Abstract
Digital pathology can efficiently assess immunohistochemistry (IHC) data on tissue microarrays (TMAs). Yet, it remains important to evaluate the comparability of the data acquired by different software applications and validate it against pathologist manual interpretation. In this study, we compared the IHC quantification of 5 clinical breast cancer biomarkers—estrogen receptor (ER), progesterone receptor (PR), human epidermal growth factor receptor 2 (HER2), epidermal growth factor receptor (EGFR), and cytokeratin 5/6 (CK5/6)—across 3 software applications (Definiens Tissue Studio, inForm, and QuPath) and benchmarked the results to pathologist manual scores.
IHC expression for each marker was evaluated across 4 TMAs consisting of 935 breast tumor tissue cores from 367 women within the Nurses’ Health Studies; each women contributing three 0.6-mm cores. The correlation and agreement between manual and software-derived results were primarily assessed using Spearman’s ρ, percentage agreement, and area under the curve (AUC).
At the TMA core-level, the correlations between manual and software-derived scores were the highest for HER2 (ρ ranging from 0.75 to 0.79), followed by ER (0.69–0.71), PR (0.67–0.72), CK5/6 (0.43–0.47), and EGFR (0.38–0.45). At the case-level, there were good correlations between manual and software-derived scores for all 5 markers (ρ ranging from 0.43 to 0.82), where QuPath had the highest correlations. Software-derived scores were highly comparable to each other (ρ ranging from 0.80 to 0.99). The average percentage agreements between manual and software-derived scores were excellent for ER (90.8%–94.5%) and PR (78.2%–85.2%), moderate for HER2 (65.4%–77.0%), highly variable for EGFR (48.2%–82.8%), and poor for CK5/6 (22.4%–45.0%). All AUCs across markers and software applications were ≥0.83.
The 3 software applications were highly comparable to each other and to manual scores in quantifying these 5 markers. QuPath consistently produced the best performance, indicating this open-source software is an excellent alternative for future use.
Keywords: Definiens, QuPath, inForm, Automation, Estrogen receptor, Progesterone receptor, Cytokeratin 5/6, Epithelium growth factor receptor
Highlights:
-
•
It is important to evaluate the comparability of the data acquired by different software applications and validate it against pathologist manual interpretation.
-
•
Definiens Tissue Studio®, inForm®, and QuPath were highly comparable to each other and to pathologist manual scores in quantifying 5 clinical breast cancer biomarkers—ER, PR, HER2, EGFR, and CK5/6—in tissue microarrays.
-
•
QuPath had the highest correlation and %agreement with manual scores.
-
•
QuPath is a reliable open-source tool to automate IHC expression quantification of breast tumor biomarkers.
Introduction
Tissue microarrays (TMAs) have enabled researchers to investigate tumor molecular characteristics in large study populations.[1], [2], [3], [4] For example, within the Nurses’ Health Studies (NHS), we, the authors, constructed TMAs of normal breast lobules to identify biomarkers associated with subsequent breast cancer development.[5], [6], [7], [8] We also utilized TMAs of breast tumors to study associations between: (1) breast cancer risk factors and tumor molecular subtypes,[9], [10], [11], [12], [13] and (2) tumor biomarkers and breast cancer prognosis.[14], [15], [16], [17]
Pathologist manual interpretation of TMA immunohistochemistry (IHC) expression is considered the current standard for epidemiological studies. Manual scoring of TMAs is, however, time-consuming, expensive, prone to subjectivity between pathologists, and semi-quantitative.18 The development of digital pathology—whole slide scanners and image analysis software applications—has enabled more quantitative and objective scoring of TMA IHC expression. Since 2010, our approach in the NHS has been to: (1) use the Definiens Tissue Studio® software to semi-automate the quantification of a marker’s IHC expression on digitized TMAs; (2) randomly select 1 TMA for manual scoring by a pathologist; and (3) benchmark the software-derived scores against manual scores for that TMA to assess the reliability of the IHC quantification by Definiens.8,13,19
It is imperative to evaluate whether IHC data acquired using newer image analysis software applications are comparable to data acquired by existing/older software applications. This is especially pertinent to prospective epidemiological cohorts such as the NHS where TMA blocks are assembled with newly diagnosed cancer cases every few years and different software applications may be used to measure IHC expression in those new TMA blocks for ongoing biomarker studies. Thus, it is important to understand potential biases in IHC data acquired using different software applications.
In this study, we compared the IHC quantification of 5 clinical breast cancer biomarkers—estrogen receptor (ER), progesterone receptor (PR), human epidermal growth factor receptor 2 (HER2), epidermal growth factor receptor (EGFR), and cytokeratin 5/6 (CK5/6)—across three software applications (Definiens, inForm®,20 and QuPath21). We benchmarked software-derived results to pathologist manual scores.
Materials and methods
Study population
The NHS, established in 1976, consists of 121,700 US female registered nurses (30–55 years). In 1989, 116,429 female nurses (25–42 years) were additionally recruited into the second cohort, NHSII. At recruitment, NHS and NHSII participants provide baseline information about their medical history and risk factors for breast cancer.22 They answer biennial follow-up questionnaires, including reporting any new breast cancer diagnosis. Breast cancer was self-reported by participants (or next of kin for decedents) and was further confirmed by NHS/NHSII medical personnel via medical records review. Participants provided written consent to obtain breast cancer pathology records and tissue specimens from the diagnosing hospital; breast cancer was additionally confirmed by central pathology review. The study protocol was approved by the institutional review boards of the Brigham and Women’s Hospital and Harvard T.H. Chan School of Public Health, and those of participating registries as required.
TMA creation
The 4 TMAs included in this study were constructed with breast tumors from 385 NHS/NHSII participants diagnosed with invasive carcinoma between 2001 and 2008. In general, for each woman, 3 x 0.6mm representative tissue cores were obtained from their primary tumors in areas annotated by a board-certified breast pathologist (GMB) and assembled into TMAs at the Specialized Histopathology Core, Dana-Farber/Harvard Cancer Center (DF/HCC), Boston, MA. A small subset of women were represented with >3 cores. These 4 TMAs consisted of a total of 1197 cores.
Tissue markers and IHC
ER, PR, HER2, EGFR, and CK5/6 IHC assays were performed using 5 μm TMA sections, standard protocols, and the appropriate positive and negative controls, at the Specialized Histopathology Core, DF/HCC.19 The antibodies used were: rabbit anti-ER clone SP at 1:40 dilution (Neomarkers #RM-9101-S1, Thermo Fisher Scientific, Waltham, MA), mouse anti-PR clone PgR 636 at 1:400 dilution (Dako #M3569, Agilent Technologies Inc, Santa Clara, CA), rabbit anti-HER2 clone SP3 at 1:40 dilution (Neomarkers #9103-SO-A), mouse anti-EGFR clone H11 at 1:200 dilution (Dako #M3563), and mouse anti-CK5/6 clone D5/16/B4 at 1:200 dilution (Dako #7237). The TMAs for each marker were stained in a single batch.
Pathologist manual scoring
A pathologist (GMB) at Beth Israel Deaconess Medical Center (BIDMC) scored the immunoreactivity for each marker. For ER and PR, nuclei of tumor cells were graded as negative (0%; complete absence of staining), low positive (estimated 1–10% tumor cells staining positive), and positive (>10% tumor cells staining positive). HER2 evaluation adhered to the 2018 American Society of Clinical Oncology/College of American Pathology guideline: 0 (no tumor membrane staining observed), 1+ (incomplete and faint membrane staining in estimated >10% of tumor cells), 2+ (weak to moderate membrane staining in >10% of tumor cells, incomplete moderate to intense membrane staining in >10% of tumor cells, or intense membrane staining in ≤10% of tumor cells), and 3+ (complete and intense membrane staining in >10% of tumor cells). For this study, HER2 scoring was reclassified as negative (0 and 1+), low positive (2+), and positive (3+). EGFR and CK5/6 were evaluated for cytoplasmic and/or membranous staining and graded as negative (0%; complete absence of staining), low positive (estimated 1–10% tumor cells staining positive), and positive (>10% tumor cells staining positive). The pathologist noted whether the cores contained tumor cells (loss of tumor region can occur due to serial sectioning); or were unevaluable (due to core loss during sectioning or staining, or tumor cells were obscured for evaluation because of staining artifacts or tissue folding; see Supplementary 1).
Digitization and semi-automated IHC quantification
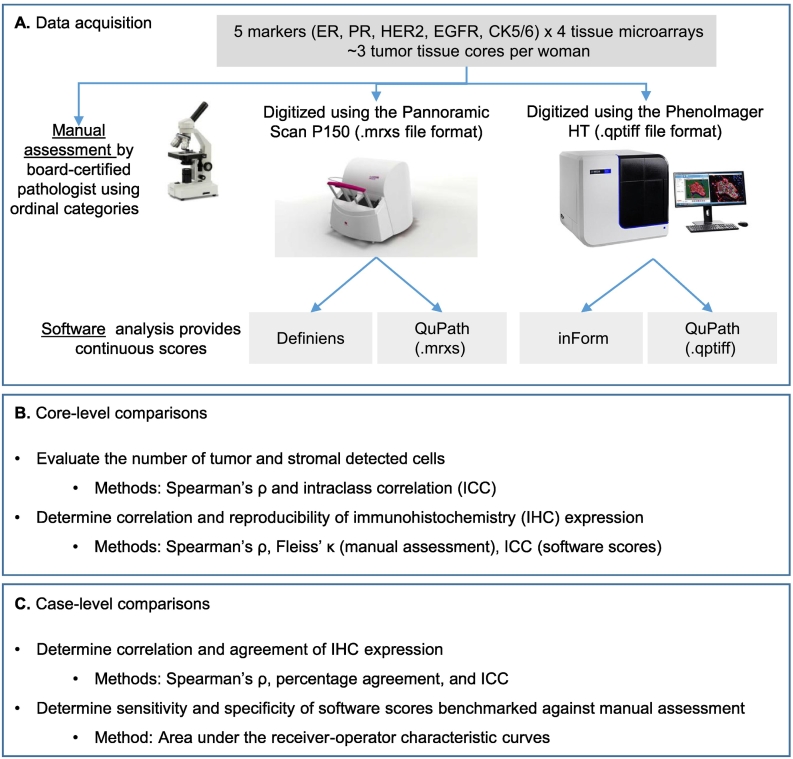
Twenty TMA sections (5 markers x 4 TMA slides) were simultaneously digitized at 20× by 2 whole-slide scanners—Pannoramic Scan P150 (3DHISTECH Ltd, Budapest, Hungary; 0.33 μm per pixel) and PhenoImager HT (formerly Vectra Polaris, Akoya Biosciences, Marlborough, MA; 0.50 μm per pixel). Since 2010, our established workflow in the NHS/NHSII involves digitizing the TMAs using the Pannoramic Scan and analyzing the images using Definiens Tissue Studio® (version 4.4.2; Definiens AG, Munich, Germany; Heng lab).8,13,19 In 2021, the Spatial Transcriptomics Unit at BIDMC acquired the PhenoImager HT scanner and its companion image analysis software, inForm® (version 2.5). The older Pannoramic Scan’s .mrxs file format was incompatible with inForm while the newer PhenoImager HT .qptiff files were incompatible with Definiens version 4.4.2. Both .mrxs and .qptiff were compatible with QuPath, an open source software.21 Hence QuPath version 0.3.0 was employed for comparison and evaluated whether images acquired from different scanners may affect IHC quantification (see Fig. 1A). Definiens, inForm, and QuPath produced 0–100% continuous estimate of immunoreactive cells for all 5 markers.
Fig. 1.
Workflow for data acquisition and analyses for this study. Twenty tissue microarray (TMA) slides were scanned by both scanners and by 3 software applications (A). A board-certified pathologist manually scored the TMA slides. Rationale and explanation of the type of analysis to be conducted at core-level (B) and when data were summarized to case-level (C).
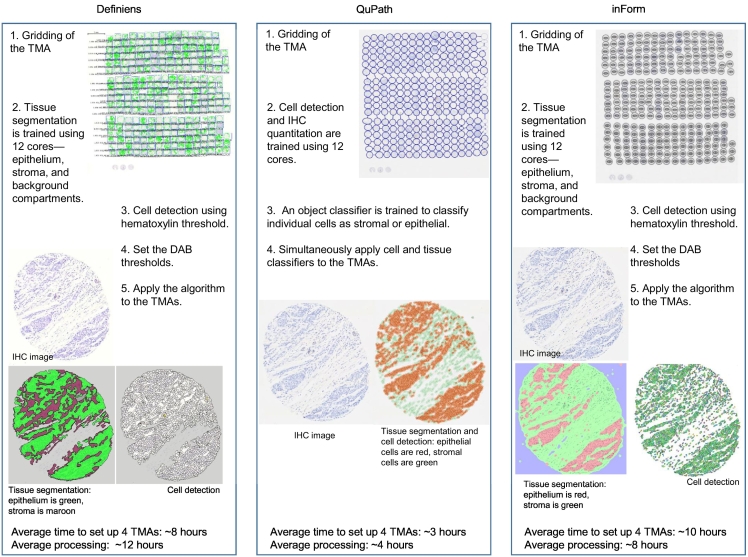
To eliminate inter-operator subjectivity, a single operator (VCB) performed all the software analyses. For each marker, we randomly selected 12 tissue cores on 1 of the 4 TMAs manually scored as negative (n=4), low positive (n=4), and positive (n=4; see Supplementary 2). These 12 cores were used across the software applications to: (1) train tissue segmentation into epithelium (i.e., tumor), stroma, and background; and (2) set the optimized IHC thresholding level (i.e., minimum intensity to score a cell as immunoreactive; see Supplementary 3). Fig. 2 is an overview of the workflow for each software.
Fig. 2.
Overview of the workflow for each software to quantify immunoreactive cells.
We used the default tissue segmentation classifier models in Definiens (unspecified) and inForm (trainable tissue segmentation with a large-scale pattern of segmentation). The best performing object classifier in QuPath was random trees. For cell detection, we used the in-built method in Definiens (unspecified) and inForm (adaptive cell segmentation). For QuPath, we used the “positive cell detection” command where we optimized the intensity thresholds for hematoxylin, nucleus/cytoplasm (mean optical density; see Supplementary 3) and used the default values for all other parameters.
Statistical analysis
We first compared the number of cells detected as tumor or stromal between the software applications for each core (Fig. 1B). This comparison was to verify that downstream IHC expression results were minimally affected by differences in the number of detected cells albeit potential misclassification by each software application. As tissue composition for each core was expected to be slightly different between serial 5 μm TMA sections, this part of the analysis was conducted on cores across 20 TMAs. We used Spearman’s ρ to correlate the number of cells detected as tumor or stromal between software. We used the intraclass correlation coefficient (ICC) to test the inter-rater reliability (ICC (C, κ); i.e., between software). ICC (A, κ) was used to test the intra-rater reliability (i.e., within each software) of the number of tumor or stromal cells detected across 2–15 cores (5 markers x 3 cores) belonging to each woman. Spearman’s ρ or ICC score of 1 indicates perfect correlation or similarity.
Next, we compared core-levelIHC expression. Since the 5 IHC markers were predominantly expressed by tumor cells, we evaluated IHC expression in regions classified as tumor/epithelium by the algorithms. For each marker, we compared the %positive score per core between manual and software (Spearman’s ρ). To evaluate the reproducibility of a marker’s IHC expression score across 2 or 3 cores pertaining to a woman, we used Fleiss’ κ for manual scores and ICC (A, κ) for software-derived scores.
For case-level comparisons, we summarized the marker’s manual and continuous scores across each woman’s cores. For manual assessment, the highest manual score was assigned to each case—the case was classified as negative if all cores were negative, low positive if at least 1 core was scored as low positive and others were negative, and positive if at least 1 core was scored as positive. For software-derived scores, the weighted average %positive was computed for each woman, i.e., the total number of positive cells/nuclei across a woman’s cores divided by the total number of tumor cells/nuclei present across the woman’s cores, and multiplied by 100. The weighted average %positive approach allows us to better capture tumor heterogeneity across a woman’s cores sampled from various areas of the tumor. As an added stringent measure, women with <100 total number of tumor cells across their cores were excluded at this stage.
For analyses at the case level, we correlated each marker’s IHC expression between manual and software results using Spearman’s ρ. Next, we categorized software-derived weighted average %positive scores into negative (<1%), low positive (1–10%), or positive (>10%) to evaluate the agreement between manual and software using %agreement and ICC (A, κ). Lastly, we created the receiver-operator characteristic (ROC) curves and utilized the area under the curve (AUC) values to determine the sensitivity and specificity of software-derived weighted average %positive scores benchmarked against binarized manual score (negative versus low positive/positive). Fig. 1C summarizes the analyses performed at the case level.
Results
IHC expression for each marker was evaluated on an average of 935 cores pertaining to 367 women. For each marker, an average of 12.3% cores had no tumor while an average of 9.6% cores were missing or not evaluable across 4 TMAs (Supplementary 1). The majority of these 367 women had Stage I disease (45.5%; Table 1). Their tumors were also mostly ER+/HER2- (64.0%) and moderately differentiated (35.4%).
Table 1.
Characteristics of the Nurses’ Health Study (NHS) and NHSII participants in this study.
| NHS | NHSII | |
|---|---|---|
| n | 192 | 175 |
| Age at breast cancer diagnosis, mean (SD) | 72 (6.7) | 52 (4.4) |
| Calendar year of breast cancer diagnosis, median (IQR) | 2008 (2007-2009) | 2006 (2005 -2007) |
| Tumor grade, n (%) | ||
| Well-differentiated | 47 (24.9) | 27 (15.4) |
| Moderately differentiated | 72 (37.5) | 58 (33.1) |
| Poorly/undifferentiated | 38 (19.8) | 35 (20.0) |
| Unknown | 35 (18.2) | 55 (31.4) |
| Tumor size, n (%) | ||
| ≤1.0 cm | 52 (27.1) | 31 (17.7) |
| 1–2 cm | 81 (42.2) | 52 (29.7) |
| 2–4 cm | 32 (16.7) | 31 (17.7) |
| >4 cm | 5 (2.6) | 8 (4.6) |
| Missing | 22 (11.5) | 53 (30.3) |
| Node status, n (%) | ||
| 0 | 155 (80.7) | 130 (74.3) |
| 1–3 | 29 (15.1) | 36 (20.6) |
| 4–9 | 4 (2.1) | 5 (2.9) |
| 10+ | 4 (2.1) | 3 (1.7) |
| Metastatic at diagnosis | 0 (0.0) | 1 (0.6) |
| Stage, n (%) | ||
| 0 (In situ) | 0 (0.0) | 0 (0.0) |
| 1 | 105 (54.7) | 62 (35.4) |
| 2 | 47 (24.5) | 46 (26.3) |
| 3 | 9 (4.7) | 12 (6.9) |
| 4 | 0 (0.0) | 1 (0.5) |
| Missing | 31 (16.2) | 54 (30.9) |
| Molecular subtype status, n (%) | ||
| ER+/PR+/HER2- or ER+/PR-/HER2- | 141 (73.4) | 94 (53.7) |
| ER+/PR+/HER2+ or ER+/PR-/HER2+ | 0 (0.0) | 2 (1.1) |
| ER-/PR-/HER2+ | 9 (4.7) | 8 (4.6) |
| ER-/PR-/HER2- | 16 (8.3) | 13 (7.4) |
| Missing | 26 (13.5) | 58 (33.1) |
Tumor and stromal cell counts: Core-level comparison
The number of detected tumor cells was highly comparable while the number of detected stromal cells was more variable between the software applications. The inter-rater reliability among software to detect the number of cells as tumor is ICC 0.92 (95% confidence interval (CI) 0.91–0.92) and stromal is 0.67 (95% CI of 0.66–0.69).
There were varying strengths of pair-wise correlations between software applications, most likely due to the variability in the classification algorithms employed by each software, notwithstanding the fact that each overall analysis algorithm was built by the same personnel. The ρ for the number of tumor cells detected ranged from 0.74 between QuPath (.mrxs) and inForm to 0.87 between Definiens and QuPath (.qptiff; all p<0.001; Supplementary 4A). There were lower pair-wise correlations for the number of stromal cell detected, ranging from ρ=0.43 (between Definiens and inForm) to ρ=0.80 (between QuPath (.mrxs) and QuPath (.qptiff); all p<0.001; Supplementary 4B). The correlation within the same file formats was moderate (.mrxs: ρ=0.78 for tumor and ρ=0.56 for stromal cells; .qptiff: ρ=0.81 for tumor and ρ=0.69 for stromal cells; Supplementary 4). Scanners had minimal effect on cell counts as correlation was very good when using QuPath to analyze both file formats (ρ=0.83 for tumor and ρ=0.80 for stromal cells; Supplementary 4).
The intra-rater reliability of the software applications to detect the number of tumor cells across cores belonging to each woman ranged from ICC 0.91 (QuPath .qptiff) to 0.94 (QuPath .mrxs; Supplementary 5). The ICC for the number of detected stromal cells ranged from 0.79 (inForm) to 0.94 (QuPath .mrxs; Supplementary 5). These excellent ICC scores reflect the high degree of similarity in tissue composition among the cores representing each woman.
IHC quantification: Core-level comparison
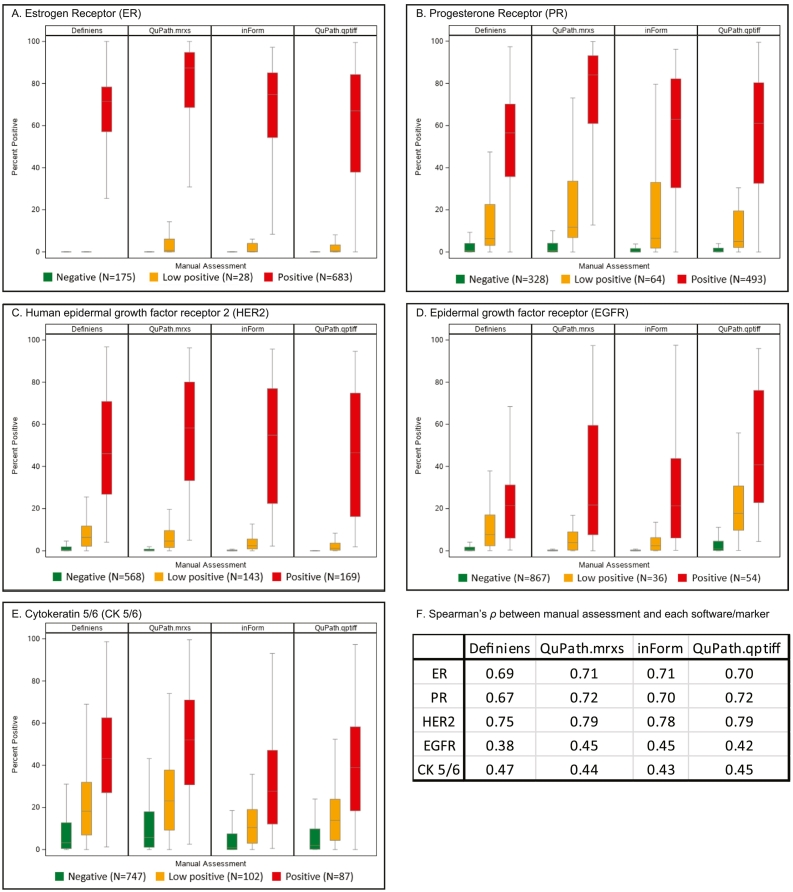
Fig. 3 displays boxplots correlating each marker’s manual scores with software-derived scores per core. The correlations were the highest for HER2 (ρ ranging from 0.75 to 0.79), followed by ER (0.69–0.71), PR (0.67–0.72), CK5/6 (0.43–0.47), and EGFR (0.38–0.45; p<0.001 for all markers).
Fig. 3.
Correlation of tumor immunohistochemistry expression between manual (ordinal categories of negative, low positive, and positive) and software-derived scores (%positivity) per core. Estrogen receptor, ER; progesterone receptor, PR; human epidermal growth factor receptor 2, HER2; epithelial growth factor receptor, EGFR; cytokeratin 5/6, CK5/6.
Across a woman’s cores, there was perfect agreement for EGFR (κ=1.00) and CK5/6 (κ=1.00), and moderate agreement for ER (κ=0.58), PR (κ=0.54), and HER2 (κ=0.50), when assessed manually (Table 2). ICC scores for each marker were similar across the software applications (Table 2). This indicates that heterogeneity in tumor marker expression was driving the variability of %positive scores between a woman’s cores (Table 2).
Table 2.
Reproducibility of each marker’s immunohistochemistry expression when manually assessed (core-level) or quantified using the software across each woman’s cores (maximum of 3).
| ER | PR | HER2 | EGFR | CK5/6 | |
|---|---|---|---|---|---|
| Manual | |||||
| Positive, n (%) | 683 (77.1) | 493 (55.7) | 169 (19.2) | 54 (5.6) | 87 (9.3) |
| Low positive, n (%) | 28 (3.2) | 64 (7.2) | 143 (16.3) | 36 (3.8) | 102 (10.9) |
| Negative, n (%) | 175 (19.7) | 328 (37.1) | 568 (64.5) | 867 (90.1) | 747 (79.8) |
| Fleiss’ κ (SE) | 0.58 (0.01) | 0.54 (0.01) | 0.50 (0.01) | 1.00 (0.02) | 1.00 (0.01) |
| Definiens (.mrxs) | |||||
| n | 766 | 790 | 783 | 802 | 778 |
| % positive, mean (SD) | 47.9 (34.4) | 33.6 (31.1) | 11.8 (21.6) | 3.0 (7.5) | 13.7 (18.5) |
| ICC (95%CI) | 0.81 (0.77, 0.84) | 0.48 (0.41, 0.56) | 0.85 (0.82, 0.87) | 0.66 (0.60, 0.71) | 0.62 (0.56, 0.68) |
| QuPath (.mrxs) | |||||
| n | 837 | 838 | 829 | 899 | 854 |
| % positive, mean (SD) | 59.6 (38.9) | 48.0 (39.7) | 13.0 (24.9) | 2.7 (10.6) | 17.5 (20.9) |
| ICC (95%CI) | 0.88 (0.86, 0.90) | 0.56 (0.49, 0.62) | 0.90 (0.88, 0.92) | 0.76 (0.72, 0.80) | 0.61 (0.54, 0.66) |
| Inform (.qptiff) | |||||
| n | 836 | 856 | 851 | 895 | 862 |
| % positive, mean (SD) | 51.9 (34.9) | 36.2 (35.2) | 11.0 (23.4) | 2.5 (9.3) | 9.7 (15.6) |
| ICC (95%CI) | 0.86 (0.84, 0.89) | 0.48 (0.41, 0.54) | 0.91 (0.89, 0.92) | 0.78 (0.74, 0.82) | 0.55 (0.49, 0.62) |
| QuPath (.qptiff) | |||||
| n | 865 | 865 | 845 | 898 | 861 |
| % positive, mean (SD) | 46.6 (35.4) | 35.7 (35.7) | 9.9 (22.7) | 8.0 (15.1) | 11.2 (16.8) |
| ICC (95%CI) | 0.85 (0.82, 0.87) | 0.50 (0.43, 0.57) | 0.91 (0.89, 0.93) | 0.75 (0.71, 0.79) | 0.61 (0.55, 0.66) |
Estrogen receptor, ER; progesterone receptor, PR; human epidermal growth factor receptor 2, HER2; epithelial growth factor receptor, EGFR; cytokeratin 5/6, CK5/6; intraclass correlation coefficient, ICC; standard error, SE; standard deviation, SD.
IHC quantification: Case-level comparison
There were good correlations between manual scores (each case was represented by the highest ordinal category across its cores, i.e., negative, low positive, and positive) and continuous software-derived scores (weighted average %positive) for all 5 markers. PR had the best correlations between manual and software (ρ ranging from 0.79 to 0.82) while correlations for EGFR and CK5/6 were lower than for ER, PR, and HER2 (0.43–0.49 for EGFR and 0.47–0.52 for CK5/6; p<0.001 for all markers; Supplementary 6). QuPath (.mrxs) had the highest ρ with manual scores for HER2, and EGFR; QuPath (.qptiff) for PR; and Definiens for ER and CK5/6. Software applications were highly comparable to each other, as demonstrated by ρ ranging from 0.80 to 0.99 across the 5 markers (Supplementary 6).
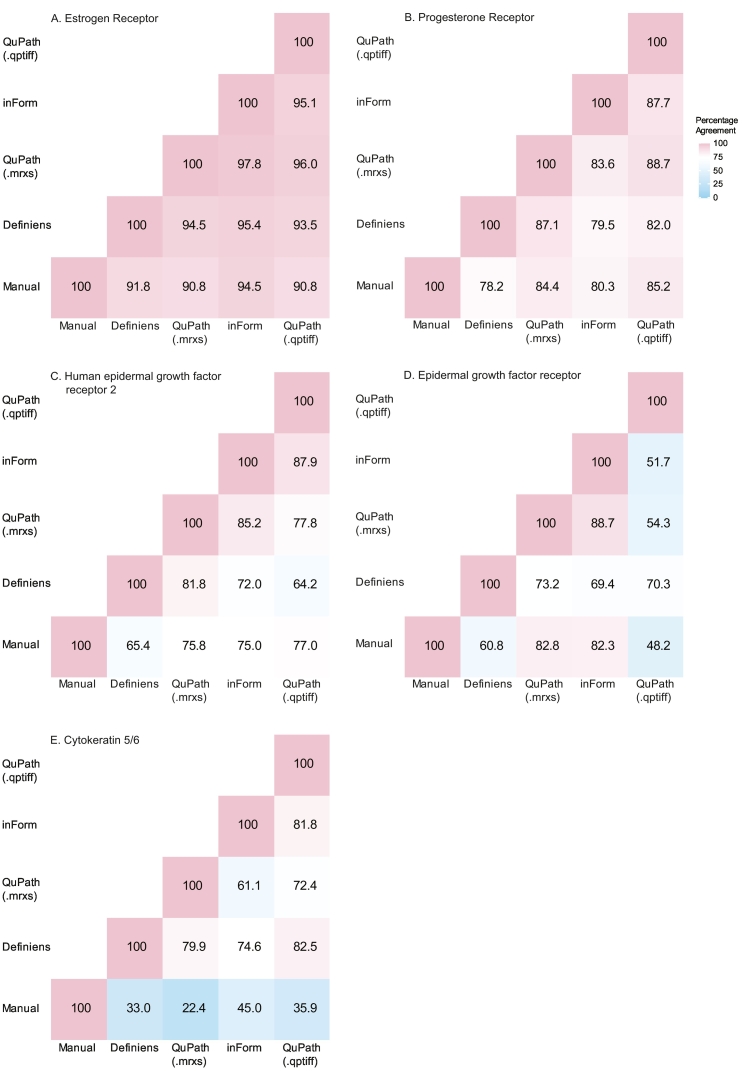
Next, we used the weighted average %positive scores approach to compare the agreement between manual and each software applications. Weighted average %positive scores were grouped into negative (<1%), low positive (1–10%), or positive (>10%) to evaluate average %agreement between manual and each software application. The average %agreements were excellent for ER (ranging from 90.8% to 94.5%) and PR (78.2%–85.2%), and moderate for HER2 (65.4%–77.0%; Fig. 4). Percentage agreements were highly variable for EGFR (48.2%–82.8%) and poor for CK5/6 (22.4%–45.0%). QuPath (.qptiff) had the highest %agreement with manual scores for PR and HER2; QuPath (.mrxs) for EGFR; and inForm for ER and CK5/6. The average %agreement between software applications were almost always better than when each software was benchmarked against manual scores (Fig. 4). ER had the highest reproducibility of the category assigned to each case across the 5 methods with an ICC score of 0.94, (95% CI 0.92–0.94), followed by PR (0.85 (0.83–0.87)), HER2 (0.81 (0.78–0.83)), EGFR (0.53 (0.48–0.58), and CK5/6 (0.49 (0.44–0.54)).
Fig. 4.
Case-level percentage agreements between manual and software-derived scores for each marker. For manual assessment, each case was represented by the highest ordinal category across its cores (negative, low positive, and positive). For software applications, the weighted average percent positive was calculated to represent each case and grouped as negative (<1%), low positive (1–10%), or positive (>10%).
The lower %agreements and ICC scores for EGFR and CK5/6 indicated that different cut-offs were needed to group their software-derived continuous scores to compare with manual scores. We dichotomized the weighted average %positive scores for EGFR and CK5/6 into ≤10% or >10%23 and compared it to dichotomized manual scores (negative/low positive versus positive). Average %agreement improved to 84.3%–97.8% for EGFR and 57.1%–75.3% for CK5/6 (Supplementary 7). InForm had the best %agreement with manual scores for both markers using dichotomized groups.
In Fig. 5, all AUCs were ≥0.83, indicating the software-derived scores were highly sensitive and specific when benchmarked against binarized manual scores (negative versus low positive/positive). Lastly, software applications were ranked based on the highest ρ or %agreement when benchmarked against manual scores. QuPath was the top-ranking software in 3 out of the 4 assessments (Supplementary 8).
Fig. 5.
Area under the receiver-operator characteristic curves (AUCs) to assess the specificity and sensitivity of software-derived weighted average %positivity benchmarked against manual assessment (negative versus low positive/positive). Estrogen receptor, ER; progesterone receptor, PR; human epidermal growth factor receptor 2, HER2; epithelial growth factor receptor, EGFR; cytokeratin 5/6, CK5/6.
Discussion
This study compared the IHC quantification of 5 breast cancer markers between 3 software applications and benchmarked their results to the current standard of pathology manually assessed scores. We showed that whilst our TMAs were well constructed—excellent ICC sores indicating highly similar tissue composition among a woman’s cores—the cores displayed heterogeneous tumor marker expression. All 3 software applications were highly comparable to each other and when compared to manual scores, especially in the quantification of ER, PR, and HER2 expression. QuPath was the overall top performing software in terms of providing the best %agreement with our manual scores, and the requiring the least time to set up and apply. Therefore, for our future work in the NHS/NHSII, QuPath is an excellent alternative software to assess tumor biomarker expression on new TMA blocks consisting of newly diagnosed breast cancer cases, and new data assessed by QuPath would be very comparable to our existing data quantified using Definiens.
CK5/6 and EGFR IHC staining are clinically used to further classify triple-negative breast cancers as basal subtypes.10,14,24 Since this study population only had 7.9% triple-negative cases, the lower ρ values for CK5/6 and EGFR comparing software-derived and manual cores at core-level, and perfect agreements across a woman’s cores when assessed manually, were driven by the high number of negative cores. As for ER, PR, and HER2, work by us19 and others[25], [26], [27] have generally demonstrated agreement between pathologist and software-derived scores. This current study further adds to the knowledge that IHC quantification of these 5 markers is highly reproducible across Definiens, inForm, and QuPath.
For IHC quantification, QuPath has previously been compared to HALO (Indica Labs) and QuantCenter (3DHISTECH) in the evaluation of Ki67;28 ImmunoRatio (ImageJ plug-in) and Visiopharm (Visiopharm) for Ki67, cleaved Poly (ADP-ribose) polymerase, and pan-cytokeratin;29 and WEKA (ImageJ plug-in) for TP53 IHC expression.30 Our study is the first to systematically compare QuPath with Definiens and inForm. One unique feature of QuPath is that it allows users to set deconvolution stains to optimize the hematoxylin, DAB (3,3'-diaminobenzidine), and residual stain vectors using red, green, and blue color channels. This allows for more specific control when defining what is and is not stained prior to detecting and classifying cells31. This additional capability allows the QuPath classifier to be better refined compared to Definiens and inForm whereby color thresholding cannot be more specifically adjusted. That, and the different classification method used in QuPath, may partly explain why the mean %positivity scores were always higher using QuPath than Definiens or inForm.
Our operator (VCB) had 2 years of experience using Definiens prior to using inForm and QuPath. VCB’s experience with Definiens allowed her to pick up inForm and QuPath relatively quickly. The usability of each of the software applications depends on the needs of the operator. Definiens and inForm are recommended for novices as the applications have fixed workflows to guide algorithm construction and data analysis. InForm (version 2.5) is more user friendly than Definiens (version 4.4.2). We were unable to update our version of Definiens as Definiens is now a privately owned software. In contrast, QuPath has many capabilities to customize the algorithms, and is geared towards advanced users. The main limitation for all 3 software applications was the detection and aligning of the cores during the TMA de-array process; and all 3 software applications required operator input to locate, edit, and label the tissue cores. For a user with TMA de-array experience, QuPath was the fastest to execute (~45 min), followed by inForm (~1 h), and Definiens (~2 h). To set thresholding, Definiens’ graphical user interface (GUI) for this task was the most straightforward. inForm GUI enables easy visualization but requires some coding experience to apply the thresholding to all tissue components. Lastly, QuPath allows for a lot of customization which can be overwhelming for a novice but welcomed by advanced users.
The strengths of our study include using a population-based, prospective collection of tumor tissue samples, analyzing a large number of cores and women across multiple TMAs, and evaluating the 5 clinically important breast cancer markers. Additional strengths of this study include our pathologist providing manual scores for every core, and having the same operator build and execute the software algorithms. The consistent ρ values and high AUCs for software-derived and manual scores indicated that IHC thresholding had been optimized for each software and reiterated the importance of using pathologists’ scores for IHC thresholding. We showed that despite potential cell misclassification by each software application, the number of detected tumor cells were very comparable across the software applications. This allowed us to interpret the variability in IHC expression as related to the different software’s classifier models and tumor heterogeneity, and not due to the number of cells detected. One limitation of our study was that we only evaluated tumor markers, hence the reliability and/or reproducibility of these software applications in evaluating stromal markers compared to manual assessment remains unknown. We previously reported that the correlations between manual and Definiens scores for some stromal markers—CD4, CD8, CD20, CD163, and glutamyl-prolyl-tRNA synthetase—ranged from ρ=0.10 for CD163 to ρ=0.72 for CD8 for 132 cases on 1 TMA19. Future work will evaluate the reproducibility of these software applications in the IHC quantification of stromal markers.
In conclusion, Definiens, inForm, and QuPath were highly comparable to each other and were reliable when benchmarked against pathology manual scores in quantifying the IHC expression of ER, PR, HER2, EGFR, and CK5/6. QuPath had the highest correlation and %agreement with manual scores and is a reliable open-source tool to automate IHC expression quantification.
Conflict of Interest
All authors do not have any conflict of interest.
Author Contributions
YJH developed the study concept and design. RMT, GMB, VCB, and HZ were involved in the creation of the tissue microarrays and acquisition of the immunohistochemistry data. TW, RMT, and AHE acquired the participant data. YJH and TW performed the data analysis. YJH, VCB, MV, and LCC provided data interpretation. All authors were involved in the writing and editing of the manuscript.
Funding
This work was supported by the National Cancer Institute at the National Institute of Health (P50CA168504, UM1CA186107, U01CA176726, and P01CA87969).
Data Availability Statement
The data that support the findings of this study are available from the Nurses’ Health Studies, however they are not publicly available. Investigators interested in using the data can request access, and feasibility will be discussed at an investigators meeting. Limits are not placed on scientific questions or methods, and there is no requirement for co-authorship. Additional data sharing information and policy details can be accessed at http://www.nurseshealthstudy.org/researchers.
Data availability
NA
Declaration of interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
The authors would like to acknowledge the contribution to this study from central cancer registries supported through the Centers for Disease Control and Prevention’s National Program of Cancer Registries (NPCR) and/or the National Cancer Institute’s Surveillance, Epidemiology, and End Results (SEER) Program. Central registries may also be supported by state agencies, universities, and cancer centers. Participating central cancer registries include the following: Alabama, Alaska, Arizona, Arkansas, California, Colorado, Connecticut, Delaware, Florida, Georgia, Hawaii, Idaho, Indiana, Iowa, Kentucky, Louisiana, Massachusetts, Maine, Maryland, Michigan, Mississippi, Montana, Nebraska, Nevada, New Hampshire, New Jersey, New Mexico, New York, North Carolina, North Dakota, Ohio, Oklahoma, Oregon, Pennsylvania, Puerto Rico, Rhode Island, Seattle SEER Registry, South Carolina, Tennessee, Texas, Utah, Virginia, West Virginia, Wyoming.
We thank Specialized Histopathology Core at DF/HCC for providing TMA construction and IHC assay services. DF/HCC is supported in part by an NCI Cancer Center Support Grant (5P30CA06516). We thank the Spatial Transcriptomics Unit at BIDMC for the use of the Akoya Biosciences PhenoImager HT scanner and inForm. We also thank Dr. Linden C. Wyatt for his inForm expertise and reading of this manuscript.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jpi.2022.100118.
Appendix A. Supplementary data
Supplementary material
References
- 1.Tamimi R.M., Baer H.J., Marotti J., Galan M., Galaburda L., Fu Y., et al. Comparison of molecular phenotypes of ductal carcinoma in situ and invasive breast cancer. Breast Cancer Res. 2008;10(4):R67. doi: 10.1186/bcr2128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Collins L.C., Wang Y., Connolly J.L., Baer H.J., Hu R., Schnitt S.J., et al. Potential role of tissue microarrays for the study of biomarker expression in benign breast disease and normal breast tissue. Appl. Immunohistochem. Mol. Morphol. 2009;17(5):438–441. doi: 10.1097/PAI.0b013e3181993d86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Horne H.N., Oh H., Sherman M.E., Palakal M., Hewitt S.M., Schmidt M.K., et al. E-cadherin breast tumor expression, risk factors and survival: Pooled analysis of 5,933 cases from 12 studies in the Breast Cancer Association Consortium. Sci. Rep. 2018;8(1):6574. doi: 10.1038/s41598-018-23733-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Abubakar M., Chang‐Claude J., Ali H.R., Chatterjee N., Coulson P., Daley F., et al. Etiology of hormone receptor positive breast cancer differs by levels of histologic grade and proliferation. Int. J. Cancer 2018;143(4):746. doi: 10.1002/IJC.31352. [DOI] [PMC free article] [PubMed]
- 5.Huh S.J., Oh H., Peterson M.A., Almendro V., Hu R., Bowden M., et al. The proliferative activity of mammary epithelial cells in normal tissue predicts breast cancer risk in premenopausal women. Cancer Res. 2016;76(7):1926–1934. doi: 10.1158/0008-5472.CAN-15-1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Oh H., Eliassen A.H., Wang M., Smith-Warner S.A., Beck A.H., Schnitt S.J., et al. Expression of estrogen receptor, progesterone receptor, and Ki67 in normal breast tissue in relation to subsequent risk of breast cancer. npj Breast Cancer 2016;2(1):16032. doi: 10.1038/npjbcancer.2016.32. [DOI] [PMC free article] [PubMed]
- 7.Tamimi R., Colditz G., Wang Y., Collins L.C., Hu R., Rosner B., et al. Expression of IGF1R in normal breast tissue and subsequent risk of breast cancer. Breast Cancer Res. Treat. 2011;128(1):243–250. doi: 10.1007/S10549-010-1313-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kensler K.H., Beca F., Baker G.M., Heng Y.J., Beck A.H., Schnitt S.J., et al. Androgen receptor expression in normal breast tissue and subsequent breast cancer risk. npj Breast Cancer 2018;4(1):33. PMCID: PMC6155011. doi:10.1038/s41523-018-0085-3. [DOI] [PMC free article] [PubMed]
- 9.Sisti J.S., Collins L.C., Beck A.H., Tamimi R.M., Rosner B.A., Eliassen A.H. Reproductive risk factors in relation to molecular subtypes of breast cancer: Results from the nurses’ health studies. Int. J. Cancer. 2016;138(10):2346–2356. doi: 10.1002/ijc.29968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tamimi R.M., Colditz G.A., Hazra A., Baer H.J., Hankinson S.E., Rosner B., et al. Traditional breast cancer risk factors in relation to molecular subtypes of breast cancer. Breast Cancer Res. Treat. 2012;131(1):159–167. doi: 10.1007/s10549-011-1702-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hirko K.A., Chen W.Y., Willett W.C., Rosner B.A., Hankinson S.E., Beck A.H., et al. Alcohol consumption and risk of breast cancer by molecular subtype: Prospective analysis of the nurses’ health study after 26 years of follow-up. Int. J. Cancer. 2016;138(5):1094–1101. doi: 10.1002/ijc.29861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang J., Zhang X., Beck A.H., Collins L.C., Chen W.Y., Tamimi R.M., et al. Alcohol Consumption and Risk of Breast Cancer by Tumor Receptor Expression. Horm. Cancer. 2015;6(5-6):237–246. doi: 10.1007/s12672-015-0235-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McGee E.E., Kim C.H., Wang M., Spiegelman D., Stover D.G., Heng Y.J., et al. Erythrocyte membrane fatty acids and breast cancer risk by tumor tissue expression of immuno-inflammatory markers and fatty acid synthase: A nested case-control study. Breast Cancer Res. 2020;22(1):78. doi: 10.1186/s13058-020-01316-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kensler K.H., Sankar V.N., Wang J., Zhang X., Rubadue C.A., Baker G.M., et al. PAM50 molecular intrinsic subtypes in the nurses’ health Study cohorts. Cancer Epidemiol. Biomark. Prev. 2019;28(4):798-806. PMCID: PMC6449178. doi:10.1158/1055-9965.EPI-18-0863. [DOI] [PMC free article] [PubMed]
- 15.Dawood S., Hu R., Homes M.D., Collins L.C., Schnitt S.J., Connolly J., et al. Defining breast cancer prognosis based on molecular phenotypes: Results from a large cohort study. Breast Cancer Res. Treat. 2011;126(1):185–192. doi: 10.1007/s10549-010-1113-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu Y., Tamimi R.M., Collins L.C., Schnitt S.J., Gilmore H.L., Connolly J.L., et al. The association between vascular endothelial growth factor expression in invasive breast cancer and survival varies with intrinsic subtypes and use of adjuvant systemic therapy: Results from the Nurses’ Health Study. Breast Cancer Res. Treat. 2011;129(1):175–184. doi: 10.1007/s10549-011-1432-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Santagata S., Hu R., Lin N.U., Mendillo M.L., Collins L.C., Hankinson S.E., et al. High levels of nuclear heat-shock factor 1 (HSF1) are associated with poor prognosis in breast cancer. Proc. Natl. Acad. Sci. U. S. A. 2011;108(45):18378–18383. doi: 10.1073/pnas.1115031108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Aeffner F., Wilson K., Martin N.T., Black J.C., Hendriks C.L.L., Bolon B., et al. The gold standard paradox in digital image analysis: Manual versus automated scoring as ground truth. Arch. Pathol. Lab. Med. 2017;141(9):1267–1275. doi: 10.5858/arpa.2016-0386-RA. [DOI] [PubMed] [Google Scholar]
- 19.Roberts M.R., Baker G.M., Heng Y.J., Pyle M.E., Astone K., Rosner B.A., et al. Reliability of a computational platform as a surrogate for manually interpreted immunohistochemical markers in breast tumor tissue microarrays. Cancer Epidemiol. 2021;74 doi: 10.1016/j.canep.2021.101999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Feng Z., Bethmann D., Kappler M., Ballesteros-Merino C., Eckert A., Bell R.B., et al. Multiparametric immune profiling in HPV- oral squamous cell cancer. JCI insight. 2017;2(14) doi: 10.1172/jci.insight.93652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bankhead P., Loughrey M.B., Fernández J.A., Dombrowski Y., McArt D.G., Dunne P.D., et al. QuPath: Open source software for digital pathology image analysis. Sci. Rep. 2017;7(1):16878. doi: 10.1038/s41598-017-17204-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Colditz G.A., Hankinson S.E. The Nurses’ Health Study: lifestyle and health among women. Nat. Rev. Cancer. 2005;5(5):388–396. doi: 10.1038/nrc1608. [DOI] [PubMed] [Google Scholar]
- 23.Howat W.J., Blows F.M., Provenzano E., Brook M.N., Morris L., Gazinska P., et al. Performance of automated scoring of ER, PR, HER2, CK5/6 and EGFR in breast cancer tissue microarrays in the Breast Cancer Association Consortium. J. Pathol. Clin. Res. 2015;1(1):18–32. doi: 10.1002/cjp2.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guiu S., Michiels S., André F., Cortes J., Denkert C., Di Leo A., et al. Molecular subclasses of breast cancer: How do we define them? The IMPAKT 2012 working group statement. Ann. Oncol. 2012;23(12):2997–3006. doi: 10.1093/annonc/mds586. [DOI] [PubMed] [Google Scholar]
- 25.Lehr H.A., Jacobs T.W., Yaziji H., Schnitt S.J., Gown A.M. Quantitative evaluation of HER-2/neu status in breast cancer by fluorescence in situ hybridization and by immunohistochemistry with image analysis. Am. J. Clin. Pathol. 2001;115(6):814–822. doi: 10.1309/AJ84-50AK-1X1B-1Q4C. [DOI] [PubMed] [Google Scholar]
- 26.Rexhepaj E., Brennan D.J., Holloway P., Kay E.W., McCann A.H., Landberg G., et al. Novel image analysis approach for quantifying expression of nuclear proteins assessed by immunohistochemistry: Application to measurement of oestrogen and progesterone receptor levels in breast cancer. Breast Cancer Res. 2008;10(5) doi: 10.1186/bcr2187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bankhead P., Fernández J.A., McArt D.G., Boyle D.P., Li G., Loughrey M.B., et al. Integrated tumor identification and automated scoring minimizes pathologist involvement and provides new insights to key biomarkers in breast cancer. Lab. Investig. 2018;98(1):15–26. doi: 10.1038/labinvest.2017.131. [DOI] [PubMed] [Google Scholar]
- 28.Acs B., Pelekanou V., Bai Y., Martinez-Morilla S., Toki M., Leung S.C.Y., et al. Ki67 reproducibility using digital image analysis: an inter-platform and inter-operator study. Lab. Investig. 2019;99(1):107–117. doi: 10.1038/s41374-018-0123-7. [DOI] [PubMed] [Google Scholar]
- 29.Miles G.J., Powley I., Mohammed S., Howells L., Pringle J.H., Hammonds T., et al. Evaluating and comparing immunostaining and computational methods for spatial profiling of drug response in patient-derived explants. Lab. Investig. 2021;101(3):396–407. doi: 10.1038/s41374-020-00511-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Prall F., Hühns M. Quantitative evaluation of TP53 immunohistochemistry to predict gene mutations: lessons learnt from a series of colorectal carcinomas. Hum. Pathol. 2019;84:246–253. doi: 10.1016/j.humpath.2018.10.012. [DOI] [PubMed] [Google Scholar]
- 31.Ruifrok A.C., Johnston D.A. Quantification of histochemical staining by color deconvolution. Anal. Quant. Cytol. Histol. 2001;23(4):291–299. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material
Data Availability Statement
The data that support the findings of this study are available from the Nurses’ Health Studies, however they are not publicly available. Investigators interested in using the data can request access, and feasibility will be discussed at an investigators meeting. Limits are not placed on scientific questions or methods, and there is no requirement for co-authorship. Additional data sharing information and policy details can be accessed at http://www.nurseshealthstudy.org/researchers.
NA