Abstract
Whole slide images contain a magnitude of quantitative information that may not be fully explored in qualitative visual assessments. We propose: (1) a novel pipeline for extracting a comprehensive set of visual features, which are detectable by a pathologist, as well as sub-visual features, which are not discernible by human experts and (2) perform detailed analyses on renal images from mice with experimental unilateral ureteral obstruction. An important criterion for these features is that they are easy to interpret, as opposed to features obtained from neural networks. We extract and compare features from pathological and healthy control kidneys to learn how the compartments (glomerulus, Bowman's capsule, tubule, interstitium, artery, and arterial lumen) are affected by the pathology. We define feature selection methods to extract the most informative and discriminative features. We perform statistical analyses to understand the relation of the extracted features, both individually, and in combinations, with tissue morphology and pathology. Particularly for the presented case-study, we highlight features that are affected in each compartment. With this, prior biological knowledge, such as the increase in interstitial nuclei, is confirmed and presented in a quantitative way, alongside with novel findings, like color and intensity changes in glomeruli and Bowman's capsule. The proposed approach is therefore an important step towards quantitative, reproducible, and rater-independent analysis in histopathology.
Keywords: Feature extraction, Pathomics, Histopathology
Introduction
Histological images, especially high-resolution Whole Slide Images (WSIs), contain a wealth of information that is not exploited to its full potential in current clinical routine. Conventionally, trained pathologists examine histopathological slides under a microscope to identify biomarkers and draw diagnostic conclusions. Through years of training, experts intuitively learn to extract visual features which aid the diagnostic process from the images. However, visual examinations are often subjective and may lack the support of concrete quantitative analyses. Furthermore, not all features are easily discernible to the human eye. Such features are often referred to as “sub-visual” features.41 Although differences in healthy and pathological cases are studied, it is difficult to obtain a thorough quantitative representation of these differences in clinical routines.
Typical tasks in histological examinations include detection,4,1,46 segmentation,12,47,5 and quantification6,13,47 of tissue structures of interest, image registration,14 and classification.2,17,15 Quantification of tissue structures involves the extraction of features characteristic to the data in consideration. Most of these tasks are extremely labor-intensive and time-demanding. However, recent advancements in the field are seeking to address the limitations presented by manual methods. In histopathology, digitization caused the field to grow rapidly, enabling the development of Artificial Intelligence (AI) techniques for automating manual tasks. Most recently, pathomics18,19 is emerging: an omics approach in digital pathology that aims at capturing quantitative features from WSIs to characterize the phenotypic features of tissues. In the context of the analysis of Glioblastoma, Lehrer et al. refer to this approach to as ‘histomics’.43 Here, they discuss the potential of discriminating between glioma subtypes with the help of morphometric features extracted from the nuclei.
Clinical analyses of histopathology images are traditionally based on descriptors obtained by domain knowledge of the experts. For instance, the presence, appearance, and number of objects like nuclei and cells form the basis of diagnosis criteria used to distinguish between healthy and pathological samples. In computer vision, traditional feature extraction methods are broadly categorized into classical image processing methods and learning-based methods. The former include feature desciptors such as color histograms, scale-invariant feature transform (SIFT),20 local binary pattern,15 and gray-level co-occurrence matrix (GLCM).21 In addition, prior domain knowledge is utilized to obtain histopathological image representations.17,22
Learning-based methods, on the other hand, involve feature extraction with the help of AI techniques, where raw image data is used to extract features with auto-encoders and Convolutional Neural Networks (CNNs).16,17 However, these features are mostly neither interpretable nor easily explainable. In this context, it is important to understand the difference between the terms “interpretability” and “explainability” in AI.23 “Interpretable” models are those where the output relates directly to the input and any changes in the input will lead to a predictable output. This is possible because the model parameters can be understood by human beings without additional aids or techniques. However, such models are often domain-specific and hence, constrained by domain knowledge. On the other hand, “explainable” models are those where the model parameters are too complicated to be understood by humans without additional aid. A second model (posthoc) is required to explain why a certain decision has been made by the model. Decision trees and linear regression models are examples of interpretable models while random forest and CNNs fall under the category of explainable models.23 These characteristics of newly proposed methods are becoming increasingly important, especially in medicine, where trustworthy AI is crucial to progress.42 According to Hasani et al,42 AI systems in healthcare should empower physicians in making informed decisions. This can, for example, be achieved through interpretable features. Hence, compared to AI systems as “black box” models, an algorithm with understandable outputs is preferable.
In this study, we will extend and follow these definitions for interpretability and explainability for features. Recently, the terms “hand-crafted pathomics” and “discovery pathomics” were proposed for feature extraction based on domain knowledge and for AI-based feature extraction, respectively.18 In contrast to most learning-based algorithms, hand-crafted pathomics produces interpretable features, hence this work focuses on this approach.
State of the art
Most studies in digital histology focus on cancer tissue,26,43,44,24,25 especially breast cancer.17,19,27 This is because cancer tissue requires annotations only on a specific tumor tissue, whereas non-tumor tissue presents several variations and combinations of normal and abnormal cells.18 Another exceptional challenge in digital histopathology is the variability of tissue types (e.g., epithelium, nervous, or connective tissue), as well the variabilites among organs under observation. This often necessitates the development of data-specific methods. Only a few research groups across the world focus on renal histopathology.18 Interestingly, a high focus is given to glomeruli detection and segmentation.1, 2, 3, 4,7,9,10,12,46,47 Specifically relevant to digital histology, the virtual translation of stains is also explored with high interest.5,8,11
Feature extraction is often performed with the aim of developing effective classification approaches, where the analyses of the extracted features themselves are limited in scope.16,28 A features-based method for the classification of pathological images showing renal cell carcinoma into different subtypes28 includes a simple color-based segmentation followed by morphological, wavelet, and texture analyses on the images. The analyses are combined to develop a robust classification system based on a simple Bayesian classifier. Although the method involves the extraction of several kinds of features, the focus is restricted to the classification task. It does not incorporate defining new features or explaining existing features in providing novel clinical insights about the tumor itself. Similarly, most current literature focuses on CNN features, which are highly effective for a given task, but due to their uninterpretability, fail to aid pathologists with novel insights on the tissue.16 Unlike the focus of this work, these features are not interpretable. One study that takes a step closer towards interpretability is presented with pathological images of uterine cervix.29 Here, the authors train a network with representative pathological features in order to understand the decisions made by the network.
In this shift of focus towards AI-based approaches, pathomics is still in its infancy. It is categorized into hand-crafted or conventional pathomics and discovery pathomics.18 While the former relies on domain knowledge to extract digital signatures from raw data, the latter uses CNNs to extract features from images. Unlike task orientated AI-based approaches, pathomics focuses on feature extraction and analysis, i.e. while such AI approaches may behave as a classifier, pathomics behaves as a feature extractor providing a structured representation of the input image data.
Yamamoto et al. presented a method that exploited features to obtain an objective measurement of pleomorphism and/or heterogeneity.13 They calculated several GLCM based texture features on nuclei segmentations and discussed their correlations with heterogeneity. Another study focusing on glioblastoma, extracted morphological features form the nuclei to demonstrate the correlation between phenotypic groups and clinical outcomes.44 They focused on understandable features and showed that nuclear circularity and eccentricity were strongly correlated with the oligodendrocyte gene expression. The authors also proposed a tool, HistomicsML, to support similar histology analyses.45 In another study,9 the authors segmented glomerular compartments and extracted features with the aim of classifying diabetic nephropathy. The authors extract color, textural, morphologic, containment, interstructural, and intrastructural distances as features. These studies are similar to our work, but with some limitations. While Yamamoto et al and Kong et al13,44 are restricted to nuclei segmentations and Ginley et al9 to a specific kidney compartment, namely the glomerulus, we extract features from several important compartments in the kidney. Moreoever, Yamamoto et al13 is limited in scope only to heterogeneity, Ginley et al9 in the classification of diabetic nephropathy, and Kong et al and Nalisnik et al44,45 in the classification of tumor regions. While we are interested in finding general differences between any two tissue types under observation. Hence, we extract several categories of features in this work.
Contribution
The contribution of this work is two-fold: (1) a novel pipeline for the extraction of features that allow meaningful analysis for pathologists and (2) a detailed case-study on kidney histopathology.
(1) We propose a pipeline to extract a large variety of interpretable features from renal WSIs. Specifically, we extract several first-order statistical (intensity), shape, color, textural, and task-specific morphological features. The features are extracted from important kidney compartments, including glomerular tuft, Bowman's capsule, tubule, interstitium, artery, and arterial lumen, of pathological as well as contralateral kidneys of mice. A comparative study enables us to highlight how individual compartments are affected by the pathology. We perform statistical analysis to understand the relation of the extracted features, both individually, and in different combinations, with the underlying tissue morphology and pathology. We also define feature selection methods to extract the most informative and discriminative features to obtain clinically relevant information about the data. As such, our contribution to the field does not lie in providing WSI-level features that are as discriminative as possible (as neural networks would do). Instead, we highlight the potential of extracting meaningful features that can aid pathologists in understanding kidney pathologies in more detail. Moreover, the pipeline is generic and can be applied to any histopathological datasets.
(2) As a case study, we evaluate the pipeline on a murine kidney disease model, Unilateral Ureteral Obstruction (UUO). Here, the induced pathological condition can be assessed in comparison with the non-manipulated healthy contralateral kidney in individual mice. We show that pathologies have a clear influence on certain features confirming expert workflows in diagnosing the condition. Particularly, we analyze how UUO affects individual compartments and provide a comprehensive list of individual features that are affected in each compartment. With our pipeline, we confirm the prior biological knowledge that in UUO, a model used to induce tubulointerstitial damage, tubules and the interstitium are the most strongly affected compartments. We also find significant differences in certain features from glomerular tuft and Bowman's capsule. Specifically, we find significant differences (p ≤ 0.005, with p denoting the Bonferroni-corrected p-value for n = 5 comparisons48) for intensity, color, and texture features in these two compartments. These findings prove that the features and analyses proposed in this study enable us to the identify pathological alterations caused by the disease, providing novel insights.
Methods
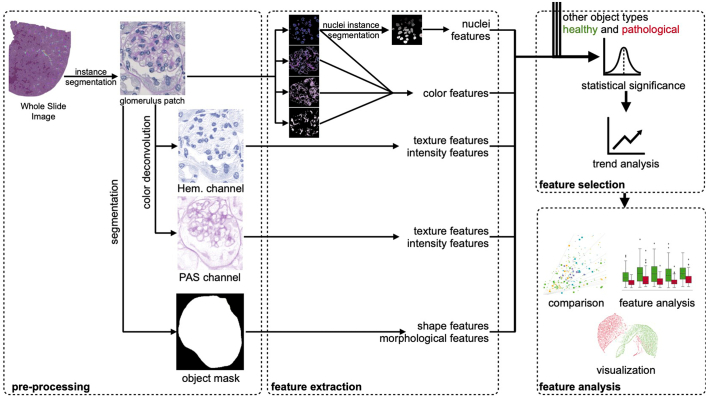
The individual processing steps of our pathomics approach comprise pre-processing, feature extraction, feature selection, and evaluation (see Fig. 1). The steps are outlined in more detail in the following subsections. Fig. 1 shows the proposed pipeline in a schematic manner with a glomerulus as an example of a segmented compartment. Henceforth, we will refer to kidney compartments as “objects”.
Fig. 1.
Schematic representation of the proposed pipeline. Pre-processing includes automatic segmentation of renal structures, color deconvolution, and instance labeling based on the labels of the WSIs. Several types of features are extracted from the segmented objects and after feature selection, feature analyses are carried out based on healthy and pathological samples.
Image data
The image data used in the study comprises 10 WSIs obtained from 5 resected mouse kidneys. The samples were taken from previously published animal experiments performed according to the regulations of the local and national ethical committee for animal welfare (Landesamt für Natur, Umwelt und Verbraucherschutz Nordrhein-Westfalen (LANUV NRW))30,31 and the study is reported in accordance with ARRIVE guidelines. LANUV NRW provided the ethical approval. In short, C57BL/6N mice (Charles River, Germany) were housed under SPF-free conditions with constant temperature and humidity under a 12-h phase light-dark cycle with free access to drinking water and food. The UUO surgery was performed under anesthesia by ketamine (14 g/kg body weight) and xylazine (8 g/kg body weight) following analgesia with Temgesic (0.05 mg/kg) until 72 h post-OP. On day 7, animals were sacrificed and perfused with saline, and kidney tissue was processed for histological examination.
Tissue was fixed in methyl Carnoy's solution and embedded in paraffin. Paraffin sections (1 μM) were stained with Periodic Acid-Schiff (PAS) reagent and counterstained with hematoxylin. For digitization of the slides, whole-slide scanners NanoZoomer HT2 with 20× objective lens (Hamamatsu Photonics, Hamamatsu, Japan) were used. Details on the tissue preservation method are as described previously by Boor et al.32 The obtained WSIs have sizes of around 50,000×40,000 pixels each.
UUO is a commonly used experimental model to cause renal fibrosis, the common pathway for most progressive renal diseases. The general progression of disease with time in case of UUO is clinically well-known,33 making this model suitable for the evaluation of novel features. Furthermore, this model affects only one kidney, allowing the other contralateral kidney to serve as the healthy control from the same subject, minimizing inter-subject variations.
Pre-processing
In the following, we describe the pre-processing steps, namely instance segmentation and labeling.
Instance segmentation
As a prerequisite to feature extraction from individual kidney compartments, we obtain a complete segmentation of the healthy and the contralateral kidneys. In particular, we segment the following kidney compartments: glomerular tuft (glomerulus), Bowman's capsule (BMC), tubule, extra-cellular matrix (interstitium), arterial blood vessels, and their lumen. Considering the large resolutions of the WSIs, it is infeasible to obtain manual segmentations. Hence, for this purpose, we use an existing deep learning-based method.6 The authors used a U-Net34-based method and report high accuracies in the segmentation of structures not only for healthy WSIs, but for UUO WSIs as well. For the former, they achieve instance segmentation accuracy of above 94% for glomerular tuft, BMC, and tubule in healthy WSIs, and around 90% in UUO WSIs. They report scores of around 88% and 80%, for arteries and arterial lumen, respectively, in case of healthy WSIs and around 75% and 82%, respectively, in case of UUO WSIs.
Instance labeling
The second step is labeling the obtained instance segmentations as ‘healthy’ or’UUO’. To keep the manual efforts minimum wherever possible, we adopt a simple approach to obtain instance labels automatically: we assign each object instance the same label as that of the WSI from which it is segmented. These instance labels serve as ground truth for all analyses. Although some objects or object instances may not reflect the global WSI label, it is expected to be the case for the majority of them. This means that not all instances of a given object in a pathological WSI may be affected and some may still be in a healthy condition. This also applies to healthy WSIs as some object instances may not be completely healthy due to natural degeneration, for instance. However, in case of UUO at day 7, the pathology affects most objects quite significantly, allowing our label assignment method possible. In the case of healthy WSIs, on the other hand, normally only a few instances can be expected to be in pathological condition (outliers).
Feature extraction
We extract several features from the objects segmented in the pre-processing step and group them into the following types. The exact number of features in each feature type is indicated in brackets.
-
1.
Intensity features (19)
-
2.
Textural features (75)
-
3.
Shape features (10)
-
4.
Morphological features (13)
-
5.
Color features (8)
-
6.
Nuclei-related features (7)
For the first 3 feature types, we rely on the PyRadiomics implementation35 for 2D images with default parameters. It consists of a comprehensive and largely standardized set of features that capture information related to the intensity distribution of Radiology images, the shape of the objects of interest, as well as their texture.
Based on expert domain knowledge, we additionally propose a set of data-specific features that capture clinically relavant characteristics, often employed in the diagnosis of renal pathologies. These comprise morphological, color, and nuclei-related features. These features are typically investigated by pathologists either qualitatively or quantitatively. A complete list and definitions of all the data-specific features are provided in Supplementary Tables 1 and 2.
Stain deconvolution
PyRadiomics is designed to extract features from radiology images, which are typically gray scale images. Hence, we adapt our images to benefit from the standardized features provided by PyRadiomics by perfoming stain deconvolution as described below.
In histological images, stains play an important role and carry relevant clinical information. To extract meaningful features for such images, we firstly perform stain deconvolution and separate the images into their constituent stain components. For this purpose, we rely on Macenko's method36 although several other methods exist, such as Ruifrok and Johnston.37 This is because Macenko's method provided excellent results in preliminary experiments. The method performs color deconvolution using the following fixed optical density matrix (Q) for separating Hematoxylin, PAS and residual color channels, respectively.
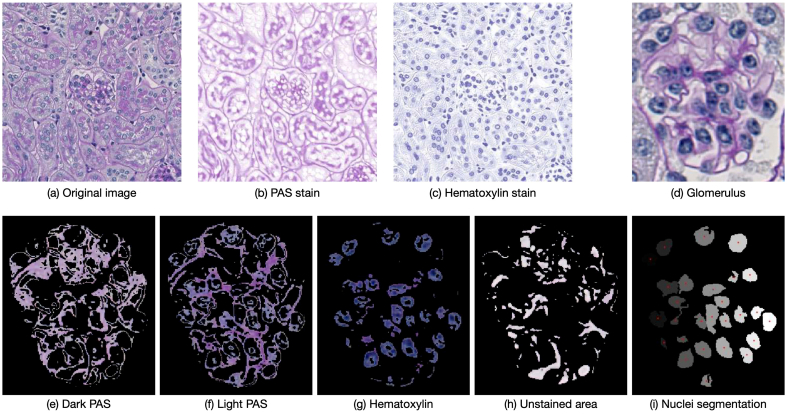
Fig. 2(a)–(c) show a small patch from a WSI deconvolved into the constituent PAS and Hematoxylin stains.
Fig. 2.
Example images showing stain deconvolution (a)–(c) and color segmentation using k-means clustering (d)–(h). An original image (a) is deconvolved into the consitituent stain components namely, PAS (b) and hematoxylin (c). Color segmentation is shown with the example of a glomerulus (d), where four color components are obtained, namely the dark PAS (e) and light magenta (f) stained regions, the regions stained with hematoxylin (g), and the unstained regions (h). Figure (g) is subsequently used in nuclei segmentation (i) using the watershed algorithm. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Intensity and textural features
Intensity and textural features are calculated on each of the two stain channels separately. Intensity features comprise first-order statistical information that describe the distribution of pixel intensities within the object. Each object instance is extracted as a bounding box image and is overlayed with the segmentation mask to calculate features within the masked region. 75 textural features are calculated from matrices including gray level co-occurrence (GLCM), neighborhood gray tone difference (NGTDM), gray level dependence (GLDM), run length (GLRLM), and size zone (GLSZM) matrices.
Shape and morphological features
As shape descriptors are independent of gray value, shape, and morphological features are extracted from the object mask. The shape and size of each object instance may vary, resulting in varied shape and sizes of the object mask itself. Hence, here the image sizes are not consistent, rather vary according to the bounding box of the object instance. We extract 10 shape and 13 morphological features. We differentiate between shape and morphological features in the way they are calculated. While shape features are based on the PyRadiomics implementation, we define the following additional morphological features: area and perimeter of the segmented contour as well as its convex hull. The calculation of convex hull of an object instance also allows us to define features including solidity and convexity, defined as the ratios between the instance area and perimeter to their respective convex area and perimeter, respectively. Furthermore, we also define features based on the minimum ellipse enclosing the segmented contour of the object mask. These include the lengths of the major and minor axes, object eccentricity, and elongation. Similarly, a feature rectangularity is defined based on the minimum enclosing bounding box as the ratio of the object area to the area of the box. Equivalent diameter is calculated as the diameter of the circle with the same area as the object. Lastly, compactness is defined as the ratio between the object area to the area of a circle with equal perimeter.
Color features
To calculate color features, we further sub-divide the PAS stain into light and dark magenta color components, as light magenta and dark magenta stain are known to dye different tissue components, such as the cytoplasm and cell membranes, respectively.
We obtain the following four colors for calculating color-related features: light magenta, dark magenta (PAS), blue (hematoxylin), and white for the unstained regions (see Fig. 2(d)–(h)). A clean separation of light magenta and dark magenta is not possible by means of stain deconvolution. For this purpose we compared two methods, a naive Otsu thresholding-based approach and a k-means clustering approach. Both methods are “trained” on a set of similar images not used in further experiments to obtain suitable thresholds or cluster centers, respectively. These are fixed and used to perform the color separation, ensuring consistent results across all images. Preliminary experiments showed superior performance with the k-means clustering approach, so we follow this approach for color segmentation. As color features, we calculate the number of pixels of a given color and the color density in the object instance. In total, this provides eight features with two features per color. Furthermore, we use the blue channel obtained as such for performing nuclei segmentation to calculate seven nuclei-related features as described in the following section.
Nuclei segmentation and related features
To extract nuclei-related features, we firstly perfom a nuclei segmentation on all the objects. For nuclei segmentation, we use the hematoxylin stain image obtained by color segmentation using k-means clustering (see Fig. 2(g) and Fig. 2(i)). The image is first pre-processed with morphological operations whereby small holes are filled and noise is removed. We use the watershed algorithm38 to ensure effective segmentation of overlapping objects, such as nuclei. For this, we compute the Euclidean distance transform to find the local peaks, followed by H-Maxima transformation and any local maxima smaller than a certain threshold (maximum of distance transformation) are suppressed. The maxima serve as markers for applying the watershed algorithm, whereby the individual nuclei, represented by the local minima, are segmented and labeled. Only in rare cases over- or undersegmentation could be observed, however, this is handled implicitly in the statistical analysis. Although this is a rather simple approach for the segmentation of nuclei, the focus in this work lies in the integration of this important sub-cellular organelle in a pathomics pipeline.
Nuclei-related features are then calculated, which include the number and density of nuclei in a given object instance. As the number of nuclei within each object varies highly, we calculate statistical features about nuclei neighbors. For a given nucleus, the neighbors of this nucleus are defined as the nuclei within a certain radius r around it (see Supplementary Fig. 2). The radius r is defined as two times the average major axis length of all nuclei in the given object instance. As the number of such neighbors is determined for each nucleus in the given object instance, statistics can be derived. In this case, the minimum, maximum, mean, medium, and standard deviation of the number of neighboring nuclei over all nuclei in the object are used.
As explained earlier, the nuclei segmentation approach deployed here does not yield a segmentation accuracy suitable for pixel-level features, such as texture and morphological features. Hence, morphological and texture features are not calculated for the nuclei.
Feature selection
Feature selection is an important step in determining the most discriminating features that highlight the differences between healthy and pathological data. We extract a number of features per object, which may only be partly useful in discriminating between the healthy and pathological samples. In fact, it may be the case that each object has a different set of discriminating features and non-discriminating ones. In such a case, the non-discriminating features may be misleading in the understanding of the effects of pathology and must be eliminated before the feature set is employed for further analysis. To select the best-suited features which carry the most discriminative information per object, we propose to use a combination of the following selection methods.
Statistical tests
With the help of statistical tests, we retain only those features which have significant differences between the healthy and the pathological WSIs. For this purpose, we compare values from individual features extracted from the pathological and the contralateral kidney of the same subject. It is important to note that we compare feature from the same subject to prevent inter-subject variations. To this end, we first determine whether our samples have equal variance using Lavene Equal Variance test. If this test shows equal variance, we perform a Student's t-test with the assumption of equal variance, otherwise with the assumption of unequal variances. We define significance level of p ≤ 0.005 to best note the differences (as compared to p ≤ 0.05, for instance). Note that p refers to the Bonferroni-corrected p-value with n = 5 comparisons, where n is the number of WSI-pairs. We follow this definition throughout this paper.
Trend analysis
In addition to statistical tests, we also analyze the relative trend between the healthy and the pathological samples. For a feature to distinguish reliably between the classes, it must consistently show the same effect. For example, if a feature is typically higher in one WSI of class “healthy” as compared to the class “pathological”, it must be higher in all other healthy WSIs as compared to the corresponding pathological ones. This ensures minimization of inter-subject variations. For this purpose, we calculate the difference in medians of the individual features from the two classes. If the difference is always positive or always negative for all WSIs of one class, this feature is selected for further analysis.
Hence, in this step, we only select those features which exhibit a consistent trend across all WSIs as well as show statistically significant differences (p ≤ 0.005) for at least three WSIs.
Experimental setting
We describe the experimental setup and several measures for the evaluation of the presented pipeline in the following:
Comparison of individual features
First of all, we evaluate the discriminativeness of individual features by comparing the distribution of feature values obtained from similar objects from individual subjects. This means we compare whether a feature shows differences between the pathological and the contralateral kidneys of the same subject with respect to an object. For visual inspection and to provide a statistical representation, we use boxplots. These plots show detailed information on the differences between individual objects which could provide important clinical insights. Since the features are interpretable, the differences can also be characterized qualitatively.
Here, we rely upon WSI pairs for comparisons. This means we compare the pathological and healthy contralateral within same subjects only. We do not aggregate the healthy WSIs into one class and the pathological into another to avoid intra-class variations, i.e. the differences in trends of feature values within the same class. A statistically representative aggregation would only be possible with higher number of WSIs per class.
We employ the feature selection techniques introduced under Section Feature selection to eliminate the least informative features. This also facilitates a more efficient analysis.
Standard score
In order to evaluate the discriminativeness of the selected features, we use the standard score, or the z-score, which is a measure of the deviation from the mean in terms of standard deviation. More specifically, here we define the z-score as follows:
where, and indicate the arithmetic means of individual features and objects for the pathological and the healthy WSIs, respectively, and σh indicates the standard deviation of the healthy WSI.
We calculate the z-score per object and per feature for each pair of WSIs, i.e. each subject. This yields 5 z-score values per feature per object. Of these 5 values, the mean and standard deviation are computed, denoted as and , respectively, and they indicate the discriminativeness of that feature for the given object. This allows us to make a comparison among all objects as well as all feature types at the same time.
Visualization of feature discriminativeness
For a visual analysis of the discriminativeness of the features, we perform dimensionality reduction on the selected features to obtain two-dimensional (2D) representations. For this purpose, we make use of the Uniform Manifold Approximation and Projection (UMAP) method.39 It is a method for dimensionality reduction that improves upon the state-of-the-art method t-Distributed Stochastic Neighbor Embedding (t-SNE)40 by preserving the global structure while having faster run time performance. Due to the limited dataset size in this study, we adopt the supervised UMAP approach, whereby the labels of the objects are available during the clustering process.
Results
In all the figures (except in Fig. 4), the healthy data is presented in green and the pathological (UUO) in red. The 5 subjects are labeled from 1 to 5.
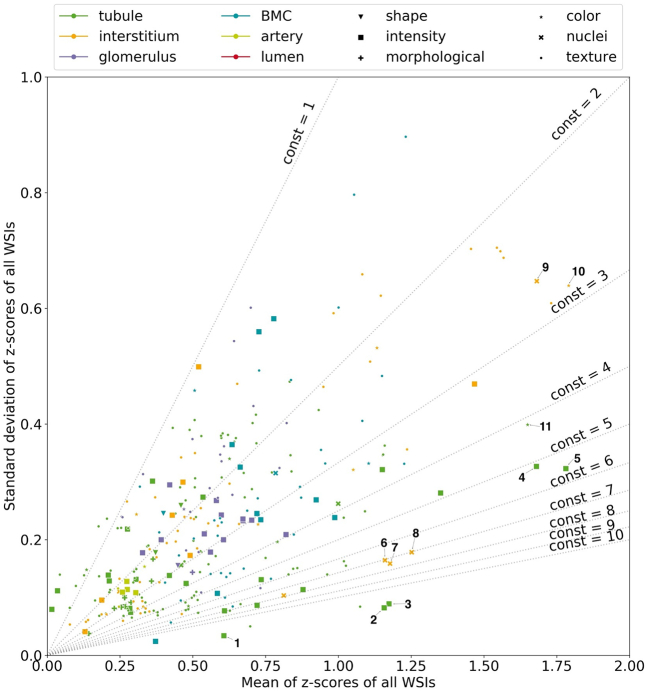
Fig. 4.
Comparison of feature types across all objects. Feature types are represented by different marker shapes and objects by different colors. Z-score of a feature is the ratio of the difference between its pathological and healthy cases to the standard deviation of the healthy feature. The X-axis shows the mean z-scores () across all WSIs and the Y-axis their standard deviation (). The dotted lines represent isolines with the relation mz/sz = const.
Selection of the most discriminative features
The most discriminative features are selected with the help of 2 criteria, namely the statistical significance and the trend between the healthy and UUO kidneys from the same subject. An example of a feature that was eliminated in the feature selection step is shown in Fig. 3(c). The figure highlights the inter-subject variation between the healthy and pathological WSIs. For instance, higher median values are observed in the healthy tissue for WSIs 1 and 4 as compared to the pathological WSIs, while a reverse trend is observed for subjects 2, 3 and 5. Features which did not exhibit the same trend across all WSIs were considered unreliable and were hence eliminated during the feature selection step. Table 1 provides an overview of the selected and the total number of features for all the objects.
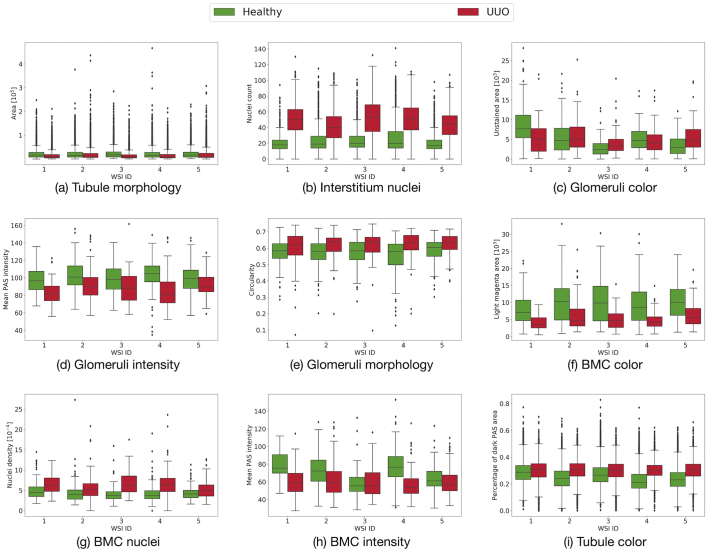
Fig. 3.
Individual features per object. The X-axis shows the 5 WSIs and the corresponding feature values are shown on the Y-Axis, with green representing healthy and red, UUO. Examples are taken of the tubule surface area as morphological feature (a), number of nuclei in the interstitium (b), unstained area in glomeruli as color feature (c), mean PAS intensity in the glomeruli (d), glomeruli circularity as morphology feature (e), area occupied by light magenta color in BMC as color feature (f), nuclei density within BMC (g), mean PAS intensity in the BMC (h), and the percentage of dark PAS area in tubule as color feature (i). Note that while all other examples show statistically significant differences (p ≤ 0.005), (c) is an example where no consistent significant differences (p ≤ 0.005) were found and was ultimately rejected in the feature selection step. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Table 1.
Overview of the selected and total number of features for all the objects. Each cell shows the number of selected/total number of features for the respective object and feature.
| Object | Features |
|||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Morphological | Shape | Color | Nuclei | PAS stain |
Hematoxylin stain |
|||||||||||
| Intensity | GLCM-based texture | GLRLM-based texture | GLSZM-based texture | GLDM-based texture | NGTDM-based texture | Intensity | GLCM-based texture | GLRLM-based texture | GLSZM-based texture | GLDM-based texture | NGTDM-based texture | |||||
| Glomerulus | 1/13 | 1/9 | 0/8 | 0/7 | 7/18 | 3/22 | 4/16 | 2/16 | 5/14 | 1/5 | 4/18 | 2/22 | 0/16 | 1/16 | 0/14 | 0/5 |
| BMC | 0/13 | 1/9 | 3/8 | 1/7 | 4/18 | 3/22 | 7/16 | 5/16 | 6/14 | 1/5 | 6/18 | 2/22 | 2/16 | 1/16 | 0/14 | 1/5 |
| Artery | 0/13 | 0/9 | 2/8 | NA | 0/18 | 0/22 | 0/16 | 0/16 | 0/14 | 0/5 | 2/18 | 0/22 | 0/16 | 0/16 | 0/14 | 0/5 |
| Lumen | 0/13 | 0/9 | 0/8 | NA | 0/18 | 0/22 | 0/16 | 0/16 | 0/14 | 0/5 | 0/18 | 0/22 | 0/16 | 0/16 | 0/14 | 0/5 |
| Tubule | 8/13 | 7/9 | 5/8 | 2/7 | 10/18 | 14/22 | 8/16 | 9/16 | 8/14 | 3/5 | 15/18 | 19/22 | 10/16 | 13/16 | 7/14 | 5/5 |
| Interstitium | NA | NA | 6/8 | 6/7 | 2/18 | 9/22 | 4/16 | 3/16 | 4/14 | 2/5 | 6/18 | 12/22 | 4/16 | 6/16 | 3/14 | 3/5 |
In the Supplementary Material, the features extracted from all objects are presented (see Supplementary Figs. 3–12). Features per object are highlighted in purple color if they are found to be significantly different (p ≤ 0.005) between the healthy and the pathological WSI. If no significant differences (p ≤ 0.005) are found between them, they are highlighted in light blue. In addition, the highlighted cells are numbered either −1 or 1 depending on the trend they exhibit. If the median of the features from a healthy WSI is higher than that of the corresponding UUO WSI, it is marked as 1, otherwise as −1. Finally, the features that are selected according to the feature selection method are outlined within black boxes. A feature is selected if it is significantly different for at least 3 of the 5 subjects as well as if all the 5 subjects exhibit the same trend for the feature, i.e. if the indicated values are either all positive, or all negative. These are considered as the most discriminating and reliable features. Hence, a detailed information about each feature per object is provided. Further observations are discussed under Section Discussion.
Object-specific assessment of individual features
Individual features from every segmented object are compared across the 5 subjects. Examples of discriminative and non-discriminative features chosen from the various feature types for different objects are shown in Fig. 3. In Fig. 3(a), the morphological feature surface area of tubules is shown. The plot shows significantly higher values and higher inter-quartile range for the healthy WSIs as compared to the pathological WSIs. Another tubule feature, namely the color feature percentage of dark PAS is shown in Fig. 3(i). The figure shows higher values of the feature in UUO WSIs compared to the healthy ones. Fig. 3(b) shows how the nuclei feature nuclei count compares in the interstitium of healthy and UUO WSIs. The plot shows a significant difference (p ≤ 0.005) with a much higher number of nuclei in UUO WSIs. Furthermore, the inter-quartile range and the standard deviation are visibly higher in UUO. In both these features, the trend comparing the two classes is the same for all WSIs, this means the median values in all the 5 WSIs in Fig. 3(a) is lower for UUO as compared to healthy WSIs and in 3(b), the median values are higher for UUO. However in Fig. 3(c), which shows the color feature unstained area for glomeruli, this is not the case.
Fig. 3 shows more examples of individual feature analysis, which include intensity, color, morphological and nuclei-related features for the objects glomeruli and BMC. The morphological feature circularity for glomeruli in UUO shows higher maximum and minimum values for UUO WSIs in comparison to healthy ones (see Fig. 3(e)). This means the former are more spherical than the healthy ones. This observation is supported by BMC shape features, which show significantly lower maximum diameters in UUO (see Supplementary Fig. 1). In UUO, glomeruli consistently have lower intensity values for the PAS stain as indicated by their first order stastistics including maximum, mean, median, 10th and 90th percentile (see Fig. 3(d) and Supplementary Fig. 1). This means that UUO glomeruli have more areas showing darker PAS stain than the healthy ones, which have comparatively more lighter PAS stain as well as unstained regions. BMC color features also show significantly smaller areas of light magenta color in UUO as compared to the healthy ones (see Fig. 3(f)). This is also evident by the lower mean values for the PAS intensity in UUO (see Fig. 3(h)). In addition, the nuclei density in healthy and UUO within BMC also changes significantly (see Fig. 3(g)) with much higher values in UUO WSIs.
Feature visualization and dimensionality reduction
In order to visualize and compare the different types of features and the objects, the standard score was defined. In Fig. 4, the absolute mean z-scores () across all WSIs are plotted on the X-axis and the standard deviation () on the Y-axis for all feature types and objects. Feature types are represented by different marker shapes and objects by different marker colors. The dotted gray lines indicate isolines with the relation mz/sz = const, where const takes the values from 1 to 10 as we move from the upper left corner to the bottom right corner of the plot. A high const value indicates that the difference between the classes is on average large compared to the standard deviation of this difference. All the features lying below a given isoline have the same relationship between their means and standard deviations. Here, only those features are shown that fulfilled the feature selection criteria. According to the definition of z-score, the most discriminating and reliable features are represented with higher values on the X-axis and lower values on the Y-axis, i.e. with higher const values.
In Fig. 4, some markers are annotated with numbers. These serve as examples to highlight differences between individual objects as well as feature types. For instance, the markers numbered 2–5 clearly show that intensity features are highly discriminative for tubules because these have high mean values with a relatively low standard deviation. Uniformity (2) and entropy (3) measure the homogeneity of the hematoxylin intensity. Both indicate strong texture variations between healthy and UUO tubules. This observation is further supported by robust mean absolute deviation (4) and inter-quartile range (5). The high mean values on the X-axis for these features indicate a high difference in the intensity distribution of the hematoxylin stain in tubules between the 2 classes. While the uniformity of the PAS intensity (1) is also useful, a lower X-axis value indicates smaller differences between healthy and pathological samples as compared to (4).
The figure further shows nuclei-related features of the interstitium with large differences between healthy and pathological samples, represented by large mean values on the X-axis. Examples are median (6), maximum (7), and mean (8) of the number of nuclei neighbors as well as the number of nuclei (9) in the region. Marker 10 represents the light magenta color area in the interstitium and marker 11, the percentage of the area of this stain in a tubule instance relative to the area of whole object instance.
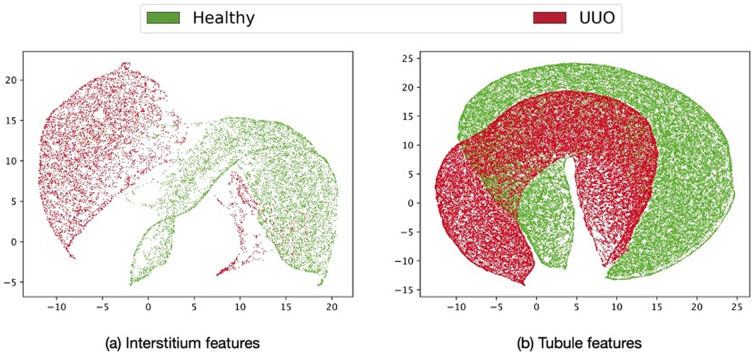
The results of dimensionality reduction using the UMAP method are shown in Fig. 5. For dimensionality reduction, features only from the two most discriminative objects, namely the interstitium (Fig. 5(a)) and the tubule (Fig. 5(b)) are considered, and only the relevant features obtained after the feature selection step are used. While the former shows 2 separable clusters for the healthy and the pathological classes, the latter shows inseparable clusters.
Fig. 5.
Visualization of feature discriminativeness via dimensionality reduction using UMAP method. Features from healthy WSIs are indicated in green and those from pathological ones in red. Clustering of relevant features from the interstitium (a) and tubule (b). Interstitium features form separable clusters while tubule features show some overlap. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Discussion
UUO is an experimental model with known pathological modifications and the study confirms the prior knowledge that tubules and interstitium are highly affected in UUO.33 We provide a comprehensive set of features that are affected in these renal structures. Interestingly, we also find some significant differences (p ≤ 0.005) in the Glomeruli and BMC which have so far not been reported in clinical literature to the best of our knowledge. These include some intensity features which show differences in light magenta or dark PAS stain, as well as in the unstained areas. Although further research is necessary to confirm the pathological causes, but an explanation for the lower unstained regions in UUO could be due to the pressure from the expanding tubulo-interstitium on these objects. In addition, in glomeruli, there is less perfusion of the glomerular capillaries since most of the filtration on day 7 of UUO occurs in the contralateral (healthy) kidney. This is also true for BMC and accounts for the smaller size of the objects in UUO as compared to the healthy WSIs. However, we also noted high standard deviations in the BMC features which justify the lack of clinical findings for this object. This could be accounted for by the possibility that not all glomeruli may be affected in pathological tissue. If only a few object instances exhibit an altered pathological condition, it is difficult to obtain reliable results without instance labelling, i.e. without the label of “healthy” or “UUO” per object instance. In such a case, further analysis based on object-level annotations is necessary.
Some commonly known effects of UUO in the renal structures are clearly represented via the extracted features. The percentage of dark PAS stain is consistently higher in the pathological tubules, which could be explained by the thickened tubular basement membrane in case of UUO. Tubular atrophy could result into prominent flattening of the tubules, which is explainable by the lower morphological area of the pathological class as compared to the healthy one. The nuclei count in the interstitium of pathological tissue is significantly higher than in healthy tissue. This is very likely due to inflammation and accumulation of interstitial fibroblasts and myofibroblasts.
A comparison of the feature types shows that while most of these categories have some highly discriminative features, the category of textural features contains the most relevant features. This is particularly the case for tubules and interstitium. For the latter, nuclei-related features and for tubule, intensity features are highly discriminating between the healthy and UUO WSIs. Surprisingly, shape features are only marginally informative. However, since they can be easily defined and calculated manually, a high focus is given to these by pathologists during manual analysis. Texture features, on the other hand, which are more relevant, are difficult to define and extract manually, and are hence neglected in manual analysis.
The proposed pipeline makes it possible to extract several types of interpretable and clinically relevant features, as well as the selection of the most discriminative features for automatic and manual analyses. The pipeline is easily applicable to new datasets and domains, such as cancer pathology. A holistic comparison of the several feature types across all objects provides additional insight into the modifications caused by pathologies. Hence, the pipeline can assist pathologists in improving diagnosis by increasing their understanding of the pathology.
An important information obtained by analyzing the individual features per WSI was the high inter-slide variability. For several features, the 5 WSIs showed varying trends in the behavior between the features extracted from the healthy and pathological kidneys. As the data in this study was limited, this behavior accounts for the lack of a classification model trained on the extracted features.
As UUO is an experimentally established model with well-known pathological changes in the tissue, we chose it to validate the effectiveness of the pipeline and the features extracted in this study. The quantitative analyses perfomed in this work confirm the prior knowledge about UUO that tubules and interstitium are the most highly affected anatomical structures in this pathological model. The majority of the features extracted from tubule and interstitium have significant differences (p ≤ 0.005) between classes with low standard deviations. This finding acts as a proof-of-principle for the extracted features as clinical studies confirm this outcome for UUO at day 7.
Limitations and future work
The pipeline could also be effectively used to study the progression of diseases by analyzing the differences among features extracted from images acquired at different disease stages. It would also be highly interesting to obtain other features, such as texture and morphological features, from the nuclei, which requires the availability of pixel level segmentations and should be the focus of future studies.
A limitation of this work is that it uses a small set of images obtained from a pre-clinical animal model. This is because it is aimed as a proof-of-concept study motivated by applications on preclinical models. Thus, the primary intention was to develop a pipeline for this use case with the aim of proving that an object level analysis has the potential to increase biological knowledge. Another limitation is the prerequisite of pixel level annotations for individual objects. For instance, due to the lack of nuclei segmentations, we were unable to extract all feature types from this object. An additional limitation is that we have not deployed and tested this approach on observational human data, which are much more variable and for which we do not have a sufficiently performing segmentation network.
The presented features could potentially also be used for classification problems. This, however, requires an aggregation of object-level features into WSI level features, or meta features, which requires higher number of WSIs. These meta-features, e.g. statistics like mean or maximum of object feature values, could then be employed to discriminatively describe an entire kidney or a biopsy section.
Conclusion
In this study, we extracted interpretable and quantitative features from renal WSIs of mice kidneys, including intensity, texture, shape, morphological, color, and nuclei-related features. While some of these features are data-specific, most of them are generic and applicable to most datasets. To the best of our knowledge, such a large-scale analysis of feature extraction has not been done before, especially on all the important compartments of renal WSIs. We also perform comprehensive feature analysis that identify novel phenotypes for improved diagnosis by increasing our understanding of pathological alterations. In addition, we provide statistical and visual comparisons of several feature types along with the comparison of the different components of the kidney. These developments could potentially serve as a framework for improving histopathological analyses of other tissues and organs as well as in other domains, such as cancer pathology.
The study clearly highlighted tubules and interstitium to be the anatomical structures that are affected in UUO, serving as a proof-of-concept for the pipeline in identifying the most discriminative features for a pathology. The dataset with paired healthy and pathological kidneys from the same subject allowed a detailed evaluation of features without influence of inter-slide and inter-subject variations. This enabled us to provide a fair evaluation on the discriminativeness of the features.
Author contributions statement
L.G and B.M.K. conceived the experiments, L.G. and N.F. conducted the experiments, B.M.K., C.S., P.B. acquired the data, N.B. generated the segmentation data, L.G., B.M.K., C.S., P.G., M.G., D.M., P.B., analysed the results, L.G. wrote the manuscript with support from P.G. All authors reviewed the manuscript.
Declaration of interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
This study was funded by the German Research Foundation (DFG) (project numbers: 233509121; 445703531; 322900939; 454024652), the “Exploratory Research Space (ERS)” of RWTH Aachen University (project number OPSF585), the German Federal Ministries of Education and Research (BMBF: STOPFSGS-01GM1901A), Health (DEEP LIVER, ZMVI1-2520DAT111) and Economic Affairs and Energy (EMPAIA), the European Research Council (ERC) under the European Union's Horizon 2020 research and innovation program (grant agreement No 101001791), and RWTH START-program (125/17 and 109/20).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jpi.2022.100097.
Contributor Information
Laxmi Gupta, Email: laxmi.gupta@lfb.rwth-aachen.de.
Barbara Mara Klinkhammer, Email: bklinkhammer@ukaachen.de.
Claudia Seikrit, Email: cseikrit@ukaachen.de.
Nassim Bouteldja, Email: nbouteldja@ukaachen.de.
Philipp Gräbel, Email: Philipp.Graebel@lfb.rwth-aachen.de.
Michael Gadermayr, Email: Michael.Gadermayr@fh-salzburg.ac.at.
Peter Boor, Email: pboor@ukaachen.de.
Dorit Merhof, Email: dorit.merhof@lfb.rwth-aachen.de.
Appendix A. Supplementary data
Supplementary material.
References
- 1.Bukowy J.D., Dayton A., Cloutier D., et al. Region-based convolutional neural nets for localization of glomeruli in trichrome-stained whole kidneysections. J Am Soc Nephrol. June 2018;29:2081–2088. doi: 10.1681/ASN.2017111210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gallego J., Pedraza A., Lopez S., et al. Glomerulus classificationand detection based on convolutional neural networks. J Imaging. Jan. 2018;4:20. [Google Scholar]
- 3.Kato T., Relator R., Ngouv H., et al. Segmental HOG: new descriptor forglomerulus detection in kidney microscopy image. BMC Bioinform. Sept. 2015;16 doi: 10.1186/s12859-015-0739-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Simon O., Yacoub R., Jain S., Tomaszewski J.E., Sarder P. Multi-radial LBP features as a tool for rapid glomerulardetection and assessment in whole slide histopathology images. Sci Rep. Feb. 2018;8 doi: 10.1038/s41598-018-20453-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.de Bel T., Hermsen M., Kers J., van der Laak J., Litjens G. In: Proceedings of the 2nd International Conference on Medical Imaging with Deep Learning. Cardoso M.J., Feragen A., Glocker B., et al., editors. vol. 102. PMLR; July 2019. Stain-transforming cycle-consistent generative adversarial networks for improved segmentation of renal histopathology; pp. 151–163. (Proceedings of Machine Learning Research,). [Google Scholar]
- 6.Bouteldja N., Klinkhammer B.M., Bülow R.D., et al. Deep learning–based segmentation and quantification in experimental kidney histopathology. J Am Soc Nephrol. Nov. 2020;32:52–68. doi: 10.1681/asn.2020050597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gadermayr M., Dombrowski A.-K., Klinkhammer B.M., Boor P., Merhof D. CNN cascades for segmenting sparseobjects in gigapixel whole slide images. Comput Med Imaging Graph. Jan. 2019;71:40–48. doi: 10.1016/j.compmedimag.2018.11.002. [DOI] [PubMed] [Google Scholar]
- 8.Gadermayr M., Gupta L., Appel V., Boor P., Klinkhammer B.M., Merhof D. Generative adversarial networks forfacilitating stain-independent supervised and unsupervised segmentation: a study on kidney histology. IEEE Trans Med Imaging. Oct. 2019;38:2293–2302. doi: 10.1109/TMI.2019.2899364. [DOI] [PubMed] [Google Scholar]
- 9.Ginley B., Lutnick B., Jen K.-Y., et al. Computational segmentation and classification of diabetic glomerulosclerosis. J Am Soc Nephrol. Sept. 2019;30:1953–1967. doi: 10.1681/ASN.2018121259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gupta L., Klinkhammer B.M., Boor P., Merhof D., Gadermayr M. International Conference on Medical Imaging with Deep Learning –Full Paper Track, (London, United Kingdom), 08–10 Jul. 2019. Iterative learning to make the most of unlabeledand quickly obtained labeled data in histology. [Google Scholar]
- 11.Gupta L., Klinkhammer B.M., Boor P., Merhof D., Gadermayr M. Lecture Notes in Computer Science. Springer International Publishing; 2019. GAN-based image enrichment in digitalpathology boosts segmentation accuracy; pp. 631–639. 13/15. [Google Scholar]
- 12.Kannan S., Morgan L.A., Liang B., et al. Segmentation of glomeruli within trichrome images using deeplearning. Kidney Int Rep. July 2019;4:955–962. doi: 10.1016/j.ekir.2019.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yamamoto Y., Saito A., Numata Y., et al. A novel method for morphological pleomorphism and heterogeneity quantitative measurement: named cell feature level co-occurrencematrix. J Pathol Inform. 2016;7(1):36. doi: 10.4103/2153-3539.189699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Borovec J., Munoz-Barrutia A., Kybic J. 2018 25th IEEE International Conference on Image Processing (ICIP) IEEE; Oct. 2018. Benchmarking of image registration methods for differently stainedhistological slides. [Google Scholar]
- 15.Ojala T., Pietikäinen M., Harwood D. A comparative study of texture measures with classification based on featureddistributions. Pattern Recogn. Jan. 1996;29:51–59. [Google Scholar]
- 16.Sari C.T., Gunduz-Demir C. Unsupervised feature extraction via deep learning for histopathological classification ofcolon tissue images. IEEE Trans Med Imaging. May 2019;38:1139–1149. doi: 10.1109/TMI.2018.2879369. [DOI] [PubMed] [Google Scholar]
- 17.Zheng Y., Jiang Z., Xie F., et al. Feature extraction from histopathological images basedon nucleus-guided convolutional neural network for breast lesion classification. Pattern Recogn. Nov. 2017;71:14–25. [Google Scholar]
- 18.Barisoni L., Lafata K.J., Hewitt S.M., Madabhushi A., Balis U.G.J. Digital pathology and computational imageanalysis in nephropathology. Nat Rev Nephrol. Aug. 2020;16:669–685. doi: 10.1038/s41581-020-0321-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tran W.T., Jerzak K., Lu F.-I., et al. Personalized breast cancer treatments using artificial intelligence in radiomics and pathomics. J Med Imag Radiat Sci. Aug 2019;50:S32–S41. doi: 10.1016/j.jmir.2019.07.010. [DOI] [PubMed] [Google Scholar]
- 20.D. Lowe. Object recognition from local scale-invariant features. In Proceedings of the Seventh IEEE International Conference on Computer Vision, IEEE, 199921.
- 21.Haralick R.M., Shanmugam K., Dinstein I. Textural features for image classification. IEEE Trans Syst Man Cybernet. Nov. 1973;SMC-3:610–621. [Google Scholar]
- 22.Kowal M., Filipczuk P., Obuchowicz A., Korbicz J., Monczak R. Computer-aided diagnosis of breast cancer basedon fine needle biopsy microscopic images. Comput Biol Med. Oct. 2013;43:1563–1572. doi: 10.1016/j.compbiomed.2013.08.003. [DOI] [PubMed] [Google Scholar]
- 23.Rudin C. Stop explaining black box machine learning models for high stakes decisions and use interpretable modelsinstead. Nat Machine Intel. May 2019;1:206–215. doi: 10.1038/s42256-019-0048-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yu K.-H., Zhang C., Berry G.J., et al. Predicting non-small cell lung cancerprognosis by fully automated microscopic pathology image features. Nat Commun. 2016;7:Aug. doi: 10.1038/ncomms12474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dabeer S., Khan M.M., Islam S. Cancer diagnosis in histopathological image: CNN based approach. Informa Med Unlocked. 2019;16 [Google Scholar]
- 26.Djuric U., Zadeh G., Aldape K., Diamandis P. Precision histology: how deep learning is poised to revitalize histomorphology for personalized cancer care. npj Precision Oncol. June 2017;1 doi: 10.1038/s41698-017-0022-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xie J., Liu R., Luttrell J., Zhang C. Deep learning based analysis of histopathological images of breast cancer. Front Genet. Feb. 2019;10 doi: 10.3389/fgene.2019.00080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Öztürk Ş., Akdemir B. Application of feature extraction and classification methods for histopathological image usingGLCM, LBP, LBGLCM. GLRLM SFTA Proc Comput Sci. 2018;132:40–46. [Google Scholar]
- 29.Uehara K., Murakawa M., Nosato H., Sakanashi H. 2020 IEEE International Conference on Image Processing (ICIP) 2020. Multi-scale explainable feature learning for pathological imageanalysis using convolutional neural networks; pp. 1931–1935. [Google Scholar]
- 30.Djudjaj S., Papasotiriou M., Bülow R.D., et al. Keratins are novel markers of renal epithelial cell injury. Kidney Int. Apr. 2016;89:792–808. doi: 10.1016/j.kint.2015.10.015. 14/15. [DOI] [PubMed] [Google Scholar]
- 31.Ehling J., Bábíčková J., Gremse F., et al. Quantitative micro-computed tomography imaging of vascular dysfunction in progressive kidney diseases. J Am Soc Nephrol. July 2015;27:520–532. doi: 10.1681/ASN.2015020204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Boor P., Bábíčková J., Steegh F., et al. Role of platelet-derived growth factor-CC in capillary rarefaction in renal fibrosis. Am J Pathol. Aug. 2015;185:2132–2142. doi: 10.1016/j.ajpath.2015.04.022. [DOI] [PubMed] [Google Scholar]
- 33.Martínez-Klimova E., Aparicio-Trejo O.E., Tapia E., Pedraza-Chaverri J. Unilateral ureteral obstruction as a modelto investigate fibrosis-attenuating treatments. Biomolecules. Apr. 2019;9:141. doi: 10.3390/biom9040141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ronneberger O., Fischer P., Brox T. U-net: convolutional networks for biomedical image segmentation. CoRR. 2015;abs/1505.04597 [Google Scholar]
- 35.van Griethuysen J.J., Fedorov A., Parmar C., et al. Computational radiomics system to decode the radiographic phenotype. Cancer Res. Oct. 2017;77:e104–e107. doi: 10.1158/0008-5472.CAN-17-0339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Macenko M., Niethammer M., Marron J.S., et al. 2009 IEEE International Symposium on Biomedical Imaging: From Nano to Macro. IEEE; June 2009. A method for normalizing histology slides for quantitative analysis. [Google Scholar]
- 37.Ruifrok A., Johnston D. Quantification of histochemical staining by color deconvolution. Anal Quant Cytol Histol. 2001;23(4):291–299. [PubMed] [Google Scholar]
- 38.Beucher S. Scanning Microscopy Supplement. 1992. The watershed transformation applied to image segmentation; pp. 299–314. 01. [Google Scholar]
- 39.McInnes L., Healy J., Melville J. UMAP: uniform manifold approximation and projection for dimension reduction. arXiv e-prints. Feb. 2018 doi: 10.48550/arXiv.1802.03426. [DOI] [Google Scholar]
- 40.van der Maaten L., Hinton G. Visualizing data using t-sne. J Mach Learn Res. 2008;9(86):2579–2605. [Google Scholar]
- 41.Madabhushi A., Lee G. Image analysis and machine learning in digital pathology: challenges and opportunities. Article Med Image Anal. 2016;33:170–175. doi: 10.1016/j.media.2016.06.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hasani N., Morris M.A., Rhamim A., et al. Trustworthy artificial intelligence in medical imaging. Article PET Clinics. 2022;17(1):1–12. doi: 10.1016/j.cpet.2021.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lehrer M., Powell R.T., Barua S., Kim D., Narang S., Rao A. Advances in Biology and Treatment of Glioblastoma. Springer; Cham: 2017. Radiogenomics and histomics in glioblastoma: The promise of linking image-derived phenotype with genomic information; pp. 143–159. [Google Scholar]
- 44.Kong J., Cooper L.A., Wang F., et al. Machine-based morphologic analysis of glioblastoma using whole-slide pathology images uncovers clinically relevant molecular correlates. PLoS One. 2013;8(11) doi: 10.1371/journal.pone.0081049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nalisnik M., Amgad M., Lee S., et al. Interactive phenotyping of large-scale histology imaging data with HistomicsML. Sci Rep. 2017;7(1) doi: 10.1038/s41598-017-15092-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Simon O., Yacoub R., Jain S., Tomaszewski J.E., Sarder P. Multi-radialLBP features as a tool for rapid glomerular detection and assessment in whole slide histopathology images. Scient Rep. 2018;8(1) doi: 10.1038/s41598-018-20453-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sarder P., Ginley B., Tomaszewski J.E. Automated renal histopathology: digital extraction and quantification of renal pathology. SPIE Proc. 2016 doi: 10.1117/12.2217329. [DOI] [Google Scholar]
- 48.Miller R.G. Springer; 1966. Simultaneous Statistical Inference. ISBN 9781461381228. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material.