Abstract
Context
Cytology is the study of whole cells in diagnostic pathology. Unlike standard histologic thinly sliced specimens, cytologic preparations consist of preparations of whole cells where cells commonly cluster and aggregate. As such, cytology preparations are generally much thicker than histologic slides, resulting in large patches of defocus when examined under the microscope. A diagnostic aggregate of cells often cannot be viewed in focus together, requiring pathologists to continually manipulate the focal plane, complicating the task of accurately assessing the entire cellular aggregate and thus in making a diagnosis. Further, it is extremely difficult to acquire useful uniformly in-focus digital images of cytology preparations for applications such as remote diagnostic evaluations and artificial intelligence models. The predominant current method to address this issue is to acquire digital images at multiple focal planes of the entire slide, which demands long scanning time, complex and expensive scanning systems, and huge storage capacity.
Aims
Here we report a unique imaging method that can acquire cytologic images efficiently and computationally render all-in-focus digital images that are highly compact.
Methods and material
This method applies a metric-based digital refocusing to microscopy data collected with a Fourier ptychographic microscope (FPM). The digitally refocused patches of images are then synthesized into an all-in-focus image.
Results
We report all-in-focus FPM results of thyroid fine needle aspiration (FNA) cytology samples, demonstrating our method’s ability to overcome the height variance of 30 μm caused by cell aggregation, and rendering images at high resolution (corresponds to a standard microscope with objective NA of 0.75) and that are all-in-focus.
Conclusions
This technology is applicable to standard microscopes, and we believe can have an impact on diagnostic accuracy as well as ease and speed of diagnosing challenging specimens. While we focus on cytology slides here, we anticipate this technology’s advantages will translate well for histology applications. This technique also addresses the issue of remote rapid evaluation of cytology preparations. Finally, we believe that by resolving the focus heterogeneity issues in standard digital images, this technique is a critical advance for applying machine learning to cytology specimens.
Keywords: Fine needle aspiration (FNA), Fourier ptychographic microscope, All-in-focus images, Aocus evaluation metric
Key messages
We develop a unique imaging method that can acquire cytologic images with a Fourier ptychographic microscope and computationally render all-in-focus digital images at high resolution (corresponds to a 0.75 NA standard microscope), overcoming height variance of 30 μm. This technology can help improve diagnostic accuracy and ease. Rendered images can be useful for artificial intelligence applications.
Introduction
Fine needle aspiration (FNA) biopsy is a safe, minimally invasive, and widely used procedure to obtain diagnostic cytologic material from multiple organs, including the thyroid gland, breast, lung and pancreas.1, 2, 3, 4, 5 In this procedure, the abnormal tissue is sampled by a thin needle, expressed onto a glass slide, stained, and then observed through a microscope. The collected sample generally contains clusters and aggregates of whole cells, the patterns of which often contain useful diagnostic information. Unlike histologic sections that are cut very thinly and generally have a uniform thickness across a slide, the aggregates of whole cells in an FNA vary in thickness. When viewed under a microscope, the cells are often distributed across multiple different focal planes requiring the pathologist to continually manipulate the focus to maintain a clear image. Importantly, it is extremely difficult to acquire useful digital images of cytologic preparations such as FNAs that are needed for remote diagnosis. Finally, cytology preparations have been very challenging to study with machine learning techniques, as out of focus areas render large areas unusable for analysis. Acquiring cytology images in which all planes can be seen in focus requires complex, time-consuming, and expensive scanning capabilities due to the fact that a cytology preparation can only be fully imaged by capturing images over multiple focal planes to constitute a volumetric image data set (z-stack).
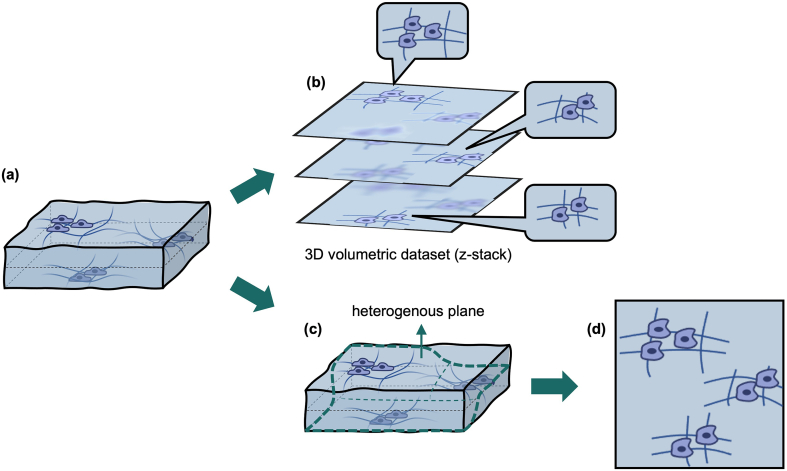
It would be advantageous if an FNA sample could be captured in a 2D image in which each of the non-overlapping cells is optically in-focus—an all-in-focus image. The 2D plane of interest is no longer flat but is a heterogenous plane that intersects each cell at its focal plane [Fig. 1]. Such an all-in-focus image is data efficient, as it is a 2D image information dataset and not a 3D volumetric dataset. Such an all-in-focus image is also highly suited to a pathologist’s workflow, as a pathologist would be able see the vast majority of cells in focus without having to scroll through a z-stack of scanned images. To be clear, such a 2D rendering would not work well for overlapping cells. Image information about overlapping cells can only be capitulated in a 3D volumetric image. Our proposed 2D strategy can work with FNA samples, because the majority of the cells are generally separated from each other by extensive fibrin clots formed during the procedures or simply by the cytoplasmic mass associated with the cells themselves.
Fig. 1.
Methods to describe a FNA sample. (a) FNA sample with cells at different planes. (b) 3D volumetric dataset. Each z-stack image has some cells in focus while other cells out of focus (c) Heterogeneous plane which intersects with each cell. (d) All non-overlapping cells are in focus in this heterogeneous plane.
To generate an all-in-focus image, we need to collect the phase and amplitude of the 2D wavefront emerging from the sample as this complete optical wavefront dataset will allow us to digitally refocus the image flexibly at different spatial points. The recently developed Fourier Ptychographic Microscopy (FPM)6, 7, 8, 9, 10, 11, 12, 13, 14 provides the requisite capability to collect such 2D wavefront data. The basic form of an FPM system is simply a standard microscope refitted with an LED array in place of the usual illumination source. During operation, the sample is illuminated in sequence by individual LEDs in an LED array and the transmission image is recorded through a low-NA objective. The collected sequence of low-resolution images can then be processed through the FPM phase-retrieval algorithm to yield both the phase and amplitude of the light field emerging from the sample. By applying a metric-based digital refocusing to the microscopy data collected with a Fourier ptychographic microscope (FPM), we can in turn render an all-in-focus image. We report all-in-focus FPM results of thyroid fine needle aspiration (FNA) cytology samples, demonstrating our method’s ability to overcome the height variance of 30 μm across the slide, as a result of cell aggregation, and rendering images at high resolution and that are all-in-focus.
Subjects and methods
Cytology material
For the Subjects and Methods section, we used an anonymized fine needle aspiration biopsy Papanicolaou smear of a thyroid gland lesion (papillary thyroid carcinoma) that was selected from the teaching files of the Department of Pathology, National University Hospital, Singapore. In accordance with the regulatory requirements at this institution, case reports involving de-identified clinical material from 2 or fewer subjects are exempt from ethics review.
For the Results section, we selected a de-identified aspirate smear slides of a thyroid FNAs from the Washington University/Barnes Jewish Hospital files. Representative diagnostic areas were selected by one of us (CB) for standard image acquisition (Aperio, see below) and for FPM imaging. Study approval was obtained from the Washington University Human Research Protection Office (IRB 202004265).
FPM imaging
General concept and experimental set up
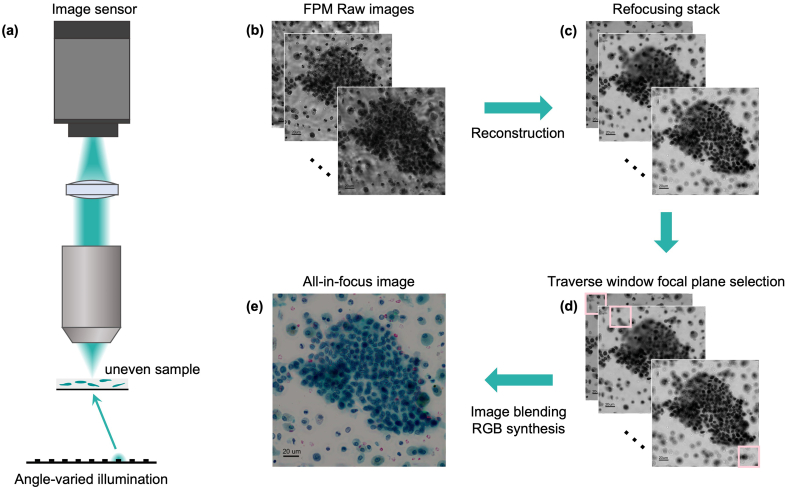
Fig. 2 shows the general process by which we accomplish all-in-focus FPM imaging. First, the uneven sample is imaged with our FPM system to collect a set of raw data. The data is then processed with the FPM algorithm to generate a stack of images at different focal planes (refocus stack). We then move a small window (vignette) across the image and select the corresponding image segment from the refocus stack that is in the sharpest focus. This selection process is repeated across the entire image and the selected segments are then fused to synthesize an all-in-focus image.
Fig. 2.
General process of all-in-focus FPM. (a) Schematic of FPM set up. (b) Raw data of FPM. (c) Refocusing stack reconstructed from raw data. (d) Focal plane selection for each part. (e) Synthesized all-in-focus color FPM image.
We implemented a transmission FPM system for this experiment. A CCD camera (ON Semi KAI-29050, 5.5 μm pixel size) served as our imaging camera. An Adafruit LED array (product ID: 607) was mounted ~66 mm below the focal plane of a 20x/0.40 objective (Plan N, Olympus), resulting in a 1.1 mm diameter field of view. The spacing between adjacent LEDs was 4 mm. During the data acquisition process, we sequentially illuminated the sample with individual LEDs of the array. Depending on the position of the lit LED, the illumination on the sample would have a specific illumination incidence angle. The transmission light was then collected through the objective and a raw image frame was recorded by the camera. In optical information terms, the collected data contains a sample angular spectrum segment. By collecting raw image frames for each LED in a 15x15 subsection of the LED array, we effectively gather image data that collectively correspond to an effective illumination NA of 0.4 and a total synthetic NA of 0.8. The central wavelengths of the full-color LED were 632.3 nm (red), 516.2 nm (green), and 471.2 nm (blue). Average power of red, green and blue LEDs were 1.7 mW, 2.1 mW and 1.6 mW, respectively. Exposure time was set to be 0.5 s for each raw image acquisition. Total acquisition time for 1 color channel was 9 min.
The data was then processed with the FPM algorithm. Broadly speaking, the algorithm combines synthetic aperture concept and phase retrieval concept to compute the expected optical wavefront at a chosen plane on the sample. The redundancy in raw image data (partially shared angular spectrum information) allows the FPM algorithm to recover phase information from the raw data that are purely intensity measurements, and subsequently, to generate the complex valued optical wavefront associated with the sample.7, 8, 9
From a user’s perspective, the operation of both raw image acquisition and FPM image processing are straightforward. The raw image acquisition procedure of our all-in-focus FPM set up is the same as that of a traditional transmission microscope: loading sample, finding the region of interest (ROI), and adjusting the focal plane. The raw images under different illumination angles will be captured automatically by the computer. Only a few inputs are required from the user to generate an optimal all-in-focus image: the exact raw images to be processed, the z-scanning range, and the z-scanning step size. With these parameters, the user will then get the all-in-focus image by a single click.
Digital refocusing by FPM
We accomplished digital refocusing in this experiment by introducing a refocus phase factor to the embedded pupil function recovery (EPRY) algorithm7,9—the component of the FPM algorithm that evaluates the imaging systems pupil function. The refocus phase factor, equivalent to defocus aberration, is given by:
| (1) |
where zr is refocus distance, k = 2π/λ is the wavenumber of the illumination light in vacuum, u, v are 2D coordinates in Fourier domain, and kmax = NA ∙ k is the cut-off frequency of the objective lens. The flowchart of a single refocusing iteration in a subregion of the sample’s Fourier domain is showed in Fig. 3a. To calculate the image at the plane with distance -zr from the focal plane, we computationally refocused the sample spectrum acquired by distance zr, which is equivalent to optically propagating the target plane to the true focal plane. We do so by performing the EPRY iteration according to our intensity observation at the focal plane. Finally, we refocused the sample Fourier spectrum by distance -zr, which is equivalent to the optical propagation back to the target defocus plane, and update the sample Fourier spectrum accordingly. In each iteration, a reconstruction error was calculated by getting the summation of each pixel’s square error between reconstruction intensity and captured intensity. Adaptive step-size strategy was adopted in our algorithm. Step-sizes for both spectrum updating and pupil updating began at 0.1 and reduced by one-half once the reconstruction error change was less than 5%. We terminated the iteration when the step-sizes were less than 0.01.
Fig. 3.
Digital refocusing ability of FPM. (a) Flowchart of FPM EPRY digital refocusing. (b) Reconstruction of USAF target at original focal plane. (c) Reconstruction of USAF target at 30 μm below the original focal plane. (d) Reconstruction of the sample at the original focal plane. (e) Reconstruction of the sample at 15 μm below the original focal plane.
We used both a USAF target slide [Fig. 3c and d] and a thyroid FNA biopsy sample slide [Fig. 3e and f] to demonstrate and verify the FPM’s digital refocusing ability. A USAF target slide 30 μm below focal plane illuminated with blue LEDs (471.2 nm) was imaged by our FPM system. The reconstruction results at the original focal plane [Fig. 3c] and the exact USAF slide plane (30 μm below the original focal plane) [Fig. 3d] demonstrate the accuracy of FPM refocusing ability.
Then we digitally refocused the FPM data from a thyroid FNA biopsy sample. RGB channels were reconstructed and then synthesized to form color images. Two focal planes, the original focal plane [Fig. 3d] and 15 μm below the original focal plane [Fig. 3e] were reconstructed. From the resulting images, we can clearly see that the cells in the sample reside in multiple focal planes—neither of the focal planes in Fig. 3d and Fig. 3e was able to place all the cells in focus simultaneously.
Focus evaluation metric
To select image segments that are in-focus, so that we can synthesis all-in-focus images, we next utilized normalized variance (NV)15,16 as a focus evaluation metric to select the sharpest image segments from a refocused image stack. As an image-contrast-based function, normalized variance is less sensitive to both noise and changes in brightness. Normalized variance of a selected part of image, denoted as NV, is given by:
| (2) |
where W and H are the width and height (unit: pixel) of the image, I(x, y) is the value of each pixel in the image and μ is the mean value of all pixels.
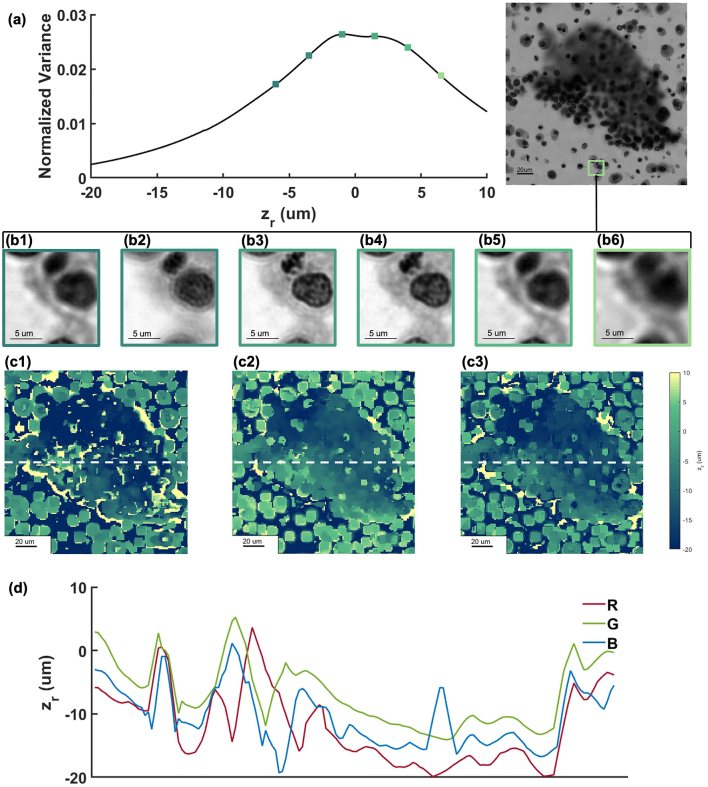
We examined the effectiveness of NV in selecting the correct focal plane for biological samples [Fig. 4a and b]. A sample was placed at the objective’s focal plane and imaged with green illumination (516.2 nm). Refocus stack ranging from -20 to 10 μm was created by FPM digital refocusing. Given the uneven nature of the thyroid sample, only a 13.75x13.75 μm2 (100 px x 100 px) region which can be regard as a flat layer was cropped. The NV curve with respect to refocusing distance is plotted in Fig. 4a. Corresponding images respectively at -5, -2.5, 0, 2.5, 5, 7.5 μm away from NV peak plane are showed in Fig. 4b. The sharpest image appears at the NV peak plane [Fig. 4b3]. As image plane shifts away from the NV peak plane, the image becomes progressively more blurred and NV value drops correspondingly.
Fig. 4.
Normal variance as a focus evaluation metric. (a) NV curve of a small region of a sample. (b) Corresponding images at selected points in (a) (-5, -2.5, 0, 2.5, 5, 7.5 μm away from NV peak plane, NV peak value at b3). Scale bar: 5 μm. (c) Depth maps of a sample. c1. Red channel. c2. Green channel. c3. Blue channel. (d) RGB depth profiles at dash-lined position in (c).
Then we generated depth maps of the sample by applying NV function to all RGB refocusing FPM stacks of the thyroid sample. The refocus range is from -20 to 10 μm. Results are showed in Fig. 4c and d. The depth map quantitively shows the height variance at different part of the sample. Fig. 4d is the exact defocus depth at dash-lined position denoted in Fig. 4c, representing RGB channels respectively. The height variance in RGB channels track each other and the residual variations can be attributed to the chromatic aberration of the objective. By separately selecting the correct focal plane for each channel, we can then synthesize a chromatic-aberration-free image.
Synthesizing an all-in-focus image
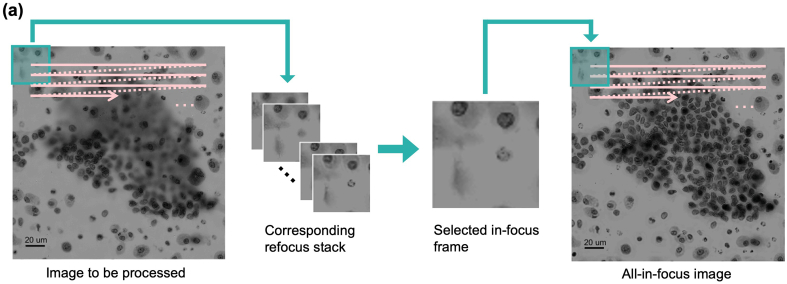
Fig. 5 summarizes the synthesis process. We shift a vignette across the entire image. At each location, the algorithm calculates the NV of the image within the vignette and selects the frame with the maximum NV as the in-focus image segment to be used. To reduce seam and mosaic, we set the traverse step size to be smaller than the vignette side length, resulting in neighbouring segments overlapping each other. At each location, the average across the segments is used as our final all-in-focus output. In our experiment, the vignette size was 100 x 100 px2 and traverse step size is 1/10 of the vignette side length. Color images are generated by repeating this process for the red, green, and blue channels.
Fig. 5.
Synthesizing an all-in-focus image. (a) Synthesis procedure. Green channel images are demonstrated. A vignette is shifted across the entire image. In focus segment at each location was selected by applying the normal variance function. All segments were then fused together to render an all-in-focus image.
Results
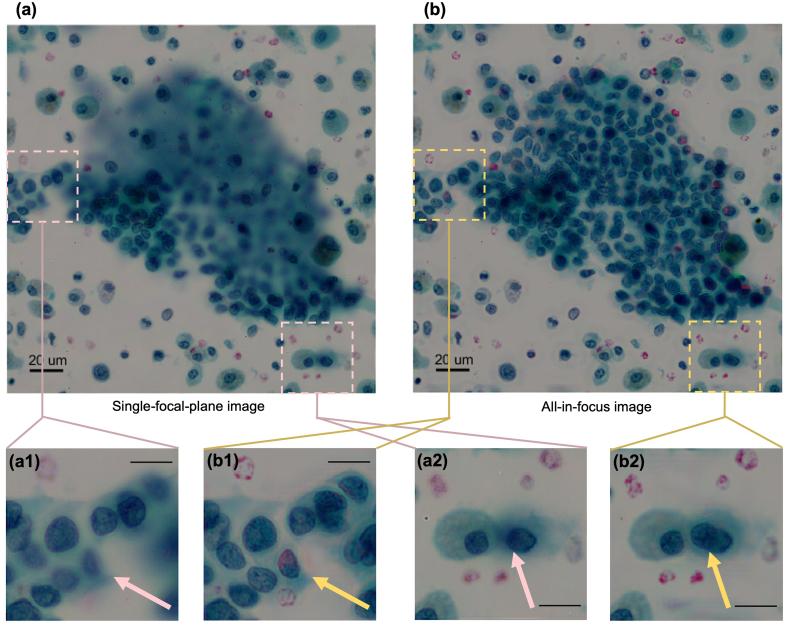
Evaluation of a single plane of focus image with an all-in-focus FPM image [Fig. 6]
Fig. 6.
Comparison between a single plane of focus image with an all-in-focus FPM image. (a) Single-focal-plane color image reconstructed from FPM. Note the large areas of out of focus image in the diagnostic cluster that results from the thickness of the preparation (b) All-in-focus color image reconstructed from FPM. Note that all non-overlapping cells in the diagnostic cluster can be seen in sharp focus, despite the thickness of the preparation. (a1,2) Corresponding details boxed out from single-focal-plane image (a). Note the out of focus cell in a1 (yellow arrow). In a2, the pink arrow points to an apparently in focus cell. Scale bar: 20 μm (b1,2) Corresponding details boxed out from all-in-focus image (b). Note that the out of focus cells in a1 can now be seen in sharp focus, with a diagnostic cell nucleus now clearly seen (yellow arrow, b1). In b2, note that the apparently in focus cell seen in a2 can now be seen to actually represent 2 cells (yellow arrow, b2). Scale bar: 20 μm
Fig. 6 shows the comparison of an image from a single focal plane [Fig. 6a] compared to an FPM all-in-focus image [Fig. 6b] on a diagnostic cell cluster in an FNA cytology preparation from a thyroid nodule. As described more fully in the figure legend, it can be easily seen that the FPM all-in-focus image allows for the precise morphologic evaluation of effectively all the cells in this diagnostic cluster, despite the thickness (and thus multiple focal planes) of the cluster.
We note that this all-in-focus FPM method only works for cells that do not overlap with other cells. In locations where cells overlap, the algorithm will still attempt to tune the effective focal plane. This will generally put a single cell in focus, but there is no guarantee it will converge correctly.
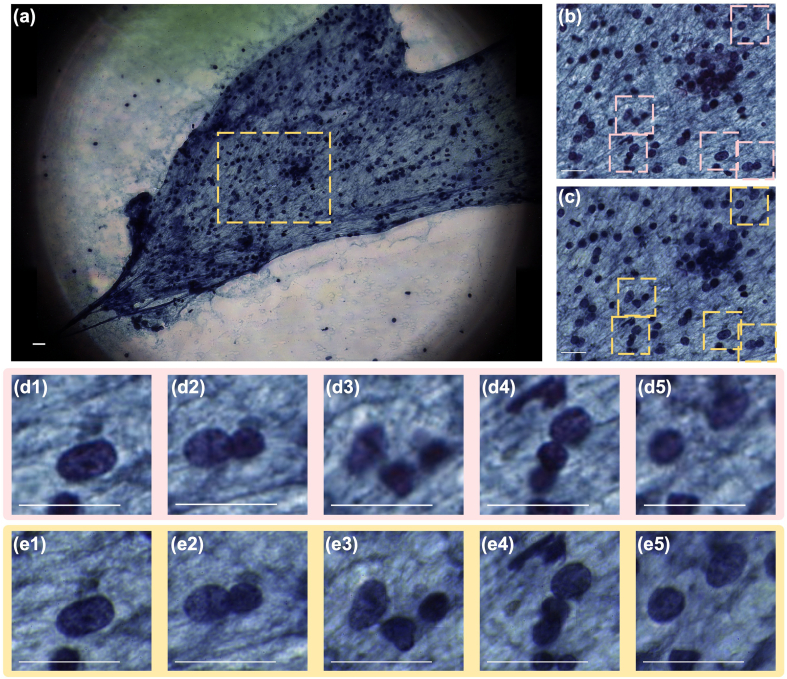
Comparison of FPM to standard digital imaging [Fig. 7]
Fig. 7.
Comparison between all-in-focus FPM and Aperio scanner. (a) All-in-focus, whole FoV, color FPM image of a thyroid FNA biopsy sample. (b) Aperio scanner result of the boxed-out region in (a). (c) FPM result of the boxed-out region in (a). (d) Corresponding details boxed-out from Aperio result (b). (e) Corresponding details boxed-out from FPM result (c). Scale bar: 20 μm. Yellow annotated figures: all-in-focus FPM results. Pink annotated figures: Aperio scanner results.
A Papanicolaou-stained thyroid FNA biopsy slide was separately scanned using the FPM system and an Aperio system AT2 digital pathology scanner to compare the performance of each imaging modality.
The Aperio scanner was equipped with an objective lens with NA of 0.75. In order to match the resolution between the Aperio scanner and our FPM system, we switched our FPM objective to a 0.75 NA objective and adjusted the illumination array to provide a maximum of 0.75 NA illumination. This larger NA illumination was achieved by setting the distance between the LED array and the objective focal plane to ~25 mm. The number of raw FPM images acquired here is 49. The FPM’s resolution dependency on NA differs from that of standard microscopy, the design choices described here were made to ensure that we had comparable resolution for the 2 systems. Interested readers are invited to read the Supplementary Information document for more information. Results of 1 ROI is presented in the following section. Additional 5 results were provided in the Supplementary document.
The acquired images from both the FPM system [Fig. 7a, c and e] and the Aperio [Fig. 7b and d] are presented. Cells that are in focus [Fig. 7d1 and d2] and out-of-focus [Fig. 7d3, d4 and d5] in the Aperio image are simultaneously in focus in the FPM image [Fig. 7e]. Comparatively, we can also observe that the nuclear details are much clearer with the all-in-focus FPM images [Fig. 7e] when compared to the images from the Aperio scanner [Fig. 7d]. Not only are more cells in focus for the all-in-focus images, the amount of cellular details is also greater. The sharper image quality of the FPM images compared to the Aperio images seems at odds with the fact that both systems were set up to have the same NA. This mismatch has also been previously observed in other high-resolution FPM experiments12 as well, and is attributable to the fact that residual presence of aberrations in commercial microscope objectives tend to degrade their achievable resolution and prevent these objectives from attaining NA-limited resolutions. In FPM imaging systems, these residual aberrations are readily corrected through the use of the component EPRY sub-routine in the FPM algorithm that is able to determine and correct pupil aberrations computationally.6,7,9 For completeness, we characterized the observed resolution for both systems and reported our findings in the Supplementary Information document.
Discussion
We have developed a method that enables all-in-focus FPM imaging of FNA biopsy slides. The all-in-focus imaging ability is realized by incorporating digital refocusing and focus evaluation into the original FPM image rendering process. In addition to inheriting original FPM’s advantage of wide-field and high resolution, all-in-focus FPM further frees user from the constraints of shallow depth-of-field in high-NA high-resolution microscopy. For color imaging, RGB channels are first processed individually and then synthesized to form an RGB all-in-focus image. Chromatic aberration associated defocus can thus be corrected through this procedure.
From a pathology perspective, the all-in-focus image is able to bring the vast majority of the cells on a slide into focus without sacrificing cellular detail. Cytopathologists use multiple different components of cellular detail to aid in making a diagnosis. These include the amount and the character of the cytoplasm, the contours of the nuclear membrane, the texture of the chromatin, and the presence or the absence of nucleoli among others. For successful evaluation of a slide, all of these different components need to be in focus and well visualized. As Fig. 6 illustrates, an all-in-focus image brings the vast majority of cells into focus when compared to a single-focal-plane image. The cellular and the nuclear detail is clearly appreciable in nearly all cells of the all-in-focus-image compared to the single-focal-plane image where some cells remain blurred and cannot be evaluated. The superiority of the all-in-focus can also be appreciated in Fig. 7, comparing images acquired by conventional scanning (Aperio) vs. the FPM imaging system. Once again, not only are almost all of the cells in focus, but they are in focus at high power, preserving diagnostic architectural, cellular, and nuclear details. The all-in-focus images are both more efficient in that more cells can be visualized without the need to focus or scan through multiple focal planes and they provide greater cellular detail which the pathologist needs to make diagnoses.
In principle, our all-in-focus algorithm can also be applied to conventionally acquired z-stack images from a standard scanner. However, there are several distinct advantages associated with using FPM data. First, FPM can acquire the requisite raw data to cover the same effective z-range without requiring the use of any mechanical actuation along the z-axis—thereby significantly simplifying the scan system. Second, commercial z-stack images are discretized along the z-axis as determined by the z-step size, while FPM is free from such conceptual limitations. For a commercial scanner, the finer the z-step size, the longer the scan duration will be and the larger the final data size will be. This trade-off necessarily constrains all z-stack imaging processes in commercial scanners. Interestingly, such trade-off does not exist for FPM. As long as the FPM is able to collect a raw image data set that allows the 2D wavefront to be accurately determined, that data can be used to virtually generate z-stack images with arbitrarily fine z-step size. From an information viewpoint, the relative efficiency of the FPM raw data set versus the conventional scanner z-stack image set is attributable to the fact that there is simply a lot of redundancies within adjacent commercial z-stack images.
The all-in-focus FPM final image is a 2D dataset. In contrast, the conventional scanner z-stack is intrinsically 3D in nature. This means that the all-in-focus FPM data set is compact in comparison. The compact data size of all-in-focus FPM images is a broadly useful advantage. In remote digital pathology applications, the smaller and more efficient data file size will greatly facilitate ease of data transmission. The comparatively smaller data file size also makes it easier to feed data into machine learning systems where the number of input nodes is always finite and limited.
The use of FPM to digitally refocus microscope slide images has previously been reported for a number of different applications.7,13,17,18 Results in neuronal cells13 demonstrated that FPM images can be digitally refocused over a range of at least ±100 μm with a synthetic NA of 0.42. In addition to light field refocusing, a new multi-slice refocusing algorithm was also proposed and 100 μm thick with 10 μm step spirogyra algae stack was reconstructed with a synthetic NA of 0.66.19 However, much of these prior works dealt with adjusting the global focus of the images, and thus cannot be directly applied to address the issue of heterogenous focal planes associated with FNA samples. To date, all-in-focus FPM imaging has only been developed and applied to blood smears.18 In that work, the segmentation-based method relied on prior knowledge of the morphology of both blood cells and plasmodium parasite, which limits its application and is not a good fit with FNA samples. Our work shows that FPM can be used to create all-in-focus images of aggregates of cells on an FNA slide without restrictive assumptions or known priors.
Slides from FNA specimens were chosen for this project because of the inherent challenges they present by their need to capture multiple focal planes. However, it should be noted that histologic sections are never completely flat, and high-power imaging and image acquisition often results in large patchy out of focus areas. Therefore, FPM is likely to have applications in remote diagnosis and image analysis applications for slides prepared from surgical pathology specimens (biopsies and resections) as well.
Looking to the future, we note that the set up reported here was not optimized for high throughput pathology slides scanning. We expect that a high throughput whole-slideall-in-focusFPM is achievable by using higher power light sources, a high-speed and high pixel-count camera, and other physical improvements to the system. Such a method can potentially facilitate better and faster pathology analysis of FNA biopsy samples.
It is also worth noting that the FPM system may have even more general applications. When scanned at high power, even “flat” thin sections in histologic slides show significant focal plane variation, hindering rapid imaging, and creating significant pitfalls for using these images in advanced applications, such as deep learning/AI, where out of focus areas are believed to be a major impediment to AI/deep learning on these preparations.20 The all-in-focus FPM method may be an ideal solution to this problem. Machine learning methods generally outright reject out-of-focus areas from consideration, and this can lead to significantly reduced analysis coverage and accuracy. As this all-in-focus FPM method can ensure that the entire image is in-focus, and therefore, it has the potential to greatly improve the performance of cell segmentation and recognition machine learning methods. In computer vision field, all-in-focus FPM images can also provide sharp edges which can greatly facilitate edge-detection-based segmentation methods. Broadly speaking, the rendering of all-in-focus pathology images has the high potential to solve a key challenge associated with machine learning based digital pathology analysis—the challenge that out-of-focus image segments are ill-suited for machine learning processing.
Funding
This work was supported by the National Institutes of Health [grant NIH- U01CA233363].
Source(s) of support
NIH-U01CA233363
Presentation at a meeting
Nil
Declaration of interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
Acknowledgement
The authors would like to thank Cheng Shen (California Institute of Technology) for imaging modality discussion, Dr. Sid Rawal and imaging team (Washington University School of Medicine) for sample preparation and imaging, and Prof. Mark Watson (Washington University School of Medicine) for pathology discussion.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jpi.2022.100119.
Contributor Information
Mingshu Liang, Email: mlliang@caltech.edu.
Cory Bernadt, Email: cbernadt@wustl.edu.
Soon Boon Justin Wong, Email: justin_sb_wong@nuhs.edu.sg.
Changsoon Choi, Email: cchoi@caltech.edu.
Richard Cote, Email: rcote@wustl.edu.
Changhuei Yang, Email: chyang@caltech.edu.
Appendix A. Supplementary data
The following are the supplementary data related to this article.
Supplementary material
References
- 1.Kline T.S., Joshi L.P., Neal H.S. Fine-needle aspiration of the breast: diagnoses and pitfalls. A review of 3545 cases. Cancer. 1979;44(4):1458–1464. doi: 10.1002/1097-0142(197910)44:4<1458::AID-CNCR2820440440>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- 2.Gharib H., Goellner J.R. Fine-needle aspiration biopsy of the thyroid: an appraisal. Ann Intern Med. 1993;118(4):282–289. doi: 10.7326/0003-4819-118-4-199302150-00007. [DOI] [PubMed] [Google Scholar]
- 3.Wiersema M.J., Vilmann P., Giovannini M., Chang K.J., Wiersema L.M. Endosonography-guided fine-needle aspiration biopsy: diagnostic accuracy and complication assessment. Gastroenterology. 1997;112(4):1087–1095. doi: 10.1016/S0016-5085(97)70164-1. [DOI] [PubMed] [Google Scholar]
- 4.Chang K.J., Nguyen P., Erickson R.A., Durbin T.E., Katz K.D. The clinical utility of endoscopic ultrasound–guided fine-needle aspiration in the diagnosisand staging of pancreatic carcinoma. Gastrointest Endosc. 1997;45(5):387–393. doi: 10.1016/S0016-5107(97)70149-4. [DOI] [PubMed] [Google Scholar]
- 5.Micames C.G., McCrory D.C., Pavey D.A., Jowell P.S., Gress F.G. Endoscopic ultrasound-guided fine-needle aspiration for non-small cell lung cancer staging: a systematic review and metaanalysis. Chest. 2007;131(2):539–548. doi: 10.1378/chest.06-1437. [DOI] [PubMed] [Google Scholar]
- 6.Guizar-Sicairos M., Fienup J.R. Phase retrieval with transverse translation diversity: a nonlinear optimization approach. Opt Express. 2008;16(10):7264–7278. doi: 10.1364/OE.16.007264. [DOI] [PubMed] [Google Scholar]
- 7.Zheng G., Horstmeyer R., Yang C. Wide-field, high-resolution Fourier ptychographic microscopy. Nat Photonics. 2013;7(9):739–745. doi: 10.1038/nphoton.2013.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ou X., Horstmeyer R., Yang C., Zheng G. Quantitative phase imaging via Fourier ptychographic microscopy. Opt Lett. 2013;38(22):4845–4848. doi: 10.1364/OL.38.004845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ou X., Zheng G., Yang C. Embedded pupil function recovery for Fourier ptychographic microscopy. Opt Express. 2014;22(5):4960–4972. doi: 10.1364/OE.22.004960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Horstmeyer R., Yang C. A phase space model of Fourier ptychographic microscopy. Opt Express. 2014;22(1):338–358. doi: 10.1364/OE.22.000338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tian L., Liu Z., Yeh L.-H., Chen M., Zhong J., Waller L. Computational illumination for high-speed in vitro Fourier ptychographic microscopy. Optica. 2015;2(10):904–911. doi: 10.1364/OPTICA.2.000904. [DOI] [Google Scholar]
- 12.Ou X., Horstmeyer R., Zheng G., Yang C. High numerical aperture Fourier ptychography: principle, implementation and characterization. Opt Express. 2015;23(3):3472–3491. doi: 10.1364/OE.23.003472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim J., Henley B.M., Kim C.H., Lester H.A., Yang C. Incubator embedded cell culture imaging system (EmSight) based on Fourier ptychographic microscopy. Biomed Opt Express. 2016;7(8):3097–3110. doi: 10.1364/BOE.7.003097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zheng G., Shen C., Jiang S., Song P., Yang C. Concept, implementations and applications of Fourier ptychography. Nat Rev Phys. 2021;3(3):207–223. doi: 10.1038/s42254-021-00280-y. [DOI] [Google Scholar]
- 15.Groen F.C.A., Young I.T., Ligthart G. A comparison of different focus functions for use in autofocus algorithms. Cytometry. 1985;6(2):81–91. doi: 10.1002/cyto.990060202. [DOI] [PubMed] [Google Scholar]
- 16.Sun Y., Duthaler S., Nelson B.J. Autofocusing in computer microscopy: selecting the optimal focus algorithm. Microsc Res Tech. 2004;65(3):139–149. doi: 10.1002/jemt.20118. [DOI] [PubMed] [Google Scholar]
- 17.Horstmeyer R., Chung J., Ou X., Zheng G., Yang C. Diffraction tomography with Fourier ptychography. Optica. 2016;3(8):827–835. doi: 10.1364/OPTICA.3.000827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Claveau R., Manescu P., Elmi M., Pawar V., Shaw M., Fernandez-Reyes D. Digital refocusing and extended depth of field reconstruction in Fourier ptychographic microscopy. Biomed Opt Express. 2020;11(1):215–226. doi: 10.1364/BOE.11.000215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tian L., Waller L. 3D intensity and phase imaging from light field measurements in an LED array microscope. Optica. 2015;2(2):104–111. doi: 10.1364/OPTICA.2.000104. [DOI] [Google Scholar]
- 20.Kohlberger T., Liu Y., Moran M., Chen P.-H., Brown T., Hipp J.D., et al. Whole-slide image focus quality: automatic assessment and impact on AI cancer detection. J Pathol Inform. 2019;10:39. doi: 10.4103/jpi.jpi_11_19. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material