Abstract
Background/Aims
We aim to evaluate the differences in the microbiome of responders and non-responders, as well as predict the response to probiotic therapy, based on fecal microbiome data in patients with diarrhea-predominant irritable bowel syndrome (IBS-D).
Methods
A multi-strain probiotics that contains Lactobacillus acidophilus (KCTC 11906BP), Lactobacillus plantarum (KCTC11867BP), Lactobacillus rhamnosus (KCTC 11868BP), Bifidobacterium breve (KCTC 11858BP), Bifidobacterium lactis (KCTC 11903BP), Bifidobacterium longum (KCTC 11860BP), and Streptococcus thermophilus (KCTC 11870BP) were used. Patients were categorized into probiotic and placebo groups, and fecal samples were collected from all patients before and at the end of 8 weeks of treatment. The probiotic group was further divided into responders and non-responders. Responders were defined as patients who experienced adequate relief of overall irritable bowel syndrome symptoms after probiotic therapy. Fecal microbiota were investigated using Illumina MiSeq and analyzed using the EzBioCloud 16S database and microbiome pipeline (https://www.EZbiocloud.net).
Results
There was no significant difference in the alpha and beta diversity between the responder and non-responder groups. The abundances of the phylum Proteobacteria and genus Bacteroides significantly decreased after probiotic treatment. Bifidobacterium bifidum, Pediococcus acidilactici, and Enterococcus faecium showed a significantly higher abundance in the probiotic group after treatment compared to the placebo group. Enterococcus faecalis and Lactococcus lactis were identified as biomarkers of non-response to probiotics. The abundance of Fusicatenibacter saccharivorans significantly increased in the responders after treatment.
Conclusions
Probiotic treatment changes some composition of fecal bacteria in patients with IBS-D. E. faecalis and L. lactis may be prediction biomarkers for non-response to probiotics. Increased abundance of F. sccharivorans is correlated to symptom improvement by probiotics in patients with IBS-D.
Keywords: Biomarkers, Irritable bowel syndrome, Microbiota, Prediction, Probiotics
Introduction
Irritable bowel syndrome (IBS) is a functional gastrointestinal disorder characterized by recurrent abdominal pain associated with defecation or a change in bowel habits.1 Approximately 5-10% of the population is affected by IBS, and its incidence has increased.2 The pathophysiology of IBS remains unknown. While various therapeutic options are available for alleviating the symptoms of IBS, the treatments are only partially effective. Patients with IBS have a poor quality of life and increased socioeconomic burdens.3,4
Increasing evidence indicates that dysbiosis, which is defined as an imbalance in the gut microbiome, plays a key role in the development of IBS. Differences in the composition of the gut microbiota have been identified between patients with IBS and healthy controls.5 In particular, patients with IBS exhibited decreased microbiome diversity and significant changes in several specific bacterial taxa compared to healthy controls.6,7 Studies that compared the gut microbiota among the IBS subtypes suggest that the microbiome may be associated with patient symptoms and disease severity.7-9 As the relationship between dysbiosis and the development of IBS gains increasing attention, therapeutic approaches targeted at the intestinal microbiota, such as dietary manipulation and antibiotic and probiotic use, are being widely investigated.10
Probiotics, defined as live microorganisms that confer health benefits to the host,11 have been extensively studied. Several studies have demonstrated the beneficial effects of probiotics in patients with IBS. Single and combination probiotic strains have been shown to reduce IBS symptoms.12-15 The possible mechanisms of action of probiotics include inhibition of pathogenic bacterial overgrowth, reduction in visceral hypersensitivity, production of short-chain fatty acids, and improvement of the gut barrier function.16,17 Despite these data, some patients with IBS still show insufficient responses to probiotic therapy. A number of randomized control trials involving patients with IBS have suggested that probiotics have no beneficial effects,18-20 whereas other studies have proposed that probiotics result in negative effects, such as abdominal pain and bloating.21,22 Thus, it is unclear whether the common probiotic strategy should be prescribed to all patients with IBS. In our previous study, we demonstrated the therapeutic effect of a probiotic mixture in patients with diarrhea-predominant IBS (IBS-D).23 In particular, 8 weeks of treatment with the probiotic mixture improved overall IBS symptoms and stool consistency. To better understand the association between probiotic therapy and treatment outcomes, we analyzed the fecal samples collected from this study. We aim to examine the differences in the microbiome based on the overall response to probiotic therapy and identify any microbial biomarkers that can predict treatment outcomes.
Materials and Methods
Study Protocol
We conducted this exploratory post hoc analysis to investigate the association between intestinal microbiome and treatment outcomes to probiotics therapy.23 A randomized, double-blind, placebo-controlled clinical trial was conducted. A short questionnaire designed to assess daily IBS symptoms was piloted during a screening period with a 1-week run-in period. Patients who had pain/discomfort for at least 2 days were included in the study according to the Design of Treatment Trials for Functional Gastrointestinal Disorders recommendations.24 After completing the screening period, the inclusion and exclusion criteria were re-evaluated, and eligible patients were randomized to receive the probiotic mixture or placebo. Randomization was performed by selecting a card from a set of identical cards from the study coordinator. All other investigators were fully blinded to the randomization until the completion of study.
Duolac7 (Cell Biotech, Co, Ltd, Seoul, Korea) is a multiple-species probiotic combination that contains 7 species of probiotic bacteria, including Lactobacillus acidophilus (KCTC 11906BP), Lactobacillus plantarum (KCTC11867BP), Lactobacillus rhamnosus (KCTC 11868BP), Bifidobacterium breve (KCTC 11858BP), Bifidobacterium lactis (KCTC 11903BP), Bifidobacterium longum (KCTC 11860BP), and Streptococcus thermophilus (KCTC 11870BP). Each capsule contains a total of 5 billion bacteria, of which 7 strains were evenly contained (7 × 108 viable cells for each strain). Placebo capsules contained excipients alone and had identical appearance, color, taste, and consistency with Duolac7 (Cell Biotech, Co).
Patients were administered oral probiotics or placebo twice a day for 8 weeks and were followed up for another 2 weeks. Over 10 weeks, patients recorded daily symptoms including abdominal pain or discomfort, urgency, bloating, passage of gas, and stool frequency and consistency using self-administered questionnaires. They were also asked weekly if their overall IBS symptoms were improved or not. They visited the clinic for an assessment of symptoms and complications every 4 weeks after the start of treatment. Adequate relief of overall IBS symptoms was assessed weekly using an interactive voice response by telephone, and responders were defined as patients who experienced adequate relief of overall IBS symptoms at least 50% of the weeks during 10-week study period. The study protocol was approved by the ethics review committee of the Chung-Ang University Hospital (C2008032 [1350]) and performed in accordance with the 1964 Declaration of Helsinki and its later amendments.
Study Population
As previously described, patients aged between 18 years and 65 years who were diagnosed with IBS-D based on the Rome III criteria were included from a university hospital.23 To exclude patients with organic abnormalities, laboratory tests such as a complete blood cell count, blood chemistry, and colonoscopy were performed during the screening period. Patients with the following clinical features were excluded: pregnancy or lactation during the study period; abnormal screening laboratory test results; severe systemic illness, such as liver disease, cardiovascular disease, renal disease, endocrine disorders, neurologic disorders, or malignant tumors; history of psychiatric disorders; previous surgery except appendectomy and abdominal wall hernia repair; and use of drugs that influence the efficacy of intestinal microbiota. In addition, patients who were judged to be ineligible by the investigators were excluded from this study. A written informed content was obtained from each patient prior to the commencement of the study.
Fecal Sample Collection
All participants provided fecal samples. Fecal samples were collected in sterile containers before and after the eighth week of treatment for microbial analyses. They were transferred to refrigerated containers in the laboratory within 12 hours of collection. We extracted the DNA from the samples and stored them at –80°C for further analysis.
Fecal Microbiota Analysis
DNA was extracted from the feces using the FastDNA SPIN kit for bacterial DNA (MP Biomedicals, Irvine, CA, USA) according to the manufacturer’s instructions. Primers targeting the V3 to V4 region of the bacterial 16S ribosomal RNA gene were used for polymerase chain reaction (PCR). The primers 341F (5’-TCGTCGGCAGCGTC-AGATGTGTATAAGAGACAG-CCTACGGGNGGCWGCAG-3’) and 805R (5’-GTCTCGTGGGCTC GG-AGATGTGTATAAGAGACAGGACTACHVGGGTATCTAATCC-3’) were used for bacterial amplification. Initial denaturation was performed at 95°C for 3 minutes, followed by 25 cycles of denaturation at 95°C for 30 seconds, primer annealing at 55°C for 30 seconds, and extension at 72°C for 30 seconds. The final elongation was performed at 72°C for 5 minutes. An i5 forward primer (5’-AATGATACGGCGACCACCGAGATCTACAC-XXXXXXXX-CGTCGGCAGCGTC-3’; X indicates the barcode region) and i7 reverse primer (5’-CAAGCAGAAG ACGGCATACGAGAT-XXXXXXXX-GTCTCGTGGGCTCGG-3’) were used for secondary amplification by attaching the Illumina NexTera barcode.
The PCR product was confirmed through 1% agarose gel electrophoresis and visualized under a Gel Doc system (BioRad, Hercules, CA, USA). Purification of the amplified products was performed with the CleanPCR (CleanNA, Waddinxveen, The Netherlands). Equal concentrations of purified products were pooled together, and short fragments (non-target products) were removed by CleanPCR (CleanNA). The quality and product size were assessed on a Bioanalyzer 2100 (Agilent, Palo Alto, CA, USA) using a DNA 7500 chip. Mixed amplicons were pooled, and sequencing was performed at ChunLab, Inc (Seoul, Korea) with the Illumina MiSeq Sequencing system (Illumina, San Diego, CA, USA) according to the manufacturer’s instructions.
Statistical Methods
As we mentioned, this study was conducted using fecal samples collected in the previous clinical trial. The sample size calculated for this study was based on the intent to detect a 25% difference in the proportion of responders between the 2 groups with 80% power at a = 0.05 while compensating for just over a 20% drop out rate.
The output data from the Illumina MiSeq sequencing system were analyzed with the EzBioCloud 16S database (ChunLab Inc)25 and 16S microbiome pipeline (ChunLab Inc, EzBioCloud 16S-based MTP app, https://www.EZbiocloud.net) for data processing, statistical analysis, and data graphing. The Chao1 estimation and Shannon diversity index were used to evaluate the richness and evenness of the samples. The overall phylogenetic distance among the groups was estimated using Bray-Curtis dissimilarity and visualized through principal coordinate analysis.
The group differences in alpha and beta diversity were tested using the Wilcoxon rank-sum test and permutational multivariate analysis of variance (PERMANOVA) with 9999 permutations, respectively. For specific taxa, the differences in the relative abundance between the groups were compared using the Kruskal-Wallis test.
Differential abundance analyses were performed using DESeq226 v1.22.2 (Bioconductor, Buffalo, NY, USA) with default parameters to investigate the enriched taxa in the placebo and probiotic groups after treatment. The taxa were considered significant if the P-value after multiple test correction was < 0.05. The potential biomarkers for probiotic response were identified with the linear discriminant analysis effect size (LEfSe)27 algorithm, which determines the features most likely to explain differences between 2 groups by coupling standard statistical tests with biological consistency and effect relevance. It was performed with default parameters. Only the taxa with logarithmic linear discriminant analysis scores > 2.0 were reported.
Results
Baseline Characteristics of Patients With Diarrhea-predominant Irritable Bowel Syndrome
As previously described, a total of 50 patients with IBS-D were enrolled and randomized into the probiotic (n = 25) and placebo (n = 25) groups.23 The proportion of responders was significantly higher in the probiotics group than in the placebo group, but change of individual symptoms were similar in the 2 groups. One and 4 patients in the probiotic and placebo groups, respectively, were excluded from this study because their fecal samples were not collected before and after treatment. In addition, the fecal samples of 2 and 3 patients in the probiotic and placebo groups, respectively, were not collected after treatment. Prior to the treatment, we analyzed the fecal samples of 24 and 21 patients in the probiotic and placebo groups, respectively. The probiotic group comprised 12 responders and 12 non-responders. After the treatment, we analyzed the fecal samples of 22 and 18 patients in the probiotic and placebo groups, respectively. The probiotic group consisted of 12 responders and 10 non-responders. The characteristics of the patients enrolled in this study are summarized in Tables 1 and 2. Patients were relatively young and had no underlying diseases. There were no significant differences in age, sex, and body mass index between the groups.
Table 1.
Baseline Characteristics of the Patients With Irritable Bowel Syndrome
| Characteristics | Probiotic group (n = 25) | Placebo group (n = 25) | P-value |
|---|---|---|---|
| Age (yr) | 37.9 ± 12.4 | 40.3 ± 11.2 | 0.492 |
| Sex | 0.156 | ||
| Male | 12 (48) | 14 (56) | |
| Female | 13 (52) | 11 (44) | |
| BMI (kg/m2) | 23 ± 3.3 | 22.9 ± 2.9 | 0.916 |
| Smoker | 2 (8) | 4 (16) | 0.416 |
| Alcohol intake | 6 (24) | 9 (36) | 0.359 |
BMI, body mass index.
Data are presented as mean ± SD or n (%).
Table 2.
Baseline Characteristics of the Responders and Non-responders to Probiotic Therapy
| Characteristics | Responders (n = 12) |
Non-responders (n = 12) |
P-value |
|---|---|---|---|
| Age (yr) | 40.4 ± 13.5 | 33.3 ± 10.9 | 0.181 |
| Sex | 1.000 | ||
| Male | 8 (66.7) | 8 (66.7) | |
| Female | 4 (33.3) | 4 (33.3) | |
| BMI (kg/m2) | 23.4 ± 3.9 | 22.6 ± 2.7 | 0.559 |
| Smoker | 0 (0.0) | 2 (16.7) | 0.140 |
| Alcohol intake | 4 (33.3) | 2 (16.7) | 0.346 |
Data are presented as mean ± SD or n (%).
Effect of Probiotic Treatment on Microbial Diversity
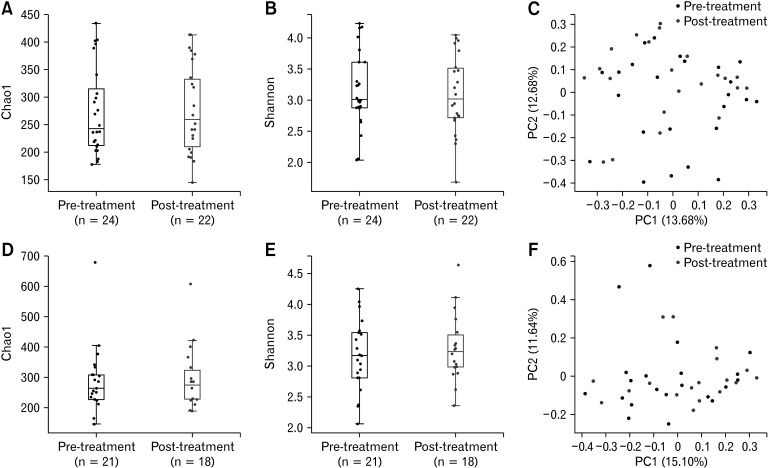
We utilized microbiome taxonomic profiles and diversity indices to determine whether the probiotic or placebo treatments resulted in changes to the diversity and composition of the gut microbiota. No significant differences in alpha diversity (Chao1, P = 0.878; Shannon, P = 0.644) were observed in the probiotic group (Fig. 1A and 1B). The principal coordinate analysis plot of the pre- and post-treatment samples demonstrated few variations (Fig. 1C). The PERMANOVA also suggested that there were no significant differences after probiotic treatment (P = 0.329). The P values of the Wilcoxon rank-sum test for the Chao1 and Shannon indices and PERMANOVA were 0.955, 0.632, and 0.686, respectively, which indicated that there were no significant differences in the diversities of the placebo group (Fig. 1D-F).
Figure 1.
Alpha and beta diversity data of the probiotic and placebo groups. The plots depict the differences in the diversity of the pre- and post-treatment samples in the (A-C) probiotic and (D-F) placebo groups. The boxplots represent the alpha diversity using the (A, D) Chao1 and (B, E) Shannon indices, and (C, F) the principal coordinate analysis plots demonstrate the beta diversity based on the Bray-Curtis dissimilarity. No significant differences in alpha diversity (Chao1, P = 0.878; Shannon, P = 0.644) were observed in the probiotic group (A, B). The permutational multivariate analysis of variance (PERMANOVA) showed that there were no significant differences after probiotic treatment (P = 0.329, C). P-values of the Wilcoxon rank-sum test for the Chao1 and Shannon indices and PERMANOVA were 0.955, 0.632, and 0.686, respectively, in the placebo group (D-F). PC, principle component.
Effect of Probiotic Treatment on Microbial Taxonomic Composition
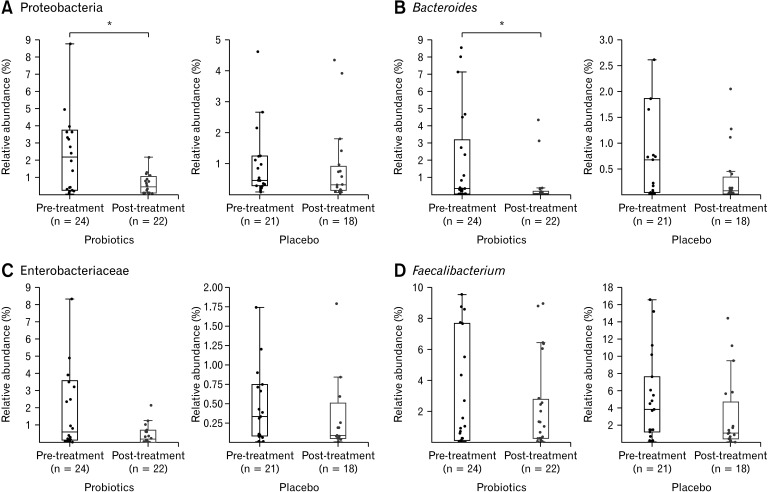
We examined the specific taxa previously associated with IBS and assessed whether their abundances changed after probiotic treatment.28 The phylum Proteobacteria (median relative abundance in the probiotics and placebo group at baseline: 1.98% vs 0.44%, P = 0.285) and genus Bacteroides (0.31% vs 0.53%, P = 0.633), which are known to be abundant in patients with IBS, significantly decreased in abundance in the probiotic group (P = 0.022 and P = 0.003, respectively), whereas no significant differences were noted in the placebo group (P = 0.215 and P = 0.063, respectively) (Fig. 2A and 2B) after treatment. In contrast, the family Enterobacteriaceae and genus Faecalibacterium, which reportedly exhibit increased and decreased abundance, respectively, in patients with IBS, showed no significant differences between the probiotic (P = 0.062 and P = 0.186, respectively) and placebo (P = 0.775 and P = 0.143, respectively) groups (Fig. 2C and 2D).
Figure 2.
Relative abundance of the taxa related to irritable bowel syndrome (IBS). The boxplots represent the relative abundance of (A) Proteobacteria, (B) Bacteroides, (C) Enterobacteriaceae, and (D) Faecalibacterium in the pre- and post-treatment samples. The abundances in the probiotic and placebo groups are shown on the left and right plots of each panel, respectively. Asterisks (*) indicate significant differences between the pre- and post-treatment samples. The phylum Proteobacteria (A) and genus Bacteroides (B) which are known to be abundant in patients with IBS, significantly decreased in abundance in the probiotic group (P = 0.022 and P = 0.003, respectively).
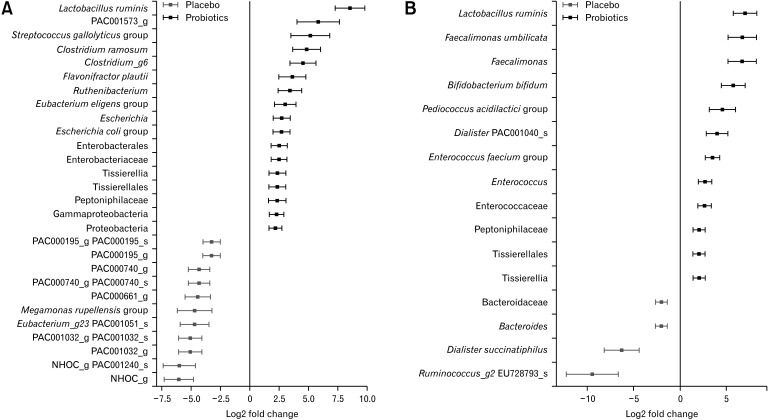
Differentially abundant taxa were identified by comparing the taxonomic profiles between the probiotic and placebo groups. Analysis of the pre-treatment fecal samples demonstrated that the probiotic group exhibited abundance of Lactobacillus ruminis, Streptococcus gallolyticus, Clostridium ramosum, Flavonifractor plautii, Eubacterium eligens, and Escherichia coli, whereas the placebo group exhibited abundance of Megamonas rupellensis (Fig. 3A). Among these species, only L. ruminis remained abundant in the probiotic group after treatment. In contrast, several taxa, including Faecalimonas umbilicata, Bifidobacterium bifidum, Pediococcus acidilactici, and Enterococcus faecium, which did not differ in abundance between the 2 groups in pre-treatment samples were abundant in the post-treatment fecal samples of the probiotic group (Fig. 3B). There were no significant differences in alpha and beta diversity between the 2 groups in pre-treatment samples (Supplementary Figure).
Figure 3.
Differentially abundant taxa in the probiotic and placebo groups. The taxa that were identified as differentially abundant by DESeq2 (A) at the baseline and (B) after probiotics treatment are listed. Several taxa, including Faecalimonas umbilicata, Bifidobacterium bifidum, Pediococcus acidilactici, and Enterococcus faecium, which did not differ in abundance between the 2 groups in pre-treatment samples (A) were abundant in the post-treatment fecal samples of the probiotic group (B).
Comparison Between the Responders and Non-responders to Probiotic Treatment
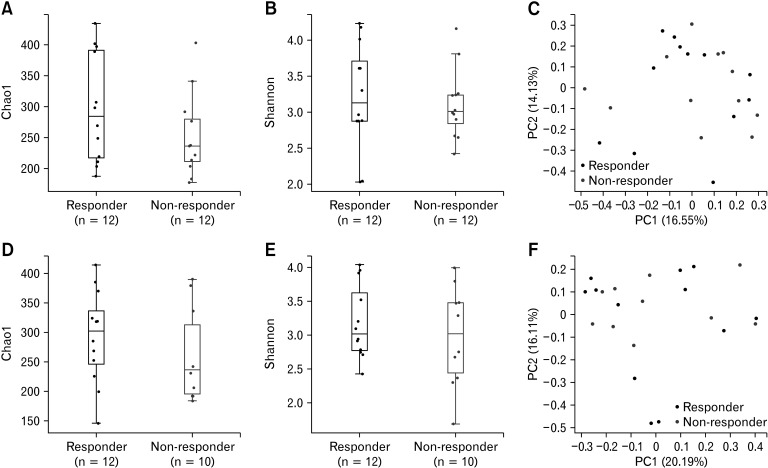
We examined the probiotic group to determine whether there were significant differences in the microbiomes of responders and non-responders. The Chao1 and Shannon indices did not demonstrate any significant differences between the responder and non-responder groups before (Fig. 4A and 4B) and after (Fig. 4D and 4E) probiotic treatment (P = 0.248, 0.729, 0.291, and 0.429). In particular, the PERMANOVA did not demonstrate any significant differences in beta diversity before and after treatment (P = 0.885 and P = 0.626, respectively) (Fig. 4C and 4F). Additionally, there were no significant differences in the alpha and beta diversity before and after probiotic treatment in the responder group (data not shown).
Figure 4.
Alpha and beta diversity of responders and non-responders. The microbial diversity of the (A-C) pre-treatment and (D-F) post-treatment samples are shown. The alpha diversity using the (A, D) Chao1 and (B, E) Shannon indices are depicted as boxplots, whereas (C, F) the beta diversity based on the Bray-Curtis dissimilarity are depicted as principal coordinate analysis plots. The Chao1 and Shannon indices did not demonstrate any significant differences between the responder and non-responder groups before (A, B) and after (D, E) probiotic treatment (P = 0.248, 0.729, 0.291, and 0.429). The permutational multivariate analysis of variance also did not demonstrate any significant differences in beta diversity before and after treatment (P = 0.885 and P = 0.626, respectively) (C, F). PC, principal component.
Potential Biomarkers Associated With the Response to Probiotics
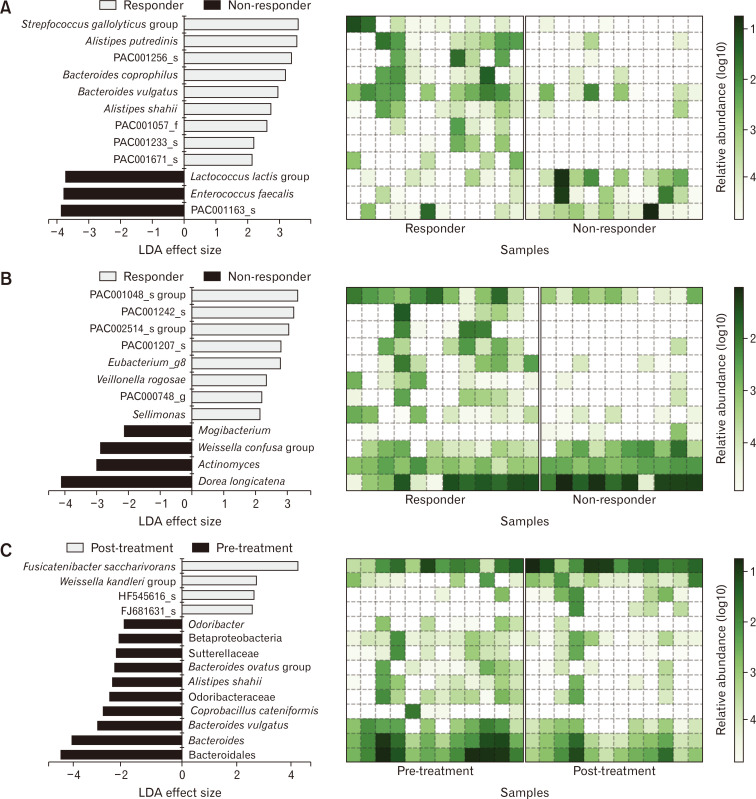
We utilized LEfSe algorithm to identify the potential biomarkers for the response to probiotic treatment. Analysis of the pre-treatment fecal samples identified S. gallolyticus, Alistipes putredinis, Bacteroides coprophilus, Bacteroides vulgatus, and Alistipes shahii as potential biomarkers for treatment response among the responders, whereas Lactococcus lactis and Enterococcus faecalis were the identified biomarkers among the non-responders (Fig. 5A). Analysis of the post-treatment fecal samples identified Vellonella rogosae as a biomarker among the responders and Dorea longicatena and Weissella confusa among the non-responders (Fig. 5B). The post-treatment samples of the responder group had a higher abundance of Fusicatenibacter saccharivorans and lower abundance of B. vulgatus, Coprobacillus cateniformis, A. shahii, and Bacteroides ovatus than the pre-treatment samples in the same group (Fig. 5C).
Figure 5.
Biomarker identification of responders to probiotic treatment. The potential biomarkers identified by the linear discriminant analysis (LDA) effect size algorithm are shown with heatmaps of the log-scaled relative abundance. Each plot represents biomarkers of the responders and non-responders in the (A) pre-treatment samples and (B) post-treatment samples and (C) biomarkers of the pre- and post-treatment samples in the responder group.
Discussion
While probiotics are widely used, its efficacy in patients with IBS remains in question, because studies have shown inconsistent results. Recent systematic reviews and meta-analyses did not provide definitive conclusions on the efficacy of probiotics for IBS.29,30
Our study aimed to determine the differences in the microbiomes of the responders and non-responders to probiotic treatment, as well as to elucidate how to predict the efficacy of probiotic therapy among patients with IBS.
Probiotic therapy did not significantly change the alpha and beta diversity, and the responder and non-responder groups demonstrated similar results. However, the relative abundance of some bacterial taxa, such as B. bifidum, P. acidilactici, and E. faecium, was significantly increased in the probiotic group compared to the placebo group. B. bifidum is a well-known and widely used probiotic. A recent study indicated that it has an anti-inflammatory effect.31 The increased abundance of B. bifidum in this study may be associated with the ingested probiotics strain. Some strains of P. acidilactici can be used as probiotics because they produce bacteriocins that have beneficial effects for the host.32-35 A randomized clinical trial demonstrated that a probiotic containing P. acidilactici improved IBS-related quality of life.36 While some safety issues remain, specific strains of E. faecium have been used as probiotics in the treatment and prevention of diarrhea or IBS. Probiotics was administered in a very small amount compared to the amount of fecal bacteria, thus it could not easily change the overall composition of fecal microbiota, but may change the abundance of some bacterial taxa into a beneficial direction in patients.
We did not identify significant differences in the relative abundance of B. bifidum, P. acidilactici, and E. faecium between the responders and non-responders; however, these species were significantly more abundant after probiotic treatment. They may have induced the response through other microbial modulatory pathway, rather than direct effect in the responder group, and it supports for the hypothesis that probiotics can modulate the gut microbial community of patients with IBS.
E. faecalis and L. lactis were identified as biomarkers that could predict non-response to probiotic treatment. Patients with IBS who did not respond to probiotic therapy demonstrated significantly higher abundances of E. faecalis and L. lactis before treatment. E. faecalis is known to have a controversial role as a commensal pathogenic bacterium in the human gut. Similar to E. faecium, specific strains of E. faecalis are used as probiotics;37 however, the poor safety profile and pathogenicity of the species is increasingly gaining attention. In addition, E. faecalis is predominantly identified in the mucosa of patients with IBD, including ulcerative colitis and Crohn’s disease. Research has proposed that the number of mucosal E. faecalis is correlated with disease activity.38,39 The proinflammatory effect of E. faecalis was identified in animal studies. E. faecalis induced inflammation in an IL-10−/− mice model.40,41 E. faecalis produces metalloprotease, which was suggested as the contributing factor that resulted in epithelial barrier dysfunction and chronic intestinal inflammation.42 The bacterial environment was shown to play a key role in E. faecalis gene expression modulation, which induces inflammatory activity.43 In contrast to E. faecalis, A. putredinis was identified as a biomarker that could predict a positive response to probiotic therapy. Alistipes is a recently isolated genus from the phylum Bacteroidetes. Although the role of this taxon in human health has not been fully understood, some species demonstrated anti-inflammatory activity in patients with liver disease. Alistipes has also demonstrated a protective role against colitis in an animal model.44-46 Low-grade intestinal inflammation is considered as one of the main mechanisms behind the development of IBS.47,48 Thus, these findings suggested that responsiveness to probiotics may be correlated with intestinal barrier function and inflammatory activity.
Dorea spp. are carbohydrate-dependent gastrointestinal bacteria that are responsible for producing much of the gas in the human intestine.49 Overproduction of gases may lead to abdominal discomfort, such as bloating. Increased bowel gas production also induces rapid colon transit, which results in diarrhea.50 Several studies have shown that Dorea spp. promote an inflammatory response by degrading intestinal mucin.51-53 Previous studies indicated that patients with IBS showed significantly higher abundance of Dorea spp.54 In our study, the non-responder group showed significantly higher abundance of D. longicatena than the responder group. It was unclear why D. longicatena was significantly more abundant in the non-responder group; however, this finding may help us understand how the microbiome interferes with the beneficial effect of probiotic therapy and results in persistent IBS symptoms. The patients who responded to probiotic therapy had a significantly increased abundance of F. saccharivorans in their post-treatment samples than their pre-treatment samples. F. saccharivorans belongs to Clostridium subcluster XIVa, which participates in maintaining the immune system homeostasis associated with regulatory T cells. F. saccharivorans produces short-chain fatty acids, such as lactic acid and acetic acid, through glucose fermentation,55 which are the key factors for improving intestinal barrier function and the host’s immune system. An animal model demonstrated that F. saccharivorans produce an anti-inflammatory effect by modulating cytokine production. In a mouse colitis model, administration of F. saccharivorans reduced the production of inflammatory cytokines, such as interleukin (IL)-13, and induced anti-inflammatory cytokines such as IL-10.56 The abundance of F. saccharivorans negatively correlated with disease activity in patients with ulcerative colitis.56 Our study results are consistent with these previous reports. Patients that responded to probiotic therapy may have experienced symptomatic improvement through anti-inflammatory effects. This finding has important implications for developing novel treatment targets against IBS.
One interesting finding was the change in the abundance of B. vulgatus, which decreased significantly after probiotic treatment in the responder group. B. vulgatus was identified as a biomarker that could predict the response to probiotic treatment. The role of B. vulgatus in the pathogenesis of IBD has been suggested.57,58 B. vulgatus has recently been shown to activate nuclear factor-κB in the epithelial enterocyte-like cell line;59 however, conflicting data exist about their relation with the inflammatory response.60,61 Further studies that account for these diverse roles need to be undertaken.
This study has several limitations. First, the relatively small sample size may be a potential for bias. Second, diet was not controlled, but the participants were recommended to continue their usual diet during the study period. Diet is considered as one of the main factors associated with symptom development and the gut microbiome. Third, it took a relatively long time from fecal sample collection to analysis. However, we extracted the DNA from the samples, and stored them at –80°C. Recent study showed that long-term storage of human fecal microbiota samples at –80°C has only limited effect on the microbial community.62 These results therefore need to be interpreted with caution. Further studies with larger sample sizes and that take lifestyle considerations, including diet, into account are required to validate our findings. To the best of our knowledge, only a few studies have evaluated the differences in the microbiome of the responders and non-responders to probiotic treatment. Moreover, only a few studies have examined the taxa that may predict the response of patients with IBS. We demonstrated significant changes in the abundance of several taxa among the responders and non-responders, as well as identified potential predictive biomarkers for probiotic treatment.
In conclusion, probiotics can alter bacterial composition in patients with IBS-D. E. faecalis and L. lactis can be considered as biomarkers for non-response to probiotic treatment among patients with IBS-D. The efficacy of probiotic treatments may be improved by reducing the abundance of D. longicatena and increasing that of F. saccharivorans. F. saccharivorans spp. can be one of the novel therapeutic targets for the treatment of IBS-D.
Supplementary Material
Note: To access the supplementary figure mentioned in this article, visit the online version of Journal of Neurogastroenterology and Motility at http://www.jnmjournal.org/, and at https://doi.org/10.5056/jnm21202.
Footnotes
Financial support: This work was supported by the Korean Society of Neurogastroenterology and Motility for 2018 under Grant KSNM 18-08, Biomedical Research Institute of Chung-Ang University Hospital for 2020 under Grant 20150921, and National Research Foundation of Korea under Grant/Award NRF-2020R1F1A1075489.
Conflicts of interest: None.
Author contributions: Seung Yong Shin, Sein Park, Jung Min Moon, Kisung Kim, Jeong Wook Kim, Jongsik Chun, Tae Hee Lee, and Chang Hwan Choi: contribution to the study conception and design, and final approval of the version to be published; Chang Hwan Choi: conception and design; Seung Yong Shin, Sein Park, Jung Min Moon, Kisung Kim, Jeong Wook Kim, and Jonsik Chun: analysis and interpretation of data; Seung Yong Shin and Sein Park: drafting the article; and Chang Hwan Choi: revising.
References
- 1.Mearin F, Lacy BE, Chang L, et al. Bowel Disorders. Gastroenterology Published Online First. 2016 Feb 18; doi: 10.1053/j.gastro.2016.02.031. doi:10.1053/j.gastro.2016.02.031. [DOI] [PubMed] [Google Scholar]
- 2.Canavan C, West J, Card T. The epidemiology of irritable bowel syndrome. Clin Epidemiol. 2014;6:71–80. doi: 10.2147/CLEP.S40245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Buono JL, Carson RT, Flores NM. Health-related quality of life, work productivity, and indirect costs among patients with irritable bowel syndrome with diarrhea. Health Qual Life Outcomes. 2017;15:35. doi: 10.1186/s12955-017-0611-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bouchoucha M, Devroede G, Girault-Lidvan N, Hejnar M, Mary F, Benamouzig R. Psychological profiles of irritable bowel syndrome patients with different phenotypes. Intest Res. 2020;18:459–468. doi: 10.5217/ir.2019.09171.8948808ab0d04f4bbeb1b255880549cd [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu HN, Wu H, Chen YZ, Chen YJ, Shen XZ, Liu TT. Altered molecular signature of intestinal microbiota in irritable bowel syndrome patients compared with healthy controls: a systematic review and meta-analysis. Dig Liver Dis. 2017;49:331–337. doi: 10.1016/j.dld.2017.01.142. [DOI] [PubMed] [Google Scholar]
- 6.Ringel Y, Carroll IM. Alterations in the intestinal microbiota and functional bowel symptoms. Gastrointest Endosc Clin N Am. 2009;19:141–150. vii. doi: 10.1016/j.giec.2008.12.004. [DOI] [PubMed] [Google Scholar]
- 7.Tap J, Derrien M, Törnblom H, et al. Identification of an intestinal microbiota signature associated with severity of irritable bowel syndrome. Gastroenterology. 2017;152:111–123. e118. doi: 10.1053/j.gastro.2016.09.049. [DOI] [PubMed] [Google Scholar]
- 8.Malinen E, Krogius-Kurikka L, Lyra A, et al. Association of symptoms with gastrointestinal microbiota in irritable bowel syndrome. World J Gastroenterol. 2010;16:4532–4540. doi: 10.3748/wjg.v16.i36.4532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhuang X, Tian Z, Li L, Zeng Z, Chen M, Xiong L. Fecal microbiota alterations associated with diarrhea-predominant irritable bowel syndrome. Front Microbiol. 2018;9:1600. doi: 10.3389/fmicb.2018.01600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Simrén M, Barbara G, Flint HJ, et al. Intestinal microbiota in functional bowel disorders: a Rome foundation report. Gut. 2013;62:159–176. doi: 10.1136/gutjnl-2012-302167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Perugorria MJ, Masyuk TV, Marin JJ, et al. Polycystic liver diseases: advanced insights into the molecular mechanisms. Nat Rev Gastroenterol Hepatol. 2014;11:750–761. doi: 10.1038/nrgastro.2014.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang Y, Li L, Guo C, et al. Effects of probiotic type, dose and treatment duration on irritable bowel syndrome diagnosed by Rome III criteria: a meta-analysis. BMC Gastroenterol. 2016;16:62. doi: 10.1186/s12876-016-0470-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Didari T, Mozaffari S, Nikfar S, Abdollahi M. Effectiveness of probiotics in irritable bowel syndrome: Updated systematic review with meta-analysis. World J Gastroenterol. 2015;21:3072–3084. doi: 10.3748/wjg.v21.i10.3072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yuan F, Ni H, Asche CV, Kim M, Walayat S, Ren J. Efficacy of Bifidobacterium infantis 35624 in patients with irritable bowel syndrome: a meta-analysis. Curr Med Res Opin. 2017;33:1191–1197. doi: 10.1080/03007995.2017.1292230. [DOI] [PubMed] [Google Scholar]
- 15.Hungin APS, Mitchell CR, Whorwell P, et al. Systematic review: probiotics in the management of lower gastrointestinal symptoms - an updated evidence-based international consensus. Aliment Pharmacol Ther. 2018;47:1054–1070. doi: 10.1111/apt.14539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cremon C, Barbaro MR, Ventura M, Barbara G. Pre- and probiotic overview. Curr Opin Pharmacol. 2018;43:87–92. doi: 10.1016/j.coph.2018.08.010. [DOI] [PubMed] [Google Scholar]
- 17.Barbara G, Feinle-Bisset C, Ghoshal UC, et al. The intestinal microenvironment and functional gastrointestinal disorders. Gastroenterology Published Online First. 2016 Feb 16; doi: 10.1053/j.gastro.2016.02.028. doi:10.1053/j.gastro.2016.02.028. [DOI] [PubMed] [Google Scholar]
- 18.Lyra A, Hillilä M, Huttunen T, et al. Irritable bowel syndrome symptom severity improves equally with probiotic and placebo. World J Gastroenterol. 2016;22:10631–10642. doi: 10.3748/wjg.v22.i48.10631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pineton de Chambrun G, Neut C, Chau A, et al. A randomized clinical trial of Saccharomyces cerevisiae versus placebo in the irritable bowel syndrome. Dig Liver Dis. 2015;47:119–124. doi: 10.1016/j.dld.2014.11.007. [DOI] [PubMed] [Google Scholar]
- 20.Ludidi S, Jonkers DM, Koning CJ, et al. Randomized clinical trial on the effect of a multispecies probiotic on visceroperception in hypersensitive IBS patients. Neurogastroenterol Motil. 2014;26:705–714. doi: 10.1111/nmo.12320. [DOI] [PubMed] [Google Scholar]
- 21.Hod K, Sperber AD, Ron Y, et al. A double-blind, placebo-controlled study to assess the effect of a probiotic mixture on symptoms and inflammatory markers in women with diarrhea-predominant IBS. Neurogastroenterol Motil Published Online First. 2017 Mar 8; doi: 10.1111/nmo.13037. doi: 10.1111/nmo.13037. [DOI] [PubMed] [Google Scholar]
- 22.Cremon C, Guglielmetti S, Gargari G, et al. Effect of Lactobacillus paracasei CNCM I-1572 on symptoms, gut microbiota, short chain fatty acids, and immune activation in patients with irritable bowel syndrome: a pilot randomized clinical trial. United European Gastroenterol J. 2018;6:604–613. doi: 10.1177/2050640617736478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ki Cha B, Mun Jung S, Hwan Choi C, et al. The effect of a multispecies probiotic mixture on the symptoms and fecal microbiota in diarrhea-dominant irritable bowel syndrome: a randomized, double-blind, placebo-controlled trial. J Clin Gastroenterol. 2012;46:220–227. doi: 10.1097/MCG.0b013e31823712b1. [DOI] [PubMed] [Google Scholar]
- 24.Irvine EJ, Whitehead WE, Chey WD, et al. Design of treatment trials for functional gastrointestinal disorders. Gastroenterology. 2006;130:1538–1551. doi: 10.1053/j.gastro.2005.11.058. [DOI] [PubMed] [Google Scholar]
- 25.Yoon SH, Ha SM, Kwon S, et al. Introducing EzBioCloud: a taxonomically united database of 16S rRNA gene sequences and whole-genome assemblies. Int J Syst Evol Microbiol. 2017;67:1613–1617. doi: 10.1099/ijsem.0.001755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Segata N, Izard J, Waldron L, et al. Metagenomic biomarker discovery and explanation. Genome Biol. 2011;12:R60. doi: 10.1186/gb-2011-12-6-r60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pittayanon R, Lau JT, Yuan Y, et al. Gut microbiota in patients with irritable bowel syndrome-a systematic review. Gastroenterology. 2019;157:97–108. doi: 10.1053/j.gastro.2019.03.049. [DOI] [PubMed] [Google Scholar]
- 29.Ford AC, Harris LA, Lacy BE, Quigley EMM, Moayyedi P. Systematic review with meta-analysis: the efficacy of prebiotics, probiotics, synbiotics and antibiotics in irritable bowel syndrome. Aliment Pharmacol Ther. 2018;48:1044–1060. doi: 10.1111/apt.15001. [DOI] [PubMed] [Google Scholar]
- 30.Dale HF, Rasmussen SH, Asiller Ö, Lied GA. Probiotics in irritable bowel syndrome: an up-to-date systematic review. Nutrients. 2019;11:2048. doi: 10.3390/nu11092048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ojima MN, Gotoh A, Takada H, et al. Bifidobacterium bifidum suppresses gut inflammation caused by repeated antibiotic disturbance without recovering gut microbiome diversity in mice. Front Microbiol. 2020;11:1349. doi: 10.3389/fmicb.2020.01349.1eaf8c2bd65c4901b3c09fdcbe01447c [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bhagat D, Raina N, Kumar A, et al. Probiotic properties of a phytase producing Pediococcus acidilactici strain SMVDUDB2 isolated from traditional fermented cheese product, Kalarei. Sci Rep. 2020;10:1926. doi: 10.1038/s41598-020-58676-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cizeikiene D, Juodeikiene G, Bartkiene E, Damasius J, Paskevicius A. Phytase activity of lactic acid bacteria and their impact on the solubility of minerals from wholemeal wheat bread. Int J Food Sci Nutr. 2015;66:736–742. doi: 10.3109/09637486.2015.1088939. [DOI] [PubMed] [Google Scholar]
- 34.Callejón S, Sendra R, Ferrer S, Pardo I. Recombinant laccase from Pediococcus acidilactici CECT 5930 with ability to degrade tyramine. PLoS One. 2017;12:e0186019. doi: 10.1371/journal.pone.0186019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yang SC, Lin CH, Sung CT, Fang JY. Antibacterial activities of bacteriocins: application in foods and pharmaceuticals. Front Microbiol. 2014;5:241. doi: 10.3389/fmicb.2014.00241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lorenzo-Zúñiga V, Llop E, Suárez C, et al. I.31, a new combination of probiotics, improves irritable bowel syndrome-related quality of life. World J Gastroenterol. 2014;20:8709–8716. doi: 10.3748/wjg.v20.i26.8709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Franz CM, Huch M, Abriouel H, Holzapfel W, Gálvez A. Enterococci as probiotics and their implications in food safety. Int J Food Microbiol. 2011;151:125–140. doi: 10.1016/j.ijfoodmicro.2011.08.014. [DOI] [PubMed] [Google Scholar]
- 38.Zhou Y, Chen H, He H, et al. Increased Enterococcus faecalis infection is associated with clinically active Crohn disease. Medicine (Baltimore) 2016;95:e5019. doi: 10.1097/MD.0000000000005019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fite A, Macfarlane S, Furrie E, et al. Longitudinal analyses of gut mucosal microbiotas in ulcerative colitis in relation to patient age and disease severity and duration. J Clin Microbiol. 2013;51:849–856. doi: 10.1128/JCM.02574-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Balish E, Warner T. Enterococcus faecalis induces inflammatory bowel disease in interleukin-10 knockout mice. Am J Pathol. 2002;160:2253–2257. doi: 10.1016/S0002-9440(10)61172-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ruiz PA, Shkoda A, Kim SC, Sartor RB, Haller D. IL-10 gene-deficient mice lack TGF-beta/Smad signaling and fail to inhibit proinflammatory gene expression in intestinal epithelial cells after the colonization with colitogenic Enterococcus faecalis. J Immunol. 2005;174:2990–2999. doi: 10.4049/jimmunol.174.5.2990. [DOI] [PubMed] [Google Scholar]
- 42.Steck N, Hoffmann M, Sava IG, et al. Enterococcus faecalis metalloprotease compromises epithelial barrier and contributes to intestinal inflammation. Gastroenterology. 2011;141:959–971. doi: 10.1053/j.gastro.2011.05.035. [DOI] [PubMed] [Google Scholar]
- 43.Lengfelder I, Sava IG, Hansen JJ, et al. Complex bacterial consortia reprogram the colitogenic activity of Enterococcus faecalis in a gnotobiotic mouse model of chronic, immune-mediated colitis. Front Immunol. 2019;10:1420. doi: 10.3389/fimmu.2019.01420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shao L, Ling Z, Chen D, Liu Y, Yang F, Li L. Disorganized gut microbiome contributed to liver cirrhosis progression: a meta-omics-based study. Front Microbiol. 2018;9:3166. doi: 10.3389/fmicb.2018.03166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sung CM, Lin YF, Chen KF, et al. Predicting clinical outcomes of cirrhosis patients with hepatic encephalopathy from the fecal microbiome. Cell Mol Gastroenterol Hepatol. 2019;8:301–318. e2. doi: 10.1016/j.jcmgh.2019.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dziarski R, Park SY, Kashyap DR, Dowd SE, Gupta D. Pglyrp-regulated gut microflora Prevotella falsenii, Parabacteroides distasonis and Bacteroides eggerthii enhance and Alistipes finegoldii attenuates colitis in mice. PLoS One. 2016;11:e0146162. doi: 10.1371/journal.pone.0146162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chey WD, Kurlander J, Eswaran S. Irritable bowel syndrome: a clinical review. JAMA. 2015;313:949–958. doi: 10.1001/jama.2015.0954. [DOI] [PubMed] [Google Scholar]
- 48.Mitselou A, Grammeniatis V, Varouktsi A, Papadatos SS, Katsanos K, Galani V. Proinflammatory cytokines in irritable bowel syndrome: a comparison with inflammatory bowel disease. Intest Res. 2020;18:115–120. doi: 10.5217/ir.2019.00125.7ad285a6240040cf9bcf723f16db1c02 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Taras D, Simmering R, Collins MD, Lawson PA, Blaut M. Reclassification of Eubacterium formicigenerans Holdeman and Moore 1974 as Dorea formicigenerans gen. nov., comb. nov., and description of Dorea longicatena sp. nov., isolated from human faeces. Int J Syst Evol Microbiol. 2002;52:423–428. doi: 10.1099/00207713-52-2-423. [DOI] [PubMed] [Google Scholar]
- 50.Pritchard SE, Marciani L, Garsed KC, et al. Fasting and postprandial volumes of the undisturbed colon: normal values and changes in diarrhea-predominant irritable bowel syndrome measured using serial MRI. Neurogastroenterol Motil. 2014;26:124–130. doi: 10.1111/nmo.12243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Crost EH, Tailford LE, Le Gall G, Fons M, Henrissat B, Juge N. Utilisation of mucin glycans by the human gut symbiont Ruminococcus gnavus is strain-dependent. PLoS One. 2013;8:e76341. doi: 10.1371/journal.pone.0076341.f6d894b98b704f8fbf268a3e5a2afe04 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Crost EH, Tailford LE, Monestier M, et al. The mucin-degradation strategy of Ruminococcus gnavus: The importance of intramolecular trans-sialidases. Gut Microbes. 2016;7:302–312. doi: 10.1080/19490976.2016.1186334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schirmer M, Smeekens SP, Vlamakis H, et al. Linking the human gut microbiome to inflammatory cytokine production capacity. Cell. 2016;167:1125–1136. e8. doi: 10.1016/j.cell.2016.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rajilić-Stojanović M, Biagi E, Heilig HG, et al. Global and deep molecular analysis of microbiota signatures in fecal samples from patients with irritable bowel syndrome. Gastroenterology. 2011;141:1792–1801. doi: 10.1053/j.gastro.2011.07.043. [DOI] [PubMed] [Google Scholar]
- 55.Takada T, Kurakawa T, Tsuji H, Nomoto K. Fusicatenibacter saccharivorans gen. nov., sp. nov., isolated from human faeces. Int J Syst Evol Microbiol. 2013;63(Pt 10):3691–3696. doi: 10.1099/ijs.0.045823-0. [DOI] [PubMed] [Google Scholar]
- 56.Takeshita K, Mizuno S, Mikami Y, et al. A single species of Clostridium subcluster XIVa decreased in ulcerative colitis patients. Inflamm Bowel Dis. 2016;22:2802–2810. doi: 10.1097/MIB.0000000000000972. [DOI] [PubMed] [Google Scholar]
- 57.Bamba T, Matsuda H, Endo M, Fujiyama Y. The pathogenic role of Bacteroides vulgatus in patients with ulcerative colitis. J Gastroenterol. 1995;30(suppl 8):45–47. [PubMed] [Google Scholar]
- 58.Shiba T, Aiba Y, Ishikawa H, et al. The suppressive effect of bifidobacteria on Bacteroides vulgatus, a putative pathogenic microbe in inflammatory bowel disease. Microbiol Immunol. 2003;47:371–378. doi: 10.1111/j.1348-0421.2003.tb03368.x. [DOI] [PubMed] [Google Scholar]
- 59.Ó Cuív P, de Wouters T, Giri R, et al. The gut bacterium and pathobiont Bacteroides vulgatus activates NF-κB in a human gut epithelial cell line in a strain and growth phase dependent manner. Anaerobe. 2017;47:209–217. doi: 10.1016/j.anaerobe.2017.06.002. [DOI] [PubMed] [Google Scholar]
- 60.Kim SC, Tonkonogy SL, Karrasch T, Jobin C, Sartor RB. Dual-association of gnotobiotic IL-10-/- mice with 2 nonpathogenic commensal bacteria induces aggressive pancolitis. Inflamm Bowel Dis. 2007;13:1457–1466. doi: 10.1002/ibd.20246. [DOI] [PubMed] [Google Scholar]
- 61.Waidmann M, Bechtold O, Frick JS, et al. Bacteroides vulgatus protects against Escherichia coli-induced colitis in gnotobiotic interleukin-2-deficient mice. Gastroenterology. 2003;125:162–177. doi: 10.1016/S0016-5085(03)00672-3. [DOI] [PubMed] [Google Scholar]
- 62.Tap J, Cools-Portier S, Pavan S, et al. Effects of the long-term storage of human fecal microbiota samples collected in RNAlater. Sci Rep. 2019;9:601. doi: 10.1038/s41598-018-36953-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.