Abstract
Identifying organs within histology images is a fundamental and non-trivial step in toxicological digital pathology workflows as multiple organs often appear on the same whole slide image (WSI). Previous works in automated tissue classification have investigated the use of single magnifications, and demonstrated limitations when attempting to identify small and contiguous organs at low magnifications. In order to overcome these shortcomings, we present a multi-magnification convolutional neural network (CNN), called MMO-Net, which employs context and cellular detail from different magnifications to facilitate the recognition of complex organs. Across N=320 WSI from 3 contract research organization (CRO) laboratories, we demonstrate state-of-the-art organ detection and segmentation performance of 7 rat organs with and without lesions: liver, kidney, thyroid gland, parathyroid gland, urinary bladder, salivary gland, and mandibular lymph node (AUROC=0.99–1.0 for all organs, Dice≥0.9 except parathyroid (0.73)). Evaluation takes place at both inter- and intra CRO levels, suggesting strong generalizability performance. Results are qualitatively reviewed using visualization masks to ensure separation of organs in close proximity (e.g., thyroid vs parathyroid glands). MMO-Net thus offers organ localization that serves as a potential quality control tool to validate WSI metadata and as a preprocessing step for subsequent organ-specific artificial intelligence (AI) use cases. To facilitate research in this area, all associated WSI and metadata used for this study are being made freely available, forming a first of its kind dataset for public use.
Keywords: Digital pathology, Convolutional neural networks, Organ identification, Multiple magnifications, Preclinical assessment, Quality control
Introduction
To advance development of drugs to cure and treat diseases, millions of histology slides are evaluated by toxicologic pathologists to assess safety of candidate therapeutic compounds prior to advancement into clinical trials. A preclinical study may include approximately 20–30 different organs for the evaluation of toxic effects of a given therapeutic compound. Given the large number of organs that are usually evaluated, it is a common practice to embed several organs within the same paraffin tissue block to save material and manual effort during tissue processing.1 As a result, there is a need to identify where and what organs are present in the slide and compare them against the expected organs to identify any discordance. Organs may be absent for a number of reasons, including: (1) insufficiently deep sectioning in the paraffin block, or (2) unexpected deviations from the study plan (e.g., tissues lost during processing). As a result, it becomes attractive to develop an automatic method for organ identification that detects and delineates the organs present in the WSI as a quality control (QC) step of the provided metadata while further enabling the development of organ-specific AI tools.
With the development of whole slide imaging technologies for digitization of glass slides, this workflow stands to be improved via the employment of machine and deep learning-based tools. Previously, in the toxicological pathology space, a set of deep learning (DL) models termed HistoNet were trained at single magnifications for identification of normal rat tissues and organs,2 showing good performance for some tissues, while noting their limited ability to differentiate morphologically similar tissues, as well as small and contiguous tissues. As discussed by Hoefling et al.,2 organ detection is non-trivial as some organs are small, anatomically connected, or embedded within other organs, thus forming one single contiguous tissue island (e.g., thyroid and parathyroid glands or lymph nodes and salivary glands). Pathologists address this challenge by examining information that is obtained from a combination of high and low magnifications during histological evaluations, thus balancing fine-grained and contextual visual information. Using a comparable computational technique, previous works in the clinical space demonstrate the advantages of using multiple magnifications combined in one single algorithm.3, 4, 5, 6, 7
When deploying such tools in the real-world settings, there is the additional challenge that these whole slide images (WSI) can originate from multiple sites, and thus show diminished model performance as a result of preanalytical variability. This variability can originate from protocol differences between contract research organization (CRO) laboratories relating to: (1) different tissue processing methods, (2) staining protocols, (3) sectioning thickness, or (4) digital image acquisition devices.8,9 Despite the existence of computer vision techniques for color normalization and augmentation to try to overcome these challenges,10,11 training and validating algorithms with heterogeneous sources of images appears to help in developing more robust and generalizable models.
Building on these works, we have constructed a preclinically deployable multi-head DL approach (MMO-Net) which uses as input a combination of magnifications of the same region to provide both context and detail. Furthermore, in line with the intended use case, we include organs with histopathological findings (i.e., lesions), as this is a common confounder in preclinical studies. These findings can be very heterogeneous and responsible for marked morphological deviations from a normal tissue, increasing the complexity of the classification task and thus warrant dedicated consideration.
In summary, our main contributions are the following:
-
●
We present MMO-Net, an organ identification network that makes use of multiple magnifications simultaneously to recognize complex organs.
-
●
We identify 7 rat organs (liver, kidney, thyroid gland, parathyroid gland, urinary bladder, salivary glands, and mandibular lymph node) with and without histopathological findings by spatially detecting and delineating the organs in a WSI through segmentation masks.
-
●
We built and are now publicly releasing a first-of-its-kind multi-centric dataset consisting of 320 WSI from 3 laboratories digitized across 2 scanners, containing over 20 different rat organs from control and treated animals.
-
●
We performed an inter/intra laboratory study design to mimic real-world workflows of identifying organs in toxicological pathology. We aim to deploy this tool in our facility to enable automatic organ metadata validation and subsequent organ-specific tool invocation in our production environment.
Materials and methods
Dataset and annotations
Nine preclinical rat studies with tissue processing performed at three different CRO laboratories (Lab A, B, and C) were selected. All studies were performed in Wistar Han rats except for study 3, performed in Sprague-Dawley rats. All experimental procedures were in accordance with the respective Swiss regulations and approved by the Cantonal Ethical Committee for Animal Research. The WSIs for this study contain organs with and without histopathological findings, and include scans from Hamamatsu (ndpi) and Aperio (svs) scanners (Table 1). Studies were selected to intentionally include significant staining variations to help develop and validate algorithms more applicable to our real-world setting (Table 1 and Fig. 1). The associated metadata of the organs included in each WSI are obtained from internal protocols that specify how the different organs are grouped per WSI.
Table 1.
Multicentric study set.
| Study | Laboratory | Format | Stain profile | Organs (n° of WSI) |
|---|---|---|---|---|
| Study 1 | Lab A | ndpi | Profile 1 | L (7), K (7), L-SG-LN (3), K-UB (3) |
| Study 2 | Lab A | ndpi | Profile 1 | L-SG-LN (10), K-UB (10), T-PT (10), NC (18) |
| Study 3 | Lab A | ndpi | Profile 1 | L (10), K (10) |
| Study 4 | Lab B | ndpi | Profile 2 | L-SG-LN (10), K-UB (10), T-PT (10), NC (18) |
| Study 5 | Lab B | svs | Profile 2 | L-SG-LN (10), K-UB (10), T-PT (10), NC (18) |
| Study 6 | Lab C | ndpi | Profile 1 | L (10), K (10) |
| Study 7 | Lab C | ndpi | Profile 1 | L-SG-LN (10), K-UB (10), T-PT (10), NC (18) |
| Study 8 | Lab C | svs | Profile 3 | L (10), K (10) |
| Study 9 | Lab C | svs | Profile 3 | L-SG-LN (10), K-UB (10), T-PT (10), NC (18) |
Liver (L), salivary glands (SG), mandibular lymph node (LN), kidney (K), urinary bladder (UB), thyroid (T) and parathyroid (PT) glands. Negative class (NC). ndpi and svs files originate from Hamamatsu and Aperio scanners, respectively. The dash symbol (-) represents organs that are grouped in the same tissue block. For each organ set in the negative class (9 in total), 2 WSIs/study were used.
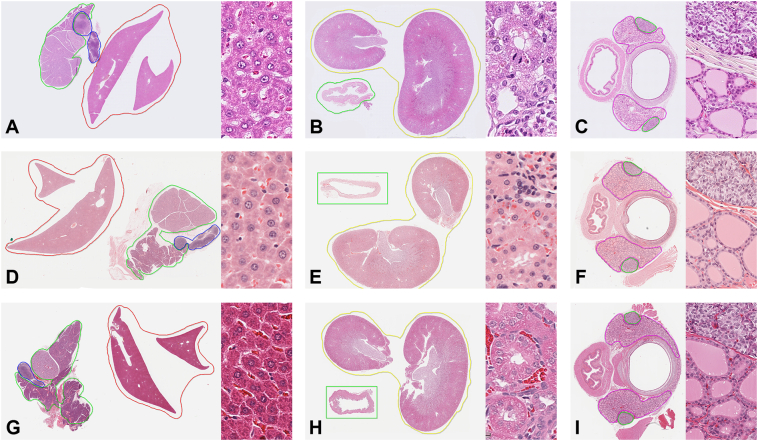
Fig. 1.
Staining variability across studies, with examples of the annotations performed. Study 2 (stain profile 1), Laboratory A (A, B, C); study 5 (stain profile 2), Laboratory B (D, E, F); study 9 (stain profile 3), Laboratory C (G, H, I). Liver, salivary glands, mandibular lymph node (A, D, G), kidney and urinary bladder (B, E, H), thyroid and parathyroid glands with trachea and esophagus (C, F, I).
The resulting dataset consists of N=320 WSI containing: liver, kidney, thyroid gland, parathyroid gland, urinary bladder, salivary gland, mandibular lymph node, and others (negative class) (see Table 2). Liver and kidney were selected due to their critical relevance in preclinical safety assessments. Organs frequently embedded with them were additionally selected, including submandibular lymph nodes and salivary glands (embedded with liver), and urinary bladder (embedded with the kidneys). The thyroid and parathyroid glands were also included as a use case to evaluate detection of small organs in close proximity. In addition, to aid in the positive selection for desired organs, various confounding organs from 9 organs sets were included as negative class for training the models (class “other”): adrenal glands, aorta, and ureters; lung and heart; stomach and intestine; skeletal muscle, sciatic nerve, mammary gland and skin; prostate and seminal vesicles; testis and epididymis; eye and harderian glands; bone with bone marrow; and spinal cord.
Table 2.
Total number of WSI used in the study.
| Organ | N° Studies | N° Laboratories | N° WSIs |
|---|---|---|---|
| Liver | 9 | 3 | 90 |
| Salivary gland, lymph node | 6 | 3 | 53 |
| Kidney | 9 | 3 | 90 |
| Urinary bladder | 6 | 3 | 53 |
| Thyroid, parathyroid | 5 | 3 | 50 |
| Other organs (Negative class) | 5 | 3 | 90 |
These WSI originating from different laboratories, and reflect the real scenario of source variability. In total, 320 WSI were used in this study. See Table 1 for more details.
Slides with histopathological findings were included for 3 organs (Fig. S1): liver, kidney, and thyroid gland. For these organs, if multiple concomitant lesions were present, the maximum lesion severity grade was considered as the organ severity grade label (see Supplemental Section 1 for additional details). Additional information with the histopathological findings can be found together with the WSI at https://doi.org/10.7303/syn30282632.12
The 7 target organs were annotated by a pathologist using HALO® v3.2 software (Indica Labs). Coarse annotations were drawn for organs which are clearly separated by background, while for touching organs, a more precise boundary delineation was performed. All annotations are exported into .xml files for subsequent data processing.
Overview of the Multi-Magnification Organ Network
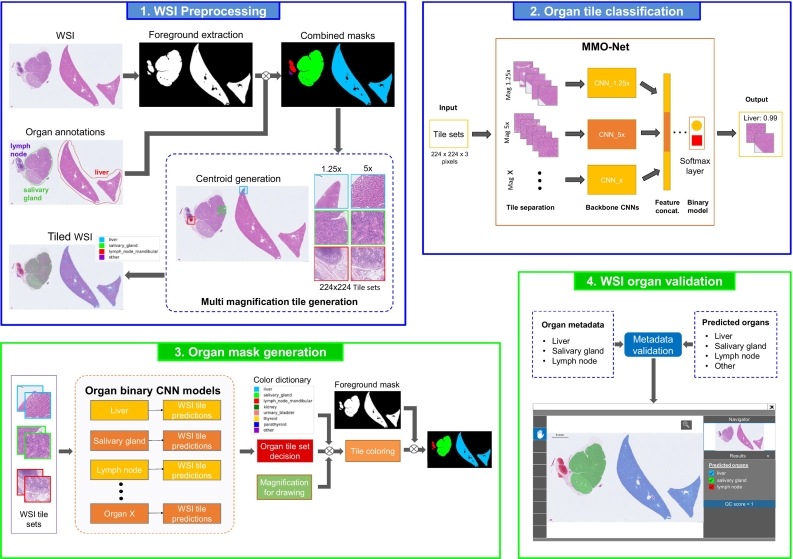
A flowchart of our approach is presented in Fig. 2 demonstrating how MMO-Net makes use of multiple magnifications simultaneously to mimic pathologist behavior during organ identification. We construct DL models per organ (one versus all) to easily enable individual model refinement and inclusion of additional organs during deployment without having to re-train and re-validate a more sophisticated multi-class classifier. Validation of the associated WSI metadata is performed after converting the classification tile predictions into segmentation masks for organ localization in the WSI space.
Fig. 2.
Summary of the proposed workflow for organ identification. Blue panels indicate Training and green panels refer to Inference. The workflow involves the generation of tiles at multiple magnifications to be used for our MMO-Net architecture to train binary models (panels 1 and 2). Subsequently, during inference, the multi-magnification tiles pass through all organ models to form the WSI level prediction mask (panel 3). Lastly, the organ metadata is validated comparing it with the organs predicted in the mask (panel 4).
WSI pre‐processing
Before model training, preprocessing steps consisted of: (1) identifying the tissue in the WSI (foreground extraction), (2) masking the tissue areas with the annotations made by the pathologist, and (3) generating, on tissue-detected areas only, the coordinates of multi-magnification tiles having the same centroid, termed tile sets (see Fig. S2).
During training, tile sets of size 224×224 pixels were dynamically extracted from the WSI at: (1) 1.25x (≈ 7.987 microns per pixel [mpp]) providing overall context and (2) 5x (≈ 1.997 mpp) to provide finer histologic details. Further details in Supplemental Section 2.
Multi-Magnification Organ Network
DenseNet-121 was chosen as the backbone CNN for MMO-Net due to its proven feature use efficiency and significantly reduced number of parameters.13,14
A visual representation of MMO-Net can be seen in Fig. 2 panel 2, where simultaneous dedicated networks learn organ feature representations from our “tile sets”. Subsequently, these dual magnification feature representations are concatenated in a fully connected layer followed by a final Softmax layer to output probability scores. For experimental comparison, DenseNet-121s were trained individually at both 1.25x and 5x. Implementation of the CNNs was done in the PyTorch framework using OpenSlide to read the WSIs and extract the image tiles. Training details are specified in Supplemental Section 3.
WSI organ mask generation
Once individual organ models are trained, WSI tile sets are passed through the models to produce the associated WSI organ segmentation mask (Fig. 2 panel 3). Organ maps are created by selecting, per pixel, the organ class with the highest SoftMax value (see Supplemental Section 4 for more information).
Assessment metrics
Classification performance
The performance of the organ classification models was assessed with the Area Under the Receiver Operating Characteristics curve (AUROC).15 A perfect model will have a value of 1.0, and a random model will have a value of 0.5.
Segmentation performance
To quantitatively assess the segmentation performance, the Dice Similarity Coefficient (DSC) was used to measure the overlap between the generated predicted organ masks and the ground truth. It is calculated as follows: where TP refers to true positives, FP to false positives, and FN to false negatives.
Experimental design
The N = 320 WSIs were divided into train, validation, and test sets, ensuring each WSI is contained only in one of the splits. The training set is used to learn the feature representations of each class and is used in updating the CNN model weights. The validation set is data used to evaluate model performance during training that does not contribute to model learning, but instead helps identify which epoch’s model weights to employ during testing. Lastly, the test set is used to assess the generalizability of our models to unseen data.
Experiment 1: Evaluate laboratory generalizability performance
The first experiment aims to assess MMO-Net’s generalizability when testing data originates from a laboratory not employed during training, i.e., its ability to recognize organs despite changes in scanners and stain variations.
Here laboratory cross-validation was performed such that one laboratory is sequentially excluded from the training set to act as the held-out unseen test set. Validation subsets are created per organ by taking 10% of the training data and stratifying by severity grades when possible. A visual representation of the splits can be seen in Fig. S3.
Results are presented averaging the 3 splits by laboratory per organ (including the standard deviation), and comparing them between the models trained at individual magnifications (1.25x, 5x, and MMO-Net at 1.25x & 5x), resulting in 63 trained binary models (7 organs x 3 laboratory-splits x 3 CNN approaches).
Experiment 2: Evaluate generalizability in real-world translatable use case
This experiment aims to build the most robust model possible by integrating data from all sites, with the intent that this model is to be deployed into our production environment.
Here the laboratories data is commingled before dividing into 70%–10%–20% for training, validation, and test splits, respectively. Stratification is performed by lesion severity grades when lesion information is available and by laboratory otherwise (see Fig. S4 for additional details).
In total, 7 binary organ-models are compared with 3 networks: 2 DenseNet-121 trained individually at 1.25x and 5x, along with a single MMO-Net trained simultaneously at 1.25x & 5x, resulting in a total of 21 models.
Experiment 3: QC WSI metadata and timing evaluation
To validate the suitability of MMO-net to act as a QC tool for validating WSI metadata: per WSI, the organ metadata information is compared with detected organs and considered correct only if all organs are present. A QC score is produced by computing the Positive Predictive Value (PPV), for all folds in Experiment 1 and the test split from Experiment 2. Lastly, the computation time needed to process each slide is recorded to determine suitability for a high-throughput preclinical environment.
Results
Experiment 1: Evaluate laboratory generalizability performance
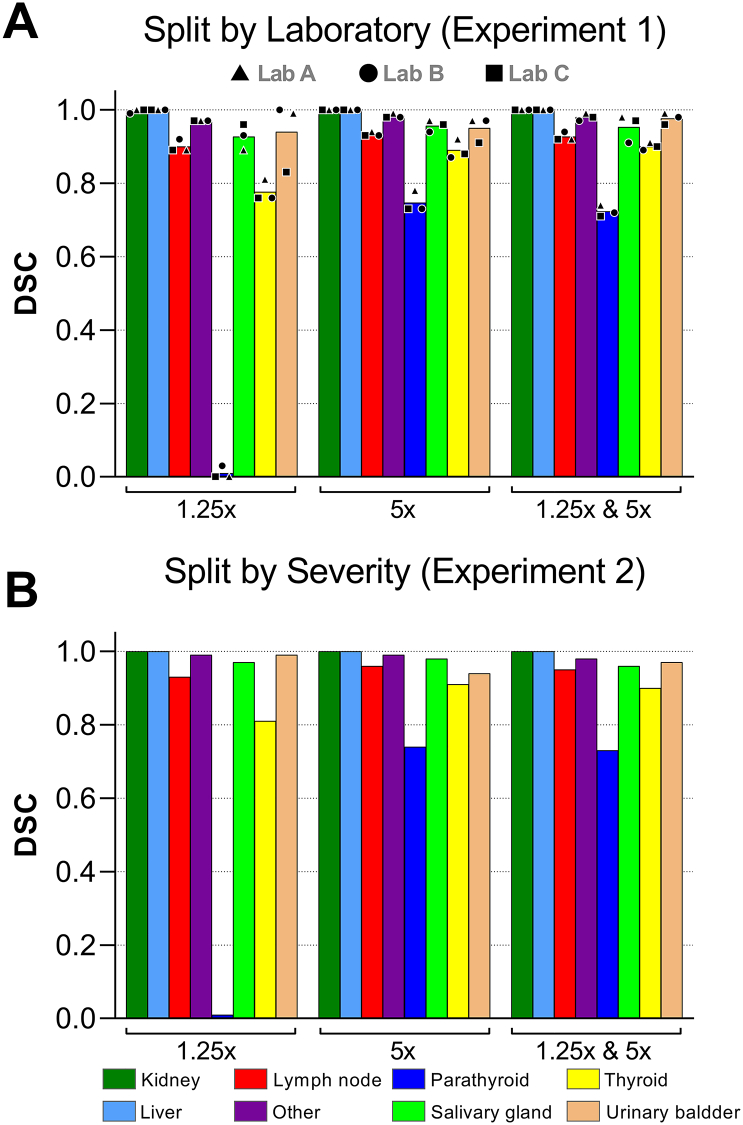
The classification results when holding out a single laboratory as a test show excellent performance for all magnifications in all experiments (upper section in Table 3). When quantitatively evaluating segmentation results (Fig. 3A), DSC values show similar behavior for all held-out laboratories. Among the 7 organs, the largest difference is observed for the parathyroid gland, with a DSC of nearly zero at 1.25x (likely due to its small size at this magnification) and a DSC > 0.7 at 5x and 1.25x & 5x. In the same manner, the thyroid gland models showed an improvement of ~0.1 from a DSC of ~0.8 (1.25x) to ~0.9 (5x and 1.25x & 5x) (Fig. 3A).
Table 3.
Model AUROC values (tile-level) per organ.
| Experiment | Magnification | Kidney | Liver | Lymph node | Para- thyroid | Salivary glands | Thyroid | Urinary bladder |
|---|---|---|---|---|---|---|---|---|
| Splits by lab (Experiment 1) | 1.25x | 0.9933 ± 0.0058 | 1.0 ± 0.000 | 0.9967 ± 0.0058 | 0.9967 ± 0.0058 | 0.9967 ± 0.0058 | 0.9933 ± 0.0116 | 0.9967 ± 0.0058 |
| 5x | 0.99 ± 0.01 | 1.0 ± 0.0 | 1.0 ± 0.0 | 1.0 ± 0.0 | 0.9967 ± 0.0058 | 0.9967 ± 0.0058 | 0.9933 ± 0.0058 | |
| 1.25-5x | 0.9967 ± 0.0058 | 1.0 ± 0.0 | 1.0 ± 0.0 | 1.0 ± 0.0 | 0.9933 ± 0.0058 | 0.9933 ± 0.0115 | 1.0 ± 0.0 | |
| Split by severity (Experiment 2) | 1.25x | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 |
| 5x | 0.99 | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | 0.99 | |
| 1.25-5x | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 |
For the experiment “Splits by laboratory”, the 3 splits results (Laboratory A, B, and C) are averaged per organ and magnification, reporting additionally the standard deviation (−/+ SD). In the column “Magnification”, 1.25x and 5x refers to DenseNet-121 models trained with the respective single magnifications while “1.25x & 5x” refers to our proposed model MMO-Net trained with both magnifications.
Fig. 3.
Macro (global) Dice Similarity Coefficient (DSC) values per organ for the organ segmentation masks for experiment 1 and 2. ‘1.25x’ and ‘5x’ refer to DenseNet-121 models trained with those single magnifications while ‘1.25x & 5x’ refers to our proposed model MMO-Net trained with both magnifications. Bar values in figure A represent the average of the 3 test splits (Laboratory A, B, and C). “Other” refers to other organs in the negative class.
Results showed that despite having lesions in the liver, kidney, and thyroid gland, MMO-net is able to identify them correctly with a DSC > 0.9 (Fig. 3A - 1.25x & 5x and Fig. 4). Focusing on the parathyroid gland, a challenging organ due to its small size, MMO-net is still able to identify it consistently (see Table 3) but due to the tile size selected, its boundaries are less refined.
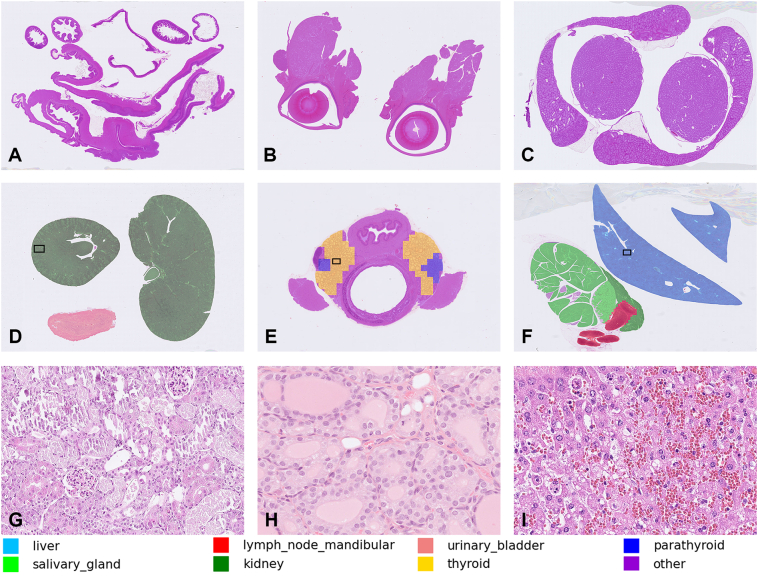
Fig. 4.
Example masks from our proposed MMO-Net (1.25x & 5x) models. Experiment 1A (Split by Laboratory A; images A, D, G); Experiment 1B (Split by Laboratory B; images B, E, H); Experiment 2 (split by lesion severity, images C, F, I). A–C, Segmentation masks from the negative class (A, stomach and intestine; B, eyes and harderian glands; C, testis and epididymis). D–F, Segmentation masks from target organs (D, kidneys and urinary bladder; E, thyroid and parathyroid glands with esophagus and trachea; F, liver, salivary gland and mandibular lymph node). G–I, H&E stains of the frames depicted in images D, E, and F, respectively, show that organs with lesions are also identified. G, kidney with tubular necrosis and mineralization. H, thyroid gland with follicular cell hypertrophy. I, liver with sinusoidal congestion and hepatocellular necrosis. The bottom legend shows the mapping color representation used for the organ masks.
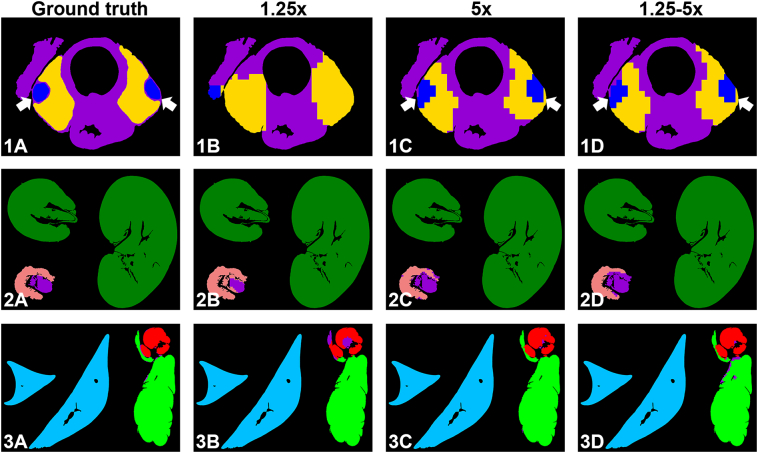
Fig. 5 provides a comparison between the ground truth and the 3 CNN models for the 7 target organs. The most notable difference can be seen in the first row with the parathyroid gland being completely undetected at 1.25x. For liver and kidney, models performed well, while for salivary gland and urinary bladder, there are small differences visible between all models.
Fig. 5.
Example WSI organ masks from the models trained at different magnifications in comparison with the ground-truth annotations. Row 1, test set split by Laboratory B (experiment 1). Rows 2 and 3, test set split by lesion severity (experiment 2). Note that for small organs like the parathyroid (arrows), 1.25x is not sufficient. For color legend see Fig. 4.
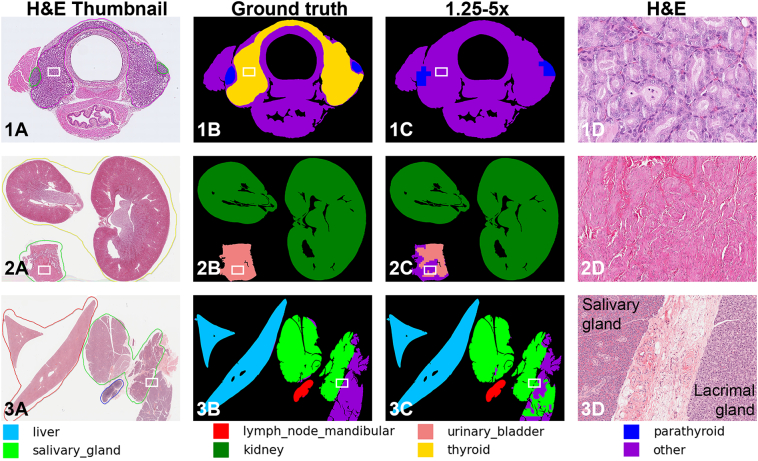
Further, visual inspection helped identify situations where our models performed poorly (Fig. 6). These situations were observed in cases of severe lesions which greatly altered tissue architecture, like marked thyroid follicular hyperplasia (Fig. 6-1). For test set Laboratory C, we observed slightly reduced DSC values for the urinary bladder (Fig. 3A), and visual inspection showed that some slides employed uncommon sectioning orientations for this organ, potentially impacting our generalizability performance (Fig. 6-2).
Fig. 6.
Examples of individual cases where our models perform poorly. Row 1, Split by Laboratory C, Thyroid is not identified in this specific case due to severe follicular hyperplasia. Row 2, Split by Laboratory C, Urinary bladder is not identified, likely due to uncommon orientation of the section and/or levels of inflation of the organ. Row 3, Split by severity, Lacrimal gland was present in this particular slide together with the salivary glands, and some regions of the lacrimal gland are incorrectly identified as salivary gland with the 1.25x & 5x model. Images on column D are H&E stains from the regions with a white frame depicted in figures from column A at 40x (1D) and 10x (2D and 3D) magnifications. (Note that the color code of the annotations on the left does not follow the color code for the masks at the bottom of the figure).
Experiment 2: Evaluate generalizability in real-world translatable use case
The quantitative results for Experiment 2 showed very similar results to Experiment 1, with excellent classification performance for all magnifications (bottom section in Table 3). The segmentation results were also very similar to Experiment 1 (Fig. 3B), showing again for the parathyroid gland a DSC > 0.7 at 5x and 1.25x & 5x compared with a DSC = 0.01 at 1.25x. Similarly, the thyroid results compared to Experiment 1 showed an improvement of ~0.1 from a DSC of 0.81 (1.25x) to ~0.9 (5x and 1.25x & 5x) (Fig. 3B).
As with Experiment 1 (Fig. 5-1), visual inspection of the masks also revealed that for small organs like the parathyroid, 1.25x is not sufficient for segmentation. Similarly, our models performed poorly in WSI containing lacrimal glands together with salivary glands (Fig. 6-3). This misclassification occurred in both experiments, likely due to the fact that both glands look alike at both magnifications despite being 2 different organs.
Experiment 3: Performance of QC metadata validation
In Experiment 1, the QC scores for the laboratories cross-validation were 1.0, 0.9896, and 1.0 (Lab A, B, and C as test set, respectively), indicating very strong performance. Importantly, it was determined that the lower QC score of Laboratory B was in fact a result of incorrect metadata and not poor performance of the model. In this context, a WSI was predicted by our model as not having a mandibular lymph node, while the metadata suggested it was present. Upon manual review, our pathologist confirmed it was in fact an error in metadata, and that our approach was able to spot this discrepancy.
This same WSI was again part of the test set for Experiment 2, wherein again it negatively affected the QC score (0.9844), yet would have successfully been flagged for manual review.
On average MMO-net takes approximately 330 s, which includes passing each WSI through the 7 organ models (30 s/model) and computing its segmentation mask (115 s). Although already suitable for our environment, it is imagined that in subsequent versions this could be highly parallelized reducing the overall time to less than 2 min.
Discussion
Organ identification with multiple magnifications (MMO-Net)
In this work, we have presented an organ identification network (MMO-Net) that makes use of multiple magnifications to simultaneously learn larger contextual information and finer morphological features.
With the use of 2 relatively low magnification levels (1.25x & 5x), MMO-net successfully recognized the 7 rat organs evaluated in this study. For comparison, models trained with low single magnification (1.25x) struggled to identify small organs like the parathyroid glands, demonstrating how detrimental suboptimal magnification selection can be. Similar issues with the thyroid and parathyroid glands were encountered by Hoefling et al.,2 where they showed higher numbers of misclassifications occur at lower magnifications (8.064 mpp ≈ 1.25x and 2.016 mpp ≈ 5x) versus higher ones (0.504 mpp ≈ 20x). As observed in our and other studies,2,6 the need for higher or lower magnifications depends on a number of factors (e.g., organ size, contact with adjacent organs, and tissue complexity), which can be challenging to balance within a single magnification. To that end, others have investigated multi-magnification approaches, enabling improved performance in classification, segmentation, and lesion detection tasks.4,5,7 Taken together, while attempting to balance context versus fine-grained detail in a single magnification is challenging, our results as well as those of others previously published suggest this can be averted by employing a multi-magnification approach.
For long-term sustainability, one-vs-all models were chosen so that in our production environment, organ models can be dynamically invoked if: (1) the WSI metadata is known a priori or (2) if identification of a specific organ is of interest. This approach has the added benefit of greater flexibility, as additional organ models can easily be dynamically added, and specific models can be individually improved if needed without requiring revalidation of all classes. Although not required here given the strong performance of individual models, as the number of additional organs grows, a secondary classifier (e.g., random forest) may be needed to learn how to appropriately weight the pre-softmax values from the individual models to produce a more robust final organ prediction.
Lesions
Identification of organs despite the presence of lesions is critical for toxicologic pathology workflows. Experiment 1 showed that MMO-net is capable of distinguishing organs independent of laboratory origin (Table 3, Fig. 3), but showed slightly diminished performance when generalizing to severe lesions. This is not unexpected as a lesion may heavily alter tissue morphology (e.g., extensive lytic necrosis). Consequently, when controlling for lesion severity during training, as done in Experiment 2, MMO-Net yielded a perfect DSC ~ 1.0 for liver and kidney organs, and a still notably improved DSC of 0.9 for thyroid glands (Fig. 3). As expected, MMO-Net appeared to heavily benefit from being exposed to wider lesion severity and variety during training, suggesting that obtaining training data similar to real-world data distributions is critical for model generalizability.
Other works identifying tissue/organs using DL approaches focused on normal histology,2,16 with no published applications to tissues with lesions. Although Hoefling et al.2 suggested that normal tissue classification could be applied to lesion detection, a formal approach has yet to be published. Segmentation of specific lesion types per organ by Kuklyte et al.6 showed that a reliable lesion detection and quantification depends on several factors including organ, type, and grading of the lesion together with magnification used. Hence, our work aims to benefit the toxicological pathology community by merging these concepts into one single approach.
An additional benefit of models like MMO-Net that can spatially localize organs, is that downstream applications for organ-specific lesion detection may be directly employed without requiring manual annotations of those organs or building additional preprocessing steps. For example, evaluating hepatocellular and follicular cell hypertrophy in rats,17,18 or even in human clinical settings,19,20 would expect to be benefited by MMO-Net.19,20
Visual organ representations
Although Hoefling et al.2 focused on embedding and tile-level validations for research and comparative histology purposes, a systematic approach for translating these results into the WSI level outputs needed for pathologist review was not mentioned. MMO-Net provides this functionality, resulting in a multi-colored mask delineating boundaries of the various organs. This map further enables the localized execution of downstream organ-specific tools, facilitating improved workflow efficiencies. MMO-Net additionally provides the needed dynamic flexibility of selecting, or adding, magnifications which can modulate the level of segmentation specificity required, regardless of organ size.
Quality control
Experiment 3 demonstrated that via these organ masks, it becomes possible to validate concordance of WSI metadata with organ WSI presence (Fig. 2 panel 4). This QC is critical for large-scale deployment, by drawing reviewer effort to only those few slides in question. As a by-product, this feedback is anticipated to help in long-term refinement of these models as challenging WSI are identified and incorporated.
Runtime considerations
Runtime per WSI is determined by the number of organ models chosen, and available computational architecture. For WSI whose metadata is known in advance, the most efficient configuration would be to perform inference with only the expected organ models. Any unlabeled tissue would be flagged and either manually reviewed or evaluated by all available models to determine its type. MMO-Net also affords the opportunity to parallelize output generation across a cluster, such that different GPUs could be assigned to different models, reducing overall computational time in a linear relationship with GPU availability. Although organ mask generation is one of the most computationally expensive parts of MMO-Net (115 s/WSI), we believe its performance could further be optimized, but remained out of scope for this proof of concept. Regardless, the current implementation of MMO-Net, with a single non-parallelized GPU approach, yields results within a timeframe deemed suitable for active usage and deployment.
Limitations and future work
Due to the nature of the mask generation approach, the boundary where organs intersect often appears block-shaped (Fig. 4). While still suitable in most cases, some uses may require higher pixel-level precision. For pixel-level boundaries, semantic segmentation approaches are ideal4,6,7 but require laborious pixel-wise annotations and a larger amount of training data.21, 22, 23 An alternative approach for providing this specificity may be to combine superpixels with deep learning24,25 hoping to benefit from superpixels’ more nuanced detection of organ/tissue boundaries.
For future work, we plan to:
-
●
Perform a more thorough validation of MMO-Net via production usage on incoming large-scale cohorts, appreciating this study represents a small fraction of the volume seen in a preclinical setting.
-
●
Focus on incorporating additional organs and animal models, enabling a more complete rollout across all our workflows.
Data release
The data release from this study includes WSI with associated metadata (study identification, organs, laboratory number, file format, color variations, severity grade label as well as the histopathological findings). Additionally, organ annotations and training–testing splits are available to help replication, benchmarking, and extension efforts.
The nature of this unique multi-site-scanner-organ dataset not only reflects, for the first time publicly available, real-world toxicological pathology studies, but also can enable additional experiments regarding color/stain normalization11,26 and impact of batch effects.8,27 Furthermore, the severity grades and histopathological findings could serve as a reference dataset for lesion detection/segmentation6 and aid in the building of models for normal vs abnormal tissue identification.
Conclusions
Our results show that it is possible to automatically identify a diverse set of organs from preclinical studies, including not only normal organs but also organs with lesions. With our MMO-Net network, we benefit from feature extraction at multiple magnifications, which provide complementary information at both context and structural detail levels. This work establishes a solid base for the identification of organs in WSI with intended usage as a first step for downstream AI-driven organ-specific models. This approach is now being deployed in our production environment for real-world evaluation. In addition, we are releasing the totality of our dataset to provide new opportunities for further algorithm development and validation.
Data availability
The dataset can be found at https://doi.org/10.7303/syn30282632.12 It contains the associated metadata, organ annotations and train-testing splits as referred in subsection Experimental Design from Materials and Methods.
Author contributions statement
All authors conceived the study design and experiments. FRP performed data collection and annotations. CGS and FA developed the computational tools used in this study. FRP, JF and VS provided histopathology expertise. CGS and FRP carried out experiments, analyzed data and created figures and tables. AJ provided additional deep learning and data analysis expertise. CGS and FRP wrote the first draft of the manuscript. AJ assisted substantially to shape the final manuscript version and all the authors were involved in reviewing the final version and gave their consent for publication.
Conflict of interest statement
All authors, except AJ are, or were, employed by F. Hoffmann-La Roche, Ltd. CGS, FRP and FA have filed the patent application “Organ Identification”, assigned to FHLR; this patent application has not been published yet. The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
This work was supported and funded by F. Hoffmann-La Roche Ltd, Basel, Switzerland, and the National Institute of Health (Award numbers U01CA239055-04, U01CA248226-02, R01DK118431). We would like to acknowledge laboratory associates in the Roche Pathology Core Labs in Basel for assistance with slide scanning. The authors thank Marco Tecilla for critical review of the manuscript and Nuno Calaim, Webster Lincoln, Monica Ge and James Cowan for providing technical expertise to release the dataset.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jpi.2022.100126.
Appendix A. Supplementary data
Supplementary material
References
- 1.Ruehl-Fehlert C., Kittel B., Morawietz G., et al. Revised guides for organ sampling and trimming in rats and mice--part 1. Exp Toxicol Pathol. 2003;55(2–3):91–106. https://www.ncbi.nlm.nih.gov/pubmed/14620530 [PubMed] [Google Scholar]
- 2.Hoefling H., Sing T., Hossain I., et al. HistoNet: a deep learning-based model of normal histology. Toxicol Pathol. 2021;49(4):784–797. doi: 10.1177/0192623321993425. https://www.ncbi.nlm.nih.gov/pubmed/33653171 [DOI] [PubMed] [Google Scholar]
- 3.Hashimoto N., Fukushima D., Koga R., et al. Multi-scale domain-adversarial multiple-instance CNN for cancer subtype classification with unannotated histopathological images. ArXiv. 2020 https://arxiv.org/abs/2001.01599 abs/2001.01599. [Google Scholar]
- 4.Ho D.J., Yarlagadda D.V.K., D’Alfonso T.M., et al. Deep Multi-Magnification Networks for multi-class breast cancer image segmentation. Comput Med Imaging Graph. 2021;88 doi: 10.1016/j.compmedimag.2021.101866. https://www.ncbi.nlm.nih.gov/pubmed/33485058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kosaraju S.C., Hao J., Koh H.M., Kang M. Deep-Hipo: multi-scale receptive field deep learning for histopathological image analysis. Methods. 2020;179:3–13. doi: 10.1016/j.ymeth.2020.05.012. https://www.ncbi.nlm.nih.gov/pubmed/32442672 [DOI] [PubMed] [Google Scholar]
- 6.Kuklyte J., Fitzgerald J., Nelissen S., et al. Evaluation of the use of single- and multi-magnification convolutional neural networks for the determination and quantitation of lesions in nonclinical pathology studies. Toxicol Pathol. 2021;49(4):815–842. doi: 10.1177/0192623320986423. https://www.ncbi.nlm.nih.gov/pubmed/33618634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.van Rijthoven M., Balkenhol M., Silina K., van der Laak J., Ciompi F. HookNet: multi-resolution convolutional neural networks for semantic segmentation in histopathology whole-slide images. Med Image Anal. 2021;68 doi: 10.1016/j.media.2020.101890. https://www.ncbi.nlm.nih.gov/pubmed/33260110 [DOI] [PubMed] [Google Scholar]
- 8.Chen Y., Zee J., Smith A., et al. Assessment of a computerized quantitative quality control tool for whole slide images of kidney biopsies. J Pathol. 2021;253(3):268–278. doi: 10.1002/path.5590. https://www.ncbi.nlm.nih.gov/pubmed/33197281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Webster J.D., Dunstan R.W. Whole-slide imaging and automated image analysis: considerations and opportunities in the practice of pathology. Vet Pathol. 2014;51(1):211–223. doi: 10.1177/0300985813503570. http://www.ncbi.nlm.nih.gov/pubmed/24091812 [DOI] [PubMed] [Google Scholar]
- 10.Janowczyk A., Basavanhally A., Madabhushi A. Stain Normalization using Sparse AutoEncoders (StaNoSA): application to digital pathology. Comput Med Imaging Graph. 2017;57:50–61. doi: 10.1016/j.compmedimag.2016.05.003. https://www.ncbi.nlm.nih.gov/pubmed/27373749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tellez D., Litjens G., Bandi P., et al. Quantifying the effects of data augmentation and stain color normalization in convolutional neural networks for computational pathology. Med Image Anal. 2019;58 doi: 10.1016/j.media.2019.101544. https://www.ncbi.nlm.nih.gov/pubmed/31466046 [DOI] [PubMed] [Google Scholar]
- 12.Gámez Serna C., Romero-Palomo F., Arcadu F., Funk J., Schumacher V., Janowczyk A. Dataset - MMO-Net (Multi Magnification Organ Network): a use case for organ identification using multiple magnifications in preclinical pathology studies. Synapse. 2022 doi: 10.7303/syn30282632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huang G., Liu Z., Maaten L.V.D., Weinberger K.Q. Densely connected convolutional networks. Proc of CVPR (Computer Vision and Pattern Recognition). 2017:2261–2269. https://ieeexplore.ieee.org/document/8099726 [Google Scholar]
- 14.Zhang C., Benz P., Argaw D.M., et al. Proc. of WACV (Winter Conference on Applications of Computer Vision) 2021. ResNet or DenseNet? Introducing dense shortcuts to ResNet; pp. 3549–3558.https://ieeexplore.ieee.org/document/9423394 [Google Scholar]
- 15.Bewick V., Cheek L., Ball J. Statistics review 13: receiver operating characteristic curves. Crit Care. 2004;8(6):508–512. doi: 10.1186/cc3000. https://www.ncbi.nlm.nih.gov/pubmed/15566624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hosseini M.S., Chan L., Tse G., et al. Proc of CVPR (Computer Vision and Pattern Recognition) 2019. Atlas of digital pathology: a generalized hierarchical histological tissue type-annotated database for deep learning; pp. 11739–11748.https://ieeexplore.ieee.org/document/8953780 [Google Scholar]
- 17.Pischon H., Mason D., Lawrenz B., et al. Artificial intelligence in toxicologic pathology: quantitative evaluation of compound-induced hepatocellular hypertrophy in rats. Toxicol Pathol. 2021;49(4):928–937. doi: 10.1177/0192623320983244. https://www.ncbi.nlm.nih.gov/pubmed/33397216 [DOI] [PubMed] [Google Scholar]
- 18.Bertani V., Blanck O., Guignard D., Schorsch F., Pischon H. Artificial intelligence in toxicological pathology: quantitative evaluation of compound-induced follicular cell hypertrophy in rat thyroid gland using deep learning models. Toxicol Pathol. 2022;50(1):23–34. doi: 10.1177/01926233211052010. https://www.ncbi.nlm.nih.gov/pubmed/34670459 [DOI] [PubMed] [Google Scholar]
- 19.Melo R.C.N., Raas M.W.D., Palazzi C., Neves V.H., Malta K.K., Silva T.P. Whole slide imaging and its applications to histopathological studies of liver disorders. Front Med (Lausanne). 2019;6:310. doi: 10.3389/fmed.2019.00310. https://www.ncbi.nlm.nih.gov/pubmed/31970160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Girolami I., Pantanowitz L., Marletta S., et al. Artificial intelligence applications for pre-implantation kidney biopsy pathology practice: a systematic review. J Nephrol. 2022;35:1–8. doi: 10.1007/s40620-022-01327-8. https://www.ncbi.nlm.nih.gov/pubmed/35441256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jayapandian C.P., Chen Y., Janowczyk A.R., et al. Development and evaluation of deep learning-based segmentation of histologic structures in the kidney cortex with multiple histologic stains. Kidney Int. 2021;99(1):86–101. doi: 10.1016/j.kint.2020.07.044. https://www.ncbi.nlm.nih.gov/pubmed/32835732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li Z., Zhang Y., Arora S. Why are convolutional nets more sample-efficient than fully-connected nets? ArXiv. 2021 https://arxiv.org/abs/2010.08515 abs/2010.08515. [Google Scholar]
- 23.Malach E., Shalev-Shwartz S. Computational separation between convolutional and fully-connected networks. ArXiv. 2021 https://arxiv.org/abs/2010.01369 abs/2010.01369. [Google Scholar]
- 24.He X., Chen K., Yang M. Proc of Collaborative Computing: Networking, Applications and Worksharing. 2021. Semi-automatic segmentation of tissue regions in digital histopathological image; pp. 678–696. [DOI] [Google Scholar]
- 25.Miao R., Toth R., Zhou Y., Madabhushi A., Janowczyk A. Quick annotator: an open-source digital pathology based rapid image annotation tool. J Pathol Clin Res. 2021;7(6):542–547. doi: 10.1002/cjp2.229. https://www.ncbi.nlm.nih.gov/pubmed/34288586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pontalba J.T., Gwynne-Timothy T., David E., Jakate K., Androutsos D., Khademi A. Assessing the impact of color normalization in convolutional neural network-based nuclei segmentation frameworks. Front Bioeng Biotechnol. 2019;7:300. doi: 10.3389/fbioe.2019.00300. https://www.ncbi.nlm.nih.gov/pubmed/31737619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Howard F.M., Dolezal J., Kochanny S., et al. The impact of site-specific digital histology signatures on deep learning model accuracy and bias. Nat Commun. 2021;12(1):4423. doi: 10.1038/s41467-021-24698-1. https://www.ncbi.nlm.nih.gov/pubmed/34285218 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material
Data Availability Statement
The dataset can be found at https://doi.org/10.7303/syn30282632.12 It contains the associated metadata, organ annotations and train-testing splits as referred in subsection Experimental Design from Materials and Methods.