Abstract
Background
The novel Artificial Intelligence-driven BestCyte® Cell Sorter Imaging System (BestCyte) enables hybrid digital screening through classification and sorting of tiles depicting cells in 8 galleries or whole slide image (WSI) reviews.
Objectives
(1) Analyze expenditures of time (minutes) for primary BestCyte cell sorter screening and adjudicative WSI rescreening of 500 blinded, randomized ThinPrep thin-layers to determine review times per Bethesda nomenclature; (2) Analyze review times for NILM qualifier diagnoses reflecting increasing interpretive complexity (i.e., Inflammation, Reactive/Repair, Bacterial cytolysis, Bacterial vaginosis, Atrophy, and Atrophic vaginitis); (3) Challenge accuracy of primary diagnoses (Downgraded, Upheld, and Upgraded) following adjudicative WSI rescreening to assess correlated review times as surrogate indicators of diagnostic confidence in BestCyte functionality (i.e., learning curve); and (4) Correlate primary and adjudicative diagnoses to calculate intra-observer reproducibility Kappa coefficients per Bethesda nomenclature.
Results
Of 500 thin-layers, the mean [primary/adjudicative rescreening review times (minutes)] were: Overall study [1.38/3.94], NILM [1.23/3.02], ASCUS [1.18/2.53], ASC-H [1.73/4.86], AGUS [1.84/6.34], LSIL [1.49/4.16], HSIL [1.52/4.10], CA [0.65/2.57]. Of 500 primary Bethesda diagnoses: 2 (0.40%) downgraded; 483 (96.6%) upheld; 15 (3.00%) upgraded after adjudicative WSI rescreening. Of 354 NILM diagnoses: 0 downgraded; 344 (97.2%) upheld; 10 (2.82%) upgraded. Of 34 ASCUS diagnoses: 2 (5.88%) downgraded; 28 (82.4%) upheld; 4 (11.8%) upgraded. Of 17 ASC-H diagnoses: 0 downgraded; 16 (94.1%) upheld; 1 (5.88%) upgraded. Of AGUS (n=1), LSIL (n=24), HSIL (n=52), CA (n=1), UNSAT (n=17): 100% upheld. Kappa coefficients with 95% (Confidence Intervals): Overall study 0.9305 (0.8983–0.9627), NILM 0.9429 (0.9110–0.9748), ASCUS 0.8378 (0.7393–0.9363), ASC-H 0.9112 (0.8113–0.9999), AGUS 1.0 (1.0–1.0), LSIL 0.9189 (0.8400–0.9978), HSIL 0.9894 (0.9685–0.9999), CA 1.0 (1.0–1.0), UNSAT 1.0 (1.0–1.0). Primary BestCyte cell image review time trends for NILM, ASCUS, LSIL, and HSIL, revealed plateaus relative to decreasing respective adjudicative WSI rescreening times.
Conclusions
Given innovative robustness, BestCyte accommodates interpretive fundamentals, enabling shorter ThinPrep thin-layer review times with optimal intra-observer concordance per Bethesda nomenclature through classifying, ranking, sorting, and displaying clinically relevant cells efficiently in galleries. BestCyte fosters continuously optimizing diagnostic confidence learning curves; may supplant manual microscopy for primary screening.
Keywords: BestCyte cell sorter imaging system, Artificial Intelligence-based cytology, Virtual primary screening, Adjudicative whole slide image rescreening, Review times, Intra-observer reproducibility
Background
With advancing scanning and computer processing technology digital platforms exploiting Artificial Intelligence (AI) have enabled impactful forays in medicine by standardizing diagnostic parameters and simplifying technical workflows.1 Equivalent systems may also be primed for Pap test cervical cancer screening and reporting given specific enhancements2,3: (i) Rapid (single or multi-plane) slide scanning to generate digital slides (i.e., whole slide images (WSI)) of cytopreparations encompassing the entire cellular milieu; (ii) Automated detection of abnormal cells (i.e., rare events) in liquid-based thin-layers exploiting dedicated AI algorithms based on object-specific parameters; (iii) Capture, mining, and web-based access to digital images displaying clinically relevant cells in high-resolution virtual high-power fields or wider fields of view (FOV); and (iv) Enabled digital WSI manipulation simulating conventional light microscopy (LM) practice. Nevertheless, diagnostic interpretations of abnormal cells identified in cervical cytopathology remain complex regardless of screening methodology as they involve subjective assessments of assemblages of cytomorphologic features depending on clinical context.4,5 Thus, digital systems that simulate LM meanwhile standardize hence optimize cytomorphologic objectivity may synchronously decrease the subjectivity and labor typically associated with cytologic screening practice.2
Using proprietary liquid-based processing technology to produce thin-layer slides, ThinPrep (Hologic, Bedford, MD, USA) and SurePath (Becton Dickinson, Burlington, NC, USA) introduced suited automated screening devices for Pap test primary screening and reporting using Bethesda nomenclature. Accordingly, Delga et al.6 investigated the BestCyte® Cell Sorter Imaging System (BestCyte) (CellSolutions, Greensboro, NC, USA) as being a novel entrant in the realm of AI-driven digital Pap test thin-layer screening given its innovative capacity to detect, classify, and sort images of cells into dedicated galleries to efficiently project cytomorphology. Given its overall design exploiting advanced scanning technology, Delga et al.6 concluded BestCyte may supplant LM when applied for Pap test screening of ThinPrep, or CellSolutions’ proprietary BestPrep, thin-layers with uniform distributions of cells and minimal 3-dimensional topography.7
Citing screening potentialities per se, Delga et al.6 raised 3 assertions that inspired the design and intent of this study: (1) Review times using BestCyte may lapse between 0.5 and 3 min per slide for the majority of cases; (2) review times may further decrease alongside increasing users’ system familiarity and perceived confidence (i.e., learning curve); and (3) BestCyte may enable efficient slide reviews utilizing less than half the time relative to LM; thus, may be a reliable primary screening device for thin-layer slides as ThinPrep.
BestCyte detects, classifies, sorts, and displays images of normal or abnormal cells, or clusters thereof, as tiles in dedicated galleries for cytologists’ assessment in 40x magnification. BestCyte also enables panning throughout the WSI using simulated 4x, 10x, 20x, or 40x objective magnifications interactively. Therefore, the system supports 2 interchangeable screening modes: Inspection of tiles sorted in galleries, or WSI analysis of the cellular milieu in toto. In either mode, BestCyte enables digital annotation of clinically relevant cells for follow-on reviews and reporting. Given this hybrid capacity, this study follows a protocol solely within the primary screening setting; that is, diagnoses rendered upon primary screening of sorted tiles in BestCyte galleries are confirmed or refuted through adjudicative WSI rescreening. This study would allow assessment of intra-observer diagnostic reproducibility of primary diagnoses, respective screening review times, and overall diagnostic confidence potentialities in BestCyte for primary screening. By limiting interpretive error to one cytologist, this study tests the BestCyte’s potential to efficiently detect and display clinically relevant cells through cell sorting; hence potentially reduce primary screening times relative to maturing learning curves as asserted by Delga et al.6
This investigation of 500 ThinPrep thin-layer slides had 5 objectives: (1) Record review time expenditures in minutes for primary BestCyte screening of sorted images and for adjudicative WSI rescreening to produce review time differentials per Bethesda nomenclature; (2) record and correlate respective review times for NILM qualifier diagnoses reflecting increasing interpretive complexity (i.e., Inflammation, Reactive/Repair, Bacterial cytolysis, Bacterial vaginosis, Atrophy, and Atrophic vaginitis); (3) challenge the accuracy of primary diagnoses rendered from review of sorted images as being Downgraded, Upheld, or Upgraded diagnoses following WSI rescreening; (4) investigate review time differentials and trends as being surrogate indicators of diagnostic confidence in BestCyte functionality (i.e., learning curve); and (5) correlate respective diagnoses to produce intra-observer reproducibility Kappa (κ) coefficients per Bethesda nomenclature.
Methods
BestCyte Cell Sorter Imaging System
The core elements of the BestCyte® Cell Sorter Imaging System developed by CellSolutions (Greensboro, NC, USA) consist of a scanner and its associated workstation connected via network (or Internet) to the BestCyte server (Fig. 1). During operation the scanner covers the entire area of the cellular milieu deposited on the cytology glass slide capturing digital images of overlapping FOV and thereafter stitching them together to compose a single, seamless, high-resolution digital slide (i.e., WSI) mirroring the clinical sample. The WSI is transferred automatically via network to the BestCyte server for proprietary ranking computations to build dedicated galleries of sorted cell images displaying diagnostically relevant cells and overall cellularity.
Fig. 1.
BestCyte Cell Sorter Imaging System.
Any given gallery may compose a maximum of 100 tiles, if as many images of cells are ranked for that gallery. BestCyte incorporates 8 galleries sequenced in a series of tabs in the display (Windows) header banner2,6: Overview, High N/C, Halos, Atypical, Elongated, Clusters, Endocervical (T-zone), and InternalCtrl. All galleries are designed to sort and display tiles of targeted cytomorphology. For instance, the Overview gallery incorporates the highest ranked tiles from the other 7 galleries to offer a composite preview of relative cellularity. The InternalCtrl gallery displays normal reference cells; it also functions as a system control indicator that the BestCyte classifier is performing at a predicted cell-display level. Cell images in galleries and corresponding WSI may be selectively displayed on interconnected review stations supporting interactive case reviews by cytologists locally or remotely to render primary, secondary, or final diagnoses.
BestCyte scanning involves configurations allowing for demarcation of the area of interest (i.e.,clinical sample) on the slide, generation of the initial focus map, and the nature of focusing algorithm (i.e.,single, or multi-plane). BestCyte optimizes WSIs based on specific configurations of optics, sensor and illumination settings, and specified pixel resolution and color settings. Also, BestCyte is optimized for robust recognition rates to accommodate potentially sub-optimal focusing so that partially defocused cells in images may be classifiable for projection and review. Ultimately, this capability enables BestCyte to generate adequate image results with superior rapidity from single focal plane scanning of uniform, thin-layer cytopreparations.
BestCyte supports digital annotation of cells in sorted images or FOV. Images of annotated (i.e., tagged) cells amass automatically in the reporting tab to compose the cytomorphology or diagnostic criteria spotted through screening defending the diagnoses raised. Selection of any one image in galleries automatically redirects the user to a full-screen WSI displaying the precise high-power field containing those cells centrally, enabling wider inspection of the adjacent cellular milieu through omnidirectional panning. Panning is made possible by use of the computer mouse, as well as selective switching between 4x, 10x, 20x, or 40x magnifications. Screening completeness may be assured through an on-screen viewfinder revealing thin-layer coverage with green color tracking.
The BestCyte configuration used for the investigations reported in this publication included a Pannoramic® P250 Flash III RX scanner using a CIS VCC-FC60FR19CL sensor (3DHISTECH Ltd. H-1141 Budapest, Öv utca 3., Hungary). The resulting WSI files had a pixel resolution of approximately 0.25 μm in a proprietary format through single plane focusing using a 20x Zeiss Plan-Apochromat objective with 0.8 numerical aperture. Simulated digital magnification was enabled by the high-resolution camera so that the cells depicted in images would be equivalent to those as reviewed through conventional 40x LM objectives. The WSI reflected the cellular sample deposited within a 20 mm diameter circular glass slide area standard for ThinPrep thin-layers (approx. 314 mm2) using a single focal plane. Each thin-layer required 1 min and 20 s (i.e., 1.33 min) for scanning, and 1 gigabyte digital storage space for the resulting WSI.
Study slide case set
Courtesy of Marlboro-Chesterfield Pathology (Pinehurst, NC, USA), 500 ThinPrep Pap test thin-layer slides (‘Pinehurst Case Set’ (PCS)) were provided to support validation research of BestCyte technology. These cases were processed using ThinPrep technology, manually screened through LM, and formally reported in 2016. The PCS consisted of non-sequentially accessioned, deidentified, and diagnostically-randomized cases enriched by LSIL cases (approx. 5% of 500 cases) and HSIL cases (approx. 10% of 500 cases) to foster investigational work with cells arising from significant cervical lesions, and for statistical confidence. The PCS slides were cleaned of all markings before digital imaging onsite at Marlboro-Chesterfield Pathology.
Study protocol
The corresponding author was trained on BestCyte functionality by CellSolutions’ staff cytologists before login authorization to the archived WSI files. As all patient clinical information and Marlboro-Chesterfield Pathology-issued diagnoses were withheld, all screening events were blinded. The PCS thin-layer slides were also withheld to sustain an exclusively virtual setting. The 500 WSI archive was accessed through wireless Internet connectivity and viewed on a 32” 4K flat-panel HP Pavilion monitor set at 100% zoom, and 9:16 ratio full-screen display.
The Bethesda diagnoses for this investigation were: NILM, ASCUS, ASC-H, AGUS, LSIL, HSIL, CA (Carcinoma), and UNSAT. For NILM cases, a series of sub-category qualifiers included: Inflammation, Reactive/Repair, Bacterial cytolysis, Atrophy, Bacterial vaginosis, and Atrophic vaginitis. These qualifiers were accordingly sequenced in data tables intending to reflect increasing interpretive complexity based on the investigator’s career experience to test for likely screening time differentials and trends between primary and adjudicative WSI rescreening.
BestCyte Cell Sorter Imaging System primary screening
All 500 PCS thin-layers were primary screened by inspecting all images of cells downloading after case selection, sorting automatically into the 8 distinct BestCyte galleries. Select tiles displaying cells of interest were electronically annotated using an on-screen Bethesda diagnosis menu as also were NILM qualifiers. BestCyte primary screening of sorted tiles facilitated: (a) Inspection of all sorted tiles and select annotation thereof to defend the Bethesda diagnoses rendered; and (b) capture of total minutes expended between case selection through to commitment to diagnosis.
Adjudicative WSI rescreening
Adjudicative rescreening involved exclusively WSI review in toto. The WSI was screened digitally using 10x magnification ensuring 20% screening overlap as in conventional LM practice. Adjudicative WSI rescreening facilitated: (a) Simulated LM rescreening to challenge the accuracy of primary BestCyte diagnoses raised through sorted images; (b) selective annotation of cells in WSI FOV to support the post-rescreening Bethesda diagnoses; (c) capture of total minutes expended to complete rescreening from WSI launch through to commitment to diagnosis; (d) recording adjudicated diagnoses as being Downgraded, Upheld, or Upgraded primary screening diagnoses; and (e) recording intra-observer correlation between respective diagnoses allowing κ and 95% Confidence Interval (CI) statistical calculations.
All data were recorded in a master Excel spreadsheet. Time expenditures recorded initially as total seconds were converted to minutes with 2 decimal points. Data converging into 5 Tables were illustrated through 7 bar plots including superimposed linear trend lines to reflect screening time trends for 4 specific Bethesda diagnoses: NILM, ASCUS, LSIL, and HSIL.
Results
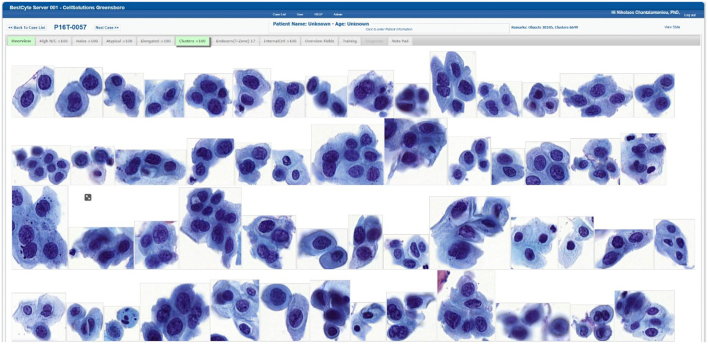
Following case selection from the on-screen registry, all tiles downloaded, sorted, and displayed into any of the 8 BestCyte galleries within 6 s uninterrupted. The cubic or rectangular dimensions and organization of sorted tiles in galleries depended on the nature of digitized cells, or clusters of cells, or their spatial orientations in 40x magnification high-power fields (Fig. 2). Small, isolated cells, as inflammatory cells, histiocytes, basal cells, endocervical cells, atrophic parabasal cells, or severely dysplastic cells were positioned centrally in smaller tiles. Larger cells, as koilocytotic squamous cells, clusters of cells, or sheets of endocervical, metaplastic, or squamous epithelial cells, were positioned randomly in either portrait or landscape layouts in proportionately larger tiles.
Fig. 2.
BestCyte Clusters gallery displaying 54 of the highest ranked of 100 sorted images of cells; 9:16 ratio full-screen display (HSIL study case 0057).
Table 1 distributes number (n) and percent (%) of primary BestCyte screening diagnoses downgraded, upheld, or upgraded following adjudicative WSI rescreening. Of all 500 primary Bethesda diagnoses: 2 (0.40%) were downgraded; 483 (96.6%) upheld, and 15 (3.00%) upgraded. Of all primary diagnoses: NILM (n=354): 0 downgraded, 344 (97.2%) upheld, and 10 (2.82%) upgraded; ASCUS (n=34): 2 (5.88%) downgraded, 28 (82.4%) upheld, and 4 (11.8%) upgraded; ASC-H (n=17): 0 downgraded, 16 (94.1%) upheld, and 1 (5.88%) upgraded; and for AGUS (n=1), LSIL (n=24), HSIL (n=52), CA (n=1), and UNSAT (n=17): 100% upheld.
Table 1.
Number (n), percent (%) of primary BestCyte screening diagnoses Downgraded, Upheld, or Upgraded following adjudicative WSI rescreening, per Bethesda diagnosis.
| BestCyte Primary Cell Sorter screening | BestCyte adjudicative WSI rescreening | Correlation with Primary BestCyte Cell Sorter diagnosis |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| Downgraded | Upheld | Upgraded | |||||||
| Diagnosis | n= | % | n= | n= | % | n= | % | n= | % |
| NILM | 354 | 70.8 | 346 | 0 | 0 | 344 | 97.2 | 10 | 2.82 |
| ASCUS | 34 | 6.80 | 28 | 2 | 5.88 | 28 | 82.4 | 4 | 11.8 |
| ASC-H | 17 | 3.40 | 18 | 0 | 0 | 16 | 94.1 | 1 | 5.88 |
| AGUS | 1 | 0.20 | 1 | 0 | 0 | 1 | 100 | 0 | 0 |
| LSIL | 24 | 4.80 | 30 | 0 | 0 | 24 | 100 | 0 | 0 |
| HSIL | 52 | 10.4 | 53 | 0 | 0 | 52 | 100 | 0 | 0 |
| CA | 1 | 0.20 | 1 | 0 | 0 | 1 | 100 | 0 | 0 |
| UNSAT | 17 | 3.40 | 17 | 0 | 0 | 17 | 100 | 0 | 0 |
| Values | 500 | 500 | 2/500 | 0.40 | 483/500 | 96.6 | 15/500 | 3.00 | |
Table 2 distributes discordant primary diagnoses either downgraded or upgraded following adjudicative WSI rescreening whereas: 2 were downgraded: (ASCUS to NILM), and 15 upgraded: (NILM to ASCUS (n=6), NILM to ASC-H (n=2), NILM to LSIL (n=2), ASCUS to LSIL (n=4), and ASC-H to HSIL (n=1)).
Table 2.
Breakdown of primary BestCyte screening diagnoses Downgraded and Upgraded following adjudicative WSI rescreening.
| Downgraded |
Upgraded |
||||
|---|---|---|---|---|---|
| Primary cell sorter diagnosis | Adjudicative WSI rescreening diagnosis | n= | Primary cell sorter diagnosis | Adjudicative WSI rescreening diagnosis | n= |
| ASCUS | NILM | 2 | NILM | ASCUS | 6 |
| NILM | ASC-H | 2 | |||
| NILM | LSIL | 2 | |||
| ASCUS | LSIL | 4 | |||
| ASC-H | HSIL | 1 | |||
| Values | 2 | 15 | |||
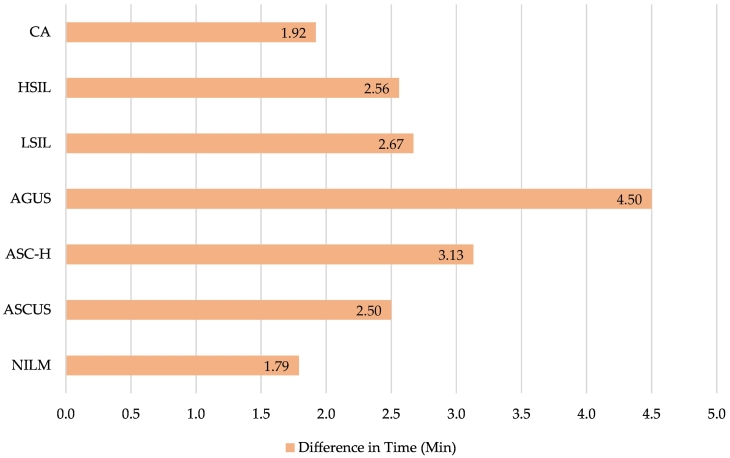
Table 3 distributes mean minute expenditures to commit to a BestCyte primary or WSI rescreening diagnosis. [Notation: UNSAT cases were excluded from this analysis. Although cells were identified in sorted tiles through BestCyte primary screening, WSI rescreening revealed unsatisfactory cases due to inadequate squamous epithelial cell component; air bubbles; dried mountant; poor technical ThinPrep cytopreparation; or (localized or global) suboptimal focusing. Screening time expenditure analysis for UNSAT was thus deemed impractical]. The overall mean time expenditure to diagnosis after primary BestCyte review for all Bethesda diagnoses (excluding UNSAT) was 1.38 min; the respective breakdowns (minutes): NILM (1.23), ASCUS (1.18), ASC-H (1.73), AGUS (1.84), LSIL (1.49), HSIL (1.52), and CA (0.65). The overall mean time expenditure to diagnosis after WSI rescreening for all Bethesda diagnoses (excluding UNSAT) was 3.94 min; the respective breakdowns (minutes): NILM (3.02), ASCUS (2.53), ASC-H (4.86), AGUS (6.34), LSIL (4.16), HSIL (4.10), and CA (2.57). Fig. 3, Fig. 4 illustrate the differences in mean review time expenditures (minutes): NILM (1.79), ASCUS (2.50), ASC-H (3.13), AGUS (4.50), LSIL (2.67), HSIL (2.56), and CA (1.92) respectively.
Table 3.
Mean time expenditures (minutes) to commit to diagnosis through primary BestCyte screening and adjudicative WSI rescreening with differences in time expenditures, per Bethesda diagnosis (excluding UNSAT).
| Primary BestCyte Cell Sorter review times |
Adjudicative WSI rescreening review times |
|||
|---|---|---|---|---|
| Diagnosis | Mean time (Min) | Diagnosis | Mean time (Min) | Difference in time (Min) |
| NILM | 1.23 | NILM | 3.02 | 1.79 |
| ASCUS | 1.18 | ASCUS | 2.53 | 2.50 |
| ASC-H | 1.73 | ASC-H | 4.86 | 3.13 |
| AGUS | 1.84 | AGUS | 6.34 | 4.50 |
| LSIL | 1.49 | LSIL | 4.16 | 2.67 |
| HSIL | 1.52 | HSIL | 4.10 | 2.56 |
| CA | 0.65 | CA | 2.57 | 1.92 |
| Mean | 1.38 | 3.94 | 2.56 | |
Fig. 3.
Mean time expenditures (minutes) for primary BestCyte Cell Sorter screening (blue) and adjudicative WSI rescreening (green), per Bethesda diagnosis.
Fig. 4.
Differences in mean time (minutes) between primary BestCyte Cell Sorter screening and adjudicative WSI rescreening, per Bethesda diagnosis.
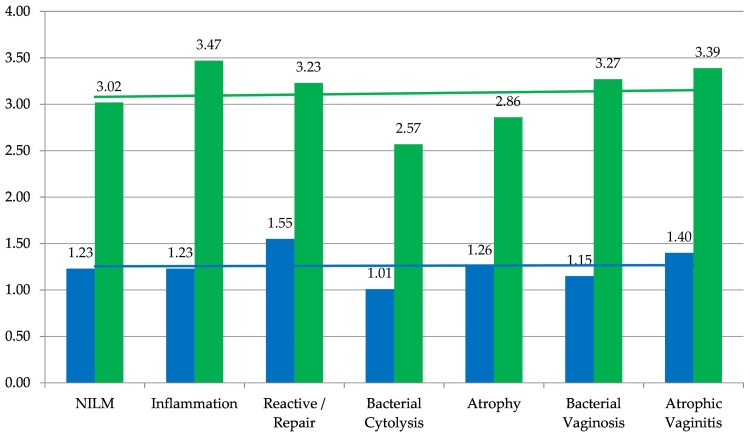
Table 4 distributes mean time expenditures to commit to NILM or NILM qualifier diagnoses with differences in mean review times. For primary and adjudicative WSI rescreening, the mean time expenditures for NILM and NILM qualifier diagnoses were 1.26 and 3.12 min, respectively; the overall difference in mean time expenditures was 1.85 min. Breakdowns of mean time expenditures for NILM and NILM qualifier diagnoses for primary BestCyte review (minutes): NILM (1.23), NILM Inflammation (1.23), NILM Reactive/Repair (1.55), NILM Bacterial cytolysis (1.01), NILM Atrophy (1.26), NILM Bacterial vaginosis (1.15), and NILM Atrophic vaginitis (1.40). The respective breakdowns for adjudicative WSI rescreening (minutes): NILM (3.02), NILM Inflammation (3.47), NILM Reactive/Repair (3.23), NILM Bacterial cytolysis (2.57), NILM Atrophy (2.86), NILM Bacterial vaginosis (3.27), and NILM Atrophic vaginitis (3.39). The differences in mean time expenditure for NILM and NILM qualifiers (minutes): NILM (1.79), NILM Inflammation (2.24), NILM Reactive/Repair (1.68), NILM Bacterial cytolysis (1.56), NILM Atrophy (1.60), NILM Bacterial vaginosis (2.12), and NILM Atrophic vaginitis (1.99). Fig. 5 illustrates these data and incorporates superimposed linear trend lines revealing a steady-state (i.e., plateau) trend for primary BestCyte review times for NILM and NILM qualifier diagnoses. In comparison, the linear trend line for adjudicative WSI rescreening times reveals a gradually increasing expenditure of time for the NILM qualifier diagnoses as sequenced.
Table 4.
Number (n) of NILM cases (and NILM qualifiers: Inflammation, Reactive/Repair, Bacterial Cytolysis, Atrophy, Bacterial vaginosis, Atrophic vaginitis) with mean time expenditures (minutes) for primary BestCyte screening and adjudicative WSI rescreening, and differences in time expenditures, per NILM and NILM qualifier diagnoses.
| Diagnosis | NILM qualifier | Adjudicative WSI rescreening diagnosis n= | BestCyte Primary Cell Sorter screening mean time (Min) | BestCyte adjudicative WSI rescreening mean time (Min) | Difference in time (Min) |
|---|---|---|---|---|---|
| NILM | NILM | 163 | 1.23 | 3.02 | 1.79 |
| NILM | Inflammation | 22 | 1.23 | 3.47 | 2.24 |
| NILM | Reactive/Repair | 65 | 1.55 | 3.23 | 1.68 |
| NILM | Bacterial cytolysis | 19 | 1.01 | 2.57 | 1.56 |
| NILM | Atrophy | 27 | 1.26 | 2.86 | 1.60 |
| NILM | Bacterial vaginosis | 45 | 1.15 | 3.27 | 2.12 |
| NILM | Atrophic vaginitis | 5 | 1.40 | 3.39 | 1.99 |
| Values | 346 | (1.26) | (3.12) | (1.85) |
Fig. 5.
Mean time expenditures (minutes) to primary BestCyte Cell Sorter screening diagnoses (blue) and adjudicative WSI rescreening diagnoses (green), for overall Bethesda NILM and NILM qualifiers: Inflammation, Reactive/Repair, Bacterial cytolysis, Atrophy, Bacterial vaginosis, Atrophic vaginitis. With superimposed linear trend lines.
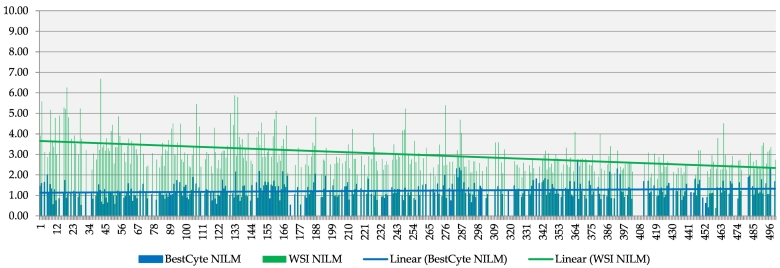
Fig. 6, Fig. 7, Fig. 8, Fig. 9 illustrate review time expenditures for all 500 PCS thin-layers for primary review (blue) and adjudicative WSI rescreening (green) with superimposed linear trend lines for: NILM (including qualifier diagnoses), ASCUS, LSIL, and HSIL respectively. Fig. 6, Fig. 7, Fig. 8, Fig. 9 reveal plateau linear trend lines for primary BestCyte review time expenditures; whereas the respective linear trend lines for adjudicative WSI rescreening reveal consistently decreasing screening time expenditures throughout the study continuum.
Fig. 6.
Time expenditures (minutes) exclusively of NILM diagnoses reflecting primary BestCyte Cell Sorter screening (blue) and adjudicative WSI rescreening (green), with superimposed linear trend lines.
Fig. 7.
Time expenditures (minutes) exclusively of ASCUS diagnoses reflecting primary BestCyte Cell Sorter screening (blue) and adjudicative WSI rescreening (green), with superimposed linear trend lines.
Fig. 8.
Time expenditures (minutes) exclusively of LSIL diagnoses reflecting primary BestCyte Cell Sorter screening (blue) and adjudicative WSI rescreening (green), with superimposed linear trend lines.
Fig. 9.
Time expenditures (minutes) exclusively of HSIL diagnoses reflecting primary BestCyte Cell Sorter screening (blue) and adjudicative WSI rescreening (green), with superimposed linear trend lines.
Table 5 distributes κ coefficients and 95% CI reflecting intra-observer reproducibility between primary BestCyte review and adjudicative WSI rescreening diagnoses for the PCS. Kappa and (95% CI): Overall 500 PCS diagnoses: 0.9305 (0.8983-0.9627); NILM including NILM qualifiers: 0.9429 (0.9110-0.9748); ASCUS: 0.8378 (0.7393-0.9363); ASC-H: 0.9112 (0.8113-0.9999); AGUS: 1.0 (1.0-1.0); LSIL: 0.9189 (0.8400-0.9978); HSIL: 0.9894 (0.9685-0.9999); CA: 1.0 (1.0-1.0); and UNSAT: 1.0 (1.0-1.0).
Table 5.
Kappa coefficients and 95% Confidence Intervals representing intra-observer reproducibility between primary BestCyte screening and adjudicative WSI rescreening diagnoses for the overall PCS, per Bethesda diagnosis.
| Overall case set/Bethesda diagnosis | Kappa coefficient | 95% confidence interval |
|---|---|---|
| Overall case set | 0.9305 | 0.8983–0.9627 |
| NILM | 0.9429 | 0.9110–0.9748 |
| ASCUS | 0.8378 | 0.7393–0.9363 |
| ASC-H | 0.9112 | 0.8113–0.9999 |
| AGUS | 1.0000 | 1.0000–1.0000 |
| LSIL | 0.9189 | 0.8400–0.9978 |
| HSIL | 0.9894 | 0.9685–0.9999 |
| CA | 1.0000 | 1.0000–1.0000 |
| UNSAT | 1.0000 | 1.0000–1.0000 |
Conclusions
The adoption of AI-driven digital technologies in diagnostic cytopathology may be understandably impeded by initial aversion.2,5 Yet as digital systems prove increasingly precise and amenable to cytologic fundamentals and workflows they seem primed for favorable application alongside increasing users’ comfortability.2,8, 9, 10, 11, 12 Furthermore, advancements that potentially simplify and minimize interpretive variance may foster wider utilization of digital technology in routine gynecological Pap test screening service.5,11, 12, 13, 14
Lew et al.2 described the chronology of digital platforms in Pap test cytopathology emphasizing the importance of domain experts.2,15 The functionality borne out of the BestCyte system is testament to input by expert cytologists, pathologists, and (computer, mechanical, optical, and biochemical) engineers in its research and development. Central to the BestCyte’s effectiveness are innumerable digital images of clinically relevant cells programmed into AI algorithms to assure their precise follow-on detection, classification, and rapid display for efficient reidentification hence appropriate interpretation and reporting through cytologic screening.
Chantziantoniou et al.5 conceptualized 5 principles involved in cytologic interpretation and their relational impact in digital technology: (1) Adequate visualization of cells; (2) completion of cytomorphologic templates; (3) evidence of diagnostic criteria; (4) assessment of cytomorphologic overlap; and (5) minimization of diagnostic pitfalls. In clinical practice, cytomorphologic overlap is reconciled through methodical screening; a skill involving multi-level mental abstractions of assemblages of cellular features against which diagnoses are raised or rejected. Screening cytologists spot then mark cells to facilitate thenceforth reidentification. This process is ultimately complex and progressively confounded by increasing subjectivity thus associated with interpretive variance depending on sample characteristics, diagnostic pitfalls, and level of expertise.4,13,14 These constructs may pose considerable diagnostic dilemmas particularly in cases revealing pronounced cytomorphologic overlap, as in those involving differential considerations for HSIL.5 Of importance therefore, and particularly in the context of cervical cancer screening, digital imaging systems as BestCyte that optimize precision by standardizing the cell selection process through algorithm-driven classification, would facilitate cytologic principles by capturing and effectively displaying rare cellular events and diagnostic criteria upon which interpretations may be raised with confidence.
The BestCyte system enabled adequate visualization of clinically relevant cells in sorted, variably sized tiles. The variety of tile sizes, dimensions, and their assortment in portrait or landscape layouts in the BestCyte galleries to optimally frame cells supported dynamic impressions of the WSI milieu upon download. This design facilitated fundamental baselines to be promptly established, such as: Relative cellularity; variety of sizes of cells or of cell clusters; squamous epithelial maturation; extent of inflammatory exudate or blood; Papanicolaou hematoxylin staining characteristics and counter-staining intensities; and euchromasia for nuclear chromatin grading. The BestCyte’s design negated potentially tedious presentation of cells in high-power fields organized as latticed equally sized cubic thumbnails juxtaposed with half-screen displays of FOV containing those cells, as in competitive digital imaging systems.2,15 BestCyte enables full-screen projection of either sorted tiles in galleries or WSI FOV in independent (Windows) displays, not simultaneously juxtaposed. This design minimized distraction or perception bias potentially arising from images of unlike cell-types, of unlike sizes, albeit in equidimensional latticed thumbnails.5,12
The data herein suggests BestCyte may efficiently support primary Pap test cervical cancer screening as 96.6% of all 500 Bethesda primary diagnoses committed through cell image sorting were confirmed following adjudicative WSI rescreening (Table 1). Through another vantage point, adjudicative rescreening, requiring an additional mean 2.56 min per case, did not reveal undetected cells of greater severity in the WSI (Table 1, Table 3). Based on the mean review times from the 500 thin-layers herein investigated, the findings equate to 21.3 total rescreening hours expended to uphold 96.6% of primary diagnoses raised throughout the Bethesda diagnostic spectrum; but also, for the inspection of significantly larger populations of cells in the WSI relative to select cells depicted in 40x images in galleries. Therefore, BestCyte may digitally optimize the practice of screening and potentially supplant LM, as was also asserted by Delga et al.6
The presentation of tiles in BestCyte galleries6 proved advantageous. The BestCyte design enabled efficient reviews of images of cells in 40x magnification for interpretive consideration given the mosaic of projected cytomorphology and relative cellularity representing the WSI. The majority of LSIL and HSIL interpretations were established from tiles populating the Overview gallery in most such cases investigated; this occurred as diagnostic criteria were displayed effectively and unequivocally. Moreover, as cells annotated in tiles or in FOV converged into the Diagnosis tab to assemble the cytomorphology spotted during screening, interpretive principles and Bethesda diagnosis considerations were simplified.5 This convergence of cytomorphology further reduced labor, hence time to diagnosis.
All AGUS, LSIL, HSIL, CA, and UNSAT primary diagnoses were upheld following adjudicative WSI rescreening (Table 1). Whereas 82.4% of ASCUS and 94.1% of ASC-H primary diagnoses were confirmed following WSI rescreening is noteworthy bearing in mind the grey zones oftentimes associated with such interpretations in clinical practice.2,12 Collectively, these cases accounted for 46 (9.20%) of the 500 diagnoses committed. Of equal importance however are 97.2% of the 354 primary NILM diagnoses confirmed upon adjudicative WSI rescreening; whereby, 183 of the 354 NILM cases (52.9%) were reported with NILM qualifiers arguably associated with interpretive complexity12 thus increased levels of subjectivity (Table 1, Table 4). However, in the context of cervical cancer screening for disease prevention, all 52 primary HSIL diagnoses were upheld after adjudicative WSI rescreening suggesting optimized capacity for BestCyte to detect, sort, and display small, severely dysplastic cells characteristic of HSIL with the required diagnostic criteria hence minimized diagnostic uncertainty (Fig. 2).5
Of the 500 PCS thin-layers investigated, 17 (3.40%) were discordant (Table 1, Table 2). Of these 17 cases, 15 (88.2%) involved ASCUS or ASC-H. The remainder 482 primary diagnoses of 500 (96.6%) were concordant. These findings are substantiated by the κ and 95% CI listed in Table 5.
Kappa coefficient ranges of 0.4–0.6, 0.6–0.8, and 0.8–1.0 reflect moderate, substantial, and nearly perfect agreement respectively.13 The κ (0.9305) arising from the overall 500 PCS study is herein noteworthy (Table 5). However, it is the investigator’s conviction that given the principles involved in cytologic interpretation,5 the κ coefficients reported for NILM (0.9429), ASCUS (0.8378), ASC-H (0.9112), and HSIL (0.9894) diagnoses are particularly remarkable (Table 5). The relatively lower κ for ASCUS agrees with studies reporting the potential diagnostic uncertainties and pitfalls oftentimes experienced in this interpretation thus introduction of objective measures as ASCUS:SIL ratios and reflex Human Papilloma Virus molecular testing to help minimize ambiguous cytologic reporting hence unfavorable clinical ramifications.12, 13, 14, 15 Yet impact of κ analysis is reportedly dependent on the prevalence of disease hence frequency of diagnosis. Sorbye et al.16 claim clinical settings with higher detection rates for disease in cervical cytology may reflect higher interpretive sensitivities for CIN2+ lesions. Thus, as the 52 HSIL diagnoses reported in this study accounted for 10.4% of the 500 PCS thin-layers (Table 1), this frequency of CIN2+ may have influenced the κ value reported for HSIL (Table 5). However, studies by Deschenes et al.17 concluded κ coefficients are more sensitive to individual cytologists’ interpretive error rates rather than to caseloads per se. Such phenomena may explain the κ coefficients herein reported since the intra-observer error rate remained constant between cell image and WSI adjudicative rescreening. Therefore, given this study’s aim to assess diagnostic confidence potentialities in BestCyte technology, diagnostic error stability and the relatively higher proportion of CIN2+ cases in the PCS may have fostered a factual evaluation.
Time to diagnosis was a critical variable in this study. Screening review time expenditures are taken to reflect a combination of screening time and technique and labor through to diagnosis commitment; thus, a meaningful surrogate predictor for diagnostic confidence in BestCyte functionality and efficiency. In part, the lower review times illustrated in Fig. 3 were facilitated by the overall functionality of the BestCyte system reflecting increasing confidence cells of greater severity are not undetected in the WSI. However, additional features contributed to lower review times. Given BestCyte’s unique projection of tiles, critical impressions formed simultaneously following download and assortment of cell images, thus minimization of WSI reviews to establish reference baselines as relative cellularity or chromasia. Likewise, the BestCyte’s Overview gallery formed adequate templates5 of high-power fields from the WSI. These compositions recreated the random slide reviews otherwise required to assess cellular milieus. Moreover, the 9:16 ratio full-screen display maximized the number of tiles sorting linearly horizontally (Fig. 2), thus minimized vertical scrolling to inspect the remainder tiles in galleries. Together, these features further reduced screening review times through to diagnosis in straight-forward cases.
When tested as a primary screening device, BestCyte produced remarkable diagnostic reproducibility despite the lower review times required to inspect tiles relative to WSI rescreening, as was surmised by Delga et al.6 (Table 3). The mean primary screening time expenditure for all 500 Bethesda diagnoses was 1.38 min (range: 0.65–1.84) compared to 3.94 min (range: 2.53–6.34) for adjudicative WSI rescreening. Also, all LSIL (n=24), HSIL (n=52), and CA (n=1) primary diagnoses were upheld following adjudicative WSI rescreening requiring an additional 2.67, 2.56, and 1.92 min, respectively. This study recorded 1 diagnosis of CA (Keratinizing squamous cell carcinoma) committed after inspecting images of isolated malignant cells amongst necrotic debris within 0.65 min. By contrast, WSI rescreening of the obscured cellular material marginating in the ThinPrep thin-layer periphery required an additional 2.57 min (Table 3, Fig. 3).
Enactment of CLIA’88 stipulated maximum workloads for screening cytologists allowing 12.5 slides per screening hour; practically, on average, 4.8 min per (conventional) slide using LM.2,4,6,18 Maximum workloads were adjusted after introduction of liquid-based technology given the smaller depositions of cellular material on glass slides relative to smears.2,18 However according to studies by Cheuvront et al.,19 whereas cytologists expended gradually less time screening thin-layers compared to smears they tripled the amount of time required to review cells per unit slide area (i.e., per mm2). Their study also revealed that cytologists’ screening times decreased from 5.9 to 2.7 min per slide alongside increasing familiarity, but only after reading greater than 500 thin-layer cases.19 Therefore, as was speculated by Delga et al.,6 the mean review time for primary cell image BestCyte screening (i.e., 1.38 min) of the 500 PCS Bethesda diagnoses (Table 3) was less than half of what would be expected for LM (i.e., 4.8 min),6,18 and within the 0.5–3.0 min range they hypothesized for thin-layers.6 These data reflect substantial potential reductions in overall screening labor.
Fig. 6, Fig. 7, Fig. 8, Fig. 9 illustrate screening time trends exclusively for NILM, ASCUS, LSIL, and HSIL diagnoses respectively. The bar plots reveal consistently decreasing WSI rescreening time expenditures throughout the study’s continuum. These findings are taken to reflect inversely increasing investigator’s confidence that cells of greater severity are not undetected in the WSI. Such diagnostic confidence potentialities in BestCyte screening of sorted images are reinforced by the plateau review time trends for these diagnoses respectively (Fig. 6, Fig. 7, Fig. 8, Fig. 9). Fig. 5 supports this assertion. Although NILM qualifier diagnoses revealed gradually increasing adjudicative WSI rescreening time expenditures arguably due to increasing subjectivity,12 this occurred despite the overall decreasing review time trend for WSI rescreening for all NILM diagnoses (Fig. 6). These findings corroborate with reported suppositions that initial aversion may diminish alongside increasing digital system familiarity and maturing learning curves, as also speculated by Delga et al.2,6,12,20 The higher WSI rescreening time expenditures at the outset of this investigation arguably reflected the concern that clinically significant cells remained possibly undetected. This skepticism is represented by the relatively higher WSI rescreening time expenditures for NILM Inflammation, Reactive/Repair, and Atrophic vaginitis qualifier diagnoses due to potential cytomorphologic overlap with HSIL or squamous cancer cells5,12 (Table 4, Fig. 5); similarly for ASC-H, AGUS, and HSIL cases, to rule out possibly isolated, small, frank malignant cells strewn within larger populations of cells in the WSI.
A meta-analysis of studies for intra-observer diagnostic concordance rates between WSI and LM screening by Girolami et al.20 in gynecological cytopathology revealed few reports since the introduction of WSI digital technology in 1999.12 Across all studies that Girolami et al.20 reviewed, the mean intra-observer κ coefficient between WSI and LM was 0.66 (relative to 0.69 for inter-observer reproducibility).20 Contributing factors cited included: Lacking preference for WSI analyses; longer review times for WSI screening relative to LM; suboptimal focusing; and the perceived superiority of LM over WSI captured through single-plane focusing.20 Nonetheless, Girolami et al.20 surmised AI algorithms may eventually overcome such limitations. Hanna et al.21 further surmised increasingly favorable perceptions for WSI screening alongside increasing users’ confidence and the facilitation of routine workflows leading to decreasing review times and labor, hence greater overall productivity. In keeping with such speculations, BestCyte recreated LM practice by adding dynamism to digital screening through AI-driven classification of cells for user’s inspection, thus may be considered non-inferior and likely significantly superior to LM for Pap test screening of thin-layers based on the data herein reported. This contention is further supported by the UNSAT cases in this study. Whereas WSI rescreening revealed thin-layers ultimately unsatisfactory for interpretation, BestCyte nonetheless projected images of cells. These findings underpin the system’s capacity to detect and display rare events. Perhaps additional studies using BestCyte technology with multi-plane scanning may lead to superior κ coefficients to those reported herein particularly for the ASCUS and ASC-H cases by reducing likely cytomorphologic overlap due to cellular 3-dimensionality, as argued by Wright et al.12,22 However, based on BestCyte’s design, digitized images from 3-dimensional cells stitched together to compose a single 2-dimensional WSI for display may be captured either through single-plane or multi-plane scanning. Accordingly, Bongaerts et al.23 investigating diagnostic concordance rates for SurePath thin-layer WSI screening relative to LM in gynecological cytopathology, using Pannoramic P250 Flash II single-plane scanning under 20x, reported a κ coefficient range of 89.4%-99.4%. Therefore, whereas near-perfect monolayer cytopreparations may be accommodated with single-plane scanning as suggested by this study, this assertion is further supported by Bongaerts et al.23 concluding that optimal diagnostic results my be achieved using WSI reviews generated through 20x single-plane scanning even if sporadic images happen to depict cells partially defocused. Nevertheless, in practice, regardless of screening technique and focal planes, cytologists’ interpretational expertise remains crucial for digital system impact.
In summary, the data reported herein validate observations reported by Delga et al.6 The time expenditures recorded between cell image primary screening and adjudicative WSI rescreening and the resulting κ coefficients from 500 PCS thin-layers reflect BestCyte’s robustness. The BestCyte cell classification platform facilitates accumulating user’s diagnostic confidence hence may confidently supplant screening practice for thin-layers through LM or pan-WSI reviews. Yet BestCyte facilitates additional advantages that may optimize diagnostic practice that ought to be considered to weigh the system’s competitive advantages.6,23 The shorter review time expenditures with reduced labor also reduce costs, increase case turn-around-times, and generate otherwise exploitable time for cytologists and organizations. This may support extended screening time expenditures for challenging cases, multi-tasking, and ancillary and prognostic marker analyses as such practices are redefining modern cytopathology. Furthermore, given its functionality, BestCyte may accommodate hybrid screening depending on nature of cytopreparations and cytologists’ preferences, multiple-users’ access thus quality assurance workflows from primary screening through to rescreening and final reporting, 5-year lookback reviews of archived images of cells or WSI, and proficiency testing and academic exercises. The ability to access WSI cases remotely through web-based connectivity adds a major dimension to BestCyte functionality and particularly in the context of global population-based cervical cancer screening. Yet another dimension is versatility, as Gelwan et al.24 investigated BestCyte technology for the detection of abnormal urothelial cells relative to LM in urinary cytopathology.
Follow-up blinded studies involving additional cytologists for the 500 PCS thin-layers may support interobserver reproducibility evaluations against reference ‘truth’ diagnoses to further evaluate diagnostic confidence potentialities in BestCyte technology.
Statement of Ethics
All 500 PCS ThinPrep thin-layers analyzed for this research were deidentified prior to digital imaging, and glass slides and respective Marlboro-Chesterfield Pathology-issued diagnoses with patient-related clinical information were withheld for blinded studies.
Funding Sources
The research work conducted for this article was supported through Consultation service to CellSolutions LLC, Greensboro, NC, USA. No other funding was received for this work.
Author Contributions
The corresponding author acquired, analyzed, and interpretated the data presented in this study and composed the manuscript.
Data Availability Statement
Data generated and analyzed are herein presented completely through Tables and Figures. Further inquiries may be directed to the corresponding author.
Conflict of Interest Statement
The corresponding author is contracted Clinical Consultant to CellSolutions LLC, Greensboro, NC, USA.
Acknowledgments
The author gratefully acknowledges support from Dr. Dell Dembosky, MD, and cytology staff at Marlboro-Chesterfield Pathology (Pinehurst, NC, USA) for availability of the 500 ThinPrep thin-layers to support this research. Likewise acknowledged is support from Dr. Thomas Gahm, PhD, President of Select Laboratory Software development, and optical engineer at CellSolutions, for providing the technical overview of the BestCyte system; and from Dr. Robert P. Hirsch, PhD, President of Stat-Aid Consulting, for calculating the reproducibility Kappa coefficients and 95% Confidence Intervals.
References
- 1.Briganti G., Le Moine O. Artificial intelligence in medicine: today and tomorrow. Front Med. 2020;7:e1–e6. doi: 10.3389/fmed.2020.00027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lew M., Wilbur D.C., Pantanowitz L. Computational cytology: lessons learned from Pap test computer-assisted screening. Acta Cytol. 2021;65:286–300. doi: 10.1159/000508629. [DOI] [PubMed] [Google Scholar]
- 3.Cui M., Zhang D.Y. Artificial intelligence and computational pathology. Lab Investig. 2021;101:412–422. doi: 10.1038/s41374-020-00514-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Clary K.M., Davey D.D., Naryshkin S., et al. The role of monitoring interpretive rates, concordance between cytotechnologist and pathologist interpretations before sign-out, and turnaround time in gynecologic cytology quality assurance. Arch Pathol Lab Med. 2013;137:164–174. doi: 10.5858/arpa.2012-0120-CC. [DOI] [PubMed] [Google Scholar]
- 5.Chantziantoniou N., Mukherjee M., Donnelly A.D., Pantanowitz L., Austin R.M. Digital applications in cytopathology: problems, rationalizations, and alternative approaches. Acta Cytol. 2018;62:68–76. doi: 10.1159/000484434. [DOI] [PubMed] [Google Scholar]
- 6.Delga A., Goffin F., Kridelka F., Maree R., Lambert C., Delvenne P. Evaluation of CellSolutions BestPrep® automated thin-layer liquid-based cytology Papanicolaou slide preparation and BestCyte® cell sorter imaging system. Acta Cytol. 2014;58(5):469–477. doi: 10.1159/000367837. [DOI] [PubMed] [Google Scholar]
- 7.Alaghehbandan R. Performance of the CellSolutions Glucyte liquid-based cytology in comparison with the ThinPrep and SurePath methods. Acta Cytol. 2013;57:189–197. doi: 10.1159/000345878. [DOI] [PubMed] [Google Scholar]
- 8.Thrall M., Pantanowitz L., Khalbuss W.E. Telecytology: clinical applications, current challenges, and future benefits. J Pathol Inform. 2011;2:51. doi: 10.4103/2153-3539.91129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pantanowitz L., Hornish M., Goulart R.A. The impact of digital imaging in the field of cytopathology. Cytojournal. 2009;6:6. doi: 10.4103/1742-6413.48606. [4103/1742-6413.48606] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Farahani N., Parwani A.V., Pantanowitz L. Whole slide imaging in pathology: advantages, limitations, and emerging perspectives. Pathol Lab Med Int. 2015;7:23–33. [Google Scholar]
- 11.Mukherjee M.S., Donnelly A.D., DeAgano V.J., Lyden E.R., Radio S.J. Utilization of virtual microscopy in cytotechnology educational programs in the United States. J Pathol Inform. 2016 Mar 1;(7):8. doi: 10.4103/2153-3539.177682. [10.4103/2153-3539.177682. PubMed PMID:27076986; PubMed Central PMCID: PMC4809110] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wright A.M., Smith D., Dhurandhar B., et al. Digital slide imaging in cervicovaginal cytology – a pilot study. Arch Pathol Lab Med. 2013;137:618–624. doi: 10.5858/arpa.2012-0430-OA. [DOI] [PubMed] [Google Scholar]
- 13.Stoler M.H., Schiffmann M. Interobserver reproducibility of cervical cytologic and histologic interpretations – realistic estimates from the ASCUS-LSIL triage study. JAMA. 2001;285:1500–1505. doi: 10.1001/jama.285.11.1500. [DOI] [PubMed] [Google Scholar]
- 14.Ko V., Nanji S., Tambouret R.H., Wilbur D.C. Testing for HPV as an objective measure for quality assurance in gynecologic cytology. Cytopathol. 2007;111:67–73. doi: 10.1002/cncr.22488. [DOI] [PubMed] [Google Scholar]
- 15.McAlpine E.D., Pantanowitz L., Michelow P.M. Challenges developing deep learning algorithms in cytology. Acta Cytol. 2021;65:301–309. doi: 10.1159/000510991. [DOI] [PubMed] [Google Scholar]
- 16.Sorbye S.W., Suhrke P., Reva B.W., Berland J., Maurseth R.J., Al-Shibli K. Accuracy of cervical cytology: Comparison of diagnoses of 100 Pap smears read by four pathologists at three hospitals in Norway. BMC Clin Pathol. 2017;17:18. doi: 10.1186/s12907-017-0058-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Deschenes M., Renshaw A.A., Auger M. Measuring the significance of workload on performance of cytotechnologists in gynecologic cytology. Cancer Cytopath. 2008;114(3):149–154. doi: 10.1002/cncr.23497. [DOI] [PubMed] [Google Scholar]
- 18.Moriarty A.T. Cytology workload calculation - Has anything really changed? Cancer Cytopathol. 2011;119(2):77–79. doi: 10.1002/cncy.20141. [DOI] [PubMed] [Google Scholar]
- 19.Cheuvront D.A., Elston R.J., Bishop J.W. Effect of a thin-layer preparation system on workload in a cytology laboratory. Lab Med. 1998;29(3):174–179. [Google Scholar]
- 20.Girolami I., Pantanowitz L., Marletta S., et al. Diagnostic concordance between whole slide imaging and conventional light microscopy in cytopathology: a systematic review. Cancer Cytopathol. 2020;128:17–28. doi: 10.1002/cncy.22195. [DOI] [PubMed] [Google Scholar]
- 21.Hanna M.G., Monaco S.E., Cuda J., Xing J., Ahmed I., Pantanowitz L. Comparison of glass slides and various digital-slide modalities for cytopathology screening and interpretation. Cancer Cytopathol. 2017;125:701–709. doi: 10.1002/cncy.21880. [DOI] [PubMed] [Google Scholar]
- 22.Evered A., Dudding N. Accuracy and perceptions of virtual microscopy compared with glass slide microscopy in cervical cytology. Cytopathology. 2011;22(2):82–87. doi: 10.1111/j.1365-2303.2010.00758.x. [DOI] [PubMed] [Google Scholar]
- 23.Bongaerts O., Clevers C., Debets M., et al. Conventional microscopical versus digital whole-slide imaging-based diagnosis of thin-layer cervical specimens: a validation study. J Pathol Inform. 2018;9:1–9. doi: 10.4103/jpi.jpi_28_18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gelwan E., Zhang M.L., Allison D.B., et al. Variability among observers utilizing the CellSolutions BestCyte Cell Sorter imaging system for the assessment of urinary tract cytology specimens. J Am Society Cytopathol. 2019;8(1):18–26. doi: 10.1016/j.jasc.2018.10.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data generated and analyzed are herein presented completely through Tables and Figures. Further inquiries may be directed to the corresponding author.