Abstract
Background
Osteoarthritis (OA) is one of the most common joint diseases and a major global public health concern. Mesenchymal stem cells (MSCs) have been widely used for the treatment of OA owing to their paracrine secretion of trophic factors, a phenomenon in which exosomes may play a major role. Here, we investigate the potential of exosomes from human umbilical cord-derived MSCs (hUC-MSCs-Exos) in alleviating OA.
Methods
The hUC-MSCs-Exos were harvested from hUC-MSC-conditioned medium using ultracentrifugation. Rats with surgically-induced OA were intra-articularly injected with hUC-MSCs-Exos. The effect of hUC-MSCs-Exos in repairing osteoarticular cartilage was assessed using hematoxylin and eosin (HE) staining, safranin-O and fast green staining and immunohistochemistry. The in vitro experiments were further carried out to verify the therapeutic effect. The effects of hUC-MSCs-Exos on the proliferation and migration of human chondrocytes were evaluated using the cell counting kit-8, EdU-555 cell proliferation kit, and transwell assays. Annexin V-FITC/PI staining were used to evaluate the effect of exosomes on chondrocyte apoptosis. An in vitro model of human articular chondrocytes treated with interleukin 1 beta (IL-1β) was used to evaluate the effects of exosomes, analyses involved using quantitative real-time polymerase chain reaction (qRT-PCR), immunofluorescence, and western blotting. The role of hUC-MSCs-Exos in macrophage polarization was examined in the monocyte cell line, Tohoku Hospital Pediatrics-1 (THP-1) by qRT-PCR and immunofluorescence.
Results
The results showed that hUC-MSCs-Exos prevented severe damage to the knee articular cartilage in the rat OA model. We confirmed the high efficacy of hUC-MSCs-Exos in promoting chondrocyte proliferation and migration and inhibiting chondrocyte apoptosis. Additionally, hUC-MSCs-Exos could reverse IL-1β-induced injury of chondrocytes and regulate the polarization of macrophages in vitro.
Conclusions
There is potential for hUC-MSCs-Exos to be used as a treatment strategy for OA.
Keywords: Human umbilical cord-derived-mesenchymal stem cell (hUC-MSCs), exosome, osteoarthritis (OA), chondrocyte, macrophage
Introduction
Osteoarthritis (OA) is one of the most common chronic joint diseases worldwide, affecting approximately 10% of men and 18% of women over 60 years of age (1). This age-related degenerative joint disease is characterized by progressive destruction of the articular cartilage and cartilage loss caused by disequilibrium between anabolism and catabolism of chondrocytes and the extracellular matrix (ECM) (2). Current treatment modalities for cartilage injuries, such as microfracture, mosaicplasty, and autologous chondrocyte implantation, are limited by donor site morbidity and inferior fibrocartilage repair (3). Furthermore, the inherent limited healing potential of cartilage tissue introduces significant challenges in its self-repair and remodeling. Therefore, the present treatment modalities for OA are limited by their inability to prevent and slow the progression of the disease (4).
Mesenchymal stem cells (MSCs), with their remarkable potential for proliferation and differentiation into various cell lineages, are recognized as a promising cell source in the treatment of various degenerative, inflammatory, and autoimmune diseases (5). Primarily, MSCs are isolated from tissues such as bone marrow, adipose tissue, and umbilical cord. These cells are widely used in cell-based therapy and tissue engineering to treat cartilage lesions and OA, and they have been extensively investigated in both animal and human studies (6-9). Additionally, human umbilical cord-derived (hUC)-MSCs are considered a better choice than MSCs because of their painless collection procedure, absence of ethical issues with respect to their use, and high proliferation potential, cell vitality, and paracrine potential (9,10). Compared to other MSCs, hUC-MSCs exert more potent anti-inflammatory effects and enable more effective cartilage regeneration (11,12). However, due to drawbacks such as the safety issues of MSCs, inconvenient storage and transportation, and differentiation for non-therapeutic purposes, there is limited research on their applications. Similar to all cell-based therapies, MSCs-based therapy is associated with significant operational costs and challenges as cell-based medicine requires stringently monitored manufacturing and storage to ensure optimal viability and vitality of the cells at all stages (harvest, expansion, storage, and delivery to patients) (3,13,14). Moreover, many studies have shown that MSCs exert therapeutic effects of reduction of cellular injury and enhanced repair through the secretion of reparative factors (13,15,16). Co-culture study has further demonstrated that MSCs secrete trophic factors to promote chondrocyte proliferation and matrix synthesis (17).
Exosomes have been identified as the principal agents mediating the therapeutic efficacy of MSCs in several diseases (15,18,19). Exosomes are extracellular vesicles with a size range of 30 to 200 nm (average ~100 nm) that have an endosomal origin. Many exosome components are derived from their cells of origin; these include DNA, RNA, lipids, metabolites, as well as cytosolic and cell-surface proteins (18,20). Repair of osteochondral defects using MSCs-derived exosomes has been characterized by increased proliferation and infiltration of chondrocytes, and enhanced ECM synthesis (21-24). The research of He et al. has shown that bone marrow MSCs-derived exosomes can effectively promote cartilage repair and ECM synthesis, as well as alleviate knee pain in the OA rats (24). Wu et al. showed that exosomes derived from infrapatellar fat pad MSCs could protect the cartilage from damage and ameliorate gait abnormalities observed in OA mice (25). Wang et al. confirmed that exosomes from embryonic MSCs exert a beneficial therapeutic effect on OA by balancing the synthesis and degradation of chondrocyte-derived ECM (26). Nevertheless, ameliorating inflammation constitutes an important aspect of strategies aimed at promoting cartilage repair (27). Zhang et al. found that exosomes derived from MSCs have an immunomodulatory effect and can induce M2 macrophages to infiltrate the defects and synovium of OA cartilage, reduce the infiltration of M1 macrophages, and downregulate the expression of interleukin 1 beta (IL-1β) and tumor necrosis factor alpha (TNF-α), thereby inhibiting the inflammatory response in OA (21). Similar immunoregulatory roles of exosomes derived from MSCs have been observed in inflammatory bowel disease (28), spinal cord injury (29), and diabetic peripheral neuropathy (30). The potential use of MSCs-derived exosomes as a novel cell-free therapy for the treatment of OA is worthy of recognition.
Therefore, in this study, we investigated the regenerative and immunomodulatory effects of hUC-MSCs-derived exosomes (hUC-MSCs-Exos) in OA. We present the following article in accordance with the ARRIVE reporting checklist (available at https://atm.amegroups.com/article/view/10.21037/atm-22-3912/rc).
Methods
Umbilical cords were obtained from mothers of healthy full-term fetuses at The Second Hospital of Jilin University, China. Informed consent was provided by all donors. This study was performed in accordance with the Guidelines for Stem Cell Research and Clinical Translation (2016, ISSCR) and Declaration of Helsinki (as revised in 2013), and approved by the Ethics Committee of The Second Hospital of Jilin University (approval No. 2019-142). Identification of exosomes was conducted individually in samples from 3 different individuals; for the subsequent experiments, the exosomes were pooled together.
Cell culture
The hUC-MSCs were isolated as described previously (10). Briefly, umbilical cords were washed with phosphate-buffered saline (PBS; Thermo Fisher Scientific, Waltham, MA, USA) containing 5% penicillin/streptomycin solution (P/S; 100 IU/mL penicillin, 100 IU/mL streptomycin; Thermo Fisher Scientific). Umbilical cord tissues were carefully dissected to remove vessels and then cut into pieces of approximately 1 mm3, followed by suspension in Dulbecco’s modified Eagle medium containing nutrient mixture F-12 (DMEM/F-12; Thermo Fisher Scientific) supplemented with 10% fetal bovine serum (FBS; Thermo Fisher Scientific), 1% P/S, and 10 ng/mL basic fibroblast growth factor (bFGF; PeproTech, Cranbury, NJ, USA). The suspension was used to coat the pre-incubated Petri dishes. The explant-seeded plates were incubated for 1–2 hours in an inverted position to ensure the proper attachment of the explants. Thereafter, 3 mL of fresh medium was gently added to the seeded plate, followed by incubation at 37 ℃ in a humidified 5% CO2 incubator. The medium was changed every 3 days. An outgrowth of adherent cells from the tissue explants were observed after 1 week of seeding. After 10–14 days, the tissue explants were removed from the Petri dishes, leaving behind the attached cells. When the hUC-MSCs had proliferated to approximately 80% confluence, cell harvesting was carried out using 0.25% ethylenediaminetetraacetic acid (EDTA)-trypsin (Thermo Fisher Scientific), followed by centrifugation at 300 ×g for 5 minutes. Harvested cells were further grown to passage 4 for all the experiments. An inverted fluorescence microscope (Olympus IX71, Tokyo, Japan) was used to monitor the phenotype and growth characteristics of the hUC-MSCs. These cells were sub-cultured and used as cell sources in subsequent experiments. Human chondrocytes (C-12710) for the knee and hip joint cartilage tissue were obtained from PromoCell’s cell culture facility and cultured in a chondrocyte growth medium (PromoCell, Heidelberg, Germany). At 24 hours after cell seeding, the medium was replaced with fresh medium supplemented with 10 ng/mL IL-1β (PeproTech, USA). The next day, the medium with or without 4×107 particles/mL hUC-MSCs-Exos was added according to the experimental design, and the cells were collected and analyzed after 48 hours. Normal cultured chondrocytes served as the control. Human monocytic Tohoku Hospital Pediatrics-1 (THP-1) cells [TIB-202; American Type Culture Collection (ATCC), Manassas, VA, USA] were cultured in Roswell Park Memorial Institute (RPMI)-1640 medium (Thermo Fisher Scientific) supplemented with 10% FBS, and 1% P/S. Based on a previous study (29), THP-1 cells were induced into M0 macrophages by incubation with 25 ng/mL phorbol 12-myristate 13-acetate (PMA; Sigma-Aldrich, St. Louis, MO, USA) for 24 hours. Once the cells became adherent, they were polarized to M1 macrophages by incubation with lipopolysaccharide (LPS; Sigma-Aldrich; 100 ng/mL) for 24 hours. M0 and M1 macrophages (5×105 cells) were exposed to hUC-MSCs-Exos (4×107 particles/mL) in RPMI-1640 medium for 48 hours.
Immunofluorescence staining
Cells were incubated with the following mouse anti-human monoclonal antibodies: anti-COL2A1 (1:100; MA5-12781, Thermo Fisher Scientific); anti-MMP-13 (1:1,000; MAB511, R&D Systems, Minneapolis, MN, USA); anti-ACAN (1:1,000; NB-600-504, Novus Biologicals, Centennial, CO, USA); and rabbit anti-human monoclonal antibodies: anti-SOX9 (1:2,000; AB5535, Sigma-Aldrich), anti-ADAMTS5 (1:1,000; NBP2-15286, Novus Biologicals), and COL1A2 [1:1,000; 95855S, Cell Signaling Technology (CST), Dancers, MA, USA]. Subsequently, cells were incubated with the corresponding goat anti-mouse secondary antibody or goat anti-rabbit secondary antibody (Alexa Fluor 488 and 555; 1:1,000; 4408S, 4409S, 4412S, 4413S, CST). Cells were then counterstained with Hoechst 33342 dye (10 µg/mL; Thermo Fisher Scientific) and visualized under an inverted fluorescence microscope.
Isolation and identification of exosomes
When hUC-MSCs reached 50–60% confluence, they were washed with PBS and cultured in conditioned medium containing DMEM/F-12, 10% non-exosomes-FBS [System Biosciences (SBI), Palo Alto, CA, USA], 1% P/S, and 10 ng/mL bFGF for an additional 48 hours at 37 ℃ in a humidified 5% CO2 incubator. Exosome isolation was carried out using ultracentrifugation (30). Collected culture suspension was transferred to conical tubes for centrifugation at 300 ×g for 10 minutes at 4 ℃ to obtain the pellet. The supernatant was centrifuged a second time at 2,000 ×g for 10 minutes at 4 ℃ to further remove cell debris, followed by a third time at 10,000 ×g for 30 minutes at 4 ℃ to further remove apoptotic bodies and other organelles. Finally, the supernatant was ultracentrifuged at 120,000 ×g for 90 minutes at 4 ℃ in a SW32Ti rotor (Beckman Coulter, Brea, CA, USA) to obtain the exosome pellet. The pellet containing hUC-MSCs-Exos was resuspended with PBS and centrifuged at 120,000 ×g for 90 minutes at 4 ℃. The supernatant was discarded, and the pellet containing hUC-MSCs-Exos was resuspended with 400 µL PBS and stored at −80 ℃. The particle number of hUC-MSCs-Exos was quantified using an exosome enzyme-linked immunosorbent assay (ELISA) complete kit (CD81 detection, EXEL-ULTRA-CD81-1, SBI, USA) following the manufacturer’s instructions. Morphology of the exosomes was observed using transmission electron microscopy (TEM). The exosomes were loaded onto copper grids and contrasted using 2% uranyl acetate (Sigma-Aldrich), dried, and observed using a TECNAI 12 TEM (FEI, Hillsboro, OR, USA). The size distribution of exosomes was measured using ZETASIZER Nano series-Nano-ZS (Malvern Instruments, Malvern, UK), and was analyzed using Zetasizer software (Malvern Instruments). Antibodies against CD63 (1:1,000; ab271286, Abcam, Cambridge, UK) and CD81 (1:1,000; ab79559, Abcam) proteins were used to analyze the incorporation of each protein into exosomes using western blotting. Exosomes were collected using ultracentrifugation. After resuspension in PBS, the exosomes were incubated with CD63-antibody-fluorescein isothiocyanate (FITC) [1:5; 561924, Becton, Dickinson, and Co. (BD), Franklin Lakes, NJ, USA] and CD81-antibody-PE (1:5; 561957, BD) for 2 hours on ice. Exosomes incubated without the antibody served as the negative control. Flow cytometry (BD Accuri C6 flow cytometer) was used to detect the surface markers of hUC-MSCs-Exos.
Animal studies
The animal experiment was performed in the Guangdong Medical Laboratory Animal Center. Male Sprague-Dawley rats (approximately 12 weeks old) weighing 300–350 g, housed in a specific-pathogen-free (SPF) animal laboratory with a 12:12-hour light/dark cycle, controlled temperature environment (20–26 ℃), and steady humidity (40–70%), were used in the experiments. All animal experiments were performed in accordance with the ethical guidelines of the National Institutes of Health Guide for the Care and Use of Laboratory Animals and approved by the Ethics Committee of the Guangdong Medical Laboratory Animal Center Care and Use Committee (B202007-15). Based on previous report, OA in our rat model was surgically induced using the anterior cruciate ligament transection in combination with medial meniscectomy (ACLT + pMMx) method, without damaging the tibial surface (31,32). Surgery and treatment of animals were performed blindly. Rats were randomly divided into 3 groups: (I) normal (without surgery; received articular cavity injection of 100 µL PBS on day 1 and 4 of every week from the 5th to 8th week; n=6); (II) OA (rats underwent surgery and received articular cavity injection of 100 µL PBS on day 1 and 4 of every week from the 5th to 8th week after surgery; n=6); (III) OA+ Exos (rats received articular cavity injection of 100 µL hUC-MSCs-Exos in PBS, 1011 particles/mL of exosomes on day 1 and 4 of every week from the 5th to 8th week after surgery; n=6) (25,33). Eight weeks after surgery, rats were sacrificed with CO2, and the knee samples were harvested for the evaluation of disease progression. The protocol was prepared before the study without registration.
Histology and immunohistochemical analysis
The tibiofemoral joints were removed, and the femoral condyles were fixed in neutral-buffered formalin (containing 4% formaldehyde) for 24 hours after the knee samples were harvested. The fixed femoral condyles were decalcified in 10% EDTA decalcification solution (Leagene, Beijing, China) for 28 days (refreshed every day) before dehydration using graded ethanol (Beijing Chemical Works, Beijing, China), vitrification using dimethyl benzene (Beijing Chemical Works), paraffin embedding, and tissue sectioning (5 µm), prior to staining with hematoxylin and eosin (H&E) for morphological evaluation. Tissue sections were stained using a safranin-O and fast green staining kit (Solarbio, Beijing, China) and examined for matrix proteoglycan and overall joint morphology. Immunohistochemistry was performed to evaluate the function of articular chondrocytes. Immunohistochemistry was performed by using Immunohistochemical staining kit (ZSGB Bio, Beijing, China) according to the manufacturer’s instructions. Briefly, endogenous peroxidase activity was quenched with 3% H2O2, followed by antigen retrieval with 0.1% trypsin and blocking with normal goat serum for 30 minutes at 4 ℃ with antibodies against the following proteins: rabbit anti-COL2A1, rabbit anti-MMP-13, mouse anti-CD86, and rabbit anti-CD163 were used as primary antibodies (1:200; ab234000, ab182422, Abcam). After washing off excess primary antibodies, samples were incubated with secondary antibodies conjugated with horseradish peroxidase (HRP): HRP-labeled goat anti-mouse IgG (1:200; A0216, Beyotime, Shanghai, China) or goat anti-rabbit IgG antibody (1:200; A0208, Beyotime). Peroxidase-conjugated antibody (1:600; Beyotime) was diluted in 1% (w/v) bovine serum albumin (BSA) solution and incubated for 1 hour. A 3,3'-diaminobenzidine (DAB) detection system (Beyotime) was used to visualize the sections. Ten sections (approximately 90 µm apart) were prepared in each knee joint sample, and the staining of pathological sections of each knee joint sample was blindly selected for 3 stained pathological sections for analysis to obtain quantitative staining results of this knee joint sample. For quantification analysis, the number of positively stained cells was counted by 3 independent pathologists from randomly selecting one field from each sample in each group using ImageJ software (National Institutes of Health, Bethesda, MD, USA). The mean number of cells per high power field (cells/HPF) was determined. Based on safranin-O and fast green staining, the Osteoarthritis Research Society International (OARSI) score, which is a well-recognized histological scoring system (31,32), was used by 3 independent pathologists in a blinded manner to evaluate the cartilage degeneration in the lateral and medial femur and tibia of rat knee joints.
Western blotting
Cells or exosomes were lysed in a radioimmunoprecipitation assay (RIPA) and phenylmethylsulfonyl fluoride (PMSF) buffer (RIPA buffer: RIPA:PMSF =100:1; Beyotime, China) for 30 minutes on ice and centrifuged at 13,000 ×g for 20 minutes at 4 ℃. The supernatant was collected, and the total protein concentration was determined using a bicinchoninic acid (BCA) kit (Beyotime). An amount of 20 µg total protein were fractionated by using 10% sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and the separated proteins were blotted onto 0.22-µm polyvinylidene fluoride (PVDF) membranes (Beyotime). Membranes were then blocked with 5% skim milk in tris-buffered saline with Tween 20 (TBST; 10 mM Tris-HCL, 150 mM NaCl, 0.25% Tween-20, pH 7.5) at 4 ℃ for 1 hour, followed by overnight incubation with the following primary antibodies: anti-COL2A1, anti-SOX9, anti-ACAN, anti-MMP-13, anti-ADAMTS5, and COL1A2; anti-GAPDH (1:1,000, 5174S, CST) served as the protein-loading control. After washing with TBST, the membranes were incubated for 1 hour at room temperature with the secondary anti-mouse or anti-rabbit IgG antibody (1:2,000, 7076S, 7074S, CST). After 3 washes with TBST, signals were detected by chemiluminescence using the ECL-Plus detection system (TransGen, Beijing, China). The relative amount of proteins on the blots was determined using ImageJ software.
Reverse transcription quantitative polymerase chain reaction (RT-qPCR)
Total RNA was extracted from cells using TRIzol (Thermo Fisher Scientific) according to the manufacturer’s instructions and reverse-transcribed into complementary DNA (cDNA) using a reverse transcription kit (Roche, Basel, Switzerland), following the manufacturer’s instructions. RT-qPCR was performed with FastStart Universal SYBR Green Master (ROX; Roche) on a Real-Time PCR System (CFX Connect Real-Time System, Bio-Rad, Munich, Germany). The fold-changes in cDNA levels of the target gene, after normalization to the levels of the reference gene GAPDH, were determined using the 2^−ΔΔCT method (32). The RT-qPCR primers for COL2A1 (NM_033150.2), SOX9 (NM_000346.3), ACAN (NM_013227.2), MMP-13 (NM_002427.3), ADAMTS5 (NM_007038.4), COL1A2 (NM_000088.3), IL-1β (NM_000576.2), TNF-α (NM_001286813.1), IL-10 (NM_000572.2), ARG1 (NM_000045.2), and GAPDH (NM_001256799.2) were purchased from Guangzhou GeneCopoeia (Guangdong, China).
Chondrocyte proliferation, migration, and apoptosis assays
Exosomes were labeled with PKH26 (Sigma-Aldrich). Briefly, 100 µL exosome suspension was mixed with 895 µL diluent C and 5 µL PKH26 for 30 minutes at room temperature; the reaction was terminated using 2 mL of 10% BSA. The exosomes were collected using ultracentrifugation (120,000 ×g for 90 min at 4 ℃). The PKH26-labeled exosomes were resuspended in PBS and centrifuged at 120,000 ×g for 90 minutes at 4 ℃. The chondrocytes were cocultured with PKH26-labeled exosomes (4×107 particles/mL) overnight. The images were recorded using a fluorescence microscope. The effect of hUC-MSCs-Exos of different concentrations (0, 0.5, 1, 2, or 4×107 particles/mL) on chondrocyte proliferation was evaluated using the Cell Counting Kit-8 (CCK-8; Dojindo, Kumamoto, Japan), as described previously (33). Cell proliferation curves were constructed by measuring the amount of formazan dye generated by the activity of cellular dehydrogenase using a microplate reader (Tecan, Salzburg, Austria) at a wavelength of 450 nm. The incorporated 5-ethynyl-2'-deoxyuridine (EdU) was detected through a reaction between the alkyne group of EdU and the fluorescent azide in a copper-catalyzed azide-alkyne cycloaddition. Chondrocytes treated with 4×107 particles/mL hUC-MSCs-Exos were measured using an EdU-555 cell proliferation kit (Ribobio, Guangzhou, China) following the manufacturer’s instructions and visualized under an inverted fluorescence microscope. The EdU-free chondrocytes served as a negative control. The migration of hUC-MSCs-Exos-treated chondrocytes was assessed using transwell chambers. In brief, after digestion and counting, approximately 5×104 cells were seeded into the upper chamber of a 24-well 8-µm-pore-size Transwell plate (Corning, Corning, NY, USA). Subsequently, 600 µL of DMEM/F-12 medium containing exosomes was added into the lower chamber and 400 µL of DMEM/F-12 medium was added into the upper chamber before incubation for 24 hours at 37 ℃. The upper chamber was then fixed with 4% paraformaldehyde (PFA) for 15 minutes, stained with 0.5% crystal violet for 10 minutes, and washed with PBS 3 times. The upper surface of the upper chamber was carefully wiped using a cotton swab to remove cells that had not migrated to the lower surface. Images were collected by using an inverted fluorescence microscope. The chondrocytes stained with crystal violet that migrated to the lower chamber were counted by using ImageJ software. Chondrocyte apoptosis was detected using a FITC-annexin V apoptosis detection kit (BD Biosciences, New Jersey, USA) according to the manufacturer’s instructions. After 24 hours of passage 3 chondrocyte seeding, the DMEM/F-12 medium was replaced with medium containing 10 ng/mL IL-1β. The next day, the medium with or without 4×107 particles/mL hUC-MSCs-Exos was added according to the experimental design, and the cells were collected and analyzed after 48 hours. Normal cultured chondrocytes served as the control. Chondrocyte apoptosis was detected by following the manufacturer’s instructions. Samples were analyzed on a FACS Calibur instrument (BD Biosciences).
Statistical analyses
All experiments were performed with at least 3 independent experiments. Data are expressed as the mean ± standard deviation. Statistical analyses were performed using GraphPad Prism 7.0 software (GraphPad Software Inc., San Diego, CA, USA). Multiple comparisons were analyzed using analysis of non-parametric or the independent-samples Student’s t-test followed by Bonferroni correction.
Results
Isolation and identification of hUC-MSCs
Cells migrated out from the UC pieces between days 7 and 14 and proliferated in the tissue culture plates (Figure S1A). Isolated cells showed the formation of colonies with fibroblast morphology. Cells at passage 4 were identified and used in subsequent experiments. After reaching 80–90% confluence, cells adopted a spindle-like shape (Figure S1B). The cells were collected, pooled, and subjected to phenotypic characterization and multipotency assays. The spindle-shaped cells that expressed the MSC differentiation markers CD44, CD73, CD90, and CD105 and possessed multipotent differentiation potentials towards chondrocytes, osteoblasts, and adipocytes were classified as hUC-MSCs (Figure S1C-S1G).
Isolation and identification of hUC-MSCs-Exos
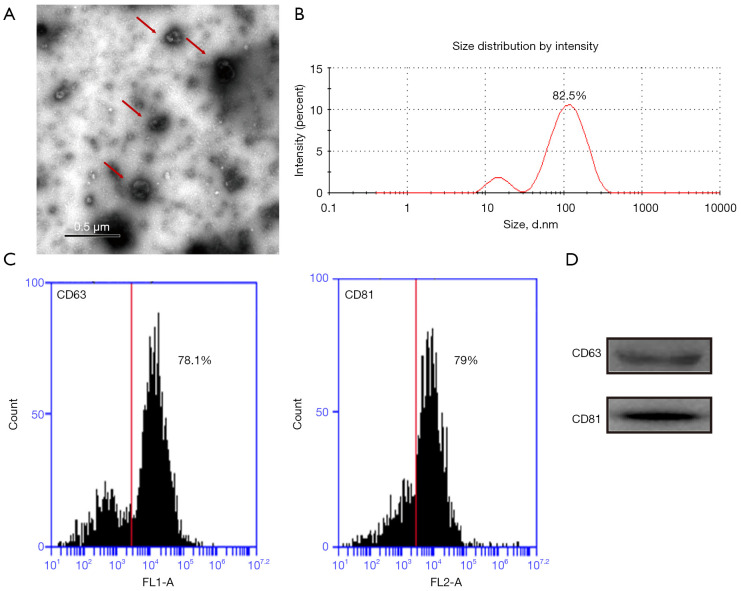
The TEM revealed that the vesicles from hUC-MSCs exhibited a round-shaped morphology with a diameter of 30–200 nm (Figure 1A). Nanoparticle tracking analysis revealed that 82.5% of vesicles displayed a particle size distribution between 30 and 200 nm (Figure 1B). Flow cytometry confirmed the expression of the vesicle differentiation markers CD63 (78.1%) and CD81 (79%) (Figure 1C). Western blotting also detected the presence of exosome marker proteins, namely CD63 and CD81 (Figure 1D). All these data indicated that hUC-MSCs-Exos were successfully isolated.
Figure 1.
Characterization of exosomes derived from hUC-MSCs (hUC-MSCs-Exos). (A) Morphology of exosomes observed by TEM. Scale bar: 0.5 µm. (B) Particle size distribution of exosomes measured by DLS. The experiment was repeated independently 3 times and representative results are shown. (C) Flow cytometry assay of the hUC-MSCs-Exos revealed expression rate of the hUC-MSCs-Exos differentiation markers, CD63 (78.1%), CD81 (79%). (D) Western blot indicated positive CD63 and CD81 protein expression in hUC-MSCs-Exos. Red arrows indicate exosomes. hUC-MSCs-Exos, hUC-MSCs-derived exosomes; hUC, human umbilical cord; MSC, mesenchymal stem cell; TEM, transmission electron microscopy; DLS, dynamic light scattering.
Exosomes attenuate OA progression
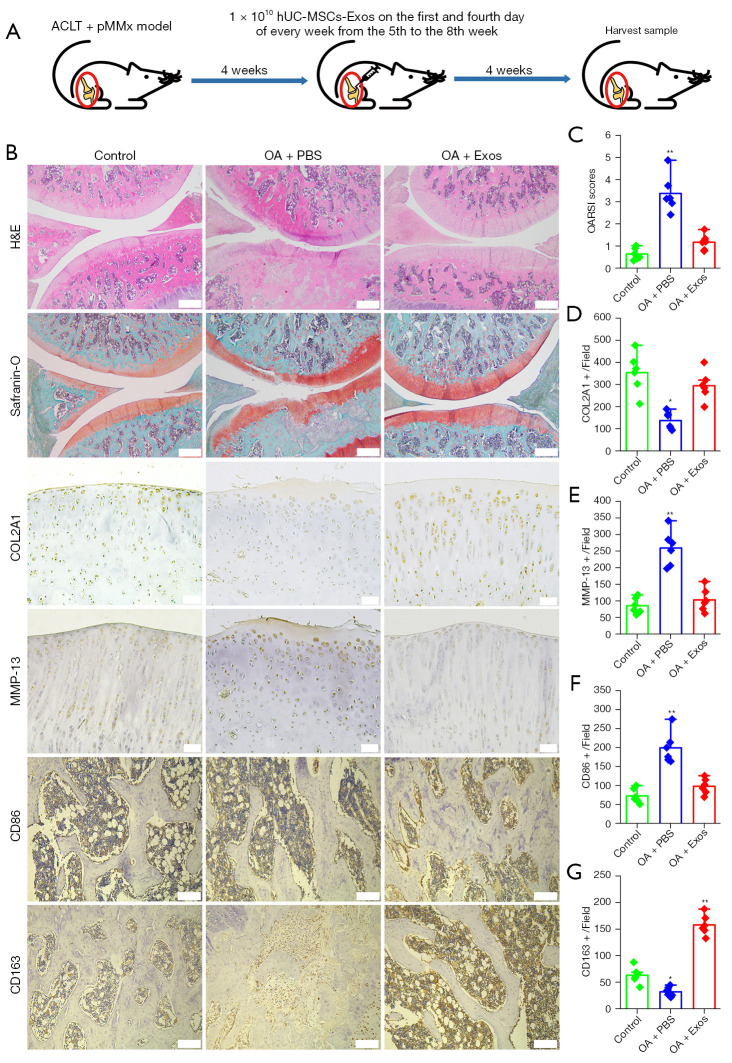
In the rat model, OA was surgically induced using the ACLT + pMMx method. Rats underwent surgery and received articular cavity injection of PBS or hUC-MSCs-Exos from the 5th to 8th week after surgery (days 1 and 4 of every week). At 8 weeks after the surgery, knee samples were harvested for the evaluation of disease progression (Figure 2A). No adverse events were observed in any of the experimental groups. We verified the potential of hUC-MSCs-Exos for OA prevention in an OA rat model (Figure 2B). In the OA + PBS group, H&E and safranin-O and fast green staining showed that the cartilage showed moderate surface irregularity, superficial fibrillation, loss of proteoglycan, and loss of cartilage in the superficial zone, but none of these effects were observed when the cartilage was treated with exosomes. Based on safranin-O and fast green staining, the OARSI score was used to evaluate OA progression. The OARSI score of group OA + Exos was 1.12±0.26 and that of group OA + PBS was 2.92±0.42 (Figure 2C). In the OA + PBS group, expression of COL2A1 in the cartilage decreased, whereas that of MMP-13 was observed on the cartilage surface. In the OA + Exos group, joint wear and cartilage matrix loss were scarcely observed. Expression of COL2A1 and MMP-13 in the cartilage was similar to that in the normal group, based on the cartilage surface (Figure 2B,2D,2E). As indicated by CD86-positive cells, M1 macrophage numbers reduced in the subchondral bone marrow cavity of rat knee joint of the OA + Exos group compared with that in the OA + PBS group (Figure 2B,2F). In contrast to the M1 macrophages, we observed an abundance of M2 macrophages, as indicated by CD163-positive cells in the subchondral bone marrow cavity of rat knee joint of the OA + Exos group compared with those in the OA + PBS group (Figure 2B,2G). These data indicated that the exosomes attenuated OA progression and prevented severe damage to the knee articular cartilage in the rat OA model, which was caused by instability of the knee joint.
Figure 2.
hhUC-MSCs-Exos prevent OA. (A) Flowchart of the in vivo experiment. (B) Staining results of H&E, safranin-O and fast green staining (scale bar: 500 μm), and immunohistochemical staining for COL2A1, MMP-13 (scale bar: 50 μm), CD86, and CD163 (scale bar: 200 μm). (C) Statistical results of chondrocytes counted in randomly selected field from each sample and the result of statistical analysis of the OARSI score in each group. Data are represented as mean ± SD. **, P<0.01 compared to the normal (n=6) by non-parametric test. (D-G) Statistical results of COL2A1+, MMP-13+, CD86+, and CD163+ cells counted in randomly selected field from each sample. Data are represented as mean ± SD. *, P<0.05; **, P<0.01 compared to the normal (n=6) by independent-samples Student’s t-test with Bonferroni correction. ACLT + pMMx, anterior cruciate ligament transection in combination with medial meniscectomy; hUC-MSCs-Exos, hUC-MSCs-derived exosomes; hUC, human umbilical cord; MSC, mesenchymal stem cell; OA, osteoarthritis; PBS, phosphate-buffered saline; OARSI, Osteoarthritis Research Society International; H&E, hematoxylin and eosin; SD, standard deviation.
Exosomes promote chondrocyte proliferation, migration, and inhibit apoptosis
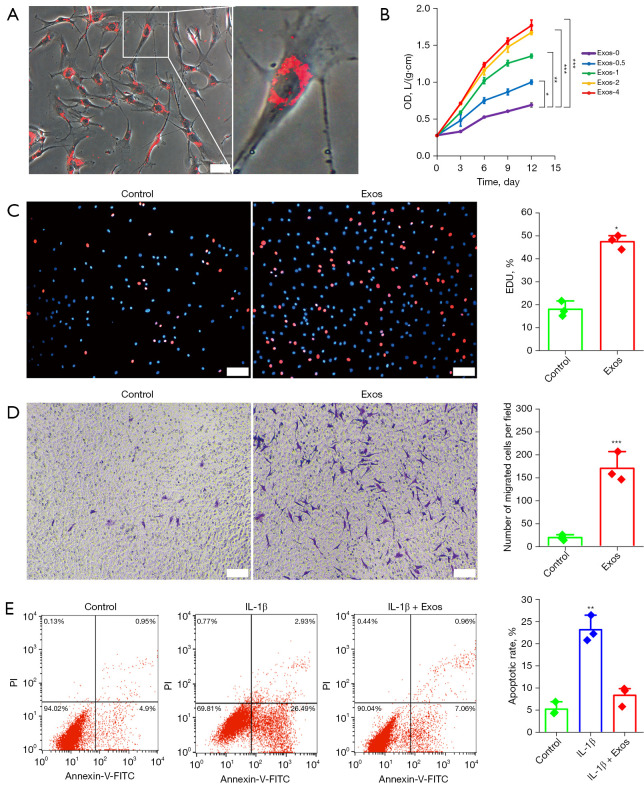
To confirm whether the chondrocytes could internalize hUC-MSCs-Exos, hUC-MSCs-Exos were labeled using red fluorescent PKH26. Chondrocytes were then incubated for 24 hours with PKH26-labeled exosomes. After washing with PBS, the PKH26-labeled exosomes were observed in the cytoplasm of the chondrocytes, confirming the internalization by chondrocytes. Almost all chondrocytes were positive for exosome internalization (Figure 3A). After quantifying the exosomes using an exosome ELISA complete kit, proliferation was evaluated following the stimulation of chondrocytes with 0, 0.5, 1, 2, or 4×107 particles/mL of hUC-MSCs-Exos. Based on our concentration range, CCK-8 assay results confirmed that all concentrations of exosomes could promote the proliferation of chondrocytes in a dose-dependent manner (Figure 3B). The results of EdU assay results further showed that the proliferative ability of chondrocytes was notably enhanced by hUC-MSCs-Exos (Figure 3C). In OA treatment, the enhancement of chondrocyte migration ability is one aspect of cartilage regeneration. The results of the transwell assay showed that the migratory ability of chondrocytes was markedly strengthened by hUC-MSCs-Exo treatment (Figure 3D).
Figure 3.
Effects of hUC-MSCs-Exos on proliferation and migration in chondrocytes. (A) Representative immunofluorescence photomicrograph of PKH26 (red)-labeled exosomes absorbed by chondrocytes (light field) on the right is an enlarged image of chondrocytes with exosomes internalized. Scale bar: 50 µm. (B) hUC-MSCs-Exos were treated with chondrocytes at different concentrations. Quantitative data are presented as means ± SD of 3 independent experiments. *, P<0.05; **, P<0.01; ***, P<0.001, independent-samples Student’s t-test with Bonferroni correction. (C) EdU-positive cells and representative images of fields showing red-colored proliferative cells. Hoechst 33342 staining was performed to detect nuclear localization (blue color). Scale bar: 100 µm. Percentage of EdU was plotted as the mean ± SD of triplicate samples from 3 independent experiments. *, P<0.05, by independent-samples Student’s t-test. (D) Migrated chondrocytes were stained by crystal violet. Scale bar: 100 µm. Migrated chondrocytes was plotted as the mean ± SD of triplicate samples from 3 independent experiments. ***, P<0.001, independent-samples Student’s t-test. (E) Annexin V-PI staining combined with flow cytometry assay to detect anti-apoptosis of hUC-MSCs-Exos in IL-1β-treated chondrocytes. **, P<0.01, independent-samples Student’s t-test with Bonferroni correction. hUC-MSCs-Exos, hUC-MSCs-derived exosomes; hUC, human umbilical cord; MSC, mesenchymal stem cell; OD, optical density; EdU, 5-ethynyl-2'-deoxyuridine; IL-1β, interleukin 1 beta; PI, propidium iodide; FITC, fluorescein isothiocyanate; SD, standard deviation.
Inhibition of apoptosis is an important factor in promoting cell proliferation. We used annexin V-FITC/propidium iodide (PI) staining and flow cytometry to detect the effect of exosomes on the inhibition of chondrocyte apoptosis (Figure 3E). We used IL-1β to induce apoptosis (29.42%) and hUC-MSCs-Exo treatment decreased the apoptotic rate of chondrocytes (8.02%). These results clearly demonstrated the superior efficacy of hUC-MSCs-Exos in promoting chondrocyte proliferation and migration and inhibiting chondrocyte apoptosis.
Exosomes reverse IL-1β-induced chondrocyte injury
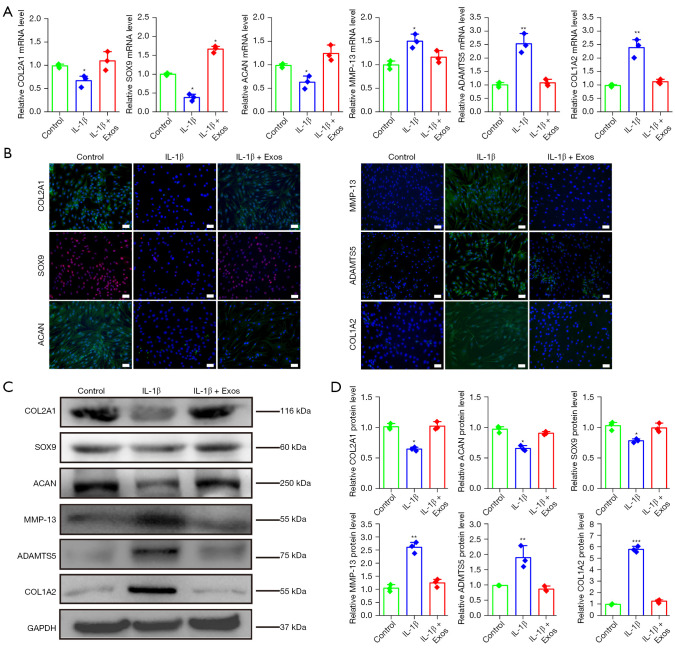
We used a model of OA-like chondrocytes reported in a previous study (34). In our experiment, qPCR results showed that IL-1β could reduce the gene expression levels of COL2A1, SOX9, and ACAN and increase those of MMP-13, ADAMTS5, and COL1A2. The OA-like chondrocyte model was successfully constructed. Interestingly, the supplementation of hUC-MSCs-Exos in chondrocytes dramatically reversed the effect of IL-1β on the gene expression of COL2A1, SOX9, ACAN, MMP-13, ADAMTS5, and COL1A2 (Figure 4A), consistent with the results obtained from the immunofluorescence staining assay (Figure 4B). Western blotting further confirmed that IL-1β could reduce protein expression of COL2A1, SOX9, and ACAN in chondrocytes and increase the expression of MMP-13, ADAMTS5, and COL1A2 at the protein level. However, more importantly, results from western blotting confirmed that hUC-MSCs-Exos could reverse the effect of IL-1β on the COL2A1, SOX9, ACAN, MMP-13, ADAMTS5, and COL1A2 proteins (Figure 4C,4D). In conclusion, exosomes could reverse the injury of chondrocytes induced by IL-1β in a model of OA-like chondrocytes in vitro.
Figure 4.
Exosomes reverse IL-1β-induced chondrocyte injury. (A) Gene expression changes in chondrocytes of COL2A1, SOX9, ACAN, MMP-13, ADAMTS5, and COL1A2 after stimulation with IL-1β and exosomes. This experiment was repeated 3 times. *, P<0.05; **, P<0.01 compared to untreated chondrocytes by independent-samples Student’s t-test with Bonferroni correction. (B) COL2A1-, ACAN-, MMP-13-, ADAMTS5-, and COL1A2-positive cells and representative images of fields showing green-colored cytoplasm of chondrocytes in each group. SOX9-positive cells and representative images of fields showing red-colored nucleus of chondrocytes in each group. Hoechst 33342 staining was performed to detect nuclear localization (blue color). Scale bar: 100 μm. (C,D) Protein expression levels of COL2A1, SOX9, ACAN, MMP-13, ADAMTS5, and COL1A2 in chondrocytes were detected using western blotting. This experiment was repeated 3 times. *, P<0.05; **, P<0.01; ***, P<0.001 compared to untreated chondrocytes by independent-samples Student’s t-test with Bonferroni correction. IL-1β, interleukin 1 beta.
Role of exosomes in macrophage polarization
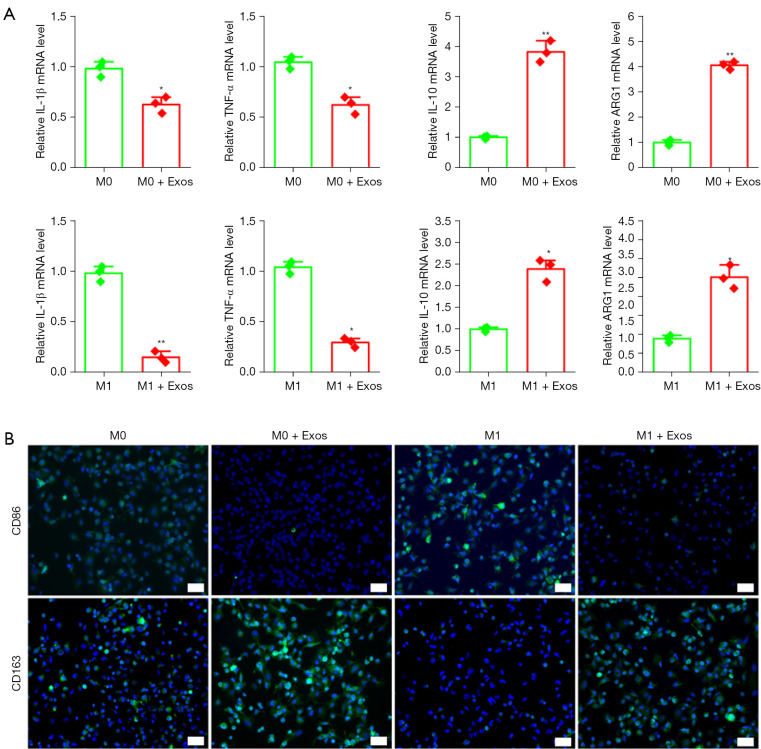
To evaluate the role of exosomes in macrophage activity, we performed an in vitro assay using a polarization protocol of the THP-1 monocyte cell line. The THP1 cells were induced to form M0 macrophages using PMA; M0 was induced to form M1 macrophages using LPS. When the M0 and M1 macrophages were treated using hUC-MSCs-Exos for 48 hours, qPCR results showed that the expression levels of anti-inflammatory factors IL-10 and ARG1 increased, whereas the expression levels of IL-1β and TNF-α decreased significantly (Figure 5A). Immunofluorescence showed that compared to the control group, the number of CD163-positive cells was significantly increased, and the number of CD86-positive cells was significantly decreased in M0 and M1 macrophages treated with exosomes (Figure 5B).
Figure 5.
Exosome effect on macrophage polarization. (A) Gene expression changes in IL-1β, TNF-α, IL-10, and ARG1 after stimulation with exosomes. This experiment was repeated 3 times. *, P<0.05; **, P<0.01 compared to macrophages without exosomes by independent-samples Student’s t-test with Bonferroni correction. (B) CD86-, CD163-positive cells and representative images of fields showing green-colored cytoplasm of macrophages in each group. Hoechst 33342 staining was performed to detect nuclear localization (blue color). Scale bar: 100 μm. IL-1β, interleukin 1 beta; TNF-α, tumor necrosis factor alpha.
Discussion
Currently, OA is a prevalent chronic joint disease (35) for which therapies are aimed at symptomatic relief, but are unable block or reverse the ongoing cartilage degeneration (2). The ideal treatment aimed at achieving optimal OA joint repair should promote the regenerative properties of chondrocytes and eliminate the destructive effects of inflammation (36). To the best of our knowledge, this is the first report on the effect of exosomes derived from hUC-MSCs in the treatment of OA. Our data demonstrated that hUC-MSCs-Exos prevented severe damage to knee articular cartilage in the rat OA model. We also demonstrated that hUC-MSCs-Exos balance the synthesis and degradation of the cartilage ECM and regulate macrophages. In the treatment of OA, exosomes derived from hUC-MSCs offer many advantages. First, studies have validated the safety of hUC-MSCs-Exos, which is crucial for the design of future clinical trials (37,38). We did not observe any adverse events in our OA model rats administered hUC-MSCs-Exos. Second, hUC-MSCs-Exos play a role in repairing tissue damage in vital organs, especially in the kidneys (39), liver (40), lung (41), and heart (42). Our previous study demonstrated that direct administration of hUC-MSCs-Exos may effectively treat cutaneous wounding (43). Third, hUC-MSCs have been presented as the ideal source of exosomes because of the absence of ethical concerns, easy availability, and non-invasive isolation with high yield (10). Fourth, the high abundance of micro RNAs (miRNAs) in hUC-MSCs-Exos plays an important role in maintaining cartilage homeostasis, promoting chondrogenesis, and regulating inflammation; these miRNAs include miRNA-21-5p (44,45), 146a-3p (46,47), 26a-5p (48), 100-5p (25,49), and let-7a (50).
Unilateral surgical ACLT + pMMx in mature rats leads to the development of progressive cartilage degeneration. In this model, exosomes were able to prevent lesion progression and exert a similar regenerative effect as exosomes from other stem cells (31). In our animal experiment, both hind limb knee joints of rats were surgically intervened for the construction of an OA model. Compared to other studies using animal models (51,52), we increased the number of exosome injections to twice a week for 4 weeks.
Previous studies have also demonstrated that exosomes derived from the human bone marrow (53), adipose (54) and embryonic (26,51) tissue-derived MSCs, and amniotic fluid stem cells (55) could cause effective cartilage repair by inducing the migration and proliferation of chondrocytes and promoting cartilage matrix synthesis. Our results also showed that hUC-MSCs-Exos could promote the proliferation and migration of chondrocytes. Tao et al. found that exosomes derived from human synovial MSCs clearly promoted chondrocyte proliferation and migration. However, there was one crucial shortcoming: the inhibition of the synthesis of ECM proteins, including ACAN and collagen II (52). Our research showed that exosomes derived from hUC-MSCs modulated chondrocytes treated with IL-1β to maintain the expression of chondrogenic markers (COL2A1, SOX9, and ACAN) and decreased MMP-13, ADAMTS5, and COL1A2 expression. Therefore, hUC-MSCs-Exos can not only promote chondrocyte proliferation and migration and inhibit IL-1β-induced chondrocyte apoptosis but also reverse IL-1β-induced chondrocyte damage by balancing the synthesis and degradation of cartilage ECM.
The most common type of arthritis, OA is a multifactorial cartilage lesion disease within a chronic inflammatory microenvironment (21). Macrophages are immune cells present in the synovial lining; their function varies with the subtype. Macrophages can polarize to either the pro-inflammatory (M1) or anti-inflammatory (M2) phenotype (56). The M1-associated cytokines, IL-1β and TNF-α, induce destructive processes in chondrocytes, such as the downregulation of ACAN and collagen II expression and upregulation of matrix metalloproteinase and aggrecanase expression (57). The M2-associated cytokines ARG1 and IL-10 are crucial for chondrogenesis (56). Macrophage-associated inflammation is a driver of OA structural damage and progression. Daghestani et al. suggested that targeting macrophages and macrophage-associated inflammatory pathways may be an effective means to treat OA (58). Anti-inflammatory intervention has been shown to promote chondrocyte survival and mitigate the risk of development to post-traumatic OA. For example, it was reported that M2 macrophage polarization in OA synovium tissues supports the survival of chondrocytes by producing anti-inflammatory IL-10 to suppress adverse inflammation, whereas M1-polarized macrophages inhibit MSCs chondrogenic differentiation via IL-6 in vitro (57). Our in vitro results also showed that hUC-MSCs-Exos can inhibit the secretion of pro-inflammatory factors, promote the expression of anti-inflammatory factors, and regulate the polarization of macrophages. Additionally, hUC-MSCs-Exos induced higher infiltration of CD163+ regenerative M2 macrophages than that of inflammatory CD86+ M1 macrophages during articular cartilage repair.
Nevertheless, the effective dosage of exosomes needs to be optimized. To date, the active ingredients in exosomes remain unknown. Further studies focusing on the efficacy of exosomes derived from a larger cohort of donors should be conducted. The observation time for animal model studies needs to be extended with an increased number of observation time points, to provide a more accurate reference for the application of exosomes in OA treatment. Since exosomes are complex carriers that contain DNA, RNA, lipids, metabolites, and various proteins, they exhibit therapeutic functions through immunomodulation, bioenergetics, or biochemical effects in various disease models. Hence, the mechanisms underlying their therapeutic effects in OA need to be investigated further.
Conclusions
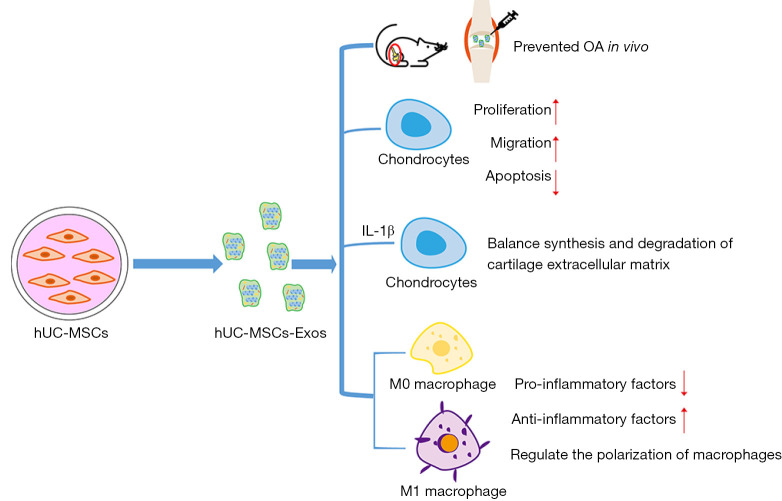
In this study, we confirmed the efficacy of exosomes derived from hUC-MSCs in treating OA. Exosomes derived from hUC-MSCs slowed the progression of early OA in vivo and prevented severe damage to the knee articular cartilage that arises as the result of the instability of the knee joint in the rat OA model. Exosomes derived from hUC-MSCs can not only promote the proliferation and migration of chondrocytes in vitro but also reverse IL-1β-associated damage. Additionally, hUC-MSCs-Exos can inhibit the secretion of pro-inflammatory factors, promote the expression of anti-inflammatory factors, and regulate the polarization of macrophages (Figure 6).
Figure 6.
Graphical abstract of the experimental design. hUC-MSCs-Exos attenuated the progression of OA and prevented severe damage to the knee articular cartilage in the rat OA model. hUC-MSCs-Exos promote chondrocytes proliferation and migration and inhibit chondrocytes apoptosis. hUC-MSCs-Exos reversed IL-1β-induced injury in vitro. hUC-MSCs-Exos could inhibit the secretion of pro-inflammatory factors, promote the expression of anti-inflammatory factors, and regulate the polarization of macrophages. hUC-MSCs-Exos, hUC-MSCs-derived exosomes; hUC, human umbilical cord; MSC, mesenchymal stem cell; OA, osteoarthritis; IL-1β, interleukin 1 beta.
Supplementary
The article’s supplementary files as
Acknowledgments
We thank the Guangdong Medical Laboratory Animal Center for providing us their equipment and service.
Funding: This work was supported by the China Natural National Science Foundation (No. 81970547), and the Science and Technology Department of Jilin Province (No. 20190101006JH).
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. This study was approved by the Ethics Committee of The Second Hospital of Jilin University (approval No. 2019-142). Informed consent was provided by all donors. This study was performed in accordance with the Guidelines for Stem Cell Research and Clinical Translation (2016, ISSCR) and Declaration of Helsinki (as revised in 2013). All animal experiments were performed in accordance with the ethical guidelines of the National Institutes of Health Guide for the Care and Use of Laboratory Animals and approved by the Ethics Committee of the Guangdong Medical Laboratory Animal Center Care and Use Committee (No. B202007-15).
Reporting Checklist: The authors have completed the ARRIVE reporting checklist. Available at https://atm.amegroups.com/article/view/10.21037/atm-22-3912/rc
Data Sharing Statement: Available at https://atm.amegroups.com/article/view/10.21037/atm-22-3912/dss
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://atm.amegroups.com/article/view/10.21037/atm-22-3912/coif). The authors have no conflicts of interest to declare.
(English Language Editor: J. Jones)
References
- 1.Nelson AE. Osteoarthritis year in review 2017: clinical. Osteoarthritis Cartilage 2018;26:319-25. 10.1016/j.joca.2017.11.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fellows CR, Matta C, Mobasheri A. Applying Proteomics to Study Crosstalk at the Cartilage-Subchondral Bone Interface in Osteoarthritis: Current Status and Future Directions. EBioMedicine 2016;11:2-4. 10.1016/j.ebiom.2016.08.047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jiang YZ, Zhang SF, Qi YY, et al. Cell transplantation for articular cartilage defects: principles of past, present, and future practice. Cell Transplant 2011;20:593-607. 10.3727/096368910X532738 [DOI] [PubMed] [Google Scholar]
- 4.Roseti L, Desando G, Cavallo C, et al. Articular Cartilage Regeneration in Osteoarthritis. Cells 2019;8:1305. 10.3390/cells8111305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pittenger MF, Mackay AM, Beck SC, et al. Multilineage potential of adult human mesenchymal stem cells. Science 1999;284:143-7. 10.1126/science.284.5411.143 [DOI] [PubMed] [Google Scholar]
- 6.Jiang Y, Jahagirdar BN, Reinhardt RL, et al. Pluripotency of mesenchymal stem cells derived from adult marrow. Nature 2002;418:41-9. 10.1038/nature00870 [DOI] [PubMed] [Google Scholar]
- 7.Jo CH, Lee YG, Shin WH, et al. Intra-articular injection of mesenchymal stem cells for the treatment of osteoarthritis of the knee: a proof-of-concept clinical trial. Stem Cells 2014;32:1254-66. 10.1002/stem.1634 [DOI] [PubMed] [Google Scholar]
- 8.Kurth TB, Dell'accio F, Crouch V, et al. Functional mesenchymal stem cell niches in adult mouse knee joint synovium in vivo. Arthritis Rheum 2011;63:1289-300. 10.1002/art.30234 [DOI] [PubMed] [Google Scholar]
- 9.Matas J, Orrego M, Amenabar D, et al. Umbilical Cord-Derived Mesenchymal Stromal Cells (MSCs) for Knee Osteoarthritis: Repeated MSC Dosing Is Superior to a Single MSC Dose and to Hyaluronic Acid in a Controlled Randomized Phase I/II Trial. Stem Cells Transl Med 2019;8:215-24. 10.1002/sctm.18-0053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhao G, Liu F, Lan S, et al. Large-scale expansion of Wharton's jelly-derived mesenchymal stem cells on gelatin microbeads, with retention of self-renewal and multipotency characteristics and the capacity for enhancing skin wound healing. Stem Cell Res Ther 2015;6:38. 10.1186/s13287-015-0031-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee NH, Na SM, Ahn HW, et al. Allogenic Human Umbilical Cord Blood-Derived Mesenchymal Stem Cells Are More Effective Than Bone Marrow Aspiration Concentrate for Cartilage Regeneration After High Tibial Osteotomy in Medial Unicompartmental Osteoarthritis of Knee. Arthroscopy 2021;37:2521-30. 10.1016/j.arthro.2021.02.022 [DOI] [PubMed] [Google Scholar]
- 12.Wang Q, Yang Q, Wang Z, et al. Comparative analysis of human mesenchymal stem cells from fetal-bone marrow, adipose tissue, and Warton's jelly as sources of cell immunomodulatory therapy. Hum Vaccin Immunother 2016;12:85-96. 10.1080/21645515.2015.1030549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Phinney DG, Pittenger MF. Concise Review: MSC-Derived Exosomes for Cell-Free Therapy. Stem Cells 2017;35:851-8. 10.1002/stem.2575 [DOI] [PubMed] [Google Scholar]
- 14.Herberts CA, Kwa MS, Hermsen HP. Risk factors in the development of stem cell therapy. J Transl Med 2011;9:29. 10.1186/1479-5876-9-29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lai RC, Yeo RW, Lim SK. Mesenchymal stem cell exosomes. Semin Cell Dev Biol 2015;40:82-8. 10.1016/j.semcdb.2015.03.001 [DOI] [PubMed] [Google Scholar]
- 16.Caplan AI, Dennis JE. Mesenchymal stem cells as trophic mediators. J Cell Biochem 2006;98:1076-84. 10.1002/jcb.20886 [DOI] [PubMed] [Google Scholar]
- 17.Wu L, Leijten JC, Georgi N, et al. Trophic effects of mesenchymal stem cells increase chondrocyte proliferation and matrix formation. Tissue Eng Part A 2011;17:1425-36. 10.1089/ten.tea.2010.0517 [DOI] [PubMed] [Google Scholar]
- 18.Kalluri R, LeBleu VS. The biology, function, and biomedical applications of exosomes. Science 2020;367:eaau6977. 10.1126/science.aau6977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hade MD, Suire CN, Suo Z. Mesenchymal Stem Cell-Derived Exosomes: Applications in Regenerative Medicine. Cells 2021;10:1959. 10.3390/cells10081959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Doyle LM, Wang MZ. Overview of Extracellular Vesicles, Their Origin, Composition, Purpose, and Methods for Exosome Isolation and Analysis. Cells 2019;8:727. 10.3390/cells8070727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang S, Chuah SJ, Lai RC, et al. MSC exosomes mediate cartilage repair by enhancing proliferation, attenuating apoptosis and modulating immune reactivity. Biomaterials 2018;156:16-27. 10.1016/j.biomaterials.2017.11.028 [DOI] [PubMed] [Google Scholar]
- 22.Mianehsaz E, Mirzaei HR, Mahjoubin-Tehran M, et al. Mesenchymal stem cell-derived exosomes: a new therapeutic approach to osteoarthritis? Stem Cell Res Ther 2019;10:340. 10.1186/s13287-019-1445-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cosenza S, Ruiz M, Toupet K, et al. Mesenchymal stem cells derived exosomes and microparticles protect cartilage and bone from degradation in osteoarthritis. Sci Rep 2017;7:16214. 10.1038/s41598-017-15376-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.He L, He T, Xing J, et al. Bone marrow mesenchymal stem cell-derived exosomes protect cartilage damage and relieve knee osteoarthritis pain in a rat model of osteoarthritis. Stem Cell Res Ther 2020;11:276. 10.1186/s13287-020-01781-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wu J, Kuang L, Chen C, et al. miR-100-5p-abundant exosomes derived from infrapatellar fat pad MSCs protect articular cartilage and ameliorate gait abnormalities via inhibition of mTOR in osteoarthritis. Biomaterials 2019;206:87-100. 10.1016/j.biomaterials.2019.03.022 [DOI] [PubMed] [Google Scholar]
- 26.Wang Y, Yu D, Liu Z, et al. Exosomes from embryonic mesenchymal stem cells alleviate osteoarthritis through balancing synthesis and degradation of cartilage extracellular matrix. Stem Cell Res Ther 2017;8:189. 10.1186/s13287-017-0632-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhao X, Zhao Y, Sun X, et al. Immunomodulation of MSCs and MSC-Derived Extracellular Vesicles in Osteoarthritis. Front Bioeng Biotechnol 2020;8:575057. 10.3389/fbioe.2020.575057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schena F, Gambini C, Gregorio A, et al. Interferon-γ-dependent inhibition of B cell activation by bone marrow-derived mesenchymal stem cells in a murine model of systemic lupus erythematosus. Arthritis Rheum 2010;62:2776-86. 10.1002/art.27560 [DOI] [PubMed] [Google Scholar]
- 29.Lankford KL, Arroyo EJ, Nazimek K, et al. Intravenously delivered mesenchymal stem cell-derived exosomes target M2-type macrophages in the injured spinal cord. PLoS One 2018;13:e0190358. 10.1371/journal.pone.0190358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fan B, Li C, Szalad A, et al. Mesenchymal stromal cell-derived exosomes ameliorate peripheral neuropathy in a mouse model of diabetes. Diabetologia 2020;63:431-43. 10.1007/s00125-019-05043-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gerwin N, Bendele AM, Glasson S, et al. The OARSI histopathology initiative - recommendations for histological assessments of osteoarthritis in the rat. Osteoarthritis Cartilage 2010;18 Suppl 3:S24-34. 10.1016/j.joca.2010.05.030 [DOI] [PubMed] [Google Scholar]
- 32.Culley KL, Dragomir CL, Chang J, et al. Mouse models of osteoarthritis: surgical model of posttraumatic osteoarthritis induced by destabilization of the medial meniscus. Methods Mol Biol 2015;1226:143-73. 10.1007/978-1-4939-1619-1_12 [DOI] [PubMed] [Google Scholar]
- 33.Zhu Y, Wang Y, Zhao B, et al. Comparison of exosomes secreted by induced pluripotent stem cell-derived mesenchymal stem cells and synovial membrane-derived mesenchymal stem cells for the treatment of osteoarthritis. Stem Cell Res Ther 2017;8:64. 10.1186/s13287-017-0510-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Meng F, Zhang Z, Chen W, et al. MicroRNA-320 regulates matrix metalloproteinase-13 expression in chondrogenesis and interleukin-1β-induced chondrocyte responses. Osteoarthritis Cartilage 2016;24:932-41. 10.1016/j.joca.2015.12.012 [DOI] [PubMed] [Google Scholar]
- 35.Hunter DJ, Bierma-Zeinstra S. Osteoarthritis. Lancet 2019;393:1745-59. [DOI] [PubMed] [Google Scholar]
- 36.Vonk LA, van Dooremalen SFJ, Liv N, et al. Mesenchymal Stromal/stem Cell-derived Extracellular Vesicles Promote Human Cartilage Regeneration In Vitro. Theranostics 2018;8:906-20. 10.7150/thno.20746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sun L, Xu R, Sun X, et al. Safety evaluation of exosomes derived from human umbilical cord mesenchymal stromal cell. Cytotherapy 2016;18:413-22. 10.1016/j.jcyt.2015.11.018 [DOI] [PubMed] [Google Scholar]
- 38.Yaghoubi Y, Movassaghpour A, Zamani M, et al. Human umbilical cord mesenchymal stem cells derived-exosomes in diseases treatment. Life Sci 2019;233:116733. 10.1016/j.lfs.2019.116733 [DOI] [PubMed] [Google Scholar]
- 39.Zhang R, Yin L, Zhang B, et al. Resveratrol improves human umbilical cord-derived mesenchymal stem cells repair for cisplatin-induced acute kidney injury. Cell Death Dis 2018;9:965. 10.1038/s41419-018-0959-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shao M, Xu Q, Wu Z, et al. Exosomes derived from human umbilical cord mesenchymal stem cells ameliorate IL-6-induced acute liver injury through miR-455-3p. Stem Cell Res Ther 2020;11:37. 10.1186/s13287-020-1550-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zheng Y, Liu J, Chen P, et al. Exosomal miR-22-3p from human umbilical cord blood-derived mesenchymal stem cells protects against lipopolysaccharid-induced acute lung injury. Life Sci 2021;269:119004. 10.1016/j.lfs.2020.119004 [DOI] [PubMed] [Google Scholar]
- 42.Ma J, Zhao Y, Sun L, et al. Exosomes Derived from Akt-Modified Human Umbilical Cord Mesenchymal Stem Cells Improve Cardiac Regeneration and Promote Angiogenesis via Activating Platelet-Derived Growth Factor D. Stem Cells Transl Med 2017;6:51-9. 10.5966/sctm.2016-0038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhao G, Liu F, Liu Z, et al. MSC-derived exosomes attenuate cell death through suppressing AIF nucleus translocation and enhance cutaneous wound healing. Stem Cell Res Ther 2020;11:174. 10.1186/s13287-020-01616-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhu H, Yan X, Zhang M, et al. miR-21-5p protects IL-1β-induced human chondrocytes from degradation. J Orthop Surg Res 2019;14:118. 10.1186/s13018-019-1160-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hu SL, Chang AC, Huang CC, et al. Myostatin Promotes Interleukin-1β Expression in Rheumatoid Arthritis Synovial Fibroblasts through Inhibition of miR-21-5p. Front Immunol 2017;8:1747. 10.3389/fimmu.2017.01747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Guan YJ, Li J, Yang X, et al. Evidence that miR-146a attenuates aging- and trauma-induced osteoarthritis by inhibiting Notch1, IL-6, and IL-1 mediated catabolism. Aging Cell 2018;17:e12752. 10.1111/acel.12752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Al-Modawi RN, Brinchmann JE, Karlsen TA. Multi-pathway Protective Effects of MicroRNAs on Human Chondrocytes in an In Vitro Model of Osteoarthritis. Mol Ther Nucleic Acids 2019;17:776-90. 10.1016/j.omtn.2019.07.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rasheed Z, Al-Shobaili HA, Rasheed N, et al. MicroRNA-26a-5p regulates the expression of inducible nitric oxide synthase via activation of NF-κB pathway in human osteoarthritis chondrocytes. Arch Biochem Biophys 2016;594:61-7. 10.1016/j.abb.2016.02.003 [DOI] [PubMed] [Google Scholar]
- 49.Luo P, Jiang C, Ji P, et al. Exosomes of stem cells from human exfoliated deciduous teeth as an anti-inflammatory agent in temporomandibular joint chondrocytes via miR-100-5p/mTOR. Stem Cell Res Ther 2019;10:216. 10.1186/s13287-019-1341-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sun F, Yang Q, Weng W, et al. Chd4 and associated proteins function as corepressors of Sox9 expression during BMP-2-induced chondrogenesis. J Bone Miner Res 2013;28:1950-61. 10.1002/jbmr.1932 [DOI] [PubMed] [Google Scholar]
- 51.Zhang S, Chu WC, Lai RC, et al. Exosomes derived from human embryonic mesenchymal stem cells promote osteochondral regeneration. Osteoarthritis Cartilage 2016;24:2135-40. 10.1016/j.joca.2016.06.022 [DOI] [PubMed] [Google Scholar]
- 52.Tao SC, Yuan T, Zhang YL, et al. Exosomes derived from miR-140-5p-overexpressing human synovial mesenchymal stem cells enhance cartilage tissue regeneration and prevent osteoarthritis of the knee in a rat model. Theranostics 2017;7:180-95. 10.7150/thno.17133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mao G, Zhang Z, Hu S, et al. Exosomes derived from miR-92a-3p-overexpressing human mesenchymal stem cells enhance chondrogenesis and suppress cartilage degradation via targeting WNT5A. Stem Cell Res Ther 2018;9:247. 10.1186/s13287-018-1004-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tofiño-Vian M, Guillén MI, Pérez Del Caz MD, et al. Microvesicles from Human Adipose Tissue-Derived Mesenchymal Stem Cells as a New Protective Strategy in Osteoarthritic Chondrocytes. Cell Physiol Biochem 2018;47:11-25. 10.1159/000489739 [DOI] [PubMed] [Google Scholar]
- 55.Zavatti M, Beretti F, Casciaro F, et al. Comparison of the therapeutic effect of amniotic fluid stem cells and their exosomes on monoiodoacetate-induced animal model of osteoarthritis. Biofactors 2020;46:106-17. 10.1002/biof.1576 [DOI] [PubMed] [Google Scholar]
- 56.Fernandes TL, Gomoll AH, Lattermann C, et al. Macrophage: A Potential Target on Cartilage Regeneration. Front Immunol 2020;11:111. 10.3389/fimmu.2020.00111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Fahy N, de Vries-van Melle ML, Lehmann J, et al. Human osteoarthritic synovium impacts chondrogenic differentiation of mesenchymal stem cells via macrophage polarisation state. Osteoarthritis Cartilage 2014;22:1167-75. 10.1016/j.joca.2014.05.021 [DOI] [PubMed] [Google Scholar]
- 58.Daghestani HN, Pieper CF, Kraus VB. Soluble macrophage biomarkers indicate inflammatory phenotypes in patients with knee osteoarthritis. Arthritis Rheumatol 2015;67:956-65. 10.1002/art.39006 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The article’s supplementary files as