1 and 3 (Fig. 1), were selected so that the amplified fragment from strain RB51 is ∼1,300 bp and the fragments from all other Brucella species (assuming an intact wboA gene) are ∼400 bp. An additional primer, primer 2 (Fig. 1), was selected manually to encompass the junction between the wboA gene and the 5′ end of IS711J. Five nanograms of purified Brucella genomic DNA was used as template for the PCR amplification. In some cases (see Table 1), a medium-sized (2 mm in diameter) bacterial colony was taken from an agar plate and resuspended in 200 μl of sterile distilled water, incubated in a boiling water bath for 15 min, and centrifuged for 5 min at 10,000 × g, and 10 μl of the supernatant was used as template. PCR amplifications were performed in a 25-μl total volume with Ready-To-Go PCR beads. Amplification was performed for 40 cycles, each cycle comprising denaturation at 95°C for 1 min, annealing at 62°C for 1 min, and extension at 72°C for 1.5 min. These parameters were selected after several trials to optimize the conditions for appropriate stringency (as determined by the absence of any undesired nonspecific bands) and better yield of the amplified product(s). Three different PCR amplifications were performed with primer combinations of primers 1 and 3; primers 2 and 3; and primers 1, 2, and 3. In reaction mixtures containing two primers, 0.5 μM (each) primer was included. Whereas in reaction mixtures containing all three primers, 0.5 μM (each) primers 1 and 2 and 1 μM primer 3 were included. Initial PCR amplifications were performed with genomic DNA from strains RB51 and 2308. As shown in Fig. 3, different sizes of fragments were amplified when primers 1 and 3 were used (∼1,300-bp fragment from RB51 and ∼400-bp fragment from 2308). Primers 2 and 3 amplified a 900-bp fragment from the RB51 genomic DNA but none from that of 2308.
TABLE 1.
Bacterial strains used in the PCR assaya
| Bacterial strain | Description | Source | PCR template |
|---|---|---|---|
| B. abortus RB51 | Rough, derived from 2308 (25) | VPI, Blacksburg, Va. | Bact,b DNA |
| B. abortus 2308 | Smooth, virulent | VPI, Blacksburg, Va. | Bact, DNAb |
| B. abortus 19 | Smooth, attenuated | VPI, Blacksburg, Va. | Bact,b DNA |
| B. abortus 45/20 | Intermediatec | VPI, Blacksburg, Va. | Bact |
| B. abortus field strains | |||
| Biovar 2 | Smooth | NADC, Ames, Iowa | DNA |
| Biovar 3 | Smooth | NADC, Ames, Iowa | DNA |
| Biovar 4 | Smooth | NADC, Ames, Iowa | DNA |
| Biovar 5 | Smooth | NADC, Ames, Iowa | DNA |
| B. melitensis 16M | Smooth | VPI, Blacksburg, Va. | Bact, DNAb |
| B. melitensis Rev1 | Smooth | VPI, Blacksburg, Va. | Bact,b DNA |
| B. melitensis B115 | Rough, O antigen in cytoplasm (11) | VPI, Blacksburg, Va. | Bact,b DNA |
| B. melitensis biovar 3 | Smooth | NADC, Ames, Iowa | DNA |
| B. suis biovar 2 | Smooth | NADC, Ames, Iowa | DNA |
| B. suis biovar 3 | Smooth | NADC, Ames, Iowa | DNA |
| B. suis biovar 4 | Smooth | NADC, Ames, Iowa | DNA |
| B. canis RM6/66 | Rough | VPI, Blacksburg, Va. | Bact |
| B. ovis | Rough | VPI, Blacksburg, Va. | Bact |
| Brucella neotomae | Smooth | NADC, Ames, Iowa | DNA |
| Ochrobactrum anthropi strains | |||
| 49237 | ATCC, Manassas, Va. | Bact, DNA | |
| 49188 | ATCC, Manassas, Va. | Bact, DNA | |
| Yersinia enterocolitica O:9 | VPI, Blacksburg, Va. | Bact |
Abbreviations: Bact, bacteria; VPI, Virginia Polytechnic Institute and State University; NADC, National Animal Disease Center; ATCC, American Type Culture Collection.
FIG. 3.
Differentiation of B. abortus RB51 from its parent strain 2308 by a wboA gene-based PCR assay. PCR amplifications with the indicated primer pairs were performed with the purified genomic DNA from strains RB51 and 2308 as templates. Negative (−ve) controls contained no template DNA. The amplified products were separated on a 0.8% agarose gel, stained with ethidium bromide, and photographed under UV light. Numbers at left indicate the 1-kb DNA ladder fragment sizes in base pairs.
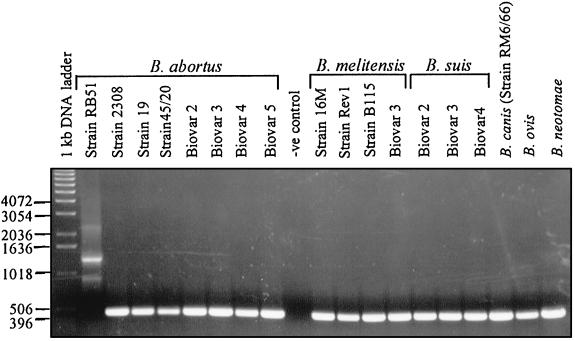
When all three primers were used in the reaction, fragments of expected sizes were amplified (400 bp from 2308 and 900 and 1,300 bp from RB51). In addition, a band of ∼2.3 kb in size was also amplified in RB51 (Fig. 3). The 900-bp and the 2.3-kb bands in strain RB51 were of lower intensity, indicating that there was some inhibition in the amplification. Adjustments of several parameters, including the concentration of Mg2+, primers, deoxynucleoside triphosphates, or changes in annealing temperature, did not result in either enhancement of the amplified products or absence of the 2.3-kb fragment. The low level of amplification of the 900-bp fragment is most probably due to the 5′→3′ exonuclease activity of Taq DNA polymerase; while extending primer 1, Taq DNA polymerase could have degraded the DNA strand that was being extended from primer 2 (primers 1 and 2 bind to the same template in strain RB51 [Fig. 1]) (15). Some of the single-stranded DNA fragments resulting from the degradation of the DNA strand that was initiated by primer 2 might have primed for the amplification of the 2.3-kb fragment. This appears likely, since the ∼2.3-kb band appeared only when all three primers were used in the amplification reaction. No such problem is encountered with strain 2308, since primer 2 cannot bind to the template; again this supports, though indirectly, the above hypothesis for the low level of amplification of the 900-bp fragment and the appearance of the 2.3-kb band in the case of RB51. We tested the specificity of this strain-specific PCR assay with various Brucella strains (Table 1). As shown in Fig. 4, when the three primers were used for the PCR assay, all the other Brucella strains tested gave an amplified product of 400 bp in size; identical results were obtained by using only primers 1 and 3 (data not shown). No amplified products were detectable when the template was genomic DNA from bacteria that are closely related to Brucella species, Ochrobactrum anthropi 49237 and 49188. Also, no products could be amplified from the genomic DNAs of Yersinia enterocolitica O:9, which synthesizes O antigen that is identical to that of Brucella (reference 7 and data not shown). Based on these results, we recommend the PCR assay with primers 1 and 3 to distinguish strain RB51 from all other Brucella strains. It should be mentioned that several attempts to clone the 2.3-kb fragment present in the amplified products of strain RB51 were unsuccessful. Amplification of the 400-bp fragment from B. ovis and Brucella canis indicates that these naturally rough species contain the wboA gene sequences. However, further studies are needed to confirm the intactness and functionality of the wboA gene in these species, since the mere presence of a gene sequence does not necessarily result in expression of a functional product.
FIG. 4.
PCR assay to differentiate B. abortus RB51 from all other Brucella species and strains. Primers 1, 2, and 3 were used in all PCR amplifications. Genomic DNA from the indicated Brucella strains was used as template. Negative (−ve) controls contained no template DNA. The amplified products were separated on a 0.8% agarose gel, stained with ethidium bromide, and photographed under UV light. Numbers at left indicate the 1-kb DNA ladder fragment sizes in base pairs.
Even though the stability of RB51 is well proven in vivo and in vitro, the actual genetic mutation(s) that contributed to the rough phenotype and attenuation of this strain has not been identified until now. This study describes the first such mutation in the wboA gene. It is clear from previous studies that deletion of the wboA gene in B. melitensis, B. abortus, and B. suis leads to the rough phenotype and attenuation (19, 29). Ongoing studies in our laboratory indicated that complementation of RB51 with a functional wboA gene resulted in O antigen production but did not result in reversion to the smooth phenotype and did not affect attenuation (unpublished data). This suggests that RB51 contains an additional genetic mutation(s) that probably affects either the export of O antigen to the bacterial surface, the coupling of O antigen to core lipopolysaccharide, or both.
Recently, several PCR assays to detect or differentiate various Brucella strains have been reported (3, 5, 6, 17, 21, 23, 26, 28). However, none of these assays could distinguish RB51 from its parent, virulent strain 2308. A PCR assay with primers 1 and 3, as described in this paper, can be used to quickly identify RB51; hence, it should be useful in studies where detecting the presence of RB51 is needed, such as those with the risk of potential abortions, which can occur if pregnant animals are vaccinated with RB51, and in studies where the fate of RB51 has to be determined after vaccination of bison and other wild as well as domestic animals. We successfully used this PCR assay to quickly verify the presence or absence of RB51 among field Brucella isolates cultured from aborted bovine fetuses (unpublished data).
REFERENCES
- 1.Acha P, Szyfres B. Zoonoses and communicable diseases common to man and animals. Washington, D.C: Pan American Health Organization; 1980. pp. 28–45. [Google Scholar]
- 2.Altschul S F, Madden T L, Schaffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baily G G, Krahn J B, Drasar B S, Stoker N G. Detection of Brucella melitensis and Brucella abortus by DNA amplification. J Trop Med Hyg. 1992;95:271–275. [PubMed] [Google Scholar]
- 4.Bricker B J, Halling S M. DNA sequence divergence of IS711: a theory on the progression of transposition events. International Congress on the E. coli. Madison, Wis: Genome; 1992. p. 1992. [Google Scholar]
- 5.Bricker B J, Halling S M. Differentiation of Brucella abortus bv. 1, 2, and 4, Brucella melitensis, Brucella ovis, and Brucella suis bv. 1 by PCR. J Clin Microbiol. 1994;32:2660–2666. doi: 10.1128/jcm.32.11.2660-2666.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bricker B J, Halling S M. Enhancement of the Brucella AMOS PCR assay for differentiation of Brucella abortus vaccine strains S19 and RB51. J Clin Microbiol. 1995;33:1640–1642. doi: 10.1128/jcm.33.6.1640-1642.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6a.Bricker, B. J., and S. M. Halling. Unpublished data.
- 7.Caroff M, Bundle D R, Perry M B, Cherwonograodzky J W, Duncan J R. Antigenic S-type lipopolysaccharide of Brucella abortus 1119-3. Infect Immun. 1984;46:384–388. doi: 10.1128/iai.46.2.384-388.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cheville N F, Jensen A E, Halling S M, Tatum F M, Morfitt D C, Hennager S G, Frerichs W M, Schurig G G. Bacterial survival, lymph node changes, and immunologic responses of cattle vaccinated with standard and mutant strains of Brucella abortus. Am J Vet Res. 1992;53:1881–1888. [PubMed] [Google Scholar]
- 9.Cheville N F, Stevens M G, Jensen A E, Tatum F M, Halling S M. Immune responses and protection against infection and abortion in cattle experimentally vaccinated with mutant strains of Brucella abortus. Am J Vet Res. 1993;54:1591–1597. [PubMed] [Google Scholar]
- 10.Chukwu C C. The instability of Brucella abortus strain 45/20 and a note on significance of using an unstable rough strain in the diagnosis of bovine brucellosis. Int J Zoonoses. 1985;12:120–125. [PubMed] [Google Scholar]
- 11.Cloeckaert A, Zygmunt M S, Nicolle J-C, Dubray G, Limet J N. O-chain expression in the rough Brucella melitensis strain B115: induction of O-polysaccharide-specific monoclonal antibodies and intracellular localization demonstrated by immunoelectron microscopy. J Gen Microbiol. 1992;138:1211–1219. doi: 10.1099/00221287-138-6-1211. [DOI] [PubMed] [Google Scholar]
- 12.Corbel M J. Brucellosis: an overview. Emerg Infect Dis. 1997;3:213–221. doi: 10.3201/eid0302.970219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Halling S M, Tatum F M, Bricker B J. Sequence and characterization of an insertion sequence, IS711, from Brucella ovis. Gene. 1993;133:123–127. doi: 10.1016/0378-1119(93)90236-v. [DOI] [PubMed] [Google Scholar]
- 14.Halling S M, Zehr E S. Polymorphism in Brucella spp. due to highly repeated DNA. J Bacteriol. 1990;172:6637–6640. doi: 10.1128/jb.172.12.6637-6640.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Holland P M, Abramson R D, Watson R, Gelfand D H. Detection of specific polymerase chain reaction product by utilizing the 5′ to 3′ exonuclease activity of Thermus aquaticus DNA polymerase. Proc Natl Acad Sci USA. 1991;88:7276–7280. doi: 10.1073/pnas.88.16.7276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jensen A E, Ewalt D R, Cheville N F, Thoen C O, Payeur J B. Determination of stability of Brucella abortus RB51 by use of genomic fingerprint, oxidative metabolism, and colonial morphology and differentiation of strain RB51 from B. abortus isolates from bison and elk. J Clin Microbiol. 1996;34:628–633. doi: 10.1128/jcm.34.3.628-633.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Leal-Klevezas D S, Martinez-Vazquez I O, Lopez-Merino A, Martinez-Soriano J P. Single-step PCR for detection of Brucella spp. from blood and milk of infected animals. J Clin Microbiol. 1995;33:3087–3090. doi: 10.1128/jcm.33.12.3087-3090.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lord V R, Schurig G G, Cherwonogrodzky J W, Marcano M J, Melendez G E. Field study of vaccination of cattle with Brucella abortus strains RB51 and 19 under high and low disease prevalence. Am J Vet Res. 1998;59:1016–1020. [PubMed] [Google Scholar]
- 19.McQuiston J R, Vemulapalli R, Inzana T J, Schurig G G, Sriranganathan N, Fritzinger D, Hadfield T L, Warren R A, Snellings N, Hoover D, Halling S M, Boyle S M. Genetic characterization of a Tn5-disrupted glycosyltransferase gene homolog in Brucella abortus and its effect on lipopolysaccharide composition and virulence. Infect Immun. 1999;67:3830–3835. doi: 10.1128/iai.67.8.3830-3835.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ouahrani S, Michaux S, Sriwidada J, Bourg G, Tournebize R, Ramuz M, Liautard J P. Identification and sequence analysis of IS6501, an insertion sequence in Brucella spp.: relationship between genomic structure and the number of IS6501 copies. J Gen Microbiol. 1993;139:3265–3273. doi: 10.1099/00221287-139-12-3265. [DOI] [PubMed] [Google Scholar]
- 21.Ouahrani-Bettache S, Soubrier M P, Liautard J P. IS6501-anchored PCR for the detection and identification of Brucella species and strains. J Appl Bacteriol. 1996;81:154–160. doi: 10.1111/j.1365-2672.1996.tb04493.x. [DOI] [PubMed] [Google Scholar]
- 22.Palmer M V, Olsen S C, Cheville N F. Safety and immunogenicity of Brucella abortus strain RB51 vaccine in pregnant cattle. Am J Vet Res. 1997;58:472–477. [PubMed] [Google Scholar]
- 23.Romeno C, Gamazo C, Pardo M, Lopez-Goni I. Specific detection of Brucella DNA by PCR. J Clin Microbiol. 1995;33:615–617. doi: 10.1128/jcm.33.3.615-617.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Roop R M, Preston-Moore D, Bagchi T, Schurig G G. Rapid identification of smooth Brucella species with a monoclonal antibody. J Clin Microbiol. 1987;25:2090–2093. doi: 10.1128/jcm.25.11.2090-2093.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schurig G G, Roop R M, Bagchi T, Boyle S, Buhrman D, Sriranganathan N. Biological properties of RB51: a stable rough strain of Brucella abortus. Vet Microbiol. 1991;28:171–188. doi: 10.1016/0378-1135(91)90091-s. [DOI] [PubMed] [Google Scholar]
- 26.Sifuentes-Rincon A M, Revol A, Barrera-Saldana H A. Detection and differentiation of the six Brucella species by polymerase chain reaction. Mol Med. 1997;3:734–739. [PMC free article] [PubMed] [Google Scholar]
- 27.Stevens M G, Hennager S G, Olsen S C, Cheville N F. Serologic responses in diagnostic tests for brucellosis in cattle vaccinated with Brucella abortus 19 or RB51. J Clin Microbiol. 1994;32:1065–1066. doi: 10.1128/jcm.32.4.1065-1066.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tcherneva E, Rijpens N, Naydensky C, Herman L. Repetitive element sequence based polymerase chain reaction for typing of Brucella strains. Vet Microbiol. 1996;51:169–178. doi: 10.1016/0378-1135(96)00036-3. [DOI] [PubMed] [Google Scholar]
- 29.Winter A J, Schurig G G, Boyle S M, Sriranganathan N, Bevins J S, Enright F M, Elzer P H, Kopec J D. Protection of BALB/c mice against homologous and heterologous species of Brucella by rough strain vaccines derived from Brucella melitensis and Brucella suis biovar 4. Am J Vet Res. 1996;57:677–683. [PubMed] [Google Scholar]
- 30.Woodard L F. Do we need another brucellosis vaccine? Mod Vet Pract. 1981;62:857–859. [PubMed] [Google Scholar]