Abstract
Histologic transformation (HT) is a major cause of drug resistance to therapy in patients with lung cancer. HTs to small-cell lung cancer (SCLC) have been reported frequently in patients with epidermal growth factor receptor (EGFR)-mutated lung cancer. Although HTs have an impact on the clinical outcomes in patients owing to a high refractoriness to treatments, there is limited data on the prevalence, causes, mechanisms, treatment efficacy, and future treatment strategies. In this review, we assess the literature regarding HTs comprehensively, including those describing EGFR-tyrosine kinase inhibitors, other molecular targeted drugs, and immune checkpoint inhibitors. Furthermore, we discuss the mechanisms of HTs and the lineage plasticity to SCLC and squamous cell carcinoma in lung cancer. In addition, we summarize the treatment efficacy and future perspectives of HTs in patients with lung cancer, and propose better management strategies for this group of patients.
Keywords: EGFR, histologic transformations, lineage plasticity, non-small-cell lung cancer, small-cell lung cancer
Introduction
Lung cancer is the leading cause of cancer-related deaths worldwide.1 According to the WHO classification of lung tumors, there are two distinctive histologic subtypes of lung cancer: non-small-cell lung cancer (NSCLC) which comprises 85% of cases and high-grade neuroendocrine carcinoma (HGNEC) which comprises 15% of cases.2 NSCLC is further categorized into various subtypes. Among these, adenocarcinoma and squamous cell carcinoma (SqCC) are common, while sarcomatoid carcinoma is rare. HGNEC is also divided into two subtypes: small-cell lung cancer (SCLC) and large-cell neuroendocrine carcinoma. Throughout the course of treatment, some patients may experience histologic transformations (HTs), resulting in anticancer drug resistance. In particular, HTs to HGNEC in patients with an epidermal growth factor receptor (EGFR) mutation-positive NSCLC on EGFR-tyrosine kinase inhibitor (TKI) treatment are most frequent. However, a comprehensive review of HTs in patients with lung cancer has been limited. In this review, we describe the characteristics, prevalence, genetic background, and future perspectives of HTs in patients with lung cancer.
HTs in patients with EGFR mutation-positive NSCLC
Transformation as a resistance mechanism
Somatic mutations in the tyrosine kinase domain of the EGFR gene are present in 15–50% of European and Asian patients with advanced NSCLC. As a result, treatment with EGFR-TKIs has extended progression-free survival (PFS) relative to chemotherapy as a first-line therapy and has been established as standard care.3,4 Although these patients demonstrate a high objective response rate to EGFR-TKIs, most develop an acquired resistance after approximately 12 months. The most common mechanism of the acquired resistance is the EGFR exon20 T790M mutation from the first or second generation (G) EGFR-TKIs.5 Currently, the third G EGFR-TKI, osimertinib, which is a potent inhibitor of active EGFR and T790M mutations, has been recognized as standard care for EGFR mutation-positive lung cancer.6 The detection of T790M mutations is important in the treatment decision after the development of resistance to the first or second G EGFR-TKIs. Therefore, as a routine clinical practice, a re-biopsy is conducted.7 The use of a repeat biopsy after clinical resistance to TKI therapy has aided the understanding of the molecular mechanisms underlying the acquired resistance to EGFR-TKIs.
In 2006, the first case of HT in a patient with lung cancer was reported by Zakowski et al.8 A 45-year-old woman with no history of smoking and EGFR del19-positive lung adenocarcinoma who had undergone erlotinib treatment for 18 months was found to have developed SCLC after a second tumor biopsy. The second biopsy of the SCLC sample retained its original EGFR mutation, which suggested that SCLC may arise from adenocarcinoma cells. Since that report, HTs have been recognized as a mechanism of resistance to EGFR-TKIs in patients with EGFR mutation-positive NSCLC. As a result, a number of case reports, systematic reviews, retrospective cohort studies, and pathological analyses of HTs in patients with lung cancer have been reported.9–13 The representative hematoxylin–eosin staining of biopsy samples from patients with HT is shown in Figure 1.
Figure 1.
Representative images of HT to SCLC from lung adenocarcinoma with an EGFR mutation. (a) hematoxylin–eosin staining of adenocarcinoma samples obtained via transbronchial lung biopsy before administering EGFR-TKI. (b) Hematoxylin–eosin staining of surgical biopsy samples of SCLC in lymph node obtained from patients exhibiting resistance to EGFR-TKI.
EGFR, epidermal growth factor receptor; HT, histologic transformation; SCLC, small-cell lung cancer; TKI, tyrosine kinase inhibitor.
Debate on HTs or de novo combined SCLC
SCLC can be categorized as pure or combined SCLC (c-SCLC). The latter features a mixed tumor histology of SCLC and NSCLC. In a previous study, c-SCLC was observed in 10% of 176 autopsied patients diagnosed with SCLC, which suggested that a small biopsy is not adequate for an accurate diagnosis.14 In practice, the presence of a c-SCLC can hinder the identification of the characteristics of HTs. The clonal selection hypothesis states that HTs may occur if the SCLC component becomes dominant when the adenocarcinoma component is killed by the EGFR inhibitors in patients with c-SCLC at the time of the initial diagnosis.15 Although this hypothesis has existed for more than a decade, strong evidence has been lacking. Generally, patients with HTs respond well to EGFR-TKIs for several months and experience greater tumor growth at the time of acquired resistance due to HT to SCLC. If the clonal selection hypothesis is true for the development of SCLC, a less dramatic response to EGFR inhibitors and earlier acquired resistance would be expected. Therefore, HTs in patients with lung cancer may be the reason for the detection of SCLC upon re-biopsy, at the time of the acquired resistance.
Prevalence of HTs in patients with EGFR mutation-positive NSCLC
The first comprehensive study using a genetic assessment was conducted by Sequist et al. in 2011, who reported a prevalence of HTs of 14% in patients with lung cancer.11 The results of large cohort studies on HTs in patients with lung cancer are summarized in Table 1. Most of the current publications have focused on HTs to HGNEC. These studies have reported that 2–15% of patients with lung cancer experience HTs to HGNEC upon the acquisition of resistance. Our largest cohort study reported among 2624 re-biopsy cases that the prevalence of HTs to HGNEC was 2.2% while that to other types of NSCLC was 0.6%.10 Overall, the time from initial diagnosis of HT to HGNEC ranged from 13 to 22 months. Four cohort studies reported that most cases included the first or second G EGFR-TKIs, and HT after the third G TKI therapy. The results indicated collectively, that a HT in patients with lung cancer is recognized as a mechanism of resistance, irrespective of the TKI generation.
Table 1.
Summary of HTs in patients with EGFR mutation-positive NSCLC.
| Author | Journal | Year | N | The number and prevalence (N, %) of HTs | HT histology | TKI (G) | 3rd G TKI (N) | Time to HT (Mo) |
|---|---|---|---|---|---|---|---|---|
| Fujimoto et al.10 | Eur J Cancer | 2022 | 2624 | 59 (2.2) 15 (0.6) |
HGNEC another NSCLC |
1st–3rd | 14 | 21.6 |
| Sequist et al.11 | Sci Transl Med | 2011 | 37 | 5 (14) | SCLC | 1st | 0 | 14.1 |
| Yu et al.16 | Clin Cancer Res | 2013 | 155 | 4 (2.6) | SCLC | 1st | 0 | 13 |
| Nosaki et al.7 | Lung Cancer | 2016 | 314 | 12 (3.8) | SCLC | 1st–2nd | 0 | N.R. |
| Zeng et al.17 | Lung Cancer | 2020 | 103 | 3 (2.9) | SCLC | 1st | 0 | N.R. |
| Piotrowska et al.18 | Cancer Discov | 2018 | 32 | 2 (6.3) 1 (3.1) |
SCLC SqCC |
3rd | 32 | N.R. |
| Lin et al.19 | Lancet Res Med | 2018 | 53 | 2 (3.8) 1 (1.9) |
SCLC SqCC |
3rd | 53 | N.R. |
| Oxnard et al.20 | JAMA Oncol | 2019 | 41 | 6 (15) | SCLC | 3rd | 41 | N.R. |
| Schoenfeld et al.21 | Clin Cancer Res | 2020 | 62 | 3 (4.8) 6 (9.7) |
SCLC SqCC |
3rd | 62 | 13.6 |
G, generation; HGNEC, high-grade neuroendocrine carcinoma; HT, histologic transformation; N.R., not reported; NSCLC, non-small-cell lung cancer; SCLC, small-cell lung cancer; TKI, tyrosine kinase inhibitor.
HTs to another subtype of NSCLC in patients with EGFR mutation-positive NSCLC
Although uncommon, HTs from one subtype of NSCLC to another have been reported in 0.6% of in patients with lung cancer.10 Most of the reports were case reports or small case series. In our previous cohort study, most TKIs that were used during HTs were first or second generation. In some reports that analyzed pre- and post-HT samples of patients treated with osimertinib, the prevalence of SqCC transformations was 2–10%; thus, larger studies on third G TKIs are needed.18,19,21 Moreover, in a systematic review of 17 patients with HTs to SqCC, most of the patients were female (82%), 41% were former smokers, and the median time to SqCC onset was 11.5 months.22
HTs to subtypes other than HGNEC or SqCC are extremely rare. For example, there have been several reports on transformations to sarcomatoid carcinoma.23,24 Specifically, six sarcomatoid HTs [five cases with EGFR mutations and one with c-ros oncogene 1 (ROS1) mutations] were reported, and the interval from initial diagnosis to HT was 9–88 (median: 31.5) months.25 Giant cells were the most common form of sarcomatoid transformations, and overall survival (OS) after HTs in patients with lung cancer was reported to be 2.5 months, which was shorter than for HTs to HGNEC.
Current problems of HTs in patients with EGFR mutation-positive NSCLC
After the FLAURA study, the mainstay first-line EGFR-TKI changed to osimertinib.26 No standard treatment has yet been developed for those with osimertinib-resistant NSCLC, and a re-biopsy after osimertinib resistance is not conducted as routine clinical practice. Therefore, no large cohort studies have been conducted and the precise incidence of EGFR mutation-positive NSCLC with HTs after osimertinib has not been determined. Currently, the major focus is on overcoming osimertinib resistance by combining new drugs, such as a mesenchymal epithelial transition (MET) inhibitor (selumetinib), a vascular endothelial growth factor receptor (VEGFR)-2 blocker (ramucirumab), and a bispecific antibody that blocks EGFR and MET receptors simultaneously (amivantamab).27–29 HTs in patients with lung cancer that occur after treatment with these agents should also be investigated in future studies.
HT induced by agents other than EGFR-TKIs
Programmed cell death 1/programmed cell death ligand 1 inhibitors
HTs have been reported mainly in patients with EGFR mutation-positive lung cancer. However, they have also been reported in patients with NSCLC undergoing treatment without EGFR-TKIs and in those with other cancers.30–33 For example, HTs to SCLC after programmed cell death 1 (PD-1) inhibitor treatment has also been reported. In addition, HTs of adenocarcinoma to SqCC and vice versa have been reported after treatment with PD-1 inhibitors such as nivolumab, pembrolizumab, and sintilimab.34–38 In contrast, HTs after programmed cell death ligand 1 (PD-L1) inhibitor administration has been reported rarely, likely due to differences in the timing of its introduction into clinical practice and its infrequent use as a single agent for advanced NSCLC.39 HTs to SCLC were detected in only one case after treatment with nivolumab followed by atezolizumab.
Among the studies on HTs in patients with lung cancer, after treatment with PD-1 inhibitors, few have examined the causes of HTs. However, several studies have reported that tumor suppressor p53 (TP53) mutations were found prior to PD-1 inhibitor administration in cases of HTs to SCLC.40,41 These findings were similar to those described below for HTs to SCLC after the use of molecular targeted agents, and no report has yet suggested a PD-1 inhibitor-specific mechanism of HT. Therefore, HTs in patients with lung cancer caused by PD-1 inhibitors may also be typical mechanisms of resistance to therapy for NSCLC. However, the rate of HTs among those patients treated with PD-1 inhibitors is unknown. It has been suggested that the frequency of HTs in patients with lung cancer may be underreported because re-biopsy after administration of immune checkpoint inhibitors (ICIs) is not common.41 Therefore, future large-scale validation is warranted.
Anaplastic lymphoma kinase-TKIs
HTs in patients with lung cancer have been reported with the use of most of the marketed anaplastic lymphoma kinase (ALK)-TKIs, such as crizotinib, alectinib, ceritinib, and lorlatinib, while no report has yet linked ALK-TKIs specifically to HTs.42–45 Moreover, in most cases, the ALK-rearrangement was retained even after the HT.42–49 As most studies have focused on the mechanisms of resistance to ALK-TKIs, such as secondary mutations, ALK-amplification, and bypass signaling pathways, the frequency of HTs as a resistance mechanism to ALK-TKIs has not yet been reported.50–52
Several studies have reported the cases of HTs to SCLC in ALK-rearranged NSCLC.42,47,48,53 Similar to HTs to SCLC caused by other drugs, HTs caused by ALK-TKIs have been implicated in the alterations in genes such as TP53 and retinoblastoma 1 gene (Rb1).45,49 Recently, the first case of a transformed SCLC from ALK-rearranged NSCLC using an analysis of the genomic and transcriptomic landscape of paired pre- and post-HT tissues was reported.54 The findings indicated that no alterations or loss of functions were detected in TP53 or Rb1, suggesting that the mechanism of HT to SCLC in ALK-rearranged NSCLC may differ from that in EGFR-mutated NSCLC. However, the number of paired-sample analyses was small, requiring further studies to examine the mechanistic differences of HTs between ALK-rearrangements and EGFR mutations. HTs to SqCC have also been reported after the use of ALK-TKIs in several cases.55–60 Two reports have demonstrated the paired pre- and post-treatment genomic landscape of the transformed ALK-rearranged SqCC; however, much remains unknown about HTs in SqCC.57,58
Other molecular targeted agents for NSCLC
HTs in patients with lung cancer are also caused by molecular targeted agents other than EGFR- and ALK-TKIs. For example, several reports have shown HTs after treatment with ROS1-TKIs or Kirsten rat sarcoma viral oncogene homolog (KRAS) G12C-inhibitors.61–63
In the case of ROS1 rearrangement-positive NSCLC, the whole exome analysis of biopsy and autopsy-derived specimens before and after crizotinib administration have provided detailed confirmation that the SCLC transformation occurred as a mechanism of resistance.61 This case showed a loss of function of TP53 and Rb1, which was similar to typical HT cases.
In terms of KRAS, mouse models and cell lines of KRAS G12D have been used frequently to study the mechanisms of oncogenesis of lung adenocarcinoma. In addition, studies using a KRAS G12D mutation-positive cell line and a mouse model have indicated that the deletion of liver kinase B1 (LKB1) may be closely related to HTs to SqCC.64,65 However, no molecular targeting agents specific for KRAS G12D are currently in clinical application, and the relationship between KRAS G12D and HT to SqCC has not yet been demonstrated in clinical practice. With respect to KRAS G12C, for which there are molecular targeted drugs are in clinical use, HTs to SqCC have been reported. In a study of KRAS G12C-positive NSCLC, the authors reported a resistance mechanism in 11 cases in which disease control was achieved for more than 12 weeks after adagrasib treatment.63 Of those 11 patients, some amplifications, oncogenic fusions, and loss of function mutations were identified. Among them, two patients exhibited HTs, both to SqCC. In one of the two cases, a circulating tumor DNA analysis of plasma samples was also performed, and no genomic resistance mechanism other than HTs to SqCC was identified (although there is no evidence that serine/threonine kinase 11 (STK11)/LKB1 was studied).
Some reports have stated that HTs in patients with lung cancer did not occur in cases where molecular targeted drugs were administered to patients with NSCLC with rare driver gene abnormalities, such as rearrangements during transfection (RET) and in the v-raf murine sarcoma viral oncogene homolog B1 (BRAF) V600E.66,67 However, the sample sizes of those studies were limited. Thus, the possibility of HTs in rare driver gene alterations in patients with lung cancer cannot be ruled out, until the resistance mechanism is studied on a large scale.
Mechanism of HTs and lineage plasticity in lung cancer
Histologic origin of lung cancer
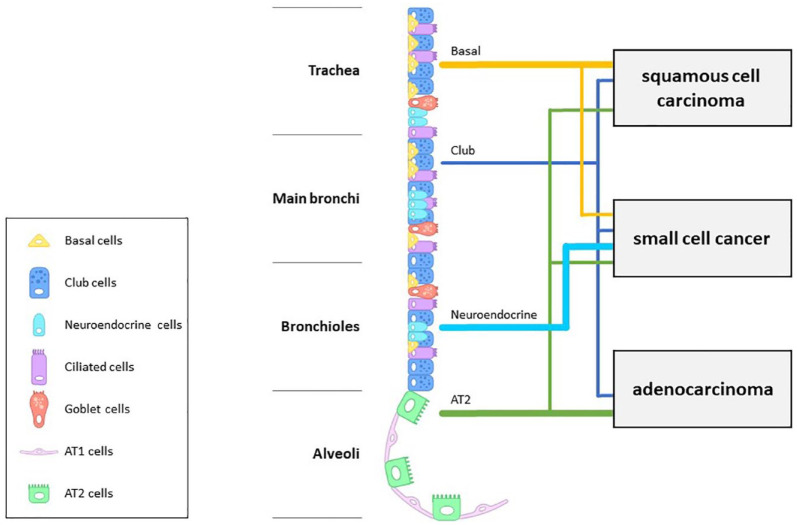
The respiratory epithelium from the trachea to the alveoli is composed of basal, club/Clara, neuroendocrine, ciliated, goblet, and alveolar type I and type II cells (Figure 2). The development of lung cancer has been studied using genetically engineered mouse models.68 Such studies have postulated that type II alveolar cells are the origin of lung adenocarcinoma,69–72 while it has been suggested recently that club cells may be responsible for this cancer subtype.73,74 Moreover, the origin of lung SqCC has been postulated to be basal cells based on the expression of p63, a marker of bronchial basal cells, and this theory is still supported as one of the origins of the disease.75–77 However, carcinogenesis of SqCC from type II alveolar cells and club cells has also been widely reported in recent years.78,79 Furthermore, because of its neuroendocrine neoplastic nature, SCLC was considered to be derived from neuroendocrine cells in the lung.80–82 Even in recent years, the development of SCLC from neuroendocrine cells is still considered one of the most promising origins.83,84 However, SCLC can arise from basal, club, and type II alveolar cells.80,85 Thus, although generally, the cell of origin of lung cancer development was presumed by histological type, each lung cancer subtype has no single histological origin.86
Figure 2.
Respiratory epithelium and the origin of lung cancers. The airway epithelium is composed of basal, club/Clara, neuroendocrine, ciliated, goblet, and alveolar type I and type II cells. The main histologic origin of each lung cancer subtype is indicated by the bold lines; however, each histological subtype has various origins.
AT1, alveolar type I cell; AT2, alveolar type II cell.
Furthermore, recent years have brought attention to the fact that the histology of lung cancer is not constant, but plastic. Cancer cell plasticity is defined as the ability of a cell to modify its identity substantially and to take on a new phenotype that resembles a distinct developmental lineage more closely.87 It is considered that some cancer cells possess plasticity and that further specific changes may cause dynamic changes, such as HTs. Herein, we review the mechanisms of HTs in patients with lung cancer, focusing on HTs to SCLC, which has been relatively well studied.
Genetic characteristics of SCLC
Inactivation of the Rb1 gene was first discovered as a characteristic genetic abnormality in retinoblastoma.88,89 Subsequently, an association with neuroendocrine tumors was suggested and it was reported to be more common in SCLC than NSCLC.90,91 However, in a mouse model, the inactivation of Rb1 alone did not cause neuroendocrine tumors in the lungs.92,93 In 2003, it was shown that loss of both TP53 and Rb1 in a wide range of lung epithelial cells in a mouse model could transform them into SCLC.94 In 2011, the inactivation of TP53 and Rb1, particularly in neuroendocrine cells and type II alveolar cells, was reported to induce small cell transformation.80 The following year, the results of a large-scale whole-genome, transcriptome, exome, and copy number analysis of SCLC were reported.95,96 As a result, mutations in TP53 and Rb1 were identified as highly frequent abnormalities in SCLC. These findings suggest that loss of function due to mutations in both TP53 and Rb1 plays a major role in the development of SCLC.
Mechanism of HT from NSCLC to SCLC
The acquired loss of function of Rb1, which occurs after the initiation of treatment for EGFR- and TP53-mutated NSCLC, was first hypothesized as the cause of SCLC transformation.15 Since the loss of TP53 and Rb1 is vital to the carcinogenesis of de novo SCLC, the Rb1 deletion was found in all 11 cases of the transformed SCLC and only in those with HTs to SCLC after resistance to EGFR-TKIs, but not in those whose cancer remained NSCLC with resistance to EGFR-TKIs.97 Therefore, it was hypothesized that the additional alterations in Rb1 during EGFR-TKI treatment would result in transformation and resistance against EGFR-TKIs. However, when the paired pre- and post-treatment tumor samples were analyzed, the alterations of Rb1 and TP53 were found to have existed before anticancer treatment.98 Furthermore, the alterations of Rb1 and TP53 were reported to be present in patients who did not develop HTs to SCLC. In other cohort studies, although EGFR/Rb1/TP53-mutant lung cancers were present in 5% of EGFR-mutant lung cancers, only approximately 20% of these cases showed HTs to SCLC after EGFR-TKI treatment.99 Moreover, EGFR-mutant lung cancers without baseline TP53 and Rb1 alterations were rarely transformed to SCLC. Taken together, the loss of TP53 and Rb1 might be a necessary, but not sufficient condition for HTs to SCLC.
Thus, a second hypothesis of the mechanisms of HTs in patients with lung cancer is that gene alterations occur before or after anticancer drug therapy in cancer cells with a baseline loss of function of TP53 and Rb1. Both of the cohort studies of the paired pre- and post-treatment tumor sample analysis reported that activation-induced cytidine deaminase (AID)/apolipoprotein B mRNA-editing enzyme (APOBEC) mutation signature was more enriched in EGFR/Rb1/TP53-mutant lung cancers which transformed to SCLC than in EGFR/Rb1/TP53-mutant lung cancers which did not transform to SCLC, suggesting that gene alterations other than TP53 and Rb1 mutations are required for HTs to SCLC.98,99 The APOBEC family is a group of DNA/RNA editing enzymes that convert cytidine (C) to uridine (U) through deamination reactions.100 APOBEC3-specific genome mutation (APOBEC3 signature) has been suggested to play an important role in the induction of genomic variations in many cancers.101–103 Furthermore, the presence of specific genomic mutations, such as the APOBEC signature, in EGFR/Rb1/TP53-mutant lung cancers prior to HTs suggests that genomic alterations other than TP53 and Rb1 may be involved in HTs to SCLC.98,99 Other genes have also been implicated as possible contributors. For example, in a mouse model of transformed human basal cells of benign prostate tissue, the addition of cellular myelocytomatosis oncogene (c-MYC) and B-cell lymphoma 2 (Bcl2) alterations to TP53, AKT, and Rb1 alterations induced SCLC. However, TP53, AKT, and Rb1 alterations without c-MYC and Bcl2 alterations induced adenocarcinoma. Furthermore, in a mouse model of transformed normal human bronchial epithelial cells, all five genes (TP53, AKT, Rb1, c-MYC, and Bcl2) had to be altered for carcinogenesis to SCLC to occur.104 Thus, not only TP53 and Rb1 loss, but also other genetic factors may be needed for HTs to SCLC. However, while involvement of AKT and c-MYC has been detected in some clinical specimens of EGFR-mutated NSCLC with HTs to SCLC, it was not as prevalent as TP53 and Rb1 alterations.98,99 Therefore, the specific set of genetic abnormalities required for SCLC transformations has not yet been identified.
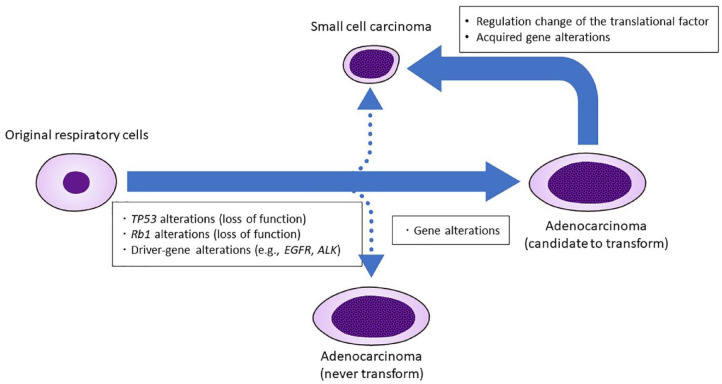
Recently, a third hypothesis of HTs in patients with lung cancer proposed that acquired transcriptional regulation occurs in cancer cells with loss of function of Rb1 and TP53. Detailed genomic, epigenomic, transcriptomic, and proteomic characterization of combined lung adenocarcinoma/SCLC, pre-transformation lung adenocarcinoma, post-transformation SCLC, never-transformed lung adenocarcinoma, and de novo SCLC have been reported.105 This study suggested that HTs in patients with lung cancer may be caused by transcriptional reprogramming rather than by acquired gene alterations. The authors identified an increased expression of genes involved in the polycomb repressive complex 2 (PRC2), phosphoinositide 3-kinases (PI3K)/AKT, and neurogenic locus notch homolog protein (NOTCH) pathways. It was also found that pre-transformed lung adenocarcinoma had an intermediate pattern between never-transformed lung adenocarcinoma and post-transformed SCLC in the methylation profiling. These findings suggested that HTs from NSCLC to SCLC may be caused by methylation-induced changes in the transcriptional regulation with a prior loss of the TP53 or Rb1 function. The current hypothesis is summarized in Figure 3.
Figure 3.
Current hypothesis of HTs to small-cell lung cancer from NSCLC. TP53, Rb1, and driver gene alterations are commonly observed before HTs, and some regulation change of translational factor or acquired gene alterations might be required for HTs.
ALK, anaplastic lymphoma kinase; EGFR, epidermal growth factor receptor; HT, histologic transformation; Rb1, retinoblastoma 1 gene; NSCLC, non-small-cell lung cancer; TP53, tumor suppressor protein p53.
Nevertheless, these hypotheses are problematic because these studies did not target only transformed SCLC and they included only a small number of patients. In addition, whether there are genetic differences between de novo SCLC and transformed SCLC remains unknown because the genomic analysis of transformed SCLC has only been performed in a small number of cases. In the few cases where the genomic analysis has been performed, no specific genetic abnormalities distinguishing the de novo SCLC from the transformed SCLC have been identified, aside from retained driver gene alterations. Therefore, in the future, more pre- and post-transformation clinical specimens of lung adenocarcinomas in which HTs to SCLC have occurred need to be collected and analyzed in detail.
Small-cell transformation of prostate cancer
Next, we discuss the small cell transformations of prostate cancer, which is helpful in considering SCLC transformations from NSCLC. HTs to small-cell carcinoma of the prostate have been known to occur after the administration of androgen receptor (AR) signaling inhibitors, such as abiraterone and enzalutamide for the treatment of metastatic castration-resistant prostate cancer.106,107 Transformations of prostate cancer into small-cell carcinoma by AR inhibition have been reported and analyzed before that of EGFR mutation-positive NSCLC to SCLC. Thus, small-cell transformations of prostate cancer have been examined in more detail than that of lung cancer in some areas. In a recent large prospective study, this condition was named treatment-emergent small-cell neuroendocrine prostate cancer (t-SCNC), and its frequency was examined. The results showed that t-SCNC as a resistance mechanism occurred in 17% of patients after the initiation of AR-targeted therapy.31 Alterations of TP53 and Rb1 were detected in several cases. In a mouse model, defects of TP53 and Rb1 depressed epigenetic initiators, such as enhancers of the zeste homolog 2 (Ezh2) and sex-determining region Y-box 2 (Sox2), were identified, which were shown to be involved in resistance to antiandrogen therapy and lineage plasticity.108 De-repression of the placental gene, paternally expressed gene 10 (PEG10) and involvement of transmembrane serine protease 2 (TMPRSS2)/endocrine/reproductive/gastrointestinal (ERG) fusion or transcription factors, such as the forkhead box protein A1 (FOXA1) and POU domain class 3 transcription factor 2 (POU3F2) have been reported to play roles in the small-cell transformation mechanisms of prostate cancer.109–112 However, although the loss of function of TP53 and Rb1 is common between NSCLC and prostate cancer in the mechanism of HTs to SCLC, some of the above-mentioned genes, especially the transcription factors, have not been identified in HTs to SCLC from NSCLC. Further studies are needed to determine whether NSCLC and prostate cancer share common or distinct mechanisms of transformation to small-cell carcinoma.
Mechanism of squamous cell transformation
Knowledge on transformations to SqCC is limited. Notably, for the de novo SqCC, there have been no reported genetic abnormalities, such as TP53 or Rb1 in SCLC, which are present in many cases and can be key to investigating the mechanisms of transformation, which may hinder the establishment of models for HTs to SqCC. However, a few studies using KRAS G12D-positive adenocarcinoma mouse models, and paired tissues before and after treatment, have suggested that the loss of the LKB1 function may play an important role in HTs to SqCC. The possibility that the loss of LKB1 function contributes to the development of de novo SqCC has been noted in several reports.113,114 In the KRAS G12D-positive adenocarcinoma mouse models, the deletion of LKB1 may have resulted in transformations to SqCC.65,115 Moreover, the administration of Adenovirus-Cre via an intranasal addition to LKB1fl/fl and phosphatase and tensin homolog (PTEN)fl/fl mice resulted in carcinogenesis of SqCC and the expression of the squamous markers keratin 5 (KRT5), p63, and Sox2, which transcriptionally resembled the human lung SqCC.116 In addition, the loss of function of LKB1 and PTEN activated AKT and the mechanistic target of rapamycin (mTOR) pathways, contributing to cell proliferation and tumorigenesis. The possibility that the loss of LKB1 or PTEN may have activated the mTOR pathway, leading to HTs to SqCC, has been suggested in reports of pre- and post-treatment clinical samples of transformed SqCC, and is a promising hypothesis.117 However, HTs to SCLC without any obvious acquired alterations of the mTOR pathway has also been reported;21 thus, the loss of LKB1 and PTEN is not considered to be the sole mechanism of SqCC transformations. Importantly, loss of LKB1 is detected in up to 2% of cases of de novo SqCC, which may make it difficult to establish a single model for HTs to SqCC.118 Further analysis of paired pre- and post-transformation tumors may reveal multiple transformation mechanisms.
Therapeutic strategies after HTs
Survival after HTs in patients with EGFR mutation-positive NSCLC
Treatment strategies after HTs in patients with lung cancer are a major problem for both patients and clinicians. Existing data from a cohort study are limited to patients with EGFR mutation-positive NSCLC. The prognosis of HTs in patients with lung cancer, obtained from cohort studies is summarized in Table 2. An OS of 9–15 months from the time of HT diagnosis has been reported in previous literature.10,119–121 Moreover, the OS and PFS of HTs to SCLC were comparable with those of de novo SCLC.122,123 From the perspective of disease behavior, one previous report suggested that HT to SCLC in patients with EGFR mutation-positive NSCLC might be more aggressive than in patients without EGFR-mutation because the median time to HT for patients with EGFR-mutation was significantly shorter than that for patients without EGFR-mutation.124 However, this might be biased by the recommendation of repeat biopsy in EGFR-mutant patients after a line of targeted therapy, whereas a repeat biopsy is not recommended in patients without EGFR mutation.125 For patients with HTs to another subtype of NSCLC, the OS was reported to be 12.1 months.10 However, the data of survival outcomes of patients with HTs to SqCC were limited compared with that of patients with HTs to SCLC. Moreover, no study has yet compared OS from the time of diagnosis between HT and non-HT cases; therefore, the clinical impact of HTs should be investigated in future studies.
Table 2.
Summary of the prognosis of patients with HGNEC or another subtype of NSCLC.
| Histology | Author | Journal | Year | N | Treatment after HT diagnosis | PFS after HT diagnosis (Mo) | OS after HT diagnosis (Mo) | OS after lung cancer diagnosis (Mo) |
|---|---|---|---|---|---|---|---|---|
| HGNEC | Fujimoto et al.10 | Eur J Cancer | 2022 | 59 | Chemo 51/ICI 12/TKI 21 | 4.1 | 12.6 | N.R. |
| Marcoux et al.119 | J Clin Oncol | 2018 | 65 | Chemo 63/ICI 17/TKI 34 | 3.4 | 10.9 | 31.5 | |
| W. Wang et al.120 | Lung Cancer | 2021 | 32 | Chemo 30/ICI 3/TKI 7 | 3.5 | 9.7 | 34.5 | |
| S. Wang et al.121 | Thorac Cancer | 2021 | 29 | Chemo 27/TKI 18 | 4.7 | 14.8 | N.R. | |
| Ferrer et al.124 | J Thorac Oncol | 2018 | 48 | Chemo 41/ICI 1/TKI 2 | N.R. | 9 | 28 | |
| Another NSCLC | Fujimoto et al.10 | Eur J Cancer | 2022 | 15 | Chemo 9/ICI 9/TKI 5 | 6.4 | 12.1 | N.R. |
HGNEC: high-grade neuroendocrine carcinoma; HT: histologic transformation; ICI: immune checkpoint inhibitor; NSCLC: non-small-cell lung cancer; N.R.: not reported; PFS: progression-free survival; OS: overall survival; TKI: tyrosine kinase inhibitor.
Efficacy of chemotherapy after HTs
The indicators of treatment efficacies such as PFS, OS, and OS after the lung cancer diagnosis are summarized in Table 2. Therapeutic strategies should be adjusted for SCLC after HTs to SCLC, since SCLC is highly malignant and develops rapidly.15 In previous studies, most patients received cytotoxic chemotherapy. Among them, SCLC-based chemotherapy (etoposide plus platinum or irinotecan plus platinum) was used most frequently.10,119 In a previous study, the PFS of etoposide plus platinum was 3.4 months. Considering that the PFS of etoposide plus platinum in de novo extensive-disease SCLC was 4–6 months, the treatment efficacy may be limited.122,123 In this clinical setting, a repeat biopsy is highly recommended to rule out SCLC transformation because platinum-etoposide chemotherapy yields responses in patients with transformed SCLC, similar to de novo SCLC.125
Efficacy of ICIs after HTs
Previous studies have shown the limited efficacy of ICIs for patients who developed HT, although recent research has suggested that HTs are associated with the increased ability to induce an immune response.105 Specifically, in our study, the total objective response rate and disease control rate of ICI therapy (PD-1/PD-L1 inhibitor monotherapy or platinum-pemetrexed combined with PD-1/PD-L1 inhibitors) were 0% and 17%, respectively, and the median PFS was 2.0 months.10 Another large-scale report showed that no responses were observed after HTs in 17 patients who received ICIs, either as a single-agent PD-1 or PD-L1 inhibitor (n = 9) or as part of a combination ipilimumab–nivolumab regimen (n = 8).119 None of the 17 patients derived clinical benefits from these therapies, as the longest time to progression was only 9 weeks.
Treatment efficacy of TKIs
Data are limited on the efficacy of TKIs after HTs in patients with lung cancer, although continuing TKI therapy after the development of HT has been used in some cases. In a retrospective study, the PFS after HTs of patients receiving chemotherapy with EGFR-TKIs was significantly longer than that of patients receiving chemotherapy without EGFR-TKIs (5.2 versus 3.0 months, p = 0.0014).121 However, the OS benefit has not been demonstrated; thus, the efficacy of continuing TKI therapy has not reached consensus. Another study has implied the beneficial efficacy of multi-kinase inhibitor therapy after HTs in patients with lung cancer.120 Anlotinib is an orally administered multi-kinase inhibitor that targets VEGFR, fibroblast growth factor receptor, platelet-derived growth factor receptors, and c-kit. Owing to its antiangiogenic effect, this agent is considered to be a potential new treatment option for SCLC.126 In a multicenter, retrospective study conducted in China, anlotinib showed good efficacy in patients with HTs to SCLC (median PFS was 4.3 months in the anlotinib group versus 0.7 months in the placebo group, HR = 0.19, p < 0.0001). From these studies, the use of TKIs after HTs in patients with lung cancer may be considered as a treatment option.
Future strategies for patients who experience HTs
Currently, there are no published prospective trials for patients with HTs. After the accumulation of data from the IMpower 133 and the CAPSIAN study, the platinum + etoposide + anti-PD-L1 inhibitor regimen has been recognized as standard care for patients with extensive-disease SCLC.127,128 In addition, experimental module 7 of the Orchard clinical trial (NCT03944772) is investigating etoposide + durvalumab + carboplatin or cisplatin for patients who develop HTs.129 Another promising strategy is the use of olaparib, a poly-ADP ribose polymerase (PARP) inhibitor. One approach to enhance the clinical activity of ICIs is to modulate the DNA damage response.130 PARP1 is highly expressed in SCLC, and PARP inhibitors have shown antitumor activity in preclinical models and in patients with SCLC. A recent study using a multiomics approach identified a greater activation of immune-related pathways in the transformed SCLC compared with the de novo SCLC.105 Therefore, olaparib is expected to be a useful treatment option for patients with EGFR-positive NSCLC that have undergone HTs to SCLC. Currently, a phase II trial of durvalumab and olaparib for patients with EGFR-mutated transformed SCLC is ongoing (NCT04538378). Furthermore, in a recent study, de novo SCLC cases were categorized into four subtypes (SCLC-A, N, P, and Y), based on the most predominant expression of the four transcription factors that are characteristic of SCLC. Achaete-scute homolog 1 (ASCL1) is predominant in SCLC-A, neurogenic differentiation 1 (NEUROD1) in SCLC-N, POU domain class 2 transcription factor 3 (POU2F3) in SCLC-P, and yes-associated protein 1 (YAP1) in SCLC-Y.131 Treatment strategies for each molecular subtype, such as Bcl2 inhibitors for SCLC-A and the Aurora inhibitors for SCLC-N are warranted.97,132 No comprehensive data have yet been compiled regarding the four subtypes of SCLC after HT. Specific treatment strategies for patients with HTs to SCLC are also warranted.
Future strategies for patients who are at high risk of HTs
Treatment strategies for patients at high risk of HTs have also been investigated. As noted previously, the loss of the TP53 and Rb1 function is not a sufficient condition for HTs to SCLC; however, it is present in most cases of HTs to SCLC. Moreover, in previous research, patients with triple-mutant (EGFR/RB1/TP53) lung cancer had a shorter time to initial EGFR-TKI discontinuation than EGFR/TP53- and EGFR-only mutant cancers.133 In patients with EGFR/RB1/TP53-mutant lung cancer, the persistent cell population after EGFR-TKI treatment may include a subclone that is at high risk of HTs to SCLC, and may benefit from the combination of EGFR-TKIs and a neuroendocrine-based chemotherapy regimen.99 Thus, the continuing use of osimertinib followed by a platinum + etoposide insertion for patients at high risk of HTs to SCLC is currently being investigated in an ongoing phase I clinical trial (NCT03567642).
In considering the treatment strategy for patients with HTs to SCLC, early detection is critical. Although a tissue biopsy is crucial for the diagnosis of HTs in patients with lung cancer, it can be difficult and invasive.134 In a previous report, elevated tumor markers, such as neuron-specific enolase and pro-gastrin-releasing peptide were reported at the time of HTs to SCLC; however, tumor markers are not precise indicators of HTs.135,136 Liquid biopsy is a recently developed, non-invasive technique used in cancer treatment. It has demonstrated an ability to detect, characterize, and monitor cancers using a serum sample. In a previous study, a tissue and plasma analysis of EGFR, TP53, and RB1 contributed to the early detection of HTs to SCLC.99,134,137 Moreover, liquid biopsy techniques have been used in the previously mentioned clinical trial (NCT03567642). Furthermore, the use of a serum digital droplet PCR (ddPCR) might be helpful in identifying high-risk patients. In a previous study, a ddPCR was conducted before osimertinib administration for previously treated NSCLC with HTs to SCLC and showed a low ratio of T790M/activating mutation.138 This study suggested that a tissue biopsy should be considered to exclude the occurrence of HTs to SCLC in cases of a low ratio of T790M/activating mutation in the blood sample. However, the number of patients included in this analysis was small; therefore, the significance of T790M mutations in HTs in patients with lung cancer should be studied in a large cohort.
Conclusions
HT is a major resistance mechanism in patients with lung cancer, not only for EGFR-TKI but also for other molecular targeted agents and ICI therapy. The most prevalent histology after HT is SCLC; however, some patients may experience HTs to another NSCLC. Recently, some reports analyzed paired pre- and post-treatment transformed-tumor samples, and several hypotheses were developed regarding the mechanism of HTs in patients with lung cancer. However, the number of cases in which such analyses have been performed remains small and the mechanisms remain unclear. Moreover, other data such as patient characteristics and treatment efficacy are scarce. Although the current standard treatment for transformed lung cancer is cytotoxic chemotherapy, the treatment efficacy has been reported to be limited. Further research is warranted for this group of patients.
Acknowledgments
The authors would like to thank Yoko Miyake (Clinical Research Center, Kobe City Medical Center General Hospital, Kobe, Japan) for figure editing. This study was supported by internal funding from Internal Medicine III, Wakayama Medical University.
Footnotes
ORCID iD: Daichi Fujimoto  https://orcid.org/0000-0003-0615-3000
https://orcid.org/0000-0003-0615-3000
Contributor Information
Yuki Sato, Department of Respiratory Medicine, Kobe City Medical Center General Hospital, Kobe, Japan.
Go Saito, Department of Respirology, Graduate School of Medicine, Chiba University, Chiba, Japan.
Daichi Fujimoto, Internal Medicine III, Wakayama Medical University, 811-1, Kimiidera, Wakayama 641-8509, Japan.
Declarations
Ethics approval and consent to participate: Not applicable
Consent to publication: Not applicable
Author contribution(s): Yuki Sato: Data curation; Visualization; Writing – original draft; Writing – review & editing.
Go Saito: Data curation; Visualization; Writing – original draft; Writing – review & editing.
Daichi Fujimoto: Conceptualization; Funding acquisition; Project administration; Supervision; Writing – original draft; Writing – review & editing.
Funding: The authors disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This study was supported by internal funding from Internal Medicine III, Wakayama Medical University.
Dr. Sato reported receiving personal fees from AstraZeneca, Chugai Pharmaceutical, MSD, Ono Pharmaceutical, Novartis, Pfizer, Taiho Pharmaceutical, Nippon Kayaku, Bristol Myers Squibb, Eli Lilly, Takeda, and Kyowa Kirin outside the submitted work.
Dr. Saito reported receiving personal fees from Ono Pharmaceutical, Chugai Pharmaceutical, AstraZeneca, Novartis Pharma, and Taiho Pharmaceutical outside the submitted work.
Dr. Fujimoto reported receiving grants from Boehringer Ingelheim and AstraZeneca; personal fees from Astra-Zeneca K.K., Ono Pharmaceutical Co., Ltd., Bristol Myers Squibb Co., Ltd., Taiho Pharmaceutical Co., Ltd., Chugai Pharmaceutical Co., Ltd., MSD K.K., Boehringer Ingelheim Japan Inc., Eli Lilly Japan K.K., and Novartis Pharma K.K. outside the submitted work. Dr. Fujimoto also served on an advisory board for AstraZeneca and Chugai Pharmaceutical.
Availability of data and materials: Not applicable.
References
- 1. Siegel RL, Miller KD, Fuchs HE, et al. Cancer statistics, 2022. CA Cancer J Clin 2022; 72: 7–33. [DOI] [PubMed] [Google Scholar]
- 2. World Health Organisation Classification of Tumours Editorial Board. Thoracic Tumours. 5th ed. Lyon, France: International Agency for Research on Cancer, 2021. [Google Scholar]
- 3. Maemondo M, Inoue A, Kobayashi K, et al. Gefitinib or chemotherapy for non-small-cell lung cancer with mutated EGFR. N Engl J Med 2010; 362: 2380–2388. [DOI] [PubMed] [Google Scholar]
- 4. Mitsudomi T, Kosaka T, Endoh H, et al. Mutations of the epidermal growth factor receptor gene predict prolonged survival after gefitinib treatment in patients with non-small-cell lung cancer with postoperative recurrence. J Clin Oncol 2005; 23: 2513–2520. [DOI] [PubMed] [Google Scholar]
- 5. Kobayashi S, Boggon TJ, Dayaram T, et al. EGFR mutation and resistance of non-small-cell lung cancer to gefitinib. N Engl J Med 2005; 352: 786–792. [DOI] [PubMed] [Google Scholar]
- 6. Mok TS, Wu YL, Ahn MJ, et al. Osimertinib or platinum-pemetrexed in EGFR T790M-positive lung cancer. N Engl J Med 2017; 376: 629–640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Nosaki K, Satouchi M, Kurata T, et al. Re-biopsy status among non-small cell lung cancer patients in Japan: a retrospective study. Lung Cancer 2016; 101: 1–8. [DOI] [PubMed] [Google Scholar]
- 8. Zakowski MF, Ladanyi M, Kris MG, et al. EGFR mutations in small-cell lung cancers in patients who have never smoked. N Engl J Med 2006; 355: 213–215. [DOI] [PubMed] [Google Scholar]
- 9. Morinaga R, Okamoto I, Furuta K, et al. Sequential occurrence of non-small cell and small cell lung cancer with the same EGFR mutation. Lung Cancer 2007; 58: 411–413. [DOI] [PubMed] [Google Scholar]
- 10. Fujimoto D, Akamatsu H, Morimoto T, et al. Histologic transformation of epidermal growth factor receptor-mutated lung cancer. Eur J Cancer 2022; 166: 41–50. [DOI] [PubMed] [Google Scholar]
- 11. Sequist LV, Waltman BA, Dias-Santagata D, et al. Genotypic and histological evolution of lung cancers acquiring resistance to EGFR inhibitors. Sci Transl Med 2011; 3: 75ra26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Bai W, Zhen C, Zhang R, et al. Clinicopathological features of patients with transformation from EGFR mutant lung adenocarcinoma to small cell lung cancer. Transl Cancer Res 2021; 10: 3694–3704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Chai X, Zhang X, Li W, et al. Small cell lung cancer transformation during antitumor therapies: a systematic review. Open Med (Wars) 2021; 16: 1160–1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Adelstein DJ, Tomashefski JF, Jr., Snow NJ, et al. Mixed small cell and non-small cell lung cancer. Chest 1986; 89: 699–704. [DOI] [PubMed] [Google Scholar]
- 15. Oser MG, Niederst MJ, Sequist LV, et al. Transformation from non-small-cell lung cancer to small-cell lung cancer: molecular drivers and cells of origin. Lancet Oncology 2015; 16: e165–e172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Yu HA, Arcila ME, Rekhtman N, et al. Analysis of tumor specimens at the time of acquired resistance to EGFR-TKI therapy in 155 patients with EGFR-mutant lung cancers. Clin Cancer Res 2013; 19: 2240–2247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Zeng L, Xiao L, Jiang W, et al. Investigation of efficacy and acquired resistance for EGFR-TKI plus bevacizumab as first-line treatment in patients with EGFR sensitive mutant non-small cell lung cancer in a real world population. Lung Cancer 2020; 141: 82–88. [DOI] [PubMed] [Google Scholar]
- 18. Piotrowska Z, Isozaki H, Lennerz JK, et al. Landscape of acquired resistance to osimertinib in EGFR-mutant NSCLC and clinical validation of combined EGFR and RET Inhibition with osimertinib and BLU-667 for acquired RET fusion. Cancer Discov 2018; 8: 1529–1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lin C-C, Shih J-Y, Yu C-J, et al. Outcomes in patients with non-small-cell lung cancer and acquired Thr790Met mutation treated with osimertinib: a genomic study. Lancet Respiratory Medicine 2018; 6: 107–116. [DOI] [PubMed] [Google Scholar]
- 20. Oxnard GR, Hu Y, Mileham KF, et al. Assessment of resistance mechanisms and clinical implications in patients With EGFR T790M-positive lung cancer and acquired resistance to osimertinib. JAMA Oncol 2018; 4: 1527–1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Schoenfeld AJ, Chan JM, Kubota D, et al. Tumor analyses reveal squamous transformation and off-target alterations as early resistance mechanisms to first-line osimertinib in EGFR-mutant lung cancer. Clin Cancer Res 2020; 26: 2654–2663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Roca E, Pozzari M, Vermi W, et al. Outcome of EGFR-mutated adenocarcinoma NSCLC patients with changed phenotype to squamous cell carcinoma after tyrosine kinase inhibitors: a pooled analysis with an additional case. Lung Cancer 2019; 127: 12–18. [DOI] [PubMed] [Google Scholar]
- 23. Nishimatsu K, Minami S, Ihara S, et al. Transformation from adenocarcinoma to pleomorphic carcinoma as an acquired resistance to epidermal growth factor receptor tyrosine kinase Inhibitors. J Med Cases 2021; 12: 310–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Toda-Ishii M, Akaike K, Kurisaki-Arakawa A, et al. Sarcomatous transformation of EGFR and TP53 mutation-positive metastatic adenocarcinoma of the lungs, masquerading as a primary pleomorphic sarcoma of the proximal femur. Int J Clin Exp Pathol 2015; 8: 3270–3278. [PMC free article] [PubMed] [Google Scholar]
- 25. Hsieh MS, Lin MW, Lee YH. Lung adenoc arcinoma with sarcomatoid transformation after tyrosine kinase inhibitor treatment and chemotherapy. Lung Cancer 2019; 137: 76–84. [DOI] [PubMed] [Google Scholar]
- 26. Soria JC, Ohe Y, Vansteenkiste J, et al. Osimertinib in untreated EGFR-mutated advanced non-small-cell lung Cancer. N Engl J Med 2018; 378: 113–125. [DOI] [PubMed] [Google Scholar]
- 27. Oxnard GR, Yang JC, Yu H, et al. TATTON: a multi-arm, phase Ib trial of osimertinib combined with selumetinib, savolitinib, or durvalumab in EGFR-mutant lung cancer. Ann Oncol 2020; 31: 507–516. [DOI] [PubMed] [Google Scholar]
- 28. Nakagawa K, Garon EB, Seto T, et al. Ramucirumab plus erlotinib in patients with untreated, EGFR-mutated, advanced non-small-cell lung cancer (RELAY): a randomised, double-blind, placebo-controlled, phase 3 trial. Lancet Oncol 2019; 20: 1655–1669. [DOI] [PubMed] [Google Scholar]
- 29. Park K, Haura EB, Leighl NB, et al. Amivantamab in EGFR exon 20 insertion-mutated non-small-cell lung cancer progressing on platinum chemotherapy: initial results from the CHRYSALIS phase I study. J Clin Oncol 2021; 39: 3391–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Woo CG, Son SM, Lee HC, et al. Histologic changes in non-small cell lung cancer under various treatments: a comparison of histology and mutation status in serial samples. Cancer Res Treat 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Aggarwal R, Huang J, Alumkal JJ, et al. Clinical and genomic characterization of treatment-emergent small-cell neuroendocrine prostate cancer: a multi-institutional prospective study. J Clin Oncol 2018; 36: 2492–2503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Morita M, Saeki H, Nakaji YU, et al. Conversion to neuroendocrine carcinoma from squamous cell carcinoma of the esophagus after definitive chemoradiotherapy. Anticancer Res 2016; 36: 4045–4049. [PubMed] [Google Scholar]
- 33. Shia J, Tickoo SK, Guillem JG, et al. Increased endocrine cells in treated rectal adenocarcinomas: a possible reflection of endocrine differentiation in tumor cells induced by chemotherapy and radiotherapy. Am J Surg Pathol 2002; 26: 863–872. [DOI] [PubMed] [Google Scholar]
- 34. Mariniello A, Righi L, Morrone A, et al. Squamous cell histological transformation in a lung adenocarcinoma patient (hyper) progressing upon immunotherapy. Tumori. Epub ahead of print March 2022. DOI: 10.1177/03008916221080487. [DOI] [PubMed] [Google Scholar]
- 35. Nagasaka M, Pansare RS, Abdulfatah E, et al. Histologic transformation in NSCLC with PD-1 therapy. J Thorac Oncol 2017; 12: e133–e134. [DOI] [PubMed] [Google Scholar]
- 36. Imakita T, Fujita K, Kanai O, et al. Small cell lung cancer transformation during immunotherapy with nivolumab: a case report. Respir Med Case Rep 2017; 21: 52–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Si X, You Y, Zhang X, et al. Histologic transformation of lung cancer during pembrolizumab therapy: a case report. Thorac Cancer 2020; 11: 793–796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Shen Q, Qu J, Sheng L, et al. Case report: transformation from non-small cell lung cancer to small cell lung cancer during anti-PD-1 therapy: a report of two cases. Front Oncol 2021; 11: 619371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Imakita T, Fujita K, Kanai O, et al. Small cell transformation of non-small cell lung cancer under immunotherapy: case series and literature review. Thorac Cancer 2021; 12: 3062–3067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Bar J, Ofek E, Barshack I, et al. Transformation to small cell lung cancer as a mechanism of resistance to immunotherapy in non-small cell lung cancer. Lung Cancer 2019; 138: 109–115. [DOI] [PubMed] [Google Scholar]
- 41. Sehgal K, Varkaris A, Viray H, et al. Small cell transformation of non-small cell lung cancer on immune checkpoint inhibitors: uncommon or under-recognized? J Immunother Cancer 2020; 8: e000697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Cha YJ, Cho BC, Kim HR, et al. A case of ALK-rearranged adenocarcinoma with small cell carcinoma-like transformation and resistance to crizotinib. J Thorac Oncol 2016; 11: e55–e58. [DOI] [PubMed] [Google Scholar]
- 43. Yamagata A, Yokoyama T, Fukuda Y, et al. Alectinib re-challenge in small cell lung cancer transformation after chemotherapy failure in a patient with ALK-positive lung cancer: a case report. Respir Med Case Rep 2021; 33: 101440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Caumont C, Veillon R, Gros A, et al. Neuroendocrine phenotype as an acquired resistance mechanism in ALK-rearranged lung adenocarcinoma. Lung Cancer 2016; 92: 15–18. [DOI] [PubMed] [Google Scholar]
- 45. Ou SI, Lee TK, Young L, et al. Dual occurrence of ALK G1202R solvent front mutation and small cell lung cancer transformation as resistance mechanisms to second generation ALK inhibitors without prior exposure to crizotinib. Pitfall of solely relying on liquid re-biopsy? Lung Cancer 2017; 106: 110–114. [DOI] [PubMed] [Google Scholar]
- 46. Fares AF, Lok BH, Zhang T, et al. ALK-rearranged lung adenocarcinoma transformation into high-grade large cell neuroendocrine carcinoma: clinical and molecular description of two cases. Lung Cancer 2020; 146: 350–354. [DOI] [PubMed] [Google Scholar]
- 47. Takegawa N, Hayashi H, Iizuka N, et al. Transformation of ALK rearrangement-positive adenocarcinoma to small-cell lung cancer in association with acquired resistance to alectinib. Ann Oncol 2016; 27: 953–955. [DOI] [PubMed] [Google Scholar]
- 48. Fujita S, Masago K, Katakami N, et al. Transformation to SCLC after treatment with the ALK Inhibitor alectinib. J Thorac Oncol 2016; 11: e67–e72. [DOI] [PubMed] [Google Scholar]
- 49. Levacq D, D’Haene N, de Wind R, et al. Histological transformation of ALK rearranged adenocarcinoma into small cell lung cancer: a new mechanism of resistance to ALK inhibitors. Lung Cancer 2016; 102: 38–41. [DOI] [PubMed] [Google Scholar]
- 50. Katayama R, Shaw AT, Khan TM, et al. Mechanisms of acquired crizotinib resistance in ALK-rearranged lung cancers. Sci Transl Med 2012; 4: 120ra117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Yanagitani N, Uchibori K, Koike S, et al. Drug resistance mechanisms in Japanese anaplastic lymphoma kinase-positive non-small cell lung cancer and the clinical responses based on the resistant mechanisms. Cancer Sci 2020; 111: 932–939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Yu Y, Ou Q, Wu X, et al. Concomitant resistance mechanisms to multiple tyrosine kinase inhibitors in ALK-positive non-small cell lung cancer. Lung Cancer 2019; 127: 19–24. [DOI] [PubMed] [Google Scholar]
- 53. Miyamoto S, Ikushima S, Ono R, et al. Transformation to small-cell lung cancer as a mechanism of acquired resistance to crizotinib and alectinib. Jpn J Clin Oncol 2016; 46: 170–173. [DOI] [PubMed] [Google Scholar]
- 54. Huang J, Zhang S-L, Zhou C, et al. Genomic and transcriptomic analysis of neuroendocrine transformation in ALK-rearranged lung adenocarcinoma after treatments with sequential ALK inhibitors: a brief report. JTO Clin Res Rep 2022; 3: 100338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Gong J, Gregg JP, Ma W, et al. Squamous cell transformation of primary lung adenocarcinoma in a patient with EML4-ALK fusion variant 5 refractory to ALK inhibitors. J Natl Compr Canc Netw 2019; 17: 297–301. [DOI] [PubMed] [Google Scholar]
- 56. Kaiho T, Nakajima T, Iwasawa S, et al. ALK rearrangement adenocarcinoma with histological transformation to squamous cell carcinoma resistant to alectinib and ceritinib. Onco Targets Ther 2020; 13: 1557–1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Park S, Han J, Sun JM. Histologic transformation of ALK-rearranged adenocarcinoma to squamous cell carcinoma after treatment with ALK inhibitor. Lung Cancer 2019; 127: 66–68. [DOI] [PubMed] [Google Scholar]
- 58. Ball M, Christopoulos P, Kirchner M, et al. Histological and molecular plasticity of ALK-positive non-small-cell lung cancer under targeted therapy: a case report. Cold Spring Harb Mol Case Stud 2022; 8: a006156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Zhang Y, Qin Y, Xu H, et al. Case report: a case report of a histological transformation of ALK-rearranged adenocarcinoma with high expression of PD-L1 to squamous cell carcinoma after treatment with alectinib. Pathol Oncol Res 2021; 27: 637745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Ueda S, Shukuya T, Hayashi T, et al. Transformation from adenocarcinoma to squamous cell lung carcinoma with MET amplification after lorlatinib resistance: a case report. Thorac Cancer 2021; 12: 715–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Lin JJ, Langenbucher A, Gupta P, et al. Small cell transformation of ROS1 fusion-positive lung cancer resistant to ROS1 inhibition. NPJ Precis Oncol 2020; 4: 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Wu CH, Su PL, Hsu CW, et al. Small cell transformation in crizotinib-resistant ROS1-rearranged non-small cell lung cancer with retention of ROS1 fusion: A case report. Thorac Cancer 2021; 12: 3068–3071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Awad MM, Liu S, Rybkin II, et al. Acquired resistance to KRAS G12C inhibition in cancer. N Engl J Med 2021; 384: 2382–2393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Ji H, Ramsey MR, Hayes DN, et al. LKB1 modulates lung cancer differentiation and metastasis. Nature 2007; 448: 807–810. [DOI] [PubMed] [Google Scholar]
- 65. Zhang H, Fillmore Brainson C, Koyama S, et al. LKB1 inactivation drives lung cancer lineage switching governed by polycomb repressive complex 2. Nat Commun 2017; 8: 14922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Facchinetti F, Lacroix L, Mezquita L, et al. Molecular mechanisms of resistance to BRAF and MEK inhibitors in BRAF V600E non-small cell lung cancer. Eur J Cancer 2020; 132: 211–223. [DOI] [PubMed] [Google Scholar]
- 67. Lin JJ, Liu SV, McCoach CE, et al. Mechanisms of resistance to selective RET tyrosine kinase inhibitors in RET fusion-positive non-small-cell lung cancer. Ann Oncol 2020; 31: 1725–1733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Kwon MC, Berns A. Mouse models for lung cancer. Mol Oncol 2013; 7: 165–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Fisher GH, Wellen SL, Klimstra D, et al. Induction and apoptotic regression of lung adenocarcinomas by regulation of a K-Ras transgene in the presence and absence of tumor suppressor genes. Genes Dev 2001; 15: 3249–3262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Mainardi S, Mijimolle N, Francoz S, et al. Identification of cancer initiating cells in K-Ras driven lung adenocarcinoma. Proc Natl Acad Sci U S A 2014; 111: 255–260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Politi K, Zakowski MF, Fan PD, et al. Lung adenocarcinomas induced in mice by mutant EGF receptors found in human lung cancers respond to a tyrosine kinase inhibitor or to down-regulation of the receptors. Genes Dev 2006; 20: 1496–1510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Lin C, Song H, Huang C, et al. Alveolar type II cells possess the capability of initiating lung tumor development. PLoS One 2012; 7: e53817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Nagaraj AS, Lahtela J, Hemmes A, et al. Cell of origin links histotype spectrum to immune microenvironment diversity in non-small-cell lung cancer driven by mutant Kras and loss of Lkb1. Cell Rep 2017; 18: 673–684. [DOI] [PubMed] [Google Scholar]
- 74. Spella M, Lilis I, Pepe MA, et al. Club cells form lung adenocarcinomas and maintain the alveoli of adult mice. Elife 2019; 8: e45571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Ferone G, Song JY, Sutherland KD, et al. SOX2 Is the determining oncogenic switch in promoting lung squamous cell carcinoma from different cells of origin. Cancer Cell 2016; 30: 519–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Ruiz EJ, Diefenbacher ME, Nelson JK, et al. LUBAC determines chemotherapy resistance in squamous cell lung cancer. J Exp Med 2019; 216: 450–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Cole BB, Smith RW, Jenkins KM, et al. Tracheal basal cells: a facultative progenitor cell pool. Am J Pathol 2010; 177: 362–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Xiao Z, Jiang Q, Willette-Brown J, et al. The pivotal role of IKKalpha in the development of spontaneous lung squamous cell carcinomas. Cancer Cell 2013; 23: 527–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Camolotto SA, Pattabiraman S, Mosbruger TL, et al. FoxA1 and FoxA2 drive gastric differentiation and suppress squamous identity in NKX2-1-negative lung cancer. Elife 2018; 7: e38579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Sutherland KD, Proost N, Brouns I, et al. Cell of origin of small cell lung cancer: inactivation of Trp53 and Rb1 in distinct cell types of adult mouse lung. Cancer Cell 2011; 19: 754–764. [DOI] [PubMed] [Google Scholar]
- 81. Park KS, Liang MC, Raiser DM, et al. Characterization of the cell of origin for small cell lung cancer. Cell Cycle 2011; 10: 2806–2815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Song H, Yao E, Lin C, et al. Functional characterization of pulmonary neuroendocrine cells in lung development, injury, and tumorigenesis. Proc Natl Acad Sci U S A 2012; 109: 17531–17536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Jia D, Augert A, Kim DW, et al. Crebbp loss drives small cell lung cancer and increases sensitivity to HDAC inhibition. Cancer Discov 2018; 8: 1422–1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Yang D, Denny SK, Greenside PG, et al. Intertumoral heterogeneity in SCLC is influenced by the cell type of origin. Cancer Discov 2018; 8: 1316–1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Ferone G, Song JY, Krijgsman O, et al. FGFR1 oncogenic activation reveals an alternative cell of origin of SCLC in Rb1/p53 mice. Cell Rep 2020; 30: 3837–3850 e3833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Ferone G, Lee MC, Sage J, et al. Cells of origin of lung cancers: lessons from mouse studies. Genes Dev 2020; 34: 1017–1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Quintanal-Villalonga A, Chan JM, Yu HA, et al. Lineage plasticity in cancer: a shared pathway of therapeutic resistance. Nat Rev Clin Oncol 2020; 17: 360–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Cavenee WK, Dryja TP, Phillips RA, et al. Expression of recessive alleles by chromosomal mechanisms in retinoblastoma. Nature 1983; 305: 779–784. [DOI] [PubMed] [Google Scholar]
- 89. Godbout R, Dryja TP, Squire J, et al. Somatic inactivation of genes on chromosome 13 is a common event in retinoblastoma. Nature 1983; 304: 451–453. [DOI] [PubMed] [Google Scholar]
- 90. Harbour JW, Lai SL, Whang-Peng J, et al. Abnormalities in structure and expression of the human retinoblastoma gene in SCLC. Science 1988; 241: 353–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Shimizu E, Coxon A, Otterson GA, et al. RB protein status and clinical correlation from 171 cell lines representing lung cancer, extrapulmonary small cell carcinoma, and mesothelioma. Oncogene 1994; 9: 2441–2448. [PubMed] [Google Scholar]
- 92. Maandag EC, van der Valk M, Vlaar M, et al. Developmental rescue of an embryonic-lethal mutation in the retinoblastoma gene in chimeric mice. EMBO J 1994; 13: 4260–4268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Jacks T, Fazeli A, Schmitt EM, et al. Effects of an Rb mutation in the mouse. Nature 1992; 359: 295–300. [DOI] [PubMed] [Google Scholar]
- 94. Meuwissen R, Linn SC, Linnoila RI, et al. Induction of small cell lung cancer by somatic inactivation of both Trp53 and Rb1 in a conditional mouse model. Cancer Cell 2003; 4: 181–189. [DOI] [PubMed] [Google Scholar]
- 95. Peifer M, Fernandez-Cuesta L, Sos ML, et al. Integrative genome analyses identify key somatic driver mutations of small-cell lung cancer. Nat Genet 2012; 44: 1104–1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Rudin CM, Durinck S, Stawiski EW, et al. Comprehensive genomic analysis identifies SOX2 as a frequently amplified gene in small-cell lung cancer. Nat Genet 2012; 44: 1111–1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Niederst MJ, Sequist LV, Poirier JT, et al. RB loss in resistant EGFR mutant lung adenocarcinomas that transform to small-cell lung cancer. Nat Commun 2015; 6: 6377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Lee JK, Lee J, Kim S, et al. Clonal history and genetic predictors of transformation into small-cell carcinomas from lung adenocarcinomas. J Clin Oncol 2017; 35: 3065–3074. [DOI] [PubMed] [Google Scholar]
- 99. Offin M, Chan JM, Tenet M, et al. Concurrent RB1 and TP53 alterations define a subset of EGFR-mutant lung cancers at risk for histologic transformation and inferior clinical outcomes. J Thorac Oncol 2019; 14: 1784–1793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Jarmuz A, Chester A, Bayliss J, et al. An anthropoid-specific locus of orphan C to U RNA-editing enzymes on chromosome 22. Genomics 2002; 79: 285–296. [DOI] [PubMed] [Google Scholar]
- 101. Roberts SA, Lawrence MS, Klimczak LJ, et al. An APOBEC cytidine deaminase mutagenesis pattern is widespread in human cancers. Nat Genet 2013; 45: 970–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Burns MB, Temiz NA, Harris RS. Evidence for APOBEC3B mutagenesis in multiple human cancers. Nat Genet 2013; 45: 977–983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Alexandrov LB, Nik-Zainal S, Wedge DC, et al. Signatures of mutational processes in human cancer. Nature 2013; 500: 415–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Park JW, Lee JK, Sheu KM, et al. Reprogramming normal human epithelial tissues to a common, lethal neuroendocrine cancer lineage. Science 2018; 362: 91–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Quintanal-Villalonga A, Taniguchi H, Zhan YA, et al. Multiomic analysis of lung tumors defines pathways activated in neuroendocrine transformation. Cancer Discov 2021; 11: 3028–3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Miyoshi Y, Uemura H, Kitami K, et al. Neuroendocrine differentiated small cell carcinoma presenting as recurrent prostate cancer after androgen deprivation therapy. BJU Int 2001; 88: 982–983. [DOI] [PubMed] [Google Scholar]
- 107. Wright ME, Tsai MJ, Aebersold R. Androgen receptor represses the neuroendocrine transdifferentiation process in prostate cancer cells. Mol Endocrinol 2003; 17: 1726–1737. [DOI] [PubMed] [Google Scholar]
- 108. Ku SY, Rosario S, Wang Y, et al. Rb1 and Trp53 cooperate to suppress prostate cancer lineage plasticity, metastasis, and antiandrogen resistance. Science 2017; 355: 78–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Bishop JL, Thaper D, Vahid S, et al. The master neural transcription factor BRN2 is an androgen receptor-suppressed driver of neuroendocrine differentiation in prostate cancer. Cancer Discov 2017; 7: 54–71. [DOI] [PubMed] [Google Scholar]
- 110. Kim J, Jin H, Zhao JC, et al. FOXA1 inhibits prostate cancer neuroendocrine differentiation. Oncogene 2017; 36: 4072–4080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Volante M, Tota D, Giorcelli J, et al. Androgen deprivation modulates gene expression profile along prostate cancer progression. Hum Pathol 2016; 56: 81–88. [DOI] [PubMed] [Google Scholar]
- 112. Akamatsu S, Wyatt AW, Lin D, et al. The placental gene PEG10 promotes progression of neuroendocrine prostate cancer. Cell Rep 2015; 12: 922–936. [DOI] [PubMed] [Google Scholar]
- 113. Mollaoglu G, Jones A, Wait SJ, et al. The lineage-defining transcription factors SOX2 and NKX2-1 determine lung cancer cell fate and shape the tumor immune microenvironment. Immunity 2018; 49: 764–779.e769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Mukhopadhyay A, Berrett KC, Kc U, et al. Sox2 cooperates with LKB1 loss in a mouse model of squamous cell lung cancer. Cell Rep 2014; 8: 40–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Han X, Li F, Fang Z, et al. Transdifferentiation of lung adenocarcinoma in mice with Lkb1 deficiency to squamous cell carcinoma. Nat Commun 2014; 5: 3261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Xu C, Fillmore CM, Koyama S, et al. Loss of LKB1 and Pten leads to lung squamous cell carcinoma with elevated PD-L1 expression. Cancer Cell 2014; 25: 590–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Park S, Shim JH, Lee B, et al. Paired genomic analysis of squamous cell carcinoma transformed from EGFR-mutated lung adenocarcinoma. Lung Cancer 2019; 134: 7–15. [DOI] [PubMed] [Google Scholar]
- 118. Cancer Genome Atlas Research Network. Comprehensive genomic characterization of squamous cell lung cancers. Nature 2012; 489: 519–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Marcoux N, Gettinger SN, O’Kane G, et al. EGFR-mutant adenocarcinomas that transform to small-cell lung cancer and other neuroendocrine carcinomas: clinical outcomes. J Clin Oncol 2019; 37: 278–285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Wang W, Xu C, Chen H, et al. Genomic alterations and clinical outcomes in patients with lung adenocarcinoma with transformation to small cell lung cancer after treatment with EGFR tyrosine kinase inhibitors: a multicenter retrospective study. Lung Cancer 2021; 155: 20–27. [DOI] [PubMed] [Google Scholar]
- 121. Wang S, Xie T, Hao X, et al. Comprehensive analysis of treatment modes and clinical outcomes of small cell lung cancer transformed from epidermal growth factor receptor mutant lung adenocarcinoma. Thorac Cancer 2021; 12: 2585–2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Noda K, Nishiwaki Y, Kawahara M, et al. Irinotecan plus cisplatin compared with etoposide plus cisplatin for extensive small-cell lung cancer. N Engl J Med 2002; 346: 85–91. [DOI] [PubMed] [Google Scholar]
- 123. Hanna N, Bunn PA, Jr., Langer C, et al. Randomized phase III trial comparing irinotecan/cisplatin with etoposide/cisplatin in patients with previously untreated extensive-stage disease small-cell lung cancer. J Clin Oncol 2006; 24: 2038–2043. [DOI] [PubMed] [Google Scholar]
- 124. Ferrer L, Giaj Levra M, Brevet M, et al. A brief report of transformation from NSCLC to SCLC: molecular and therapeutic characteristics. J Thorac Oncol 2019; 14: 130–134. [DOI] [PubMed] [Google Scholar]
- 125. Park CK, Oh IJ, Kim YC. Is transformed small cell lung cancer (SCLC) different from de novo SCLC? Transl Cancer Res 2019; 8: 346–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Cheng Y, Wang Q, Li K, et al. Anlotinib vs placebo as third- or further-line treatment for patients with small cell lung cancer: a randomised, double-blind, placebo-controlled Phase 2 study. Br J Cancer 2021; 125: 366–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Horn L, Mansfield AS, Szczesna A, et al. First-line atezolizumab plus chemotherapy in extensive-stage small-cell lung cancer. N Engl J Med 2018; 379: 2220–2229. [DOI] [PubMed] [Google Scholar]
- 128. Paz-Ares L, Dvorkin M, Chen Y, et al. Durvalumab plus platinum-etoposide versus platinum-etoposide in first-line treatment of extensive-stage small-cell lung cancer (CASPIAN): a randomised, controlled, open-label, phase 3 trial. Lancet 2019; 394: 1929–1939. [DOI] [PubMed] [Google Scholar]
- 129. Yu HA, Goldberg SB, Le X, et al. Biomarker-directed phase II platform study in patients with EGFR sensitizing mutation-positive advanced/metastatic non-small cell lung cancer whose disease has progressed on first-line osimertinib therapy (ORCHARD). Clin Lung Cancer 2021; 22: 601–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Brown JS, Sundar R, Lopez J. Combining DNA damaging therapeutics with immunotherapy: more haste, less speed. Br J Cancer 2018; 118: 312–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Rudin CM, Poirier JT, Byers LA, et al. Molecular subtypes of small cell lung cancer: a synthesis of human and mouse model data. Nat Rev Cancer 2019; 19: 289–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Gong X, Du J, Parsons SH, et al. Aurora a kinase inhibition is synthetic lethal with loss of the RB1 tumor suppressor gene. Cancer Discov 2019; 9: 248–263. [DOI] [PubMed] [Google Scholar]
- 133. Vokes NI, Chambers E, Nguyen T, et al. Concurrent TP53 mutations facilitate resistance evolution in EGFR-mutant lung adenocarcinoma. J Thorac Oncol 2022; 17: 779–792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Pizzutilo EG, Pedrani M, Amatu A, et al. Liquid biopsy for small cell lung cancer either de novo or transformed: systematic review of different applications and meta-analysis. Cancers (Basel) 2021; 13: 2265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Zhang Y, Li XY, Tang Y, et al. Rapid increase of serum neuron specific enolase level and tachyphylaxis of EGFR-tyrosine kinase inhibitor indicate small cell lung cancer transformation from EGFR positive lung adenocarcinoma? Lung Cancer 2013; 81: 302–305. [DOI] [PubMed] [Google Scholar]
- 136. Oya Y, Yoshida T, Uemura T, et al. Serum ProGRP and NSE levels predicting small cell lung cancer transformation in a patient with ALK rearrangement-positive non-small cell lung cancer: a case report. Oncol Lett 2018; 16: 4219–4222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Vendrell JA, Quantin X, Serre I, et al. Combination of tissue and liquid biopsy molecular profiling to detect transformation to small cell lung carcinoma during osimertinib treatment. Ther Adv Med Oncol 2020; 12: 1758835920974192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. Minari R, Bordi P, Del Re M, et al. Primary resistance to osimertinib due to SCLC transformation: issue of T790M determination on liquid re-biopsy. Lung Cancer 2018; 115: 21–27. [DOI] [PubMed] [Google Scholar]