Abstract
Background:
The tumor suppressor genes play a critical role in cellular and molecular mechanisms such as cell cycle processes, cell differentiation and apoptosis. Aberrant DNA methylation of tumor suppressor genes and subsequent gene expression changes have shown to be involved in the initiation and progression of various malignancies including thyroid malignancies. In this review, we investigated what is known about the impact of promoter hypermethylation on the key tumor suppressor genes known to be involved in cell growth and/or apoptosis of thyroid cancer.
Methods:
The most important databases were searched for research articles until June 2020 to identify reported tumor suppressor genes that are modulated by methylation modulation changes in thyroid carcinoma. Following the inclusion and exclusion criteria, 26 studies were reviewed using the full text to meet the inclusion and exclusion criteria.
Results:
The tumor suppressor genes reviewed here are suggestive biomarkers and potential targetable drugs. Inactivation of RASSF1A, DAPK1, SLCFA8, and TSHR through aberrant epigenetic methylation could activate BRAF/MEK/ERK kinase pathways with potential clinical implications in thyroid cancer patients. RARβ2 and RUNX3 could suppress cell cycle and induce apoptosis in malignant cells. TIMP3 and PTEN could prevent angiogenesis and invasion through PIP3 pathway and arrest VEFG activity.
Conclusion:
The methylation status of key genes in various types of thyroid malignancies could be used in early diagnosis as well as differentiation of malignant and benign thyroid. This is valuable in drug repurposing and discovering alternative treatments or preventions in thyroid cancer.
Keywords: DNA methylation, Tumor suppressor gene, Epigenetic, Thyroid cancer, Neoplasm
Introduction
Thyroid cancer has known as a common malignancy among endocrine neoplasms. It is estimated to reach the fourth common cancer by 2030 (1). There are four main types of thyroid cancer including Papillary Thyroid Carcinoma (PTC), Follicular Thyroid Carcinoma (FTC), Medullary Thyroid Carcinoma (MTC) and Anaplastic Thyroid Carcinoma (ATC). The differentiated thyroid carcinomas that are papillary and follicular types consist more than 90% of all types of thyroid carcinoma. Both MTC and ATC are account for 5%-3% and less than 3% of the thyroid carcinoma, respectively (2–4).
Most thyroid cancer cases have a desirable prognosis and are successfully treated with surgery and radio-iodinated treatment (5). Early cancer diagnosis before metastasis is crucial because it can improve prognosis, patient’s quality of life and provide more therapeutical options. Biological biomarkers, such as DNA, mRNA, proteins and metabolites can be used for early cancer diagnosis. These biomarkers may be presented or produced by the tumor itself and/or other tissues found in all kinds of body fluids, tissues, or cell lines (6). These biomarkers can be categorized as active subgroups of proteins (i.e. enzymes, glycoproteins, and receptors), genetic markers (mutations, amplifications, or translocations) and epigenetic markers (such as tumor suppressor gene hyper-methylation). The biomarkers are been utilized in diagnostic and screenings areas of the clinic such as estimating tumor volume, evaluating the response to therapy, assessing recurrence and prognostic factors in illness progression (3).
DNA methylation, as one of the main epigenetic mechanisms, plays an important role in pathological conditions such as tumorigenesis including thyroid tumor development. A methyl (the chemical group here) group is added to the regions that are rich in CpG dinucleotides (CpG islands) in the’ 5’-flanking regions of the promoter of a given gene. In particular, methylation occurrence within the promoter of the tumor suppressor genes leads to decreased gene expression (also known as gene silencing). Consequently, the decreased levels of expression result in modified function of those tumor suppressor genes. This changed function is loss of control of cell cycle process and consequently cell growth (7, 8). Silencing of tumor suppressors is vital in initiating and progressing cancer. Increased levels of methylation (hypermethylation) in promoter tumor suppressors as an important hallmark of development and progression of different human malignancies particularly thyroid carcinogenesis (9). It occurs at early stages of tumor development leading to transcriptionally inactive tumor suppressor genes (7).
Inactivation of cancer-related genes may offer a selective benefit to cells subjected to transformation or progressing to a more malignant pheno-type, and therefore, may significantly influence the tumor promotion and progression in carcinomas. Thus, promoter hypermethylation has been widely assessed in biomarker utilization to early diagnosis and detection of cancers that is been useful as a prognostic indicator in malignancies (10).
Here, a comprehensive overview of the key genes, known as tumor suppressor genes involved in thyroid tumorigenesis, is provided. The mechanisms of actions of promoter hypermethylation in preventing cell growth or apoptosis induction in thyroid malignancies have been discussed.
Methods
Research strategy
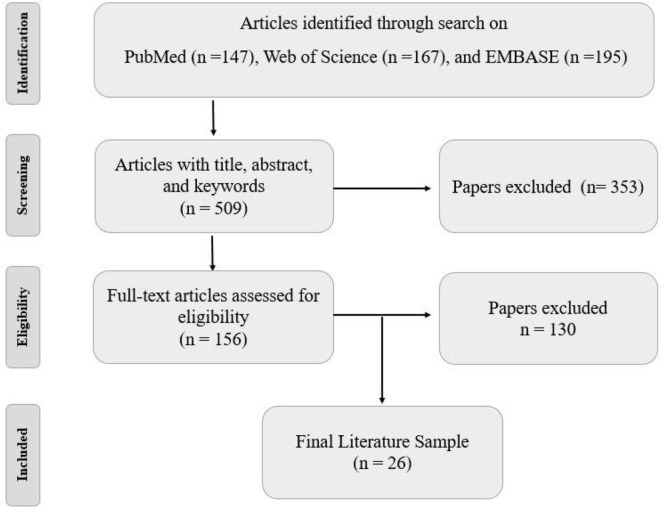
PubMed, EMBASE, and Web of Science databases were searched for research articles until June 2020 to identify reported tumor suppressor genes that are modulated by methylation modulation changes in thyroid carcinoma. In these datasets, we looked for the following keywords: “thyroid” and “cancer or tumor or neoplasm or carcinoma”, and “suppressor or anti-oncogene” and “gene” and “methylation”. One hundred and thirteen, 195, and 145 studies were selected from PubMed, EMBASE, and Web of Science according to their titles and abstracts. The references of the selected articles were manually reviewed to avoid missing any additional studies (Fig. 1).
Fig. 1:
The workflow that was used in the present review
Final literatures were selected based on inclusion and exclusion criteria. As a final point, twenty-six articles were choose for this review
Selection criteria
The characteristic criteria, followed in this review, were 1) original research article 2) studies that reported correlations with known tumor suppressor genes, hyper-methylation, clinic-pathological parameters and prognosis of thyroid cancer, 3) tumor suppressor genes that represent hypermethylation within their promoter regions that were assessed with strong evidence. To limit this review to higher-quality studies we excluded letters, case report studies, expert opinion, conference abstracts, editorials, ex-vivo studies, human xenografts, and studies with no report on tumor suppressor hypermethylation. Moreover, we filtered out the studies with similar samples and overlapped information.
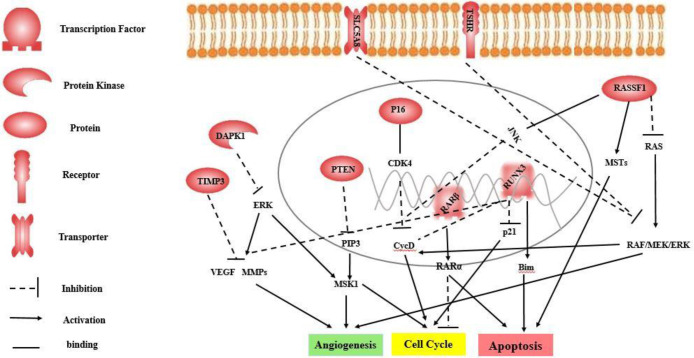
Following the above-mentioned criteria, 26 studies were reviewed using the full text to meet the inclusion and exclusion criteria. Review articles, in-vitro studies, Kyoto Encyclopedia of Gene and Genomes (KEGG) and pathway studio website were used to gain more information explaining the probable tumor suppressors pathway in preventing thyroid cancer (Fig. 2) (11, 12).
Fig. 2:
The effect of tumor suppressor genes during suppression of pathways involved in thyroid cancer tumorigenesis and progress (Source: The authors of this paper).
SLC5A8 and TSHR inhibit the RAF/MEK/ERK pathway, involved cell cycle through cyclin D1 and angiogenesis.
RASSF1 acts through three ways. First, inhibiting the RAS oncogene and consequently preventing RAF/MEK/ERK pathway, cell cycle and angiogenesis. Second, motivating pro-apoptotic kinases (MSTs) and then inducing apoptosis. Third, activating c-Jun N-terminal kinase (JNK) pathway and then stopping cell cycle through cyclin D1.
RUNX3 interrupts Wnt/β-catenin signaling pathway, involved in cell proliferation and invasion. It also forms the complex with TCF4-b-catenin complex and consequently inhibiting the cyclinD1 activity. RUNX3 cooperates to activating the Transforming growth factor-β (TGF-β) pathway by induction of p21 and Bim and regulates apoptosis. RARβ forms complex with RARα and induced apoptosis and inhibit cell cycle.
P16 forms complex with Cyclin-dependent kinase 4 (CDK4) and then inhibits cyclin D1, prevents cell growth and stop cell cycle.
PTEN interrupts in the phosphoinositol 3-kinase (PI3K) AKT pathway through effects on common genetic alterations in this pathway, which plays an important role in tumorigenesis of thyroid cancer.
DAPK1 inhibit the RAF/MEK/ERK pathway, involved cell cycle and angiogenesis through Mitogen- and stress-activated kinase 1 (MSK1). In addition,
TIMP3 inhibits angiogenesis through effects on Vascular endothelial growth factor (VEGF) signaling and matrix metallopeptidase 9 (MMPs)
Results and Discussion
Tumor suppressor genes
Cycling-dependent kinase inhibitor 2A (P16); or multiple tumor suppressor 1 and or p16INK4a, belongs to cyclin-dependent kinases (CDK) and plays a vital inhibitory role in regulating cell cycle by slowing down the cell progression during the G1 to S phase. Nowadays, P16 possess tumor suppressor activity by regulating cell proliferation thus contributing to the pathogenesis (13–15). P16 promoter methylation and consequently gene silencing causes a lack of protein expression and therefore impacts on its function leading to increased risk of cancer. The p16 gene inactivation is been detected in colon, lung, brain, esophagus, stomach, pancreas, and lymphomas cancers (16). Aberrant hypermethylation of P16 promoter may affect the loss of function causing the progression in thyroid carcinoma cell lines (17). The hypermethylation of P16 promoter has been reported in 44% and 50% of PTC and FTC cases, respectively, compared to 13% of non-tumorous thyroid tissues (18). Another study showed significant differences between PTC tissues vs. adjacent normal thyroid tissues (24% vs. 0%) (14). Lam et al. revealed silencing of P16 promoter in 41% PTC tissues while none of the nodular was negative for p16 hypermethylation (19).
Promoter hypermethylation was significantly associated with lymph node metastasis and extra thyroid invasion (18). A remarkable frequency of aberrant promoter methylation was detected in aggressive undifferentiated thyroid carcinoma, invasive PTC with lymph node and distant metastasis (10, 14, 19). P16 is involved in the transformation of differentiated tumor to undifferentiated phenotype (anaplastic) (18). However, the methylation status of P16 was not associated with clinicopathological characteristics in PTC cases (20). An exvivo study confirmed that silencing of P16 occurred in C643 and Hth74 cell lines result in a remarkable frequency of aberrant promoter methylation was detected in aggressive undifferentiated thyroid carcinoma. This data revealed that 100% promoter methylation of P16 occurred in undifferentiated thyroid cancer cell lines (10). A meta-analysis confirmed that p16 gene promoter methylation is vital for the PTC risk (21). Promoter methylation of P16 plays a major role in the process of neoplastic progression and tumor dedifferentiation in thyroids. Notably, the methylation status of P16 promoter was shown to be a common and most likely an early event during this process.
Ras Association domain Family members 1 A (RASSF1A); is an isoform of Ras Association Domain Family that is located on chromosome 3p21.3 and associated with preserving genomic stability, stabilization of microtubules. Based on KEEG pathway analysis, shown by several studies (ref), RASSF1A is contributed to cell cycle, proliferation and apoptosis (22). The RASSF1A gene silencing has been shown to occur via promoter hypermethylation and has been linked to the pathogenesis of different human cancer types. These events suggest the tumor suppressor effect of this gene might be presented via cell proliferation and suppress apoptosis process (3, 13). In particular, hypermethylation of RASSF1A occurs in 75% of FTC cases, 20% of PTC cases, 44% of benign adenoma, and 0% of normal tissues (23). The aberrant promoter methylation of RASSF1 has been identified in FTC and PTC (13, 23–25). Even though the frequency of promoter methylation of RASSF1A in malignant tissues was less than benign tissues (76% vs. 93%), the mean of methylation in malignant tissues was higher than benign tissues (28.3 vs. 11.4) (20). This finding is in line with an in-vitro study that revealed RASSF1A hypermethylation in nine PTC, ATC, and FTC-related cell lines (24). BRAF mutation and RASSF1A deactivation were related to each other inversely. In other words, RASSF1A deactivation occurred in PTC patients who have BRAF mutation (26).
RASSF1A promoter methylation is strongly associated with thyroid cancer risk and pathogenesis of PTC, however, there is no correlation between the risk and stage of the disease or distant metastasis (13, 27). Therefore, inactivation of RASSF1A promoter was not shown to be correlated with the severity of thyroid malignancies. In addition, RASSF1A inactivation occurs at an early stage of thyroid tumorigenesis. Based on KEGG pathway by previous studies, RASSF1A causes apoptosis by regulating pro-apoptotic kinases (MSTs) (22). Similar to P16, RASSF1A plays a vital role in the cell cycle at the G1-S checkpoint by connecting to c-Jun N-terminal kinase (JNK) pathway and modulating the levels of cyclin D1 (22). RASSF1A inhibits the RAS proto-oncogene activity, involved in cell cycle progression and angiogenesis via RAF-MEK-ERK pathway (28).
In 2009, the epigenetic inactivation of RASSF10, a novel RASSF member, was identified as a new target for the pathogenesis of thyroid cancer (in 66% of the primary tumor in thyroid tissues) (29). RASSF2, a third family member of RASSFs, was inactivated in 63% of primary thyroid tumor (ref). RASSF2 promoter hypermethylation was associated with a decreased expression of?. This study used a DNA methylation inhibitor to reactivate RASSF2 transcription. RASSF2 could induce apoptosis in thyroid cancer cells (30). However, more studies are needed to accept these RASSF members which act as tumor suppressor genes.
Solute Carrier Family 5 Member 8 (SLC5A8); is a member of the sodium solute symporter family, and transports iodide via a passive mechanism (elaborate passive mechanism?). The family members are expressed in various tissues such as thyroid gland and could play both roles as a transporter and a symporter (7, 8). They are also played as sodium-independent facilitative transferring iodide via the basal membrane of thyroid follicular cells (31). SLC5A8 is frequently methylated in various types of cancer (32–34). Aberrant methylation of the SLC5A8 promoter was assessed in various populations and sample sizes. Significant hyper-methylation was observed in 76/231 (33%) by Hu et al. (7), 36/138 (26.1%) by Kiseljak-Vassiliades et al. (35), 34/86 (39.5%) by Zane et al. (36), and. In addition, promoter silencing of SLC5A8 was detected in circular DNA in PTC patients by high-resolution melting analysis by Khatami et al (25). The SLC5A8 methylation status in thyroid gland is shown to be associated with pathological features in thyroid cancer such as extra-thyroidal invasion, multifocality and disease aggressiveness (particularly in PTC) and BRAF mutations (7, 25, 32). The association between down-regulating and promoter hypermethylation of SLC5A8 provides evidence of tumor suppressor role of SLC5A8 gene in thyroid cancer. Similar to SLC5A8, several studies demonstrated aberrant methylation in SLC5A5 gene in PTC cases compared to normal thyroid tissue and hyperthyroid samples (2, 3). No evidence supported the exact pathway, which involved in tumor suppressor activity of SLC5A8. However, because of the association between BRAF and SLC5A8, this gene is probably contributed to modulate MAPK pathway in thyroid cancer especially in PTC.
Retinoic Acid Receptor, Beta2 (RARβ2); is a member of the thyroid-steroid hormone receptor superfamily that encodes retinoic acid receptor beta (a nuclear transcriptional regulator). This receptor is involved in epithelial cell tumorigenesis by binding to the active form of vitamin A, retinoic acid and ERK signaling pathway. RARβ2 gene methylation is seen frequently in human cancers (7). RARβ2 promoter was remarkably hypermethylated in thyroid cancers compared with benign thyroid tumors representing a positive correlation with silencing of RARβ2 and BRAF mutations (26). However, the frequency of RARβ2 methylation occurrence was less than another tumor suppressor such as TIMP3, SLC5A8 and DAPK1 genes. RARβ2 gene promoter methylation was assessed in 50/231 (22%) (7), 23/138 (16.7%) of PTC cases (35). Even though RARβ2 gene silencing was associated with thyroid tumorigenesis, there are a handful of studies that showed no significant difference in RARβ2 methylation frequencies between thyroid cancer and non-malignant cases (20, 37). This suggests that RARβ2 inactivation is associated with BRAF mutation (7, 10, 20). Overall, RARβ2 might act as a tumor suppressor gene. However, RARβ and RARα prevented cell growth and induced apoptosis (38) but this remains to be confirmes by in-vitro studies.
Death-associated protein kinase 1 (DAPK1); is a calcium calmodulin-regulated serine/threonine-protein kinase. DAPK1 mediates the gamma-interferon-related programmed cell death mechanism (39). DAPK1 exerts gene silencing via hypermethylation in many human cancer types including thyroid cancer (7, 40, 41). Promoter hypermethylation-induced silencing of DAPK1 occurred in 78/231 (34%) (7), 40/138 (29%) (35), and (32%) of PTC cases (2). Aberrant methylation of DAPK1 was significantly higher in PTC cases than in FTC, MTC and goiter cases (10). However, one study revealed no significant differences of the frequencies of DAPK1 methylation among thyroid cancer tissues and normal/benign tissues (37). DAPK1 methylation is significantly linked to tumor multi-focality and BRAF mutations? In aggressive PTC (7). So far, the effect of DAPK1 as a tumor suppressor is exerted by pro-apoptosis which occurs in cell survival and apoptosis-related pathways and prevents cell proliferation by inhibiting the ERK in MAPK signaling pathway.
Tissue inhibitor of metalloproteinase-3 (TIMP3); belongs to TIMP gene family, known as tissue inhibitor of metalloproteinase. This gene acts as a mitogenic stimulator in cell growth, angiogenesis, invasion and metastasis inhibitor in several cancer types (7). This gene presents hyper-methylation in some cancers, such as thyroid cancer, esophageal adenocarcinoma, and uveal melanoma (7, 26, 42–44). Promoter methylation status of TIMP3 was detected in 53% of PTC cases using Real-time quantitative methylation-specific PCR. Notably, in this research, the association between promoter methylations of TIMP3 and BRAF mutations, aggressive pathological PTC features, such as extrathyroidal invasion, lymph node metastasis, and tumor multifocality were also found (7). The frequencies of TIMP3 methylation was 67/138 (48%) in PTC cases (35). Another study assessed the TIMP3 methylation rate 27%, 42% and 51% in normal, benign stages of thyroid malignancies and thyroid cancer, respectively. (37). The significantly positive association of promoter methylation as a result of TIMP3 and BRAF mutations in PTC samples have an important role in PTC development. Some pathways are shown related to TIMP3, such as ERK signaling and Akt signaling. VEGF signaling pathway and Angiogenesis are also two pathways, which TIPM3 is involved. TIMP3 could inhibit matrix metallopeptidase 9 (MMP9). TIMP3 is downregulated as a result of promoter methylation and modulates the MMP9 binding to receptors and consequently, the angiogenesis and vessel information are initiated (45). TIMP3 inhibits vascular endothelial growth factor receptor2 (VEGFR2) in VEGF pathways. Therefore, it can prevent angiogenesis via activation of a kinase cascade that includes RAS and MAPK (46, 47). The studies abovementioned suggest that
TIMP3 may act as a tumor suppressor gene. Thyrotropin Receptor (TSHR); gene encodes a receptor that binds to thyroid-stimulating hormone (TSH). This protein is located on the membrane of thyroid follicular cells. In addition, TSHR plays a crucial role in iodide uptake by sodium/iodide symporter (48). The epigenetic silencing of TSHR gene has been observed in thyroid tumor-igenesis. A significantly higher prevalence of promoter methylation for TSHR has been detected in a higher aggressive ATC compared to other thyroid cancer types (10). Khan et al. found a higher frequency of TSHR methylation in thyroid tumors, and epigenetic silencing was significantly associated with BRAF mutations (49). One study revealed aberrant methylation in TSHR gene in 11 out of 32 (34%) PTC tissues, and 2/27 (7%) control tissues (2). The findings were similar using thyroid tissues samples or fine-needle aspiration cytology samples. The methylation status was significantly higher in malignant cases than in non-malignant cases, particularly in PTC cases (71% vs. 46%) (50). In contrast, TSHR is hypermethylated in 20/44 (45%) thyroid cancer cases, 24/44 (55%) benign adenoma cases and 9/15 (60%) normal thyroid tissues. The TSHR hypermethylation levels were not significant between groups (37). Finally, recent meta-analyses have covered all reports focused on the methylation of TSHR in thyroid cancer. Based on this study, the gene methylation level is higher in PTC cases significantly associated with age, lymph node metastasis, clinical stage and tumor size (51, 52). TSHR potentially suppresses the RAF-MEK-ERK pathway in thyroid cancer (53). Therefore, TSHR gene may be an applicant marker for PTC diagnosis.
Runt-related transcription factor 3 (RUNX3); is expressed into different isoforms. The isoforms regulate the gene expression by an impact on promoters and play an important role as a tumor suppressor gene such as p53, p21, and p27. These target genes are vital in the regulation of epithelial proliferation and apoptosis. Similarly, the core-binding factor (CBF) complex is regulated by RUNX genes. The CBF complex activates several regulators of growth, survival and differentiation pathways. RUNX3 promoter silencing is reported in several cancers such as gastric, colorectal and lung cancer (50, 54). Ko et al. conducted the first study based on an assessment of the methylation level of RUNX3 in thyroid cancer (55). There is one study conducted by Yin et al, in Chinese language not included in this review. Aberrant methylation of RUNX3 promoter was significantly detected in thyroid cancer higher than in noncancerous controls, particularly in PTC subtype (55, 56). Hypermethylation of promoter CpG islands of RUNX3 in thyroid cell lines was confirmed using in-vitro methods (55). Subsequently, the relation between RUNX3 silencing and the risk of tumor recurrence was detected (14). RUNX3 might act as a tumor suppressor by interrupting Wnt/β-catenin signaling pathway, involved in cell proliferation and invasion. RUNX3 acts through forming the complex with TCF4-b-catenin complex and consequently inhibiting the cyclinD1 activity. RUNX3 cooperates in activating the Transforming growth factor- β (TGF-β) pathway by induction of p21 and Bim (57, 58). Notably, these mechanisms must be confirmed in thyroid cancer pathway too.
Even though the epigenetic modifications have been confirmed by several studies in thyroid cancer cases but the relationship between methylation changes and gene expression and clinic-pathological characteristics has not yet been investigated.
Phosphatase and tensin homolog (PTEN); is a tumor suppressor gene located in 10q23.31 chromosomal region and negative regulator of the phosphoinositol 3-kinase (PI3K) AKT pathway in multiple cancers. Common genetic alterations in AKT pathway plays an important role in tumor-igenesis of thyroid cancer (59). Enhanced signaling activity of PI3K/AKT pathway contributes to cell growth, proliferation, differentiation and DNA repair (ref). On the other hand, PTEN inactivation affects cell cycle regulation via decreasing cell cycle inhibitor activity of p27 and p50 (59–62). PTEN inactivation occurs through genetic and epigenetic modifications including deletion, mutation and aberrant promoter methylation in thyroid cancer (59, 62). Interestingly, the prevalence of PTEN methylation is higher than mutations or loss of heterozygosity in thyroid neoplasms (63). Aberrant promoter silencing of PTEN was detected in PTC, FTC and ATC cases associated with the progression of thyroid cancer, however, the promoter hypermethylation was low in cell lines (10, 59, 63). Although the PTEN silencing is shown to be associated with thyroid tumorigenesis it is not associated with aggressive lymph node metastasis and extrathyroid extension (62). More studies on different tumor types are needed to explore whether PTEN can be a candidate marker for thyroid cancer diagnosis.
The recent finding of frequent methylation of PTEN promoter in thyroid tumors explains aberrant silencing of this gene in thyroid cancer, as promoter methylation is an important epigenetic mechanism in silencing of genes. In summary, our data are consistent with an interesting model that signaling of the PI3K/AKT pathway activated by its activating genetic alterations can be enhanced by coexisting aberrant methylation and hence epi-genetic down-regulation of the PTEN gene, which normally antagonizes the signaling of this pathway. Further studies are needed to explore the molecular mechanisms underlying the link between such genetic and epigenetic changes. As we exected, hypermethylation of PTEN promoter was significantly detected in Fine Needle Aspiration (FNA) samples (PTC cases), but it could not improve the specificity of FNA diagnosis (62).
Conclusion
Promoter silencing of certain genes appears to be associated with various stages of thyroid cancer and invasive tumor behavior and clinical implications. The tumor suppressor genes reviewed here are suggestive biomarkers and potential targetable drugs. Inactivation of RASSF1A, DAPK1, SLCFA8, and TSHR through aberrant epigenetic methylation could activate BRAF/MEK/ERK kinase pathways with potential clinical implications in thyroid cancer patients. In particular, MAPK pathway is one the important process in thyroid pathogenesis. RARβ2 and RUNX3 could suppress cell cycle and induce apoptosis in malignant cells. TIMP3 and PTEN could prevent angiogenesis and invasion through PIP3 pathway and arrest VEFG activity.
Aberrant methylation is known as an important epigenetic modification that results in tumor suppressor inactivation in human cancers. The methylation status of key genes in various types of thyroid malignancies could be used in early diagnosis as well as differentiation of malignant and benign thyroid. This is valuable in drug repurposing and discovering alternative treatments or preventions in thyroid cancer. For instance, demethylating of tumor suppressors genes involved in a given pathway may contribute to suppressing that pathway consequently enhancing tumor suppressor activity that might improve the treatment process. Given the importance of inactivation of tumor suppressor genes, more studies are needed to identify and examine new molecules with tumor suppressor activity. To design molecular panels improving diagnosis and treatment of thyroid cancer.
Ethical considerations
Ethical issues (Including plagiarism, informed consent, misconduct, data fabrication and/or falsification, double publication and/or submission, redundancy, etc.) have been completely observed by the authors.
Footnotes
Conflict of interest
The authors declare that there is no conflict of interest.
References
- 1.Rahib L, Smith BD, Aizenberg R, Rosenzweig AB, Fleshman JM, Matrisian LM. (2014). Projecting cancer incidence and deaths to 2030: the unexpected burden of thyroid, liver, and pancreas cancers in the United States. Cancer Res, 74(11):2913–21. [DOI] [PubMed] [Google Scholar]
- 2.Smith JA, Fan CY, Zou C, Bodenner D, Kokoska MS. (2007). Methylation status of genes in papillary thyroid carcinoma. Arch Otolaryngol Head Neck Surg, 133(10):1006–11. [DOI] [PubMed] [Google Scholar]
- 3.Stephen JK, Chitale D, Narra V, Chen KM, Sawhney R, Worsham MJ. (2011). DNA methylation in thyroid tumorigenesis. Cancers (Basel), 3(2):1732–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hedayati M, Nozhat Z, Hannani M. (2016). Can the Serum Level of Myostatin be Considered as an Informative Factor for Cachexia Prevention in Patients with Medullary Thyroid Cancer? Asian Pac J Cancer Prev, 17(S3):119–23. [DOI] [PubMed] [Google Scholar]
- 5.Qiang W, Zhao Y, Yang Q, et al. (2014). ZIC1 is a putative tumor suppressor in thyroid cancer by modulating major signaling pathways and transcription factor FOXO3a. J Clin Endocrinol Metab, 99(7):E1163–72. [DOI] [PubMed] [Google Scholar]
- 6.Stephen JK, Chen KM, Merritt J, Chitale D, Divine G, Worsham MJ. (2018). Methylation markers differentiate thyroid cancer from benign nodules. J Endocrinol Invest, 41(2):163–70. [DOI] [PubMed] [Google Scholar]
- 7.Hu S, Liu D, Tufano RP, et al. (2006). Association of aberrant methylation of tumor suppressor genes with tumor aggressiveness and BRAF mutation in papillary thyroid cancer. Int J Cancer, 119(10):2322–9. [DOI] [PubMed] [Google Scholar]
- 8.Xing M. (2007). Gene methylation in thyroid tumorigenesis. Endocrinology, 148(3):948–53. [DOI] [PubMed] [Google Scholar]
- 9.Lal G, Padmanabha L, Smith BJ, et al. (2006). RIZ1 is epigenetically inactivated by promoter hypermethylation in thyroid carcinoma. Cancer, 107(12):2752–9. [DOI] [PubMed] [Google Scholar]
- 10.Schagdarsurengin U, Gimm O, Dralle H, Hoang-Vu C, Dammann R. (2006). CpG island methylation of tumor-related promoters occurs preferentially in undifferentiated carcinoma. Thyroid, 16(7):633–42. [DOI] [PubMed] [Google Scholar]
- 11.Kanehisa M, Sato Y, Furumichi M, Morishima K, Tanabe M. (2019). New approach for understanding genome variations in KEGG. Nucleic Acids Res, 47(D1):D590–d5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nikitin A, Egorov S, Daraselia N, Mazo I. (2003). Pathway studio--the analysis and navigation of molecular networks. Bioinformatics, 19(16):2155–7. [DOI] [PubMed] [Google Scholar]
- 13.Jiang JL, Tian GL, Chen SJ, Xu LI, Wang HQ. (2015). Promoter methylation of p16 and RASSF1A genes may contribute to the risk of papillary thyroid cancer: A meta-analysis. Exp Ther Med, 10(4):1549–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang P, Pei R, Lu Z, Rao X, Liu B. (2013). Methylation of p16 CpG islands correlated with metastasis and aggressiveness in papillary thyroid carcinoma. J Chin Med Assoc, 76(3):135–9. [DOI] [PubMed] [Google Scholar]
- 15.Ben-Ari Fuchs S, Lieder I, Stelzer G, et al. (2016). GeneAnalytics: An Integrative Gene Set Analysis Tool for Next Generation Sequencing, RNAseq and Microarray Data. Omics, 20(3):139–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kamb A, Gruis NA, Weaver-Feldhaus J, et al. (1994). A cell cycle regulator potentially involved in genesis of many tumor types. Science, 264(5157):436–40. [DOI] [PubMed] [Google Scholar]
- 17.Elisei R, Shiohara M, Koeffler HP, Fagin JA. (1998). Genetic and epigenetic alterations of the cyclin-dependent kinase inhibitors p15INK4b and p16INK4a in human thyroid carcinoma cell lines and primary thyroid carcinomas. Cancer, 83(10):2185–93. [PubMed] [Google Scholar]
- 18.Boltze C, Zack S, Quednow C, Bettge S, Roessner A, Schneider-Stock R. (2003). Hypermethylation of the CDKN2/p16INK4A promotor in thyroid carcinogenesis. Pathol Res Pract, 199(6):399–404. [DOI] [PubMed] [Google Scholar]
- 19.Lam AK, Lo CY, Leung P, Lang BH, Chan WF, Luk JM. (2007). Clinicopathological roles of alterations of tumor suppressor gene p16 in papillary thyroid carcinoma. Ann Surg Oncol, 14(5):1772–9. [DOI] [PubMed] [Google Scholar]
- 20.Mohammadi-asl J, Larijani B, Khorgami Z, et al. (2011). Qualitative and quantitative promoter hypermethylation patterns of the P16, TSHR, RASSF1A and RARβ2 genes in papillary thyroid carcinoma. Med Oncol, 28(4):1123–8. [DOI] [PubMed] [Google Scholar]
- 21.Wu W, Yang SF, Liu FF, Zhang JH. (2015). Association between p16 Promoter Methylation and Thyroid Cancer Risk: A Meta-analysis. Asian Pac J Cancer Prev, 16(16):7111–5. [DOI] [PubMed] [Google Scholar]
- 22.Donninger H, Vos MD, Clark GJ. (2007). The RASSF1A tumor suppressor. J Cell Sci, 120(Pt 18):3163–72. [DOI] [PubMed] [Google Scholar]
- 23.Xing M, Cohen Y, Mambo E, et al. (2004). Early occurrence of RASSF1A hypermethylation and its mutual exclusion with BRAF mutation in thyroid tumorigenesis. Cancer Res, 64(5):1664–8. [DOI] [PubMed] [Google Scholar]
- 24.Schagdarsurengin U, Gimm O, Hoang-Vu C, Dralle H, Pfeifer GP, Dammann R. (2002). Frequent epigenetic silencing of the CpG island promoter of RASSF1A in thyroid carcinoma. Cancer Res, 62(13):3698–701. [PubMed] [Google Scholar]
- 25.Khatami F, Larijani B, Heshmat R, Nasiri S, Haddadi-Aghdam M, Teimoori-Toolabi L. (2020). Hypermethylated RASSF1 and SLC5A8 promoters alongside BRAF(V600E) mutation as biomarkers for papillary thyroid carcinoma, J Cell Physiol, 235(10):6954–6968. [DOI] [PubMed] [Google Scholar]
- 26.Hoque MO, Rosenbaum E, Westra WH, et al. (2005). Quantitative assessment of promoter methylation profiles in thyroid neoplasms. J Clin Endocrinol Metab, 90(7):4011–8. [DOI] [PubMed] [Google Scholar]
- 27.Shou F, Xu F, Li G, et al. (2017). RASSF1A promoter methylation is associated with increased risk of thyroid cancer: a meta-analysis. Onco Targets Ther, 10:247–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Saavedra HI, Knauf JA, Shirokawa JM, et al. (2000). The RAS oncogene induces genomic instability in thyroid PCCL3 cells via the MAPK pathway. Oncogene, 19(34):3948–54. [DOI] [PubMed] [Google Scholar]
- 29.Schagdarsurengin U, Richter AM, Wohler C, Dammann RH. (2009). Frequent epigenetic inactivation of RASSF10 in thyroid cancer. Epigenetics, 4(8):571–6. [DOI] [PubMed] [Google Scholar]
- 30.Schagdarsurengin U, Richter AM, Hornung J, Lange C, Steinmann K, Dammann RH. (2010). Frequent epigenetic inactivation of RASSF2 in thyroid cancer and functional consequences. Mol Cancer, 9:264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ganapathy V, Gopal E, Miyauchi S, Prasad PD. (2005). Biological functions of SLC5A8, a candidate tumour suppressor. Biochem Soc Trans, 33(Pt 1):237–40. [DOI] [PubMed] [Google Scholar]
- 32.Li H, Myeroff L, Smiraglia D, et al. (003). SLC5A8, a sodium transporter, is a tumor suppressor gene silenced by methylation in human colon aberrant crypt foci and cancers. Proc Natl Acad Sci U S A, 100(14):8412–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Park JY, Kim D, Yang M, et al. (2013). Gene silencing of SLC5A8 identified by genome-wide methylation profiling in lung cancer. Lung Cancer, 79(3):198–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Babu E, Ramachandran S, Coothan Kandaswamy V, et al. (2011). Role of SLC5A8, a plasma membrane transporter and a tumor suppressor, in the antitumor activity of dichloroacetate. Oncogene, 30(38):4026–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kiseljak-Vassiliades K, Xing M. (2011). Association of Cigarette Smoking with Aberrant Methylation of the Tumor Suppressor Gene RARbeta2 in Papillary Thyroid Cancer. Front Endocrinol (Lausanne), 2:99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zane M, Agostini M, Enzo MV, et al. (2013). Circulating cell-free DNA, SLC5A8 and SLC26A4 hypermethylation, BRAF(V600E): A non-invasive tool panel for early detection of thyroid cancer. Biomed Pharmacother, 67(8):723–30. [DOI] [PubMed] [Google Scholar]
- 37.Brait M, Loyo M, Rosenbaum E, et al. (2012). Correlation between BRAF mutation and promoter methylation of TIMP3, RARbeta2 and RASSF1A in thyroid cancer. Epigenetics, 7(7):710–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Connolly RM, Nguyen NK, Sukumar S. (2013). Molecular pathways: current role and future directions of the retinoic acid pathway in cancer prevention and treatment. Clin Cancer Res, 19(7):1651–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kanehisa M, Furumichi M, Tanabe M, Sato Y, Morishima K. (2017). KEGG: new perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res, 45(D1):D353–d61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Qi JH, Ebrahem Q, Moore N, et al. (2003). A novel function for tissue inhibitor of metalloproteinases-3 (TIMP3): inhibition of angiogenesis by blockage of VEGF binding to VEGF receptor-2. Nat Med, 9(4):407–15. [DOI] [PubMed] [Google Scholar]
- 41.Schneider-Stock R, Roessner A, Ullrich O. (2005). DAP-kinase--protector or enemy in apoptotic cell death. Int J Biochem Cell Biol, 37(9):1763–7. [DOI] [PubMed] [Google Scholar]
- 42.Brueckl WM, Grombach J, Wein A, et al. (2005). Alterations in the tissue inhibitor of metalloproteinase-3 (TIMP-3) are found frequently in human colorectal tumours displaying either microsatellite stability (MSS) or instability (MSI). Cancer Lett, 223(1):137–42. [DOI] [PubMed] [Google Scholar]
- 43.Darnton SJ, Hardie LJ, Muc RS, Wild CP, Casson AG. (2005). Tissue inhibitor of metalloproteinase-3 (TIMP-3) gene is methylated in the development of esophageal adenocarcinoma: loss of expression correlates with poor prognosis. Int J Cancer, 115(3):351–8. [DOI] [PubMed] [Google Scholar]
- 44.van der Velden PA, Zuidervaart W, Hurks MH, et al. (2003). Expression profiling reveals that methylation of TIMP3 is involved in uveal melanoma development. Int J Cancer, 106(4):472–9. [DOI] [PubMed] [Google Scholar]
- 45.Slenter DN, Kutmon M, Hanspers K, et al. (2018). WikiPathways: a multifaceted pathway database bridging metabolomics to other omics research. Nucleic Acids Res, 46(D1):D661–d7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Maitland ML, Lou XJ, Ramirez J, et al. (2010). Vascular endothelial growth factor pathway. Pharmacogenet Genomics, 20(5):346–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Whirl-Carrillo M, McDonagh EM, Hebert JM, et al. (2012). Pharmacogenomics knowledge for personalized medicine. Clin Pharmacol Ther, 92(4):414–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Szkudlinski MW, Fremont V, Ronin C, Weintraub BD. (2002). Thyroid-stimulating hormone and thyroid-stimulating hormone receptor structure-function relationships. Physiol Rev, 82(2):473–502. [DOI] [PubMed] [Google Scholar]
- 49.Khan MS, Pandith AA, Masoodi SR, Wani KA, Ul Hussain M, Mudassar S. (2014). Epigenetic silencing of TSHR gene in thyroid cancer patients in relation to their BRAF V600E mutation status. Endocrine, 47(2):449–55. [DOI] [PubMed] [Google Scholar]
- 50.Kartal K, Onder S, Kosemehmetoglu K, Kilickap S, Tezel YG, Kaynaroglu V. (2015). Methylation status of TSHr in well-differentiated thyroid cancer by using cytologic material. BMC Cancer, 15:824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Qu M, Wan S, Ren B, Wu H, Liu L, Shen H. (2020). Association between TSHR gene methylation and papillary thyroid cancer: a meta-analysis. Endocrine, 69(3):508–515. [DOI] [PubMed] [Google Scholar]
- 52.Khatami F, Larijani B, Heshmat R, et al. (2017). Meta-analysis of promoter methylation in eight tumor-suppressor genes and its association with the risk of thyroid cancer. PLoS One, 12(9):e0184892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Liu D, Hu S, Hou P, Jiang D, Condouris S, Xing M. (2007). Suppression of BRAF/MEK/MAP kinase pathway restores expression of iodidemetabolizing genes in thyroid cells expressing the V600E BRAF mutant. Clin Cancer Res, 13(4):1341–9. [DOI] [PubMed] [Google Scholar]
- 54.Joung KH, Shong M. (2012). Epigenetic regulation of RUNX3 in thyroid carcinoma. Korean J Intern Med, 27(4):391–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ko HJ, Kim BY, Jung CH, et al. (2012). DNA Methylation of RUNX3 in Papillary Thyroid Cancer. Korean J Intern Med, 27(4):407–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Botezatu A, Iancu IV, Plesa A, et al. (2019). Methylation of tumour suppressor genes associated with thyroid cancer. Cancer Biomark, 25(1):53–65. [DOI] [PubMed] [Google Scholar]
- 57.Chuang LS, Ito Y. (2010). RUNX3 is multifunctional in carcinogenesis of multiple solid tumors. Oncogene, 29(18):2605–15. [DOI] [PubMed] [Google Scholar]
- 58.Chen F, Liu X, Bai J, Pei D, Zheng J. (2016). The emerging role of RUNX3 in cancer metastasis (Review). Oncol Rep, 35(3):1227–36. [DOI] [PubMed] [Google Scholar]
- 59.Hou P, Ji M, Xing M. (2008). Association of PTEN gene methylation with genetic alterations in the phosphatidylinositol 3-kinase/AKT signaling pathway in thyroid tumors. Cancer, 113(9):2440–7. [DOI] [PubMed] [Google Scholar]
- 60.Lu YM, Cheng F, Teng LS. (2016). The association between phosphatase and tensin homolog hypermethylation and patients with breast cancer, a meta-analysis and literature review. Sci Rep, 6:32723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Georgescu MM. (2010). PTEN Tumor Suppressor Network in PI3K-Akt Pathway Control. Genes & cancer, 1(12):1170–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Beg S, Siraj AK, Jehan Z, et al. (2015). PTEN loss is associated with follicular variant of Middle Eastern papillary thyroid carcinoma. Br J Cancer, 112(12):1938–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Alvarez-Nuñez F, Bussaglia E, Mauricio D, et al. (2006). PTEN promoter methylation in sporadic thyroid carcinomas. Thyroid, 16(1):17–23. [DOI] [PubMed] [Google Scholar]