Abstract
Anti-hepatitis B core antigen (HBcAg)-positive hepatitis B surface antigen (HBsAg)-negative plasma samples from blood donors were tested by nested PCR. DNA positivity was more significantly associated with high levels of anti-HBcAg than with low levels of anti-HBsAg antibodies. Analysis of a dilution of anti-HBcAg antibodies might result in a more rational exclusion of anti-HBcAg-positive HBsAg-negative samples, reducing the number of donations discarded and enabling more countries to incorporate anti-HBcAg testing.
The incidence of posttransfusion hepatitis B virus (HBV) infection has been significantly reduced with the development of serological tests for the detection of HBV surface antigen (HBsAg). It is known, however, that some blood derivatives, negative for HBsAg but positive for antibodies against hepatitis B core antigen (anti-HBcAg), are able to transmit the infection, both after transfusion and after transplantation of organs (6, 17). The presence of anti-HBcAg antibodies without any other HBV serological marker is frequently found in different population groups. Different situations may account for this result: (i) a false-positive anti-HBcAg result; (ii) low levels of HBV replication inside the hepatocyte, without detectable production of HBsAg; (iii) the window phase of acute HBV infection; (iv) the loss of anti-HBsAg with time or failure to develop an antibody response against the antigen after infection; or (v) the presence of a vaccine escape mutant, not detected by most of the currently available HBsAg detection tests (3, 9, 11, 16).
In some countries with a high prevalence of HBV infection, such as Japan, exclusion of all anti-HBcAg-positive plasma units would result in a drastic reduction in the number of units available for transfusion. Additional testing, like determination of the level of anti-HBcAg antibody (9), has been included to enable the transfusion of some of the anti-HBcAg-positive plasma units. On the other hand, anti-HBcAg testing is not mandatory in some countries. More rational criteria for discarding HBcAg-positive blood units may help these countries to adopt this additional testing. The aim of this study was the molecular and serological characterization of anti-HBcAg-positive, HBsAg-negative plasma units from blood donors, in order to evaluate whether the measures practiced in some Asiatic countries could be adopted in other settings with a lower prevalence of HBV infection.
The study comprised 171 plasma samples, screened as positive for anti-HBcAg antibodies and negative for HBsAg, from voluntary blood donors from Banco Municipal de Sangre, Caracas, Venezuela, and from Planta Procesadora de Derivados Sanguíneos Quimbiotec, Caracas, Venezuela. Anti-HBcAg positivity was corroborated, by a monoclonal inhibition enzyme immunoassay (EIA) (14), in 167 of the 171 samples (97.7%) originally referred as positive by blood banks. Discrepant samples were also found to be negative by a commercial EIA (Hepanostika; Organon-Teknika). Anti-HBcAg antibody titers were determined by diluting samples in anti-HBcAg-negative plasma. Immunoglobulin M (IgM) anti-HBcAg antibodies were determined in PCR-positive samples by a commercial EIA (Corzyme-M; Abbott Laboratories, Diagnostics Division). Absence of HBsAg in plasma samples was confirmed by a double sandwich EIA (13). Anti-HBsAg antibody levels were determined by a commercial EIA (Roche Diagnostic GmbH).
The presence of a conserved region of the HBsAg gene was assayed by nested PCR (2). Two DNA extraction methods were used. In the first one, 10 μl of plasma was treated with 10 μl of NaOH and then neutralized with 20 μl of HCl according to a previously reported procedure (7). Ten microliters of extracted material (equivalent to 2.5 μl of starting material) was amplified by nested PCR. In the second method, 550 μl of plasma was treated with 25 mg of proteinase K per ml–70 mM Tris-HCl–35 mM EDTA–3.5% sodium dodecyl sulfate for 3 h at 50°C. After addition of 10 mg of bovine serum albumin per ml (8), DNA was extracted with phenol-chloroform and precipitated by ethanol. Half of the original input of plasma (10 μl of extracted material, equivalent to 275 μl of starting material) was used for amplification. Positive controls were HBsAg-positive samples infected with the most divergent genotype (F) of HBV, highly prevalent in Venezuela (1). A sample was considered positive when repeatedly found positive after amplification of newly extracted material.
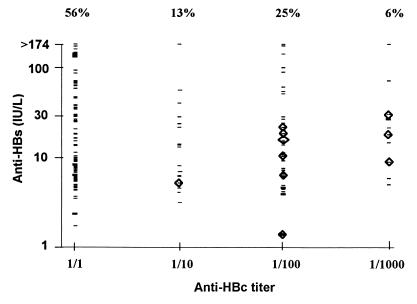
Two populations of anti-HBcAg-positive plasma units were observed: one with low antibody titers in the monoclonal inhibition assay (positive only undiluted, 56% of the total plasma) and another with titers equal to or greater than 1/100 (31% [Fig. 1]). The same bimodal distribution of anti-HBcAg antibodies was observed in a Japanese population of blood donors (9). About half of the anti-HBcAg-positive plasma units (45%) exhibited anti-HBsAg levels below the limit established as protective (10 IU/liter) (Fig. 1). It has been reported elsewhere that about half of the anti-HBcAg-positive blood donor samples from Brazil are not positive for another HBV marker (19). This prevalence of isolated anti-HBcAg antibodies is higher than the one found among U.S. blood donors (20 to 30%) (15). It is not known whether some HBV genotypes induce lower degrees of antibody response after a natural infection. If so, this could account for the low levels of anti-HBsAg observed among several Venezuelan and Brazilian blood plasma units, where the predominant infecting genotype is the most divergent genotype, F (1, 10). On the other hand, even if anti-HBcAg false positivity was reduced by an additional immunoassay, false-positive anti-HBcAg results may still account for some of these low-anti-HBcAg and low-anti-HBsAg plasma units, as suggested earlier (15, 16).
FIG. 1.
Anti-HBcAg and anti-HBsAg levels in anti-HBcAg-positive plasma. HBV DNA-positive samples are inside diamonds.
HBV DNA was detected in a low number of anti-HBcAg-positive samples, in agreement with previous reports (4, 16, 20). Viral DNA was detected in 8 of 167 (4.8%) plasma samples by the low-volume extraction procedure. Two additional positive plasma samples (10 of 167 [6%]) were detected by a procedure which allowed the analysis of a higher volume of plasma and reduced the risk of false-negative results by inhibition of PCR (8). The difference in the results obtained with the two extraction procedures stresses the importance of an adequate PCR procedure to detect potentially infective blood and derivatives. HBV DNA has been more frequently found among anti-HBcAg-positive specimens (5, 21). These differences may be due to the analysis of anti-HBcAg-selected sera with high levels of antibody and/or to use of a small sample size. Sample volume tested in PCR is even more critical when testing viremia in plasma units because of the high volume of blood and derivatives usually administered to the patient (12). A good correlation, however, has been found previously between PCR determination and infectivity testing in chimpanzees (18).
No HBV DNA-positive sample was found among the 94 of 167 plasma samples with low levels of anti-HBcAg antibodies. A significantly higher prevalence of HBV DNA was found among the samples with anti-HBcAg antibody titers equal to or higher than 1/10 (10 of 73 [14%] (Table 1). Viremia was also associated with anti-HBsAg antibody levels lower than 30 IU/liter. Low anti-HBsAg levels were also present in viremic samples from blood donors associated with posttransfusion hepatitis in a previous study (12). The difference in frequency of HBV DNA positivity was, however, of borderline significance among plasma samples with low and high levels of anti-HBsAg antibodies (Table 1). Applying a quantitative criterion for exclusion of anti-HBcAg-positive plasma and discarding samples with anti-HBcAg antibody titers equal to or higher than 1/10 would result in the exclusion of only 73 of 167 plasma samples (44%). In contrast, discarding plasma samples with anti-HBsAg levels lower than 30 IU/liter would result in the exclusion of 122 of 167 plasma samples (73%). The apparent bimodal distribution of anti-HBcAg-positive plasma suggests that it may be relatively easy to incorporate an anti-HBcAg dilution, or a higher cutoff for the percent inhibition in the anti-HBcAg inhibition assays, depending on the serological test used, that could discriminate between these two populations. Only 1 of the 10 HBV DNA-positive samples was positive for the presence of IgM anti-HBcAg antibodies, and this sample was positive for anti-HBcAg antibodies only at a 1/10 dilution (Fig. 1). Although IgM antibodies are not frequent among anti-HBcAg-positive blood donor plasma samples (12a), additional studies are needed to evaluate the possible usefulness of detecting this acute-phase marker among anti-HBcAg-positive samples in blood donors, to discard possible donations from patients during the window of infection. In Venezuela, about 4% of blood donors are positive for anti-HBcAg and negative for HBsAg, and this marker accounts for at least 50% of the total units discarded (14a). Upon confirmation in a larger sample size, these preliminary results suggest that a screening based upon anti-HBcAg level detection might then result in up to 2.2% more units available for transfusion in Venezuela. On the other hand, a more rational exclusion of anti-HBcAg-positive samples might enable additional countries to incorporate this testing.
TABLE 1.
Viremia in plasma samples positive for anti-HBcAg antibodies and negative for HBsAg according to the levels of anti-HBcAg and anti-HBsAg antibodiesa
| Anti-HBc titer | No. positive/no. tested (%) for sample group:
|
||
|---|---|---|---|
| 0–30 IU of anti HBsAg/liter (n = 122) | >30 IU of anti-HBsAg/liter (n = 45) | All (n = 167) | |
| 1/1 (n = 94) | 0/64 | 0/30 | 0/94 (0) |
| ≥1/10 (n = 73) | 10/58 | 0/15 | 10/73 (14) |
| All (n = 167) | 10/122 (8) | 0/45 (0) | 10/167 (6) |
Statistical differences were evaluated by the Fisher exact test according to the Epi Info program, version 5.01b (Centers for Disease Control and Prevention, Atlanta, Ga.). Significance of column 1 compared with column 2 is P = 0.064; significance of row 1 compared with row 2 is P = 0.002.
Acknowledgments
Grant S1-98002701 from CONICIT, Venezuela, supported this work.
We are grateful to Jose Gregorio Quijada for his comments and suggestions. We thank Firelei Sirit from Quimbiotec, Venezuela, for providing some of the anti-HBcAg-positive plasma samples; Roche Diagnostics GmbH Molecular Systems R&D, Tutzing, Germany, for providing the kits for anti-HBsAg testing; and Emilia Ortegano from Instituto Nacional de Higiene, Venezuela, for IgM anti-HBcAg testing.
REFERENCES
- 1.Blitz L, Pujol F H, Swenson P D, Porto L, Atencio R, Araujo M, Costa L, Callejas D, Torres J, Fields H A, Lambert S, Van Geyt C, Norder H, Magnius L O, Echeverría J M, Stuyver L. Antigenic diversity of hepatitis B virus strains of genotype F in Amerindians and other population groups from Venezuela. J Clin Microbiol. 1998;36:1–4. doi: 10.1128/jcm.36.3.648-651.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Carman W F, Zanetti A R, Karayiannis P, Waters J, Manzillo G, Tanzi E, Zuckerman A J, Thomas H C. Vaccine-induced escape mutant of hepatitis B virus. Lancet. 1990;336:325–329. doi: 10.1016/0140-6736(90)91874-a. [DOI] [PubMed] [Google Scholar]
- 3.Carman W F, Korula J, Wallace L, MacPhee R, Mimms L, Decker R. Fulminant reactivation of hepatitis B due to envelope protein mutant that escaped detection by monoclonal HBsAg ELISA. Lancet. 1995;345:1406–1407. doi: 10.1016/s0140-6736(95)92599-6. [DOI] [PubMed] [Google Scholar]
- 4.Douglas D D, Taswell H F, Rakela J, Rabe D. Absence of hepatitis B virus DNA detected by polymerase chain reaction in blood donors who are hepatitis B surface antigen negative and antibody to hepatitis B core antigen positive from a United States population with a low prevalence of hepatitis B serologic markers. Transfusion. 1992;33:212–216. doi: 10.1046/j.1537-2995.1993.33393174446.x. [DOI] [PubMed] [Google Scholar]
- 5.Gomes S A, Yoshida C F T, Niel C. Detection of hepatitis B virus DNA in hepatitis B surface antigen-negative serum by polymerase chain reaction: evaluation of different primer pairs and conditions. Acta Virol. 1996;40:133–138. [PubMed] [Google Scholar]
- 6.Hoofnagle J H, Seeff L B, Bales Z B, Zimmerman H J. Type B hepatitis after transfusion with blood containing antibody to hepatitis B core antigen. N Engl J Med. 1978;298:1379–1383. doi: 10.1056/NEJM197806222982502. [DOI] [PubMed] [Google Scholar]
- 7.Kaneko S, Feinstone S M, Miller R H. Rapid and sensitive method for the detection of serum hepatitis B virus using the polymerase chain reaction technique. J Clin Microbiol. 1989;27:1930–1933. doi: 10.1128/jcm.27.9.1930-1933.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Klein A, Barsuk R, Shlomo D, Nusbaum O, Shouval D, Galun E. Comparison of methods for extraction of nucleic acid from hemolytic serum for PCR amplification of hepatitis B virus DNA sequences. J Clin Microbiol. 1997;35:1897–1899. doi: 10.1128/jcm.35.7.1897-1899.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lisuka H, Ohmura K, Ishijima A, Satoh K, Tanaka T, Okamoto H, Miyakawa Y, Mayumi M. Correlation between anti-HBc titers and HBV DNA in blood units without detectable HBsAg. Vox Sang. 1992;63:107–111. doi: 10.1111/j.1423-0410.1992.tb02495.x. [DOI] [PubMed] [Google Scholar]
- 10.Magnius L O, Norder H. Subtypes, genotypes and molecular epidemiology of the hepatitis B virus as reflected by sequence variability of the S-gene. Intervirology. 1995;38:24–34. doi: 10.1159/000150411. [DOI] [PubMed] [Google Scholar]
- 11.Mason A L, Xu L, Guo L, Kuhns M, Perrillo R P. Molecular basis for persitent hepatitis B virus infection in the liver after clearance of serum hepatitis B surface antigen. Hepatology. 1998;27:1736–1742. doi: 10.1002/hep.510270638. [DOI] [PubMed] [Google Scholar]
- 12.Mosley J W, Stevens C E, Aach R D, Hollinger F B, Mimms L T, Solomon L R, Barbosa L H, Nemo G J. Donor screening for antibody to hepatitis B core antigen and hepatitis B virus infection in transfusion recipients. Transfusion. 1995;35:5–12. doi: 10.1046/j.1537-2995.1995.35195090661.x. [DOI] [PubMed] [Google Scholar]
- 12a.Pujol, F. H. Unpublished results.
- 13.Pujol F H, Rodriguez I, Devesa M, Rangel-Aldeo R, Liprandi F. A double sandwich monoclonal enzyme immunoassay for detection of hepatitis B surface antigen. J Immunoass. 1993;14:21–31. doi: 10.1080/15321819308019838. [DOI] [PubMed] [Google Scholar]
- 14.Pujol F H, Bertolotti A, Fields H A, Khudyakov Y E, Kalinina T H, Liprandi F. A monoclonal inhibition enzyme immunoassay for detection of antibodies against hepatitis B core antigen: confirmation of an immunodominant epitope. J Immunoass. 1994;15:239–249. doi: 10.1080/15321819408009575. [DOI] [PubMed] [Google Scholar]
- 14a.Salazar, M. Personal communication.
- 15.Schifman R B, Rivers S L, Sampliner R E, Krammes J E. Significance of isolated hepatitis B core antibody in blood donors. Arch Intern Med. 1993;153:2261–2266. [PubMed] [Google Scholar]
- 16.Silva A E, McMahon B J, Parkinson A J, Sjogren M H, Hoofnagle J H, Di Bisceglie A M. Hepatitis B virus DNA in persons with isolated antibody to hepatitis B core antigen who subsequently received hepatitis B vaccine. Clin Infect Dis. 1998;26:895–897. doi: 10.1086/513918. [DOI] [PubMed] [Google Scholar]
- 17.Uemoto S, Sugiyama K, Marusawa H, Inomata Y, Asonuma K, Egawa H, Kiuchi T, Miyake Y, Tanaka K, Chiba T. Transmission of hepatitis B virus from hepatitis B core antibody-positive donors in living related liver transplants. Transplantation. 1998;65:494–499. doi: 10.1097/00007890-199802270-00007. [DOI] [PubMed] [Google Scholar]
- 18.Ulrich P P, Bhat R A, Seto B, Mack D, Sninsky J, Vyas G N. Enzymatic amplification of hepatitis B virus DNA in serum compared with infectivity testing in chimpanzees. J Infect Dis. 1989;160:37–43. doi: 10.1093/infdis/160.1.37. [DOI] [PubMed] [Google Scholar]
- 19.Vasconcelos H C, Yoshida C F, Vanderborght B O, Schatzmayr H G. Hepatitis B and C prevalences among blood donors in the south region of Brazil. Mem Inst Oswaldo Cruz. 1994;89:503–507. doi: 10.1590/s0074-02761994000400002. [DOI] [PubMed] [Google Scholar]
- 20.Wang J-T, Wang T-H, Sheu L-N, Lin J-T, Chen D-S. Detection of hepatitis B virus DNA by polymerase chain reaction in plasma of volunteer blood donors negative for hepatitis B surface antigen. J Infect Dis. 1991;163:397–399. doi: 10.1093/infdis/163.2.397. [DOI] [PubMed] [Google Scholar]
- 21.Zabaleta M E, Toro F L, Ruiz M E, Colmenares C J, Bianco N E, Machado I V. Assessment of former and newly developed HBV assays in a third world setting. J Med Virol. 1992;38:240–245. doi: 10.1002/jmv.1890380403. [DOI] [PubMed] [Google Scholar]