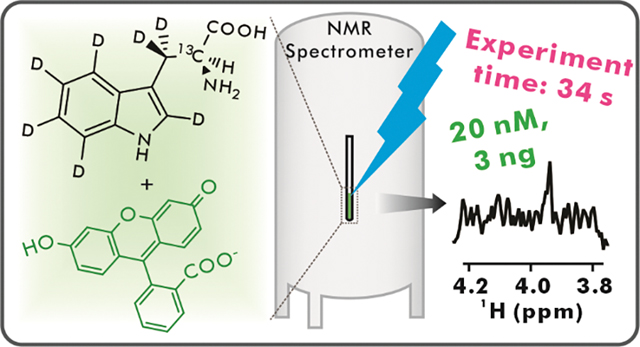
Abstract
NMR spectroscopy is a powerful tool to investigate molecular structure and dynamics. The poor sensitivity of this technique, however, limits its ability to tackle questions requiring dilute samples. Low-concentration photo-chemically induced dynamic nuclear polarization (LC-photo-CIDNP) is an optically-enhanced NMR technology capable of addressing the above challenge by increasing the detection limit of aromatic amino acids in solution up to 1,000-fold, either in isolation or within proteins. Here, we show that the absence of NMR-active nuclei close to a magnetically active site of interest (e.g., the structurally diagnostic Hα-Cα pair of amino acids) is expected to significantly increase LC-photo-CIDNP hyperpolarization. Then, we exploit the spin-diluted tryptophan isotopolog Trp-α-13C-β,β,2,4,5,6,7-d7 and take advantage of the above prediction to experimentally achieve a ca 4-fold enhancement in NMR sensitivity over regular LC-photo-CIDNP. This advance enables the rapid (within seconds) detection of 20 nM concentrations or the molecule of interest, corresponding to a remarkable 3 ng detection limit. Finally, the above Trp isotopolog is amenable to incorporation within proteins and is readily detectable at 1 μM concentration in complex cell-like media, including E. coli cell-free extracts.
Keywords: Hyperpolarization, photo-CIDNP, NMR, isotope labeling
Graphical Abstract
Introduction
Nuclear magnetic resonance (NMR) spectroscopy is a non-invasive technique capable of yielding invaluable information on molecular structure and dynamics at atomic resolution. On the other hand, NMR is intrinsically insensitive due to small differences in population between nuclear spin eigenstates, even at the highest commercially available magnetic field of 28.2 T (1, 2). Although low sensitivity can be compensated by increased sample concentration, this is often not a feasible option due to limited solubility, small amounts of material and high aggregation propensities. The above challenges are particularly severe in the case of biological samples in aqueous media. Numerous hyperpolarization methods were developed to increase the sensitivity of liquid-state NMR by altering nuclear spin populations at thermal equilibrium. Major developments comprise Overhauser dynamic nuclear polarization (ODNP) (3–5), dissolution dynamic nuclear polarization (DDNP) (6–8), and parahydrogen-induced polarization (PHIP) (9–15), including signal amplification by reversible exchange (SABRE) (16–33).
On the other hand, a few challenges have hampered the wide applicability of the above hyperpolarization technologies, to date. These include expensive instrumentation, harsh sample conditions (e.g., high/low temperatures, freeze-thaw cycles), long hyperpolarization times, applicability to a limited number of molecules of interest, safety considerations, inability to perform in situ hyperpolarization, requirement of high sample concentration (50 μM or more) and unfeasible data collection in physiologically relevant environments.
Earlier identification of chemically induced dynamic nuclear polarization (CIDNP) (34, 35) in organic reactions led to the development of a photochemically enhanced NMR approach known as photo-CIDNP. While originally employed to assess solvent exposure (36), this technique has more recently been employed to significantly increase NMR sensitivity in solution at high applied field (37–43). Photo-CIDNP is devoid of most of the drawbacks of other approaches listed in the previous paragraph. This technique requires aromatic moieties and benefits from an O2-free environment.
Photo-CIDNP exploits spin-selective processes between a molecule of interest and a photosensitizer dye. The phenomenon proceeds via a radical pair mechanism, as shown in Figure 1A. Photo-CIDNP can be employed for NMR sensitivity enhancement purposes (44) and, when combined with heteronuclear correlation spectroscopy, it enables highly sensitive detection of hyperpolarized 13C and 15N (38, 39, 45). More recently, trace amounts of commercially available glucose oxidase (GO) and catalase (CAT) were employed to remove dissolved molecular oxygen, thus preventing excited-state dye quenching and further increasing NMR sensitivity (37). The use of fluorescein (Fig. 1B) as photosensitizer, in place of flavin mononucleotide (FMN), led to extension of this technique to low sample concentration (low- to sub-μM) (40). The use of photosensitizers characterized by a long triplet-state lifetime (e.g., 20 ms for fluorescein (40, 46)) gave rise to a branch of photo-CIDNP known as low-concentration photo-CIDNP (LC-photo-CIDNP). Replacement of costly and hard-to-maintain high-power laser with LEDs (Fig. 1C) with no sensitivity losses led to enhanced ease of access, portability and safety (42).
Figure 1. Basic aspects of low-concentration photo-chemically induced dynamic nuclear polarization (LC-photo-CIDNP).
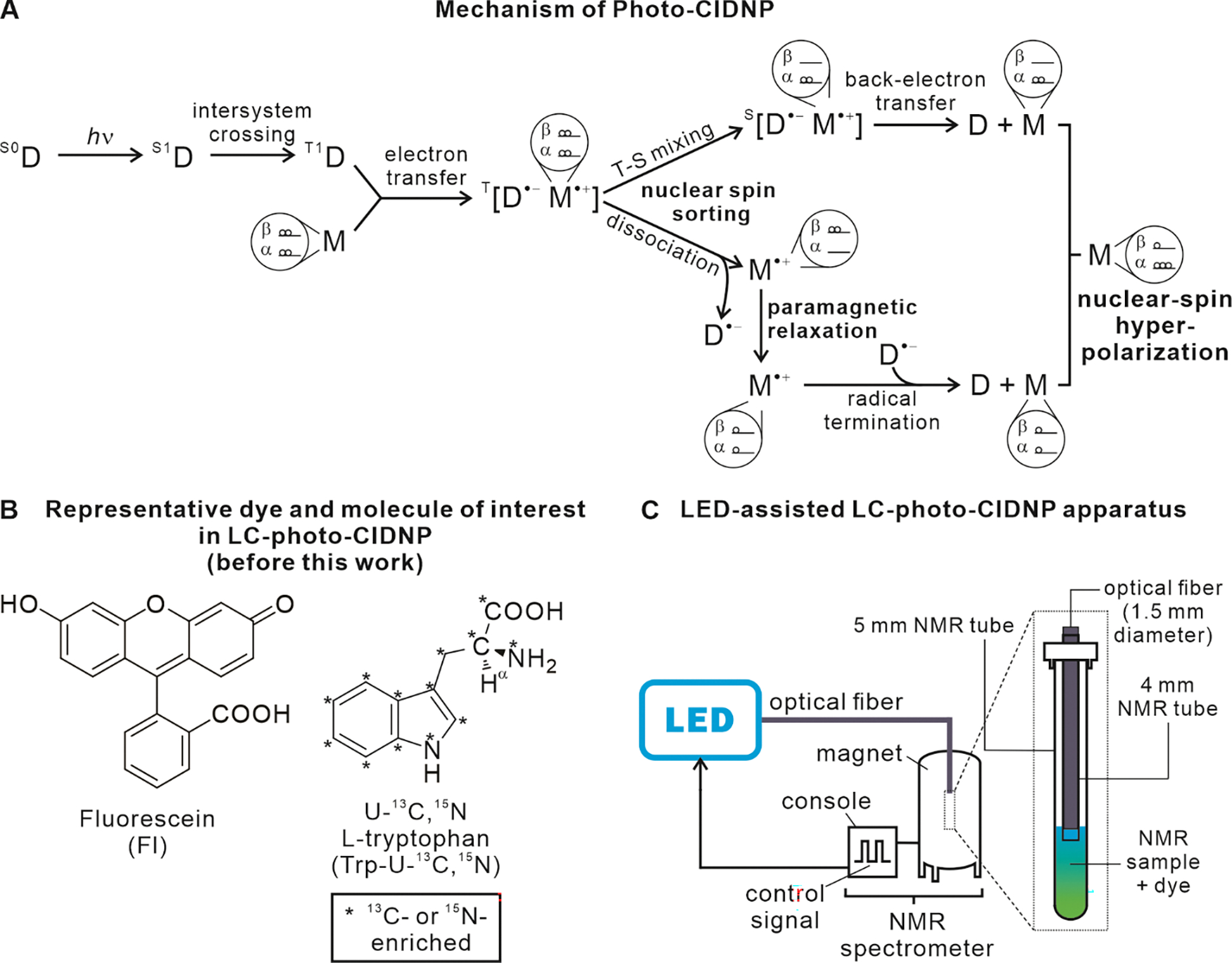
A. Schematic illustration of the radical-pair mechanism of photo-CIDNP in liquids. D and M denote the dye and molecule of interest, respectively. S0, S1 and T1 refer to the ground singlet, 1st excited singlet and 1st excited electronic triplet states, respectively. The T-S acronym denotes triplet-singlet mixing. The frequency of T-S mixing of the T[D•−M•+] radical pair depends on the nuclear spin states of the molecule of interest M•+, due to hyperfine coupling between unpaired electron and nuclear spins. Therefore, depending on the value of nuclear spin states, molecules can either undergo fast or slow T-S mixing. The population with nuclear spin states undergoing slow T-S mixing experiences radical-pair dissociation followed by paramagnetic relaxation, resulting in thermal nuclear-spin polarization. Conversely, the population with nuclear spin states experiencing fast T-S mixing undergoes T-S mixing followed by fast back electron transfer. Importantly, this population experiences nuclear-spin hyperpolarization. In practice, one detects the sum of the two populations, in a photo-CIDNP experiment. B. Structure of a typical small molecule of interest and photosensitizer dye employed in LC-photo-CIDNP. C. Scheme illustrating typical LC-photo-CDINP instrumentation.
LC-photo-CIDNP performs optimally at low-to-sub μM sample concentration (40, 41), and it provides fast (0.2 s) in situ nuclear hyperpolarization. The latter features enable long-term signal-averaging and multi-dimensional NMR of extremely dilute samples. This technology leads to enhanced signal-to-noise ratio (S/N) of tryptophan (Trp) and tyrosine (Tyr), either as free amino acids or within polypeptides and proteins (38, 41–43). Remarkably, high LC-photo-CIDNP enhancements are observed for Cα nuclei of aromatic amino acids, likely due to their high hyperfine coupling constants (38, 47), enabling the efficient detection of Hα-Cα pairs. The Cα chemical shifts are particularly useful because they are diagnostic of backbone secondary structure in proteins (48). To take advantage of proton’s higher sensitivity, 13C-to-1H polarization transfer via reverse INEPT is typically employed (38). In addition, 2D data collection is carried out to preserve information on 13Cα chemical shifts (38, 41). The recently developed 13C RASPRINT pulse sequence is optimized for ultra-fast data collection in aqueous solvent (42).This sequence exploits minimal recycle delays, given that most nuclear-spin polarization is generated during the optical irradiation time, thus abrogating the need for lengthy inter-scan intervals (42). The 13C RASPRINT sequence is particularly useful for the ultra-rapid detection of Hα-Cα pairs at low sample concentration, down to ca. 500 nM (42). More recent studies showed that LC-photo-CIDNP hyperpolarization can be further enhanced in the presence of low-μM concentrations of reductive radical quenchers including ascorbic acid, a.k.a. vitamin C, leading to detection of 200 nM aromatic amino acids (43). Despite the above advances, it is highly desirable to render liquid-state NMR spectroscopy even more sensitive so that it approaches detection limits of other leading techniques, e.g., fluorescence and mass spectrometry.
In this work, we provide theoretical and experimental evidence illustrating that nearby NMR-active nuclei (e.g., 1H’s, 13C and 15N) significantly decrease the extent of observable 13C LC-photo-CIDNP polarization. Upon replacement of these nuclei with either NMR-inactive (12C) or low-gyromagnetic-ratio (2H) nuclei by selective isotope labeling, we were able to achieve an unprecedented improvement in NMR detection level in solution, down to 20 nM Trp, in only a few seconds. In addition, we showed that the above isotopolog-assisted optically enhanced technology is also effective in complex cell-like media, enabling the analysis of aromatic amino acids in physiologically relevant milieu at atomic resolution and extremely high sensitivity.
RESULTS AND DISCUSSION
Theoretical predictions on the dependence of LC-photo-CIDNP-hyperpolarization on isotope substitution patterns and g-factor values.
LC-photo-CIDNP polarization depends on the g-factor of the radical form of the reduced photosensitizer dye. Fluorescein is typically employed as a dye. The g-factor of its reduced radical form in solution, however, is not known. Note that different protonation states of fluorescein in solution are expected to affect g-factor values. We carried out density functional theory (DFT) calculations to estimate the pKa and g-factor of the fluorescein dye radical at 25 °C and pH 7.0. Water solvation effects were taken into account via the integral equation formalism polarizable continuum model (IEFPCM). Predicted g-factor values were determined for the fully protonated state of the Fl• dye radical and for seven variably deprotonated states, as listed in Figure 2A. The pKa values of the three titratable protons were employed to determine the population of each of the eight variably protonated species listed in Figure 2A via the Henderson-Hasselbalch equation. The resulting individual populations and corresponding g-factors were employed to predict the overall weighted-average g-factor of the fluorescein dye radical at 25 °C and pH 7.0. This g-factor value was mostly contributed by Fl•−, and was found to be 2.003077.
Figure 2. Computational predictions.
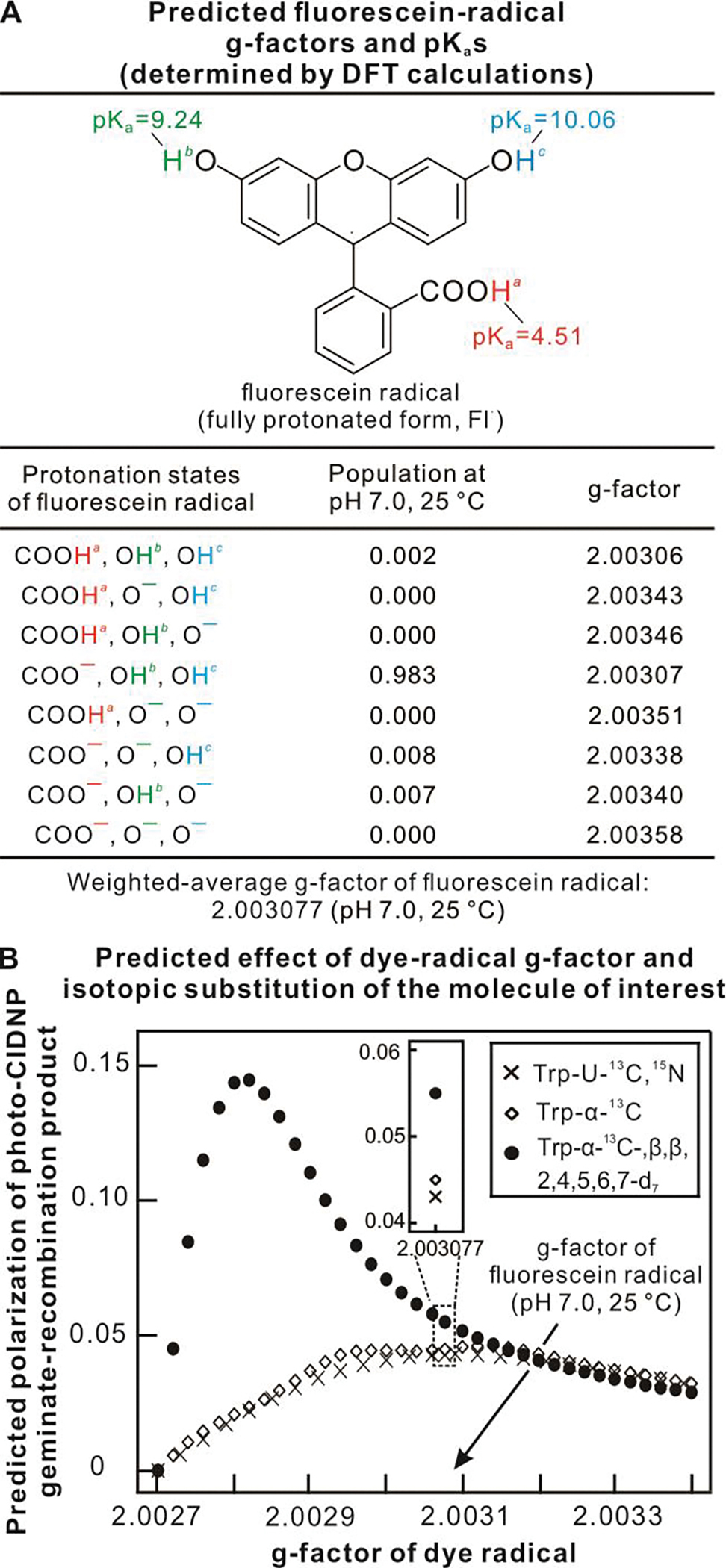
A. Predicted pKa and isotropic g-factor values for fluorescein radicals of different protonation states. B. Predicted photo-CIDNP polarization of three Trp isotopologs as a function of dye g-factor. Simulations were performed using g-factor of Trp• = 2.0027, and hyperfine coupling constant of Trp and fluorescein radicals from literature sources (see details in Suppl. Inf.).
The known g-factor of Trp•+ (47) and the weighted-average g-factor of the fluorescein dye radical, determined as described in the previous paragraph, were used as input for computational predictions of LC-photo-CIDNP polarization as a function of g-factor of the dye radical. We followed known theoretical and practical guidelines (49–51), with some modifications, as detailed below.
For freely-diffusing radical pairs, the population difference between any two nuclear-spin configurations 1 and 2 of the geminate-recombination product is
| [1] |
where p1, p2 are the populations with nuclear-spin configuration 1 and 2, respectively; p is a normalization factor (see Supplementary Information), ωTS1 and ωTS2 are the triplet-singlet mixing frequencies of the nuclear-spin configurations, and τd is the average time the radical-pair components remain closely associated (beyond orbital-overlap distances) before long-term dissociation. The triplet-singlet mixing frequency of each of any possible nuclear-spin configurations is
| [2] |
where the i and j subscripts denote individual nuclei of the dye and molecule of interest, respectively. In addition, Δg is the difference between the g-factors of the radical-pair components (i.e., the dye and molecule of interest), μB denotes the Bohr’s magneton B0 is the applied magnetic field, and m and A are the magnetic nuclear-spin quantum number and hyperfine coupling constant, respectively.
For different isotope labeling schemes of the molecule of interest, corresponding to different isotopologs, the third term of equation [2] is expected to vary depending on specific Aj values. Therefore, we reasoned that different isotopic substitutions ought to give rise to variable populations of geminate-pair products, resulting into different levels of photo-CIDNP hyperpolarization (52). Hence we proceeded to explore how the g-factor of dye radicals is expected to affect the theoretically achievable geminate-pair recombination products, in the case of different Trp isotopologs. We focused on three representative labeling schemes of the molecule of interest: namely, (i) uniformly 13C-labeled Trp (Trp-U-13C), (ii) a hypothetical Trp isotopolog bearing a 13C-1H pair at the α carbon (Cα) and extensive deuteration at non-exchangeable sites (Trp-α-13C-β,β,2,4,5,6,7-d7, Fig. 3A), and, finally, (iii) a reference Trp isotopolog enriched in 13C at the Cα and bearing natural-abundance nuclides at all other sites (Trp-α-13C, Fig. 3B). Importantly, all of the above isotopologs have a 13C at the α position, honoring the fact that the LC-photo-CIDNP hyperpolarization approach is particularly effective at enhancing the 13Cα of the Trp and Tyr amino acids, either alone or within proteins (41). Additional details about the computations, including assessment of overall probabilities of geminate recombination, proportional to nuclear-spin polarization at the Trp α carbon, are provided in the Supplementary Information.
Figure 3. Synthesis and characterization of Trp isotopologs.
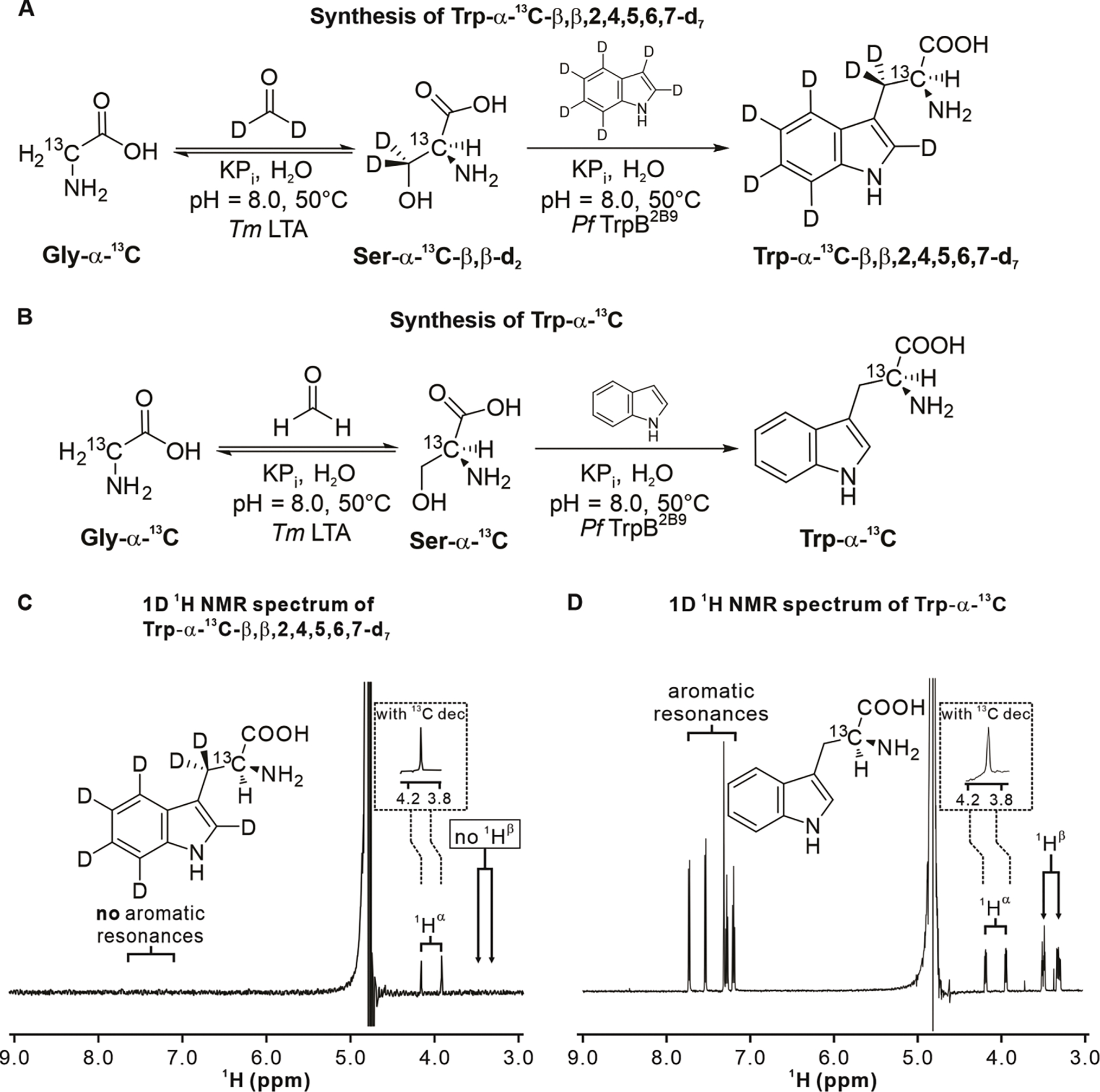
A. Scheme illustrating the enzyme-cascade synthesis of Trp-α-13C-β,β,2,4,5,6,7-d7. B. Scheme illustrating the enzyme-cascade synthesis of Trp-α-13C. C. 1D 1H NMR characterization of Trp-α-13C-β,β,2,4,5,6,7-d7. A pulse-acquire data-collection scheme with a 121.5 Hz solvent presaturation scheme during the entire 5 s recycle delay was employed. 8 scans were acquired. The sample (ca. 100 μM) was prepared in 90% H2O, 10% D2O with traces of sodium trimethylsilylpropanesulfonate (DSS) used as an internal reference. The inset shows the 1Hα region of a spectrum including 13C decoupling during acquisition using bi_p5m4sp_4sp.2 composite pulse decoupling (see Exp. Section in Suppl. Inf.). D. 1D 1H NMR characterization of HPLC-purified and lyophilized Trp-α-13C (ca. 200 μM). The inset shows the 1Hα region of the 13C-decoupled spectrum. The same acquisition and processing parameters as in panel C were used.
The simulation results for the above three Trp isotopologs are shown in Figure 2B. Even at optimal values of g-factor of the dye radical, the two compounds bearing no deuteration are predicted to afford only limited sensitivity enhancement. On the other hand, the Trp-α-13C-β,β,2,4,5,6,7-d7 deuterated isotopolog (solid circles in the plot of Fig. 2B) is expected to undergo drastic polarization enhancements that significantly vary in magnitude depending on the g-factor of the dye radical. These enhancements extend up to nearly 14.5% for dyes that bear an optimal g-factor value.
Under typical LC-photo-CIDNP conditions, fluorescein is employed as photosensitizer (41). As detailed elsewhere (40), this dye bears a conveniently long triplet-state lifetime that persists across the infrequent collisions between dye and molecule of interest, at moderate sample concentrations (low μM and lower). Therefore, despite its sub-optimal g-factor value in the plot of Figure 2B, fluorescein is the current photosensitizer of choice in LC-photo-CIDNP experiments. When fluorescein is employed as a photosensitizer at room temperature and 25 °C, the predicted probability of geminate recombination (proportional to nuclear-spin hyperpolarization (52)) of uniformly 13C-labeled Trp (Trp-U-13C) is the lowest (ca. 5%). On the other hand, Trp-α-13C is predicted to experience a 4.4% more geminate-recombination than Trp-U-13C. More strikingly, the selectively labeled isotopolog Trp-α-13C-β,β,2,4,5,6,7-d7, which includes extensive deuteration at Cβ and aromatic carbon sites, is predicted to experience 28.2% more geminate recombination than Trp-U-13C.
Trp analogs bearing variable isotopic substitution patterns were prepared and characterized.
Encouraged by the computational predictions of Figure 2B, we proceeded to prepare some relevant isotopologs and experimentally test their performance in terms of achievable LC-photo-CIDNP hyperpolarization.
Previous selective labeling schemes were based on supplementing bacterial growth media with [4-13C]-δ-aminolevulinic acid (53) or with different singly labeled 13C-glucose isotopologs (54). These methods, however, suffer from low isotope-incorporation yields and are not easily customizable to the desired labeling patterns. Here, we employed an alternative recently developed enzyme-cascade approach (55) to readily achieve high levels of isotope enrichment starting from affordable and readily available precursors. This approach is particularly convenient due to its modular nature (55). For instance, it enables the facile synthesis of a wide variety of desired isotopologs by simply changing the labeling pattern of the starting materials (i.e., glycine and indole) and solvent (water).
The synthetic schemes of Figure 3A and 3B detail the route that led to the generation of Trp-α-13C-β,β,2,4,5,6,7-d7 and Trp-α-13C with 44.7% and 31.8% yield, respectively. Given that the amounts of purified products were more than adequate for our purposes, no further optimization was performed. The identity of purified Trp-α-13C-β,β,2,4,5,6,7-d7 and Trp-α-13C was confirmed by ESI MS and 1H NMR. The pulse-acquire 1H NMR spectra of the two isotopologs are shown in Figure 3C and 3D and confirm the presence of the expected 13C-1H scalar coupling. The latter disappears upon 13C decoupling. In addition, no 1H aromatic resonances are observed for Trp-α-13C-β,β,2,4,5,6,7-d7, consistent with expectations.
The Trp-α-13C-β,β,2,4,5,6,7-d7 isotopolog leads to large LC-photo-CIDNP sensitivity enhancements.
The two Trp isotopologs described in the previous section and the commercially available uniformly labeled Trp-U-13C,15N were initially analyzed under dark (i.e., LED-off) conditions. The 13C RASPRINT pulse sequence (42) was employed, except that the typically short recycle delays (50 ms) (42) were replaced by long ones (5 s) to allow for nearly complete longitudinal relaxation. This pulse sequence enables the detection of 13C-bound protons and is routinely employed in LC-photo-CIDNP (42).
Figure 4A, which focuses on Hα protons under dark conditions, shows that the selectively labeled Trp-α-13C and Trp-α-13C-β,β,2,4,5,6,7-d7 isotopologs display a larger S/N than uniformly labeled Trp (Trp-U-13C,15N). While Figure 4A provides a useful qualitative assessment, the different sample concentrations prevent quantitative comparisons. A more quantitative analysis of NMR behavior under dark conditions is provided in Figure 4B. The plot in this figure examines both intensities at the resonance maximum and areas, and normalizes the data so that they are independent of sample concentration. Here we note that, relative to Trp-U-13C,15N, the Trp-α-13C isotopolog displays 20% increases in resonance area and intensity at the maximum. Conversely, Trp-α-13C-β,β,2,4,5,6,7-d7 displays a 42% increase in area and an even more dramatic 192% increase in intensity at the maximum. The latter enhancement results from narrower 1H linewidths, as confirmed by T2 measurements (Supplementary Information). In summary, perhaps not surprisingly, detection of Trp Hα resonances under dark conditions is rendered more facile by the extensive deuteration of the Trp-α-13C-β,β,2,4,5,6,7-d7 isotopolog. It is worth noting that Hα resonances are important in biology because they are diagnostic of structure, especially when analyzed in the context of protein backbone.
Figure 4. LED-off and LED-on data show enhancements due to both dark and light contributions.
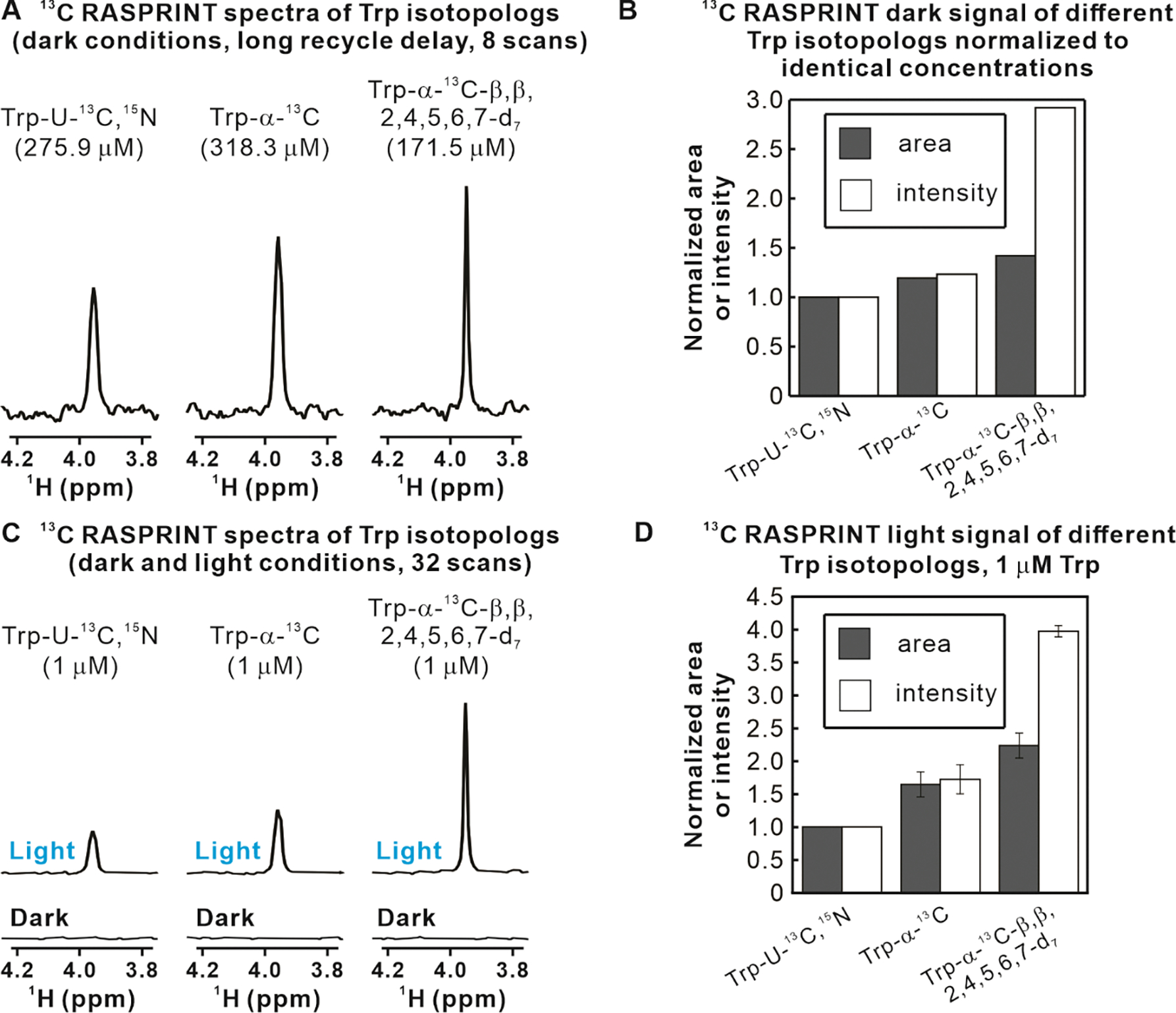
A. NMR spectra showing the Hα resonance of different Trp isotopologs. Data were collected with the 1D 13C RASPRINT pulse sequence under dark (i.e., LED-off) conditions at the concentrations listed above the spectra. A 5 s recycle delay was employed. B. Quantitative analysis of Hα-resonance areas and intensities-at-the-maximum relative to Trp-U-13C,15N, after normalization to eliminate differences due to variable concentrations. C. LC-photo-CIDNP spectra acquired with the 1D 13C RASPRINT pulse sequence under light (LED-on) conditions for three Trp isotopologs of equal concentration. D. Quantitative analysis of Hα resonance area and intensity-at-the-maximum under light (i.e., LED-on) conditions relative to Trp-U-13C,15N. Data in panels B and D are shown as avg ± S.E. (n=2). All spectra were acquired at 24 °C with 64 scans, 4 dummy scans, and an optical irradiation time of 0.2 s.
Next, we performed 1D LC-photo-CIDNP experiments on all three Trp isotopologs under light (i.e., LED-on) conditions. Data were collected at very low sample concentration (1μM). The NMR spectra under light conditions, shown in Figure 4C, display considerable increases in S/N upon going from Trp-U-13C,15N to Trp-α-13C to Trp-α-13C-β,β,2,4,5,6,7-d7. Remarkably, the latter isotopolog displays a ca. 4-fold increase in S/N relative to the already intense signal of uniformly labeled Trp (Trp-U-13C,15N). More quantitative studies, displayed in Figure 4D, show that Trp-α-13C displays a 70% increase in resonance area and intensity relative to Trp-U-13C,15N, while Trp-α-13C-β,β,2,4,5,6,7-d7 features a 120% increase in area and a 300% increase in resonance intensity.
At this juncture, it is important to consider a few simple items. First, NMR sensitivity (1, 56) is defined as S/N divided by the square root of experiment time t, i.e.,. Second, the NMR S/N ratio depends on the amplitude of both the resonance of interest and the noise. Third, all data were collected with identical acquisition and processing parameters. Taking all of the above considerations into account, the results of Figure 4C and 4D clearly show that the Trp-α-13C-β,β,2,4,5,6,7-d7 isotopolog displays a remarkable 4-fold increase in sensitivity relative to uniformly labeled Trp (Trp-U-13C,15N). Note that all experiments under light conditions must bear equivalent noise values. This result is even more significant if one notices that (a) all the Figure 4C spectra acquired under dark conditions display no detectable signal, and (b) all the Figure 4 light and dark spectra included only 32 scans. In conclusion, the overall sensitivity of the 13C RASPRINT experiment for the Trp-α-13C-β,β,2,4,5,6,7-d7 isotopolog must be very high. More details on specific enhancements factors and polarization values are provided in a later section.
The Trp-α-13C-β,β,2,4,5,6,7-d7 isotopolog enables optically-enhanced NMR at low nanomolar concentration.
Encouraged by the results described in the previous sections, we further explored the LC-photo-CIDNP detection limit of the Trp-α-13C-β,β,2,4,5,6,7-d7 isotopolog. Towards this end, we took a few deliberate simple steps. First, to optimize detection of the target isotopolog, we modified the 13C RASPRINT pulse sequence by removing its constant-time evolution. Note that the absence of 13Cs at the β and carbonyl positions (13Cβ and 13C’) of the Trp-α-13C-β,β,2,4,5,6,7-d7 isotopolog renders constant-time evolution unnecessary, due to lack of 13C-13C J couplings (57). Second, we converted all the on-resonance selective pulses into hard pulses and removed the 13C’ off-resonance pulse. These modifications are justified by the fact that the Trp-α-13C-β,β,2,4,5,6,7-d7 isotopolog only bears one 13C and lacks 13C-13C J couplings. Therefore, there is no need for 13Cα selective excitation or for selective C’ pulses to refocus 13Cα−13C’ J couplings.
Importantly, all the above modifications are beneficial because they lead to shorter experiment times and reduce signal losses arising from pulse imperfections and transverse relaxation during the pulse scheme. The resulting pulse sequence, displayed in Figure 5A, was denoted as 13C isotopolog-optimized RASPRINT or 13C iso-RASPRINT.
Figure 5. LC-photo-CIDNP in combination with selective isotope labeling enables the detection of Trp at low-nanomolar concentration.
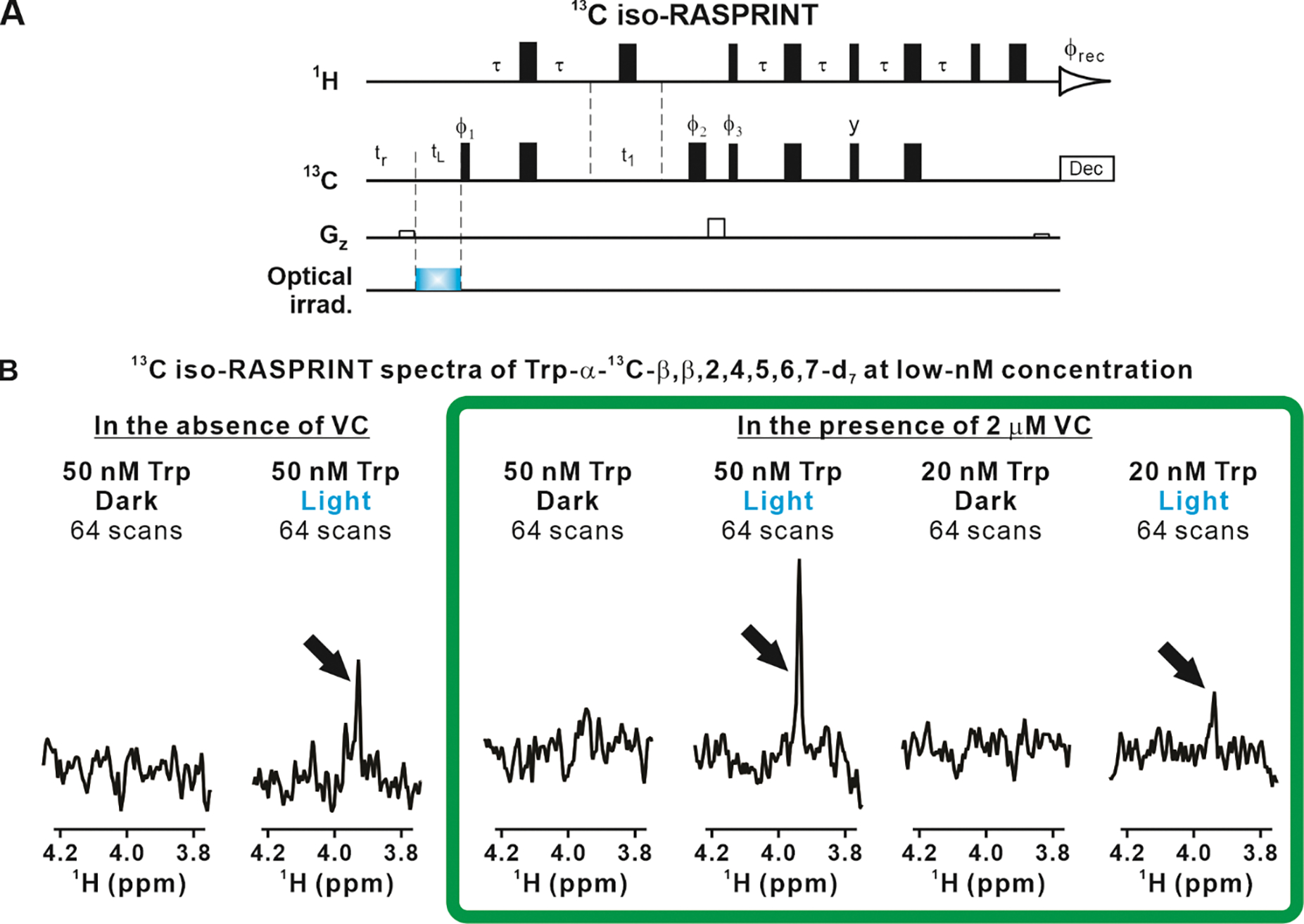
A. Scheme illustrating the 13C iso-RASPRINT pulse sequence, optimized for the detection of the Trp-α-13C-β,β,2,4,5,6,7-d7 isotopolog. The 13C iso-RASPRINT sequence differs from the 13C RASPRINT sequence in that on-resonance selective pulses are replaced by hard pulses and off-resonance selective pulses are removed. In addition, the constant-time evolution was removed, yet a short spin-echo was preserved to compensate for the effect of pulse field gradients. B. LC-photo-CIDNP (1D 13C iso-RASPRINT) spectra of 50 and 20 nM Trp-α-13C-β,β,2,4,5,6,7-d7. Data were acquired under dark and light conditions, in the absence and presence of 2 μM vitamin C (VC). In each spectrum, signal-averaging was carried out across 64-scans preceded by 4 dummy scans. The optical irradiation time was 0.2 s. Data were collected at 24 °C. The spectra enclosed within the green frame show the highest degree of nuclear-spin hyperpolarization.
Then, we tested the LC-photo-CIDNP behavior of extremely dilute (50 and 20 nM) Trp-α-13C-β,β,2,4,5,6,7-d7 upon data collection with the 13C iso-RASPRINT pulse sequence. The results (two outmost-left spectra in Fig. 5B) show that the 50 nM sample yields a detectable resonance of moderate intensity after 64 scans (Fig. 5B). On the other hand, the 20 nM sample displayed no observable signal (data not shown). Given that reductive radical quenchers including ascorbic acid, also known as vitamin C (VC) is known to protect LC-photo-CIDNP molecules of interest from photodamage (43), potentially extending detection limit, we prepared an additional set of samples containing a small concentration of VC (2 μM). The results, shown in the four righthandside panels of Figure 5B, prove that in the presence of the VC reductive radical quencher even 20 nM Trp-α-13C-β,β,2,4,5,6,7-d7 can be readily detectable, within only 64 scans. This is a remarkably small concentration, corresponding to an extremely favorable detection limit of 3 ng.
Quantitative assessment of LC-photo-CIDNP enhancements and polarization values.
To more quantitatively evaluate the extent of polarization enhancement generated via LC-photo-CIDNP for each of the isotopologs analyzed in this work, we compared resonance areas under dark and light conditions according to the relation below
| [3] |
where ε is the polarization enhancement factor, defined as the ratio between nuclear spin polarization under light and dark conditions. Note that we acquired the dark (LED-off) spectra at much higher concentrations (ca. 100 μM) than the LED-on experiments, and then adjusted for concentration differences according to equation [3]. In addition, we employed long (5 s) recycle delays to ensure nearly complete relaxation between scans. We collected the same number of scans under dark and light conditions. Clearly, if the shorter delays employed in the light experiments had been employed for data collection under dark conditions, the apparent polarization enhancement factors would have been much larger. We refrained from doing this so that we could provide the highest possible reference S/N values under dark conditions.
From percent polarization enhancements ε and from thermal-equilibrium percent polarization P%,te, assessed from the Boltzmann distribution in the presence of a 600 MHz applied field (1), we also determined the percent polarization P% according to
| [4] |
The resulting ε and P% values of all three isotopologs examined in this work are shown in Figure 6.
Figure 6. The Trp-α-13C-β,β,2,4,5,6,7-d7 isotopolog displays unprecedented LC-photo-CIDNP enhancement factors ε and percent polarization in solution.
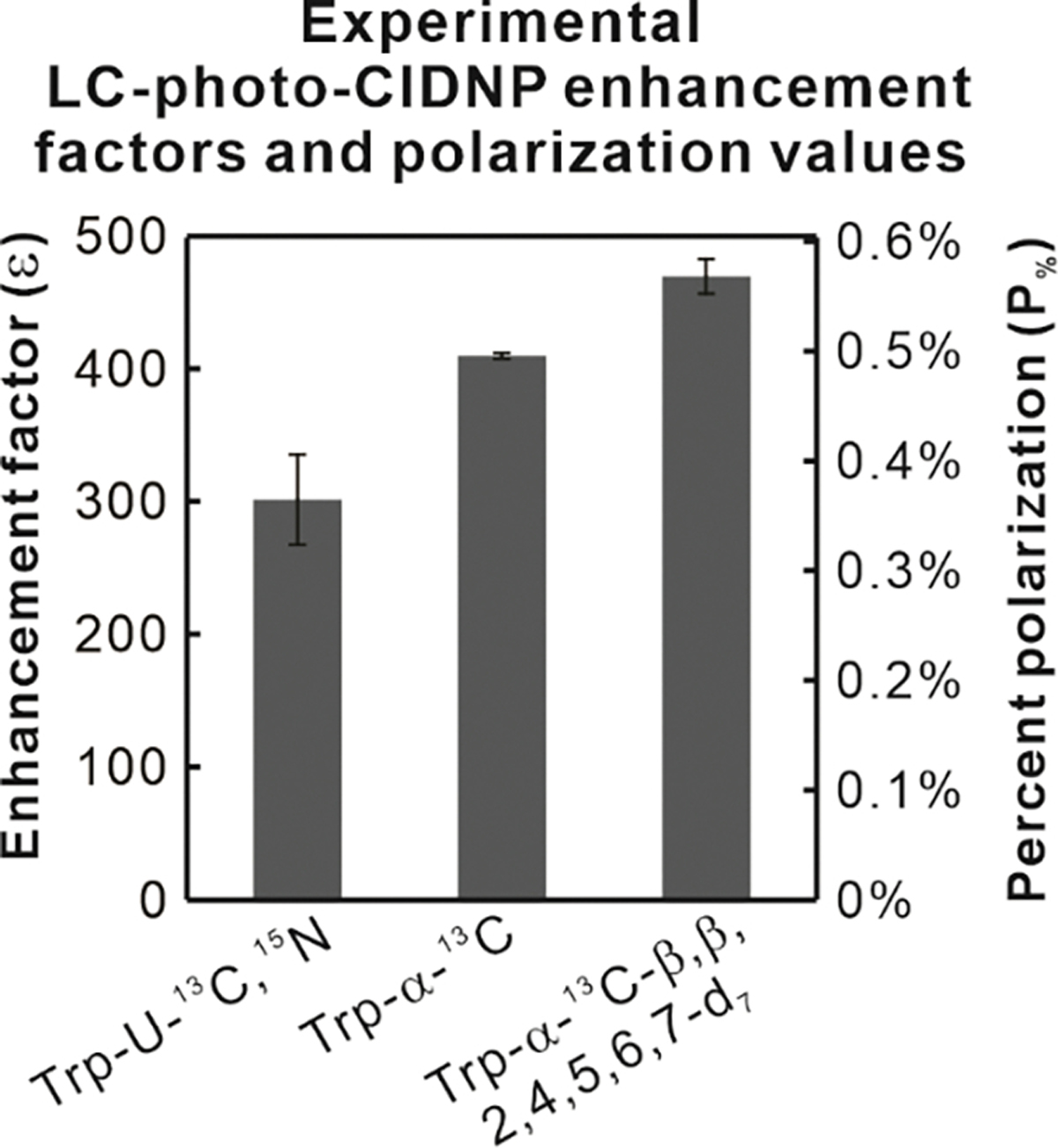
Enhancement factors (ε) were determined from experimental data according to equation [3]. Percent polarization values were assessed from enhancement factors and thermal-equilibrium percent polarizations according to equation [4]. Thermal-equilibrium percent polarizations were estimated at room temperature at 600 MHz applied field. Sample concentrations were 1 μM and 100 μM in LED-on and LED-off experiments, respectively. All values are shown as avg ± S.E., with n=2, where n denotes the number of independent experiments.
The enhancement factor and percent polarization of the two best isotopologs are discussed next. As shown in Figure 6, the ε and P% values of Trp-α-13C are 410 ± 2 and 0.496 ± 0.003%, respectively. The Trp-α-13C-β,β,2,4,5,6,7-d7 displays the highest values; namely ε and P% of 470 ± 13 and 0.568 ± 0.016%. The Trp-α-13C and Trp-α-13C-β,β,2,4,5,6,7-d7 isotopologs performed considerably better than uniformly labeled Trp (Trp-U-13C,15N), with increases of 38% and 57%, respectively.
The observed sensitivity enhancements arise from both LED-on and LED-off effects.
The data of Figure 4B, collected under dark (LED-off) conditions, show that selective 13C labeling as well as introduction of deuteration at Cβ and across the aromatic ring induces some sensitivity enhancement even in the absence of optical irradiation. Therefore, some non-photo-CIDNP-related factors must contribute to the overall observed effects.
We were able to identify two specific dark-state contributions. First, there was an increase in 1Hα T2 arising from either the removal of some nearby NMR-active nuclei and(or) from their replacement with low-gyromagnetic-ratio (low-γ) nuclei due to selective labeling. The results of 1Hα T2 measurements are presented in the Supplementary Information. Clearly, this effect accounts for the sharper resonances, and longer 1Hα T2 displayed by the Trp-α-13C-β,β,2,4,5,6,7-d7 isotopolog even under dark conditions. Second, the elimination of many 13C-13C, multi-bond 1H-13C and 1H-1H J couplings arising from selective labeling prevents loss of coherence via J coupling during the pulse sequence. This effect was quantified upon comparing the area/concentration ratios for the 1Hα resonance of each isotopolog in LC-photo-CIDNP measurements carried out under dark conditions.
In addition, there were also contributions due to LC-photo-CIDNP hyperpolarization effects under light (LED-on) conditions expected from the theoretical arguments discussed in the previous sections. The contributions arising from all three factors (linewidth reduction, J-coupling effects and LC-photo-CIDNP hyperpolarization) were quantified as described in the Supplementary Text and Supplementary Figure S4. The results are summarized in Table 1 for each of the isotopologs examined in this work. For easier-evaluation purposes, the data were normalized relative to Trp-U-13C,15N. The product of all three separately-assessed contributions agrees well with the overall enhancement factor in sensitivity. Overall, it is intriguing to note that the sensitivity enhancements described in this work were achieved by a combination of “dark and light effects”.
Table 1.
Relative sensitivity of LC-photo-CIDNP NMR experiments (1D 13C RASPRINT pulse sequence) carried out on different Trp isotopologs. The role of individual light (LED-on) and dark (LED-off) contributions is listed. Uniformly labeled Trp (Trp- U-13C,15N) is regarded as reference compound.
| topolog | Relative sensitivity partitioned into individual contributions |
Product of individual contributions d | Overall relative sensitivity (assessed via 13C RASPRINT, light cond.) e | ||
|---|---|---|---|---|---|
| LC-photo-CIDNP hyperpolarization a(light effect) | Elimination of J-coupling b(dark effect) | Linewidth reduction c(dark effect) | |||
|
| |||||
| Trp-U-13C,15N | 100% | 100% | 100% | 100% | 100% |
| Trp-α-13C | 138±4% | 119% | 126 ± 2% | 207 ± 7% | 172 ± 20% |
| Trp-α-13C- β,β, 2,4,5,6,7-d7 | 157±4% | 142% | 190 ± 5% | 424 ± 16% | 397 ± 9% |
Determined from data in Fig. 6, including assessment of errors.
Determined from data in Fig. 4B. Only one experiment was carried out.
Assessed from T2 experiments in Supplementary Fig. S2. Values are shown as avg ± S.E., with n=3, where n denotes the number of independent experiments.
Errors were assessed by error propagation of the individual contributions.
Three 13C RASPRINT experiments were carried out for each isotopolog under LED-on conditions. Relative sensitivities were assessed from resonance intensities of experiments run with identical parameters, including number of scans and duration. Relative sensitivities are shown as avg ± S.E.
LC-photo-CIDNP is effective in complex biologically relevant media.
In addition to the simple environments discussed so far, including only few purified components, LC-photo-CIDNP can also be applied to samples within extremely complex biological milieux. To demonstrate this capability in the case of the LC-photo-CIDNP technology presented here, we prepared an E. coli S30 bacterial cell extract according to known procedures (58). This extract includes most E. coli soluble proteins. The composition of the corresponding NMR sample is schematically illustrated in Figure 7A and the procedure followed to generate the cell extract is outlined in Figure 7B. Importantly, this complex biological sample included 1 μM Trp-α-13C-β,β,2,4,5,6,7-d7. Figure 7C illustrates the fact that no signal was observable for this extremely dilute Trp isotopolog under dark conditions. In contrast, 1 μM Trp-α-13C-β,β,2,4,5,6,7-d7 became readily detected after only 8 scans under light conditions. Remarkably, due to the nearly absent recycle delay of 13C iso-RASPRINT under light conditions, the entire data collection, including the hyperpolarization time, only took a total of ca. 4 s (excluding the time for the 4 dummy scans, i.e., 2 additional seconds).
Figure 7. LC-photo-CIDNP in combination with selective isotope labeling enables hypersensitive data collection in a complex cell-like medium.
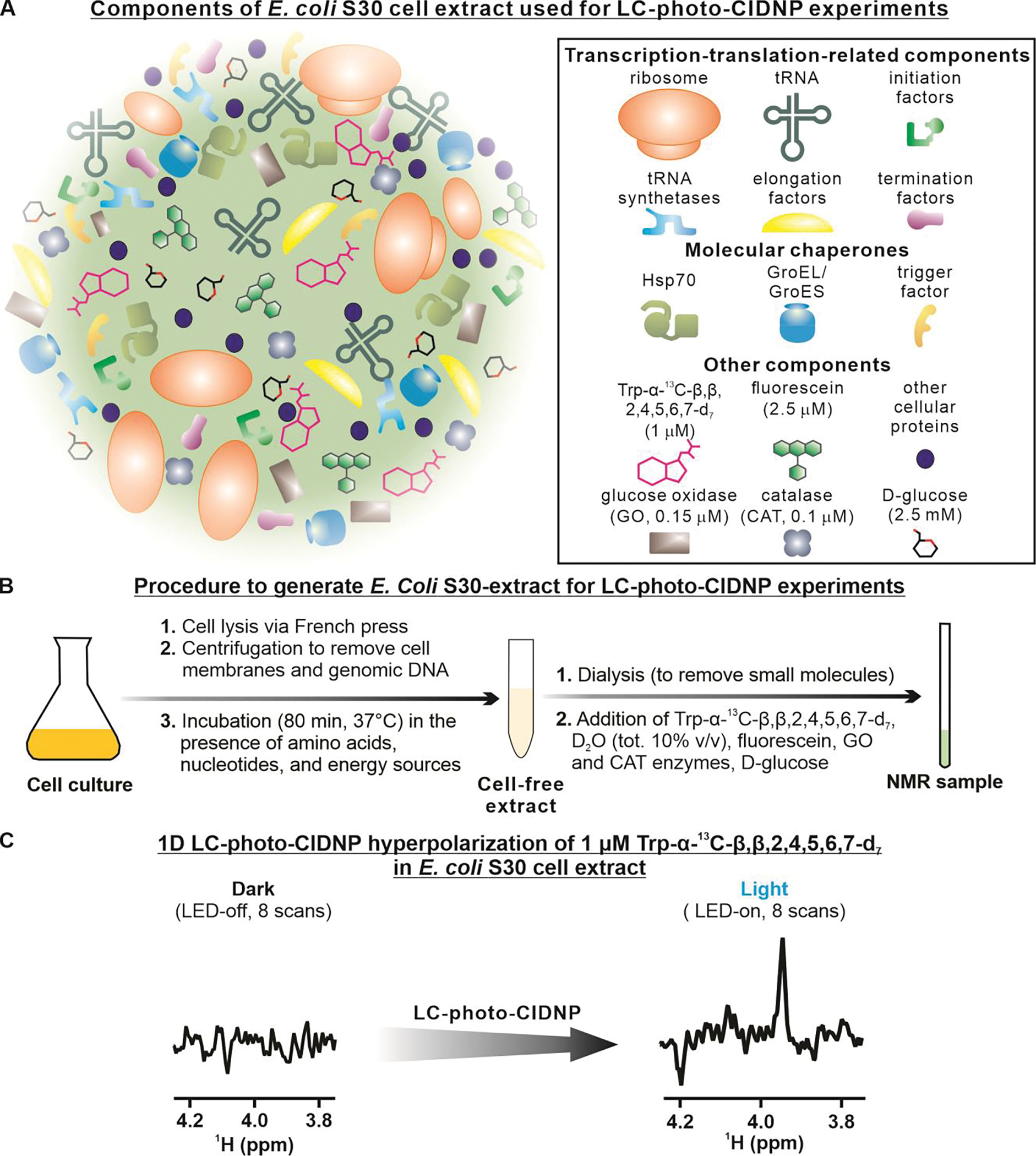
A. Schematic illustration of the components of the bacterial cell-like medium, i.e., an E. Coli S30 extract, employed in this work. B. Summary of the adopted procedures to generate the E. Coli S30 extract. C. LC-photo-CIDNP spectrum of 1 μM Trp-α-13C-β,β,2,4,5,6,7-d7 in an E. coli S30 cell extract under dark (i.e., LED-off) and light (i.e., LED-on) conditions. Each spectrum was acquired at 24 °C with 8 scans, 4 dummy scans, and 0.2 s of optical irradiation.
Comparisons with other hyperpolarization approaches in liquids.
As a comparison, two representative examples from other leading nuclear-spin hyperpolarization approaches will be listed next. The well-known dissolution DNP (dDNP) technology enabled detection of 20 μM protein samples (59). This technology has also been used in the context of protein-ligand interactions (6, 60–65). In addition, Kouřil et al. (66) recently showed that 32% 13C polarization was rendered possible by bullet-DNP, an interesting variation of dDNP employing small sample-dilution factors during the dissolution process. In both the above studies, considerably higher sample concentrations were required than in the present work. Moreover, unlike LC-photo-CIDNP, the dDNP approach requires a freeze-thaw process that is not always desirable in the case of biomolecules.
Very recent studies showed that parahydrogen-induced polarization (PHIP), especially via the SABRE approach, can be employed for the detection of amino acids in solution (18, 67, 68). While this methodology is promising, identification of suitable amino-acid precursors for SABRE remains difficult, and extension of this technique to proteins has not been feasible.
CONCLUSIONS AND OUTLOOK
In this study we introduce a combination of LC-photo-CIDNP and selective isotope labeling to enable the NMR detection of low-nM biomolecular concentrations by liquid-state NMR. The use of selective isotope labeling enables an additional ca. 4-fold increase in sensitivity relative to a uniformly isotopically enriched reference molecule (Trp-U-13C,15N). This gain in sensitivity was crucial to access the low-nanomolar (20 nM) concentration range, providing a remarkable detection limit of only 3 ng in a few seconds. To the best of our knowledge, this is the lowest concentration achieved by high-resolution biomolecular NMR in liquids to date. Our methodology works well even in extremely complex physiologically relevant environments, including an E. coli S30 cell extract. The capability to assess Trp levels (69–71), metabolism and transport in biological media is important because of the emerging role of this amino acid and its metabolites in health and disease (72–78).
Importantly, the LC-photo-CIDNP technology, including the advances presented here, is also readily amenable to extensions to Trp-containing macromolecules. For instance, LC-photo-CIDNP of polypeptides and proteins has already been established (38, 41, 43), and it yields comparable enhancements to free Trp. Further, given that Trp analogs can be routinely incorporated into proteins via auxotroph strains (79), one can envisage that the same strains can also be employed to incorporate Trp isotopologs like Trp-α-13C-β,β,2,4,5,6,7-d7 into proteins. Ultrasensitive detection of Hα-Cα pairs in proteins is highly desirable because it provides access to site-specific information on backbone secondary structure (48). Further, the 13C iso-RASPRINT pulse sequence can readily be run in 2D mode to resolve overlapping resonances within mixtures of aromatic amino acids or in the context of large biomolecules.
In all, the advances described in this work pave the way to a variety of biomolecular NMR applications requiring high sensitivity and fast data collection. It will be exciting to follow the forthcoming progress of this technology in the years to come.
Supplementary Material
ACKNOWLEDGMENTS
We thank Alexandra Yurkovskaya and Andrew Buller for insightful discussions, and Greg Sabat for assistance with mass spectrometry. We are grateful to Charles G. Fry, Heike Hofstetter and Cathy Clewett for technical assistance with NMR. Some of this research was performed using the computer resources and assistance of the UW-Madison Center for High Throughput Computing (CHTC) in the Department of Computer Sciences (80, 81). The CHTC is supported by UW-Madison, the Advanced Computing Initiative, the Wisconsin Alumni Research Foundation, the Wisconsin Institutes for Discovery, and the National Science Foundation, and is an active member of the OSG Consortium, which is supported by the National Science Foundation and the U.S. Department of Energy’s Office of Science.
FUNDING SOURCES
This work was funded by the National Institutes of Health (grants R01GM125995 and S10OD012245), from the National Science Foundation (grant MCB 2124672) and from the University of Wisconsin-Madison (Bridge Funds from the College of Letters & Science).
Footnotes
ASSOCIATED CONTENT
The Supporting Information is available free of charge at [URL TBD].
Description of theoretical predictions on photo-CIDNP polarization, additional experimental details on NMR transvers relaxation measurements, description of assessment of LED-on and LED-off effects used to generate Table 1, experimental section.
REFERENCES
- 1.Lee JH, Okuno Y, Cavagnero S, Sensitivity enhancement in solution NMR: emerging ideas and new frontiers. J. Magn. Reson. 241, 18–31 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ardenkjaer-Larsen JH et al. , Facing and overcoming sensitivity challenges in biomolecular NMR spectroscopy. Angew. Chem. Int. Ed. 54, 9162–9185 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schops P, Spindler PE, Prisner TF, Multi-frequency pulsed Overhauser DNP at 1.2 Tesla. Zeit. Phys. Chem. Intl. J. Res. Phys. Chem. Chem. Phys. 231, 561–573 (2017). [Google Scholar]
- 4.Denysenkov VP, Prisner TF, Liquid-state Overhauser DNP at high magnetic fields. Emagres 8, 41–54 (2019). [Google Scholar]
- 5.Bennati M, Orlando T, Overhauser DNP in liquids on 13C nuclei. Emagres 8, 11–18 (2019). [Google Scholar]
- 6.Kim Y, Hilty C, “Applications of Dissolution-DNP for NMR Screening” in Biological Nmr, Pt B, Wand AJ, Ed. (2019), vol. 615, pp. 501–526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kagawa A et al. , High-field NMR with dissolution triplet-DNP. J. Magn. Reson. 309, 106623 (2019). [DOI] [PubMed] [Google Scholar]
- 8.Jahnig F et al. , A spin-thermodynamic approach to characterize spin dynamics in TEMPO-based samples for dissolution DNP at 7 T field. J. Magn. Reson. 303, 91–104 (2019). [DOI] [PubMed] [Google Scholar]
- 9.Bowers CR, Weitekamp DP, Transformation of symmetrization order to nuclear-spin magnetization by chemical reaction and nuclear magnetic resonance. Phys. Rev. Lett. 57, 2645–2648 (1986). [DOI] [PubMed] [Google Scholar]
- 10.Bowers CR, Weitekamp DP, Parahydrogen and synthesis allow dramatically enhanced nuclear alignment. J. Am. Chem. Soc. 109, 5541–5542 (1987). [Google Scholar]
- 11.Carson PJ, Bowers CR, Weitekamp DP, The PASADENA effect at a solid surface: High-sensitivity nuclear magnetic resonance of hydrogen chemisorption. J. Am. Chem. Soc. 123, 11821–11822 (2001). [DOI] [PubMed] [Google Scholar]
- 12.Pravica MG, Weitekamp DP, Net NMR alignment by adiabatic transport of parahydrogen addition products to high magnetic field. Chem. Phys. Lett. 145, 255–258 (1988). [Google Scholar]
- 13.Wang WY et al. , Pairwise stereoselective hydrogenation of propyne on supported Pd-Ag catalysts investigated by parahydrogen-induced polarization. J. Phys. Chem. C 125, 17144–17154 (2021). [Google Scholar]
- 14.Salnikov OG et al. , Heterogeneous 1H and 13C parahydrogen-Induced polarization of acetate and pyruvate esters. ChemPhysChem 22, 1389–1396 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pokochueva EV, Burueva DB, Salnikov OG, Koptyug IV, Heterogeneous catalysis and parahydrogen-induced polarization. ChemPhysChem 22, 1421–1440 (2021). [DOI] [PubMed] [Google Scholar]
- 16.Adams RW et al. , Reversible interactions with para-hydrogen enhance NMR sensitivity by polarization transfer. Science 323, 1708–1711 (2009). [DOI] [PubMed] [Google Scholar]
- 17.Duckett SB, Mewis RE, Application of Parahydrogen Induced Polarization Techniques in NMR Spectroscopy and Imaging. Acc. Chem. Res. 45, 1247–1257 (2012). [DOI] [PubMed] [Google Scholar]
- 18.Pravdivtsev AN, Buntkowsky G, Duckett SB, Koptyug IV, Hovener JB, Parahydrogen-induced polarization of amino acids. Angew. Chem. 60, 23496–23507 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Taylor DA, Natrajan LS, Nilsson M, Adams RW, SABRE-enhanced real-time pure shift NMR spectroscopy. Magn. Reson. Chem. 59, 1244–1252 (2021). [DOI] [PubMed] [Google Scholar]
- 20.Spiridonov KA et al. , Phosphite-containing iridium polarization transfer catalysts for NMR signal amplification by reversible exchange. Mendeleev Commun. 31, 475–477 (2021). [Google Scholar]
- 21.Kiryutin AS, Yurkovskaya AV, Petrov PA, Ivanov KL, Simultaneous 15N polarization of several biocompatible substrates in ethanol-water mixtures by signal amplification by reversible exchange (SABRE) method. Magn. Reson. Chem. 59, 1216–1224 (2021). [DOI] [PubMed] [Google Scholar]
- 22.Barskiy DA et al. , The Feasibility of Formation and Kinetics of NMR Signal Amplification by Reversible Exchange (SABRE) at High Magnetic Field (9.4 T). J. Am. Chem. Soc. 136, 3322–3325 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Barskiy DA et al. , In Situ and Ex Situ Low-Field NMR Spectroscopy and MRI Endowed by SABRE Hyperpolarization. ChemPhysChem 15, 4100–4107 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Feng BB, Coffey AM, Colon RD, Chekmenev EY, Waddell KW, A pulsed injection parahydrogen generator and techniques for quantifying enrichment. J. Magn. Reson. 214, 258–262 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shchepin RV et al. , N-15 Hyperpolarization of Imidazole-N-15(2) for Magnetic Resonance pH Sensing via SABRE-SHEATH. Acs Sensors 1, 640–644 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shchepin RV et al. , Hyperpolarization of “Neat” Liquids by NMR Signal Amplification by Reversible Exchange. Journal of Physical Chemistry Letters 6, 1961–1967 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shi F, Coffey AM, Waddell KW, Chekmenev EY, Goodson BM, Heterogeneous Solution NMR Signal Amplification by Reversible Exchange. Angewandte Chemie-International Edition 53, 7495–7498 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shi F, Coffey AM, Waddell KW, Chekmenev EY, Goodson BM, Nanoscale Catalysts for NMR Signal Enhancement by Reversible Exchange. J. Phys. Chem. C 119, 7525–7533 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shi F et al. , Aqueous NMR Signal Enhancement by Reversible Exchange in a Single Step Using Water-Soluble Catalysts. J. Phys. Chem. C 120, 12149–12156 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Theis T, Truong M, Coffey AM, Chekmenev EY, Warren WS, LIGHT-SABRE enables efficient in-magnet catalytic hyperpolarization. J. Magn. Reson. 248, 23–26 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Theis T et al. , Microtesla SABRE Enables 10% Nitrogen-15 Nuclear Spin Polarization. J. Am. Chem. Soc. 137, 1404–1407 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Truong ML et al. , Irreversible Catalyst Activation Enables Hyperpolarization and Water Solubility for NMR Signal Amplification by Reversible Exchange. J. Phys. Chem. B 118, 13882–13889 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Truong ML et al. , N-15 Hyperpolarization by Reversible Exchange Using SABRE-SHEATH. J. Phys. Chem. C 119, 8786–8797 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bargon J, Fischer H, Johnsen U, Kernresonanz-emissionslinien während rascher radikalreaktionen. Zeitschrift für Naturforschung A 22, 1551–1555 (1967). [Google Scholar]
- 35.Ward HR, Lawler RG, Nuclear magnetic resonance emission and enhanced absorption in rapid organometallic reactions. J. Am. Chem. Soc. 89, 5518–5519 (1967). [Google Scholar]
- 36.Kaptein R, Dijkstra K, Nicolay K, Laser photo-CIDNP as a surface probe for proteins in solution. Nature 274, 293–294 (1978). [DOI] [PubMed] [Google Scholar]
- 37.Lee JH, Cavagnero S, A novel tri-enzyme system in combination with laser-driven NMR enables efficient nuclear polarization of biomolecules in solution. J. Phys. Chem. B 117, 6069–6081 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lee JH, Sekhar A, Cavagnero S, 1H-Detected 13C photo-CIDNP as a sensitivity enhancement tool in solution NMR. J. Am. Chem. Soc. 133, 8062–8065 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lyon CE, Jones JA, Redfield C, Dobson CM, Hore PJ, Two-dimensional 15N-1H photo-CIDNP as a surface probe of native and partially structured proteins. J. Am. Chem. Soc. 121, 6505–6506 (1999). [Google Scholar]
- 40.Okuno Y, Cavagnero S, Fluorescein: a photo-CIDNP sensitizer enabling hypersensitive NMR data collection in liquids at low micromolar concentration. J. Phys. Chem. B 120, 715–723 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Okuno Y et al. , Laser- and cryogenic probe-assisted NMR enables hypersensitive analysis of biomolecules at submicromolar concentration. Proc. Natl. Acad. Sci. USA 116, 11602–11611 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yang H, Hofstetter H, Cavagnero S, Fast-pulsing LED-enhanced NMR: A convenient and inexpensive approach to increase NMR sensitivity. J. Chem. Phys. 151, 245102 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yang H, Mecha MF, Goebel CP, Cavagnero S, Enhanced nuclear-spin hyperpolarization of amino acids and proteins via reductive radical quenchers. J. Magn. Reson. 324, 106912 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kaptein R, Dijkstra K, Müller F, Van Schagen CG, Visser AJWG, 360-MHz laser-induced phot-CIDNP in photoreaction of flavins. J. Magn. Reson. 31, 171–176 (1978). [Google Scholar]
- 45.Hore PJ, Broadhurst RW, Photo-CIDNP of biopolymers. Prog. Nucl. Magn. Reson. Spectrosc. 25, 345–402 (1993). [Google Scholar]
- 46.Lindqvist L, A flash photolysis study of fluorescein. Arkiv for Kemi 16, 79–138 (1961). [Google Scholar]
- 47.Kiryutin AS, Morozova OB, Kuhn LT, Yurkovskaya AV, Hore PJ, 1H and 13C Hyperfine coupling constants of the tryptophanyl cation radical in aqueous solution from microsecond time-resolved CIDNP. J. Phys. Chem. B 111, 11221–11227 (2007). [DOI] [PubMed] [Google Scholar]
- 48.Spera S, Bax A, Empirical correlation between protein backbone conformation and Cα. and Cβ. 13C nuclear magnetic resonance chemical shifts. J. Am. Chem. Soc. 113, 5490–5492 (1991). [Google Scholar]
- 49.Adrian FJ, Singlet-triplet splitting in diffusing radical pairs and magnitude of chemically induced electron-spin polarization. J. Chem. Phys. 57, 5107 (1972). [Google Scholar]
- 50.Ivanov KL, Pravdivtsev AN, Yurkovskaya AV, Vieth HM, Kaptein R, The role of level anti-crossings in nuclear spin hyperpolarization. Prog. Nucl. Magn. Reson. Spectrosc. 81, 1–36 (2014). [DOI] [PubMed] [Google Scholar]
- 51.Lyon CE, Lopez JJ, Cho BM, Hore PJ, Low field CIDNP of amino acids and proteins: characterization of transient radicals and NMR sensitivity enhancement. Mol. Phys. 100, 1261–1269 (2002). [Google Scholar]
- 52.Okuno Y, Cavagnero S, Photochemically induced dynamic nuclear polarization: basic principles and applications. eMagRes 6, 283–313 (2017). [Google Scholar]
- 53.Janssen GJ et al. , Observation of the solid-state photo-CIDNP effect in entire cells of cyanobacteria synechocystis. Photosynth. Res. 104, 275–282 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Eisenreich W et al. , Strategy for enhancement of 13C-photo-CIDNP NMR spectra by exploiting fractional 13C-labeling of tryptophan. J. Phys. Chem. B 119, 13934–13943 (2015). [DOI] [PubMed] [Google Scholar]
- 55.Thompson CM, McDonald AD, Yang H, Cavagnero S, Buller AR, Modular control of L-tryptophan isotopic substitution via an efficient biosynthetic cascade. Org. Biomol. Chem. 18, 4189–4192 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ernst RR, Bodenhausen G, Wokaun A, Principles of nuclear magnetic resonance in one and two dimensions (Oxford University Press, New York, 1989). [Google Scholar]
- 57.Cavanagh J, Fairbrother W, Palmer AI, R. M, NJ S, Protein NMR Spectroscopy Principles and Practice (Elsevier Academic Press, ed. 2nd, 2007). [Google Scholar]
- 58.Bakke CK, Jungbauer LM, Cavagnero S, In vitro expression and characterization of native apomyoglobin under low molecular crowding conditions. Prot. Eng. Expr. Purif. 45, 381–392 (2006). [DOI] [PubMed] [Google Scholar]
- 59.Ragavan M, Iconaru LI, Park C-G, Kriwacki RW, Hilty C, Real-time analysis of folding upon binding of a disordered protein by using dissolution DNP NMR spectroscopy. Angew. Chem. 56, 7070–7073 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kim J, Mandal R, Hilty C, Characterization of Membrane Protein-Lipid Interactions in Unfolded OmpX with Enhanced Time Resolution by Hyperpolarized NMR. Chembiochem 21, 2861–2867 (2020). [DOI] [PubMed] [Google Scholar]
- 61.Kim Y, Liu MX, Hilty C, Parallelized Ligand Screening Using Dissolution Dynamic Nuclear Polarization. Anal. Chem. 88, 11178–11183 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kim Y, Liu MX, Hilty C, Determination of binding affinities using hyperpolarized NMR with simultaneous 4-channel detection. J. Magn. Reson. 295, 80–86 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kim Y, Wang YY, Chen HY, Hilty C, “In-Vitro Dissolution Dynamic Nuclear Polarization for Sensitivity Enhancement of NMR with Biological Molecules” in Protein NMR: Methods and Protocols, Ghose R, Ed. (2018), vol. 1688, pp. 155–168. [DOI] [PubMed] [Google Scholar]
- 64.Liu MX, Kim Y, Hilty C, Characterization of Chemical Exchange Using Relaxation Dispersion of Hyperpolarized Nuclear Spins. Anal. Chem. 89, 9154–9158 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wang YY, Kim J, Hilty C, Determination of protein-ligand binding modes using fast multi-dimensional NMR with hyperpolarization. Chemical Science 11, 5935–5943 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kouřil K, Kouřilová H, Bartram S, Levitt MH, Meier B, Scalable dissolution-dynamic nuclear polarization with rapid transfer of a polarized solid. Nature Comm. 10, 1733 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kaltschnee L et al. , Hyperpolarization of amino acids in water utilizing parahydrogen on a rhodium nanocatalyst. Chem. Eur. J. 25, 11031–11035 (2019). [DOI] [PubMed] [Google Scholar]
- 68.Sellies L, Aspers R, Feiters MC, Rutjes F, Tessari M, Parahydrogen hyperpolarization allows direct NMR detection of alpha-amino acids in complex (bio)mixtures. Angew. Chem. 60, 26954–26959 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kositz C et al. , High serum tryptophan concentration in pollinosis patients is associated with unresponsiveness to pollen extract therapy. Int. Arch. All. Immun. 147, 35–40 (2008). [DOI] [PubMed] [Google Scholar]
- 70.Menta R et al. , Tryptophan concentration is the main mediator of the capacity of adipose mesenchymal stromal cells to inhibit T-lymphocyte proliferation in vitro. Cytother. 16, 1679–1691 (2014). [DOI] [PubMed] [Google Scholar]
- 71.Weinlich G, Murr C, Richardsen L, Winkler C, Fuchs D, Decreased serum tryptophan concentration predicts poor prognosis in malignant melanoma patients. Dermatol. 214, 8–14 (2007). [DOI] [PubMed] [Google Scholar]
- 72.Agus A, Planchais J, Sokol H, Gut Microbiota Regulation of Tryptophan Metabolism in Health and Disease. Cell Host & Microbe 23, 716–724 (2018). [DOI] [PubMed] [Google Scholar]
- 73.Bosi A, Banfi D, Bistoletti M, Giaroni C, Baj A, Tryptophan Metabolites Along the Microbiota-Gut-Brain Axis: An Interkingdom Communication System Influencing the Gut in Health and Disease. Int. J. Tryp. Res. 13, 1–25 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Gao K, Mu CL, Farzi A, Zhu WY, Tryptophan Metabolism: A Link Between the Gut Microbiota and Brain. Adv. Nutr. 11, 709–723 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Gostner JM et al. , Tryptophan Metabolism and Related Pathways in Psychoneuroimmunology: The Impact of Nutrition and Lifestyle. Neuropsychobiol. 79, 89–99 (2020). [DOI] [PubMed] [Google Scholar]
- 76.Kokturk O, Kanbay A, “Tryptophan Metabolism and Sleep” in Tryptophan Metabolism: Implications for Biological Processes, Health and Disease, Engin A, Engin AB, Eds. (2015), pp. 239–252. [Google Scholar]
- 77.Konopelski P, Ufnal M, Indoles - Gut Bacteria Metabolites of Tryptophan with Pharmacotherapeutic Potential. Curr. Drug Met. 19, 883–890 (2018). [DOI] [PubMed] [Google Scholar]
- 78.Roth W, Zadeh K, Vekariya R, Ge Y, Mohamadzadeh M, Tryptophan Metabolism and Gut-Brain Homeostasis. International Journal of Molecular Sciences 22, 1877–2013 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Broos J, “Biosynthetic incorporation of tryptophan analogs in proteins” in Fluorescence spectroscopy and microscopy - methods and protocols, Engelborghs Y, Visser AJWG, Eds. (Humana Press, New York, 2014), chap. 15, pp. 359–370. [DOI] [PubMed] [Google Scholar]
- 80.Pordes R et al. (2007) The open science grid. in 3rd Annual Scientific Discovery through Advanced Computing Conference (SciDAC 2007) (Boston, MA). [Google Scholar]
- 81.Sfiligoi I et al. , The Pilot Way to Grid Resources Using glideinWMS. WRI World Congress on Computer Science and Information Engineering 2, 428–432 (2009). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


