Abstract
Molecular subtypes of medulloblastoma [Sonic Hedgehog (SHH), Wingless/INT (WNT), Group 3, and Group 4] are defined by common patterns of gene expression. These differential gene expression patterns appear to result in different histomorphology and prognosis. Quantitative histomorphometry is a well-known method of computer-aided pathology image analysis. The hypotheses we sought to examine in this preliminary proof of concept study were whether computer extracted nuclear morphological features of medulloblastomas from digitized tissue slide images could independently: (1) distinguish between molecularly determined subgroups and (2) identify patterns within these subgroups that correspond with clinical outcome. Our dataset was composed of 46 medulloblastoma patients: 16 SHH (5 dead, 11 survived), 3 WNT (0 dead, 3 survived), 12 Group 3 (4 dead, 8 survived), and 15 were Group 4 (5 dead, 10 survived). A watershed-based thresholding scheme was used to automatically identify individual nuclei within digitized whole slide hematoxylin and eosin tissue images. Quantitative histomorphometric features corresponding to the texture (variation in pixel intensity), shape (variations in size, roundness), and architectural rearrangement (distances between, and number of connected neighbors) of nuclei were subsequently extracted. These features were ranked using feature selection schemes and these top-ranked features were then used to train machine-learning classifiers via threefold cross-validation to separate patients based on: (1) molecular subtype and (2) disease-specific outcomes within the individual molecular subtype groups. SHH and WNT tumors were separated from Groups 3 and 4 tumors with a maximum area under the receiver operating characteristic curve (AUC) of 0.7, survival within Group 3 tumors was predicted with an AUC of 0.92, and Group 3 and 4 patients were separated into high- and low-risk groups with p = 0.002. Model prediction was quantitatively compared with age, stage, and histological subtype using univariate and multivariate Cox hazard ratio models. Age was the most statistically significant variable for predicting survival in Group 3 and 4 tumors, but model predictions had the highest hazard ratio value. Quantitative nuclear histomorphometry can be used to study medulloblastoma genetic expression phenotypes as it may distinguish meaningful features of disease pathology.
Introduction
Medulloblastoma is the most common malignant central nervous system tumor in pediatric patients, resulting in an overall cure rate of around 70%, and a five-year survival rate of about 75%,1 and significant neurological morbidity to many survivors as a result of both the disease and treatment.2 Survival rates also vary between subgroups, e.g., Wingless/INT (WNT) typically has a >95% five-year survival rate, whereas Sonic Hedgehog (SHH) may be divided amongst SHHα, SHHβ, SHHɣ, and SHHδ, of those the majority have intermediate risk, whereas SHHβ has the worst relative outcome, Group 4 tumors can be split into low- and high-risk subgroups with 72% and 36% ten-year survival rates respectively, and Group 3 tumors have the overall worst outcomes with <60% five-year survival rates.3 There is considerable evidence that patterns of gene expression can potentially help identify disease subtypes with better and worse prognosis.4,5 Recent work in molecular classification of medulloblastomas has resulted in identification of four distinct subgroups—SHH, WNT, Group 3, and Group 4—with “distinct epidemiology, clinical presentation”, molecular phenotypes, and outcomes.2 These four molecular subtypes were identified using unsupervised clustering of mRNA expression,2,6,7 and the identified subgroups have differences in the signaling pathways and driver genes, such as MYC, MYCN, KDM6A, GFI1, and GFI1B.8 Similar variations in genetic expression phenotypes have been used to characterize SHH, WNT, and non-SHH/WNT medulloblastoma tumors specifically.9 In addition, there is evidence that within these groups there may be even smaller subgroups with distinct phenotypes and molecular signatures, especially among the less well-defined Groups 3 and 4.6
Quantitative histomorphometry (QH) is the use of computer-aided image analysis of digitized pathology images to find disease specific patterns that may be predictive of underlying biology,10, 11, 12 molecular subtype, mutational status, disease outcome, or treatment response. Nuclei color,13 texture,14 shape,15 and architecture16,17 features have been used to predict cancer outcome, aggressiveness, and grade. Quantitative nuclei color and shape features were found to correlate strongly with poor prognosis,13,18 and molecular subtypes13 in work relating to brain tumors. In medulloblastomas, the level of anaplasia (variation in nuclear size, chromatin staining, with highly round, pleomorphic, or angular nuclei19, 20, 21) has been shown to be prognostic. Our group has previously demonstrated that algorithms can distinguish anaplastic from non-anaplastic medulloblastoma,22,23 and other groups have demonstrated QH feature differences in medulloblastoma tumors with different types of molecular expression,24 and biological and clinical factors that are predictive of survival.25 However, to date QH approaches have not been applied to explicitly predicting disease-specific survival in medulloblastomas.
In this work, we sought to identify quantifiable nuclear histomorphometric features that may be able to distinguish between molecular subgroups in medulloblastoma, as well as identifying histomorphometric associations with patient outcome within molecular subgroups.
Methodology
We extracted features from digitized whole-slide images to predict: (1) the molecular subtype (Experiment 1), and (2) patient outcome (Experiment 2).
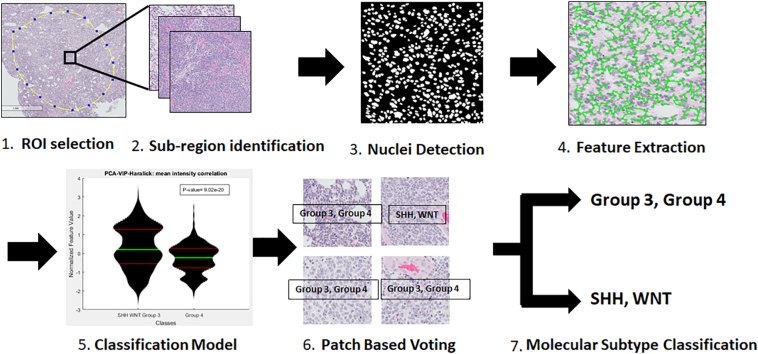
We used the following steps (Fig. 1). First, hematoxylin and eosin (H&E) slides were scanned and digitized. A pathologist identified regions of interest (ROIs) that contained primarily tumor nuclei (Fig. 1A). Second, 2000×2000 pixel subregions were extracted from these ROIs (Fig. 1B). Third, nuclear segmentation was performed using watershed separation26 (Fig. 1C). Fourth, we extracted features selected due to their effectiveness at detection and grading in prostate and breast cancer17,27,28 from segmented nuclei (Fig. 1D). Fifth, we selected top-performing features and used them in supervised machine learning classifiers (Fig. 1E). Finally, classifier predictions generated per-patch class predictions, and the majority of the predictions in each patient were used in a patch-based voting scheme to create a final prediction for each patient (Fig. 1E and F). In the first experiment, this feature-based per-patch and per-patient voting schemes were applied to molecular subtype classification (Fig. 1G), whereas in experiment 2, these features were used to predict survival within molecular subgroups.
Figure 1.
Workflow for molecular subtype classification. (1) Pathologist selection of suitable regions of interest (ROIs) in whole slides that are suitable for analysis. (2) Extraction of patches within each ROI for analysis. (3) Deep learning nuclei detection and segmentation. (4) Feature extraction. (5) Feature ranking and classification using patient-separated 3-fold cross-validation. (6) Patch-based voting to predict the molecular subgroup of each patient. (7) Per-patch and per-patient classification.
Our dataset comprised 69 H&E medulloblastoma formalin-fixed paraffin embedded whole-slide images obtained from patients under the age of 18 from the Children’s Hospital Los Angeles and digitized at University Hospital.1 Further description of dataset selection criteria can be found in the supplemental section (Fig. 2, Table 1, Table 2 and Supplemental Table 4).
Figure 2.

Flow chart of patient inclusion criteria resulting in a final cohort of 46 patients.
Table 1.
Dataset characteristics––demographic and cancer subtype distribution for the data used in this study.
| Database composition | Subtype | ||
|---|---|---|---|
| Total medulloblastoma | 69 | SHH | 16 |
| Frozen | 16 | Survived | 11 |
| Bad samples | 7 | Died | 5 |
| Usable samples | 46 | WNT | 3 |
| Age range | 0–17 | Survived | 3 |
| Female | 20 | Died | 0 |
| Male | 26 | Group 3 | 12 |
| Race | Survived | 8 | |
| Hispanic Latino | 27 | Died | 4 |
| Asian | 7 | Group 4 | 15 |
| White | 8 | Survived | 10 |
| Other | 4 | Died | 5 |
Table 2.
Demographic and clinical variable distribution, with Cox hazard ratio p-values with respect to survival.
| Subtype | Number | Lived | Died | %Survival |
|---|---|---|---|---|
| SHH | ||||
| Classic | 5 | 3 | 2 | 60 |
| Anaplastic | 1 | 0 | 1 | 0 |
| Desmoplastic | 10 | 8 | 2 | 80 |
| WNT | ||||
| Classic | 3 | 3 | 0 | 100 |
| Anaplastic | 0 | 0 | 0 | NA |
| Desmoplastic | 0 | 0 | 0 | NA |
| Group 3 | ||||
| Classic | 7 | 4 | 3 | 57.14 |
| Anaplastic | 5 | 4 | 1 | 80 |
| Desmoplastic | 0 | 0 | 0 | NA |
| Group 4 | ||||
| Classic | 12 | 8 | 4 | 66.67 |
| Anaplastic | 3 | 2 | 1 | 66.67 |
| Desmoplastic | 0 | 0 | 0 | NA |
Both experiments used the same approach for feature extraction, feature ranking, and classification. ROIs from each whole-slide image were verified by a pathologist to ensure that only ROIs containing invasive cancer were used in analysis. These ROIs from each of the 46 patients were divided into 1464 subregions, each comprising 2000×2000 pixels, and representing an area of 0.2 mm2 each. Subregions were identified by imposing a grid upon the ROI and finding all of the non-overlapping 2000×2000 pixel subregions which were entirely inside the ROI. These subregions were then processed using a watershed nuclei segmentation algorithm.26 This method was selected due to the fact that the nuclei interrogated were consistently darker than their surroundings, and in the case of nuclei expressing prominent nucleoli, the edges of nuclei were dark enough to allow for robust watershed nuclei segmentation.
From the segmented nuclei, a total of 227 features were extracted, corresponding to one of three categories: architectural, shape, and texture. These features measure histological characteristics that are relevant for stratifying tumor grade.19 Architectural features which measure the distance between nuclei can reflect qualities such as nodularity, anaplasiastic cells are identified by their large size, and texture features can capture variations in chromatin staining found in prominent nucleoli and apoptotic cells.19 Architectural features were obtained by performing quantitative analysis of nuclear graphs, such as Delaunay Triangles, Voronoi Diagrams, Minimum Spanning Trees (MST), and Cell Cluster Graphs (CCG).29 These nuclear graphs were constructed using the individual nuclei as the vertices of the graph. The choice of vertex connectivity determined the type of nuclear graph (i.e., Delaunay, Voronoi, MST, CCG) constructed. Features extracted from the graphs included changes in the lengths of edges and distance between nearest vertices. Cellular disorder was measured using Cell Orientation Graphs, previously described in Lee et al.27 Nuclear shape features included Invariant Moment, Fourier Descriptor, and Length/Width ratios. A comprehensive enumeration of all the image features extracted is presented in the Supplemental Material. Texture features included Haralick features, which involve statistics relating to frequency of specific pixel values being adjacent to each other.30
Feature ranking was used to identify the most relevant image features for each of the two experiments. The most relevant features identified were subsequently used in conjunction with machine learning classifiers. A number of popular feature ranking methods were evaluated including Wilcoxon rank-sum,31 PCA-VIP,32 and Maximum-Relevance Minimum-Redundancy (MRMR)33 with two variants—Mutual Information Difference, and Mutual Information Quotient (MRMR-MID and MRMR-MIQ).34 In each machine learning model, the number of features used was equal to the number of total patches used in cross-validation, divided by 10. Those numbers are listed in the supplemental section (Supplemental Table 5).
In each experiment, four different types of classifiers (Random Forest, Neural Network, Support Vector Machine, and Linear Discriminant Analysis), and four different feature ranking methods [Ranksum, Primary Component Analysis with Variable Importance Projection (PCA-VIP), MRMR MID, and MRMR MIQ] were used. These classifiers have varying strengths and weaknesses. For instance, neural networks are apt at identifying non-linear relationships, but are prone to overfitting, whereas other models such as Linear Discriminant Analysis are less likely to overfit, but only consider simpler combinations of features to generate predictions. By evaluating multiple feature ranking and classification methods, we attempted to identify the optimal combination of feature ranking and machine learning model for each of the two experiments.
For each of the two experiments described below, we identified the corresponding set of most highly ranked and predictive nuclear morphological features.
In experiment 1, we investigated the ability of histomorphometric features to distinguish between a single molecular subtype and all other subtypes. Three such molecular subtypes were investigated: SHH, Group 3, and Group 4 medulloblastoma tumors. First, histomorphometric features were used to separate each molecular subtype from all other molecular subtypes. Second, histomorphometric features were used to distinguish between two groups of molecular subtypes put together: SHH and WNT vs. Group 3 and Group 4.
In experiment 2, we investigated the ability of nuclear histomorphometric features to predict long-term survival within selected molecular subgroups. This experiment comprised three sub-parts: (1) identifying histomorphometric features that were predictive of disease specific survival, (2) identify whether specific histomorphometric features were more prognostic of survival within each molecular group, and (3) identify whether specific histomorphometric features were more prognostic of survival within two groups: SHH and WNT combined, and Groups 3 and 4 combined due to the similarity of genetic expression between Groups 3 and 4, and between the SHH and WNT groups.2 For each experiment, the top-performing feature ranking and machine learning classification methods were then employed with 100 iterations of per-patient 3-fold cross-validation. For each run of cross-validation, the machine classifier helped identify which patients in the hold out fold were high- or low-risk. In order to determine the final predicted risk category for each patient for Kaplan–Meier survival curves, the prediction for each patient when placed in the testing fold was recorded over 100 iterations, and the majority vote determined the disease-specific outcome for that patient.
Results
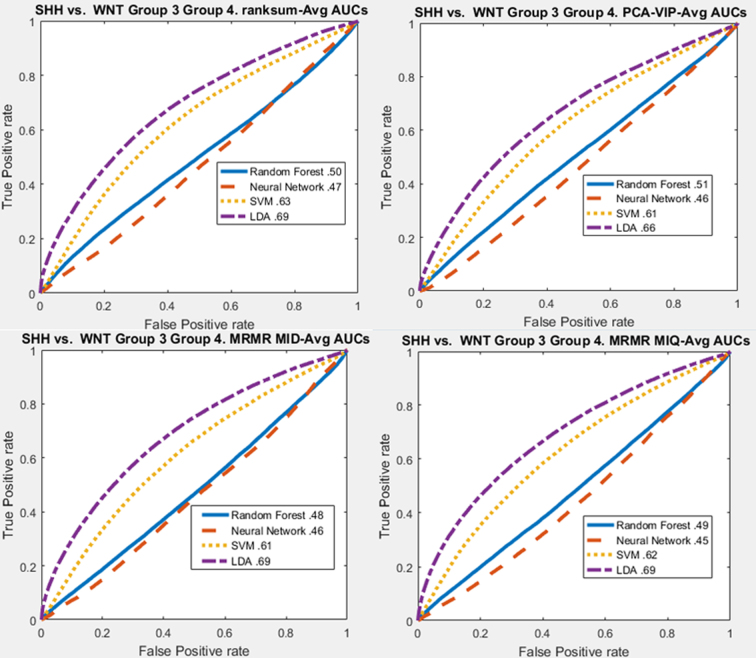
When separating SHH from all other subtypes combined, the top-performing features were measurements of the Haralick intensity, disorder of cell orientation, standard deviation of fractal dimension, and perimeter to area ratio features (Supplemental Table 6). Across the four feature ranking methods, Haralick, nuclei shape, and cell orientation entropy features were agreed upon as the top 10 most significant (Supplemental Table 9). The highest AUC for indicating whether or not subregions were SHH was 0.69 using LDA (Table 3, Supplemental Figure 7).
Table 3.
AUC curves for molecular subgroup separation based on nuclear histomorphometric features using rank-sum feature Ranking. Top left: SHH vs. WNT, Group 3, and Group 4. Top right: Group 3 vs. SHH, WNT, and Group 4. Bottom left: Group 4 vs. SHH, WNT, and Group 3. Bottom right: SHH and WNT vs. Groups 3 and 4.
| Molecular subgroup prediction AUC | Random forest | Neural network | SVM | LDA |
|---|---|---|---|---|
| SHH vs. WNT, Group 3, Group 4 | 0.5 | 0.47 | 0.63 | 0.69 |
| Group 3 vs. SHH, WNT, Group 4 | 0.55 | 0.49 | 0.67 | 0.63 |
| Group 4 vs. SHH, WNT, Group 3 | 0.52 | 0.46 | 0.55 | 0.59 |
| SHH, WNT vs. Group 3, Group 4 | 0.5 | 0.48 | 0.64 | 0.7 |
Supplemental figure 7.
Group separation performance using histologic features: SHH vs. WNT, Group 3, and Group 4
The top-performing features for Group 3 separation from other subgroups were the variation in the distances between nuclei and each of their neighbors, standard deviation of Haralick covariance matrix, and disorder of cell orientation (Fig. 3, Supplemental Table 6). Haralick texture features were the only features agreed upon as the top 10 most significant for all feature ranking methods (Supplemental Table 10). The highest AUC for separating subregions of Group 3 from all other subtypes was 0.67 using an SVM (Table 3, Supplemental Figure 8).
Figure 3.
Visualization of top-performing feature (architecture: disorder of distance between nearest seven neighbors) for separating Group 3 from all other groups. Left to right: Tissue at varying degrees of magnification within the region of interest (ROI) with a group: Low magnification ROI, subregion, and subregion zoom in to clearly display nuclei architecture. Far right: Nuclei are colored by the disorder of the distances between each nucleus and their seven closest neighbors (normalized). Top and bottom rows display Group 3 and 4 samples, respectively.
The top-performing nuclear features for separating Group 4 from other molecular subtypes were nuclei shape (particularly the median invariant moment), and Haralick standard deviation of intensity variance (Supplemental Table 6). Nuclei shape features were the only features agreed upon as the top 10 most significant for all feature ranking methods (Supplemental Table 7). The highest AUC for separating subregions of Group 4 from all other subtypes was 0.59 using LDA (Table 1, Supplemental Figure 9).
Supplemental figure 9.
Group 4 vs. SHH, WNT, and Group 3
The top-performing features for predicting whether subregions were SHH or WNT vs. Group 3 or Group 4 were Haralick intensity correlations, Haralick standard deviation of contrast entropy, and the standard deviation of the fractal dimension of nuclei shape (Supplemental Table 12). The highest AUC for predicting whether subregions were SHH or WNT vs. Group 3 or Group 4 was 0.7 using LDA (Table 3, Supplemental Figure 10).
Supplemental figure 10.
SHH and WNT vs. Groups 3 and 4
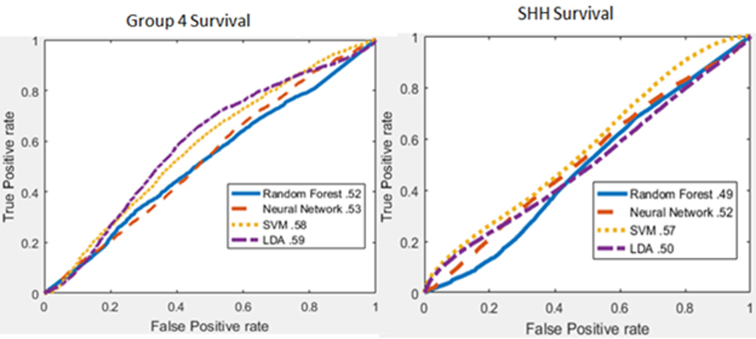
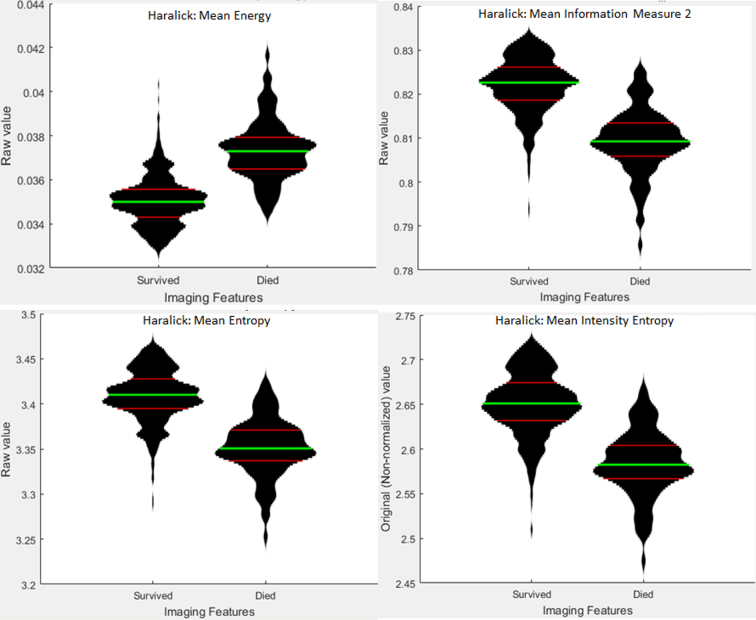
Rank-sum results are displayed here, whereas the results from other feature ranking methods are in the supplemental section. Of the three parts of this experiment listed above, the survival predictions for Group 3 and Groups 3 and 4 had significant results, and are thus shown below. The top-performing features for predicting which Group 3 subregions were associated with survival were Haralick mean and standard deviation of contrast and intensity texture features (Supplemental Table 6). The highest AUC for predicting which Group 3 subregions were associated with survival was 0.92 using a Neural Network (Fig. 4, left). Features such as Haralick texture features were useful for distinguishing between long- and short-term survivors in groups 3 and 4 (Fig. 5). The top-performing features for predicting which Group 3 and 4 subregions were associated with survival were Haralick mean and standard deviation of intensity energy and entropy texture features (Supplemental table 8, Supplementary material), whereas the AUC performance of Group 3, Group 4, and SHH survival models can be found in Supplemental figure 11, Supplemental figure 12, Supplemental figure 13. The features identified as most relevant to the survival for each of the groups are listed in supplemental tables. Features most significant to the survival for each group are listed in Supplemental table 14, Supplemental table 15, Supplemental table 16, Supplementary material. Haralick texture features were the only features that were represented in the top 10 most significant features for each feature ranking method (supplemental material). The highest AUC for predicting which Groups 3 and 4 subregions were associated with survival was 0.83 using SVM (Fig. 4, right), and the KM survival curves had a p-value of 0.002 (Fig. 5, right), and their feature distributions can be found in Supplemental Figure 14.
Figure 4.
Survival area under the curve curves within individual and paired molecular subgroups based on nuclear histomorphometric features. Left: Group 3. Right: Groups 3 and Group 4 combined.
Figure 5.
Feature filter maps, displaying a zoomed in sample from 1 of 4 cases left to right: Groups 3 and 4, short- and long-term survival. Top row: hematoxylin and eosin image of tissue. Bottom row: normalized colormap intensity of the top-performing feature for separating survival and non-survival in Groups 3 and 4 (Haralick mean energy) of nuclei overlaid on top of original image. Dark blue pixels correspond with low Haralick values, whereas yellow and red pixels correspond with high Haralick values
Supplemental figure 11.
Group 3 survival AUC curves
Supplemental figure 12.
Group 4 and SHH survival AUC curves
Supplemental figure 13.
Group 4 survival AUC curves
Supplemental figure 14.
Feature distributions for Groups 3 and 4 Survival. Feature Distributions for the top four ranked features for determining survival for Groups 3 and 4 combined using Rank-sum feature ranking. Green lines indicate the mean of each population, and red lines indicate the 25th and 75th percentiles of the distribution. Width of the plot indicates the relative number of data points at each non-normalized feature value along the y-axis.
The distinction between high- and low-risk groups is clearly visible in the highest performing features. The features themselves can be used to visualize differences between high- and low-risk patients. Figure 6 shows normalized Haralick values overlaid onto images of H&E stained tissue to show the difference between high- and low-risk patients. Statistical significance was measured using p-values derived via the Cox hazard ratio test, and the severity of each feature tested for survival was measured using the hazard ratio, as well as the statistical significance of each variable. A 95% confidence interval of the hazard ratio was calculated for each feature (Supplemental Table 8).
Figure 6.
Kaplan–Meier survival curves within individual and paired molecular subgroups based on nuclear histomorphometric features. Blue lines indicate predicted high survival groups, whereas red lines indicate predicted low survival groups. Left: Group 3. Right: Group 3 and Group 4 combined.
Discussion
Medulloblastoma molecular subtypes have been identified using immunohistochemical data, including protein expression, mutation, and methylation data,2,6,7,35,36 resulting in the identification of four distinct subtypes: SHH, WNT, Group 3, and Group 4. SHH is defined by changes in expression in SHH, basal cell, WNT/β, and axonal guidance signaling, and it primarily affects children younger than 5 and older than 11.37 WNT tumors are defined by axonal guidance, WNT/β, O-glycan biosynthesis, and basal cell carcinoma signaling, and affects primarily children between the ages of 6 and 16.37 Group 3 is defined by changes in expression in MYC, phototransduction, WNT/β, and glutamate signaling, and primarily affects children between the ages of 3 and 6.37 Group 4 is defined by changes in expression in semaphorin, cAMP, G protein-coupled receptor, and p53 signaling, and primarily affects children between the ages of 6 and 16.37
Both histology as well as molecular subgroups are important sources of information when identifying subgroups and predicting prognosis. Variations in both histological phenotype and genetic expression correlate with prognostic changes; e.g., MBEN has comparatively good prognosis, whereas c-myc gene expression and LCA each result in poor prognosis.2,38,39 However, histological and molecular subtypes have dramatically different phenotypical expression, prognosis, and rates of incidence.2,37 MYC and MYCN overexpression are an example of possible correlation between molecular subtype and histomorphometric features, as MYC amplification could not be directly linked to a histological subtype but is absent in Group 4, whereas found in WNT and Group 3,2 and it is strongly correlated with prominent nucleoli in histological slides.40 MYC and MYCN expression have even been shown to be “more effective than conventional clinical (histological) criteria at predicting patient survival” and was considered in initial determination of molecular subgroup categories.37 Considered together, molecular subtype and histological features of medulloblastoma may provide further utility in subgroup-specific treatment investigation as well as at clinical presentation, potentially providing further ability to stratify patient risk and inform treatment decisions.
Our goal in this work was to investigate the relationship between quantifiable nuclear features and molecular subtypes and survival. In this work, we used nuclear histological features obtained through digital image analysis to: (1) predict the molecular subgroups of medulloblastoma samples, and (2) make statistically significant predictions of survival risk within medulloblastoma subgroups.
In experiment 1, QH features were used to predict molecular subtype. Haralick texture features, cell orientation, and nuclei shape features featured prominently in separating molecular subgroups. Nuclear size, shape, and texture features have previously been identified as significant to medulloblastoma, due to the role that those characteristics play in identifying anaplasia, which is characterized with darker chromatin staining; nuclei size, which can fluctuate by up to 50%; and nuclei shape, which can shift to either being extremely round or angular.19,20
In experiment 2, QH features were used to try to predict short- vs. long-term survival within molecular subgroups. The most dominant features for predicting survival in Group 3 and Groups 3 and 4 were Haralick texture features (Supplemental Table 7); in Group 4 were primarily haralick and shape features (Supplemental Table 15); and in SSH tumors entirely shape features (Supplemental Table 13). Haralick texture features measure the repetition of similar pixel values in geometric patterns, and may detect uneven distributions of hematoxylin staining, multinucleation, or other pleomorphic patterns, which are an independent prognostic indicator for medulloblastoma.41 In particular, chromatin staining patterns have been found to strongly correlate with both MYC and MYCN expression, and is positively correlated with poor prognosis.40
We acknowledge study limitations, such as limited dataset size due to rarity of the disease, as well as the recent introduction of medulloblastoma molecular subtyping. Secondly, we focused solely on the role of listed nuclear morphology metrics in this work. Third, this analysis was performed within selected ROIs. Additional work is clearly needed to evaluate whether these regions were truly representative of the morphological landscape of the tumor. In the future, we hope to address these limitations to more precisely define subgroups and aid in prognosis, treatment, and targeted therapies.
The following are the supplementary data related to this article.
Group 3 vs. SHH, WNT, and Group 4
Demographic and clinical variable distribution, with Cox Hazard Ratio p-values with respect to survival.
Number of features used in experiments by subgroups investigated.
Each group Individually vs all feature ranking
Group 3 and Group 3+4 survival top 10 features
Hazard ratio and P value for model predictions vs. clinical variables
SHH vs. All top features for each feature ranking method
Group 3 vs. All top features for each feature ranking method
Group 4 vs. All top features for each feature ranking method
SHH and WNT vs Groups 3+4 top features for each feature ranking method
SHH Survival top 10 features for each feature ranking method.
Group 3 Survival top 10 features for each feature ranking method.
Group 4 Survival top 10 features for each feature ranking method.
Groups 3 & 4 Survival top 10 features for each feature ranking method.
Ethical policy and institutional review board statement
The datasets used and/or analyzed during this study are available from the corresponding author on reasonable request.
Financial support and sponsorship
Research reported in this publication was supported by the National Cancer Institute (award numbers 01CA249992-01A1, R01CA202752-01A1, R01CA208236-01A1, R01CA216579-01A1, R01CA220581-01A1, R01CA257612-01A1, 1U01CA239055-01, 1U01CA248226-01, and 1U54CA254566-01); National Heart, Lung and Blood Institute (award numbers 1R01HL15127701A1 and R01HL15807101A1); National Institute of Biomedical Imaging and Bioengineering (award number 1R43EB028736-01); National Center for Research Resources (award number 1 C06 RR12463-01); VA Merit Review Award from the United States Department of Veterans Affairs Biomedical Laboratory Research and Development Service (award number IBX004121A); the Office of the Assistant Secretary of Defense for Health Affairs through the Breast Cancer Research Program (award number W81XWH-19-1-0668); the Prostate Cancer Research Program (award numbers W81XWH-15-1-0558 and W81XWH-20-1-0851); the Lung Cancer Research Program (award numbers W81XWH-18-1-0440 and W81XWH-20-1-0595); the Peer Reviewed Cancer Research Program (award numbers W81XWH-18-1-0404 and W81XWH-21-1-0345); the Kidney Precision Medicine Project (KPMP) Glue Grant; the Ohio Third Frontier Technology Validation Fund; the Clinical and Translational Science Collaborative of Cleveland from the National Center for Advancing Translational Sciences (NCATS) component of the National Institutes of Health and NIH roadmap for Medical Research (award number UL1TR0002548); the Wallace H.! Coulter Foundation Program in the Department of Biomedical Engineering at Case Western Reserve University; and sponsored research agreements from Bristol Myers-Squibb, Boehringer-Ingelheim, and Astrazeneca. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health, the U.S. Department of Veterans Affairs, the Department of Defense, or the United States Government. Fellowship from the Hartwell Foundation.
Conflicts of interest
Dr. Madabhushi is an equity holder in Elucid Bioimaging and in Inspirata Inc. In addition, he has served as a scientific advisory board member for Inspirata Inc, Astrazeneca, Bristol Meyers-Squibb and Merck. Currently, he serves on the advisory board of Aiforia Inc and currently consults for Caris, Roche, Cernostics, and Aiforia. He also has sponsored research agreements with Philips, AstraZeneca, Boehringer-Ingelheim, and Bristol Meyers-Squibb. His technology has been licensed to Elucid Bioimaging. He is also involved in three different R01 grants with Inspirata Inc.
Declaration of patient consent
The above-named study was reviewed by a member of the CHLA IRB. The study does not meet the definition of human subject research per 45 CFR 46.102(f), as the project does not include data obtained through intervention or interaction with the individual, or identifiable private information.
References
- 1.Orr B.A. Pathology, diagnostics, and classification of medulloblastoma. Brain Pathol. 2020 May;30(3):664–678. doi: 10.1111/bpa.12837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Northcott P.A., Korshunov A., Pfister S.M., Taylor M.D. The clinical implications of medulloblastoma subgroups. Nat Rev Neurol. 2012 May 8;8(6):340–351. doi: 10.1038/nrneurol.2012.78. [DOI] [PubMed] [Google Scholar]
- 3.Juraschka K., Taylor M.D. Medulloblastoma in the age of molecular subgroups: a review: JNSPG 75th Anniversary Invited Review Article. J Neurosurg Pediatr. 2019 Oct;24(4):353–363. doi: 10.3171/2019.5.PEDS18381. [DOI] [PubMed] [Google Scholar]
- 4.Castellino R.C., Barwick B.G., Schniederjan M., et al. Heterozygosity for Pten promotes tumorigenesis in a mouse model of medulloblastoma. Gelovani JG, editor. PLoS One. 2010 May 26;5(5):e10849. doi: 10.1371/journal.pone.0010849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.MacDonald T., Brown K., LaFleur B., et al. Erratum: Expression profiling of medulloblastoma: PDGFRA and the RAS/MAPK pathway as therapeutic targets for metastatic disease. Nat Genet. 2003;35:287. doi: 10.1038/ng1103-287a. [DOI] [PubMed] [Google Scholar]
- 6.Cho Y.-J., Tsherniak A., Tamayo P., et al. Integrative genomic analysis of medulloblastoma identifies a molecular subgroup that drives poor clinical outcome. J Clin Oncol. 2011 Apr 10;29(11):1424–1430. doi: 10.1200/JCO.2010.28.5148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Taylor M.D., Northcott P.A., Korshunov A., et al. Molecular subgroups of medulloblastoma: the current consensus. Acta Neuropathol (Berl). 2012 Apr;123(4):465–472. doi: 10.1007/s00401-011-0922-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lin C.Y., Erkek S., Tong Y., et al. Active medulloblastoma enhancers reveal subgroup-specific cellular origins. Nature. 2016 Jan 27;530(7588):57–62. doi: 10.1038/nature16546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ellison D.W., Dalton J., Kocak M., et al. Medulloblastoma: clinicopathological correlates of SHH, WNT, and non-SHH/WNT molecular subgroups. Acta Neuropathol (Berl). 2011 Mar;121(3):381–396. doi: 10.1007/s00401-011-0800-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Romo-Bucheli D., Janowczyk A., Gilmore H., Romero E., Madabhushi A. A deep learning based strategy for identifying and associating mitotic activity with gene expression derived risk categories in estrogen receptor positive breast cancers. Cytom Part J Int Soc Anal Cytol. 2017 Feb 13;91(6):566–573. doi: 10.1002/cyto.a.23065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Romo-Bucheli D., Janowczyk A., Romero E., Gilmore H., Madabhushi A. In: Automated tubule nuclei quantification and correlation with oncotype DX risk categories in ER+ breast cancer whole slide images. Gurcan M.N., Madabhushi A., editors. 2016. p. 979106.http://proceedings.spiedigitallibrary.org/proceeding.aspx?doi=10.1117/12.2211368 [cited 2016 Aug 3]. Available from. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Basavanhally A., Xu Jun, Madabhushi A., Ganesan S. IEEE. 2009. Computer-aided prognosis of ER+ breast cancer histopathology and correlating survival outcome with Oncotype DX assay; pp. 851–854.http://ieeexplore.ieee.org/document/5193186/ [cited 2017 Apr 26]. Available from. [Google Scholar]
- 13.Hang Chang Ju, Han Borowsky A., Loss L., et al. Invariant delineation of nuclear architecture in glioblastoma multiforme for clinical and molecular association. IEEE Trans Med Imaging. 2013 Apr;32(4):670–682. doi: 10.1109/TMI.2012.2231420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nielsen B., Albregtsen F., Kildal W., Danielsen H.E. Prognostic classification of early ovarian cancer based on very low dimensionality adaptive texture feature vectors from cell nuclei from monolayers and histological sections. Anal Cell Pathol J Eur Soc Anal Cell Pathol. 2001;23(2):75–88. doi: 10.1155/2001/683747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Diamond D.A., Berry S.J., Umbricht C., Jewett H.J., Coffey D.S. Computerized image analysis of nuclear shape as a prognostic factor for prostatic cancer. The Prostate. 1982;3(4):321–332. doi: 10.1002/pros.2990030402. [DOI] [PubMed] [Google Scholar]
- 16.Doyle S., Agner S., Madabhushi A., Feldman M., Tomaszewski J. IEEE. 2008. Automated grading of breast cancer histopathology using spectral clusteringwith textural and architectural image features; pp. 496–499.http://ieeexplore.ieee.org/lpdocs/epic03/wrapper.htm?arnumber=4541041 [cited 2016 Sep 6]. Available from. [Google Scholar]
- 17.Doyle S., Hwang M., Shah K., Madabhushi A., Feldman M., Tomaszeweski J. IEEE. 2007. Automated grading of prostate cancer using architectural and textural image features; pp. 1284–1287.http://ieeexplore.ieee.org/lpdocs/epic03/wrapper.htm?arnumber=4193528 [cited 2016 Mar 18]. Available from. [Google Scholar]
- 18.Han J., Wang Y., Cai W., Borowsky A., Parvin B., Chang H. In: Medical Image Computing and Computer-Assisted Intervention – MICCAI 2016 [Internet] Ourselin S., Joskowicz L., Sabuncu M.R., Unal G., Wells W., editors. Springer International Publishing; Cham: 2016. Integrative analysis of cellular morphometric context reveals clinically relevant signatures in lower grade glioma; pp. 72–80.http://link.springer.com/10.1007/978-3-319-46720-7_9 [cited 2017 Nov 14]. Available from. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Eberhart C.G., Kepner J.L., Goldthwaite P.T., et al. Histopathologic grading of medulloblastomas: A Pediatric Oncology Group Study. Cancer. 2002 Jan 15;94(2):552–560. doi: 10.1002/cncr.10189. [DOI] [PubMed] [Google Scholar]
- 20.Giangaspero F., Wellek S., Masuoka J., Gessi M., Kleihues P., Ohgaki H. Stratification of medulloblastoma on the basis of histopathological grading. Acta Neuropathol (Berl). 2006 Jul;112(1):5–12. doi: 10.1007/s00401-006-0064-x. [DOI] [PubMed] [Google Scholar]
- 21.Eberhart C.G., Burger P.C. Anaplasia and grading in medulloblastomas. Brain Pathol. 2006 Apr 5;13(3):376–385. doi: 10.1111/j.1750-3639.2003.tb00037.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cruz-Roa A., Arevalo J., Basavanhally A., Madabhushi A., González F. In: A comparative evaluation of supervised and unsupervised representation learning approaches for anaplastic medulloblastoma differentiation. Romero E., Lepore N., editors. 2015. p. 92870G.http://proceedings.spiedigitallibrary.org/proceeding.aspx?doi=10.1117/12.2073849 [cited 2017 Apr 20]. Available from. [Google Scholar]
- 23.Cruz-Roa A., Gonzalez F., Galaro J., et al. A visual latent semantic approach for automatic analysis and interpretation of anaplastic medulloblastoma virtual slides. Med Image Comput Comput-Assist Interv MICCAI Int Conf Med Image Comput Comput-Assist Interv. 2012;15(Pt 1):157–164. doi: 10.1007/978-3-642-33415-3_20. [DOI] [PubMed] [Google Scholar]
- 24.Nafe R., Schlote W. Histomorphometry of brain tumours: histomorphometry of brain tumours. Neuropathol Appl Neurobiol. 2004 Aug 5;30(4):315–328. doi: 10.1111/j.1365-2990.2004.00596.x. [DOI] [PubMed] [Google Scholar]
- 25.Ray A. A clinicobiological model predicting survival in medulloblastoma. Clin Cancer Res. 2004 Nov 15;10(22):7613–7620. doi: 10.1158/1078-0432.CCR-04-0499. [DOI] [PubMed] [Google Scholar]
- 26.Veta M., van Diest P.J., Kornegoor R., Huisman A., Viergever M.A., Pluim J.P.W. Automatic nuclei segmentation in H&E stained breast cancer histopathology images. PLoS One. 2013;8(7) doi: 10.1371/journal.pone.0070221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee G., Ali S., Veltri R., Epstein J.I., Christudass C., Madabhushi A. Cell orientation entropy (COrE): predicting biochemical recurrence from prostate cancer tissue microarrays. Med Image Comput Comput-Assist Interv MICCAI Int Conf Med Image Comput Comput-Assist Interv. 2013;16(Pt 3):396–403. doi: 10.1007/978-3-642-40760-4_50. [DOI] [PubMed] [Google Scholar]
- 28.Basavanhally A., Feldman M., Shih N., et al. Multi-field-of-view strategy for image-based outcome prediction of multi-parametric estrogen receptor-positive breast cancer histopathology: comparison to Oncotype DX. J Pathol Inform. 2011;2:S1. doi: 10.4103/2153-3539.92027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ali S., Veltri R., Epstein J.A., Christudass C., Madabhushi A. In: Cell cluster graph for prediction of biochemical recurrence in prostate cancer patients from tissue microarrays. Gurcan M.N., Madabhushi A., editors. 2013. p. 86760H.http://proceedings.spiedigitallibrary.org/proceeding.aspx?doi=10.1117/12.2008695 [cited 2016 Mar 18]. Available from. [Google Scholar]
- 30.Haralick R.M., Shanmugam K., Dinstein I. Textural features for image classification. IEEE Trans Syst Man Cybern. 1973 Nov;3(6):610–621. [Google Scholar]
- 31.Devore J. 2015. Probability and Statistics for Engineering and the Sciences. Brooks Cole (1/1/2015) Cengage Learning. [Google Scholar]
- 32.Ginsburg S.B., Viswanath S.E., Bloch B.N., et al. Novel PCA-VIP scheme for ranking MRI protocols and identifying computer-extracted MRI measurements associated with central gland and peripheral zone prostate tumors. J Magn Reson Imaging JMRI. 2015 May;41(5):1383–1393. doi: 10.1002/jmri.24676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Peng H., Long F., Ding C. Feature selection based on mutual information: criteria of max-dependency, max-relevance, and min-redundancy. IEEE Trans Pattern Anal Mach Intell. 2005 Aug;27(8):1226–1238. doi: 10.1109/TPAMI.2005.159. [DOI] [PubMed] [Google Scholar]
- 34.Ding C., Peng H. Minimum redundancy feature selection from microarray gene expression data. J Bioinform Comput Biol. 2005 Apr;03(02):185–205. doi: 10.1142/s0219720005001004. [DOI] [PubMed] [Google Scholar]
- 35.Korshunov A., Chavez L., Northcott P.A., et al. DNA-methylation profiling discloses significant advantages over NanoString method for molecular classification of medulloblastoma. Acta Neuropathol (Berl). 2017 Dec;134(6):965–967. doi: 10.1007/s00401-017-1776-9. [DOI] [PubMed] [Google Scholar]
- 36.Fattet S., Haberler C., Legoix P., et al. Beta-catenin status in paediatric medulloblastomas: correlation of immunohistochemical expression with mutational status, genetic profiles, and clinical characteristics: β-catenin status in paediatric medulloblastomas. J Pathol. 2009 May;218(1):86–94. doi: 10.1002/path.2514. [DOI] [PubMed] [Google Scholar]
- 37.Northcott P.A., Korshunov A., Witt H., et al. Medulloblastoma comprises four distinct molecular variants. J Clin Oncol. 2011 Apr 10;29(11):1408–1414. doi: 10.1200/JCO.2009.27.4324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Garre M.L., Cama A., Bagnasco F., et al. Medulloblastoma variants: age-dependent occurrence and relation to gorlin syndrome--a new clinical perspective. Clin Cancer Res. 2009 Mar 10;15(7):2463–2471. doi: 10.1158/1078-0432.CCR-08-2023. [DOI] [PubMed] [Google Scholar]
- 39.Stearns D. c-myc overexpression causes anaplasia in medulloblastoma. Cancer Res. 2006 Jan 15;66(2):673–681. doi: 10.1158/0008-5472.CAN-05-1580. [DOI] [PubMed] [Google Scholar]
- 40.Wang L.L., Teshiba R., Ikegaki N., et al. Augmented expression of MYC and/or MYCN protein defines highly aggressive MYC-driven neuroblastoma: a Children’s Oncology Group study. Br J Cancer. 2015 Jun 30;113(1):57–63. doi: 10.1038/bjc.2015.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lamont J.M. Combined histopathological and molecular cytogenetic stratification of medulloblastoma patients. Clin Cancer Res. 2004 Aug 15;10(16):5482–5493. doi: 10.1158/1078-0432.CCR-03-0721. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Group 3 vs. SHH, WNT, and Group 4
Demographic and clinical variable distribution, with Cox Hazard Ratio p-values with respect to survival.
Number of features used in experiments by subgroups investigated.
Each group Individually vs all feature ranking
Group 3 and Group 3+4 survival top 10 features
Hazard ratio and P value for model predictions vs. clinical variables
SHH vs. All top features for each feature ranking method
Group 3 vs. All top features for each feature ranking method
Group 4 vs. All top features for each feature ranking method
SHH and WNT vs Groups 3+4 top features for each feature ranking method
SHH Survival top 10 features for each feature ranking method.
Group 3 Survival top 10 features for each feature ranking method.
Group 4 Survival top 10 features for each feature ranking method.
Groups 3 & 4 Survival top 10 features for each feature ranking method.